Abstract
We propose a new method and optical instrumentation for mouse brain imaging based on extended-focus optical coherence microscopy. This in vivo imaging technique allows the evaluation of the cytoarchitecture at cellular level and the circulation system dynamics in three dimensions. This minimally invasive and non-contact approach is performed without the application of contrasting agents. The optical design achieved a resolution of 2.2 μm over a distance of 800 μm, which was sufficient to obtain a detailed three-dimensional image of a wild-type mouse’s brain down to the layer III of the cortex. Intrinsically contrasted microvessels and structures similar to the bodies of neurons were distinguishable.
OCIS codes: (110.4500) Optical coherence tomography, (110.3175) Interferometric imaging, (110.2945) Illumination design, (170.3880) Medical and biological imaging
1. Introduction
The recent application of optical imaging techniques to determine the structure and function of brain has provided new perspectives for the better understanding of the neural function, pathogenesis of disorders, and cognitive functions of the mammalian central nervous system [1, 2]. These methods offer unique possibilities when compared to other modalities recognized as gold standards in neuroimaging, like magnetic resonance imaging, positron emission tomography, or X-ray-based computer tomography. One of their main advantages is the broad range of contrast mechanisms that result from the detectable interactions of light with tissue, including those based on the intrinsic optical properties of brain tissue, like wavelength-dependent absorption, scattering, or autofluorescence [3, 4]. The use of exogenous fluorescent agents provides even more possibilities at the expense of the higher level of invasiveness of the procedure [5, 6]. Nevertheless, they are still significantly less invasive than those using ionizing radiation. Last, but not least, optical methods are relatively inexpensive in terms of infrastructure and utilization and therefore easily accessible and available for multimodal use.
Rodent models are the basic platform for brain research. A rat or mouse brain is very challenging in terms of effective optical three-dimensional (3D) imaging. Usually, only the cortex (the superficial 1.2 mm of the brain in mice) is optically accessible owing to the limited light penetration. In cerebral cortex research, two important interacting systems are of particular interest: the neuronal cytoarchitecture and the circulatory system. The physiological mutual dependence of these two systems is very strong and establishes the correct functioning of the brain [7]. Blood vessels occupy mostly the arachnoid layer and penetrate into the cortex. Neurons of different types are located within the cortex and show a diverse body structure and varying distribution according to depth, which allows the distinction between particular layers of the cortex. Comprehensive non-invasive simultaneous studies of the two systems still remain a challenge. An optical modality suitable for this task must address the following concerns:
-
1.
Both neurons and vessels in the brain are distributed over a wide range; therefore, a method must allow 3D imaging in significantly large volumes (at least 1 mm × 1 mm × 1 mm in the case of the mouse brain).
-
2.
The method must be applicable in vivo and it should preferably not require the use of exogenous contrast agents, which can perturb the physiological equilibrium and therefore make the interpretation of the results unclear. Intrinsic optical contrasting is preferable.
-
3.
The spatial resolution of the method must be of the order of micrometers to resolve both neurons and the smallest blood vessels.
-
4.
Owing to the exceptionally dynamic metabolism of the brain (especially in rodents), the speed of the setup must be sufficiently high to ensure an appropriate time resolution for the effective tracing of observable changes.
-
5.
The method must be non-contact for the comfort of the animal (reduction of extraneous uncontrollable stimuli) and support high ergonomics in the imaging system—especially by maintaining a sufficiently long working distance to manipulate the imaging subject and thus perform multiple imaging sessions using the same animal.
Previous literature shows that optical coherence microscopy (OCM) has a considerable potential in comprehensive rodent brain imaging, can deliver extensive useful information about its structure [8] and function [9, 10], and fulfills most of the mentioned prerequisites. Additionally, it has been successfully utilized in many diverse aspects of brain research [11].
OCM has been successfully applied for structural imaging of the murine cortex both in vitro and in vivo [12]. The precise design of the scanning protocols allows the effective measurement of blood dynamics in cortical vessels. Furthermore, Doppler techniques provide valuable quantitative information on the blood flow in particular vessels [13]. The use of a variety of techniques based on the correlation of spatially oversampled data allows the visualization of the net of vessels and provides detailed angiographic maps [14]. OCM has been applied to monitor changes in the brain due to conditions including stroke [15] and cancer [16].
One of the most interesting conclusions drawn from in situ studies is that OCM can provide the contrast of the somas of single neurons. Owing to their optical properties, the bodies of the neurons manifest themselves as darker regions in the bright background of the cortex. Extraordinarily detailed images of the cortex, which even allowed the analysis of the geometry of single pyramidal neurons or the distinction of myelinated axons, were provided by Srinivasan et al. [17] and Leahy et al. [18]. However, a high image quality required advanced averaging for effective speckle noise reduction. This was obtained by acquiring multiple 3D sets of data for different on-axis positions of the animal. This approach was very effective but required i) a greatly extended measurement time (assimilating the OCM measurement to an ordinary confocal microscopy study) and ii) a significant limitation to the comfort of the animal and a possibility of its manipulation by using a high-numerical-aperture immersion microscopic objective. It provoked our considerations on whether the two disadvantages can be effectively overcome.
In contrast to optical coherence tomography (OCT), the axial sectioning in OCM is limited by confocal gating, similarly to all other confocal microscopic systems. This fundamental limitation of standard OCM results from the application of illumination and detection of the fundamental spatial laser mode (the so-called Gaussian beam). In this case, the depth of focus (defined as twice the Rayleigh range) is very short; for example, for a scanning beam of the focus diameter (measured at 1/e2 of the peak intensity) of 2 µm corresponding depth of focus is only approximately 8 µm for a wavelength of 800 nm. Therefore, confocal gating requires mechanical scanning of the sample in the axial direction, which significantly increases the total acquisition time. To address this problem, the concept of extended focus has been successfully introduced in various applications of OCM. The focus extension has been provided by the use of a multimode waveguide for multi-focus illumination [19, 20], smart numerical refocusing using a synthetic aperture [21], aperture apodization [22], and a Bessel beam [23]. The Bessel beam can be produced using an axicon lens, a circular aperture, or a phase mask. In this case, extending the length of focus while preserving the axial sectioning by coherence gating is possible by a combination of several factors, including a Bessel-like beam cross section with a strong central peak of constant shape along the optical axis, a very low signal from the sample that eliminates the influence of side lobs, and high optical powers illuminating the sample to counterbalance the sensitivity loss owing to the Gaussian confocal gating. This concept has been already presented [24] and applied for quantitative cerebral blood flow imaging [25] and imaging of cerebral beta-amyloidosis [26].
The motivation for the work presented here is the validation of the extended focus approach using Bessel beam illumination for effective, fast, and high-resolution imaging of the murine brain that addresses simultaneously two aspects: cytoarchitecture visualization at the level of single neurons and circulation system dynamics analysis. We present high-resolution 3D images of the mouse cortex that contain characteristic details whose distribution and optical properties show substantial similarity to the bodies of neurons [18]. To the best of our knowledge, this is the first report of a non-contact method and instrumentation that can achieve the high-resolution imaging of the cortex microvessels of wild-type mouse and can provide images that allows the analysis of the 3D distribution of the structures that presumably are the bodies of neurons without the application of contrasting agents.
2. Materials and methods
2.1 Experimental setup
The experimental setup was designed as a platform for comprehensive structural and functional measurements of the mouse brain in vivo. It consisted of two optical modalities: an optical coherence scanning microscope and a brightfield microscope (Fig. 1). The two channels were coupled with the use of a dichroic mirror and employed the same objective lens.
Fig. 1.
(a) Experimental setup; (b) example of image obtained in the brightfield preview mode; (c) anesthetized animal during experimentation (distance between the tip of the microscopic objective and the cranial window: 10 mm).
Optical coherence microscope
The extended-focus Fourier domain OCM apparatus was based on the Mach-Zehnder interferometer. The general concept was derived from the previously reported work of Leitgeb et al. [24]. The light was divided into the object (70% of total power) and reference (30% of total power) arms using a fiber beam splitter (AC Photonics, USA). The reference arm was equipped with a gradient filter for controlling the intensity of the light, glass for matching the dispersion in both arms of the interferometer, and a combination of a beam splitter and a translational stage for adjusting the optical path length difference between the two arms.
In the object arm, the Bessel beam was created by passing a collimated beam of diameter 3.47 mm through an axicon lens (Thorlabs, USA, apex angle 176°). The beam was scaled and transferred by two telescopes to the focal plane of the tube lens. The tube lens transformed the beam to a light ring positioned at the aperture of the objective lens (Olympus RMS10X-PF, 10x, NA 0.3), which then created the actual Bessel beam at the object position. Between the lenses of the second telescope, a pair of galvo scanners was mounted to enable scanning in the two axes perpendicular to the optical axis and thus provided the advantage of 3D imaging. The working distance of the optical setup was as long as 10 mm, which significantly facilitated the manipulation of the animal. The microscope operated in the epi mode with standard Gaussian detection. The backscattered light was recombined with the light propagating in the reference arm and coupled to the spectrometer (diffraction grating: 1200 lpmm, Wasatch Photonics, camera: Basler Sprint, 70 kHz, 2048 pixels). The OCM instrument was designed to operate with a femtosecond laser (Fusion, Femtolasers; central wavelength: 795 nm, spectrum FWHM: 131 nm), resulting in an axial resolution of 2.4 μm (theoretical value: 2.17 μm). The power of the illumination beam at the sample was 11 mW. The sensitivity of the setup was approximately 92 dB.
Brightfield microscope
The brightfield channel was used for the precise positioning of the animal with respect to the OCM scanning area. The optical path included a telescope and a CCD web camera (LifeCam HD Cinema, Microsoft) with an adjustable focusing objective lens, which allowed the independent adjusting of the imaging plane for preview. Thus, the extended OCM focus could penetrate more deeply into the brain structure while the preview ensured the proper positioning of the animal based on the larger vessels located in the arachnoid. The accessible area of the brightfield preview was approximately twice as wide as the available OCM scanning area. The cold mirror was chosen to let the short-wavelength tail of the laser spectrum of the source be partly visible in the brightfield preview. Therefore, the actual area covered during the OCM measurement could be controlled on-line.
2.2 Bessel beam
The Bessel beam was generated by the axicon lens and transferred to the object using a set of telescopes. The annular aperture was inserted in the first telescope to block unwanted residual rays coming from the imperfect axicon tip. In the proposed geometry, the measured value of the width of the central peak of the Bessel beam was smaller than 2.2 μm along a path of 800 µm. The point-spread function (PSF) of the system was characterized by the analysis of 3D images of a dedicated phantom (National Physics Laboratory, United Kingdom). The device consisted of iron dioxide particles embedded in a transparent solidified polymer. The mean diameter of the particles (200 nm) was significantly smaller than the expected resolution. The object was raster-scanned with an interval of 0.275 µm to obtain adequately dense spatial sampling. The Z-projection of this set of data provided the lateral profile of the imaging beam and revealed a characteristic Bessel pattern. The effective spatial resolution was defined by the FWHM of the central intensity peak and it was measured to have a value <2.2 µm over the distance of 800 µm, reaching a minimum of 1.75 µm (theoretical value: 1.64 µm).
The phantom used for the PSF measurement is shown in Fig. 2(a), while the schematic of Fig. 2(b) shows the area of convergence of the conical beam and how it was observed inside the phantom. The ring is a cross section of the conical beam projected at the bottom of the phantom. The position of the actual focus of the Bessel beam is evident. Figure 2(c) presents an example of a B-scan of the phantom with color-coded depth, in which single scatterers are localized. In Fig. 2(d), the Z-projection of the full set of B-scans is shown with the same color-coding of the depth.
Fig. 2.
(a) The phantom used for the PSF measurement; (b) schematic of the Bessel beam geometry and photograph of the Bessel beam scattered in the phantom used for the measurement of PSF; (c) structural B-scan (logarithmic scale) of the phantom with color-coded depth; (d) en face image of the artifact, depth coded as in (c) Scale bar: 20 μm.
Figure 3 contains the reconstructed profiles of the Bessel beam measured at different depths. The intensity is presented in logarithmic scale. The width of the central peak does not exceed 2.2 µm throughout the distance of approximately 800 µm. On the other hand, the intensity of the central peak varies significantly over this distance. The images are not intensity-normalized for a reliable presentation of this effect. The brightness of the central peak reaches a maximum at a depth of 400 µm and decreases slightly asymmetrically on both sides of this level, as expected from the theory of the formation of the Bessel beam. However, the reduction of the signal is mostly caused by the detection rather than the illumination optics. The use of an axicon lens results in the need of decoupling the illumination and detection paths; this was obtained by using the Mach-Zehnder interferometer. This necessity is caused by the significant loss of signal light intensity in the back-propagation path of the light through the axicon lens and the poor coupling efficiency of the light to the fiber in case of the Bessel beam [24]. In effect, the detection path was consistent with Gaussian optics. The sensitivity depends on the product of the illumination and detection 3D point spread functions, which results in the combination of the advantages of using a Bessel beam and the drawbacks of Gaussian detection optics, which provides a reasonable compromise.
Fig. 3.
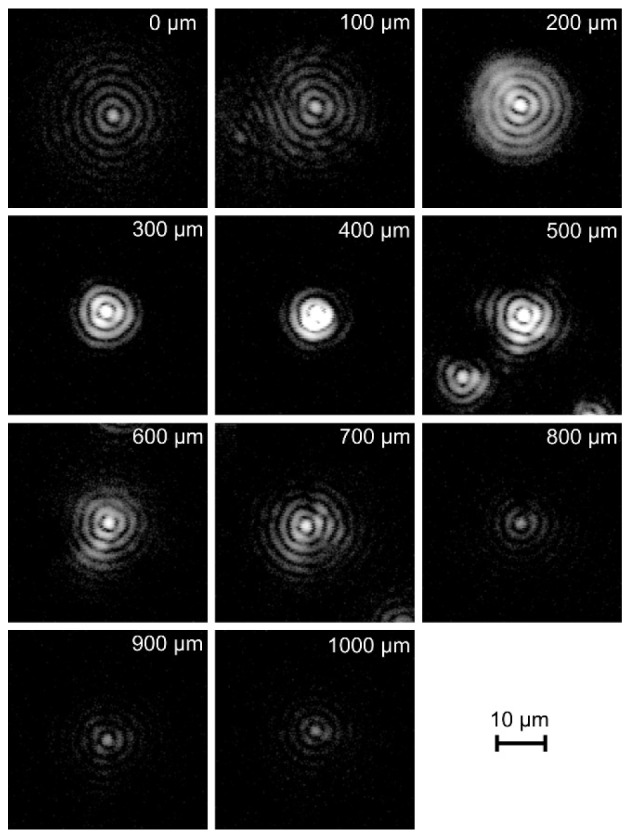
PSF cross sections of the optical setup reconstructed at variable depths (logarithmic intensity scale).
2.3 Scanning protocols
The scanning protocol was designed to simultaneously provide high-resolution spatial imaging and flow mapping. The flow measurements were performed with the use of differential algorithms, which required multiple acquisitions of B-scans for every slow-scanner-axis position and further pointwise analysis of the changes in the complex signal [27]. In consecutively acquired B-scans, the phase variance for each point was calculated, providing contrast for the dynamic changes due to the blood flow. The B-scans in the analyzed set required pre-alignment, which was based on the signal from an automatically detected static structure.
The scanning protocol was 300 A-scans × 300 B-scans (corresponding to 1.1 mm × 1.1 mm). Six B-scans were acquired before the scanner deflected the beam to the next slow-axis-scanner position. Thus, a four-dimensional data set was acquired for every measurement, allowing pointwise speckle decorrelation analysis for angiographic imaging. The repetition time for every A-scan was 20 μs. The total time of scanning of the sample was 9.7 s, while the total measurement time, including data transfer and saving to disk, was 18 s.
2.4 Animal preparation
Young adult mice (6–10 weeks) were used in the experiments. They were prepared for imaging by implanting a cranial window centered on the barrel cortex. The mice were placed in a stereotaxic frame and were deeply anaesthetized with isoflurane (4% for induction, 1.5–2% for surgery). Dexamethasone (0.2 mg/kg) and Carprofen (5 mg/kg) were administered subcutaneously to prevent swelling of the brain and inflammatory response. A warming pad with an anal probe was used for monitoring the body temperature. After removing the skin over the top of the skull, a drop of lidocaine (1%)-epinephryne (1:100000) solution was applied onto the periosteum to avoid excessive bleeding or pain. A circle of approximately 4 mm diameter was removed from the skull and the dura was exposed. An optical chamber was then constructed by covering the intact dura with 1.2% low-melting-point agarose (Sigma-Aldrich, USA/Germany) and a 5-mm-diameter cover glass glued with cyanoacrylate-based glue and sealed with dental acrylic.
The actual OCM measurement was performed after a recovery of at least one week after the cranial window installation. The animal was placed in a stereotaxic frame with the warming pad and was deeply anaesthetized with isoflurane (4% for induction, 1.5–2% for actual procedure). The stereotaxic frame was positioned with the use of a custom-made 5-axis platform. The windowed part of the animal’s brain was accessed by the extended focus of the imaging system. The long working distance of the optical setup allowed the precise and comfortable manipulation of the animal.
The animal procedures were approved by the National Bioethical Committee for Research on Animals of the Nencki Institute of Experimental Biology.
3. Results
3.1 Structural imaging
The extended focus provided detailed images of the brain structure. The light penetrated effectively through the highly scattering region of the dura, reaching layer II/III of the cortex. The images reveal a distinct network of dark spots clearly contrasting with the bright background of the cortex. We consider that these structures correspond to neuron bodies and we hereafter refer to them as such; we discuss the validity of this assumption in the Conclusions section. To extract these fine structures from the speckle noise, we used a speckle contrast reduction algorithm based on averaging of the registered data. The selected scanning protocol delivered highly oversampled data that could be used for effective spatial and temporal speckle averaging.
Temporal averaging was effective owing to the bulk movements of the animal’s head caused by breathing and the heartbeat. The amplitude of those movements appeared to be sufficient to decorrelate speckles between the frames. Even the most careful fastening of the animal’s head in the stereotactic frame did not prevent this motion. Although this was obviously problematic for angiographic analysis, as a positive side-effect, it allowed the averaging of the speckles. The resolution loss due to these movements is difficult to determine because of its random nature; however, it is significantly smaller than the resolution loss due to the spatial averaging. Time averaging was possible in the short and long scale. In the short scale, the averaging of the intensity images was performed using data from one set of six repeated B-scans (time interval of the order of 6 ms). However, the time between averaged frames was not sufficiently long to provide any substantial decorrelation of the speckles. Averaging between 3D sets of data obtained the expected improvement. In this case, the time interval between averaged frames was approximately 18 s. The structural images presented in this manuscript are the result of averaging five consecutive 3D data sets.
On the other hand, the enhancement of the contrast of the cortex cytoarchitecture details could be obtained by averaging the data in the spatial dimension. In this case, the discrete nature of the acquired data set implied a minimum resolution loss of the order of the distance between the measurement points, which was approximately 3.67 µm in the chosen scanning protocol. The averaging of five adjacent B-scans increased the decorrelation distance to 14.7 µm, which corresponds to an effective lateral resolution in the slow-scanner axis. We determined that this number was optimal in terms of a satisfactory trade-off between the resolution and the gain in the signal-to-noise ratio.
The criterion used to determine the parameters applied in the averaging procedure was the visibility of the objects of interest, which allowed the visual inspection of their number for quantitative analysis. We determined that none of the two averaging strategies alone provided satisfactory results. Therefore, the final averaging scheme included both approaches.
Figure 4 presents the effect of the averaging of the intensity images. We selected to apply the mean-intensity projection method. The resulting cross-sectional images reveal the stratified structure of the brain in the proximity of a big cerebral artery, with its cross section shown on the right side of the image. The bottom surface of the glass coverslip, dura, arachnoid, and cortex are easily visible. As an example of the effect of averaging overlapping frames within a single 3D cubic volume, a single B-scan is presented in Fig. 4(a). In Fig. 4(b), we display the same image smoothened by spatial averaging of five adjacent B-scans. Figure 4(c) presents the effect of averaging of the B-scans in time, which resulted in further speckle noise decrease and the enhancement of the contrast of the structures of interest. The dynamic fly-through projection along the slow-scanner axis of the despeckled 3D data set allows even better perception of the details of the structure (see Visualization 1 (13.4MB, MP4) ).
Fig. 4.
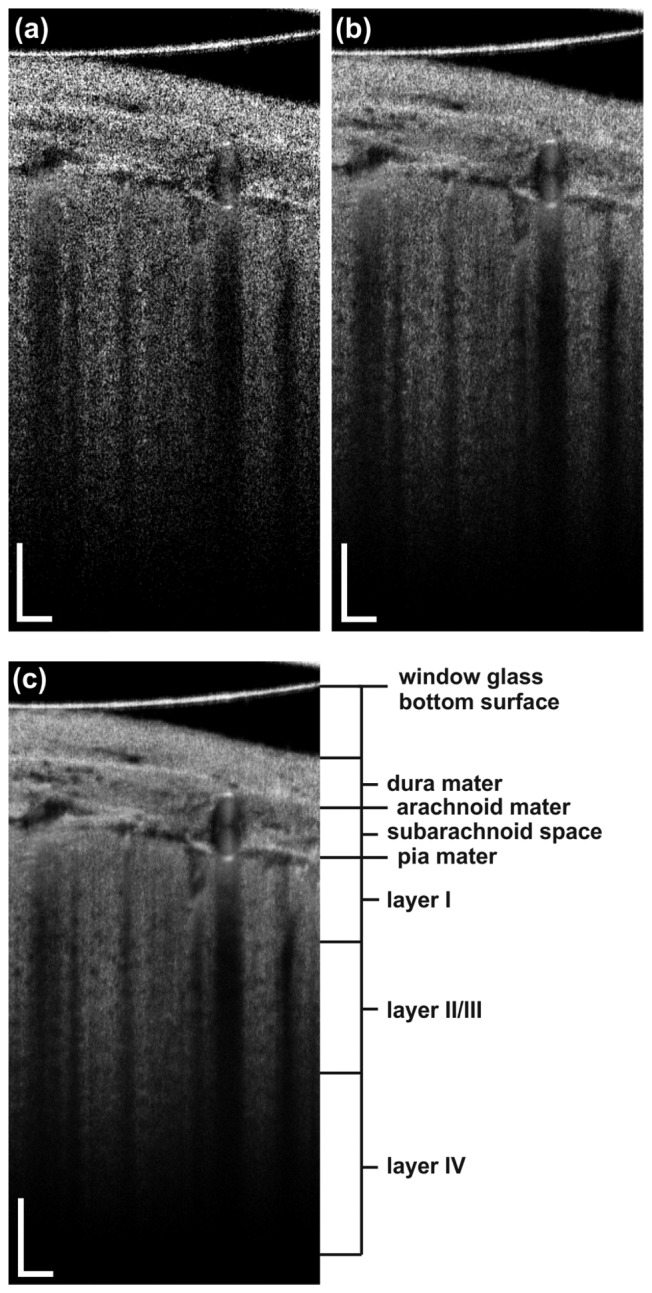
(a) Structural B-scan of the mouse brain (b) after spatial averaging and (c) after spatio-temporal averaging. Scale bars: 100 µm. A fly-through projection across the whole 3D structural data set represented by (c) is presented in Visualization 1 (13.4MB, MP4) .
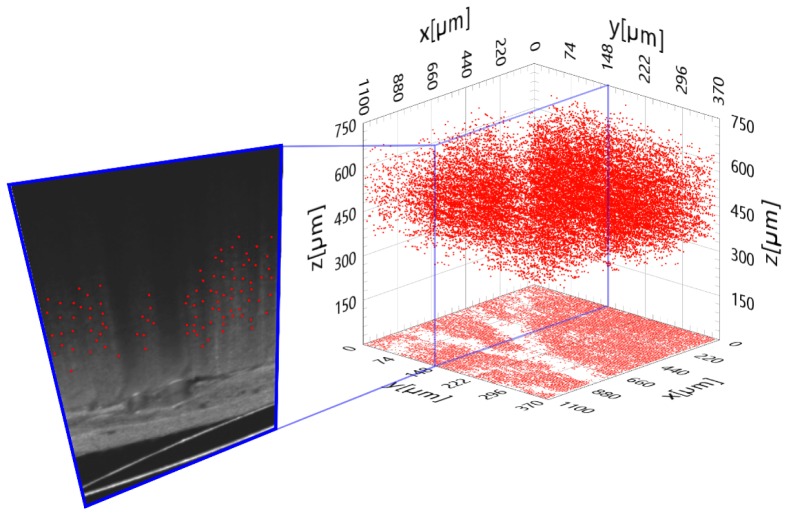
The quality of the obtained images was sufficient for statistical analyses of the cytoarchitecture of the II/III layer of the mouse cortex. The dimensions of the supposed single neuron body could only be estimated approximately and were of the order of 10 μm, which is consistent with their known values. We performed a manual localization of the centers of those objects as the obtained contrast was not sufficiently high to apply effectively any automatic segmentation algorithm. Additionally, the significant number of blood vessels casting shadows on the structure beneath them further impeded this task. The localization allowed the 3D visualization of the distribution of the objects within the accessible region of the cortex. In Fig. 5, we present the result of this procedure performed on a cuboid of 1.10 mm × 0.37 mm × 0.75 mm.
Fig. 5.
Spatial distribution of the neuron-like structures detected in the mouse cerebral cortex.
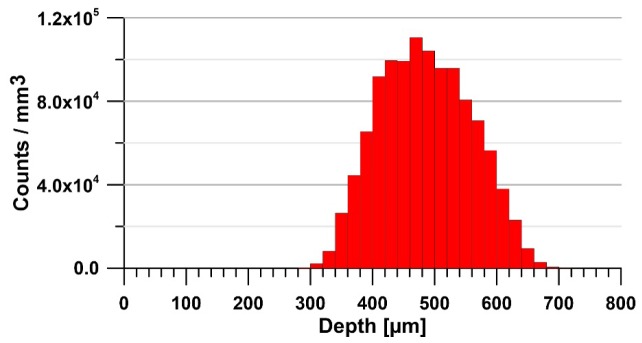
The localized counts were used to calculate their average density at different depths. It was obtained by integrating the counts in the XY plane for a 20-μm-thick slice. The value was corrected by considering the ratio of the shadowed (by blood vessels) part of the structure to the well-exposed one. The resulting plot is shown in Fig. 6. The presumed neurons were found starting at a depth of approximately 280 μm from the surface of the dura, which corresponds to the border of the II/III cortical layer, where the pyramidal neurons are normally localized. At a depth of approximately 480 μm, the average density reaches its maximum of approximately 1.1 × 105/mm3, which matches quite well the neuron density in this layer reported in the literature (approximately 1.37 × 105/mm3) [28]. The decrease of the density in deeper regions (layer IV of the cortex) disagrees with the expected increase to approximately 1.8 × 105/mm3. The observed decrease is caused by two main factors. First, the average size of the neurons normally decreases at this depth [17], which makes them less detectable at a given resolution. Second, high scattering within the entire cerebral column causes a significant OCM signal drop and the structures of interest lose contrast and can no longer be reliably recognized. In effect, the signal-to-noise ratio in the deeper cortex regions decreases to a level where the proposed quantitative analysis becomes unreliable. In fact, the histogram in Fig. 6 shows that the calculated values of this parameter can be considered as close to the values expected for neurons distribution only within the range of approximately 200 µm. In the deeper sectors, it is underestimated.
Fig. 6.
Mean density of the counted neuron-like structures versus depth.
A more detailed analysis of the data also allows the production of two-dimensional maps of the densities of the discussed dark object revealed in the cortex. The number of counts in voxels of 20 μm × 20 μm × 20 μm at different depths is presented in Fig. 7. Here, the effects of shadowing of the blood vessels have not been corrected. The figure demonstrates the variation of the number of the detected structures at the examined interfaces of the cortex and their distribution in the XY plane.
Fig. 7.
Two-dimensional neuron density maps for various depths (indicated in the top structural B-scan).
The quantitative analysis of the neuron density in vivo is undeniably a very challenging task and the discussed weaknesses of the data provided by our setup obtains it with limited precision and in a limited range. Nevertheless, the ability to provide this level of quality for this type of analysis in a non-invasive manner and with the achieved time resolution remains unique. We believe that the advantages of the method can be more easily appreciated in the quantitative regime. If the assumption that the detected structures are in fact neuron bodies holds, the analysis of the relative changes of the number of neurons as a function of time or of the localized flow is feasible, e.g., for monitoring the progress of brain necrosis in the case of a stroke or any other injury involving neuron death.
3.2 Flow mapping
The result of the contrasting of the blood flow by analyzing the phase variance is presented in Fig. 8(a). In this example B-scan, the detected dynamic signal is depicted in red. It consists of one part corresponding to the actual movement of the blood and one originating from the static structure below the vessel. Figure 8(b) presents the combination of the angiographic B-scan and the corresponding structural image, which facilitates the distinction between the two parts.
Fig. 8.
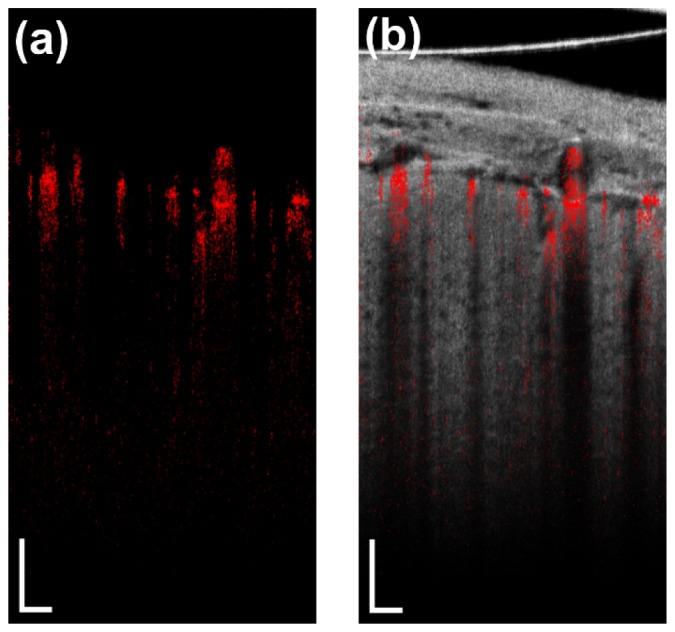
a) Angiographic B-scan; b) angiographic B-scan overlaid on the corresponding structural image.
It is especially informative to examine the flow maps in wide-angle en face projections. To provide a wide-field projection of the brain while maintaining high lateral resolution, the image was built with a set of tiles corresponding to 3D data sets acquired separately. The tiles had lateral dimensions 1.1 mm × 1.1 mm and partially overlapped with each other to facilitate alignment and stitching. For each tile, the animal was mechanically positioned with the use of a translational platform on which the stereotactic frame was mounted. The en face projection was produced by integrating the signal along the Z-axis for every A-scan, which resulted in a single point of the final projection. Representative maps—structural and angiographic—are shown in Fig. 9. The extended focus allowed the detailed projection of the vessels from the entire volume of the covered region at different depths. The use of the extended-focus setup provided the fast acquisition of the required series of 3D data sets without readjusting the axial position of the mouse’s brain. Moreover, as Fig. 4 suggests, the head of the animal was tilted with reference to the horizontal line and the surface of the brain showed some curvature. In the case of short focus, the focal plane would cause uneven sharpness at different depths from the surface of the brain, which would make the interpretation of the images difficult.
Fig. 9.

Wide-field maps of a) the structure and b) the flow of the mouse brain. Scale bar: 200 μm.
4. Conclusions
In this manuscript, we report on the performance of a custom-made extended-focus OCM setup for mouse brain imaging. This approach maintained a resolution of 2.2 μm over a distance of 800 µm, which is sufficient to penetrate layer II/III of the mouse cerebral cortex. It is particularly worth noting that the Bessel beam in our setup could penetrate the highly scattering and relatively thick dura to provide a signal from the cortex at a reasonable sensitivity level. The non-removal of the dura during animal preparation is much less invasive than the opposite case and is therefore highly desirable, particularly in long-term studies of one animal. The investigation of the changes in a mouse cerebral cortex in a period of weeks seems to be feasible in terms of survival rate of the animals by using the procedure of cranial window insertion.
Additionally, we demonstrate that the time resolution of the method provides 3D sets of data covering 1.1 mm × 1.1 mm brain areas every 18 s and allows to trace dynamic changes in the brain with this time resolution. The method seems very well suited for application to systematic real-time studies of the brain e.g. during and after ischemic stroke. As we showed, obtained 3D data sets may contain detailed information on both: the structure and the dynamics of the cortex. The method allowed the detection of the objects that we assume to be single neuron bodies in layer II/III of the cortex. These objects can be statistically analyzed. The observed structures showed substantial similarity to what has been reported in the literature as the manifestation of neuron bodies in terms of distribution statistics and optical properties. The effective resolution of the averaged structural images was not high enough for detailed cytoarchitecture visualization and analysis so as to allow a direct validation of our assumption by e.g. a single-neuron-geometry study. However, in contrast to the cited studies [17, 18] our solution i) does not require mechanical scanning in the axial direction and is therefore much faster, allowing the dynamic study of the properties of the cortex cytoarchitecture and blood flow, ii) ensures a much higher working distance (10 mm, no immersion needed), which is favorable for measurements requiring more advanced animal handling, and iii) allows imaging while preserving the dura. To the best of our knowledge, the instrument and methodology presented here is the first one exhibiting this particular set of properties and fills a significant gap in the research on the structure and function of the murine brain.
We do not provide a direct validation of our assumption regarding the correspondence between the dark-spot field in the cortex with the bodies of neurons. We assume the results provided by the group of V. Srinivasan [17, 18] to be representative in terms of the optical properties of neuron bodies. The cited research was performed using rats, while we examined mice. The neuron distributions in the volume of the cortex differ between the two species but the optical properties remain very similar and the same appearance can be expected in optically registered images. Because the observed structure details show very similar contrast to what was presented in the previous work, we postulate per analogiam that the details in the cortex observed in our images are in fact neuron bodies. We invite the reader to interpret the presented images and analysis that produced our arguments which, although formulated in the subjunctive mood, we consider as relatively strong. We believe that such discussion is valuable and provides a significant contribution to the field.
The validation of the results could be performed with the use of a fluorescence microscope. The unique properties of the extended-focus setup result from its specific optical design, which cannot be easily transformed to one allowing fluorescence imaging. Fundamentally, to observe cell bodies in vivo, a fluorescence imaging system with high resolution and penetration depth would require the use of an immerse objective, which is impossible to incorporate in the current Bessel beam setup. Any other solution, including histology or correlative imaging by two instruments, would not ensure the imaging of identical volumes and the validation of the method would also be questionable.
The quality of the neuron density maps highly depends on the performance of the system, especially its characteristic sensitivity variation in the axial direction. It is significantly affected by the limitations resulting from Gaussian optics detection. The challenge of overcoming this limitation seems to be one of the most significant ones in terms of taking full advantage of the use of Bessel beam illumination and will be examined in our future studies.
Acknowledgments
We would like to thank Danuta Bukowska-Sampson for her involvement in the early stage of the project, fruitful discussions, and valuable suggestions.
Funding
Polish National Center for Research and Development (NCBR) (PBS1/A9/20/2013); Polish Ministry of Science and Higher Education (“Iuventus Plus” program for years 2015–2017 grant IP 2014 045973). Polish National Science Center (NCN) (MAESTRO Program DEC-2011/02/A/ST2/00302).
References and links
- 1.Hillman E. M. C., “Optical brain imaging in vivo: techniques and applications from animal to man,” J. Biomed. Opt. 12(5), 051402 (2007). 10.1117/1.2789693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.In Vivo Optical Imaging of Brain Function (CRC Press, 2009). [PubMed] [Google Scholar]
- 3.Grinvald A., Lieke E., Frostig R. D., Gilbert C. D., Wiesel T. N., “Functional architecture of cortex revealed by optical imaging of intrinsic signals,” Nature 324(6095), 361–364 (1986). 10.1038/324361a0 [DOI] [PubMed] [Google Scholar]
- 4.Optical Methods and Instrumentation in Brain Imaging and Therapy (Springer, New York, 2013). [Google Scholar]
- 5.Hong G., Diao S., Chang J., Antaris A. L., Chen C., Zhang B., Zhao S., Atochin D. N., Huang P. L., Andreasson K. I., Kuo C. J., Dai H., “Through-skull fluorescence imaging of the brain in a new near-infrared window,” Nat. Photonics 8(9), 723–730 (2014). 10.1038/nphoton.2014.166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Susaki E. A., Tainaka K., Perrin D., Kishino F., Tawara T., Watanabe T. M., Yokoyama C., Onoe H., Eguchi M., Yamaguchi S., Abe T., Kiyonari H., Shimizu Y., Miyawaki A., Yokota H., Ueda H. R., “Whole-Brain Imaging with Single-Cell Resolution Using Chemical Cocktails and Computational Analysis,” Cell 157(3), 726–739 (2014). 10.1016/j.cell.2014.03.042 [DOI] [PubMed] [Google Scholar]
- 7.Liao L.-D., Tsytsarev V., Delgado-Martínez I., Li M.-L., Erzurumlu R., Vipin A., Orellana J., Lin Y.-R., Lai H.-Y., Chen Y.-Y., Thakor N. V., “Neurovascular coupling: in vivo optical techniques for functional brain imaging,” Biomed. Eng. Online 12(1), 38 (2013). 10.1186/1475-925X-12-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choi W. J., Wang R. K., “Swept-source optical coherence tomography powered by a 1.3-μm vertical cavity surface emitting laser enables 2.3-mm-deep brain imaging in mice in vivo,” J. Biomed. Opt. 20(10), 106004 (2015). 10.1117/1.JBO.20.10.106004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Watanabe H., Rajagopalan U. M., Nakamichi Y., Igarashi K. M., Kadono H., Tanifuji M., “Functional optical coherence tomography of rat olfactory bulb with periodic odor stimulation,” Biomed. Opt. Express 7(3), 841–854 (2016). 10.1364/BOE.7.000841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Devor A., Sakadžić S., Srinivasan V. J., Yaseen M. A., Nizar K., Saisan P. A., Tian P., Dale A. M., Vinogradov S. A., Franceschini M. A., Boas D. A., “Frontiers in optical imaging of cerebral blood flow and metabolism,” J. Cereb. Blood Flow Metab. 32(7), 1259–1276 (2012). 10.1038/jcbfm.2011.195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu G., Chen Z., “Optical Coherence Tomography for Brain Imaging,” in Optical Methods and Instrumentation in Brain Imaging and Therapy, Madsen J. S., ed. (Springer; New York, New York, NY, 2013), pp. 157–172. [Google Scholar]
- 12.Chong S. P., Merkle C. W., Cooke D. F., Zhang T., Radhakrishnan H., Krubitzer L., Srinivasan V. J., “Noninvasive, in vivo imaging of subcortical mouse brain regions with 1.7 μm optical coherence tomography,” Opt. Lett. 40(21), 4911–4914 (2015). 10.1364/OL.40.004911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.You J., Du C., Volkow N. D., Pan Y., “Optical coherence Doppler tomography for quantitative cerebral blood flow imaging,” Biomed. Opt. Express 5(9), 3217–3230 (2014). 10.1364/BOE.5.003217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baran U., Wang R. K., “Review of optical coherence tomography based angiography in neuroscience,” Neurophotonics 3(1), 010902 (2016). 10.1117/1.NPh.3.1.010902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baran U., Li Y., Wang R. K., “Vasodynamics of pial and penetrating arterioles in relation to arteriolo-arteriolar anastomosis after focal stroke,” Neurophotonics 2(2), 025006 (2015). 10.1117/1.NPh.2.2.025006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vakoc B. J., Lanning R. M., Tyrrell J. A., Padera T. P., Bartlett L. A., Stylianopoulos T., Munn L. L., Tearney G. J., Fukumura D., Jain R. K., Bouma B. E., “Three-dimensional microscopy of the tumor microenvironment in vivo using optical frequency domain imaging,” Nat. Med. 15(10), 1219–1223 (2009). 10.1038/nm.1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Srinivasan V. J., Radhakrishnan H., Jiang J. Y., Barry S., Cable A. E., “Optical coherence microscopy for deep tissue imaging of the cerebral cortex with intrinsic contrast,” Opt. Express 20(3), 2220–2239 (2012). 10.1364/OE.20.002220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leahy C., Radhakrishnan H., Srinivasan V. J., “Volumetric imaging and quantification of cytoarchitecture and myeloarchitecture with intrinsic scattering contrast,” Biomed. Opt. Express 4(10), 1978–1990 (2013). 10.1364/BOE.4.001978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lorenser D., Yang X., Sampson D. D., “Ultrathin fiber probes with extended depth of focus for optical coherence tomography,” Opt. Lett. 37(10), 1616–1618 (2012). 10.1364/OL.37.001616 [DOI] [PubMed] [Google Scholar]
- 20.Yin B., Chu K. K., Liang C.-P., Singh K., Reddy R., Tearney G. J., “μOCT imaging using depth of focus extension by self-imaging wavefront division in a common-path fiber optic probe,” Opt. Express 24(5), 5555–5564 (2016). 10.1364/OE.24.005555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ralston T. S., Marks D. L., Carney P. S., Boppart S. A., “Interferometric synthetic aperture microscopy,” Nat. Phys. 3(2), 129–134 (2007). 10.1038/nphys514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu L., Gardecki J. A., Nadkarni S. K., Toussaint J. D., Yagi Y., Bouma B. E., Tearney G. J., “Imaging the subcellular structure of human coronary atherosclerosis using micro-optical coherence tomography,” Nat. Med. 17(8), 1010–1014 (2011). 10.1038/nm.2409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McGloin D., Dholakia K., “Bessel Beams: Diffraction in a new light,” (2005).
- 24.Leitgeb R. A., Villiger M., Bachmann A. H., Steinmann L., Lasser T., “Extended focus depth for Fourier domain optical coherence microscopy,” Opt. Lett. 31(16), 2450–2452 (2006). 10.1364/OL.31.002450 [DOI] [PubMed] [Google Scholar]
- 25.Bouwens A., Bolmont T., Szlag D., Berclaz C., Lasser T., “Quantitative cerebral blood flow imaging with extended-focus optical coherence microscopy,” Opt. Lett. 39(1), 37–40 (2014). 10.1364/OL.39.000037 [DOI] [PubMed] [Google Scholar]
- 26.Bolmont T., Bouwens A., Pache C., Dimitrov M., Berclaz C., Villiger M., Wegenast-Braun B. M., Lasser T., Fraering P. C., “Label-free imaging of cerebral β-amyloidosis with extended-focus optical coherence microscopy,” J. Neurosci. 32(42), 14548–14556 (2012). 10.1523/JNEUROSCI.0925-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fingler J., Schwartz D., Yang C., Fraser S. E., “Mobility and transverse flow visualization using phase variance contrast with spectral domain optical coherence tomography,” Opt. Express 15(20), 12636–12653 (2007). 10.1364/OE.15.012636 [DOI] [PubMed] [Google Scholar]
- 28.DeFelipe J., Alonso-Nanclares L., Arellano J. I., “Microstructure of the neocortex: comparative aspects,” J. Neurocytol. 31(3-5), 299–316 (2002). 10.1023/A:1024130211265 [DOI] [PubMed] [Google Scholar]