Abstract
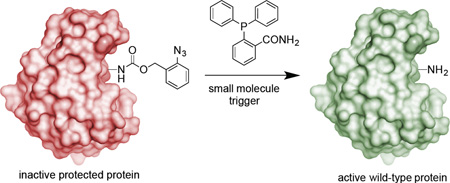
Using small molecules to control the function of proteins in live cells with complete specificity is highly desirable, but challenging. Here we report a small molecule switch that can be used to control protein activity. The approach uses a phosphine-mediated Staudinger reduction to activate protein function. Genetic encoding of an ortho-azidobenzyloxycarbonyl amino acid using a pyrrolysyl tRNA synthetase/tRNACUA pair in mammalian cells enables the site-specific introduction of a small molecule-removable protecting group into the protein of interest. Strategic placement of this group renders the protein inactive until deprotection through a bioorthogonal Staudinger reduction delivers the active, wild-type protein. This developed methodology was applied to the conditional control of several cellular processes, including bioluminescence (luciferase), fluorescence (EGFP), protein translocation (nuclear localization sequence), DNA recombination (Cre), and gene editing (Cas9).
Graphical abstract
Introduction
Conditional control of protein function in live cells is essential for studies of the molecular details of biological processes. Several inhibitor-based loss-of-function strategies have been developed to reduce protein function and levels, for example, selective inhibition of kinase function using a bump-and-hole strategy,1 selective induction of target protein degradation with small molecule probes that recruit the ubiquitin proteasome system,2,3 and selective small molecule-protein bioconjugations leading to the blocking of allosteric and active sites.4 In contrast, the design and discovery of small molecule switches for activation of protein function has been more challenging,5,6 with the rapamycin-mediated dimerization of FKBP12 and FRB protein domains being the most dominant example.7–9 This small molecule switch has been successfully applied to the activation of transcription factors, kinases, split enzyme systems such as TEV protease, cellular protein translocation, and many others.10–13 However, limitations include the need for the expression of two fusion proteins and the engineering of protein function to be controlled through FKBP12-FRB dimerization. Limited examples exist were single protein-rapamycin complex formations have been developed to control protein stability and kinase function; however, extensive protein engineering was required for the generation of these switches.14,15 Other small molecule switches have been identified in nature and engineered for conditional control of protein function,16 but examples of have been limited. In addition to small molecule activation of protein function, tools for optical activation of protein function have received significant interest in recent years.17,18 Small molecule control and light control complement each other and can be synergistically employed in a powerful fashion.19,20
We designed a small molecule switch for activation of protein function through the site-specific incorporation of an ortho-azidobenzyloxycarbonyl lysine (OABK, Figure 1a). The azidobenzyloxycarbonyl protecting group blocks protein function, until removed through bioorthogonal deprotection via a Staudinger reduction. The Staudinger reduction-induced deprotection of amino acids adds an additional tool to the expanding set of chemical and photochemical peptide and protein activation approaches.21–26 Related reactions have been extensively used in bioconjugations in vitro and in vivo, and several types of Staudinger ligations have been reported.27–29 Azide groups installed on biological molecules are non-toxic and fully orthogonal to all cellular chemistries as demonstrated by numerous examples of bioconjugation reactions in cells and whole organisms.
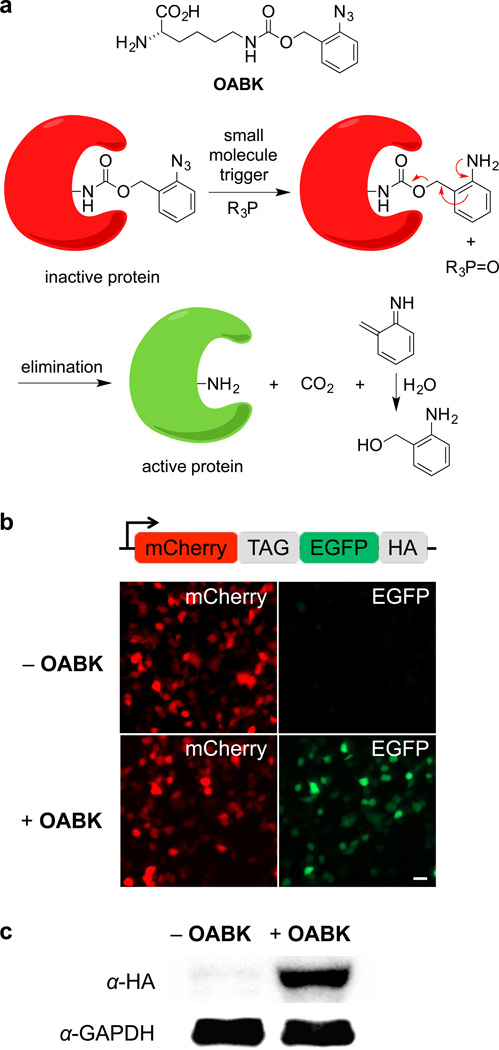
Figure 1. Staudinger reduction-based protein activation through genetic encoding of OABK in living cells.
(a) Structure of OABK and schematic of the phosphine-triggered protein activation through protecting group removal via a Staudinger reduction. OABK is site-specifically incorporated into a protein of interest via genetic code expansion, and then the protein is deprotected and activated through a phosphine-induced Staudinger reduction followed by a 1,4-elimination of the azidobenzyl group. (b) Micrographs confirming amino acid-dependent incorporation of OABK into mCherry-TAG-EGFP-HA in HEK293T cells. EGFP fluorescence was observed only in the presence of OABK, due to suppression of the TAG stop codon. Scale bar represents 20 µm. (c) Confirmation of OABK-dependent full-length protein expression through an anti-HA Western blot.
Results and Discussion
Genetic encoding of OABK and deprotection in vitro
The amino acid OABK was readily synthesized in three steps from 2-azidobenzyl alcohol via a succinimidyl carbonate (Supplementary Scheme 1). Phosphine-induced Staudinger reduction of OABK yields an aniline derivative that undergoes a 1,4-elimination and decarboxylation, as confirmed by LC/MS analysis (Figure 1a, Supplementary Figure S1), including time-course experiments (Supplementary Figure S2).30 Deprotection results in the formation of lysine and, when OABK is incorporated into a protein, the formation of active, wild-type protein. Several unnatural lysine derivatives have been genetically encoded through the addition of engineered pyrrolysyl tRNA/tRNA synthetase pairs from archaea to the translational machinery of bacterial, yeast, and human cells.31–35 Based on a previously reported Y271A/Y349F PylRS mutant (termed OABKRS),36 we demonstrated the first incorporation of OABK into proteins in mammalian cells (Figure 1b–c). A plasmid encoding the synthetase and an amber-suppressor reporter (pOABKRS-mCherry-TAG-EGFP-HA, Supplementary Figure S4), together with a plasmid encoding the tRNA (p4CMVE-U6-PylT), was transfected into human embryonic kidney (HEK) 293T cells. The cells were incubated for 24 hrs in the absence or presence of OABK (0.5 mM) and fluorescence imaging revealed EGFP expression only in the presence of the unnatural amino acid (Figure 1b). This was further confirmed by Western blot analysis with an anti-HA antibody (Figure 1c, Supplementary Figure S6). These results are in agreement with OABK-dependent protein expression in bacterial cells and confirm the high fidelity of the OABKRS in both pro- and eukaryotic systems (Supplementary Figure S5). The efficient incorporation, a sfGFP-OABK protein yield of 12 mg/L was obtained in E. coli, was further confirmed by electrospray ionization mass spectrometry (ESI-MS), revealing an experimentally obtained mass of 28402.75 Da, in agreement with the expected mass of 28403.03 Da (Supplementary Figure S5). Furthermore, efficient in vitro protein deprotection through Staudinger reduction was confirmed by mass spec analysis of sfGFP-OABK after treatment with the small molecule trigger (the observed mass of 28226.41 Da for deprotected sfGFP matches the expected mass of 28227.89 Da; deprotection was quantitative and no protected sfGFP-OABK was detected (Supplementary Figure S5).
Small molecule-triggered fluorescent protein activation
Enhanced green fluorescent protein (EGFP) was selected as an initial target protein in order to test if this activation strategy would allow triggering of protein function in cells, since the activity of EGFP can be measured in real time using live cell imaging.37 The lysine K85 in EGFP was targeted for OABK introduction, since it undergoes crucial electrostatic and H-bonding interactions with a nearby aspartate, serine, and cysteine, all key residues for chromophore maturation and fluorescence (Figure 2a). HEK293T cells were co-transfected with pEGPF-K85TAG-mCherry and pOABKRS-4PylT in the presence of OABK. The mCherry serves as a transfection and expression control. After 24 h incubation, no EGFP fluorescence was observed in the absence of any small molecule trigger. However, addition of 2-(diphenylphosphino)benzoic acid to the cells led to EGFP activation (Figure 2b). Time-lapse imaging and quantification of cellular fluorescence revealed a t½ of 98 min for activation of EGFP (Figure 2c and Supplementary Movie 1). Individual steps contributing to the kinetics of this activation include permeation of the phosphine into the cell, Staudinger reduction, 1,4-elimination of the lysine protecting group, EGFP folding, and fluorophore maturation through oxidation. Since individual rate constants of several of these steps are unknown, we decided to compare the overall kinetics to the optical activation of an EGFP-K85 mutant containing a caged coumarin-lysine (Supplementary Figure S8). Here, in agreement with previously observed EGFP maturation kinetics,37,38 a t½ of 35 min was observed, indicating that, as expected, the fluorescent protein activation with the small molecule trigger is slightly slower than the light-activation. Thus, genetically encoded OABK in conjunction with small molecule activation allows for the conditional regulation of intracellular protein maturation.
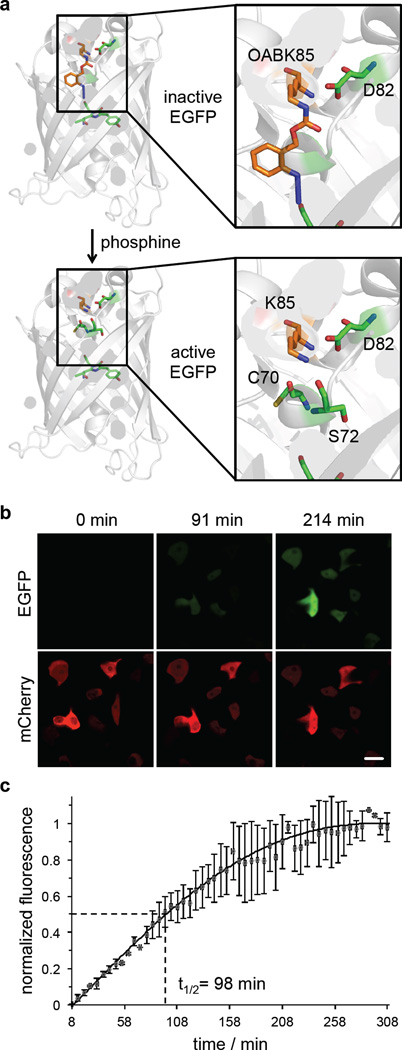
Figure 2. Small molecule-induced activation of EGFP fluorescence.
(a) Incorporation of OABK (0.5 mM) at position K85 of EGFP inhibits fluorophore formation until the native lysine is generated through small molecule activation (model based on PDB 4EUL). (b) Micrographs of HEK293T cells showing EGFP fluorescence only in the presence of the small molecule 2-(diphenylphosphino)benzoic acid (2DPBA, 500 µM), but not in its absence. Scale bar represents 20 µm. (c) Time-lapse imaging and fluorescence quantification indicates a combined t1/2 of deprotection and EGFP maturation of 98 min. Error bars represent standard deviations from three cells.
Small molecule-triggered enzyme activation
To demonstrate broader applicability of this approach after these initial encouraging results and to obtain an easily quantified readout that can be used to screen a variety of different phosphines for their ability to activate protein function in cells and in cell lysates, we chose firefly luciferase (FLuc) as a second target protein. FLuc contains a catalytic site lysine, K206, which has previously shown to block catalytic activity when substituted with a sterically demanding hydroxycoumarin lysine.37 Similarly, introducing OABK at position K206 inhibits FLuc enzymatic activity by restricting access of ATP to the active site (Figure 3b), until the enzyme is deprotected and activated through phosphine treatment. Several different phosphine derivatives were selected, based on their availability, solubility, and previous use in Staudinger reductions (Figure 3a). These include tris(2-carboxyethyl)phosphine (TCEP, 1), a TCEP methylester (2), triphenylphosphine (TPP, 3), phosphanetriyltris(benzenesulfonic acid) trisodium salt (TPPTS, 4), 2-(diphenylphosphino) benzoic acid (2DPBA, 5), and 2-(diphenylphosphino)benzamide (2DPBM, 6). All phosphines were commercially available, besides 2 and 6 which were synthesized as previously described.39,40 To screen these small molecule triggers, HEK293T cells were co-transfected with the mutated firefly luciferase plasmid (pGL3-K206TAG) and the MbOABKRS/PyltRNACUA pair (pOABKRS-4PylT) in the presence of OABK (0.5 mM). After 24 h incubation, the cells were either treated with one of the six phosphine derivatives 1–6 or were continued to be incubated in the absence of the small molecule trigger. The incorporation of OABK into FLuc blocked luciferase activity in the absence of small molecule activation, as determined by a Bright-Glo luciferase assay (Figure 3C). However, after only a 3 h incubation with the phosphines 2 (500 µM), 3 (50 µM solubility limit in media), 5 (500 µM), or 6 (50 µM) firefly luciferase activity was increased by 43-fold, 28-fold, 44-fold, and 46-fold, respectively – showing excellent OFF to ON switching of enzymatic function. Not unexpected, the reactions with 1 and 4 did not show luciferase activation in live cells due to their diminished cell permeability (Figure 3c).41 The pronounced differential activity of TCEP (1) in lysate versus cells (Figure 3c–d) may provide an opportunity to selectively activate surface displayed or secreted proteins while not affecting intracellular proteins. A dose-dependent luciferase assay in live cells was conducted with the most effective triarylphosphines 5 (5–500 µM) and 6 (5–100 µM), indicating a maximal activation plateau starting at concentrations as low as 50 µM (5) and 25 µM (6) (Figure 3e). Thus, among the six phosphines, 5 and 6 were used for all subsequent experiments, including a time-course of cellular luciferase function (Supplementary Figure S7; the chemiluminescence intensity gradually increased over time, reaching a plateau at 150 min with a t½ of 55 min for activation of FLuc).
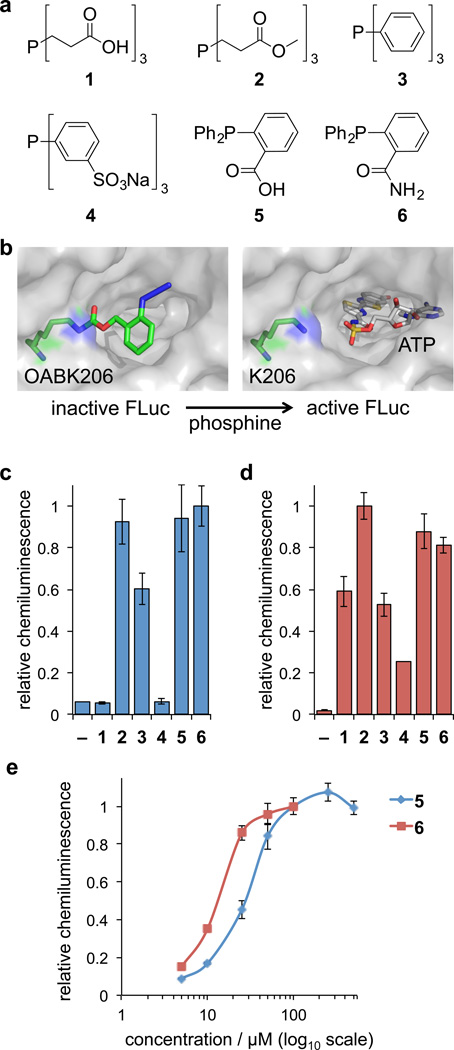
Figure 3. Phosphine screening for Staudinger reduction-mediated deprotection and activation of firefly luciferase in cells and cell lysates.
(a) Structures of the six phosphine derivatives TCEP (1), TCEP ester (2), TPP (3), TPPTS (4), 2DPBA (5), and 2DPBM (6). (b) Model of FLuc containing OABK at position K206, thereby blocking access of ATP to the catalytic site until the azidobenzyloxycarbonyl group is cleaved by the small molecule trigger (based on PDB 2D1S). (c) Initial screening of phosphines 1–6 (at maximum concentrations based on the solubility and cell viability, see Supplementary Figure 3) in order to identify efficient activators of OABK-FLuc in HEK293T cells and in (d) cell lysates. Error bars represent standard deviations from three independent experiments. (e) Live cell dose-dependent luciferase assays were conducted for the most efficient small molecule triggers 5 (5, 10, 25, 50, 100, 250, 500 µM) and 6 (5, 10, 25, 50, 100 µM), revealing 6 as the most active phosphine. The solubility of 6 in an aqueous environment limits its application to ≤100 µM. Error bars represent standard deviations from three independent experiments.
Based on these results, the site-specific incorporation of OABK into proteins provides a novel approach to the small molecule activation of enzymatic function in live cells, complementing other protecting group strategies to conditionally control cellular processes, including light-removable caging groups.42,43 Small molecule-activation of protein function can be employed in cases where optical stimulation is not feasible, such as opaque, thick, or structurally complex specimens (most metazoans) and in systems that are not amenable to stimulation through irradiation, e.g., due to conflicting fluorescent reporters or UV toxicity. Moreover, bulk activation of large numbers of cells can be readily achieved, and, intriguingly, sequential activation of cellular processes is possible by using both light and small molecules as triggers, as shown in the engineering of dual-input mammalian genetic circuits with extremely stringent regulation of gene expression in vivo.19 Thus, the Staudinger-reduction-induced protein activation expands the toolbox for conditionally controlled protein control in live cells. It compares favorably to Diels-Alder-mediated21 and palladium-mediated24 protein deprotection methods, since benign triarylphosphine reagents have been extensively used in biological studies, since low concentrations (as low as 25 µM) of the small molecule trigger were found to efficiently activate protein function, and since the synthesis of OABK and its incorporation into proteins in both bacterial and mammalian systems is highly efficient and high yielding.
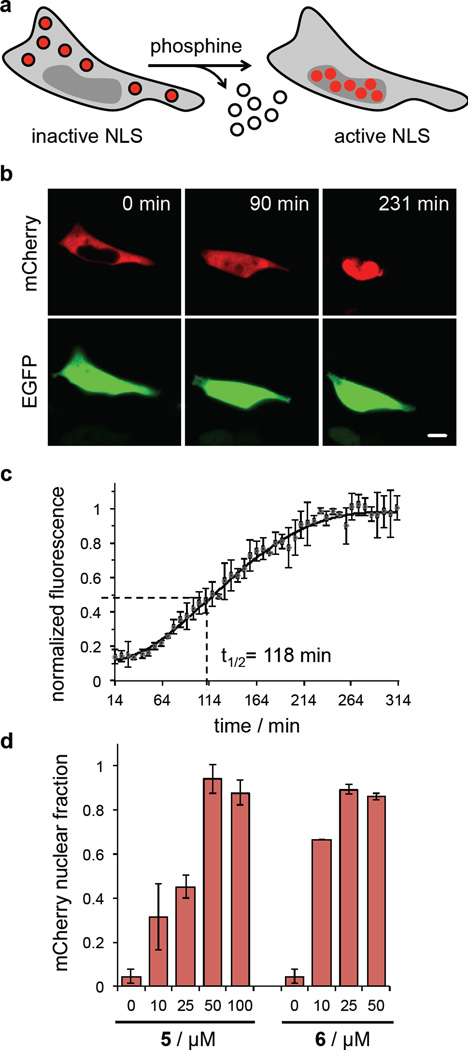
Small molecule-triggered protein translocation
In addition to the small molecule triggering of enzymatic function (FLuc) and protein maturation (EGFP), we wanted to demonstrate the small molecule control of a protein substrate. In addition, we wanted to measure in live cells if quantitative deprotection can be achieved. Thus, we investigated small molecule-activated nuclear translocation by genetically incorporating OABK into the nuclear localization signal (NLS) of the transcription factor SATB1 (Figure 4a).44 The presence of the azidobenzyl group at position K29 completely inhibited translocation of an mCherry reporter fused to the NLS, as observed in HEK 293T cells expressing SATB1K29OABK-mCherry. After 24 hr of incubation, the cells were treated with 5 (500 µM) and nuclear translocation of mCherry was triggered through Staudinger reduction-induced formation of the native SATB1-NLS (Figure 4b and Supplementary Movie 2). Translocation was completed within 231 min, as shown by time-lapse imaging, and fluorescence quantification over time revealed a t½ of 118 min (Figure 4c). When translocation rates were compared to the optical triggering of a caged SATB1-NLS (t½ of 54 min) (Supplementary Figure S9), the small molecule activated process occurred within a similar timeframe (approximately two vs. one hour). Importantly, complete protein translocation from the cytoplasm to the nucleus was observed, demonstrating complete OABK-SATB1 deprotection in live cells through small molecule triggering. Furthermore, dose-dependent assays showed that concentrations as low as 50 µM (5) and 25 µM (6) were sufficient for full deprotection and activation of the nuclear translocation signal, as all cytoplasmic mCherry quantitatively shuttled to the nucleus. The dose-response also indicated a correlation between the switch-on ratio and the phosphine concentration in the intracellular environment, allowing for precise titration of the extend of cellular action (Figure 4d).
Figure 4. Small molecule-triggered protein translocation.
(a) Site-specific incorporation of OABK at K29 in the SATB1 nuclear localization sequence leads to complete exclusion of mCherry from the nucleus, until the wild-type NLS is generated through treatment with the phosphine 5. (b) Micrographs of HEK293T cells showing no nuclear localization of OABK-SATB1-mCherry and complete activation of translocation from the cytoplasm to the nucleus after activation with 5. EGFP expression was used as a transfection and imaging control. Scale bar represents 10 µm. (c) In order to determine the kinetics of the phosphine deprotection coupled nuclear translocation, time-lapse imaging was conducted and normalized mCherry fluorescence in the nucleus indicates a combined t1/2 of 118 min. Error bars represent standard deviations from three cells. (d) Dose-dependent mCherry nuclear import through activation of SatB1 translocation with different concentrations of 5 and 6 revealed the ability to titrate the biological effect and the high triggering efficiency of the phosphine 6 at concentrations as low as 25 µM. Fluorescent cells were imaged in four randomly selected fields of view per well, and fluorescence intensities were determined and normalized to the maximum fluorescence intensity in (c).Data was analyzed using ImageJ. Error bars represent standard deviations from four cells.
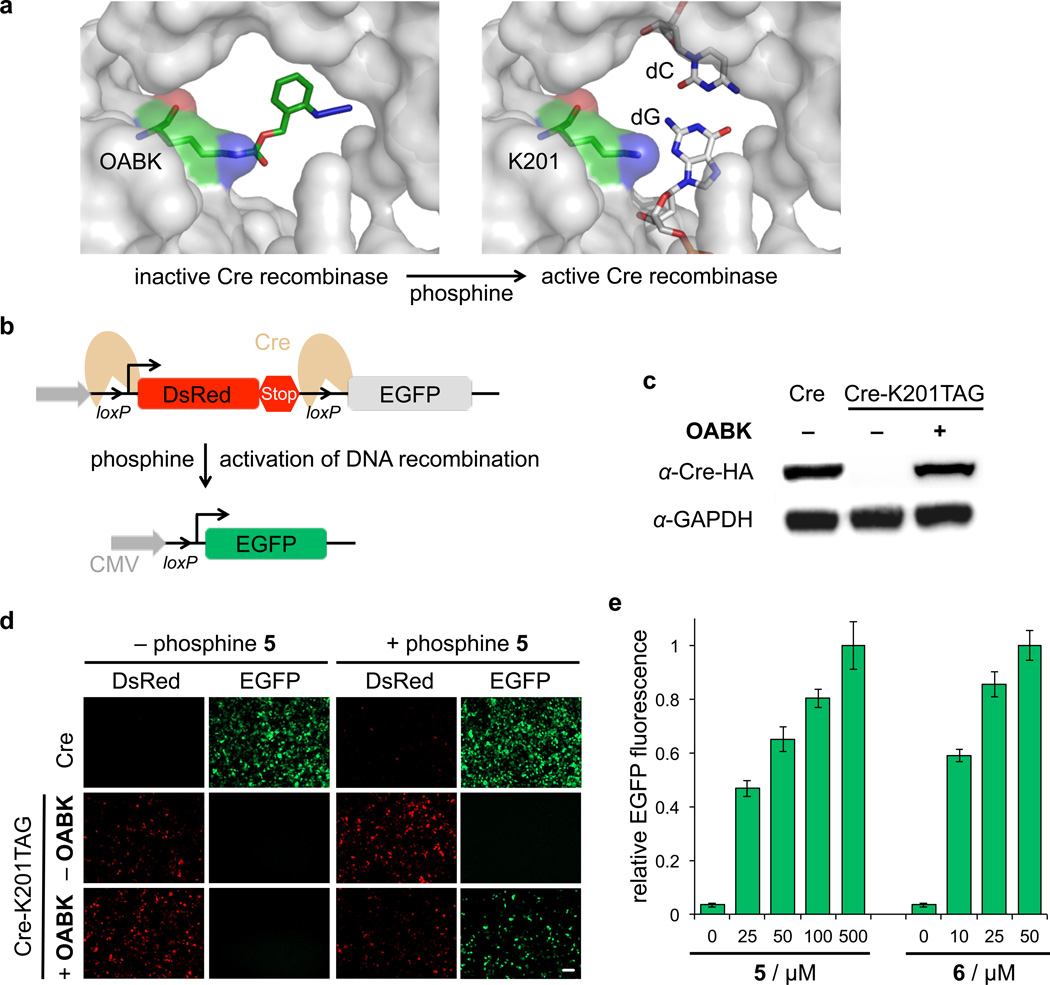
Small molecule-triggered DNA recombination
To further explore the general applicability of the developed small molecule switch of protein function, we investigated its function in two additional proteins that act upon DNA as their substrate. Bacteriophage P1 Cre recombinase recognizes loxP sites on double-stranded DNA, induces phosphodiester cleavage, and either inserts or excises sequences through a double recombination event that proceeds via a holiday junction.45 The Cre-loxP recombination system has been developed as a versatile genome manipulation tool and has been extensively applied in gene knock-out and knock-in studies.46 To apply our phosphine-triggered switch, we incorporated OABK at position K201. A lysine in position 201 is essential for catalytic activity of Cre recombinase, because it is located in a loop extending into the DNA minor groove near the cleavage site of the DNA, and has interactions with the N3 of the +1 guanine base and the 5’-O and 4’-O of the −1 sugar, as well as a tightly bound water molecule within hydrogen bonding distance (Figure 5a).47 The expression level of Cre-K201OABK is comparable to wild-type Cre recombinase expression in HEK293T cells (Figure 5c, Supplementary Figure S10). Activation of Cre-OABK with 5 in live cells was explored using a Cre stoplight reporter assay (Figure 5b–c).48 Importantly, cells expressing Cre-OABK exclusively showed DsRed expression in the absence of the small molecule trigger 5, demonstrating the complete catalytic inactivity of the recombinase enzyme containing the K201→OABK mutation. Through 5-induced deprotection, the protein was activated very effectively and the functional Cre catalyzed DNA recombination to excise DsRed and a transcriptional terminator, switching on EGFP expression (Figure 5b and 5d). To identify the optimal phosphine activation conditions, the Cre recombination event was triggered with varying concentrations of 5 and 6. Overall, activation of DNA recombination can be efficiently achieved at high levels through triggering with 25 µM of 6 (Figure 5e). This is in full agreement with the previous luciferase activation and the activation of nuclear translocation, which indicated virtually complete intracellular protein deprotection at 25 µM of 6. These results demonstrate that conditional activation of DNA recombination was successfully accomplished through a small molecule-mediated Staudinger reduction.
Figure 5. Small molecule-triggered DNA recombination.
(a) Model of the regulation of Cre recombinase activity through the K201→OABK mutation based on PDB 1CRX. Before addition of the phosphine trigger, the protecting group blocks the active site lysine from interacting with the DNA. Upon activation with phosphines, the caging group is removed and Cre recombination is restored. (b) Schematic design of the Cre recombinase activity assay using a Stoplight dual reporter. In the absence of activated Cre, DsRed is exclusively expressed under control of the CMV promoter. When Cre-OABK is activated by the phosphine 5 or 6, functional Cre excises the DsRed-terminator cassette between the loxP sites, and EGFP is placed under control of the CMV promoter and is expressed. (c) Anti-HA-tag Western blot showing amino acid-dependent expression of modified Cre recombinase in HEK 293T cells in the absence or presence of OABK (0.25 mM), as well as a GAPDH loading control. (d) EGFP fluorescence imaging shows small molecule activation of Cre recombinase-catalyzed DNA recombination in HEK 293T cells using the dual fluorescence reporter. Scale bar represents 100 µm. (e) Analysis of EGFP expression using imaging cytometry reveals dose-dependent activation of Cre recombinase with increasing concentrations of 5 and 6. The number of both DsRed and EGFP fluorescent cells were counted in four randomly selected fields of view per well and normalized to the total number of fluorescent cells. Data was analyzed using ImageJ. Error bars represent standard deviations from three independent experiments.
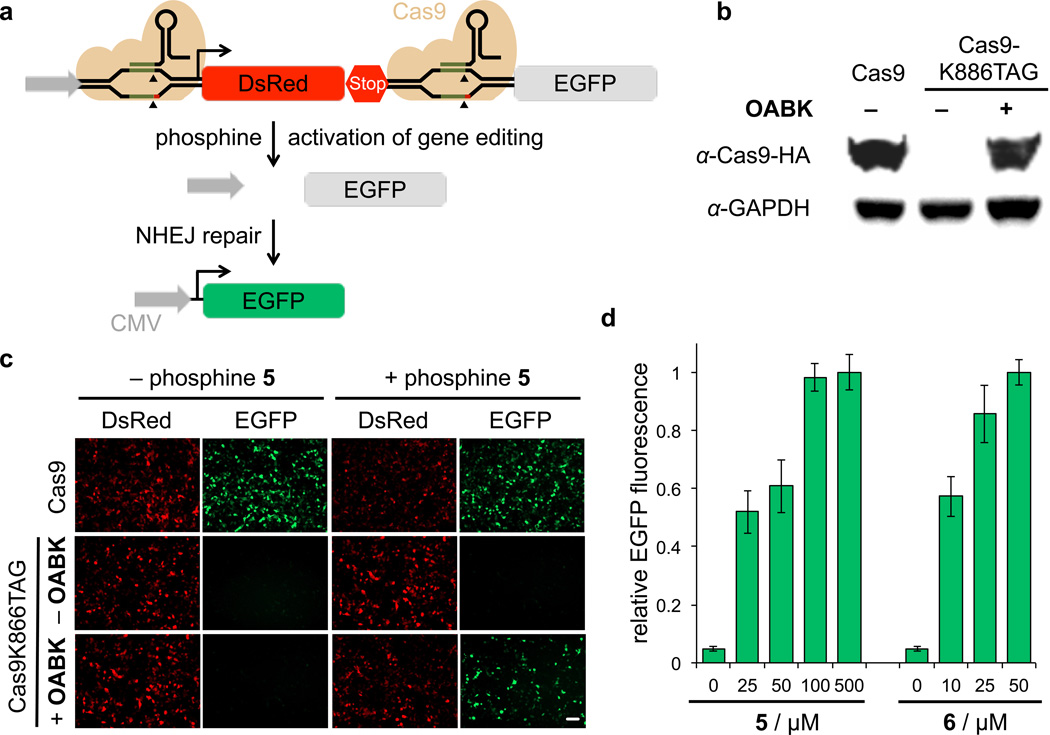
Small molecule-triggered CRISPR/Cas9 gene editing
The second DNA-editing enzyme we applied the engineered small molecule switch to was Cas9. The CRISPR/Cas9 system has been established as an extremely valuable tool for gene editing in cells and model organisms.49 Most recently, conditional regulation of Cas9 enzymatic activity has been shown to provide spatial and temporal control over gene editing and to mitigate off-target effects observed with constitutively active Cas9.50,51 To implement the Staudinger reduction switch, we incorporated OABK at position K866, a lysine residue that undergoes a large conformational change upon gRNA binding, thereby becoming exposed to the surface and possibly facilitating orientation of the DNA for nuclease cleavage.52 This site has been successfully used for the optical control of Cas9 function.53 The function of Cas9-OABK in the absence and presence of 5 was investigated using a DsRed/EGFP dual reporter assay (Figure 6a; encoded on pIRG). Cells expressing Cas9-OABK only showed DsRed fluorescence, because transcription terminates after DsRed in the absence of activated Cas9, indicating complete inhibition of enzymatic function. Once Cas9-OABK is activated by the small molecule phosphine, the complex of functional Cas9 and gRNAs induces excision of the DsRed-terminator cassette through DNA double-strand break, followed by NHEJ repair, and leading to EGFP expression. The expression level of Cas9-K866OABK is similar to wild-type Cas9 expression in mammalian cells (Figure 6b, Supplementary Figure S11), and the activity after small molecule triggering almost reached native levels (Figure 6c, Supplementary Figure S12). To optimize the concentration of phosphines for activation of gene editing in live cells, varying concentrations of 5 and 6 were investigated. Consistent with the other small molecule-triggered processes, dose-dependent results were obtained, and 25 µM was identified as the universally effective concentration of 6 for activation of Cas9-OABK and other proteins in live cells (Figure 6d). These experiments successfully demonstrate that conditional activation of CRISPR/Cas9 gene editing can be accomplished via a Staudinger reduction.
Figure 6. Small molecule-triggered CRISPR/Cas9 gene editing.
(a) Schematic design of the CRISPR/Cas9 activity assay using the pIRG dual reporter. In the absence of activated Cas9, transcription terminates after the DsRed gene and no EGFP is expressed. When Cas9-OABK is activated by the phosphine 5 or 6, the complex of functional Cas9 and gRNAs induces excision of the DsRed-terminator cassette, followed by DNA repair and EGFP expression. (b) Anti-HA western blot showing amino acid-dependent expression of modified Cas9 in HEK 293T cells without or with OABK (0.25 mM), as well as a GAPDH loading control. (c) Small-molecule control of CRISPR/Cas9 gene editing in HEK 293T cells using the pIRG reporter shows that EGFP fluorescence is only observed only in the presence of 5. Residual DsRed fluorescence was still observed, due to reporter expression during the 24 h incubation before small molecule activation. Scale bar represents 100 µm. (d) EGFP expression, and thus Cas9 function, can be titrated in response to increasing concentrations of 5 and 6. EGFP expression was analyzed using imaging cytometry by counting fluorescent cells in four randomly selected fields of view per well. Data was analyzed using ImageJ. Error bars represent standard deviations from three independent experiments.
Conclusions
In conclusion, the developed Staudinger reduction-based switch may provide a general method for the small molecule activation of proteins – as demonstrated in the conditional activation of protein functions as diverse as enzymatic activity (luciferase, Cre recombinase, and Cas9), protein maturation (EGFP), protein-protein interactions (SatB1 nuclear localization sequence), and protein-nucleic acid interactions (Cre recombinase and Cas9). While numerous small molecule inhibitors of protein function have been discovered through classical screening approaches (or have been engineered through bump & hole and targeted degradation strategies) and have been applied to the study of numerous biological processes, small molecule activators of specific protein function are rare. The small molecule activation approach reported here is highly specific since only a single, selected amino acid in only the protein of interest is modified. It is easily implemented based on rational design following structural or mechanistic data, and, compared to existing methodologies that typically rely on the expression of fusion proteins or the conditional activation of protein expression, the site-specific introduction of a small protecting group at an essential site of the protein produces minimal structural perturbations while very effectively blocking protein function. Removal of the protecting group through a bioorthogonal Staudinger reduction activates protein function by generating the wild-type protein without any appended fusion domains. The timing of activation can be controlled on the minute time scale and thus is faster than small molecule activation of transcription, as, for example, in case of the well-established Tet-On system. In contrast to optical control of protein function, no specialized photostimulation equipment is required and the developed small molecule switch is orthogonal to optical switches in the visible spectrum, potentially enabling precisely staged protein activation events. If necessary, depending on the study that is being conducted, the endogenous, non-phosphine controlled copy of the protein could be deleted through gene editing (CRISPR/Cas9 or TALENs) or be knocked-down (RNAi or antisense).
METHODS
Chemical synthesis protocols are included in the Supplementary Information.
Incorporation of OABK into proteins in bacterial cells
The plasmid pBAD-sfGFP-Y151TAG-pylT was co-transformed with pBK-OABKRS into chemically competent E. coli Top10 cells. A single colony was used to inoculate LB media overnight and 500 µL of the overnight culture was added to 25 mL of LB media, supplemented with 1 mM of OABK and 25 µg/mL of tetracycline and 50 µg/mL of kanamycin. Cells were grown at 37 °C, 250 rpm, and protein expression was induced with 0.2% arabinose when the OD600 reached ~0.5. After overnight expression at 37 °C, cells were pelleted and washed once with PBS. The cell pellet was re-suspended in 6 mL of phosphate lysis buffer (50 mM, pH 8.0) and Triton X-100 (60 µL, 10%), gently mixed, and incubated for an hour at 4 °C. The cell suspensions were sonicated with 6 short burst of 30 s followed by intervals of 30 s for cooling, and then the cell lysates were centrifuged at 4 °C, 13000 g, for 10 minutes. The supernatant was transferred to a 15 mL conical tube and 100 µL of Ni-NTA resin (Qiagen) was added. The mixture was incubated at 4 °C for 2 h under shaking. The resin was collected by centrifugation (500 g, 10 min), washed twice with 400 µL of lysis buffer, followed by two washes with 400 µL of wash buffer containing 20 mM imidazole. The protein was eluted with 400 µL of elution buffer containing 250 mM imidazole. The purified sfGFP-OABK was analyzed by ESI-MS, revealing a mass of 28402.75 Da, in agreement with the expected mass of 28403.03 Da.
Incorporation of OABK into protein in mammalian cells
HEK293T cells were seeded at ~50,000 cells per well (200 µL) in DMEM (Dulbecco's Modified Eagle Medium, Gibco) supplemented with 10% FBS (Gibco), 1% Pen-Strep (Corning Cellgro) and 2 mM L-glutamin (Alfa Aesar) in 96-well black plates (Greiner) in a humidified atmosphere with 5% CO2 at 37 °C. At ~80% confluency, cells were transfected with pOABKRS-mCherry-TAG-EGFP-HA (Supplementary Figure 3) and p4CMVE-U6-PylT (100 ng of each plasmid) using linear PEI (1.5 µL, 0.323 mg/mL). After an overnight incubation at 37 °C in the presence or absence of OABK (0.5 mM), the cells were washed once with PBS (200 µL) and imaged on a Zeiss Axio Observer Z1 Microscope (10X objective, NA 0.8 plan-apochromat) with mCherry (Ex: BP550/25; Em: BP605/70) and EGFP (Ex: BP470/40; Em: BP525/50) filter cubes. To confirm the expression of the fusion protein and also differentiate between expression levels, a Western blot was performed. HEK293T cells were seeded at ~900,000 cells per well in DMEM media (2 mL) in 6-well plates and co-transfected with pOABKRS-mCherry-TAG-EGFP-HA and p4CMVE-U6-PylT (1.5 µg of each plasmid) using linear PEI (10 µL, 0.323 mg/mL) in the presence or absence of OABK (0.5 mM) at ~80% cell confluency. After 24 h incubation, the media was removed and the cells were washed once with chilled PBS (2 mL), lysed with mammalian protein extraction buffer (200 µL, GE Healthcare) containing complete protease inhibitor cocktail (Sigma) on ice, and the cell lysates were cleared at 14,000 g (4 °C, 20 min). The supernatant (12 µL) was added to 4X SDS sample loading buffer and the protein lysates were boiled at 95 °C, and then analyzed by SDS-PAGE (10%). After gel electrophoresis and transfer to a nitrocellulose membrane (GE Healthcare), the membrane was blocked in TBS with 0.1% Tween 20 (Fisher Scientific) and 5% milk powder for 1 h. The blots were probed and incubated in TBST with the primary antibody, α-HA-probe (Y-11) rabbit polyclonal lgG, (1:1000 dilution, sc-805, Santa Cruz) overnight at 4 °C, followed by a goat anti-rabbit IgG-HRP secondary antibody (1:500,000 dilution, sc-2004, Santa Cruz) for 1 h at room temperature. As a protein loading control, Western blotting was also carried out to detect the endogenous levels of total GAPDH protein using a mouse anti-GAPDH antibody (Santa Cruz) visualized with an HRP-conjugated anti-mouse secondary antibody (sc-2005, Santa Cruz). The blots were washed by TBST for three times, and then incubated in working solution (0.5 mL, Thermo Scientific SuperSignal West Femto Maximum Sensitivity Substrate) for 1 min. The blots were placed in a clear plastic wrap and imaged on a ChemiDoc (Bio-Rad).
Luciferase-based screen of phosphines for protein activation in cells
Plasmids pGL3-K206TAG and pOABKRS-PylT were co-transfected in the presence of OABK into HEK293T cells in 96-well plates (using the same protocol as above). After 24 h incubation, cells were washed three times and incubated in fresh medium (200 µL) for 2 h to remove excess OABK. For the luciferase activation in live cells, media (200 µL) containing the phosphines 1–6 was added and incubation was continued for 3 h. Luciferase activities were measured using a BrightGlo luciferase assay kit (Promega) and a microplate reader (Tecan M1000) with an integration time of 1 s. For luciferase assays in cell lysates, cells were lysed through addition of 20 µL of lysis buffer, followed by addition of the phosphines 1–6 (in 80 µL media) and the mixtures were incubated for 3 h. Substrate solution (100 µL, Promega) was added and luminescence was measured as described above.
Live-cell fluorescence imaging and fluorescence measurements
HEK 293T cells were plated at ~100,000 cells per well (400 µL) into a poly-D-lysine coated 8-well chamber slide (Lab-Tek). At ~75% confluency, cells were co-transfected with pEGFP-K85TAG-mCherry or pEGFP-K29TAG-SatB1-mCherry and pOABKRS-4PylT (200 ng of each plasmid) using linear PEI (3 µL, 0.323 mg/mL). After a 20 h incubation at 37 °C and 5% CO2 in DMEM with 10% FBS in the presence of OABK (0.25 mM), cells were washed three times with phenol red-free DMEM (200 µL), followed by a 3 h incubation in order to remove any non-incorporated OABK. Before small molecule activation, cells were focused using the TXRED channel, and imaged with a Nikon A1 confocal microscope (40X oil objective, 2X zoom, FITC (Ex = 488 nm) and TXRED (Ex = 560 nm) channels). Subsequently, media was changed to media (400 µL) containing 5 or 6, and EGFP and mCherry fluorescence was imaged every 5 min for 5 h (scan resolution 512X512, 2X scan zoom, dwell time 1.9 ms). The mean fluorescence intensities in the nucleus, in case of activation of SatB1-mCherry, or the cytoplasm, in the case of EGFP activation, were quantified using NIS Elements software to calculate Fnt/Fnmax or Fct/Fcmax ratios followed by normalized.
Supplementary Material
Acknowledgments
This research was supported in part by the National Institutes of Health (1R01GM112728), the National Science Foundation (MCB-1330746), and the Charles E. Kaufman Foundation of The Pittsburgh Foundation. K.M. is grateful for a Japan Society for the Promotion of Science Postdoctoral Fellowship for Research Abroad. We thank the Chin lab (MRC) for plasmids encoding the PylRS and PylT genes, the Asokan lab (UNC) for the pgRNA and pIRG plasmids, and the Hughes lab (MSU) for the Cre Stoplight (pC-SL) plasmid.
Footnotes
Author contributions. A.D. and J.L. conceived and designed the experiments; J.L, Q.L., and K.M. performed the experiments and analyzed the data; A.D. and J.L. co-wrote the paper.
References
- 1.Bishop AC, et al. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature. 2000;407:395–401. doi: 10.1038/35030148. [DOI] [PubMed] [Google Scholar]
- 2.Buckley DL, Crews CM. Small-molecule control of intracellular protein levels through modulation of the ubiquitin proteasome system. Angew. Chem. Int. Ed. 2014;53:2312–2330. doi: 10.1002/anie.201307761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Winter GE, et al. Phthalimide conjugation as a strategy for in vivo target protein degradation. Science. 2015;348:1376–1381. doi: 10.1126/science.aab1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Warner JB, Muthusamy AK, Petersson EJ. Specific modulation of protein activity by using a bioorthogonal reaction. Chembiochem. 2014;15:2508–2514. doi: 10.1002/cbic.201402423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buskirk AR, Liu DR. Creating small-molecule-dependent switches to modulate biological functions. Chem. Biol. 2005;12:151–161. doi: 10.1016/j.chembiol.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 6.Zorn JA, Wells JA. Turning enzymes ON with small molecules. Nat. Chem. Biol. 2010;6:179–188. doi: 10.1038/nchembio.318. [DOI] [PubMed] [Google Scholar]
- 7.Putyrski M, Schultz C. Protein translocation as a tool: The current rapamycin story. FEBS Lett. 2012;586:2097–2105. doi: 10.1016/j.febslet.2012.04.061. [DOI] [PubMed] [Google Scholar]
- 8.Fegan A, White B, Carlson JC, Wagner CR. Chemically controlled protein assembly: techniques and applications. Chem. Rev. 2010;110:3315–3336. doi: 10.1021/cr8002888. [DOI] [PubMed] [Google Scholar]
- 9.Rakhit R, Navarro R, Wandless TJ. Chemical biology strategies for posttranslational control of protein function. Chem. Biol. 2014;21:1238–1252. doi: 10.1016/j.chembiol.2014.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ho SN, Biggar SR, Spencer DM, Schreiber SL, Crabtree GR. Dimeric ligands define a role for transcriptional activation domains in reinitiation. Nature. 1996;382:822–826. doi: 10.1038/382822a0. [DOI] [PubMed] [Google Scholar]
- 11.Gray DC, Mahrus S, Wells JA. Activation of specific apoptotic caspases with an engineered small-molecule-activated protease. Cell. 2010;142:637–646. doi: 10.1016/j.cell.2010.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karginov AV, Ding F, Kota P, Dokholyan NV, Hahn KM. Engineered allosteric activation of kinases in living cells. Nat. Biotechnol. 2010;28:743–747. doi: 10.1038/nbt.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jullien N, Sampieri F, Enjalbert A, Herman JP. Regulation of Cre recombinase by ligand-induced complementation of inactive fragments. Nucleic Acids Res. 2003;31:e131. doi: 10.1093/nar/gng131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bonger KM, Chen LC, Liu CW, Wandless TJ. Small-molecule displacement of a cryptic degron causes conditional protein degradation. Nat. Chem. Biol. 2011;7:531–537. doi: 10.1038/nchembio.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chu PH, et al. Engineered kinase activation reveals unique morphodynamic phenotypes and associated trafficking for Src family isoforms. Proc. Natl. Acad. Sci. U. S. A. 2014;111:12420–12425. doi: 10.1073/pnas.1404487111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miyamoto T, et al. Rapid and orthogonal logic gating with a gibberellin-induced dimerization system. Nat. Chem. Biol. 2012;8:465–470. doi: 10.1038/nchembio.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gautier A, et al. How to control proteins with light in living systems. Nat. Chem. Biol. 2014;10:533–541. doi: 10.1038/nchembio.1534. [DOI] [PubMed] [Google Scholar]
- 18.Weitzman M, Hahn KM. Optogenetic approaches to cell migration and beyond. Curr. Opin. Cell Biol. 2014;30:112–120. doi: 10.1016/j.ceb.2014.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen X, et al. Synthetic dual-input mammalian genetic circuits enable tunable and stringent transcription control by chemical and light. Nucleic Acids Res. 2016;44:2677–2690. doi: 10.1093/nar/gkv1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jeong JW, et al. Wireless optofluidic systems for programmable in vivo pharmacology and optogenetics. Cell. 2015;162:662–674. doi: 10.1016/j.cell.2015.06.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li J, Jia S, Chen PR. Diels-Alder reaction-triggered bioorthogonal protein decaging in living cells. Nat. Chem. Biol. 2014;10:1003–1005. doi: 10.1038/nchembio.1656. [DOI] [PubMed] [Google Scholar]
- 22.Hemphill J, Chou CJ, Chin JW, Deiters A. Genetically encoded light-activated transcription for spatiotemporal control of gene expression and gene silencing in mammalian cells. J. Am. Chem. Soc. 2013;135:13433–13439. doi: 10.1021/ja4051026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li H, Hah JM, Lawrence DS. Light-mediated liberation of enzymatic activity: "Small Molecule" caged protein equivalents. J. Am. Chem. Soc. 2008;130:10474–10475. doi: 10.1021/ja803395d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li J, et al. Palladium-triggered deprotection chemistry for protein activation in living cells. Nat. Chem. 2014;6:352–361. doi: 10.1038/nchem.1887. [DOI] [PubMed] [Google Scholar]
- 25.Vila-Perello M, Hori Y, Ribo M, Muir TW. Activation of protein splicing by protease- or light-triggered O to N acyl migration. Angew. Chem. Int. Ed. 2008;47:7764–7767. doi: 10.1002/anie.200802502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sainlos M, Iskenderian-Epps WS, Olivier NB, Choquet D, Imperiali B. Caged mono- and divalent ligands for light-assisted disruption of PDZ domain-mediated interactions. J. Am. Chem. Soc. 2013;135:4580–4583. doi: 10.1021/ja309870q. [DOI] [PubMed] [Google Scholar]
- 27.Kohn M, Breinbauer R. The Staudinger ligation-a gift to chemical biology. Angew. Chem. Int. Ed. 2004;43:3106–3116. doi: 10.1002/anie.200401744. [DOI] [PubMed] [Google Scholar]
- 28.Van Berkel SS, Van Eldijk MB, Van Hest JC. Staudinger ligation as a method for bioconjugation. Angew. Chem. Int. Ed. 2011;50:8806–8827. doi: 10.1002/anie.201008102. [DOI] [PubMed] [Google Scholar]
- 29.Sletten EM, Bertozzi CR. From mechanism to mouse: a tale of two bioorthogonal reactions. Acc. Chem. Res. 2011;44:666–676. doi: 10.1021/ar200148z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alouane A, Labruere R, Le Saux T, Schmidt F, Jullien L. Self-immolative spacers: kinetic aspects, structure-property relationships, and applications. Angew. Chem. Int. Ed. 2015;54:7492–7509. doi: 10.1002/anie.201500088. [DOI] [PubMed] [Google Scholar]
- 31.Chen PR, et al. A facile system for encoding unnatural amino acids in mammalian cells. Angew. Chem. Int. Ed. 2009;48:4052–4055. doi: 10.1002/anie.200900683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takimoto JK, Dellas N, Noel JP, Wang L. Stereochemical basis for engineered pyrrolysyl-tRNA synthetase and the efficient in vivo incorporation of structurally divergent non-native amino acids. ACS Chem. Biol. 2011;6:733–743. doi: 10.1021/cb200057a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hancock SM, Uprety R, Deiters A, Chin JW. Expanding the genetic code of yeast for incorporation of diverse unnatural amino acids via a pyrrolysyl-tRNA synthetase/tRNA pair. J. Am. Chem. Soc. 2010;132:14819–14824. doi: 10.1021/ja104609m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Plass T, Milles S, Koehler C, Schultz C, Lemke EA. Genetically encoded copper-free click chemistry. Angew. Chem. Int. Ed. 2011;50:3878–3881. doi: 10.1002/anie.201008178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang YS, Fang XQ, Wallace AL, Wu B, Liu WSR. A rationally designed pyrrolysyl-tRNA synthetase mutant with a broad substrate spectrum. J. Am. Chem. Soc. 2012;134:2950–2953. doi: 10.1021/ja211972x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yanagisawa T, et al. Multistep engineering of pyrrolysyl-tRNA synthetase to genetically encode N(epsilon)-(o-azidobenzyloxycarbonyl) lysine for site-specific protein modification. Chem. Biol. 2008;15:1187–1197. doi: 10.1016/j.chembiol.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 37.Luo J, et al. Genetically encoded optochemical probes for simultaneous fluorescence reporting and light activation of protein function with two-photon excitation. J. Am. Chem. Soc. 2014;136:15551–15558. doi: 10.1021/ja5055862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Iizuka R, Yamagishi-Shirasaki M, Funatsu T. Kinetic study of de novo chromophore maturation of fluorescent proteins. Anal. Biochem. 2011;414:173–178. doi: 10.1016/j.ab.2011.03.036. [DOI] [PubMed] [Google Scholar]
- 39.Kawai K, et al. A reductant-resistant and metal-free fluorescent probe for nitroxyl applicable to living cells. J. Am. Chem. Soc. 2013;135:12690–12696. doi: 10.1021/ja404757s. [DOI] [PubMed] [Google Scholar]
- 40.Saneyoshi H, et al. Triphenylphosphinecarboxamide: an effective reagent for the reduction of azides and its application to nucleic acid detection. Org. Lett. 2014;16:30–33. doi: 10.1021/ol402832w. [DOI] [PubMed] [Google Scholar]
- 41.Cline DJ, et al. New water-soluble phosphines as reductants of peptide and protein disulfide bonds: Reactivity and membrane permeability. Biochemistry. 2004;43:15195–15203. doi: 10.1021/bi048329a. [DOI] [PubMed] [Google Scholar]
- 42.Baker AS, Deiters A. Optical control of protein function through unnatural amino acid mutagenesis and other optogenetic approaches. ACS Chem. Biol. 2014;9:1398–1407. doi: 10.1021/cb500176x. [DOI] [PubMed] [Google Scholar]
- 43.Brieke C, Rohrbach F, Gottschalk A, Mayer G, Heckel A. Light-controlled tools. Angew. Chem. Int. Ed. 2012;51:8446–8476. doi: 10.1002/anie.201202134. [DOI] [PubMed] [Google Scholar]
- 44.Engelke H, Chou C, Uprety R, Jess P, Deiters A. Control of protein function through optochemical translocation. ACS Synth. Biol. 2014;3:731–736. doi: 10.1021/sb400192a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ho PS, Eichman BF. The crystal structures of DNA Holliday junctions. Curr. Opin. Struct. Biol. 2001;11:302–308. doi: 10.1016/s0959-440x(00)00219-0. [DOI] [PubMed] [Google Scholar]
- 46.Nagy A. Cre recombinase: The universal reagent for genome tailoring. Genesis. 2000;26:99–109. [PubMed] [Google Scholar]
- 47.Gibb B, et al. Requirements for catalysis in the Cre recombinase active site. Nucleic Acids Res. 2010;38:5817–5832. doi: 10.1093/nar/gkq384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang YS, Hughes TE. Cre stoplight: A red/green fluorescent reporter of Cre recombinase expression in living cells. Biotechniques. 2001;31:1036, 1038, 1040–1041. doi: 10.2144/01315st03. [DOI] [PubMed] [Google Scholar]
- 49.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Davis KM, Pattanayak V, Thompson DB, Zuris JA, Liu DR. Small molecule-triggered Cas9 protein with improved genome-editing specificity. Nat. Chem. Biol. 2015;11:316–318. doi: 10.1038/nchembio.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zetsche B, Volz SE, Zhang F. A split-Cas9 architecture for inducible genome editing and transcription modulation. Nat. Biotechnol. 2015;33:139–142. doi: 10.1038/nbt.3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nishimasu H, et al. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 2014;156:935–949. doi: 10.1016/j.cell.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hemphill J, Borchardt EK, Brown K, Asokan A, Deiters A. Optical control of CRISPR/Cas9 gene editing. J. Am. Chem. Soc. 2015;137:5642–5645. doi: 10.1021/ja512664v. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.