Abstract
African trypanosomiasis, caused by parasites of the genus Trypanosoma, is a complex of devastating vector‐borne diseases of humans and livestock in sub‐Saharan Africa. Central to the pathogenesis of African trypanosomes is their transmission by the arthropod vector, Glossina spp. (tsetse fly). Intriguingly, the efficiency of parasite transmission through the vector is reduced following depletion of Trypanosoma brucei Procyclic‐Specific Surface Antigen‐2 (TbPSSA‐2). To investigate the underlying molecular mechanism of TbPSSA‐2, we determined the crystal structures of its ectodomain and that of its homolog T. congolense Insect Stage Antigen (TcISA) to resolutions of 1.65 Å and 2.45 Å, respectively using single wavelength anomalous dispersion. Both proteins adopt a novel bilobed architecture with the individual lobes displaying rotational flexibility around the central tether that suggest a potential mechanism for coordinating a binding partner. In support of this hypothesis, electron density consistent with a bound peptide was observed in the inter‐lob cleft of a TcISA monomer. These first reported structures of insect stage transmembrane proteins expressed by African trypanosomes provide potentially valuable insight into the interface between parasite and tsetse vector.
Keywords: Trypanosoma brucei, Trypanosoma congolense, tsetse, ectodomain, X‐ray crystallography, bi‐lobed architecture, conformational flexibility
Short abstract
Interactive Figure 1; Interactive Figure 2 | PDB Code(s): 5KLH; 5KMX
Introduction
African trypanosomiasis is a devastating complex of diseases caused by parasites of the genus Trypanosoma. These parasites, transmitted by the infamous tsetse fly (Glossina spp.), affect both humans (T. brucei rhodesiense, T. b. gambiense) and livestock (T. b. brucei, T. simiae, T. congolense, T. vivax) in sub‐Saharan Africa. In addition to, and often overshadowed by the variant surface glycoproteins responsible for antigenic variation of bloodstream forms, are surface proteins expressed by the parasites during transit through the tsetse vector. These latter proteins do not exhibit the extreme antigenic variation observed with the bloodstream stage molecules and thus potentially offer more options as therapeutic targets for parasite control.
African trypanosomes rely on a multi‐stage life cycle for survival. At each stage, these extracellular parasites express an array of surface proteins which enable their survival in the tsetse and allow their ultimate transmission to a new mammalian host.1 Although much has been learned about the predominant insect stage surface molecules, very little is known about the role of low abundant surface transmembrane (TM) proteins. Characterization of these TM proteins offers the potential to define the nuances of the molecular networks at the interface between parasite and vector that can be potentially targeted for transmission blocking therapies.
More than two decades ago, Jackson et al. identified a TM protein in the insect stages of the T. brucei sspp. called Procyclic Stage Specific Antigen‐2 (TbPSSA‐2).2 Fragoso et al. subsequently showed that a TbPSSA‐2 knock‐out reduced the efficiency of trypanosome migration from the tsetse midgut to the salivary glands highlighting the importance of this protein for parasite transmission.3 Recently, we4 identified a homolog of this molecule in T. congolense and determined its expression profile throughout the parasite's life cycle. This homolog was subsequently named Congolense Insect Stage Specific Antigen (CISSA).5 To be consistent with current nomenclature, we have renamed this molecule T. congolense Insect Stage antigen (TcISA). While TbPSSA‐2 and TcISA are likely functionally related, the basis of this function is unknown and their primary sequences do not align with those of any structurally or functionally characterized protein.
Towards establishing structure–function relationships for these trypanosome insect stage‐specific molecules, we determined the crystal structures of the TbPSSA‐2 and TcISA ectodomains. Intriguingly, both proteins adopt a previously uncharacterized bilobed architecture with intriguing implications for ligand coordination. These first structures of insect stage TM proteins expressed by T. brucei and T. congolense are discussed with respect to their potential role in molecular crosstalk between African trypanosomes and their insect vectors.
Results and Discussion
TbPSSA‐2 and TcISA ectodomains adopt a novel architecture with conformational flexibility
Analysis of the TbPSSA‐2 and TcISA ectodomain structures revealed an asymmetric, bi‐lobed architecture formed predominantly by β strands with the addition of three short helices (α‐1, 2, 4) positioned at the lobe periphery. The two lobes are linked together by an elongated β‐strand (β8) that spans the entire length of the protein and a short loop connecting β5 and β6 [Fig. 1(B)—red]. The larger lobe 1 (L1) consists of two polypeptide segments from the amino terminus (TbPSSA‐2: S40‐S123; TcISA: P44‐T123) and carboxyl terminus (TbPSSA‐2: L189‐V258; TcISA: L189‐E255). The smaller lobe 2 (L2) consists of P124‐Y188 in both TbPSSA‐2 and TcISA. L1 is comprised of nearly orthogonally stacked β sheets with the larger sheet consisting of β1, 4, 5, 8, 9, 10 and the smaller sheet β2, 3, and 11. The smaller L2 is stabilized by four intramolecular disulfide linkages characterized by a β‐α‐β motif with β‐7 and β‐6 coordinating the β‐8 linker [Fig. 1(C)]. A structural homology search using the DaliLite server6 using the entire ectodomain or the individual lobes revealed no significant hits indicating that TbPSSA‐2 and TcISA adopt a previously uncharacterized domain architecture.
Figure 1.
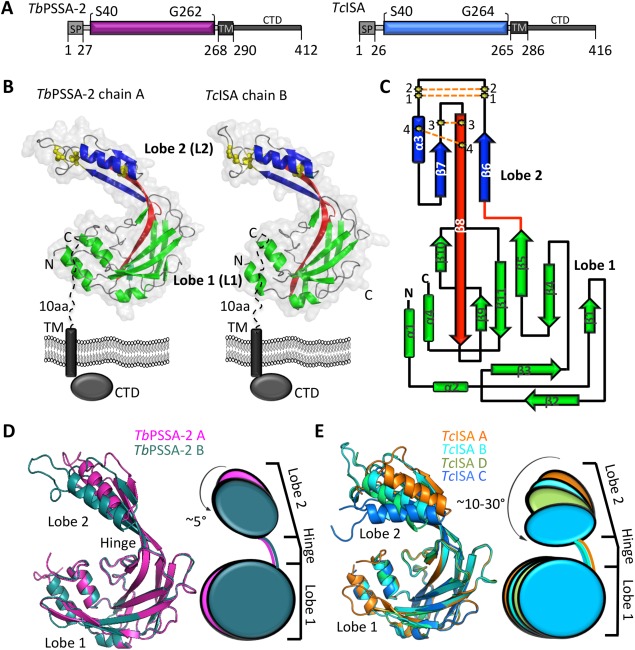
TbPSSA‐2 and TcISA ectodomains adopt a novel architecture and exhibit conformational flexibility. (A) Schematic representation of TbPSSA‐2 and TcISA with predicted structural domains, TM regions and signal peptides. (B) Secondary structure depictions of TbPSSA‐2 (left) and TcISA (right) highlighting lobe 1 (green) and lobe 2 (blue). The beta strand and the loop connecting the two lobes are shown in red. Four disulfide bonds in lobe 2 are shown as yellow ball and sticks. (C) Topology diagram of TbPSSA‐2 and TcISA colored as in (B). Rotational flexibility between lobes L1 and L2 in (D) TbPSSA‐2 (chain A‐pink; B‐dark teal) and (E) TcISA (chain A‐orange; B‐cyan; C‐limegreen; D‐marine). An interactive view is available in the electronic version of the article.
To investigate the potential for inter‐lobe flexibility, we took advantage of the fact that both TbPSSA‐2 and TcISA structures contain multiple copies in the asymmetric unit (two copies of TbPSSA‐2 and four copies of TcISA) [Fig. 1(D,E)]. While individual domain rmsd values showed only subtle differences in L1 (TbPSSA‐2: 0.25 Å over 125 Cα; TcISA: 0.27‐0.33 Å over 103‐118 Cα) and L2 (TbPSSA‐2: 0.43 Å over 47 Cα; TcISA: 0.42‐0.72 Å over 42‐47 Cα), the lobes showed significant rotational displacement relative to each other. In TbPSSA‐2, L2 rotates ∼5° (into the plane) relative to L1 for a total displacement of 6 Å [Fig. 1(D)]. In TcISA L2 rotates 30° (into the plane) relative to L1 with an 18 Å displacement [Fig. 1(D)]. Further, the DynDom protein domain motion analysis server7 classified the β8 strand residues E130, R131, W132, K184, Q185, and E186 in TbPSSA‐2, and R126, V127, K128, R129, E130, K131, K184, Q185, and N186 in TcISA as high probability candidates for participating in a hinge‐like structure. Consistent with the observed inter‐lobe flexibility, the ectodomains of TbPSSA‐2 and TcISA elute as monomers from a size exclusion column. However, it has been previously reported that TbPSSA‐2 may form multimers in vivo,3 reflecting a possible role for the cytoplasmic domain in promoting higher order assembly.
Hinge region in TcISA reveals ligand coordinating potential
One possible functional outcome of the conformational flexibility of the hinge region might be to enable positioning of the individual lobes to support ligand coordination. Consistent with this possibility, one of the TcISA monomers in the AU (chain A) formed a complex with a short peptide that bound at the base of the inter‐lobe hinge region [Fig. 2(A)]. While the backbone of the bound peptide showed clear electron density, the side chains could not be definitively registered possibly due to reduced occupancy [Fig. 2(A)]. Analysis of the remaining monomers in the AU reveal that the bound peptide is likely derived from the neighbouring chain C molecule for which the 15 residue C‐terminal tail could not be directly modelled. Notably, the last modelled residue in chain C is positioned ∼11 Å from the where the peptide is bound in chain A. Overall, the bound peptide is stabilized through a combination of shape complementarity and intermolecular hydrogen bonds including two backbone‐backbone hydrogen bonds with K128 and Y224. Additionally, the bound peptide coordinates two water molecules (HOH29 and HOH45). A closer inspection of the hinge region revealed a pair of basic residues (R129 and K131 pair) that, in chain A, coordinate a bound sulfate molecule and, as a result, are directed away from the peptide‐binding region [Fig. 2(B)—middle panel]. In the three other chains where no sulfate is bound the side‐chains of R129 and K131 sterically clash with the peptide overlaid from chain A [Fig. 2(B)—right panel]. Thus, R129 and K131 may regulate access to the inter‐lobe cleft.
Figure 2.
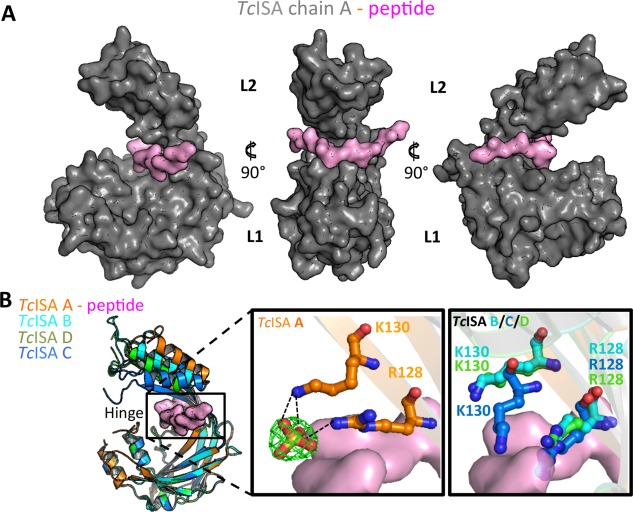
Hinge region in TcISA reveals the potential for ligand binding. (A) TcISA chain A surface (grey) highlighting the bound peptide (pink). (B) Superimposition of TcISA monomers (left) with cleft indicated by black box. Middle panel: R127 and K130 in the hinge region of chain A coordinating a sulfate group which is depicted as stick model with a 2Fo‐Fc electron density map (green mesh) contoured at 1.5σ. Right panel: R128 and K130 of chains B, C, and D showing steric clash with overlaid peptide from chain A. An interactive view is available in the electronic version of the article.
Although identifying the binding partners of these proteins is key to fully elucidating their function(s), these ectodomain structures provide important insight into the molecular architecture of Trypanosoma surface proteins expressed during insect transmission. Combined with functional data that show TbPSSA‐2 plays an important role in parasite survival in the tsetse, the novel domain architecture of TbPSSA‐2 and TcISA with their potential for ligand coordination may contribute to designing transmission‐blocking therapies.
Materials and Methods
Cloning, expression, and purification
TbPSSA‐2 (Genbank8 accession number: AAA30244.1) and TcISA (TritrypDB9 accession number: TcIL3000_10_9440) construct boundaries are shown in Figure 1(A). Signal peptides were predicted using SignalP 4.010 and TM domains were predicted using TMHMM 2.0.11 Synthetic genes (Genscript) were codon optimized for E. coli and cloned into pET28a (Novagen) with a modified N‐terminal Trx‐His6 tag. Both proteins were expressed in E. coli Rosetta‐gami 2 (DE3) and purified by nickel affinity, size exclusion (Superdex 16/600 75) in 20 mM HEPES, 150 mM NaCl, pH 6.8, and cation‐exchange chromatography (HiTrap SP FF column in 20 mM Tris‐HCl, 10 mM NaCl, pH 6.2 with NaCl gradient).
Protein crystallization and data collection
Crystals of TbPSSA‐2 were grown in 0.2 M (NH4)2SO4, 0.1 M Bis‐tris pH 6.5, and 25% PEG 3350 while TcISA crystallized in 20% PEG 3350 with 0.2 M (NH4)2SO4 using sitting drops. For phasing purposes, TbPSSA‐2 crystals were soaked in mother liquor supplemented with NaI to a final concentration of 0.5 M for 45 s. Crystals were cryoprotected in mother liquor supplemented with 12.5% glycerol. Diffraction data were collected on beamline 9‐2 at the Stanford Synchrotron Radiation Laboratory (SSRL).
Data processing, structure solution, and refinement
Diffraction data for TbPSSA‐2 native and iodide soaked crystals were processed to 1.65 and 1.85 Å resolution, respectively using Imosflm12 and Scala13 in CCP4.14 A total of 28 iodide sites were identified in TbPSSA‐2 and refined using SHELX C/D/E.15 High quality phases were obtained after density modification in dm and enabled building 60% of the sequence using buccaneer.16 The remaining structure was built manually and used as a molecular replacement model for the higher resolution native data using Phaser.17 Solvent atoms were selected using COOT18 and refined in Phenix Refine.19 Diffraction data for TcISA crystals were processed to 2.45 Å resolution using the methods described for TbPSSA‐2. Initial phases were obtained by molecular replacement using Phaser20 with TbPSSA‐2 as a search model. Overall, 5% of the reflections were set aside for calculation of R free. Stereo‐chemical analysis performed with PROCHECK and SFCHECK in CCP414 showed at least 96% of the residues in the favoured conformations and no residues in disallowed orientations. Data collection and refinement statistics of TbPSSA‐2 and TcISA are presented in Table 1. All the images of the structures were prepared with PYMOL.22 Atomic coordinates and structure factors of TbPSSA‐2 and TcISA have been deposited in the Protein Data Bank with accession codes 5KLH and 5KMX.
Table 1.
Data Collection and Refinement Statistics
| TbPSSA‐2_I SAD | TbPSSA‐2 | TcISA | |
|---|---|---|---|
| A. Data collection statistics | |||
| Space group | P1 | P1 | P21 |
| a, b, c (Å) | 32.13, 57.82, 60.61 | 31.82, 57.81, 61.24 | 75.81, 87.65, 98.89 |
| α, β, γ (deg.) | 74.24, 76.45, 89.76 | 73.55, 76.43, 89.98 | 90, 113.27, 90 |
| Wavelength | 1.1807 | 1.1807 | 0.979 |
| Resolution range (Å) | 56.59–1.80 (1.84–1.80) | 56.95–1.65 (1.67–1.65) | 39.47–2.45 (2.49–2.45) |
| Measured reflections | 184,718 (10,800) | 171,475 (8353) | 146,837 (11,384) |
| Unique reflections | 35,093 (2079) | 45,400 (2236) | 42,734 (2110) |
| Redundancy | 5.3 (5.2) | 3.8 (3.7) | 3.4 (3.5) |
| Completeness (%) | 93.2 (91.9) | 92.3 (90.7) | 97.6 (98.3) |
| I/σ(I) | 11.3 (2.4) | 10.3 (2.0) | 17.8 (1.2) |
| R merge a | 0.087 (0.822) | 0.066 (0.542) | 0.044 (0.517) |
| CC1/2 | 0.997 (0.771) | 0.995 (0.739) | 0.960 (0.821) |
| Number of I sites | 28 | N/A | N/A |
| B. Refinement statistics | |||
| Resolution (Å) | 47.12–1.65 | 39.48–2.45 | |
| R crys T b/R free c | 0.177/0.214 | 0.257/0.289 | |
| No. of atoms | |||
| Protein (A/B/C/D/U) | 1728/1692 | 1630/1568/1468/1486/74 | |
| Sulfate/glycerol | 5 | 40/12 | |
| Solvent | 462 | 68 | |
| B‐values (Å2) | |||
| Protein (A/B/C/D/U) | 21.2/24.8 | 65.8/70.0/87.1/84.8/68.4 | |
| Sulfate/glycerol | 28.5 | 75.3/62.4 | |
| Solvent | 28.1 | 51.9 | |
| r.m.s. deviation from ideality | |||
| Bond lengths (Å) | 0.007 | 0.004 | |
| Bond angles (deg.) | 1.07 | 0.78 | |
| Ramachandran statistics (%) | |||
| Most favoured | 98.6 | 96.0 | |
| Allowed | 1.2 | 4.0 | |
| Disallowed | 0.0 | 0.0 | |
Values in parentheses are for the highest resolution shell
R merge = ∑hkl∑i|Ihkl,i − [Ihkl]|/∑hkl∑iIhkl,i, where [Ihkl] is the average of symmetry related observations of a unique reflection
R cryst = ∑|F obs − F calc|/∑F obs, where F obs and F calc are the observed and the calculated structure factors, respectively.
R free is R using 5% of reflections randomly chosen and omitted from refinement*.
Acknowledgments
The authors thank the staff at the Stanford Synchrotron Radiation Laboratory for their expert contributions. The name TcISA was derived in part to honour Dr. Isabel Roditi (Isa) who has made major seminal contributions to the study of trypanosome insect stage molecules over the past few decades.
References
- 1. Roditi I, Liniger M (2002) Dressed for success: the surface coats of insect‐borne protozoan parasites. Trends Microbiol 10:128–134. [DOI] [PubMed] [Google Scholar]
- 2. Jackson DG, Smith DK, Luo C, Elliott JF (1993) Cloning of a novel surface antigen from the insect stages of Trypanosoma brucei by expression in COS cells. J Biol Chem 268:1894–1900. [PubMed] [Google Scholar]
- 3. Fragoso CM, Schumann Burkard G, Oberle M, Renggli CK, Hilzinger K, Roditi I (2009) PSSA‐2, a membrane‐spanning phosphoprotein of Trypanosoma brucei, is required for efficient maturation of infection. PLoS One 4:e7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Eyford BA, Sakurai T, Smith D, Loveless B, Hertz‐Fowler C, Donelson JE, Inoue N, Pearson TW (2011) Differential protein expression throughout the life cycle of Trypanosoma congolense, a major parasite of cattle in Africa. Mol Biochem Parasitol 177:116–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tonkin ML, Workman SD, Eyford BA, Loveless BC, Fudge JL, Pearson TW, Boulanger MJ (2012) Purification, crystallization and X‐ray diffraction analysis of Trypanosoma congolense insect‐stage surface antigen (TcCISSA). Acta Cryst F68:1503–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Holm L, Rosenstrom P (2010) Dali server: conservation mapping in 3D. Nucleic Acids Res 38:W545–W549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Girdlestone C, Hayward S (2016) The DynDom3D webserver for the analysis of domain movements in multimeric proteins. J Comput Biol 23:21–26. [DOI] [PubMed] [Google Scholar]
- 8. Benson DA, Cavanaugh M, Clark K, Karsch‐Mizrachi I, Lipman DJ, Ostell J, Sayers EW (2013) GenBank. Nucleic Acids Res 41:D36–D42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Aslett M, Aurrecoechea C, Berriman M, Brestelli J, Brunk BP, Carrington M, Depledge DP, Fischer S, Gajria B, Gao X, Gardner MJ, Gingle A, Grant G, Harb OS, Heiges M, Hertz‐Fowler C, Houston R, Innamorato F, Iodice J, Kissinger JC, Kraemer E, Li W, Logan FJ, Miller JA, Mitra S, Myler PJ, Nayak V, Pennington C, Phan I, Pinney DF, Ramasamy G, Rogers MB, Roos DS, Ross C, Sivam D, Smith DF, Srinivasamoorthy G, Stoeckert CJ Jr., Subramanian S, Thibodeau R, Tivey A, Treatman C, Velarde G, Wang H (2010) TriTrypDB: a functional genomic resource for the Trypanosomatidae. Nucleic Acids Res 38:D457–D462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Petersen TN, Brunak S, von Heijne G, Nielsen H (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8:785–786. [DOI] [PubMed] [Google Scholar]
- 11. Krogh A, Larsson B, Von Heijne G, Sonnhammer EL (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305:567–580. [DOI] [PubMed] [Google Scholar]
- 12. Battye TG, Kontogiannis L, Johnson O, Powell HR, Leslie AG (2011) iMOSFLM: a new graphical interface for diffraction‐image processing with MOSFLM. Acta Cryst D67:271–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Evans P (2006) Scaling and assessment of data quality. Acta Cryst D62:72–82. [DOI] [PubMed] [Google Scholar]
- 14. Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS (2011) Overview of the CCP4 suite and current developments. Acta Cryst D67:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sheldrick GM (2010) Experimental phasing with SHELXC/D/E: combining chain tracing with density modification. Acta Cryst D66:479–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cowtan K (2007) Fitting molecular fragments into electron density. Acta Cryst D64:83–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. McCoy AJ (2007) Solving structures of protein complexes by molecular replacement with Phaser. Acta Cryst D63:32–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Emsley P, Cowtan K (2004) Coot: model‐building tools for molecular graphics. Acta Cryst D60:2126–2132. [DOI] [PubMed] [Google Scholar]
- 19. Afonine PV, Grosse‐Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, Adams PD (2012) Towards automated crystallographic structure refinement with phenix.refine. Acta Cryst D68:352–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. McCoy AJ, Grosse‐Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ (2007) Phaser crystallographic software. J Appl Cryst 40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Collaborative Computational Project N (1994) Acta Cryst D50:760–763. [Google Scholar]
- 22. Schrodinger LL (2015) The PyMOL Molecular Graphics System, Version 1.8.


