Abstract
Escherichia coli and other Gram-negative and -positive bacteria employ type IV secretion systems (T4SSs) to translocate DNA and protein substrates, generally by contact-dependent mechanisms, to other cells. The T4SSs functionally encompass two major subfamilies, the conjugation systems and the effector translocators. The conjugation systems are responsible for interbacterial transfer of antibiotic resistance genes, virulence determinants, and genes encoding other traits of potential benefit to the bacterial host. The effector translocators are used by many Gram-negative pathogens for delivery of potentially hundreds of virulence proteins termed effectors to eukaryotic cells during infection. In E. coli and other species of Enterobacteriaceae, T4SSs identified to date function exclusively in conjugative DNA transfer. In these species, the plasmid-encoded systems can be classified as the P, F, and I types. The P-type systems are the simplest in terms of subunit composition and architecture, and members of this subfamily share features in common with the paradigmatic Agrobacterium tumefaciens VirB/VirD4 T4SS. This review will summarize our current knowledge of the E. coli systems and the A. tumefaciens P-type system, with emphasis on the structural diversity of the T4SSs. Ancestral P-, F-, and I-type systems were adapted throughout evolution to yield the extant effector translocators, and information about well-characterized effector translocators also is included to further illustrate the adaptive and mosaic nature of these highly versatile machines.
INTRODUCTION
Secretion of DNA and protein macromolecules across the Gram-negative cell envelope requires elaboration of dedicated translocation systems and the coupling in space and time of substrate recruitment, processing, and translocation reactions. Translocation of substrates to another cell by contact-dependent mechanisms adds more complexity through the requisite establishment of productive cell-cell junctions. The type IV secretion systems (T4SSs) deliver various macromolecular substrates, including DNA and monomeric and multimeric proteins, to a diversity of bacterial, fungal, plant, and human cell types (1, 2). Some T4SSs also function as contact-independent exporters or importers of substrates to or from the extracellular milieu. This functional versatility makes the T4SSs unique among the known bacterial translocation systems. In Escherichia coli, the known T4SSs are restricted in their substrate repertoires to mobile genetic elements (MGEs), although these systems also are capable of translocating certain proteins associated with the DNA transfer process. Detailed investigations of conjugation machines have established the importance of T4SSs over evolutionary time in the shaping of bacterial genomes and, on a more immediate time scale, for dissemination of virulence and antibiotic resistance genes in clinical settings (3, 4). These studies also have contributed greatly to our current understanding of T4SS machine architecture, mechanism of action, and evolution (3, 5, 6, 7).
The T4SSs are functionally grouped as (i) conjugation systems, (ii) effector translocators, or (iii) contact-independent DNA/protein exchange systems (1). The conjugation systems are the largest subfamily, present in nearly all bacterial species and some archaeal species (5). These systems are specifically employed for dissemination of the mobile elements that encode them, and they also can mediate transfer of some genetically unlinked, non-self-transmissible mobile elements. The effector translocators, so far shown to function only in Gram-negative pathogens or symbionts, deliver effector proteins to eukaryotic cells (8, 9, 10). The translocated substrates disrupt host cell physiological processes, enabling bacterial colonization and spread. The contact-independent exchange systems, consisting of only a few members, function in release of DNA or protein substrates to the milieu or, alternatively, uptake of exogenous DNA (11, 12, 13).
In E. coli, several conjugation systems have been characterized in detail (see Fig. 1). These systems elaborate two surface structures, a cell-envelope-spanning translocation channel and an extracellular pilus (6, 7). A classification scheme that will be used in this review groups the E. coli conjugation systems by the type of conjugative pilus produced. Accordingly, F-type systems elaborate long, flexible pili that dynamically extend and retract (14, 15, 16). These pili support equally efficient mating in liquid or solid surfaces. The P-type systems, by contrast, elaborate short, rigid pili for which no evidence exists of dynamic extension and retraction. Well-characterized conjugation systems encoded by E. coli plasmids RP4, R388, and pKM101, as well as the Agrobacterium tumefaciens VirB/VirD4 system, are P-type systems (17). A third group of plasmids represented by R64 and ColIb-P9 encode two types of pilus structures: a thick, rigid conjugative pilus and a long, flexible type IV pilus. Despite their name, type IV pili are ancestrally unrelated to conjugative pili produced by T4SSs and instead are phylogenetically and functionally related to type II secretion systems (T2SSs) (18). Nevertheless, type IV pili do play a role in enhancing conjugative transfer of these plasmids, especially in liquid media, by virtue of their ability to extend and retract, reminiscent of F pili. The T4SSs encoded by this group of plasmids are designated as I type, and they have a number of physical and functional features that distinguish them from the F- and P-type systems (19, 20). A major aim of this review is to summarize structure-function relationships of paradigmatic F-, P-, and I-type conjugation systems of E. coli and the VirB/VirD4 system of A. tumefaciens.
Figure 1.
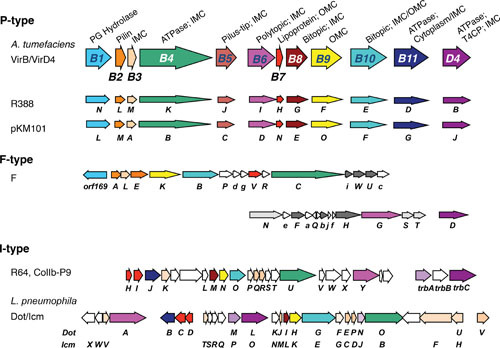
Conservation of type IV secretion genes among P-, F-, and I-type T4SSs. (P type) The A. tumefaciens VirB/VirD4 reference system with subunit enzymatic functions and associations with inner membrane complex (IMC), outer membrane complex (OMC), or pilus. PG Hydrolase, peptidoglycan hydrolase; T4CP, type IV coupling protein. (F type) Genes related to the virB/virD4 genes are color coded. Genes encoding functions required for F pilus/retraction are shaded in dark gray, and for mating pair stabilization (Mps) or surface exclusion in light gray (VirB6-like TraG also functions in mating pair stabilization). Uppercase letters are tra genes, lowercase letters are trb genes. (I type) virB/virD4-like genes are color coded. Genes unique to the I-type systems encoding inner membrane proteins are in beige; genes encoding subunits that functionally interact with the DotL T4CP are in light-shaded purple. P- and I-type systems employ three ATPases related to VirD4, VirB4, and VirB11; F-type systems employ only homologs of VirD4 and VirB4. Unless otherwise indicated, the systems shown are functional in E. coli.
On the basis of detailed phylogenetic studies, it is postulated that the present-day effector translocators evolved from P-, F-, and I-type conjugation systems (5). In general, this process involved the appropriation by ancestral conjugation systems of structural motifs from unknown ancestries that enabled diversification of substrate repertoires and the capacity to deliver substrates to eukaryotic host cells. A second goal of this review is to compare well-characterized conjugation and effector translocator systems to shed light on this evolutionary process and to illustrate the striking structural diversity and functional versatility of this fascinating translocator superfamily.
OVERVIEW OF T4SS ARCHITECTURE AND TRANSLOCATION PATHWAY
The paradigmatic A. tumefaciens VirB/VirD4 T4SS provides a unifying genetic nomenclature for a discussion of the T4SSs (Fig. 1) (21). The T4SSs of Gram-negative bacteria can be viewed as compilations of four distinct proteins or subassemblies: (i) the VirD4 type IV coupling protein (T4CP); (ii) the inner membrane complex (IMC) composed of VirB4 ATPase, VirB11 ATPase (when present), polytopic VirB3 and VirB6, and bitopic VirB8; (iii) the outer membrane complex (OMC) composed of outer membrane-associated VirB7 and VirB9, and a cell-envelope-spanning subunit VirB10; (iv) the conjugative pilus composed of VirB2 pilin, a proteolytic fragment of the VirB1 transglycosylase (VirB1*) and the pilus-tip protein VirB5 (Fig. 2A) (6, 7). The VirB/VirD4 T4SS and related P-type systems in E. coli are among the simplest T4SSs functioning in Gram-negative species in terms of subunit composition and machine architecture. Other P-type systems as well as the F- and I-type systems encode homologs or orthologs of most or all of the VirB subunits, but also possess additional domains, subunits, or protein subcomplexes that endow these systems with specialized functions and confer greater structural complexity (9, 14, 22).
Figure 2.
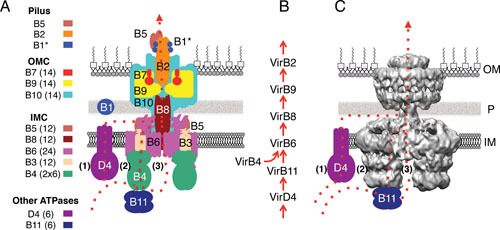
Architecture of a prototypical P-type T4SS. The E. coli R388 T4SS3–10 structure (26), presented schematically using the A. tumefaciens VirB/VirD4 reference nomenclature and as visualized by transmission electron microscopy. (A) The T4SS is composed of an extracellular pilus, an outer membrane complex (OMC), and inner membrane complex (IMC), the VirD4-like T4CP (substrate receptor) and, for most systems, a VirB11 ATPase. Substructures of the T4SS: The conjugative pilus was not part of the solved T4SS3–10 structure, but is postulated to associate with the structure as depicted. In the A. tumefaciens VirB/VirD4 T4SS, the pilus is composed of a fragment of the VirB1 transglycosylase (VirB1*), VirB2 pilin, and VirB5 tip protein (126, 138). The OMC is composed of the VirB7, VirB9, and VirB10 subunits in copy numbers listed in parentheses (26). The VirB1 transglycosylase is required for pilus assembly but not for elaboration of the translocation channel. The IMC is composed of the VirB5, VirB8, VirB6, VirB3, and VirB4 subunits with copy numbers listed in parentheses (26). Other ATPases, including VirD4 and VirB11, were not part of the solved T4SS3–10 structure, but are postulated to associate with the IMC as depicted. Color-coding of the subunits matches that for the corresponding virB/virD4 genes in Fig. 1. (B) The A. tumefaciens subunits shown to form formaldehyde cross-links with the T-DNA substrate during transfer through the VirB/VirD4 T4SS with the TrIP assay. Red arrows denote the proposed translocation pathway (23). (C) The R3883–10 structure solved by transmission electron microscopy, reproduced with permission by reference 26. The OMC is postulated to house the translocation channel that extends through the periplasm and across the outer membrane. The OMC is connected to the IMC by a narrow stalk. Two hexamers of VirB4 extend into the cytoplasm and are postulated to establish contacts with the VirD4 T4CP and the VirB11 ATPase. (A and C) For both the schematic and solved T4SS3–10 structure, three possible routes of substrate translocation across the inner membrane are depicted in red dashed lines. A single route is postulated, in a red dashed line, through the OMC for substrate passage to the cell exterior (see text for details). IM, inner membrane; P, periplasm; OM, outer membrane.
Two recent lines of investigation have shaped our current view of T4SS architecture and the substrate translocation pathway (Fig. 2). Although investigations commencing in the 1940s established the importance of so-called mating pair formation (Mpf) proteins for conjugative DNA transfer, it was not until the early 2000s that the first definitive evidence was obtained that these proteins assemble as a mating channel. By use of a ChIP-based, formaldehyde (FA)-crosslinking assay termed transfer DNA immunoprecipitation (TrIP), a translocation pathway was described for the A. tumefaciens transfer-DNA (T-DNA) substrate through the VirB/VirD4 T4SS (23). With the TrIP assay, FA-cross-linkable substrate contacts were detected with the VirD4 T4CP, VirB11 ATPase, inner membrane proteins VirB6 and VirB8, and periplasmic/outer membrane subunits VirB2 and VirB9 (Fig. 2B). These subunits were thus postulated to constitute the cell-envelope-spanning “mating” channel. Other components of the T4SS that did not form FA cross-links with the T-DNA substrates were proposed to function as structural scaffolds or stabilizing elements for the channel (23).
Second, structures of T4SS subunits or soluble domains and of larger subassemblies have been solved by X-ray crystallography or electron microscopy. Most notably, structures recently were presented for the OMC and the IMC/OMC subassemblies of P-type systems encoded by E. coli plasmids pKM101 and R388 (24, 25, 26). The R388-encoded IMC/OMC subassembly is the largest T4SS structure solved to date and is composed of homologs of the A. tumefaciens VirB3 through VirB10 subunits, lacking only homologs of VirD4, VirB11, VirB1, and VirB2 (26). This structure, here referred to as R388 T4SS3–10, is the current architectural blueprint for the T4SSs (Fig. 2C) and is described in more detail below.
By integrating results of the in vivo TrIP and ultrastructural studies, a current model for translocation of substrates through a P-type T4SS postulates that substrates first dock with the VirD4 T4CP and then are transferred to VirB11 for further processing. All three ATPases, VirD4, VirB11, and VirB4, then coordinate substrate transfer across the inner membrane via a channel composed minimally of VirB6 and VirB8. The substrate is then delivered into a channel, composed of VirB2 and VirB9, that is housed within the OMC for translocation across the periplasm and outer membrane (Fig. 2) (7, 23, 27). The following sections describe this P-type translocation pathway and interesting structural or mechanistic variations among the F- and I-type systems.
SUBSTRATES AND SUBSTRATE RECOGNITION
In the initiating steps of type IV secretion, DNA and protein substrates are processed and recruited to cognate T4SSs for translocation. The early DNA substrate-processing reactions have been extensively reviewed elsewhere (3, 28). In brief, these are conserved reactions in most bacteria, carried out by proteins termed DNA transfer and replication (Dtr) factors. Dtr proteins bind specific origin-of-transfer (oriT) sequences associated with mobile elements, resulting in formation of the catalytically active relaxosome. One Dtr protein, the relaxase, cleaves the DNA strand destined for transfer (the T strand) at the nic site through a DNA phosphodiesterase reaction that results in covalent linkage of a catalytic Tyr residue with the 5′ end of the cleaved DNA. Accessory Dtr factors play critical roles in guiding the relaxase to the oriT sequence and facilitating the nicking reaction on supercoiled DNA substrates. Upon nicking, the relaxase serves to pilot the covalently associated T strand through the conjugation or “mating” channel. The early DNA-processing reactions are spatially positioned at or near the entrance to the conjugation channel, specifically mediated by contacts formed between relaxosome components and the VirD4-like T4CP (28, 29, 30). Accordingly, the T4CP functions as the receptor and docking site for cognate DNA substrates. This receptor activity, however, is not restricted to DNA substrates. T4CPs are associated with nearly all effector translocator systems, where they also function as receptors for protein substrates (2, 31).
DNA Substrate Recognition Signals
The nature of DNA substrate-T4CP contacts is not yet fully defined, but studies have identified motifs associated with relaxases that are required for DNA transfer. These motifs are postulated to specify the docking of DNA substrates with cognate T4CPs and are designated as translocation signals (TSs). Among the relaxases characterized to date, two types of TSs have been identified. The F plasmid-encoded TraI relaxase, for example, is a large protein with relaxase and helicase domains in its two halves. TSs termed TSA and TSB were mapped, respectively, in the two halves of the protein (32). Both TSs have a consensus sequence (G[E/D]R[L/M]R[V/F]T), and a structure of the TSA region of TraI was solved recently by X-ray crystallography (33). It consists of three domains, each structurally similar to SF1B helicase family domains. A putative T4CP interaction surface was mapped to one of the helicase domains through mutational analysis. Other relaxases, including R388-encoded TrwC and RSF1010-encoded MobA, carry TSs similar to those identified in TraI, suggestive of a common mode of relaxase binding to cognate T4CPs (34, 35, 36).
Relaxases alternatively have C-terminal signals that can contribute to DNA substrate-T4CP docking. In A. tumefaciens, for example, the VirD2 relaxase carries such a TS, which is composed of a high proportion of positively charged residues. This motif is critical for the VirD2-VirD4 T4CP interaction and for transfer of the T-DNA substrate through the VirB/VirD4 T4SS to plant cells (see below). Interestingly, the VirB/VirD4 T4SS also translocates several protein substrates to plant cells, and these substrates also carry C-terminal, positively charged TSs (37, 38). Finally, relaxases can carry a combination of internal and C-terminal TSs for docking with cognate receptors. In some cases, such relaxases have been shown to employ these signals to mediate transfer of DNA substrates through distinct T4SSs. RSF1010 plasmids, for example, are promiscuous in the sense that they can translocate through many different T4SSs. RSF1010’s MobA relaxase carries internal TSs required for RSF1010 transfer through P-type T4SSs in E. coli. However, the relaxase also carries a C-terminal, positively charged TS that promotes RSF1010 transfer through the A. tumefaciens VirB/VirD4 T4SS (34, 36, 38). Similarly, the TrwC relaxase employs internal TSs for substrate transfer through the R388-encoded T4SS, and a C-terminal motif for substrate passage through a Bartonella henselae VirB/VirD4 T4SS (35).
Relaxases are only partly responsible for specifying the docking of DNA substrates with cognate T4CPs. Other Dtr accessory factors can confer substrate specificity, as best documented for the F-type systems. For F plasmids, substrate docking relies on the formation of highly specific contacts between the accessory factor TraM and the TraD receptor. In fact, the TraM-TraD interaction is the only structural interface solved to date by X-ray crystallography, as described in further detail in the next section.
T4CPs and the DNA Substrate-Docking Reaction
The T4CPs are so named because they link secretion substrates with cognate T4SS channels (23, 39, 40, 41). They typically are composed of an N-terminal transmembrane domain (NTD) joined to a large cytoplasmic moiety that consists of a nucleotide binding domain (NBD) and an α-helical bundle termed the all-alpha-domain (AAD) (Fig. 3A, B). An X-ray structure showed that a soluble fragment of the TrwB T4CP assembles as a globular homohexamer of ∼110 Å in diameter and 90 Å in height, with a ∼20-Å-wide channel in the center. This channel forms an 8-Å-wide constriction at the cytoplasmic pole of the molecule. The NTD extends from the opposite end of the globular assembly, as shown by electron microscopy, giving rise to an overall F1-F0-like ball-stem structure (Fig. 3A) (41, 42). These findings, coupled with evidence that T4CPs exhibit sequence and structural similarities with the FtsK and SpoIIIE DNA translocases, prompted a model in which the T4CP serves as the translocase for delivery of DNA substrates across the inner membrane (See Fig. 2, route 1) (43).
Figure 3.
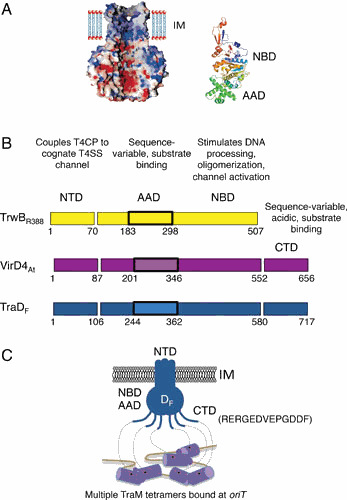
Domain architecture of type IV coupling proteins (T4CPs). (A) The structural prototype of the T4CPs, R388-encoded TrwB, is a hexamer with a stem composed of the N-terminal transmembrane domains (NTDs) of the 6 protomers and a ball composed of the nucleotide binding domains (NBDs) and all-alpha-domains (AADs). (Left) A space-filling model of the TrwB hexamer showing the Connolly surface (red, negatively charged; blue, positively charged) and central channel connecting the cytoplasm to the periplasm. IM, inner membrane. (Right) A ribbon diagram of the NBD (multicolored) and AAD (green). In the assembled hexamer, the AAD sits at the entrance of the hexamer lumen. Images reproduced with permission by reference 43. (B) Schematics display the domain architectures of P-types TrwB and A. tumefaciens VirD4 and F-type TraD; numbers represent domain junctions with residue numbers relative to the start codon. The AAD is boxed. Known or predicted properties are listed above each domain. TrwB lacks a C-terminal domain (CTD), but such domains are carried by other VirD4 homologs including F-type TraD. (C) The TraD T4CP in the inner membrane with the various domains listed. The extreme C terminus of the CTD (residues in parentheses) is negatively charged and forms specific contacts with C-terminal α-helical domains (purple cylinders) of the TraM Dtr accessory protein. TraM’s N-terminal ribbon-helix-helix domain (RHH; purple dots) mediates binding to sbmA sites located within the F plasmid oriT sequence. The TraD-TraM interaction specifies F plasmid transfer through the F-encoded T4SS. IM, inner membrane.
Two features of the TrwB structure point to a role for the AAD in DNA substrate docking: (i) the AAD is positioned at the cytoplasmic pole of the TrwB homohexamer (Fig. 3A) and (ii) the AAD structurally resembles the DNA binding domain of XerD recombinase (41). Mutational analyses confirmed the functional importance of AADs associated with TrwB as well as a homolog encoded by plasmid RP4 (44, 45). A recent study further showed that the AADs of the A. tumefaciens VirD4 T4CP and of a T4CP encoded by Enterococcus faecalis pCF10 are essential for translocation of cognate DNA substrates in vivo (46). Purified forms of these domains bind DNA substrates and, more importantly, display specificity for cognate relaxases and other Dtr factors (46). As mentioned above, the A. tumefaciens VirD2 relaxase possesses a C-terminal TS required for T-DNA transfer to plants (38, 47, 48, 49). A VirD2 mutant deleted of this TS fails to bind VirD4’s AAD in vitro, suggesting that a VirD2 TS-VirD4 AAD interaction constitutes a basis for docking of the T-DNA substrate with the VirB/VirD4 channel in A. tumefaciens (49).
T4CPs also can carry C-terminal domains (CTDs) of variable lengths and sequence compositions (Fig. 3B) (2). Contributions of these CTDs to substrate transfer are unspecified, with one notable exception. For F-type systems, the CTD of the TraD receptor plays a critical role in docking of the F plasmid substrate (Fig. 3C) (50). An acidic motif at the extreme C terminus of the CTD was shown by X-ray crystallography to mediate specific contacts with the Dtr accessory factor TraM (51, 52). Mutational analyses further confirmed that the TraD hexamer-TraM interaction forms the basis of a highly specific relaxosome-T4CP interaction in vivo (52, 53). This interaction both mediates highly efficient F plasmid transfer and blocks access of the RSF1010 plasmid substrate to the T4CP, resulting in strong inhibition of RSF1010 plasmid transfer (50). TraD’s CTD thus functions as a specificity checkpoint by ensuring efficient docking of the cognate F plasmid substrate at the exclusion of alternative substrates. At this juncture, therefore, the available data suggest that T4CPs recruit cognate DNA substrates through interactions mediated by AADs and, when present, CTDs. Further work is needed to define substrate-T4CP interactions at higher resolutions and identify factors controlling the dynamics of these contacts.
The “coupling” function of T4CPs is only partly defined by its receptor activities. T4CPs also interact with cognate T4SS channel subunits to mediate delivery of docked substrates through the channel. The NTDs of T4CPs are required for these contacts, as shown by two-hybrid screens and mutational analyses (54, 55). These domains are implicated in formation of contacts with VirB10-like subunits (54, 55, 56), and it has been proposed that such contacts are responsible for the coupling of T4CPs with cognate channels. On purely stoichiometric grounds, however, it is difficult to envision how a T4CP hexamer(s) forms specific contacts with VirB10 subunits, which, according to the R388 T4SS3–10 structure, exist in 14 copies in the translocation channel (Fig. 2) (26, 41). Another complicating aspect of this interaction is that T4CPs have been shown to undergo profound changes both in conformation and oligomeric state in response to substrate docking and ATP binding signals (57, 58; see below). Accordingly, it seems more reasonable that, rather than forming stable, high-affinity contacts with T4SS channel subunits, T4CPs associate dynamically with T4SSs, possibly in response to activation by intracellular signals. That the T4CP is not a fixed, structural component of the T4SS is further supported by the fact that, in its absence, the T4SS is fully functional in its ability to elaborate conjugative pili (2, 22).
STRUCTURE AND FUNCTION OF TYPE IV MACHINES
The T4CPs deliver secretion substrates to the translocation channel composed of the VirB homologs. As mentioned above, the R388 T4SS3–10 structure currently represents the architectural blueprint for the T4SS superfamily (Fig. 2C). It is dominated by two large subassemblies, the IMC and OMC, which are connected by a narrow stalk. The following sections summarize current structure-function information about these subassemblies and the respective components, with emphasis on their contributions to the dynamics of substrate transfer across the inner and outer membranes.
Contributions of ATPases to Substrate Trafficking
In the R388 T4SS3–10 structure, two hexamers of the VirB4-like ATPase form the “legs” of the IMC substructure, but the VirD4- and VirB11-like ATPases were absent (Fig. 2). The ATPase interaction network is thus not yet described in molecular detail, nor is the specific contribution(s) of ATP energy to the overall process of substrate transfer. There is, however, compelling evidence for the P-type systems that all three ATPases coordinate their activities to drive substrate transfer across the inner membrane (54, 59). Interestingly, while the P- and I-type conjugation systems employ homologs of the VirD4, VirB4, and VirB11 ATPases (19, 20, 60, 61), in striking contrast, the F-type systems as well as conjugation systems functioning in Gram-positive species employ only homologs VirD4 and VirB4 (8, 22).
The role of the T4CP as a substrate receptor is described above. Importantly, receptor activity does not require the ATP hydrolysis by the T4CP, suggesting that this catalytic activity is required for subsequent steps of the transfer process. One such postulated function is that the T4CP itself catalyzes the delivery of secretion substrates across the inner membrane (Fig. 2, route 1). According to this model, the DNA substrate binds the T4CP via its AAD and then enters the central lumen of the hexamer. The substrate is then pumped through the lumen by rounds of ATP hydrolysis across the membrane (3). At this time, the only support for this model is that the T4CP is phylogenetically and structurally similar to the DNA translocases SpoIIIE and FtsK (43). It is also difficult to reconcile this model with results of the TrIP studies showing that all three ATPases, VirD4, VirB4, and VirB11, are required for delivery of the DNA substrate to the inner membrane subunits VirB6 and VirB8 (23). Thus, alternative models have postulated that the three ATPases form an energy center that coordinates early-stage DNA-processing reactions as well as delivery of the DNA transfer intermediate across the membrane (Fig. 2, routes 2 and 3).
Specific contributions of the VirB-like ATPases to substrate transfer have not been defined, but results of mutational analyses as well as structural information about these subunits have supplied important clues regarding their functions. For the VirB11-like ATPases, results of the TrIP studies showed that VirB11 interacts with the DNA substrate only in cells producing the VirD4 receptor, implying that VirD4 delivers the substrate to VirB11 (23). Mutational studies further supplied evidence that VirB11 also engages with effector protein substrates (62). Structural studies of VirB11 homologs revealed that these proteins belong to a secretion ATPase superfamily, whose other members include GspE subunits associated with T2SSs and type IV pilus assembly systems and InvC associated with a Salmonella enterica type III secretion system (T3SS) (63–65). These ATPases undergo large nucleotide-dependent conformational changes thought to be important for providing the mechanical leverage necessary for driving machine assembly processes or substrate unfolding. Indeed, studies confirmed that InvC catalyzes both the release of bound secretion chaperones and unfolding of effector proteins in an ATP-dependent manner (66). By analogy, VirB11 subunits might catalyze the unfolding of relaxases and effector proteins prior to translocation through the T4SS.
The VirB4 ATPases are large (>70 kDa) proteins that bind tightly or integrally with the cytoplasmic membrane. Interestingly, these proteins are phylogenetically related to and exhibit structural similarities with T4CPs (5, 67). VirB4-like subunits have been shown to form multiple contacts with other IMC subunits, as well as with the VirB10 subunit (54, 59, 68). In A. tumefaciens, cross-linking of the T-DNA substrate with VirB4 was not detected by TrIP, but production of VirB4 was necessary for detectable substrate cross-linking with the VirB6 and VirB8 channel components (23). In more recent studies, evidence was presented for DNA substrate binding in vivo by a VirB4 homolog associated with a E. faecalis T4SS (69) and for DNA binding in vitro by VirB4 homologs associated with the E. coli pKM101- and R388-encoded T4SSs (70, 71). These findings raise the possibility that VirB4 plays a more direct role in mediating the transfer of DNA substrates across the inner membrane than originally envisioned (23). Accordingly, it can be proposed that, upon docking of the DNA substrate with the VirD4-like receptor, the substrate is delivered to the VirB11 ATPase for further processing. In turn, VirB11 might deliver the substrate to the VirB4 ATPase for transfer to or across the inner membrane (Fig. 2, routes 2 and 3).
The Inner Membrane Complex and Its Contribution to Substrate Transfer across the Inner Membrane
In early studies, the IMC was envisioned as a subassembly consisting of the ATPases and one or a few copies each of the membrane proteins VirB3, VirB6, and VirB8 (17). Structural definition of the R388 T4SS3–10 subassembly, however, now establishes that the IMC is considerably larger than previously thought (Fig. 2C) (26). This substructure presents as an arch of 255 Å in width sitting on top of the two VirB4 hexamer barrel structures each with dimensions of 105 Å in width and 135 Å in length. The arch, composed of 12 copies each of VirB8 and VirB3, and 24 copies of VirB6, sits in the inner membrane. Intriguingly, the arch also is composed of 12 copies of VirB5, which also localizes at the tip of the conjugative pilus (26). Finally, the IMC is composed of 14 copies of VirB10, which also is a major structural component of the OMC (see below). In view of known topologies of these subunits, the IMCs of P-type systems can be estimated to consist of at least 200 transmembrane helices.
VirB6 channel activity?
Results of the TrIP studies in A. tumefaciens favored a translocation pathway whereby the three ATPases coordinate delivery of DNA substrates to a membrane channel composed of VirB6 and VirB8 (Fig. 2, route 3) (23, 54). Various VirB6 mutations also were found to selectively block formation of T-DNA contacts with VirB8, suggesting that VirB6 and VirB8 act sequentially in the substrate transfer pathway (Fig. 2B) (72). Other mutations in VirB6 selectively inhibited formation of substrate contacts with the VirB2 and VirB9 subunits, further indicating that VirB6 also might form contacts with OMC components necessary for passage of the DNA substrate through the distal portion of the channel (see below) (72).
VirB6 subunits minimally are ∼300 residues with 5 or more membrane-spanning domains and a large central, periplasmic domain (2, 72). In view of the recent T4SS3–10 structure, it is intriguing to consider how 24 copies of VirB6 might be configured as part of the translocation channel. One possibility warranting further investigation is that VirB6 oligomerizes to form more than one functional channel across the inner membrane. This could, for example, enable the simultaneous passage of multiple substrates through a given T4SS in response to environmental cues. For conjugation machines mediating the transfer of a mobile genetic element to a bacterial recipient, such redundancy of function might not seem necessary. However, as noted earlier, conjugation systems translocate not only DNA substrates but also certain proteins, e.g., primases, single-stranded DNA binding proteins (SSBs), that function in establishment of the mobile element upon transfer to recipient cells (73, 74). Successful transfer of a DNA substrate therefore could rely on the coordinated and simultaneous transfer of DNA-metabolizing proteins, sometimes in many copies as in the case of SSBs, through a given T4SS. It is also noteworthy that effector translocator systems typically deliver multiple substrates to eukaryotic target cells. While this could be achieved through reiterative rounds of transfer through a single channel or different T4SS machines, a potentially more efficient mechanism would employ multiple channels within the framework of a single T4SS machine for the simultaneous export of multiple substrates. This could explain how, in Legionella pneumophila, the Dot/Icm system is able to rapidly deliver as many as several hundred effectors to eukaryotic cells during an infection (75, 76).
With these considerations in mind, the various translocation routes depicted in Fig. 2 might not be mutually exclusive. For example, the T4CP or VirB4-like ATPase might suffice to translocate DNA substrates (routes 1 and 2), while the VirB6/VirB8 channel mediates protein trafficking (route 3). Even for DNA transfer, a model was proposed that the T4CP functions as the translocase for the DNA component of a DNA substrate and the VirB6/8 subunits mediate transfer of the relaxase bound to the 5′ end of the DNA substrate (3). Such a model is intriguing as it invokes two distinct translocation pathways for the delivery of a single DNA substrate, the relaxase-T-strand transfer intermediate, across the inner membrane. It also has been envisioned that large periplasmic loops of VirB6 monomers might assemble to form an entry portal for DNA substrates delivered to the periplasm by the T4CP ATPase. This proposal was based on findings in A. tumefaciens that mutations in a central periplasmic loop of VirB6 blocked formation of formaldehyde cross-links with the DNA substrate (72). In fact, the existence of an entry portal in the periplasm for feeding substrates into the channel housed by the OMC is appealing in view of evidence that some effector translocator systems recruit and export substrates that are first delivered to the periplasm via the general secretory pathway (GSP; see below).
VirB8, an IMC-OMC connector?
The VirB8 subunits are bitopic proteins with a short cytoplasmic N-terminal domain, a membrane-spanning domain, and a large C-terminal periplasmic domain (77). VirB8 subunits are signatures of T4SSs carried by Gram-negative and -positive species (8), and recent work has established that their periplasmic domains adopt a common structural fold (78, 79, 80, 81, 82). This fold presents as large extended β-sheets with five α-helices, giving rise to an overall globular fold. VirB8 subunits pack as dimers or trimers in the crystal structures, and results of mutational analyses suggest that oligomerization is physiologically relevant. Indeed, the dimer interface has served as a target for a high-throughput screen that resulted in the identification of small-molecule inhibitors of T4SSs (83).
Recently, crystal structures were solved for the periplasmic domains of VirB8-like DotI and TraM associated with the I-type L. pneumophila Dot/Icm and plasmid R64 T4SSs (82). Interestingly, the VirB8-like domain of DotI forms a stack of two octameric rings in the crystal structure, and results of mutational analyses suggest that the contacts between the two octameric rings are biologically important in vivo. The octameric structure distinguishes DotI of the I-type systems from P-type homologs, which are predicted to assemble as a dodecameric ring in the IMC (26, 82). Nevertheless, in view of the earlier TrIP data generated with the A. tumefaciens system, it is reasonable to propose that these subunits assemble as ring-like structures at the IMC/OMC junction for conveyance of substrates from the IMC into the channel housed by the OMC.
The Outer Membrane Complex and Its Contribution to Substrate Transfer across the Outer Membrane
In A. tumefaciens, the outer membrane lipoprotein VirB7 and VirB9 interact with bitopic inner membrane protein VirB10, forming a subassembly originally termed the core complex (see Fig. 2A) (17, 21). This complex, renamed the OMC, is intrinsically stable and stabilizing for most of the other VirB subunits (17). A. tumefaciens ring-shaped OMC complexes were visualized by transmission electron microscopy (TEM) (84), and corresponding OMCs associated with P-type T4SSs encoded by E. coli plasmids pKM101 and R388 have been structurally solved by X-ray crystallography, cryoelectron microscopy, and TEM (24, 25, 26).
The pKM101-encoded OMC is composed of 14 copies each of the VirB7, VirB9, and VirB10 homologs (24, 25). This is a large, 1.05 MDa barrel-shaped structure of 185 Å in width and length. It is composed of two layers (I and O layers) forming a double-walled structure. The I layer, composed of the N-terminal domains of VirB9 and VirB10, is anchored in the inner membrane via the N-terminal transmembrane domain of VirB10. The N-terminal domains of VirB10 form a ring of 55 Å in diameter when the OMC is produced in the absence of the IMC. The O layer, composed of VirB7 and domains of VirB9 and VirB10, has a main body and cap that is thought to span the outer membrane. The cap, with a narrow hole of 10 Å in diameter, consists of 14 copies of a domain of VirB10 termed the antennae projection (AP). The OMC thus forms a large barrel with openings at both ends (24, 25). VirB10-like proteins are highly unusual bacterial membrane proteins in extending across the entire cell envelope (24, 85). This feature lends itself well to roles for these subunits not only as structural scaffolds for the T4SS (27) but also as transducers of activating signals from within or outside the cell across the T4SS (see below).
In the R388 T4SS3–10 structure, the OMC is linked to the IMC by a thin structure termed the stalk (Fig. 2B) (26). The composition of the stalk is not defined but must consist in part of the 14 copies of VirB10’s N-proximal region since the N terminus of VirB10 extends across the inner membrane. The biological importance of the stalk, or even its existence in the context of the T4SS when embedded in the cell envelope of an intact cell, is not yet established. However, a gap between the IMC and OMC junction may enable access of substrates delivered into the periplasm via the GSP to the OMC for translocation across the outer membrane.
The interior chamber of the OMC is sufficiently large to house a translocation channel consisting of VirB2 and VirB9, which were shown by TrIP to cross-link with the translocating T-DNA substrate (24, 25, 86). Interestingly, VirB10 did not detectably interact with the DNA substrate (23), even though VirB10’s AP presumably forms the outer membrane-spanning pore (24, 25). OMC structures solved to date lack VirB2 pilin homologs, and it is conceivable that VirB2-like subunits assemble as part of the channel at the distal end of the OMC to add structural complexity to the outer membrane pore (Fig. 2A) (27).
The Translocation Route
With an emerging knowledge of T4SS architecture, it is interesting to return to the question of how substrates are routed through the T4SS across the cell envelope. Translocation systems of Gram-negative bacteria deliver their cargoes either in one or in two steps. One-step pathways employ a single channel that spans the entire cell envelope, and two-step pathways employ an inner membrane translocase and a second system that recruits the substrate from the periplasm for delivery across the outer membrane (see reference 18). Most T4SSs are thought to employ a one-step route, as depicted in Fig. 2, routes 2 and 3. For conjugation systems, such a pathway is highly likely given the prevalence of nucleases in the periplasm. Indeed, in uninterrupted matings over periods of many minutes, E. coli Hfr strains can translocate single-stranded forms of nearly entire chromosomes through F-type T4SSs without apparent degradation. A one-step pathway does not exclude the use of a T4CP for delivery of DNA cargoes across the inner membrane (Fig. 2, route 1), however, because the physical disposition of the T4CP relative to the T4SS3–10 subassembly is not known presently. Thus, the T4CP could fit within the body of the IMC, possibly even substituting for one or both of the VirB4 “legs,” to enable DNA delivery through a T4CP-channel contact without substrate exposure to the periplasm (see Fig. 2).
It is simplest to envision that a one-step pathway also is employed for translocation of protein substrates, but this is not invariably the case, because at least two systems lack a T4CP and instead rely on the GSP or another inner membrane translocase for delivery of the protein substrate to the periplasm. The Bordetella pertussis Ptl system was the first described T4SS utilizing a two-step pathway. The sole substrate of the Ptl system is the pertussis toxin (PT) of the A/B family of toxins. The A and B subunits of PT carry canonical sec signal sequences for secretion to the periplasm by the GSP. Once in the periplasm, the PT subunits assemble as the holotoxin, which is then recruited and exported by the Ptl system to the extracellular milieu (11). The Ptl system resembles other P-type systems in subunit composition and most likely in overall architecture (see Fig. 1 and 2) (87). With the R388 T4SS3–10 subassembly as a structural reference (Fig. 2C), it is enticing to suggest that the holotoxin enters the Ptl T4SS through a portal positioned at the IMC/OMC junction. The VirB T4SSs elaborated by Brucella spp., also likely employ a two-step translocation route for delivery of effectors into eukaryotic host cells. Interestingly, the Brucella T4SSs also lack associated T4CPs, yet only some effectors carry canonical sec signal sequences, while others do not. Precisely how the latter are translocated across the inner membrane remains an intriguing question for further study (88, 89, 90).
Signal-Mediated Activation of T4SSs
The T4SSs generally translocate substrates only upon establishment of direct contact with target cells. This indicates that the channels are gated and activated by propagation of a signal from recipient to donor cells. Studies in the 1970s of the F transfer system supplied strong experimental support for such a signal, generated upon contact of the F pilus with a recipient cell (91). This signal is transduced to the interior of the donor cell where it stimulates early F plasmid-processing reactions. More recent work supplied further evidence for signal communication from the F pilus to the cell interior (92). The F-like R1-encoded pilus is the receptor for bacteriophage R17, which enters cells through undefined mechanisms involving pilus retraction or penetration of the pilus lumen. It was discovered that R17 binds and enters only cells in which the TraD T4CP has already engaged with the F relaxosome. Phage R17 binding appears to supply a requisite extracellular signal, possibly by mimicking recipient cell contact, which together with the DNA substrate-T4CP docking signal activates the channel to allow phage uptake.
Studies in A. tumefaciens also have provided evidence for signal transduction along the VirB/VirD4 T4SS. In this system, VirB10 was shown to undergo a conformational switch in response to sensing of two intracellular signals mediated through the VirD4 T4CP. The first signal is conveyed through a two-step reaction involving the docking of the T-DNA substrate with VirD4 and its subsequent transfer to the VirB11 ATPase (49). The second signal corresponds to the ATP hydrolysis activities of VirD4 and VirB11 (93). Integration of these signals results in a conformational change in the structural scaffold VirB10, as shown by a change in protease susceptibility. This conformational change is necessary for passage of the DNA substrate through the distal portion of the VirB channel composed of the VirB2 pilin and VirB9 subunits, as shown with the TrIP assay (49, 93). The nature of the conformational change is not yet known, but a role for VirB10 in the coupling of intracellular signals to gating of an outer membrane pore is suggested by isolation of a mutation near the VirB10 AP pore that “locks” VirB10 in the energized conformation and allows for release of secretion substrates to the cell surface independently of target cell contact (94). Therefore, as shown for a number of small-molecule and macromolecular transport systems, the VirB/VirD4 T4SS appears to be activated by a combination of ligand and energy signals.
It is tempting to suggest that intra- and extracellular signals generally activate T4SS channels through the envelope-spanning VirD4-VirB10 transduction network. For type IV secretion, signal activation would open channels enabling substrate passage to target cells. In a reverse reaction, exogenous DNA or phage binding to pilus or other T4SS receptors might activate channels for uptake of nucleic acids across the cell envelope.
MODULATION OF DONOR-TARGET CELL INTERACTIONS BY ADAPTED FORMS OF CHANNEL SUBUNITS
Besides their functions as structural components of the translocation channel, several T4SS subunits have been adapted for novel purposes through acquisition of novel domains or motifs. Most notably, the IMC component VirB6 and the OMC subunits VirB7 and VirB10 of several T4SSs carry domains of demonstrated or postulated importance for modulation of donor cell contacts with bacterial or eukaryotic target cells. The next sections describe some of these specialized functions.
Extended VirB6s
VirB6 subunits are minimally ∼300 kDa and have at least 5 or 6 membrane-spanning helices (2, 72). A subset additionally carries a large hydrophilic domain of functional importance. These subunits were designated “extended VirB6,” and they are found in conjugation systems and many effector translocator systems (2). For the F-type T4SSs, the polytopic motif of VirB6-like TraG subunits is followed by a large ∼600 residue C-terminal domain (Fig. 4) (22). This domain is extensively α-helical in its predicted secondary structure, and has been suggested to functionally substitute for VirB8, on the basis of its periplasmic location and the absence of a VirB8 ortholog in the F-type systems (14, 22). Interestingly, TraG also is involved in entry exclusion, a process that blocks redundant DNA transfer between identical donor cells. When a donor cell contacts another donor cell, the C-terminal domain of TraG is thought to extend across the outer membranes of the paired cells to form a specific contact with TraS, a protein also encoded by F-carrying donor cells (Fig. 4) (95, 96). This contact signals a nonproductive donor-donor cell junction, and blocks DNA transfer. Similar findings were reported for homologs of TraG and TraS encoded by the SXT ICE (integrative and conjugative element) of Vibrio cholera (97). Precisely how the TraG-TraS interaction blocks DNA transfer is not yet known, but might involve transduction of a signal across the donor-donor cell junction resulting in a conformational change in the T4SS that blocks channel function (Fig. 4).
Figure 4.
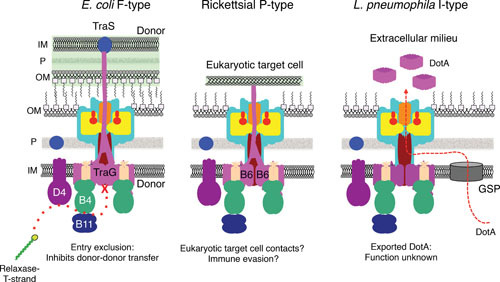
Adaptations of the IMC: VirB6-like subunits. Polytopic VirB6 subunits with lengths of ∼300 residues are components of the IMC. Many T4SSs have larger forms of these subunits, designated extended-VirB6. These forms of VirB6 are composed of a polytopic VirB6-like domain and a hydrophilic domain as large as ∼1,000 residues. (Left) F-type F plasmids of E. coli encode large VirB6-like TraG subunits. These subunits participate in entry exclusion, which prevents nonproductive plasmid transfer between donor cells. TraG’s C-terminal domain extends or is delivered via the T4SS across the donor-donor cell junction, where it binds the entry exclusion protein TraS located in the inner membrane of the paired donor. This interaction might transduce a signal resulting in a conformational change in the T4SS that blocks nonproductive F plasmid transfer to other F-carrying cells (96). (Middle) Rickettsial P-type T4SSs are composed of 4 or more large VirB6 subunits whose hydrophilic domains are unrelated in sequence and which might be surface-displayed for target cell binding or immune evasion (98, 99, 100). (Right) The I-type Dot/Icm system of L. pneumophila secretes the highly hydrophobic DotA (∼1,000 residues) to the milieu where it assembles as large ring-shaped complexes. DotA has a canonical signal sequence, which is thought to mediate DotA delivery through the General Secretory Pathway (GSP) across the inner membrane. In the periplasm, DotA is then recruited to the Dot/Icm T4SS for delivery across the outer membrane to the cell exterior (101).
The Rickettsia spp. P-type T4SSs have multiple copies of variant forms of “extended-VirB6” subunits (98, 99). They range in size between ∼600 and 1,400 residues and the polytopic VirB6 motif can be located in one or both terminal regions or centrally. There is evidence for surface display of VirB6 domains in Wolbachia, supporting the notion that such exported domains are involved in establishment of endosymbiotic relationships (Fig. 4) (100). Whether these domains protrude through the OMC chamber as depicted in Fig. 4, or are proteolytically cleaved from the rest of the protein prior to export to the cell surface is presently not known.
I-type systems also code for “extended-VirB6” subunits. E. coli plasmid R64 encodes TraY, a 745-residue protein with an unusual hydropathy profile (19). The N- and C-terminal thirds of the protein are highly hydrophobic with between 4 and 6 TM motifs, and the central third is hydrophilic and predicted to reside in the periplasm. In the related L. pneumophila Dot/Icm system, the TraY ortholog is DotA. DotA is ∼300 residues larger than TraY, and possesses the same general hydropathy profile with multiple N- and C-terminal TM domains flanking a central hydrophilic domain. Strikingly, DotA is released in a Dot/Icm-dependent manner to the extracellular milieu where it forms ring-shaped complexes (Fig. 4) (101). In contrast to R64-encoded TraY, DotA has an N-terminal signal sequence, leading to a proposal that it is secreted across the membrane by the GSP (101). If true, DotA would be delivered to the cell surface in two steps: first, across the inner membrane by the GSP and, second, across the outer membrane by the Dot/Icm T4SS (Fig. 4). No function has been ascribed to the extracellular form of DotA, and no follow-up studies have defined its export route in further detail.
VirB7 Adaptations
VirB7 subunits are typically small lipoproteins that play an important role in stabilizing the OMC at the outer membrane. However, larger forms of VirB7-like lipoproteins functioning in P- and I-type systems have novel features, including surface variable regions, as shown for Helicobacter pylori CagT (102, 103), or N0 domains as shown for Xanthomonas citri VirB7 and L. pneumophila DotD (Fig. 5) (104, 105). Studies have shown that surface-variable CagT is required for CagA translocation and pilus biogenesis (106, 107), and also might contribute to immune evasion by H. pylori (102). By contrast, N0 domains are structural modules thought to serve a variety of functions relating to transport processes. They are features of transport-associated proteins, including TonB, secretins of the types II and III secretion systems and related type IV pilus systems, bacteriophages, and type VI secretion systems (see reference 104). The N0 domains of X. citri VirB7 and L. pneumophila DotD are envisioned to form additional rings at the base of the OMC that contribute to channel gating or contacts with the IMC (104, 105).
Figure 5.
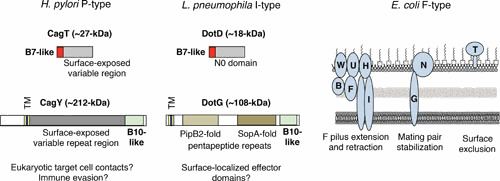
Adaptations of the OMC: VirB7- and VirB10-like subunits. The outer membrane complexes (OMCs) of paradigmatic T4SSs are composed of the small (∼5-kDa) lipoprotein VirB7, outer membrane-associated VirB9, and envelope-spanning VirB10 (∼50-kDa). Many T4SSs have larger forms of the VirB7- and VirB10-like subunits that carry surface-displayed domains of functional importance. (Left) H. pylori P-type T4SSs carry larger forms of VirB7-like CagT and VirB10-like CagY with repeat domains that are exposed on the cell surface. TM, transmembrane domain. (Middle) L. pneumophila I-type T4SSs also carry larger forms of the VirB7- and VirB10-like proteins. VirB7-like DotD has an N0 domain that is thought to form an extra ring of structural importance for the OMC. DotG has an N-terminal transmembrane domain (TM), a C-terminal VirB10 structural fold (20), and internal structural folds similar to that of Salmonella T3SS effector PipB2 and the Salmonella effector SopA (114, 116), as determined by Phyre2 structural modeling (113). It is proposed that these domains of DotG protrude through the OMC or are proteolytically released from the VirB10 scaffold domain for delivery into the eukaryotic cell where they exert effector activities. (Right) E. coli F-type T4SSs carry novel subcomplexes that may or may not be physically associated with the T4SS at the cell surface. The TraW/U/F/B/F/TrbI subunits mediate F pilus extension and retraction, TraN and VirB10-like TraG stabilize mating pairs and the TraT lipoprotein prevents redundant F plasmid transfer among donor cells through surface exclusion (see reference 22).
VirB10 Adaptations
The VirB10-like subunits are among the most sequence- and structurally variable subunits of the T4SSs. In the Cag T4SS of H. pylori, only a small C-terminal region of CagY is similar to VirB10, whereas a large central region is composed of multiple repeats (Fig. 5) (108). This central region is surface displayed and associates with a pilus structure (109, 110). Furthermore, during infection this region undergoes extensive genetic rearrangements that disrupt or activate the Cag T4SS (108, 110). Through host immune-driven recombination, CagY is postulated to function as a sensor of the host immune response and, in turn, regulate Cag T4SS function to maximize persistent infection (110).
A second example of an interesting but as yet unexplored OMC adaptation is found in the I-type systems. In E. coli, plasmid R64 codes for TraO, which closely resembles VirB10 in size and predicted overall structure (19). In striking contrast, in the related L. pneumophila Dot/Icm system, “VirB10-like” DotG is over 1,000 residues and only the extreme C-terminal region resembles VirB10 (111, 112). DotG possesses central variable domains consisting in part of multiple sets of pentapeptide repeats (111). Although this region of DotG has been postulated to reside in the periplasm, it is noteworthy that DotG homologs from various Legionella species carry structural motifs of known bacterial effector proteins, as shown by Phyre 2 modeling (Fig. 5) (113). Most strikingly, L. pneumophila DotG (accession no. AF026534.1) has structural folds highly similar to PipB2, a T3SS effector of Salmonella enterica serovar Typhimurium (Phyre2 modeling: c2leza; 99.2% confidence; 18% sequence identity) During Salmonella infection, secreted PipB2 participates in reorganization of late endosome/lysosome compartments by linking kinesin-1 onto the Salmonella-containing vacuole (SCV) membrane. The pentapeptide motif is required to efficiently recruit kinesin-1, whereas the N-terminal domain suffices for type III translocation and association with SCVs (114, 115). A C-proximal pentapeptide motif of DotG (accession no. AF026534.1) also has a structural fold similar to the Salmonella T3SS effector SopA (Phyre2 modeling: c2qzaA, 99.7% confidence; 12% sequence identity), which structurally mimics eukaryotic ubiquitin ligase (Fig. 5) (116). The central regions of DotG subunits from different L. pneumophila species are highly variable, and a search among 11 other DotG/IcmE subunits identified structural folds similar to other effector domains that were mainly embedded within pentapeptide motifs (P. J. Christie, unpublished findings). Thus, it can be proposed that DotG subunits of Dot/Icm T4SSs might elaborate effector domains that, reminiscent of passenger domains of type V autotransporters (117), are either tethered to the L. pneumophila cell surface or released by proteolysis for delivery into eukaryotic cells.
CONJUGATIVE PILI AND PILUS BIOGENESIS
All conjugation systems and probably most effector translocator systems of Gram-negative bacteria elaborate pili or other surface appendages. Although there is compelling evidence that these structures promote attachment of donor cells to potential bacterial or eukaryotic target cells, the contributions of these organelles to the process of substrate transfer is somewhat controversial. On the one hand, studies of E. coli F-type pili showed that donor cells can inefficiently deliver the F plasmid to distantly located recipient cells, suggestive of a role for extended F pili as conduits for the DNA substrate (118, 119). On the other hand, E. coli and A. tumefaciens mutants lacking detectable pili can still efficiently transfer DNA substrates, establishing that extended pili are not obligatory features of T4SS channels (61, 62, 120, 121). In natural settings, it is likely these surface organelles enhance DNA transfer efficiencies, not as conduits for substrates, but rather through their ability to promote aggregation of donor and recipient cells and development of robust biofilms on biotic and abiotic surfaces. Through these adhesive activities, conjugative pili promote cellular contacts favoring formation of mating junctions that resist shear forces and other environmental perturbations.
E. coli F pili are presently the best characterized group of conjugative pili. These pili are typically long, ranging from 1 to 20 μm in length, and flexible (14). Among their most noteworthy properties, they stochastically and dynamically extend and retract. This is thought to enable donor cells to scan the local environment for viable recipient cells as a means of enhancing F plasmid transfer efficiencies, particularly in low-cell-density, aqueous environments (14, 15, 16). In contrast to the F pili, the P pili are thicker, more rigid, and typically much shorter, although length measurements are complicated by the fact that isolated pili are often broken (122, 123, 124). These pili do not appear to undergo cycles of extension and retraction, but instead accumulate in the milieu, either through breakage or an active sloughing mechanism. They tend to aggregate as a mesh of polymers, which might promote nonspecific clumping of donor and recipient cells resulting in formation of productive mating pairs.
The biogenesis of pili elaborated by E. coli conjugation systems has been a subject of study for over 50 years. The general requirements for pilus assembly are the same as those for elaboration of the translocation channel, with two exceptions. First, the T4CP is not required for pilus production (14). Second, the VirB1 transglycosylase is important for pilus assembly but not a functional translocation channel (60, 125). In fact, the contribution of VirB1 to pilus assembly appears not to be associated with its cell wall hydrolase activity but rather with the export of a proteolytic fragment termed VirB1* to the cell surface. At this location, VirB1* interacts with VirB2 pilin and VirB5 and, by an undefined mechanism, promotes pilus polymerization (126). The biogenesis of conjugative pili can be divided into early- and later-stage reactions, as summarized below.
Early-Stage Pilus Assembly Reactions
F and P pili are synthesized as pro-proteins with unusually long (∼30 to 50 residues) leader peptides that are cleaved upon insertion into the inner membrane (127). The F plasmid-encoded TraA pro-pilin inserts into the membrane by a mechanism dependent on the proton motive force, but not the GSP (128). The F plasmid-encoded membrane protein TraQ also is required for correct orientation and stabilization of TraA pro-pilin in the membrane (129). Membrane-embedded pro-pilins are processed further by proteases and other posttranslational modifications. Following signal sequence cleavage by LepB, F pili are acetylated at their N termini, although this modification is not required for F pilus assembly (14). Pilin subunits of P pili undergo a novel head-to-tail cyclization required for stabilization of the pilin in the membrane and for pilus assembly (130, 131). Processed forms of the conjugative pilins typically are composed of alternating stretches of hydrophilic and hydrophobic residues along the length of the protein, giving a characteristic membrane topology such that the N and C termini are in the periplasm and a small hydrophilic domain of only a few residues is in the cytoplasm (128, 132, 133, 134). Approximately ∼100,000 monomers of F pilin assemble in the membrane for use in building the pilus upon receipt of an unknown signal (135).
F pili assemble by sequential addition of pilin monomers to the base of the growing pilus (15). These pili are composed of a single type of pilin, although the composition and nature of the pilus tip is currently unknown. F pili are hollow cylinders of ∼9 nm in diameter with an axial lumen of ∼3 nm. Recent studies showed that a single F pilus is composed of distinct 1-start and 4-start helical symmetries, a property thought to enable these pili to withstand considerable extension and contraction forces as might be encountered upon engagement of the pilus with recipient cells (136). E. coli cells typically possess 1 to 5 F pili per cell, although variant F systems can encode up to 20 pili per cell. The pili are randomly distributed around the cell, and it is thought that there are more pilus assembly sites on the cell surface than extended pili (14). The structures of P pili have not yet been solved. They are difficult to visualize when attached to the cell surface, but indirect detection through decoration with pilus-specific phage or green fluorescent protein labeling of pilus assembly proteins yielded estimates as high as 20 pilus assembly sites per cell (14, 137).
The mechanism by which pilins are extracted from the hydrophobic membrane environment was evaluated by monitoring accessibility of Cys residues engineered along the length of A. tumefaciens VirB2 to a membrane-impermeable thiol-reactive reagent. This study defined the inner membrane topology of VirB2 and also presented evidence that the VirB4 ATPase, with a contribution by the VirB11 ATPase, catalyzes extraction of the pilin from the membrane (134). Consistent with these findings, VirB4 also stabilizes subunits required for pilus assembly and is essential for interaction of VirB2 with another pilus-associated protein, VirB5 (68). VirB5 associates with the tip of pili elaborated by the A. tumefaciens VirB/VirD4 system, and the VirB2-VirB5 interaction is thought to be essential for pilus nucleation (138). VirB4 thus is postulated to function in dislocation of mature pilins from the inner membrane to mediate formation of the VirB2-VirB5 nucleation complex for subsequent pilus polymerization (134). F-type systems encode VirB5-like TraE subunits, but no evidence exists for a pilus tip association. Rather, these subunits are thought to form part of the T4SS in the periplasm, which, interestingly, agrees with recent evidence for association of VirB5-like TrwJ with the IMC of the R388 T4SS3–10 structure (22, 26). VirB5 subunits might have multiple functions in relation to assembly of the IMC as well as the conjugative pilus.
Later-Stage Pilus Assembly and F Pilus Dynamics
The membrane pool of pilin monomers is recruited upon receipt of an unknown signal(s) to build the pilus, presumably within or on top of the T4SS. In view of the R388 T4SS3–10 structure showing that two VirB4 hexamers are located at the base of the IMC (Fig. 2) (26), it is reasonable to suggest that VirB4 subunits catalyze extraction of pilin monomers from the membrane and then feed them into the chamber of the OMC. Once in the OMC chamber, pilins would undergo polymerization to build the conjugative pilus. The central chamber of the pKM101-encoded OMC is approximately 100 Å in diameter, sufficiently large to house the conjugative pilus, but the diameter of the outer membrane cap is at most only 32 Å and clearly not large enough to accommodate the pilus. These observations suggest at least two alternative models for pilus assembly. First, the pilus assembles from an IMC platform, and, as it extends through the OMC chamber, it induces profound structural changes in the distal portion of OMC. Second, pilin monomers are delivered through the OMC chamber where polymerization initiates on an outer membrane platform formed by VirB10’s AP (cap) domain (see Fig. 2C). At this time, it is not possible to discriminate between these models.
As mentioned above, the F pili are so far the only group of conjugative pili for which a dynamic mode of extension and retraction has been unequivocally established. F pilus assembly requires a putative TraV/TraK/TraB OMC, as well as IMC subunits VirB3-like TraL, VirB4-like ATPase TraC, and VirB6-like TraG (14). These systems code for a unique set of proteins, TraF, -H, -U, -W and TrbB, -I (Fig. 5) (16, 139, 140). These proteins form an interaction network distinct from the presumptive TraV/K/B OMC (141). The TraF/H/U/W-TrbB/I complex is thought to regulate the dynamics of F pilus extension/retraction, although the underlying mechanism is not yet defined. TraF, -H, -U, and -W localize at the outer membrane (14). TrbI is of special interest as a bitopic inner membrane protein that contributes specifically to the process of pilus retraction through contacts with the outer membrane-associated TraF/H/U/W protein complex (Fig. 5) (142).
F pilus extension and retraction has been visualized by use of fluorescently labeled phage R17, which binds along the sides of F pili. Intriguingly, F pili were shown to stochastically extend and retract regardless of the presence of recipient cells in the vicinity (15). Pilus polymerization and retraction requires ∼5 min for completion, and new pili are elaborated prior to retraction of older pili, suggesting that initiation and retraction events are randomly timed and not coordinated. These studies also presented evidence supporting earlier work showing that F pili rotate along their longitudinal axis during retraction, causing a bound recipient cell to spin as it is drawn closer to the donor cell (15). Such a rotary motion of flexible pili during extension and retraction is thought to allow pili to “sweep” a large volume around the donor cell in order to enhance the probability of a productive encounter with a recipient cell, which would be of particular benefit for cells growing in liquid environments (16).
MATING PAIR FORMATION AND THE MATING JUNCTION
In E. coli, conjugative pili first form loose contacts with potential recipients. At this stage, mating by F- and P-type plasmids can be disrupted by physical means or treatment with 0.01% SDS. Donor-recipient cell contacts soon stabilize and become difficult to break apart (143, 144, 145, 146). F-type systems encode two proteins, TraN and TraG, that specify this function (Fig. 5). These proteins, termed mating pair stabilization proteins, are unique to the F-type systems and promote formation of tight mating junctions (145).
TraN family proteins are large (∼600 to 1200 residues) cysteine-rich proteins thought to function as adhesins for formation of stable donor-recipient cell-mating pairs. TraN also was shown to coordinate its functions through interactions with the TraV subunit of the putative TraV/K/B OMC (145). TraN binds both OmpA and LPS on recipient cells, which likely explains the basis for F-specific mating-pair stabilization. This was evidenced by the identification of mutations in recipients that disrupt formation of tight mating junctions in genes encoding OmpA and constituents of the lipopolysaccharide (LPS) biosynthesis pathway (145, 147).
The second mating-pair stabilization protein, TraG, was discussed above as a VirB6-like protein with an α-helical C terminus that can extend or be delivered into recipient cells where it binds the inner membrane protein TraS to block DNA transfer in nonproductive donor-donor pairs (96). In matings with F-minus recipients, however, the C-terminal domain of TraG is thought to alternatively form contacts with another, unidentified inner membrane protein to stabilize the mating junction (96). TraG also has been implicated as a binding partner of TraN (148) and, through this interaction, promote formation of mating pairs (Fig. 5).
The architecture of conjugative mating junctions is presently not defined in E. coli or any other species. E. coli donor cells carrying plasmid RP4 form mating junctions at poles and along the lengths of cells, as shown by electron microscopy. Junctions at cell poles were estimated to be ∼150 to 200 nm in length, whereas those along the cell body extended up to 1,500 nm in length or up to half the lengths of the cells (143). Intriguingly, no structures resembling the R388 T4SS3–10 or the OMC or IMC subassemblies, or any other structures, have been detected at the mating junction (149). This suggests the intriguing possibility that intact channels form only transiently upon channel activation by substrate-docking signals and establishment of donor-recipient cell contacts.
Discerning the assembly mechanism and architecture of the mating junction is further complicated by evidence that E. coli conjugation systems are exceedingly promiscuous in their capacity to deliver cargos to a wide range of Gram-negative and -positive bacteria. In fact, this promiscuity extends to eukaryotic cells, as both F- and P-type systems have been shown to deliver substrates to yeast and even human cells (150, 151, 152, 153).
CONCLUSIONS AND FUTURE DIRECTIONS
In recent years, mechanistic and structural features of E. coli conjugation systems have been described in unprecedented detail. Most noteworthy, the recent ultrastructural studies have generated an architectural blueprint of a nearly entire T4SS that, when combined with results of the earlier TrIP studies, enhances our understanding of the substrate transfer route. Despite this exciting progress, long-term studies of E. coli conjugation systems have described many features of the F-, P-, and I-type systems whose underlying mechanisms remain poorly understood. Answers to the following “big picture” questions surrounding type IV secretion will require sustained efforts and implementation of creative state-of-the-art technologies and approaches.
What is the architecture of the translocation channel and the pathway(s) by which substrates are delivered through the channel? In this review, three possible routes across the inner membrane were described involving the T4CP, VirB ATPases, and integral membrane components of the IMC. Refined studies are needed to discriminate between these possibilities and to define specific contributions of the ATPases and ATP energy consumption to the early-stage translocation reactions. We also now have an OMC structure in atomic detail and, based on this structure and results of the TrIP studies, it is proposed that the translocation channel responsible for conveying substrates to the cell surface is located within the OMC. Visualization of this channel and the nature of its interaction with the IMC channel is critical for answering the larger question of whether T4SSs employ one- or two-step translocation pathways.
Donor and recipient cells are known to form tight mating junctions, but the architecture of this junction remains poorly defined. Indeed, the mechanisms by which T4SSs establish productive target cell contacts and then presumably dissociate after mating are perhaps the greatest mysteries surrounding type IV secretion. What are the physical and functional relationships of the conjugative pilus, the translocation channel, and the mating junction? With recent advances in superresolution and cryoelectron microscopy, it should be feasible using the highly efficient F- and P-type systems in E. coli to visualize mating junction formation in real time and the junction itself at near atomic resolution. Equally important is the goal of visualizing the T4SS within the mating junction and specific T4SS machine contacts with the recipient cell envelope. Of broader biological interest, since E. coli also can deliver DNA substrates to yeast and human cells, we should be able to use this model bacterium to define the architecture of junctions formed during interkingdom mating.
Intracellular and extracellular signals are known to activate T4SSs, but what are the molecular and structural details of signal activation? Also of interest, conjugative DNA transfer must be coordinated with DNA replication and cell division to avoid replication of a mobile element from occurring at the same time as transfer. A stabilization system (Stb) has been identified as one mechanism functioning to reconcile the maintenance mode with the propagation mode a mobile element (154). What other types of molecular signals and pathways coordinate these potentially conflicting processes?
Finally, while there is clear evidence for the structural diversification of P-, F-, and I-type systems during evolution, what novel functions do these structural acquisitions specify? An especially interesting avenue of research for the coming years is to decipher how T4SS machine adaptations and host-derived or other environmental signals confer spatiotemporal control over substrate translocation. The E. coli conjugation systems remain the best subjects for refined structure-function studies of T4SSs. Yet, a full understanding of T4SS diversity also will require detailed investigations of effector translocators with particular attention to specialized features acquired during the evolution of these machines.
ACKNOWLEDGMENTS
I thank members of the Christie laboratory for helpful discussions. This work was supported by NIH R01GM48476 and R21AI105454.
Conflicts of interest: The author declares no conflicts.
REFERENCES
- 1.Cascales E, Christie PJ. 2003. The versatile bacterial type IV secretion systems. Nat Rev Microbiol 1:137–149. [PubMed] 10.1038/nrmicro753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alvarez-Martinez CE, Christie PJ. 2009. Biological diversity of prokaryotic type IV secretion systems. Microbiol Mol Biol Rev 73:775–808. [PubMed] 10.1128/MMBR.00023-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cabezón E, Ripoll-Rozada J, Peña A, de la Cruz F, Arechaga I. 2015. Towards an integrated model of bacterial conjugation. FEMS Microbiol Rev 39:81–95. [PubMed] [DOI] [PubMed] [Google Scholar]
- 4.Juhas M. 2015. Horizontal gene transfer in human pathogens. Crit Rev Microbiol 41:101–108. [PubMed] 10.3109/1040841X.2013.804031 [DOI] [PubMed] [Google Scholar]
- 5.Guglielmini J, de la Cruz F, Rocha EP. 2013. Evolution of conjugation and type IV secretion systems. Mol Biol Evol 30:315–331. [PubMed] 10.1093/molbev/mss221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christie PJ, Whitaker N, González-Rivera C. 2014. Mechanism and structure of the bacterial type IV secretion systems. Biochim Biophys Acta 1843:1578–1591. [PubMed] 10.1016/j.bbamcr.2013.12.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chandran Darbari V, Waksman G. 2015. Structural biology of bacterial type IV secretion systems. Annu Rev Biochem 84:603–629. [PubMed] 10.1146/annurev-biochem-062911-102821 [DOI] [PubMed] [Google Scholar]
- 8.Bhatty M, Laverde Gomez JA, Christie PJ. 2013. The expanding bacterial type IV secretion lexicon. Res Microbiol 164:620–639. [PubMed] 10.1016/j.resmic.2013.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kubori T, Nagai H. 2016. The Type IVB secretion system: an enigmatic chimera. Curr Opin Microbiol 29:22–29. [PubMed] 10.1016/j.mib.2015.10.001 [DOI] [PubMed] [Google Scholar]
- 10.Asrat S, Davis KM, Isberg RR. 2015. Modulation of the host innate immune and inflammatory response by translocated bacterial proteins. Cell Microbiol 17:785–795. [PubMed] 10.1111/cmi.12445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Locht C, Coutte L, Mielcarek N. 2011. The ins and outs of pertussis toxin. FEBS J 278:4668–4682. [PubMed] 10.1111/j.1742-4658.2011.08237.x [DOI] [PubMed] [Google Scholar]
- 12.Ramsey ME, Woodhams KL, Dillard JP. 2011. The gonococcal genetic island and type IV secretion in the pathogenic Neisseria. Front Microbiol 2:61. [PubMed] 10.3389/fmicb.2011.00061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stingl K, Müller S, Scheidgen-Kleyboldt G, Clausen M, Maier B. 2010. Composite system mediates two-step DNA uptake into Helicobacter pylori. Proc Natl Acad Sci USA 107:1184–1189. [PubMed] 10.1073/pnas.0909955107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arutyunov D, Frost LS. 2013. F conjugation: back to the beginning. Plasmid 70:18–32. [PubMed] 10.1016/j.plasmid.2013.03.010 [DOI] [PubMed] [Google Scholar]
- 15.Clarke M, Maddera L, Harris RL, Silverman PM. 2008. F-pili dynamics by live-cell imaging. Proc Natl Acad Sci USA 105:17978–17981. [PubMed] 10.1073/pnas.0806786105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Silverman PM, Clarke MB. 2010. New insights into F-pilus structure, dynamics, and function. Integr Biol Camb 2:25–31. [PubMed] 10.1039/B917761B [DOI] [PubMed] [Google Scholar]
- 17.Christie PJ, Atmakuri K, Krishnamoorthy V, Jakubowski S, Cascales E. 2005. Biogenesis, architecture, and function of bacterial type IV secretion systems. Annu Rev Microbiol 59:451–485. [PubMed] 10.1146/annurev.micro.58.030603.123630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Costa TR, Felisberto-Rodrigues C, Meir A, Prevost MS, Redzej A, Trokter M, Waksman G. 2015. Secretion systems in Gram-negative bacteria: structural and mechanistic insights. Nat Rev Microbiol 13:343–359. [PubMed] 10.1038/nrmicro3456 [DOI] [PubMed] [Google Scholar]
- 19.Sampei G, Furuya N, Tachibana K, Saitou Y, Suzuki T, Mizobuchi K, Komano T. 2010. Complete genome sequence of the incompatibility group I1 plasmid R64. Plasmid 64:92–103. [PubMed] 10.1016/j.plasmid.2010.05.005 [DOI] [PubMed] [Google Scholar]
- 20.Nagai H, Kubori T. 2011. Type IVB secretion systems of Legionella and other Gram-negative bacteria. Front Microbiol 2:136. 10.3389/fmicb.2011.00136. [PubMed] 10.3389/fmicb.2011.00136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Christie PJ. 2004. Type IV secretion: the Agrobacterium VirB/D4 and related conjugation systems. Biochim Biophys Acta 1694:219–234. [PubMed] 10.1016/j.bbamcr.2004.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lawley TD, Klimke WA, Gubbins MJ, Frost LS. 2003. F factor conjugation is a true type IV secretion system. FEMS Microbiol Lett 224:1–15. [PubMed] 10.1016/S0378-1097(03)00430-0 [DOI] [PubMed] [Google Scholar]
- 23.Cascales E, Christie PJ. 2004. Definition of a bacterial type IV secretion pathway for a DNA substrate. Science 304:1170–1173. [PubMed] 10.1126/science.1095211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fronzes R, Schäfer E, Wang L, Saibil HR, Orlova EV, Waksman G. 2009. Structure of a type IV secretion system core complex. Science 323:266–268. [PubMed] 10.1126/science.1166101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chandran V, Fronzes R, Duquerroy S, Cronin N, Navaza J, Waksman G. 2009. Structure of the outer membrane complex of a type IV secretion system. Nature 462:1011–1015. [PubMed] 10.1038/nature08588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Low HH, Gubellini F, Rivera-Calzada A, Braun N, Connery S, Dujeancourt A, Lu F, Redzej A, Fronzes R, Orlova EV, Waksman G. 2014. Structure of a type IV secretion system. Nature 508:550–553. [PubMed] 10.1038/nature13081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trokter M, Felisberto-Rodrigues C, Christie PJ, Waksman G. 2014. Recent advances in the structural and molecular biology of type IV secretion systems. Curr Opin Struct Biol 27:16–23. [PubMed] 10.1016/j.sbi.2014.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zechner EL, Lang S, Schildbach JF. 2012. Assembly and mechanisms of bacterial type IV secretion machines. Philos Trans R Soc Lond B Biol Sci 367:1073–1087. [PubMed] 10.1098/rstb.2011.0207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mihajlovic S, Lang S, Sut MV, Strohmaier H, Gruber CJ, Koraimann G, Cabezón E, Moncalián G, de la Cruz F, Zechner EL. 2009. Plasmid R1 conjugative DNA processing is regulated at the coupling protein interface. J Bacteriol 191:6877–6887. [PubMed] 10.1128/JB.00918-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sut MV, Mihajlovic S, Lang S, Gruber CJ, Zechner EL. 2009. Protein and DNA effectors control the TraI conjugative helicase of plasmid R1. J Bacteriol 191:6888–6899. [PubMed] 10.1128/JB.00920-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vincent CD, Friedman JR, Jeong KC, Sutherland MC, Vogel JP. 2012. Identification of the DotL coupling protein subcomplex of the Legionella Dot/Icm type IV secretion system. Mol Microbiol 85:378–391. [PubMed] 10.1111/j.1365-2958.2012.08118.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lang S, Gruber K, Mihajlovic S, Arnold R, Gruber CJ, Steinlechner S, Jehl MA, Rattei T, Fröhlich KU, Zechner EL. 2010. Molecular recognition determinants for type IV secretion of diverse families of conjugative relaxases. Mol Microbiol 78:1539–1555. [PubMed] 10.1111/j.1365-2958.2010.07423.x [DOI] [PubMed] [Google Scholar]
- 33.Redzej A, Ilangovan A, Lang S, Gruber CJ, Topf M, Zangger K, Zechner EL, Waksman G. 2013. Structure of a translocation signal domain mediating conjugative transfer by type IV secretion systems. Mol Microbiol 89:324–333. [PubMed] 10.1111/mmi.12275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parker C, Meyer RJ. 2007. The R1162 relaxase/primase contains two, type IV transport signals that require the small plasmid protein MobB. Mol Microbiol 66:252–261. [PubMed] 10.1111/j.1365-2958.2007.05925.x [DOI] [PubMed] [Google Scholar]
- 35.Alperi A, Larrea D, Fernández-González E, Dehio C, Zechner EL, Llosa M. 2013. A translocation motif in relaxase TrwC specifically affects recruitment by its conjugative type IV secretion system. J Bacteriol 195:4999–5006. [PubMed] 10.1128/JB.00367-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meyer R. 2015. Mapping type IV secretion signals on the primase encoded by the broad-host-range plasmid R1162 (RSF1010). J Bacteriol 197:3245–3254. [PubMed] 10.1128/JB.00443-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vergunst AC, Schrammeijer B, den Dulk-Ras A, de Vlaam CM, Regensburg-Tuïnk TJ, Hooykaas PJ. 2000. VirB/D4-dependent protein translocation from Agrobacterium into plant cells. Science 290:979–982. [PubMed] 10.1126/science.290.5493.979 [DOI] [PubMed] [Google Scholar]
- 38.Vergunst AC, van Lier MC, den Dulk-Ras A, Stüve TA, Ouwehand A, Hooykaas PJ. 2005. Positive charge is an important feature of the C-terminal transport signal of the VirB/D4-translocated proteins of Agrobacterium. Proc Natl Acad Sci USA 102:832–837. [PubMed] 10.1073/pnas.0406241102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cabezón E, Sastre JI, de la Cruz F. 1997. Genetic evidence of a coupling role for the TraG protein family in bacterial conjugation. Mol Gen Genet 254:400–406. [PubMed] 10.1007/s004380050432 [DOI] [PubMed] [Google Scholar]
- 40.Schröder G, Lanka E. 2003. TraG-like proteins of type IV secretion systems: functional dissection of the multiple activities of TraG (RP4) and TrwB (R388). J Bacteriol 185:4371–4381. [PubMed] 10.1128/JB.185.15.4371-4381.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gomis-Rüth FX, Moncalián G, Pérez-Luque R, González A, Cabezón E, de la Cruz F, Coll M. 2001. The bacterial conjugation protein TrwB resembles ring helicases and F1-ATPase. Nature 409:637–641. [PubMed] 10.1038/35054586 [DOI] [PubMed] [Google Scholar]
- 42.Hormaeche I, Alkorta I, Moro F, Valpuesta JM, Goni FM, De La Cruz F. 2002. Purification and properties of TrwB, a hexameric, ATP-binding integral membrane protein essential for R388 plasmid conjugation. J Biol Chem 277:46456–46462. [PubMed] 10.1074/jbc.M207250200 [DOI] [PubMed] [Google Scholar]
- 43.Gomis-Rüth FX, Coll M. 2001. Structure of TrwB, a gatekeeper in bacterial conjugation. Int J Biochem Cell Biol 33:839–843. [PubMed] 10.1016/S1357-2725(01)00060-7 [DOI] [PubMed] [Google Scholar]
- 44.Schröder G, Krause S, Zechner EL, Traxler B, Yeo HJ, Lurz R, Waksman G, Lanka E. 2002. TraG-like proteins of DNA transfer systems and of the Helicobacter pylori type IV secretion system: inner membrane gate for exported substrates? J Bacteriol 184:2767–2779. [PubMed] 10.1128/JB.184.10.2767-2779.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.de Paz HD, Larrea D, Zunzunegui S, Dehio C, de la Cruz F, Llosa M. 2010. Functional dissection of the conjugative coupling protein TrwB. J Bacteriol 192:2655–2669. [PubMed] 10.1128/JB.01692-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Whitaker N, Chen Y, Jakubowski SJ, Sarkar MK, Li F, Christie PJ. 2015. The all-alpha domains of coupling proteins from the Agrobacterium tumefaciens VirB/VirD4 and Enterococcus faecalis pCF10-encoded type IV secretion systems confer specificity to binding of cognate DNA substrates. J Bacteriol 197:2335–2349. [PubMed] 10.1128/JB.00189-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van Kregten M, Lindhout BI, Hooykaas PJ, van der Zaal BJ. 2009. Agrobacterium-mediated T-DNA transfer and integration by minimal VirD2 consisting of the relaxase domain and a type IV secretion system translocation signal. Mol Plant Microbe Interact 22:1356–1365. [PubMed] 10.1094/MPMI-22-11-1356 [DOI] [PubMed] [Google Scholar]
- 48.Atmakuri K, Cascales E, Burton OT, Banta LM, Christie PJ. 2007. Agrobacterium ParA/MinD-like VirC1 spatially coordinates early conjugative DNA transfer reactions. EMBO J 26:2540–2551. [PubMed] 10.1038/sj.emboj.7601696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cascales E, Atmakuri K, Sarkar MK, Christie PJ. 2013. DNA substrate-induced activation of the Agrobacterium VirB/VirD4 type IV secretion system. J Bacteriol 195:2691–2704. [PubMed] 10.1128/JB.00114-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sastre JI, Cabezón E, de la Cruz F. 1998. The carboxyl terminus of protein TraD adds specificity and efficiency to F-plasmid conjugative transfer. J Bacteriol 180:6039–6042. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Beranek A, Zettl M, Lorenzoni K, Schauer A, Manhart M, Koraimann G. 2004. Thirty-eight C-terminal amino acids of the coupling protein TraD of the F-like conjugative resistance plasmid R1 are required and sufficient to confer binding to the substrate selector protein TraM. J Bacteriol 186:6999–7006. [PubMed] 10.1128/JB.186.20.6999-7006.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lu J, Wong JJ, Edwards RA, Manchak J, Frost LS, Glover JN. 2008. Structural basis of specific TraD-TraM recognition during F plasmid-mediated bacterial conjugation. Mol Microbiol 70:89–99. [PubMed] 10.1111/j.1365-2958.2008.06391.x [DOI] [PubMed] [Google Scholar]
- 53.Lu J, Frost LS. 2005. Mutations in the C-terminal region of TraM provide evidence for in vivo TraM-TraD interactions during F-plasmid conjugation. J Bacteriol 187:4767–4773. [PubMed] 10.1128/JB.187.14.4767-4773.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Atmakuri K, Cascales E, Christie PJ. 2004. Energetic components VirD4, VirB11 and VirB4 mediate early DNA transfer reactions required for bacterial type IV secretion. Mol Microbiol 54:1199–1211. [PubMed] 10.1111/j.1365-2958.2004.04345.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Llosa M, Zunzunegui S, de la Cruz F. 2003. Conjugative coupling proteins interact with cognate and heterologous VirB10-like proteins while exhibiting specificity for cognate relaxosomes. Proc Natl Acad Sci USA 100:10465–10470. [PubMed] 10.1073/pnas.1830264100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.de Paz HD, Sangari FJ, Bolland S, García-Lobo JM, Dehio C, de la Cruz F, Llosa M. 2005. Functional interactions between type IV secretion systems involved in DNA transfer and virulence. Microbiology 151:3505–3516. [PubMed] 10.1099/mic.0.28410-0 [DOI] [PubMed] [Google Scholar]
- 57.Tato I, Zunzunegui S, de la Cruz F, Cabezon E. 2005. TrwB, the coupling protein involved in DNA transport during bacterial conjugation, is a DNA-dependent ATPase. Proc Natl Acad Sci USA 102:8156–8161. [PubMed] 10.1073/pnas.0503402102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tato I, Matilla I, Arechaga I, Zunzunegui S, de la Cruz F, Cabezon E. 2007. The ATPase activity of the DNA transporter TrwB is modulated by protein TrwA: implications for a common assembly mechanism of DNA translocating motors. J Biol Chem 282:25569–25576. [PubMed] 10.1074/jbc.M703464200 [DOI] [PubMed] [Google Scholar]
- 59.Ripoll-Rozada J, Zunzunegui S, de la Cruz F, Arechaga I, Cabezón E. 2013. Functional interactions of VirB11 traffic ATPases with VirB4 and VirD4 molecular motors in type IV secretion systems. J Bacteriol 195:4195–4201. [PubMed] 10.1128/JB.00437-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Berger BR, Christie PJ. 1994. Genetic complementation analysis of the Agrobacterium tumefaciens virB operon: virB2 through virB11 are essential virulence genes. J Bacteriol 176:3646–3660. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Haase J, Lurz R, Grahn AM, Bamford DH, Lanka E. 1995. Bacterial conjugation mediated by plasmid RP4: RSF1010 mobilization, donor-specific phage propagation, and pilus production require the same Tra2 core components of a proposed DNA transport complex. J Bacteriol 177:4779–4791. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sagulenko E, Sagulenko V, Chen J, Christie PJ. 2001. Role of Agrobacterium VirB11 ATPase in T-pilus assembly and substrate selection. J Bacteriol 183:5813–5825. [PubMed] 10.1128/JB.183.20.5813-5825.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Savvides SN, Yeo HJ, Beck MR, Blaesing F, Lurz R, Lanka E, Buhrdorf R, Fischer W, Haas R, Waksman G. 2003. VirB11 ATPases are dynamic hexameric assemblies: new insights into bacterial type IV secretion. EMBO J 22:1969–1980. [PubMed] 10.1093/emboj/cdg223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hare S, Bayliss R, Baron C, Waksman G. 2006. A large domain swap in the VirB11 ATPase of Brucella suis leaves the hexameric assembly intact. J Mol Biol 360:56–66. [PubMed] 10.1016/j.jmb.2006.04.060 [DOI] [PubMed] [Google Scholar]
- 65.Savvides SN. 2007. Secretion superfamily ATPases swing big. Structure 15:255–257. [PubMed] 10.1016/j.str.2007.02.003 [DOI] [PubMed] [Google Scholar]
- 66.Akeda Y, Galán JE. 2005. Chaperone release and unfolding of substrates in type III secretion. Nature 437:911–915. [PubMed] 10.1038/nature03992 [DOI] [PubMed] [Google Scholar]
- 67.Middleton R, Sjölander K, Krishnamurthy N, Foley J, Zambryski P. 2005. Predicted hexameric structure of the Agrobacterium VirB4 C terminus suggests VirB4 acts as a docking site during type IV secretion. Proc Natl Acad Sci USA 102:1685–1690. [PubMed] 10.1073/pnas.0409399102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yuan Q, Carle A, Gao C, Sivanesan D, Aly KA, Höppner C, Krall L, Domke N, Baron C. 2005. Identification of the VirB4-VirB8-VirB5-VirB2 pilus assembly sequence of type IV secretion systems. J Biol Chem 280:26349–26359. [PubMed] 10.1074/jbc.M502347200 [DOI] [PubMed] [Google Scholar]
- 69.Li F, Alvarez-Martinez C, Chen Y, Choi KJ, Yeo HJ, Christie PJ. 2012. Enterococcus faecalis PrgJ, a VirB4-like ATPase, mediates pCF10 conjugative transfer through substrate binding. J Bacteriol 194:4041–4051. [PubMed] 10.1128/JB.00648-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Peña A, Matilla I, Martín-Benito J, Valpuesta JM, Carrascosa JL, de la Cruz F, Cabezón E, Arechaga I. 2012. The hexameric structure of a conjugative VirB4 protein ATPase provides new insights for a functional and phylogenetic relationship with DNA translocases. J Biol Chem 287:39925–39932. [PubMed] 10.1074/jbc.M112.413849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Durand E, Oomen C, Waksman G. 2010. Biochemical dissection of the ATPase TraB, the VirB4 homologue of the Escherichia coli pKM101 conjugation machinery. J Bacteriol 192:2315–2323. [PubMed] 10.1128/JB.01384-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jakubowski SJ, Krishnamoorthy V, Cascales E, Christie PJ. 2004. Agrobacterium tumefaciens VirB6 domains direct the ordered export of a DNA substrate through a type IV secretion system. J Mol Biol 341:961–977. [PubMed] 10.1016/j.jmb.2004.06.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rees CE, Wilkins BM. 1989. Transfer of Tra proteins into the recipient cell during bacterial conjugation mediated by plasmid ColIb-P9. J Bacteriol 171:3152–3157. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rees CED, Wilkins BM. 1990. Protein transfer into the recipient cell during bacterial conjugation: studies with F and RP4. Mol Microbiol 4:1199–1205. [PubMed] 10.1111/j.1365-2958.1990.tb00695.x [DOI] [PubMed] [Google Scholar]
- 75.Ninio S, Roy CR. 2007. Effector proteins translocated by Legionella pneumophila: strength in numbers. Trends Microbiol 15:372–380. [PubMed] 10.1016/j.tim.2007.06.006 [DOI] [PubMed] [Google Scholar]
- 76.Zhu W, Banga S, Tan Y, Zheng C, Stephenson R, Gately J, Luo ZQ. 2011. Comprehensive identification of protein substrates of the Dot/Icm type IV transporter of Legionella pneumophila. PLoS One 6:e17638. 10.1371/journal.pone.0017638. 10.1371/journal.pone.0017638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kumar RB, Das A. 2001. Functional analysis of the Agrobacterium tumefaciens T-DNA transport pore protein VirB8. J Bacteriol 183:3636–3641. [PubMed] 10.1128/JB.183.12.3636-3641.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Terradot L, Bayliss R, Oomen C, Leonard GA, Baron C, Waksman G. 2005. Structures of two core subunits of the bacterial type IV secretion system, VirB8 from Brucella suis and ComB10 from Helicobacter pylori. Proc Natl Acad Sci USA 102:4596–4601. [PubMed] 10.1073/pnas.0408927102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bailey S, Ward D, Middleton R, Grossmann JG, Zambryski PC. 2006. Agrobacterium tumefaciens VirB8 structure reveals potential protein-protein interaction sites. Proc Natl Acad Sci USA 103:2582–2587. [PubMed] 10.1073/pnas.0511216103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Porter CJ, Bantwal R, Bannam TL, Rosado CJ, Pearce MC, Adams V, Lyras D, Whisstock JC, Rood JI. 2012. The conjugation protein TcpC from Clostridium perfringens is structurally related to the type IV secretion system protein VirB8 from Gram-negative bacteria. Mol Microbiol 83:275–288. [PubMed] 10.1111/j.1365-2958.2011.07930.x [DOI] [PubMed] [Google Scholar]
- 81.Goessweiner-Mohr N, Grumet L, Arends K, Pavkov-Keller T, Gruber CC, Gruber K, Birner-Gruenberger R, Kropec-Huebner A, Huebner J, Grohmann E, Keller W. 2013. The 2.5 Å structure of the Enterococcus conjugation protein TraM resembles VirB8 type IV secretion proteins. J Biol Chem 288:2018–2028. [PubMed] 10.1074/jbc.M112.428847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kuroda T, Kubori T, Thanh Bui X, Hyakutake A, Uchida Y, Imada K, Nagai H. 2015. Molecular and structural analysis of Legionella DotI gives insights into an inner membrane complex essential for type IV secretion. Sci Rep 5:10912. 10.1038/srep10912. 10.1038/srep10912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Paschos A, den Hartigh A, Smith MA, Atluri VL, Sivanesan D, Tsolis RM, Baron C. 2011. An in vivo high-throughput screening approach targeting the type IV secretion system component VirB8 identified inhibitors of Brucella abortus 2308 proliferation. Infect Immun 79:1033–1043. [PubMed] 10.1128/IAI.00993-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sarkar MK, Husnain SI, Jakubowski SJ, Christie PJ. 2013. Isolation of bacterial type IV machine subassemblies. Methods Mol Biol 966:187–204. [PubMed] 10.1007/978-1-62703-245-2_12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jakubowski SJ, Kerr JE, Garza I, Krishnamoorthy V, Bayliss R, Waksman G, Christie PJ. 2009. Agrobacterium VirB10 domain requirements for type IV secretion and T pilus biogenesis. Mol Microbiol 71:779–794. [PubMed] 10.1111/j.1365-2958.2008.06565.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fronzes R, Christie PJ, Waksman G. 2009. The structural biology of type IV secretion systems. Nat Rev Microbiol 7:703–714. [PubMed] 10.1038/nrmicro2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Winans SC, Burns DL, Christie PJ. 1996. Adaptation of a conjugal transfer system for the export of pathogenic macromolecules. Trends Microbiol 4:64–68. [PubMed] 10.1016/0966-842X(96)81513-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.de Jong MF, Sun YH, den Hartigh AB, van Dijl JM, Tsolis RM. 2008. Identification of VceA and VceC, two members of the VjbR regulon that are translocated into macrophages by the Brucella type IV secretion system. Mol Microbiol 70:1378–1396. [PubMed] 10.1111/j.1365-2958.2008.06487.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Myeni S, Child R, Ng TW, Kupko JJ III, Wehrly TD, Porcella SF, Knodler LA, Celli J. 2013. Brucella modulates secretory trafficking via multiple type IV secretion effector proteins. PLoS Pathog 9:e1003556. 10.1371/journal.ppat.1003556. 10.1371/journal.ppat.1003556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ke Y, Wang Y, Li W, Chen Z. 2015. Type IV secretion system of Brucella spp. and its effectors. Front Cell Infect Microbiol 5:72. 10.3389/fcimb.2015.00072. [PubMed] 10.3389/fcimb.2015.00072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ou JT. 1975. Mating signal and DNA penetration deficiency in conjugation between male Escherichia coli and minicells. Proc Natl Acad Sci USA 72:3721–3725. [PubMed] 10.1073/pnas.72.9.3721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lang S, Kirchberger PC, Gruber CJ, Redzej A, Raffl S, Zellnig G, Zangger K, Zechner EL. 2011. An activation domain of plasmid R1 TraI protein delineates stages of gene transfer initiation. Mol Microbiol 82:1071–1085. [PubMed] 10.1111/j.1365-2958.2011.07872.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cascales E, Christie PJ. 2004. Agrobacterium VirB10, an ATP energy sensor required for type IV secretion. Proc Natl Acad Sci USA 101:17228–17233. [PubMed] 10.1073/pnas.0405843101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Banta LM, Kerr JE, Cascales E, Giuliano ME, Bailey ME, McKay C, Chandran V, Waksman G, Christie PJ. 2011. An Agrobacterium VirB10 mutation conferring a type IV secretion system gating defect. J Bacteriol 193:2566–2574. [PubMed] 10.1128/JB.00038-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Anthony KG, Klimke WA, Manchak J, Frost LS. 1999. Comparison of proteins involved in pilus synthesis and mating pair stabilization from the related plasmids F and R100-1: insights into the mechanism of conjugation. J Bacteriol 181:5149–5159. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Audette GF, Manchak J, Beatty P, Klimke WA, Frost LS. 2007. Entry exclusion in F-like plasmids requires intact TraG in the donor that recognizes its cognate TraS in the recipient. Microbiology 153:442–451. [PubMed] 10.1099/mic.0.2006/001917-0 [DOI] [PubMed] [Google Scholar]
- 97.Marrero J, Waldor MK. 2007. Determinants of entry exclusion within Eex and TraG are cytoplasmic. J Bacteriol 189:6469–6473. [PubMed] 10.1128/JB.00522-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gillespie JJ, Ammerman NC, Dreher-Lesnick SM, Rahman MS, Worley MJ, Setubal JC, Sobral BS, Azad AF. 2009. An anomalous type IV secretion system in Rickettsia is evolutionarily conserved. PLoS One 4:e4833. 10.1371/journal.pone.0004833. [PubMed] 10.1371/journal.pone.0004833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gillespie JJ, Brayton KA, Williams KP, Diaz MA, Brown WC, Azad AF, Sobral BW. 2010. Phylogenomics reveals a diverse Rickettsiales type IV secretion system. Infect Immun 78:1809–1823. [PubMed] 10.1128/IAI.01384-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rancès E, Voronin D, Tran-Van V, Mavingui P. 2008. Genetic and functional characterization of the type IV secretion system in Wolbachia. J Bacteriol 190:5020–5030. [PubMed] 10.1128/JB.00377-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Nagai H, Roy CR. 2001. The DotA protein from Legionella pneumophila is secreted by a novel process that requires the Dot/Icm transporter. EMBO J 20:5962–5970. [PubMed] 10.1093/emboj/20.21.5962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Fischer W. 2011. Assembly and molecular mode of action of the Helicobacter pylori Cag type IV secretion apparatus. FEBS J 278:1203–1212. [PubMed] 10.1111/j.1742-4658.2011.08036.x [DOI] [PubMed] [Google Scholar]
- 103.Terradot L, Waksman G. 2011. Architecture of the Helicobacter pylori Cag-type IV secretion system. FEBS J 278:1213–1222. [PubMed] 10.1111/j.1742-4658.2011.08037.x [DOI] [PubMed] [Google Scholar]
- 104.Nakano N, Kubori T, Kinoshita M, Imada K, Nagai H. 2010. Crystal structure of Legionella DotD: insights into the relationship between type IVB and type II/III secretion systems. PLoS Pathog 6:e1001129. 10.1371/journal.ppat.1001129. [PubMed] 10.1371/journal.ppat.1001129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Souza DP, Andrade MO, Alvarez-Martinez CE, Arantes GM, Farah CS, Salinas RK. 2011. A component of the Xanthomonadaceae type IV secretion system combines a VirB7 motif with a N0 domain found in outer membrane transport proteins. PLoS Pathog 7:e1002031. 10.1371/journal.ppat.1002031. 10.1371/journal.ppat.1002031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ding H, Zeng H, Huang L, Dong Y, Duan Y, Mao X, Guo G, Zou Q. 2012. Helicobacter pylori chaperone-like protein CagT plays an essential role in the translocation of CagA into host cells. J Microbiol Biotechnol 22:1343–1349. [PubMed] 10.4014/jmb.1202.02025 [DOI] [PubMed] [Google Scholar]
- 107.Johnson EM, Gaddy JA, Voss BJ, Hennig EE, Cover TL. 2014. Genes required for assembly of pili associated with the Helicobacter pylori cag type IV secretion system. Infect Immun 82:3457–3470. [PubMed] 10.1128/IAI.01640-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Aras RA, Fischer W, Perez-Perez GI, Crosatti M, Ando T, Haas R, Blaser MJ. 2003. Plasticity of repetitive DNA sequences within a bacterial (Type IV) secretion system component. J Exp Med 198:1349–1360. [PubMed] 10.1084/jem.20030381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Rohde M, Püls J, Buhrdorf R, Fischer W, Haas R. 2003. A novel sheathed surface organelle of the Helicobacter pylori cag type IV secretion system. Mol Microbiol 49:219–234. [PubMed] 10.1046/j.1365-2958.2003.03549.x [DOI] [PubMed] [Google Scholar]
- 110.Barrozo RM, Cooke CL, Hansen LM, Lam AM, Gaddy JA, Johnson EM, Cariaga TA, Suarez G, Peek RM Jr, Cover TL, Solnick JV. 2013. Functional plasticity in the type IV secretion system of Helicobacter pylori. PLoS Pathog 9:e1003189. 10.1371/journal.ppat.1003189. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Segal G, Purcell M, Shuman HA. 1998. Host cell killing and bacterial conjugation require overlapping sets of genes within a 22-kb region of the Legionella pneumophila genome. Proc Natl Acad Sci USA 95:1669–1674. [PubMed] 10.1073/pnas.95.4.1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Vogel JP, Andrews HL, Wong SK, Isberg RR. 1998. Conjugative transfer by the virulence system of Legionella pneumophila. Science 279:873–876. [PubMed] 10.1126/science.279.5352.873 [DOI] [PubMed] [Google Scholar]
- 113.Kelley LA, Sternberg MJ. 2009. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc 4:363–371. [PubMed] 10.1038/nprot.2009.2 [DOI] [PubMed] [Google Scholar]
- 114.Henry T, Couillault C, Rockenfeller P, Boucrot E, Dumont A, Schroeder N, Hermant A, Knodler LA, Lecine P, Steele-Mortimer O, Borg JP, Gorvel JP, Méresse S. 2006. The Salmonella effector protein PipB2 is a linker for kinesin-1. Proc Natl Acad Sci USA 103:13497–13502. [PubMed] 10.1073/pnas.0605443103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Baisón-Olmo F, Cardenal-Muñoz E, Ramos-Morales F. 2012. PipB2 is a substrate of the Salmonella pathogenicity island 1-encoded type III secretion system. Biochem Biophys Res Commun 423:240–246. [PubMed] 10.1016/j.bbrc.2012.05.095 [DOI] [PubMed] [Google Scholar]
- 116.Diao J, Zhang Y, Huibregtse JM, Zhou D, Chen J. 2008. Crystal structure of SopA, a Salmonella effector protein mimicking a eukaryotic ubiquitin ligase. Nat Struct Mol Biol 15:65–70. [PubMed] 10.1038/nsmb1346 [DOI] [PubMed] [Google Scholar]
- 117.Bernstein HD. 2010. Type V secretion: the autotransporter and two-partner secretion pathways. Ecosal Plus 4. 10.1128/ecosalplus.4.3.6.. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Harrington LC, Rogerson AC. 1990. The F pilus of Escherichia coli appears to support stable DNA transfer in the absence of wall-to-wall contact between cells. J Bacteriol 172:7263–7264. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Babic A, Lindner AB, Vulic M, Stewart EJ, Radman M. 2008. Direct visualization of horizontal gene transfer. Science 319:1533–1536. [PubMed] 10.1126/science.1153498 [DOI] [PubMed] [Google Scholar]
- 120.Lawley TD, Gordon GS, Wright A, Taylor DE. 2002. Bacterial conjugative transfer: visualization of successful mating pairs and plasmid establishment in live Escherichia coli. Mol Microbiol 44:947–956. [PubMed] 10.1046/j.1365-2958.2002.02938.x [DOI] [PubMed] [Google Scholar]
- 121.Jakubowski SJ, Cascales E, Krishnamoorthy V, Christie PJ. 2005. Agrobacterium tumefaciens VirB9, an outer-membrane-associated component of a type IV secretion system, regulates substrate selection and T-pilus biogenesis. J Bacteriol 187:3486–3495. [PubMed] 10.1128/JB.187.10.3486-3495.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bradley DE. 1980. Morphological and serological relationships of conjugative pili. Plasmid 4:155–169. [PubMed] 10.1016/0147-619X(80)90005-0 [DOI] [PubMed] [Google Scholar]
- 123.Bradley DE, Taylor DE, Cohen DR. 1980. Specification of surface mating systems among conjugative drug resistance plasmids in Escherichia coli K-12. J Bacteriol 143:1466–1470. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Paranchych W, Frost LS. 1988. The physiology and biochemistry of pili. Adv Microb Physiol 29:53–114. [PubMed] 10.1016/S0065-2911(08)60346-X [DOI] [PubMed] [Google Scholar]
- 125.Bayer M, Eferl R, Zellnig G, Teferle K, Dijkstra A, Koraimann G, Högenauer G. 1995. Gene 19 of plasmid R1 is required for both efficient conjugative DNA transfer and bacteriophage R17 infection. J Bacteriol 177:4279–4288. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zupan J, Hackworth CA, Aguilar J, Ward D, Zambryski P. 2007. VirB1* promotes T-pilus formation in the vir-Type IV secretion system of Agrobacterium tumefaciens. J Bacteriol 189:6551–6563. [PubMed] 10.1128/JB.00480-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Majdalani N, Moore D, Maneewannakul S, Ippen-Ihler K. 1996. Role of the propilin leader peptide in the maturation of F pilin. J Bacteriol 178:3748–3754. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Majdalani N, Ippen-Ihler K. 1996. Membrane insertion of the F-pilin subunit is Sec independent but requires leader peptidase B and the proton motive force. J Bacteriol 178:3742–3747. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Maneewannakul K, Maneewannakul S, Ippen-Ihler K. 1993. Synthesis of F pilin. J Bacteriol 175:1384–1391. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Eisenbrandt R, Kalkum M, Lai EM, Lurz R, Kado CI, Lanka E. 1999. Conjugative pili of IncP plasmids, and the Ti plasmid T pilus are composed of cyclic subunits. J Biol Chem 274:22548–22555. [PubMed] 10.1074/jbc.274.32.22548 [DOI] [PubMed] [Google Scholar]
- 131.Eisenbrandt R, Kalkum M, Lurz R, Lanka E. 2000. Maturation of IncP pilin precursors resembles the catalytic Dyad-like mechanism of leader peptidases. J Bacteriol 182:6751–6761. [PubMed] 10.1128/JB.182.23.6751-6761.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Paiva WD, Grossman T, Silverman PM. 1992. Characterization of F-pilin as an inner membrane component of Escherichia coli K12. J Biol Chem 267:26191–26197. [PubMed] [PubMed] [Google Scholar]
- 133.Paiva WD, Silverman PM. 1996. Effects of F-encoded components and F-pilin domains on the synthesis and membrane insertion of TraA-'PhoA fusion proteins. Mol Microbiol 19:1277–1286. [PubMed] 10.1111/j.1365-2958.1996.tb02472.x [DOI] [PubMed] [Google Scholar]
- 134.Kerr JE, Christie PJ. 2010. Evidence for VirB4-mediated dislocation of membrane-integrated VirB2 pilin during biogenesis of the Agrobacterium VirB/VirD4 type IV secretion system. J Bacteriol 192:4923–4934. [PubMed] 10.1128/JB.00557-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Frost LS. 2009. Conjugation, bacterial, p 294–308. In Schaechter M (ed), Desk Encyclopedia of Microbiology, 2nd ed, Academic Press, San Diego, CA. 10.1016/B978-012373944-5.00007-9 [DOI] [Google Scholar]
- 136.Wang YA, Yu X, Silverman PM, Harris RL, Egelman EH. 2009. The structure of F-pili. J Mol Biol 385:22–29. [PubMed] 10.1016/j.jmb.2008.10.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Gilmour MW, Lawley TD, Rooker MM, Newnham PJ, Taylor DE. 2001. Cellular location and temperature-dependent assembly of IncHI1 plasmid R27-encoded TrhC-associated conjugative transfer protein complexes. Mol Microbiol 42:705–715. [PubMed] 10.1046/j.1365-2958.2001.02682.x [DOI] [PubMed] [Google Scholar]
- 138.Aly KA, Baron C. 2007. The VirB5 protein localizes to the T-pilus tips in Agrobacterium tumefaciens. Microbiology 153:3766–3775. [PubMed] 10.1099/mic.0.2007/010462-0 [DOI] [PubMed] [Google Scholar]
- 139.Harris RL, Silverman PM. 2004. Tra proteins characteristic of F-like type IV secretion systems constitute an interaction group by yeast two-hybrid analysis. J Bacteriol 186:5480–5485. [PubMed] 10.1128/JB.186.16.5480-5485.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Arutyunov D, Arenson B, Manchak J, Frost LS. 2010. F plasmid TraF and TraH are components of an outer membrane complex involved in conjugation. J Bacteriol 192:1730–1734. [PubMed] 10.1128/JB.00726-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Harris RL, Hombs V, Silverman PM. 2001. Evidence that F-plasmid proteins TraV, TraK and TraB assemble into an envelope-spanning structure in Escherichia coli. Mol Microbiol 42:757–766. [PubMed] 10.1046/j.1365-2958.2001.02667.x [DOI] [PubMed] [Google Scholar]
- 142.Maneewannakul S, Maneewannakul K, Ippen-Ihler K. 1992. Characterization, localization, and sequence of F transfer region products: the pilus assembly gene product TraW and a new product, TrbI. J Bacteriol 174:5567–5574. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Dürrenberger MB, Villiger W, Bächi T. 1991. Conjugational junctions: morphology of specific contacts in conjugating Escherichia coli bacteria. J Struct Biol 107:146–156. [PubMed] 10.1016/1047-8477(91)90018-R [DOI] [PubMed] [Google Scholar]
- 144.Achtman M, Kennedy N, Skurray R. 1977. Cell–cell interactions in conjugating Escherichia coli: role of traT protein in surface exclusion. Proc Natl Acad Sci USA 74:5104–5108. [PubMed] 10.1073/pnas.74.11.5104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Klimke WA, Rypien CD, Klinger B, Kennedy RA, Rodriguez-Maillard JM, Frost LS. 2005. The mating pair stabilization protein, TraN, of the F plasmid is an outer-membrane protein with two regions that are important for its function in conjugation. Microbiology 151:3527–3540. [PubMed] 10.1099/mic.0.28025-0 [DOI] [PubMed] [Google Scholar]
- 146.Low B, Wood TH. 1965. A quick and efficient method for interruption of bacterial conjugation. Genet Res 6:300–303. [PubMed] 10.1017/S001667230000416X [DOI] [PubMed] [Google Scholar]
- 147.Anthony KG, Sherburne C, Sherburne R, Frost LS. 1994. The role of the pilus in recipient cell recognition during bacterial conjugation mediated by F-like plasmids. Mol Microbiol 13:939–953. [PubMed] 10.1111/j.1365-2958.1994.tb00486.x [DOI] [PubMed] [Google Scholar]
- 148.Firth N, Skurray R. 1992. Characterization of the F plasmid bifunctional conjugation gene, traG. Mol Gen Genet 232:145–153. [PubMed] 10.1007/BF00299147 [DOI] [PubMed] [Google Scholar]
- 149.Samuels AL, Lanka E, Davies JE. 2000. Conjugative junctions in RP4-mediated mating of Escherichia coli. J Bacteriol 182:2709–2715. [PubMed] 10.1128/JB.182.10.2709-2715.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Heinemann JA, Sprague GFJ Jr. 1989. Bacterial conjugative plasmids mobilize DNA transfer between bacteria and yeast. Nature 340:205–209. [PubMed] 10.1038/340205a0 [DOI] [PubMed] [Google Scholar]
- 151.Bates S, Cashmore AM, Wilkins BM. 1998. IncP plasmids are unusually effective in mediating conjugation of Escherichia coli and Saccharomyces cerevisiae: involvement of the tra2 mating system. J Bacteriol 180:6538–6543. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Waters VL. 2001. Conjugation between bacterial and mammalian cells. Nat Genet 29:375–376. [PubMed] 10.1038/ng779 [DOI] [PubMed] [Google Scholar]
- 153.Silby MW, Ferguson GC, Billington C, Heinemann JA. 2007. Localization of the plasmid-encoded proteins TraI and MobA in eukaryotic cells. Plasmid 57:118–130. [PubMed] 10.1016/j.plasmid.2006.08.006 [DOI] [PubMed] [Google Scholar]
- 154.Guynet C, Cuevas A, Moncalián G, de la Cruz F. 2011. The stb operon balances the requirements for vegetative stability and conjugative transfer of plasmid R388. PLoS Genet 7:e1002073. 10.1371/journal.pgen.1002073. [PubMed] 10.1371/journal.pgen.1002073 [DOI] [PMC free article] [PubMed] [Google Scholar]


