Abstract
Emergence of antibiotic resistance is an example of the incredible plasticity of bacteria to survive in all environments. The search for new antibiotics active against traditional targets is more challenging due not only to the lack of novel natural products to fulfill the current clinical needs against multidrug-resistant (MDR) bacteria, but also for the possible “collateral” effects on the human microbiota. Thus, non-traditional approaches to combat MDR bacteria have been proposed. Here, we discuss the possibility of targeting the membrane response to the antibiotic attack (cell membrane adaptation) as a viable strategy to increase the activity of current antimicrobials, enhance the activity of the innate immune system and prevent development of resistance during therapy using the three-component regulatory system LiaFSR of enterococci as a model.
Introduction
It has long been recognized that the bacterial response to antibiotics is one of the most important evolutionary phenomena with major clinical consequences in the history of medicine. Antibiotics are rapidly disappearing from the clinical armamentarium posing immense challenges in the care of patients. The dwindling pipeline of new antimicrobial compounds available to treat multidrug-resistant (MDR) bacteria suggests that the current approaches to develop antibacterial agents are not sustainable [1, 2]. In recent years, the use of next-generation sequencing, combined with rational drug design, has broadened the search for novel compounds and targets [3, 4]. This approach could potentially open the door to new therapeutic strategies by targeting novel bacterial systems involved in essential cellular processes such as cell division or maintaining membrane integrity. Here, we review the rationale to target a signal transduction system involved in protecting the cell envelope against the antimicrobial attack. We postulate that this approach may restore the activity of currently used antibiotics and also likely favor clearance of the organism by the innate immune system.
LiaFSR system as a master regulator of the cell membrane stress response to the antimicrobial peptide attack
The LiaFSR system is a major player in the response against lipid II interfering antibiotics (particularly bacitracin and vancomycin) in Bacillus subtilis [5]. This system has subsequently been shown to orchestrate the response to membrane active agents including organic solvents, detergents and cationic antimicrobial peptides (e.g. daptomycin [DAP]) [6–9]. In general, LiaFSR is organized into an operon with at least three genes, i) liaS, which encodes a bifunctional sensor histidine-kinase [HK]/phosphatase, ii) liaR, encoding the response regulator of the system and iii) liaF, which encodes a transmembrane domain protein that negatively regulates the transcriptional effects of LiaR, presumably by locking LiaS into a “phosphatase mode” [10, 11]. Of interest, the LiaFSR system is highly conserved across the low G+C bacteria, a group that contains many relevant human pathogens including Staphylococcus aureus, enterococci, streptococci and Listeria monocytogenes (Table 1).
Table 1.
Amino acid identity of Bacillus subtilis LiaR homologs in clinically relevant Gram-positive organisms
| Organism | Identity (%) |
|---|---|
| Enterococcus faecalis | 62 |
| Enterococcus faecium | 64 |
| Lactococcus lactis | 53 |
| Staphylococcus aureus | 53 |
| Staphylococcus epidermidis | 51 |
| Staphylococcus lugdunensis | 52 |
| Streptococcus mutans | 55 |
| Streptococcus pneumoniae | 63 |
| Streptococcus pyogenes | 56 |
VraSR is the homolog of LiaSR in S. aureus, and is part of a 4 gene operon, vraUTSR [12], that is induced by exposure to cell-acting antibiotics such as β-lactams, bacitracin, DAP, glycopeptides, d-cycloserine and antimicrobial peptides [13, 14]. It was first observed that VraSR was upregulated in vancomycin-intermediate S. aureus (VISA) strains [15], and subsequent work has shown that this system upregulates targets including penicillin binding protein 2, teichoic acid synthesis, prsA (a chaperone), and murZ (UDP-N-acetylglucosamine enolpyruvyl transferase), presumably to repair cell wall damage [13, 16–20]. Interestingly, serial passage of S. aureus in sub-inhibitory concentrations of DAP generated a derivative in which VraSR was upregulated and exhibited both VISA and DAP-nonsusceptible (DNS) phenotypes, further suggesting that VraSR responds to broad perturbations of the cell wall and cell membrane [21]. Similarly, mutations in vraSR can influence susceptibility to both vancomycin and β-lactams [22]. Moreover, dysregulation of the vraSR operon have been shown to affect methicillin and DAP susceptibility in clinical isolates of methicillin-resistant S. aureus (MRSA) [12, 23]. Collectively, VraSR is an important mediator of cell response to a variety of antibiotics in S. aureus and its dysregulation could lead to a MDR phenotype.
LiaSR is also implicated as a major determinant in biofilm formation and acid resistance in S. mutans [24] and two other streptococcal species, S. gordonii and S. uberis [25, 26]. A study by McCormick et al. demonstrated that expression of the dlt operon is influenced by LiaSR in S. gordonii [26]. Mutants defective of LiaS or LiaR were unable to upregulate expression of the dlt operon to counteract envelope stress when exposed to polymyxin B. These observations suggest that LiaR plays a critical role in responding to global stressors to maintain membrane integrity.
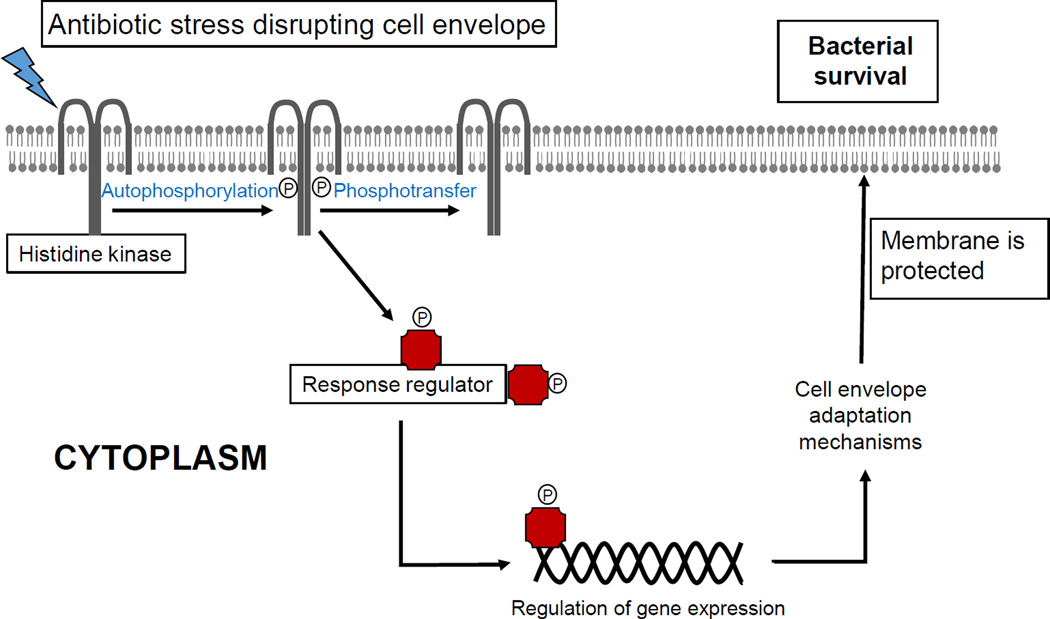
The role of LiaFSR in enterococci has only been described recently in the context of DNS. A substitution in the transmembrane protein LiaF was first encountered in a clinical strain of DNS E. faecalis [27] and subsequent investigations identified mutations in LiaFSR as the most frequent genetic changes associated with DNS in E. faecium [28, 29]. A quantitative experimental evolutionary approach also demonstrated that alterations in liaFSR were the initial pivotal step leading to development of DNS in E. faecalis [30]. Mutation in liaF, presumably resulting in activation of the LiaSR system, can decrease DAP susceptibility against E. faecalis and abolished DAP bactericidal activity in time-kill assays [27, 31]. Interestingly, this mutation also contributed to remodeling of the cell membrane by redistributing cell membrane cardiolipin microdomains away from the division septa in E. faecalis [32]. In E. faecium, alterations in LiaFSR are associated with DNS and loss of bactericidal activity in “susceptible” strains [28, 29]. These data provide evidence to suggest that the LiaFSR system is important for cell membrane homeostasis and antibiotic resistance [32]. Recent structural and biochemical studies indicate that substitutions affecting LiaR are associated with DNS and constitutively activate the response regulator by mimicking phosphorylation (independent of LiaS), locking the protein in an “on” state by altering the oligomerization state [33] (Figure 1).
Figure 1. Bacterial cell membrane adaptation via two-component signal transduction systems.
Protection of cell membrane is of paramount importance for bacterial survival. Several mechanism to protect the cell membrane against environmental stressors (including antibiotics) are available. The participation of signal transduction systems (two-component regulatory systems) is one of the most important strategies to mediate the cell membrane adaptive response.
LiaR as a novel target for anti-adaptation antibiotics
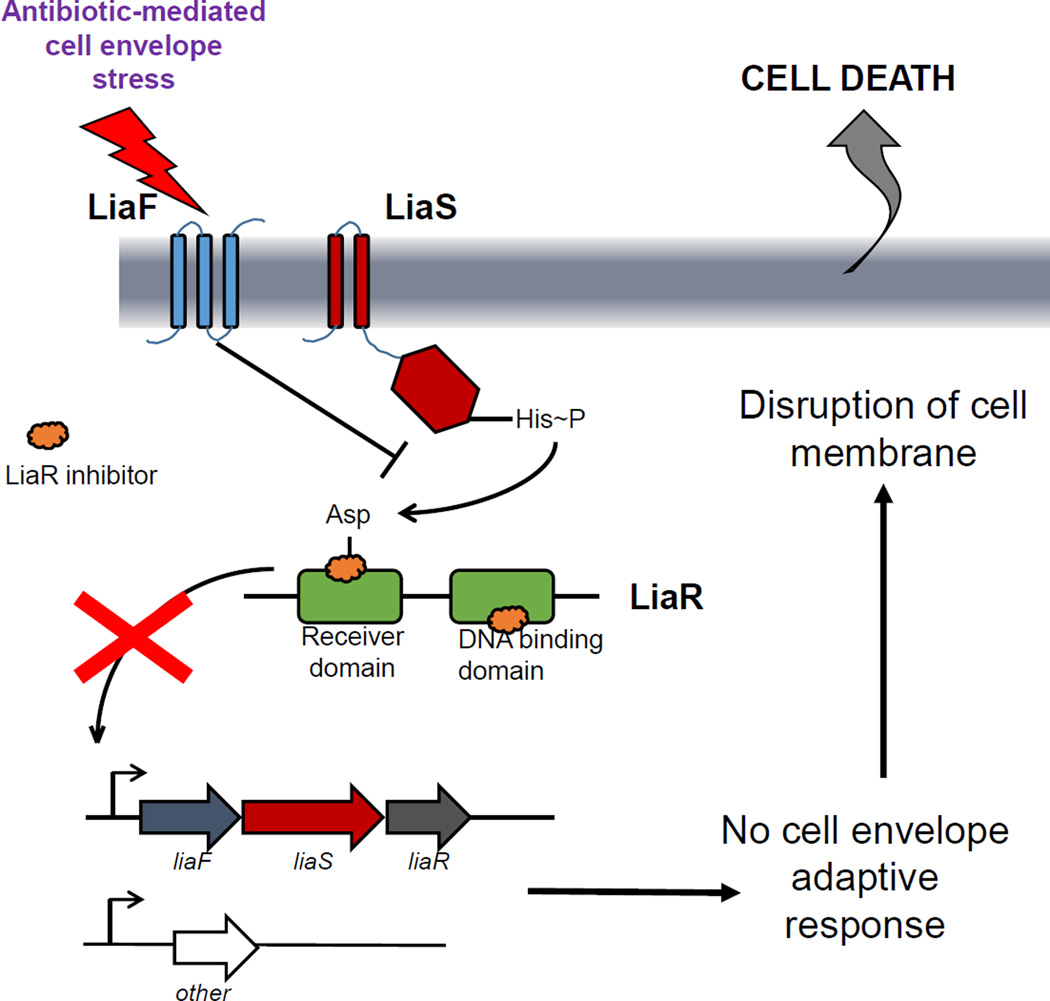
Since the LiaFSR system seems to play a prominent role in the mechanism of bacterial protection against the antibiotic attack in Gram-positive bacteria, a logical hypothesis is that inactivation of this system may make bacteria more vulnerable to antibiotics that target the cell envelope (Figure 2). Moreover, since one of the mechanisms by which the innate immune system responds to invading bacteria is through the production of antimicrobial peptides by immune cells (which also target the cell membrane), disruption of the mechanism of cell membrane protection is likely to enhance the ability of the host to clear bacteria in vivo.
Figure 2. A model of LiaR inhibitors.
Inhibiting the LiaR response regulator (green) would prevent the activation of the cell membrane adaptive response triggered by antibiotics targeting the cell envelope.
In order to support these hypotheses, a series of liaR deletion mutants in MDR enterococci were tested for their ability to survive the antimicrobial peptide challenge. A non-polar deletion of liaR in both DAP-resistant and –susceptible E. faecalis markedly increased susceptibility of the mutants to DAP, decreasing the MIC beyond wild-type levels (hypersusceptibility). This effect was seen independently of the presence of mutations in other genes that have been associated with DNS [34], confirming that LiaR plays a major role against the antibiotic attack. Interestingly, loss of functional LiaR also resulted in a marked increase in activity of other antibiotics and cationic antimicrobial peptides such as LL-37 (produced by human neutrophils), strengthening the concept that targeting LiaR could positively modulate the innate immune system making bacteria more vulnerable to clearance. Furthermore, the DAP hypersusceptibilty that occurs after deletion of liaR was also observed in strains of E. faecium regardless of the presence of mutations in liaFSR or the genetic background [35], indicating that alterations of LiaR have a “universal” effect in all species of enterococci. Of note, null vraR mutants of clinical DNS MRSA also exhibited increased susceptibility to DAP [23]. Moreover, deletion of vraR reversed methicillin and non-β-lactam [36] resistance and had better outcomes with β-lactam therapy (namely oxacillin) in mouse infection models [12, 37].
Novel target: the pathway forward
The current era of MDR presents clinicians with the challenges of limited therapeutic efficacy and drug toxicity where safe, effective therapies once existed. To be accepted as “novel”, the design of compounds should consider the following characteristics: i) novel chemical class, ii) new target, iii) a novel mechanism against an existing target and iv) low likelihood of developing resistance [38]. As discussed previously, search for compounds that are directed against unconventional targets are at the frontline of the war against antibiotic resistance. An understanding of the molecular events that lead to bacterial cell adaptation after antibiotic challenge is likely to open new avenues of targets with molecules that enhance the activity of current antimicrobials, reverse resistance and also enhance the immune system. While traditional antibiotics target proteins with essential functions, a focus on signal transduction systems can disrupt many cellular processes and even lead to a generalized arrest of cellular function. It is interesting to note that the modification of signaling pathways has been a remarkable success in anticancer therapies but has not been exploited in the development of antimicrobial strategies. As mentioned, LiaSR TCS is activated by a variety of stimuli, including many clinically useful antibiotics with different mechanisms of action (Figure 2). However, each of these signals converges to the response regulator LiaR, which serves as the courier to activate expression of downstream genes. Thus, by inhibiting the response regulator we could prevent the expression of a resistance phenotype, even in the presence of the resistance genes (Figure 2).
LiaR or its homologs emerge as an appealing target for novel antimicrobial development due to several characteristics. First, the LiaFSR system is conserved and specific to many low G+C Gram-positive bacteria, including many clinically relevant pathogens (Table 1). The high degree of identity between LiaR response regulators in these species suggests that development of an inhibitor targeting LiaR with broad range of activity against pathogenic organisms is feasible. Second, while LiaFSR does not appear to be essential in all organisms studied [10, 17, 34, 35, 39, 40, antimicrobial activity from the compound itself is not expected. Rather, LiaR has proven to be the “Achilles heel” of the membrane adaptive response and inactivation results in susceptibility to several antimicrobials and possibly enhancement of the innate immune system (see below) (Figure 2). Inhibition of LiaR can “resurrect” antimicrobial agents that are bactericidal, less toxic and more cost effective. Third, deletion of LiaR demonstrated parallel enhancement of the activity of host derived cationic antimicrobial peptides. Thus, we can augment the organism’s susceptibility to the patient’s own immune system by weakening the ability of bacteria to defend against antimicrobial peptide produced by immune cells. This concept will potentially allow those with weaker immune response to mount a more robust mucosal defense and possibly decrease colonization with MDR organisms that are the harbingers of infection [34, 35]. Lastly, emergence of resistance in LiaR-deficient mutants has been evaluated in two clinically important enterococcal species. In one study, E. faecalis was shown to incorporate exogenous fatty acids which circumvented the loss of LiaR and improved survival to a variety of membrane stressors, including DAP [41]. In a serial passage, the same hypersusceptible isolate developed resistance to DAP after 19 days of exposure to sub-inhibitory concentrations. In E. faecium, the species that is by far a greater clinical challenge, resistance was not found after 19 days of adaptation [42]. Of note, response to antibiotics via LiaSR has been shown to begin within 10 minutes of induction [13]. Thus, with appropriate dosing and prompt management of the infectious source, the opportunities to develop resistance can be limited.
The anti-adaptive concept may have several limitations. The effectiveness of these nonconventional agents has not been demonstrated yet. The activity of conventional antibiotics is predicted by bacterial growth inhibition assays. However, anti-adaption agents are not expected to affect growth suggesting that standard minimum inhibitory concentrations would not be practical. Additional assays such as time-kill assay and animal infection models will be necessary to determine activity of these agents, which are time consuming and costly. As stated, anti-adaptation drugs are expected to synergize with existing antimicrobials. Thus, combinations of anti-adaptive and antimicrobial agents are needed for treatment. Yet, different genus of Gram-positive bacteria display heterogenoeus phenotype(s) in LiaR-depleted mutants (see above) and the optimal combinations are not known. Moreover, the potential for pathogens to acquire tolerance/resistance to anti-adaptive agents or become more virulent by bypassing LiaR-dependent responses needs to be evaluated. Thus, careful characterization of this approach to increase our understanding of the evolutionary risks for anti-adaptive agents will require a multidisciplinary approach.
Concluding remarks
The alarming increase in resistance to antibiotics in pathogenic bacteria urges researchers to identify innovative and unconventional strategies for antibiotic discovery. We provide the rationale for targeting cell membrane adaptation systems which could be an interesting approach to deal with MDR organisms. Based on data derived from targeting the LiaR response regulator in clinical strains of MDR enterococci, a LiaR inhibitor may be used to disarm clinically relevant Gram-positive organisms and restore activity of currently available antibiotics and we believe that pursuing this avenue should yield novel insights into bacterial physiology and drug development.
Highlights.
The Lia system mediates response to cell membrane stressors in Gram-positive bacteria
Disruption of the Lia system rendered bacteria susceptible to a variety of antibiotics
LiaR or its homologs emerge as an appealing target for novel antimicrobial development
Acknowledgments
This work was supported by the National Institute of Allergy and Infectious Diseases, National Institutes of Health (grants R01 AI093749, R21 AI114961, R21/R33 AI121519 and K24-AI114818 to C.A.A., K08 AI113317 to T.T.T, and R01 AI080714 to Y.S.)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, Scheld M, Spellberg B, Bartlett J. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin Infect Dis. 2009;48:1–12. doi: 10.1086/595011. [DOI] [PubMed] [Google Scholar]
- 2.Bassetti M, Righi E. Development of novel antibacterial drugs to combat multiple resistant organisms. Langenbecks Arch Surg. 2015;400:153–165. doi: 10.1007/s00423-015-1280-4. [DOI] [PubMed] [Google Scholar]
- 3.Padmanabhan R, Mishra AK, Raoult D, Fournier PE. Genomics and metagenomics in medical microbiology. J Microbiol Methods. 2013;95:415–424. doi: 10.1016/j.mimet.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 4.Klemm E, Dougan G. Advances in understanding bacterial pathogenesis gained from whole-genome sequencing and phylogenetics. Cell Host Microbe. 2016;19:599–610. doi: 10.1016/j.chom.2016.04.015. [DOI] [PubMed] [Google Scholar]
- 5.Mascher T, Margulis NG, Wang T, Ye RW, Helmann JD. Cell wall stress responses in Bacillus subtilis: the regulatory network of the bacitracin stimulon. Mol Microbiol. 2003;50:1591–1604. doi: 10.1046/j.1365-2958.2003.03786.x. [DOI] [PubMed] [Google Scholar]
- 6.Mascher T, Zimmer SL, Smith TA, Helmann JD. Antibiotic-inducible promoter regulated by the cell envelope stress-sensing two-component system LiaRS of Bacillus subtilis. Antimicrob Agents Chemother. 2004;48:2888–2896. doi: 10.1128/AAC.48.8.2888-2896.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pietiainen M, Gardemeister M, Mecklin M, Leskela S, Sarvas M, Kontinen VP. Cationic antimicrobial peptides elicit a complex stress response in Bacillus subtilis that involves ECF-type sigma factors and two-component signal transduction systems. Microbiology. 2005;151:1577–1592. doi: 10.1099/mic.0.27761-0. [DOI] [PubMed] [Google Scholar]
- 8.Hachmann AB, Angert ER, Helmann JD. Genetic analysis of factors affecting susceptibility of Bacillus subtilis to daptomycin. Antimicrob Agents Chemother. 2009;53:1598–1609. doi: 10.1128/AAC.01329-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wecke T, Zuhlke D, Mader U, Jordan S, Voigt B, Pelzer S, Labischinski H, Homuth G, Hecker M, Mascher T. Daptomycin versus friulimicin B: in-depth profiling of Bacillus subtilis cell envelope stress responses. Antimicrob Agents Chemother. 2009;53:1619–1623. doi: 10.1128/AAC.01046-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jordan S, Junker A, Helmann JD, Mascher T. Regulation of LiaRS-dependent gene expression in Bacillus subtilis: identification of inhibitor proteins, regulator binding sites, and target genes of a conserved cell envelope stress-sensing twocomponent system. J Bacteriol. 2006;188:5153–5166. doi: 10.1128/JB.00310-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schrecke K, Jordan S, Mascher T. Stoichiometry and perturbation studies of the LiaFSR system of Bacillus subtilis. Mol Microbiol. 2013;87:769–788. doi: 10.1111/mmi.12130.. • The native stoichiometry of LiaFSR is 18:4:1 and LiaR is a bifunctional histidine kinase that has phosphatase activity in Bacillus subtilis.
- 12.Boyle-Vavra S, Yin S, Jo DS, Montgomery CP, Daum RS. VraT/YvqF is required for methicillin resistance and activation of the VraSR regulon in Staphylococcus aureus. Antimicrob Agents Chemother. 2013;57:83–95. doi: 10.1128/AAC.01651-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuroda M, Kuroda H, Oshima T, Takeuchi F, Mori H, Hiramatsu K. Two-component system VraSR positively modulates the regulation of cell-wall biosynthesis pathway in Staphylococcus aureus. Mol Microbiol. 2003;49:807–821. doi: 10.1046/j.1365-2958.2003.03599.x. [DOI] [PubMed] [Google Scholar]
- 14.Mensa B, Howell GL, Scott R, DeGrado WF. Comparative mechanistic studies of brilacidin, daptomycin, and the antimicrobial peptide LL16. Antimicrob Agents Chemother. 2014;58:5136–5145. doi: 10.1128/AAC.02955-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuroda M, Kuwahara-Arai K, Hiramatsu K. Identification of the up- and down-regulated genes in vancomycin-resistant Staphylococcus aureus strains Mu3 and Mu50 by cDNA differential hybridization method. Biochem Biophys Res Commun. 2000;269:485–490. doi: 10.1006/bbrc.2000.2277. [DOI] [PubMed] [Google Scholar]
- 16.Utaida S, Dunman PM, Macapagal D, Murphy E, Projan SJ, Singh VK, Jayaswal RK, Wilkinson BJ. Genome-wide transcriptional profiling of the response of Staphylococcus aureus to cell-wall-active antibiotics reveals a cell-wall-stress stimulon. Microbiology. 2003;149:2719–2732. doi: 10.1099/mic.0.26426-0. [DOI] [PubMed] [Google Scholar]
- 17.Yin S, Daum RS, Boyle-Vavra S. VraSR two-component regulatory system and its role in induction of pbp2 and vraSR expression by cell wall antimicrobials in Staphylococcus aureus. Antimicrob Agents Chemother. 2006;50:336–343. doi: 10.1128/AAC.50.1.336-343.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sobral RG, Jones AE, Des Etages SG, Dougherty TJ, Peitzsch RM, Gaasterland T, Ludovice AM, de Lencastre H, Tomasz A. Extensive and genome-wide changes in the transcription profile of Staphylococcus aureus induced by modulating the transcription of the cell wall synthesis gene murF. J Bacteriol. 2007;189:2376–2391. doi: 10.1128/JB.01439-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dengler V, Meier PS, Heusser R, Berger-Bachi B, McCallum N. Induction kinetics of the Staphylococcus aureus cell wall stress stimulon in response to different cell wall active antibiotics. BMC Microbiol. 2011;11:16. doi: 10.1186/1471-2180-11-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gardete S, Kim C, Hartmann BM, Mwangi M, Roux CM, Dunman PM, Chambers HF, Tomasz A. Genetic pathway in acquisition and loss of vancomycin resistance in a methicillin resistant Staphylococcus aureus (MRSA) strain of clonal type USA300. PLoS Pathog. 2012;8:e1002505. doi: 10.1371/journal.ppat.1002505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Camargo IL, Neoh HM, Cui L, Hiramatsu K. Serial daptomycin selection generates daptomycin-nonsusceptible Staphylococcus aureus strains with a heterogeneous vancomycin-intermediate phenotype. Antimicrob Agents Chemother. 2008;52:4289–4299. doi: 10.1128/AAC.00417-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoo JI, Kim JW, Kang GS, Kim HS, Yoo JS, Lee YS. Prevalence of amino acid changes in the yvqF, vraSR, graSR, and tcaRAB genes from vancomycin intermediate resistant Staphylococcus aureus. J Microbiol. 2013;51:160–165. doi: 10.1007/s12275-013-3088-7. [DOI] [PubMed] [Google Scholar]
- 23.Mehta S, Cuirolo AX, Plata KB, Riosa S, Silverman JA, Rubio A, Rosato RR, Rosato AE. VraSR two-component regulatory system contributes to mprF-mediated decreased susceptibility to daptomycin in in vivo-selected clinical strains of methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2012;56:92–102. doi: 10.1128/AAC.00432-10. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li YH, Lau PC, Tang N, Svensater G, Ellen RP, Cvitkovitch DG. Novel two-component regulatory system involved in biofilm formation and acid resistance in Streptococcus mutans. J Bacteriol. 2002;184:6333–6342. doi: 10.1128/JB.184.22.6333-6342.2002. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCormick NE, Halperin SA, Lee SF. Regulation of D-alanylation of lipoteichoic acid in Streptococcus gordonii. Microbiology. 2011;157:2248–2256. doi: 10.1099/mic.0.048140-0. [PubMed] [DOI] [PubMed] [Google Scholar]
- 26.Salomaki T, Karonen T, Siljamaki P, Savijoki K, Nyman TA, Varmanen P, Iivanainen A. A Streptococcus uberis transposon mutant screen reveals a negative role for LiaR homologue in biofilm formation. J Appl Microbiol. 2015;118:1–10. doi: 10.1111/jam.12664. [DOI] [PubMed] [Google Scholar]
- 27.Arias CA, Panesso D, McGrath DM, Qin X, Mojica MF, Miller C, Diaz L, Tran TT, Rincon S, Barbu EM, et al. Genetic basis for in vivo daptomycin resistance in enterococci. New England Journal of Medicine. 2011;365:892–900. doi: 10.1056/NEJMoa1011138. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Munita JM, Panesso D, Diaz L, Truc TT, Reyes J, Wanger A, Murray BE, Arias CA. Correlation between mutations in liaFSR of Enterococcus faecium and minimal inhibitory concentration of daptomycin: revisiting daptomycin breakpoints. Antimicrob Agents Chemother. 2012;56:4354–4359. doi: 10.1128/AAC.00509-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Diaz L, Tran TT, Munita JM, Miller WR, Rincon S, Carvajal LP, Wollam A, Reyes J, Panesso D, Rojas NL, et al. Whole-genome analyses of Enterococcus faecium isolates with diverse daptomycin MICs. Antimicrob Agents Chemother. 2014;58:4527–4534. doi: 10.1128/AAC.02686-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller C, Kong J, Tran TT, Arias CA, Saxer G, Shamoo Y. Adaptation of Enterococcus faecalis to daptomycin reveals an ordered progression to resistance. Antimicrob Agents Chemother. 2013;57:5373–5383. doi: 10.1128/AAC.01473-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Munita JM, Tran TT, Diaz L, Panesso D, Reyes J, Murray BE, Arias CA. A liaF codon deletion abolishes daptomycin bactericidal activity against vancomycin-resistant Enterococcus faecalis. Antimicrob Agents Chemother. 2013 doi: 10.1128/AAC.00021-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tran TT, Panesso D, Mishra NN, Mileykovskaya E, Guan Z, Munita JM, Reyes J, Diaz L, Weinstock GM, Murray BE, et al. : Daptomycin-resistant Enterococcus faecalis diverts the antibiotic molecule from the division septum and remodels cell membrane phospholipids. MBio. 2013;4 doi: 10.1128/mBio.00281-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Davlieva M, Shi Y, Leonard PG, Johnson TA, Zianni MR, Arias CA, Ladbury JE, Shamoo Y. A variable DNA recognition site organization establishes the LiaR-mediated cell envelope stress response of enterococci to daptomycin. Nucleic Acids Res. 2015;43:4758–4773. doi: 10.1093/nar/gkv321.. • An adaptive mutation that constitutively activate LiaR in the absence of phosphorylation is correlated with increased oligomerization and associated with daptomycin resistance.
- 34. Reyes J, Panesso D, Tran TT, Mishra NN, Cruz MR, Munita JM, Singh KV, Yeaman MR, Murray BE, Shamoo Y, et al. A liaR deletion restores susceptibility to daptomycin and antimicrobial peptides in multidrug-resistant Enterococcus faecalis. J Infect Dis. 2015;211:1317–1325. doi: 10.1093/infdis/jiu602.. •• Deletion of LiaR demonstrated increase susceptibility to antibiotics and cationic antimicrobial peptides in Enterococcus faecalis.
- 35. Panesso D, Reyes J, Gaston EP, Deal M, Londono A, Nigo M, Munita JM, Miller WR, Shamoo Y, Tran TT, et al. Deletion of liaR reverses daptomycin resistance in Enterococcus faecium independent of the genetic background. Antimicrob Agents Chemother. 2015;59:7327–7334. doi: 10.1128/AAC.01073-15.. •• Deletion of LiaR produced hypersusceptibility to daptomycin regardless of genetic background in Enterococcus faecium.
- 36.McCallum N, Meier PS, Heusser R, Berger-Bachi B. Mutational analyses of open reading frames within the vraSR operon and their roles in the cell wall stress response of Staphylococcus aureus. Antimicrob Agents Chemother. 2011;55:1391–1402. doi: 10.1128/AAC.01213-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jo DS, Montgomery CP, Yin S, Boyle-Vavra S, Daum RS. Improved oxacillin treatment outcomes in experimental skin and lung infection by a methicillin-resistant Staphylococcus aureus isolate with a vraSR operon deletion. Antimicrob Agents Chemother. 2011;55:2818–2823. doi: 10.1128/AAC.01704-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gwynn MN, Portnoy A, Rittenhouse SF, Payne DJ. Challenges of antibacterial discovery revisited. Ann N Y Acad Sci. 2010;1213:5–19. doi: 10.1111/j.1749-6632.2010.05828.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 39.Suntharalingam P, Senadheera MD, Mair RW, Levesque CM, Cvitkovitch DG. The LiaFSR system regulates the cell envelope stress response in Streptococcus mutans. J Bacteriol. 2009;191:2973–2984. doi: 10.1128/JB.01563-08. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eldholm V, Gutt B, Johnsborg O, Bruckner R, Maurer P, Hakenbeck R, Mascher T, Havarstein LS. The pneumococcal cell envelope stress-sensing system LiaFSR is activated by murein hydrolases and lipid II-interacting antibiotics. J Bacteriol. 2010;192:1761–1773. doi: 10.1128/JB.01489-09. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harp JR, Saito HE, Bourdon AK, Reyes J, Arias CA, Campagna SR, Fozo EM. Exogenous fatty acids protect Enterococcus faecalis from daptomycin induced membrane stress independent of the response regulator LiaR. Appl Environ Microbiol. 2016 doi: 10.1128/AEM.00933-16. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miller WR, editor. abstract 1390, IDWeek 2015. San Diego, CA: 2015. Oct, [Google Scholar]