Abstract
RcsB, the transcription‐associated response regulator of the Rcs phosphorelay two‐component signal transduction system, activates cell stress responses associated with desiccation, cell wall biosynthesis, cell division, virulence, biofilm formation, and antibiotic resistance in enteric bacterial pathogens. RcsB belongs to the FixJ/NarL family of transcriptional regulators, which are characterized by a highly conserved C‐terminal DNA‐binding domain. The N‐terminal domain of RcsB belongs to the family of two‐component receiver domains. This receiver domain contains the phosphoacceptor site and participates in RcsB dimer formation; it also contributes to dimer formation with other transcription factor partners. Here, we describe the crystal structure of the Escherichia coli RcsB receiver domain in its nonphosphorylated state. The structure reveals important molecular details of phosphorylation‐independent dimerization of RcsB and has implication for the formation of heterodimers.
Keywords: transcriptional regulator, Rcs phosphorelay, two‐component signal transduction system, phosphorylation domain, FixJ/NarL family
Short abstract
PDB Code(s): 5I4C
Abbreviations
- HTH
helix‐turn‐helix
- SEC‐MALS
size exclusion chromatography with multi‐angle light scattering
Introduction
Escherichia coli and related organisms employ the Rcs phosphorelay system to modulate intracellular responses to environmental stresses associated with desiccation, antibiotic exposure, biofilm formation and virulence.1, 2, 3, 4, 5 A complex two‐component signal transduction system, the Rcs phosphorelay consists of five proteins: the outer membrane lipoprotein RcsF, the inner membrane protein Iga, the inner membrane hybrid sensor kinase RcsC, the histidine phosphotransferase RcsD and the response regulator RcsB.1, 2, 3, 4, 5, 6 The latter controls the transcription of 5% of E. coli genes.7
Like many other two‐component response regulators, RcsB can become active when it is phosphorylated on a conserved aspartyl residue (D56) located within its N‐terminal receiver domain.8 Phosphorylation of RcsB can occur via two distinct mechanisms. In response to an extracytoplasmic stress, it can receive a phosphoryl group via an ATP‐dependent phosphorylation cascade that involves RcsC and RcsD; alternatively, it can receive a phosphoryl group from the central metabolite acetyl phosphate in response to certain metabolic stresses.9, 10, 11
RcsB forms both homo and heterodimers.12, 13, 14 As a homodimer, RcsB regulates the small non‐encoding RNA rprA,15 the osmoregulated gene osmC,16 and the master regulator of flagellar biosynthesis flhDC.9, 17 RcsB also partners with other transcriptional factors to form heterodimers. These partners include RcsA, GadE, BglJ, RmpA, PhoP, MatA, and RflM.12, 13, 14, 17, 18, 19, 20, 21, 22, 23, 24 The formation of RcsB heterodimers extends the regulatory activity of the Rcs phosphorelay system to modulate virulence, facilitate biofilm formation, and regulate cell envelope stress responses that contribute to antibiotic resistance in pathogens.5, 22, 25, 26
RcsB is a 216 amino acid protein that belongs to the FixJ/NarL family of transcriptional regulators, defined by a conserved DNA‐binding helix‐turn‐helix (HTH) domain.27 Like many other FixJ/NarL members, RcsB is a two‐domain protein that contains this C‐terminal DNA‐binding domain linked to an N‐terminal regulatory domain. A long flexible linker connects the two RcsB domains [Fig. 1(A)]. Others previously determined the NMR structure of the RcsB C‐terminal DNA‐binding domain from Erwinia amylovora, a close relative of E. coli.28 This study revealed the DNA‐binding HTH domain as an all‐helical structure comprised of four α‐helices: α7, α8, α9, and α10 [Fig. 1(B)]. Helices α8 and α9 comprise the HTH DNA‐binding motif, whereas helix α6 is a part of the interdomain linker in the structure of the RcsB DNA‐binding domain [Fig. 1(B)]. The larger, RcsB N‐terminal regulatory receiver domain, whose structure remains unknown, contains the phosphoacceptor site (residue D56).
Figure 1.
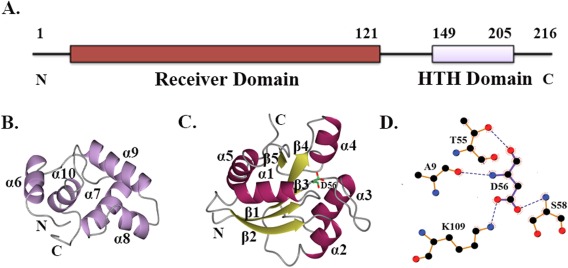
RcsB domain composition. A. Functional domains of RcsB protein as annotated in Pfam database (pfam.xfam.org/). B. Ribbon diagram of the C‐terminal DNA‐binding domain of RcsB from E. amylovora (PDB ID 1P4W) determined by NMR. C. Ribbon diagram of the N‐terminal receiver domain of RcsB from E. coli. β‐strands and α‐helices of the RcsB receiver domain are shown in yellow and dark purple, respectively. Phosphoacceptor residue D56 is shown in cylinder model, where carbon atoms are in green, nitrogen atom in blue, and oxygen atoms in red. Secondary structure elements on C‐ and N‐terminal domain of RcsB are labeled. D. LigPlot+ diagram of phosphorylation site. Hydrogen bonds for D56 residue are shown as dark blue dashes. In the diagram, oxygen atoms are colored in red, carbon atoms in black, and nitrogen atoms in blue.
Phosphorylation of the RcsB N‐terminal receiver domain is thought to induce structural changes allowing homodimerization of RcsB and heterodimerization of RcsB‐RcsA, and modulating binding to their DNA sites.21, 28 In contrast, a recent reports provide evidence that the activities of the RcsB‐BglJ, RcsB‐MatA and RcsB‐RflM complexes do not depend on the RcsB phosphorylation.14, 17 It has been shown that a dimerization interface in both RcsB homodimers and heterodimers resides within the RcsB N‐terminal receiver domain.14 However, it is not known if phosphorylated or nonphosphorylated RcsB form homodimers, heterodimers with its auxiliary partners, or other oligomers in solution or upon binding to their DNA‐binding sites.
In this study, we focus on the structural details of phosphorylation‐independent RcsB dimerization. We present the X‐ray structure at 2.0 Å resolution of the nonphosphorylated receiver domain of RcsB from E. coli. We characterize the RcsB receiver domain phosphorylation site, and perform a structural comparison between the RcsB receiver domain and structures of other FixJ/NarL family members. Importantly, our data provide structural insights into the dimerization of the RcsB receiver domain in both RcsB homo and heterodimeric forms.
Results
SEC‐MALS analysis of the RcsB receiver domain
Size exclusion chromatography with multi‐angle light scattering (SEC‐MALS) analysis of the RcsB receiver domain (present at a concentration of 3 mg/mL) detected two peaks that correspond to the dimeric (average MW of 31 kDa) and monomeric (average MW of 19 kDa) states of the RcsB receiver domain in solution [Fig. 2(A), S1]. To determine if protein concentration and/or phosphorylation mediate dimer formation, we also performed SEC‐MALS analysis of the RcsB receiver domain at a lower concentration (1 mg/mL) in the absence or presence of 50 mM acetylphosphate (AcP). In the absence of AcP, SEC‐MALS analysis detected two peaks that correspond to the dimeric and monomeric states with an average MW of 30 and 19 kDa, respectively [Fig. 2(A), S1]. A similar mass distribution was observed in the presence of AcP; the major fractions in peak 1 and 2 had average MWs of 31 and 20 kDa, respectively [Fig. 2(A), S1]. The MALS analysis indicates that the eluted peaks of the RcsB receiver domain at high and low concentration, in the absence or presence of AcP, contain a dimer‐monomer mixture; however, at high concentration, the fraction of dimer is larger than that of the monomer.
Figure 2.
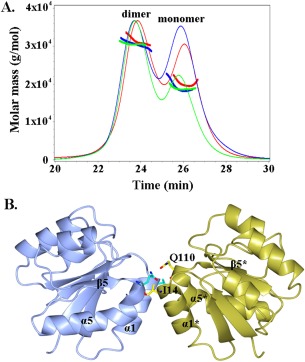
SEC‐MALS elution profile and dimer of the RcsB receiver domain. A. The elution profile of the RcsB N‐terminal domain at concentration 3 mg/mL and at concentration 1 mg/mL in absence and presence of AcP is shown as solid line in green, blue, and red, respectively. Peaks that correspond to dimeric and monomeric state of the RcsB receiver domain are marked. Molecular mass distribution is shown as bold solid lines within peaks. B. Ribbon diagram of the dimer of RcsB receiver domain. α‐helices, β‐strand and residues (cylinder model in cyan) involved in hydrogen bonds within dimerization contact area are labeled. In the symmetry‐related subunit of the crystallographic dimer, the α‐helices and β‐strand are labeled with asterisk and residues are shown in yellow. The oxygen atoms of residues are colored in red and nitrogen in blue.
Structure of the RcsB receiver domain
The crystal structure of the RcsB receiver domain in its nonphosphorylated form was determined at 2.0 Å resolution by the molecular replacement method [Fig. 1(C)]. The structure of the crystallized RcsB receiver domain contains one protein molecule (residues 2‐132) in the asymmetric unit. The first N‐terminal residue and 15 C‐terminal residues comprising part of the inter‐domain linker were not ordered in the electron density map. The RcsB receiver domain has an α/β structural fold with a central five‐stranded β‐sheet (β1 (residues 4‐10), β2 (28‐35), β3 (51‐57), β4 (82‐87) and β5 (105‐109)) surrounded by five α helices (α1 (12‐24), α2 (37‐45), α3 (66‐78), α4 (93‐98), and α5 (111‐125)) and two short helical fragments containing residues 46‐48 and 99‐102.
Within the receiver domain, the conserved phosphoacceptor residue D56 is located on the surface loop at the C‐terminus of strand β3 [Fig. 1(C)]. The phosphorylation site is surrounded by the following residues: A9, D10, D11, H12, T55, L57, S58, M59, P60, L86, T87 and K109. In the structure, the N and O atoms of the D56 main chain are hydrogen bonded with the O atom of the A9 main chain and the OG1 atom of the T55 side chain, respectively [Fig. 1(D)]. The OD1 and OD2 atoms of the D56 side chain are hydrogen bonded with the NZ atom of the K109 side chain and the N atom of the S58 main chain, respectively [Fig. 1(D)].
To identify if a relevant dimer of the RcsB receiver domain is present in the crystal lattice, we ran the PISA (Protein Interface, Surfaces and Assemblies) server.29 Its analysis of quaternary structure of the RcsB receiver domain, applying packing arrangements of protein molecules in the crystal, identified a dimer of the RcsB receiver domain subunits [Fig. 2(B)] with a favorable interaction energy ΔG of −10.5 kcal/mol. This dimerization interface has a buried surface area of 530 Å2. The small size of the dimerization interface is consistent with the observation of a dimer‐monomer mixture detected by the SEC‐MALS analysis [Fig. 2(A)]. The dimer interface is formed by 14 residues located primarily on helix α1 (residues 12‐15, 17‐19, 21, and 22), loop β5‐α5 (residues 109 and 110) and helix α5 (residues 112 and 113). Within the dimerization contact area, residues I14 and Q110 form four hydrogen bonds and residues P13, I14, V15, F17, G18, I19, M88, A112, and P113 form hydrophobic interactions.
Structural homology between E. Coli RcsB and FixJ/NarL family regulators
Structural comparison using the secondary structure alignment program in the ProFunc server30 identified a number of regulatory domains with a structural fold similar to that of the E. coli RcsB receiver domain and with RMSD values of 1.6 Å. The list of characterized homologous structures includes the receiver domain of YycF from Streptococcus pneumoniae (PDB ID 1NXW, Z score 9.7 31), the regulatory domain of PmrA from Klebsiella pneumoniae (PDB ID 3W9S, Z score 10.8 32), the receiver domain of transcription regulator RR468 from Thermotoga maritima (PDB ID 4JA2, Z score 9.5 33), and the receiver domain of the CheY regulator from T. maritima (PDB ID 4TMY, Z score 9.6 34). Despite low sequence similarity between the listed proteins, the structural fold of each of their receiver domains is well conserved. Conformational differences are localized in the loops that connect β‐strands and α‐helixes [Fig. 3(A,B)]. In the receiver domain of structurally characterized response regulators, phosphorylation of the aspartyl residue mainly alters reorganization of the loops between β1‐α1, β3‐α3, β4‐α4, and helix α4.35, 36
Figure 3.
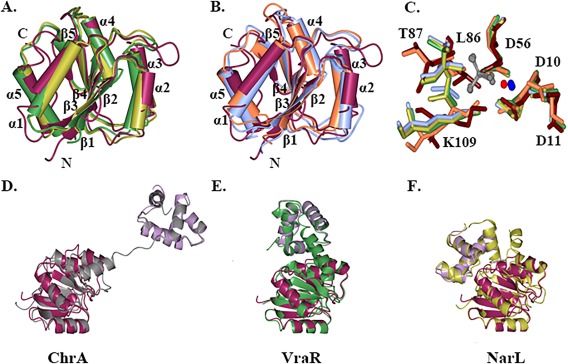
Structural comparison of RcsB with FixJ/NarL family regulators. A. Superposition of the nonphosphorylated RcsB receiver domain (maroon) with homologous receiver domain of YycF in inactive form (green) and PmrA in active form (gold). B. Superposition of the nonphosphorylated RcsB receiver domain with homologous receiver domain of CheY in inactive form (coral) and RR468 in active form (light blue). C. Superposition of conserved residues (D10, D11, D56, L86, T87, K109) at phosphorylation site of the RcsB receiver domain (tan) and active site residues of the receiver domain of RR468 with aspartate beryllium trifluoride residue (green) and magnesium ion, PmrA in complex with beryllium fluoride ion (carbons in light blue) and magnesium ion, CheY (orange) in complex with magnesium ion (red) and PhoP from E. coli (PDB ID 2PL1) in presence of beryllium fluoride ion (gold) and magnesium ion. Residues are shown as cylinder model. The beryllium fluoride (grey) and magnesium (blue) ions of the receiver domain response regulators in active state are shown as ball‐and‐stick model. D., E., and F. Superposition of RcsB C‐terminal DNA‐binding (lilac) and N‐terminal receiver domain (dark purple) with ChrA (grey), VraR (green), and NarL (gold) regulator. The average RMSD between superimposed RcsB C‐terminal and N‐terminal domains and homologous domains of full‐length FixJ/NarL regulators are ∼ 1.8 and 1.6 Å, respectively. Structures were superimposed in program CCP4mg.
Structures have been determined for homologous regulatory receiver domains in complex with beryllium trifluoride ion ( ) or with a modified aspartate residue (L‐peptide linking) that mimics the phosphorylated state.32, 33, 35, 36 A comparison of the conserved active site residues of the nonphosphorylated RcsB receiver domain with the active site residues of receiver domain response regulators (RR468, PmrA, and PhoP) in their active phosphorylated state and CheY in its inactive nonphosphorylated state shows identical positioning of the D10, D11, and D56 residues involved in interactions with the or with the phosphoryl group of the modified aspartyl residue and Mg2+ [Fig. 3(C)]. The superposition of the active site residues between receiver domains in their inactive (RcsB, CheY) and active state (RR468, PmrA, and PhoP) shows rearrangement of the L86, T87, and K109 residues located on the strand β4, the loop β4‐α4, and β5‐α5, respectively [Fig. 3(C)]. These results support the contention that residues D10, D11, D56, L86, T87, and K109 of the RcsB receiver domain play essential roles for the phosphotransfer reaction and that phosphorylation will likely induce conformational changes of loops (β1‐α1, β3‐α3, β4‐α4) and helixes (α4, α6) in the structure of the full‐length RcsB protein or its isolated receiver domain. These changes should affect dimer‐monomer equilibrium and/or alter domain orientations in the full‐length RcsB as seen in the structure of characterized response regulators and their isolated receiver domains.35, 36, 37
A search for homologous structures based on sequence conservation revealed existing PDB structures of known full‐length FixJ/NarL regulators that are similar to E. coli RcsB. The highest ranked proteins based on sequence identity are the response regulator ChrA in a haem‐sensing two‐component system from Corynebacterium diphtheria (PDB ID 4YN838), the vancomycin‐resistance‐associated response regulator VraR from Staphylococcus aureus (PDB ID 4GVP37), and the nitrate/nitrite response regulator NarL from E. coli (PDB ID 1RNL39). All these response regulators contain DNA‐binding and receiver domains connected by a long inter‐domain linker. Structural comparison of the RcsB domains with homologous regulators in their non‐phosphorylated state shows analogous fold and structural topology between individual domains [Fig. 3(D‐F)]. However, the relative orientation of domains and both the lengths and conformations of the inter‐domain linkers differ in these response regulators. These differences could affect the dimerization interfaces of these response regulators and thus contribute to their unique function.
Discussion
In this study, we report the first crystal structure of the N‐terminal receiver domain of the stress response regulator RcsB from the two‐component Rcs phosphorelay system of E. coli. The structural fold and phosphorylation site of the E. coli RcsB receiver domain are similar to other known receiver domains of homologous regulators from the FixJ/NarL family of proteins. Our SEC‐MALS data showed that the RcsB receiver domain can exist in solution as a dimer and our structural analysis revealed that it can form a dimer in the crystal lattice. We suggest that this dimeric state is physiologically relevant and might correspond to the non‐phosphorylated dimeric state of the RcsB receiver domain. In addition, the SEC‐MALS results indicate that this dimer is not very stable in solution such that the protein exists as a dimer‐monomer equilibrium system. Furthermore, we found that phosphorylation, by addition of AcP, does not have a major impact on dimerization of the RcsB receiver domains; in contrast, increased protein concentration strongly favors dimer formation [Fig. 2(A)]. It is possible that the absence of the interdomain helix α6 and/or the lack of the C‐terminal DNA‐binding domain make the dimer of the RcsB receiver domains less stable and more prone to dissociation. In the structure of the RcsB receiver domain, the dimer interface is formed by helices α1, α5 and loop β5‐α5. Similar dimerization interfaces between receiver domains have been described in the structure of homologous regulators, including the nitrate/nitrite response regulator NarL from E. coli, the vancomycin‐resistance‐associated response regulator VraR from S. aureus and the response regulator DesR, which controls fluidity of membrane phospholipids.37, 39, 40, 41, 42 We previously demonstrated that full‐length RcsB can form tetramers and dimers in solution.10 Thus, the data suggest that the observed dimer likely has functional importance.
RcsB can form an RcsB‐RcsB homodimer or RcsB heterodimers with other transcriptional regulators, including RcsA, BglJ, GadE, MatA, DctR, RmpA, RflM, and PhoP.12, 13, 14, 17, 18, 19, 20, 21, 22, 23, 24 The formation of some of these dimers can depend on phosphorylation state of the RcsB monomer. For example, it has been reported that the RcsB homodimer and the RcsB‐RcsA heterodimer form when RcsB is phosphorylated.14, 21, 43, 44 In contrast, formation of RcsB‐MatA, RcsB‐GadE, RcsB‐RflM or RcsB‐BglJ heterodimers appears to be unaffected by phosphorylation.14, 17 The data also suggest that both phosphorylated and nonphosphorylated RcsB in their homodimeric and heterodimeric states would form analogous dimerization interfaces comprised of helices α1, α5 and loop β5‐α5 of the RcsB receiver domain.14 This study also analyzed the impact of residues important for RcsB activity and located on α4‐β5‐α5 and α1 of the presumed dimerization surface (I14, D62, Y64, D66, R76, H77, L95, S96, L99, D100, E104, I106, L108, T114, D115, and K118).14 Mutation of I14 located on helix α1 had the highest impact on activity in all tested RcsB protein dimers. Thus, the dimeric structure of the nonphosphorylated RcsB receiver domains obtained in this study is in agreement with this previously published mutagenesis study.14
Therefore, we hypothesize that the non‐phosphorylated and phosphorylated states of the RcsB homodimer would have a similar dimerization interface formed through helices α1, α5 and loop β5‐α5 as seen in the structure of the determined RcsB receiver domain. This dimerization interface is mostly formed by hydrophobic interactions of the type that usually play a key role in protein‐protein complexes. In the heterodimeric state, the RcsB monomer could form a similar dimer interface (through helices α1/α5) with a monomer of the RcsB auxiliary partner. Confirmation of this hypothesis will require further analysis of interactions between RcsB and its auxiliary transcriptional regulators in the homo‐ and heterodimeric states.
Materials and Methods
Cloning, expression and purification
The gene sequence of the truncated N‐terminal receiver domain (residues 1‐147) of RcsB (NCBI accession code AAC75277, GI: 1788546) from E. coli str. K‐12 substr. MG1655 was synthesized (GenScript, USA) and subcloned into the IPTG (Isopropyl β‐D‐1‐Thiogalactopyranoside)‐inducible pMCSG7 vector with a short N‐terminal 6‐His tag without TEV cleavage site.45 This truncated protein was prepared at the Recombinant Protein Production Core (rPPC) facility at Northwestern University (Evanston, IL). For purification, the plasmid containing the truncated RcsB receiver domain was transformed by electroporation into electrocompetent BL21(DE3)‐magic cells (E. coli) on Gene Pulser Xcell electroporation System (Bio‐Rad, Hercules, CA). Transformed cells were inoculated in LB (Luria Broth), supplemented with 400 μg/mL ampicillin and 25 μg/mL kanamycin and grown overnight in Innova‐44 Incubator‐Shaker (Eppendorf Int., Hamburg, Germany) at 200 rpm and 37°C to provide the starter culture. The main culture was inoculated from the starter culture and grown in TB media (Terrific Broth), supplemented with 400 μg/mL ampicillin and 25 μg/mL kanamycin and incubated in a BIOSTAT Bplus Bioreactor (Sartorius Stedim Biotech, Goettingen, Germany) at stirrer rate 300 rpm, pH 7.2, gas flow rate 3 L/min, O2 setpoint: 20% and 37°C. The cell culture was induced by 0.6 mM IPTG when OD600 reached 0.8 and then incubated overnight at 25°C. Cells were harvested and centrifuged in an Avanti J‐E high‐speed centrifuge (Beckman Coulter Life Sciences, Brea, CA) at 8000 g for 15 minutes at 4°C. Cells were resuspended in lysis buffer (1.5 mM magnesium acetate, 1 mM calcium chloride, 250 mM sodium chloride, 100 mM ammonium sulfate, 40 mM disodium phosphate, 3.25 mM citric acid, 5% glycerol, 5 mM imidazole, 5 mM 2‐mercaptoethanol (BME), 0.08% n‐dodecyl‐beta‐maltoside (DDM), 1 mM phenylmethylsulfonyl fluoride (PMSF), 20 μM leupeptin), homogenized in an EmulsiFlex‐C5 High Pressure Homogenizer (Avestin, Ottawa, ON, Canada) and clarified in the centrifuge at 36000 g 4°C for 40 min. The solution containing expressed truncated RcsB N‐terminal domain was purified on an ÄKTAxpress™ (GE Healthcare Life Science, Piscataway, NJ) FPLC purification system by Ni‐NTA chromatography on a His Trap FF 5 mL column followed by size exclusion chromatography on a HiLoad 26/600 Superdex 200 prep grade column at 4°C. Impurities were washed on a His Trap FF column with loading buffer (10 mM Tris‐HCl, 500 mM sodium chloride, 5 mM BME, pH 8.3) and the protein construct was eluted in elution buffer (10 mM Tris‐HCl, 500 mM sodium chloride, 500 mM imidazole, pH 8.3). Before elution, an extra wash step with 25 mM imidazole in loading buffer was applied to remove other proteins. Finally, the purified truncated RcsB N‐terminal domain was collected in loading buffer and concentrated in Vivaspin Protein Concentrator Spin Columns with MWCO 10000 (GE Healthcare Life Science, Piscataway, NJ). Molecular weight and protein purity were analyzed by SDS‐PAGE (Bio‐Rad, Hercules, CA).
Crystallization and data collection
Crystallization experiments were set up using the crystallization robotic system Phoenix (Art Robbins Instruments, Sunnyvale, CA) on Corning 96‐well crystallization plates for the sitting‐drop vapor‐diffusion method at 19°C. Drops were composed of a 1:1 mixture of reservoir solution from the crystal screens Classics II, Classics Lite, Protein Complex, JCSG+, PACT, and PEGs II (Qiagen, Inc), as well as protein solution at a concentration of 6 mg/mL in loading buffer. A crystal of truncated RcsB N‐terminal receiver domain was obtained from the drop containing 3.5 M sodium formate pH 7.0. For data collection, the crystal was transferred into 4 M sodium formate for cryo‐protection and flash cooled in the liquid nitrogen. X‐ray diffraction data were collected at LS‐CAT 21ID‐F beamline of the Advance Photon Source (Argonne National Laboratory, Argonne, IL) with 0.97872 Å wavelength radiation. The diffraction data were integrated and merged with the HKL‐2000 program suite.46 The crystal space group, cell parameters, and X‐ray data collection statistics are summarized in Table 1.
Table 1.
The Crystal Space Group, Cell Parameters, X‐Ray Data Collection, Structure Refinement Statistics of the Atomic Model for the RcsB Receiver Domain
| Crystal parameters | |
| Resolution (last shell) (Å) | 50.0‐2.0 (2.03‐2.0) |
| Space group | P3221 |
| Unit cell parameters | |
| a, b, c (Å) | 61.2, 61.2, 65.2 |
| α, β, γ (°) | 90, 90, 120 |
| Matthews coefficient (Å3/Da) | 2.4 |
| Solvent content (%) | 49 |
| Data Collection | |
| Completeness (%) | 100 (100) |
| No of unique reflections | 9939 |
| I/σ(I) | 20.1 (2.7) |
| R merge (%) | 0.09 (0.6) |
| CC1/2 | 0.95 (0.82) |
| Redundancy | 8.6 (5.8) |
| Wilson B‐factor (Å2) | 33.1 |
| Refinement | |
| R (%)/R free (%) | 17.3/23.8 |
| RMSD bond length (Å) | 0.019 |
| RMSD bond angle (°) | 1.6 |
| Average B value (Å2) | 38 |
| Number of molecules in AU No of atoms | 1 |
| Protein | 1101 |
| Water molecules | 81 |
| Ramachandran analysis a | |
| Favored (%)/n | 97.7/129 |
| Allowed (%)/n | 2.3/2 |
| Outlier (%)/n | 1 |
RMSD stands for root‐mean‐square deviation. AU stands for asymmetric unit.
Defined by validation program MolProbity.
Structure determination and refinement
The structure of the RcsB N‐terminal receiver domain was determined by the molecular replacement method using BALBES, a molecular replacement pipeline.47 The best structure solution obtained from BALBES was rebuilt with the AutoBuild model‐building program in the PHENIX program suite.48 The structure was refined with REFMAC,49 followed by manual model refinement with Coot.50 The structure was validated using MolProbity 51 and the PDB ADIT validation server (http://deposit.pdb.org/validate/). Refinement and characterization of the atomic model for the RcsB receiver domain are summarized in Table 1. The figures were generated with CCP4mg52 and LigPlot+.53
PDB accession codes
The atomic coordinates and structure factors for the RcsB receiver domain are deposited into the Protein Data Bank (www.rcsb.org)54 with accession code 5I4C.
SEC‐MALS analysis
The size exclusion chromatography with multi‐angle light scattering (SEC‐MALS) experiment to obtain an absolute molecular weight of RcsB N‐terminal receiver domain was performed at Northwestern University (Evanston, IL) at the Keck Biophysics Facility, using a Waytt Dawn Heleos II multi‐angle scattering (MALS) detector (Wyatt Technology Europe GmbH, Dernbach, Germany) coupled with Agilent Technologies’ 1100 LC HPLC system (Agilent Technologies, Santa Clara, CA). Two samples of the purified RcsB receiver domain, at 1 and 3 mg/mL were prepared in a buffer containing 10 mM Tris‐HCl, 500 mM sodium chloride, 10 mM magnesium chloride, 0.5 mM DTT, pH 8.3. To prepare phosphorylated RcsB receiver domain, 1 mg/mL of the receiver domain was incubated for 2 h in the presence of 50 mM lithium potassium acetylphosphate (prepared freshly before use) and 10 mM magnesium chloride in a loading buffer following protocols described in 42. A total of 300 µL of RcsB receiver domain at 3 mg/mL or 1 mg/mL, or 250 µL of the phosphorylated RcsB receiver domain at 1 mg/mL were applied to a Superdex 75 10/300 GL column and equilibrated in 10 mM Tris‐HCl, 500 mM sodium chloride, 10 mM magnesium chloride, 0.5 mM DTT, pH 8.3 buffer (GE Healthcare, Piscataway, NJ) at a flow rate of 0.5 mL/min at room temperature. For a reference, lysozyme protein (Sigma‐Aldrich Corp., St. Louis, MO) was used. The data were processed with the program ASTRA (Wyatt Technology Europe GmbH, Dernbach, Germany).
Supporting information
Supporting Information Figure 1.
Acknowledgments
The data collection was performed at the LS‐CAT at the Advanced Photon Source. We would like to thank Thient Aung and the Keck Biophysics Facility at Northwestern University (Evanston, IL) for assistance with SEC‐MALS data collection.
References
- 1. Gottesman S, Trisler P, Torres‐Cabassa A (1985) Regulation of capsular polysaccharide synthesis in Escherichia coli K‐12: characterization of three regulatory genes. J Bacteriol 162:1111–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Majdalani N, Gottesman S (2005) The Rcs phosphorelay: a complex signal transduction system. Annl Rev Microbiol 59:379–405. [DOI] [PubMed] [Google Scholar]
- 3. Clarke DJ (2010) The Rcs phosphorelay: more than just a two‐component pathway. Future Microbiol 5:1173–1184. [DOI] [PubMed] [Google Scholar]
- 4. Evans KL, Kannan S, Li G, de Pedro M, Young KD (2013) Eliminating a set of four penicillin binding proteins triggers the Rcs phosphorelay and Cpx stress responses in Escherichia coli . J Bacteriol 195:4415–4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Laubacher ME, Ades SE (2008) The Rcs phosphorelay is a cell envelope stress response activated by peptidoglycan stress and contributes to intrinsic antibiotic resistance. J Bacteriol 190:2065–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cho SH, Szewczyk J, Pesavento C, Zietek M, Banzhaf M, Roszczenko P, Asmar A, Laloux G, Hov AK, Leverrier P, Van der Henst C, Vertommen D, Typas A, Collet JF (2014) Detecting envelope stress by monitoring β‐barrel assembly. Cell 159:1652–1664. [DOI] [PubMed] [Google Scholar]
- 7. Howery KE, Clemmer KM, Simsek E, Kim M, Philip N (2015) Regulation of the min cell division inhibition complex by the Rcs phosphorelay in Proteus mirabilis. J Bacteriol 197:2499–2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ferrières L, Clarke DJ (2003) The RcsC sensor kinase is required for normal biofilm formation in Escherichia coli K‐12 and controls the expression of a regulon in response to growth on a solid surface. Mol Microbiol 50:1665–1682. [DOI] [PubMed] [Google Scholar]
- 9. Fredericks CE, Shibata S, Aizawa S, Reimann SA, Wolfe AJ (2006) Acetyl phosphate‐sensitive regulation of flagellar biogenesis and capsular biosynthesis depends on the Rcs phosphorelay. Mol Microbiol 61:734–747. [DOI] [PubMed] [Google Scholar]
- 10. Hu LI, Chi BK, Kuhn ML, Filippova EV, Walker‐Peddakotla AJ, Bäsell K, Becher D, Anderson WF, Antelmann H, Wolfe AJ (2013) Acetylation of the response regulator RcsB controls transcription from a small RNA promoter. J Bacteriol 195:4174–4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wolfe AJ (2010) Physiologically relevant small phosphodonors link metabolism to signal transduction. Cur Opinion Microbiol 13:204–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Castanié‐Cornet MP, Cam K, Bastiat B, Cros A, Bordes P, Gutierrez C (2010) Acid stress response in Escherichia coli: mechanism of regulation of gadA transcription by RcsB and GadE. Nucleic Acid Res 38:3546–3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Salcheider SL, Jahn A, Schnetz K (2014) Transcriptional regulation by BglJ‐RcsB, a pleiotropic heteromeric activator in Escherichia coli . Nucleic Acid Res 42:2999–3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pannen D, Fabisch M, Gausling L, Schnetz K (2016) Interaction of the RcsB response regulator with auxiliary transcription regulators in Escherichia coli . J Biol Chem 291:2357–2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Majdalani N, Hernandez D, Gottesman S (2002) Regulation and mode of action of the second small RNA activator of RpoS translation, RprA. Mol Microbiol 46:813–826. [DOI] [PubMed] [Google Scholar]
- 16. Davalos‐Garcia M, Conter A, Toesca I, Gutierrez C, Cam K (2001) Regulation of osmC gene expression by the two‐component system rcsB‐rcsC in Escherichia coli. J Bacteriol 183:5870–5876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kuhne C, Singer HM, Grabisch E, Codutti L, Carlomango T, Scrima A, Erhardt M (2016) RflM mediates target specificity of the RcsCDB phosphorelay system for transcriptional repression of flagellar synthesis in Salmonella enterica. Mol Microbiol 101:841–855. [DOI] [PubMed] [Google Scholar]
- 18. Castanié‐Cornet MP, Treffandier H, Francez‐Charlot A, Gutierrez C, Cam K (2007) The glutamate‐dependent acid resistance system in Escherichia coli: essential and dual role of the His‐Asp phosphorelay RcsCDB/AF. Microbiol 153:238–246. [DOI] [PubMed] [Google Scholar]
- 19. Mouslim C, Latifi T, Groisman EA (2003) Signal‐dependent requirement for the co‐activator protein RcsA in transcription of the RcsB‐regulated ugd gene. J Biol Chem 278:50588–50595. [DOI] [PubMed] [Google Scholar]
- 20. Nassif X, Honoré N, Vasselon T, Cole ST, Sansonetti PJ (1989) Positive control of colanic acid synthesis in Escherichia coli by rmpA and rmpB, two virulence‐plasmid genes of Klebsiella pneumoniae. Mol Microbiol 3:1349–1359. [DOI] [PubMed] [Google Scholar]
- 21. Kelm O, Kiecker C, Geider K, Bernhard F (1997) Interaction of the regulator proteins RcsA and RcsB with the promoter of the operon for amylovoran biosynthesis in Erwinia amylovora. Mol Gen Genet 256:72–83. [DOI] [PubMed] [Google Scholar]
- 22. Krin E, Danchin A, Soutourina O (2010) RcsB plays a central role in H‐NS‐dependent regulation of motility and acid stress resistance in Escherichia coli . Res Microbiol 161:363–371. [DOI] [PubMed] [Google Scholar]
- 23. Brill JA, Quinlan‐Walshe C, Gottesman S (1988) Fine‐structure mapping and identification of two regulators of capsule synthesis in Escherichia coli K‐12. J Bacteriol 170:2599–2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Virlogeux I, Waxin H, Ecobichon C, Lee JO, Popoff MY (1996) Characterization of the rcsA and rcsB genes from Salmonella typhi: rcsB through tviA is involved in regulation of Vi antigen synthesis. J Bacteriol 178:1691–1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Latasa C, García B, Echeverz M, Toledo‐Arana A, Valle J, Campoy S, García‐del Portillo F, Solano C, Lasa I (2012) Salmonella biofilm development depends on the phosphorylation status of RcsB. J Bacteriol 194:3708–3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sun YC, Guo XP, Hinnebusch BJ, Darby C (2012) The Yersinia pestis Rcs phosphorelay inhibits biofilm formation by repressing transcription of the diguanylate cyclase gene hmsT. J Bacteriol 194:2020–2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chen J, Xie J (2011) Role and regulation of bacterial LuxR‐like regulators. J Cell Biochem 112:2694–2702. [DOI] [PubMed] [Google Scholar]
- 28. Pristovsek P, Sengupta K, Lohr F, Schafer B, von Trebra MW, Ruterjans H, Bernhard F (2003) Structural analysis of the DNA‐binding domain of the Erwinia amylovora RcsB protein and its interaction with the RcsAB box. J Biol Chem 278:17752–17759. [DOI] [PubMed] [Google Scholar]
- 29. Krissinel E (2010) Crystal contacts as nature's docking solutions. J Comput Chem 31:133–143. [DOI] [PubMed] [Google Scholar]
- 30. Laskowski RA, Macarthur MW, Moss DS, Thornton JM (1993) Procheck—a program to check the stereochemical quality of protein structures. J Appl Cryst 26:283–291. [Google Scholar]
- 31. Bent CJ, Isaacs NW, Mitchell TJ, Riboldi‐Tunnicliffe A (2004) Crystal structure of the response regulator 02 receiver domain, the essential YycF two‐component system of Streptococcus pneumoniae in both complexed and native states. J Bacteriol 186:2872–2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Luo SC, Lou YC, Rajasekaran M, Chang YW, Hsiao CD, Chen C (2013) Structural basis of a physical blockage mechanism for the interaction of response regulator PmrA with connector protein PmrD from Klebsiella pneumoniae. J Biol Chem 288:25551–25561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Podgornaia AI, Casino P, Marina A, Laub MT (2013) Structural basis of a rationally rewired protein‐protein interface critical to bacterial signaling. Structure 21:1636–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Usher KC, de la Cruz AF, Dahlquist FW, Swanson RV, Simon MI, Remington SJ (1998) Crystal structures of CheY from Thermotoga maritima do not support conventional explanations for the structural basis of enhanced thermostability. Protein Sci 7:403–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bachhawat P, Stock AM (2007) Crystal structures of the receiver domain of the response regulator PhoP from Escherichia coli in the absence and presence of the phosphoryl analog beryllofluoride. J Bacteriol 189:5987–5995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ahn DR, Song H, Kim J, Lee S, Park SY (2013) The crystal structure of an activated Thermotoga maritima CheY with N‐terminal region of FliM. Int J Biol Macromol 54:76–83. [DOI] [PubMed] [Google Scholar]
- 37. Leonard PG, Golemi‐Kotra D, Stock AM (2013) Phosphorylation‐dependent conformational changes and domain rearrangements in Staphylococcus aureus VraR activation. Proc Natl Acad Sci USA 110:8525–8530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Doi A, Nakamura H, Shiro Y, Sugimoto H (2015) Structure of the response regulator ChrA in the haem‐sensing two‐component system of Corynebacterium diphtheriae. Acta Cryst F71:966–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Baikalov I, Schröder I, Kaczor‐Grzeskowiak M, Grzeskowiak K, Gunsalus RP, Dickerson RE (1996) Structure of the Escherichia coli response regulator NarL. Biochemistry 35:11053–11061. [DOI] [PubMed] [Google Scholar]
- 40. Schnell R, Agren D, Schneider G (2008) 1.9 A structure of the signal receiver domain of the putative response regulator NarL from Mycobacterium tuberculosis. Acta Cryst F64:1096–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Park AK, Moon JH, Chi YM (2013) Crystal structure of the response regulator spr1814 from Streptococcus pneumoniae reveals unique interdomain contacts among NarL family proteins. Biochem Biophys Res Commun 434:65–69. [DOI] [PubMed] [Google Scholar]
- 42. Trajtenberg F, Albanesi D, Ruetalo N, Botti H, Mechaly AE, Nieves M, Aguilar PS, Cybulski L, Larrieux N, Mendoza D, Buschiazzo A (2014) Allosteric activation of bacterial response regulators: the role of the cognate histidine kinase beyond phosphorylation. Mbio 5:e02105–e02114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wehland M, Bernhard F (2000) The RcsAB box. Characterization of a new operator essential for the regulation of exopolysaccharide biosynthesis in enteric bacteria. J Biol Chem 275:7013–7020. [DOI] [PubMed] [Google Scholar]
- 44. Zavilgersky GB, Kotova YuV M IV (2003) The effects of regulatory proteins RcsA and RcsB on the expression of the Vibrio fischeri lux operon in Escherichia coli . Mol Biol 37:598–604. [PubMed] [Google Scholar]
- 45. Makowska‐Grzyska M, Kim Y, Maltseva N, Li H, Zhou M, Joachimiak G, Babnigg G, Joachimiak A (2014) Protein production for structural genomics using E. coli expression. Meth Mol Biol 1140:89–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Otwinowski Z, Minor W (1997) Processing of X‐ray diffraction data collected in oscillation mode. Methods Enzymol 276:307–326. [DOI] [PubMed] [Google Scholar]
- 47. Long F, Vagin AA, Young P, Murshudov GN (2008) BALBES: a molecular‐replacement pipeline. Acta Cryst D64:125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L‐W, Kapral GJ, Grosse‐Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH (2010) PHENIX: a comprehensive Python‐based system for macromolecular structure solution. Acta Cryst D66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA (2011) REFMAC5 for the refinement of macromolecular crystal structures. Acta Cryst D67:355–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Emsley P, Cowtan K (2004) Coot: model‐building tools for molecular graphics. Acta Cryst D60:2126–2132. [DOI] [PubMed] [Google Scholar]
- 51. Chen VB, Arendall WB, III , Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC (2010) MolProbity: all‐atom structure validation for macromolecular crystallography. Acta Cryst D66:12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Potterton L, McNicholas S, Krissinel E, Gruber J, Cowtan K, Emsley P, Murshudov GN, Cohen S, Perrakis A, Noble M (2004) Developments in the CCP4 molecular‐graphics project. Acta Cryst D60:2288–2294. [DOI] [PubMed] [Google Scholar]
- 53. Wallace AC, Laskowski RA, Thornton JM (1996) LIGPLOT: a program to generate schematic diagrams of protein‐ligand interactions. Protein Eng 8:127–134. [DOI] [PubMed] [Google Scholar]
- 54. Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE (2000) The Protein Data Bank. Nucleic Acid Res 28:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information Figure 1.


