Abstract
Objectives
There is a need for validated predictive markers of gemcitabine response to guide precision medicine treatment in pancreatic cancer. We previously validated human Equilibrative Nucleoside Transporter 1 (hENT1) as a predictive marker of gemcitabine treatment response using Radiation Therapy Oncology Group (RTOG) 9704. Controversy exists about the predictive value of gemcitabine metabolism pathway biomarkers: deoxycytidine kinase (DCK), ribonucleotide reductase 1 (RRM1), ribonucleotide reductase 2 (RRM2) and p53R2.
Methods
RTOG 9704 prospectively randomized 538 patients after pancreatic resection to receive either 5-fluorouracil (5-FU) or gemcitabine. Tumor DCK, RRM1, RRM2 and p53R protein expression were analyzed using a Tissue Microarray (TMA) and Immunohistochemistry (IHC) and correlated with treatment outcome (overall survival and disease free survival) by unconditional logistic regression analysis.
Results
229 patients were eligible for analysis from both the 5-FU and Gemcitabine arm. Only RRM2 proteins expression, and not DCK, RRM1, or p53R2 protein expression was associated with survival in the gemcitabine treatment arm.
Conclusions
Despite limited data from other nonrandomized treatment data our data does not support the predictive value of DCK, RRM1, or p53R2. Efforts should focus on hENT1, and possibly RRM2 as a valid predictive marker of treatment response of gemcitabine in pancreas cancer.
Keywords: Pancreatic Cancer, Precision Medicine, hENT1, DCK, RRM1, RRM2
INTRODUCTION
The availability of both gemcitabine-based and non-gemcitabine-based treatment regimens for pancreatic cancer and the overall poor dismal prognosis associated with this disease necessitates the validation of predictive markers of treatment response in this disease, to facilitate precision medicine options. (1–3) RTOG 9704 is a phase III randomized post-operative adjuvant study in patients with pancreatic cancer comparing pre- and post-chemoradiation 5-fluorouracil with pre- and post- chemoradiation gemcitabine, and represents an ideal study for evaluating predictive markers of gemcitabine treatment response.(4) Using it, we previously showed protein expression of hENT1, the predominant transmembrane transporter for gemcitabine, as a possible treatment predictive marker in the adjuvant setting for pancreatic cancer(5), which was further validated by several subsequent studies(Supplementary Table 1).(6–13)
There are several reasons for why there is conflicting data about the predictive value of other markers involved with gemcitabine metabolism (DCK,(7, 14, 15) RRM1(7, 9, 16–18), RRM2(17, 19) and p53R2) (20) (21) (22), some of which are being used in routine clinical practice to make decisions about precision medicine options for patients. Firstly, several gene expression studies have been performed without the appropriate microdissection of the tumor tissue to separate it from the stromal tissue. This suggests the protein-based markers by immunohistochemistry (IHC) represent the most reproducible methodology for these types of studies. Secondly, several studies used heterogeneous collection of data and disease stages, non-prospective and non-randomized treatment regimens often involving multiple drug regimens. Finally, different antibodies and IHC scoring systems have been used across several studies.
In view of the appropriate design and prospective tissue collection of the RTOG 9704 study, and its initial use in validating hENT1 as a promising predictive marker, we decided to use this study to analyze DCK, RRM1, RRM2 and p53R2 protein expression, correlate it with treatment outcome, and to compare our results with current existing published data to assess the predictive value of these markers for guiding precision medicine options with gemcitabine treatment.
METHODS
Patient Selection and Consent
The RTOG tissue bank received tumor blocks from a total of 229 of the 538 patients who had undergone surgical resection and were entered in the RTOG 9704 prospective adjuvant treatment trial. Permission to perform this study was obtained by the Institutional Review Board. (23) A tissue micro-array (TMA), which was also used for the original hENT1 study, was constructed using tissue core samples from the patients enrolled in the RTOG 9704 study, using three cores from different areas of a patient’s tumor.
Immunohistochemistry and Scoring
Rabbit polyclonal antibodies were raised against a synthetic human DCK peptide, and tissue immunohistochemistry was performed per prior study.(24) Semi-quantitative scoring of tumoral cytoplasmic staining was used for the evaluation of DCK protein expression by a single pathologist (R.L.). Staining of DCK protein was assigned a score from 0 to 2 based on staining intensity (no staining = 0, weakly positive staining = 1, and strongly positive staining = 2). A final score (0–200) was determined by multiplying the intensity score and the percentage of the positive cells in the specimen, as described previously.(15) The mean score of triplicate tissues from each patient was used for analysis, and the scores were dichotomized into low DCK and high DCK based on the median score of all the cytoplasmic means per patient data in the three TMAs.
Immunofluorescence combined with AQUA, using antiserum to RRM1 was generated from rabbits was used to assess in situ expression of nuclear RRM1 as described previously.(25) The final slides were scanned with SpotGrabber (HistoRx, New Haven, CT, USA), and images were analyzed with AQUA (version 1.6, PM-2000, HistoRx) by a pathologist with experience with RRM1 staining (ZZ). The AQUA scores ranged from 0 (no expression) to 3000 (maximal observed expression) for the triplicate TMA tissues for each patient. The median value of the RRM1 expression levels were used to divided the patients into high RRM1 and low RRM1 levels.
Tumoral protein expression of hRRM2 was performed using a mouse polyclonal antibody against hRRM2 (Convance, Princeton, NJ) as previously described.(26). Two independent scorers (YY, XL) experienced with RRM2 immunohistochemistry, assessed and scored the cytoplasmic RRM2 immunostaining intensities, ranging from 0 (no staining), to 3 (maximum staining), with a consensus score being agreed upon if there was initial discordance between the two scorers.(26) The mean cytoplasmic RRM2 score was assessed per patient, and the RRM2 cytoplasmic score was also categorized as “Low RRM2” expressing (mean cytoplasmic score <1.5 (<mean + 1 S.D.) and “High RRM2” expressing (mean cytoplasmic score ≥1.5 (≥mean + 1 S.D.).
Due to non-specific targeting of commercial p53R2 antibody, a new highly specific p53R2 anti-rabbit antibody was used.(27) Details of the de-paraffinization protocol and IHC were described in a previous publication.(28). Each sample was scored by two independent investigators in a double-blind manner (XL, YY), with a consensus score being agreed upon if there was initial discordance between the two scorer. Cytoplasmic staining of p53R2 protein was assigned a score from 0 to 2 based on staining intensity (no staining = 0, weakly positive staining = 1, and strongly positive staining = 2) per prior study.(27) The percentage of adenocarcinoma cells stained at each intensity level was recorded for each, resulting in a weighted scores ranged between 0 and 200. For the purposes of statistical analysis, the scores were dichotomized into low p53R2 and high p53R2 based on the median score.
IHC Biomarkers Statistical Analysis
The DCK, RRM1, RRM2, p53R2 (Gemcitabine pathway biomarkers) immunohistochemistry scores were submitted to the RTOG Statistical Department for analysis without knowledge of patient demographics, treatment arm randomization or outcome. A statistical comparison to assess whether missing data were associated with dichotomized baseline characteristics (pathological t-stage (T1, T2 vs. T3, T4), AJCC stage (I, II vs. III, IV), primary tumor location (head vs. everything else)). was carried out using the chi-square test for each of the markers.
Association between the gemcitabine pathway biomarkers protein expression, dichotomized (“Low” vs. “High”) with tumor demographic details and treatment outcome (Overall survival (OS), Disease Free Survival (DFS)) were sought by the chi-square test and the Cox proportional hazards model. Both treatment arms of the study were analyzed. OS and DFS were estimated using the Kaplan-Meier method and the biomarkers were compared using the log-rank test. The following variables were included in the multivariate analyses based on analysis of the original trial: age, gender, race, treatment arm (when appropriate), nodal involvement (no vs. yes), tumor diameter (< 3 vs. ≥ 3 cm), KPS (100, 90 vs. 60, 70, 80), and surgical margin status (negative vs. positive and negative vs. unknown). Results were expressed as Hazard Ratio (H.R), (H.R. > 1 denoting increased risk of death) and considered significant at p-value ≤0.05.
Literature Search
A systematic literature search upto December 2014 was performed in MEDLINE and EMBASE to identify relevant studies. An initial search study using recognized search terms [hENT1 or DCK or RRM1 or RRM2 or p53R2] and [Pancreatic Cancer or Pancreatic Carcinoma] and [Gemcitabine] was conducted. Studies were considered eligible for comparison with the current study if there were performed using immunohistochemistry and in the adjuvant setting.
RESULTS
DCK Protein Expression
Of the 229 patient tumor samples evaluated, 186 had analyzable DCK immunostaining. There were no positive statistical associations between baseline characteristics and missing and determined DCK protein expression (Supplementary Table 2 and Supplementary Table 3). For cytoplasm mean across three TMAs, the median score was 125.00 (min-max: 0.00–200.00) to dichotomize the results in to “Low DCK” and “High DCK”. “High DCK” expression was seen in 50 of 88 patients in the gemcitabine treatment arm, and 44 of 98 patients randomized to the 5-FU treatment arm.
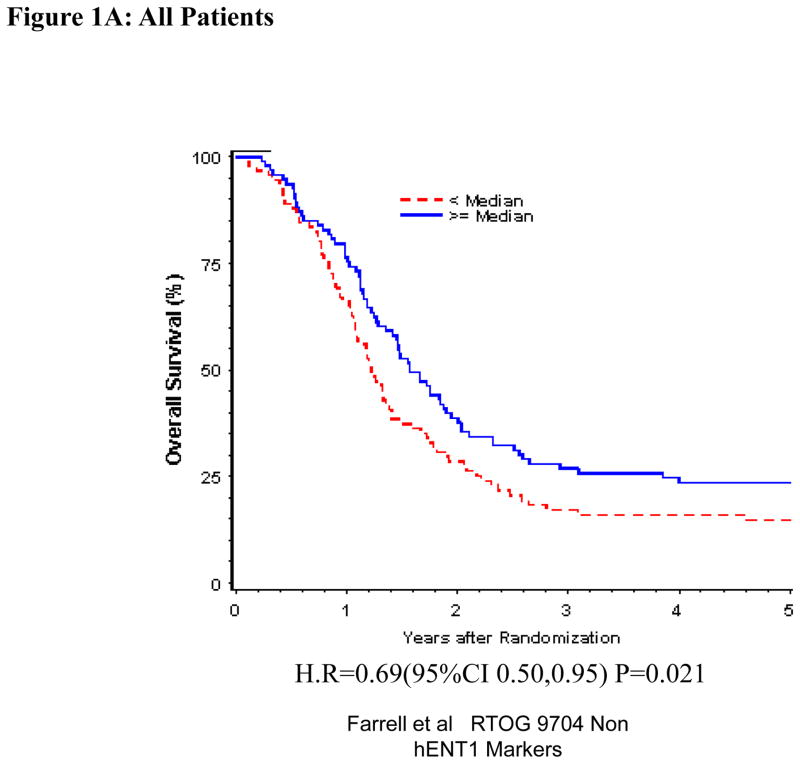
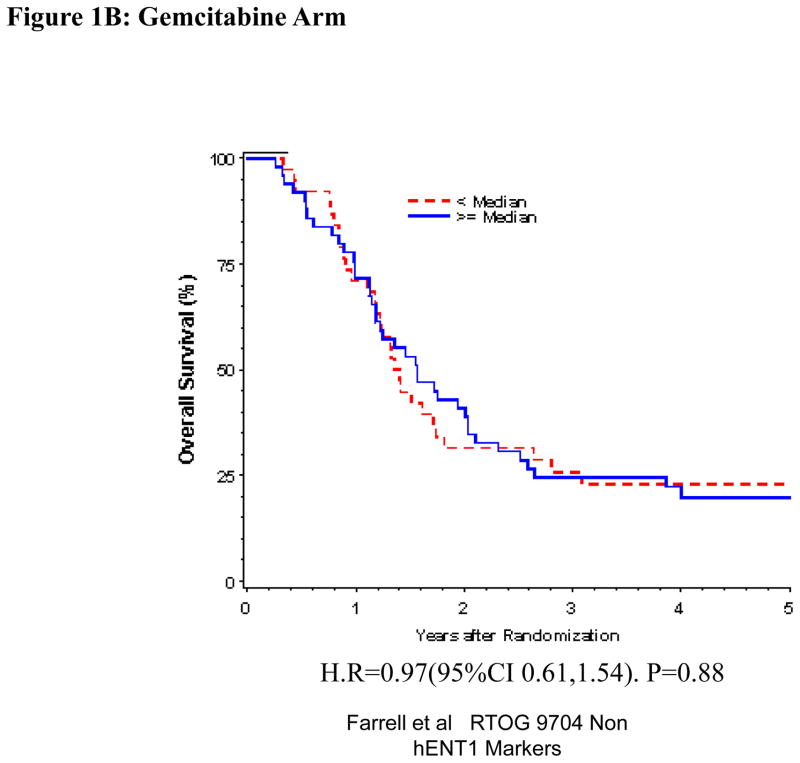
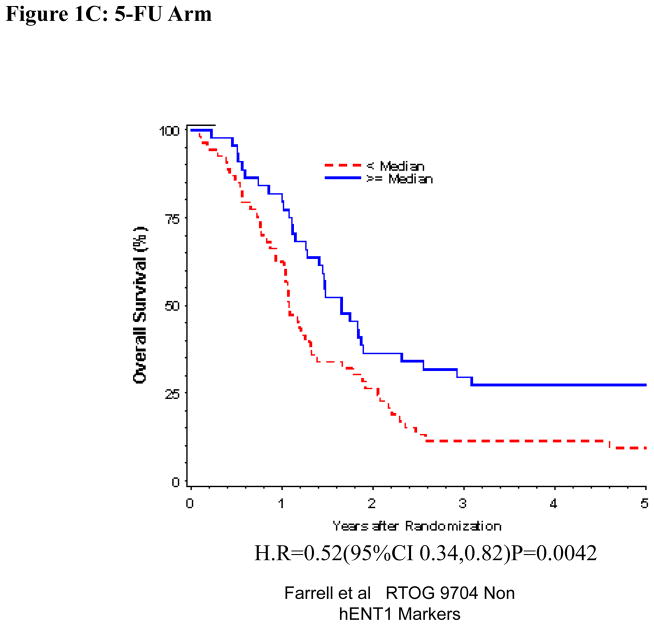
Univariate and multivariate analyses for dichotomized DCK: “Low DCK” vs. “High DCK” was performed for all patients (Table 1). By multivariate analysis, DCK expression was independently and significantly associated with OS despite adjusting for baseline characteristics in these multivariate models for the overall treatment group (HR = 0.71, p=0.044) (Table 1, Figure 1 A), but this was not seen with DFS. DCK proteins expression was not significantly associated with OS or DFS among those patients randomized to the gemcitabine treatment arm on multivariate analysis (Table 1, Figure 1B). However, multivariate analysis confirmed improved OS when comparing all patients with “High DCK” to patients with “Low DCK” (H.R. = 0.50, p=0.0026) (Table 1, Figure 1C), but no association was seen for DFS in the 5-FU treatment arm.
Table 1.
Overall survival and Disease-Free Survival for hENT1, dCK, RRM1, RRM2 and p53R2 (univariate and multivariate analysis)
| hENT1(ref 5) | dCK | RRM1 | RRM2 | P53R2 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Low/High vs No hENT1 | High vs Low DCK | High vs Low RRM1 | High vs Low RRM2 | Low vs High p53R2 | ||||||
| Univariate | HR | p | HR | p | HR | p | HR | p-value | HR | p |
| Overall Survival | ||||||||||
| Both Groups | 0.73 | 0.09 | 0.69 | 0.022 | 0.72 | 0.04 | 1.42 | 0.17 | 1.05 | 0.80 |
| RT+5-FU | 0.93 | 0.75 | 0.52 | 0.0048 | 0.53 | 0.0039 | 1.72 | 0.10 | 0.90 | 0.64 |
| RT+GEM | 0.51 | 0.02 | 0.97 | 0.88 | 1.01 | 0.97 | 1.10 | 0.81 | 1.28 | 0.35 |
| Disease Free Survival | ||||||||||
| Both Groups | 0.74 | 0.09 | 0.84 | 0.25 | 0.82 | 0.19 | 1.62 | 0.045 | 1.05 | 0.79 |
| RT+5FU | 0.88 | 0.60 | 0.67 | 0.061 | 0.65 | 0.036 | 1.50 | 0.21 | 0.76 | 0.23 |
| RT+GEM | 0.57 | 0.05 | 1.11 | 0.66 | 1.06 | 0.80 | 1.74 | 0.12 | 1.50 | 0.11 |
| Multivariate | ||||||||||
| Overall Survival | ||||||||||
| Both Groups | 0.66 | 0.03 | 0.71 | 0.044 | 0.71 | 0.031 | 1.58 | 0.069 | 1.07 | 0.71 |
| RT+5-FU | 0.78 | 0.31 | 0.50 | 0.0026 | 0.51 | 0.0021 | 1.78 | 0.078 | 0.98 | 0.92 |
| RT+GEM | 0.40 | 0.004 | 0.92 | 0.72 | 1.20 | 0.44 | 1.42 | 0.39 | 1.36 | 0.25 |
| Disease Free Survival | ||||||||||
| Both Groups | 0.61 | 0.009 | 0.88 | 0.40 | 0.84 | 0.26 | 1.62 | 0.0045 | 1.01 | 0.97 |
| RT+5FU | 0.72 | 0.18 | 0.67 | 0.06 | 0.65 | 0.036 | 1.78 | 0.21 | 0.76 | 0.23 |
| RT+GEM | 0.39 | 0.003 | 1.37 | 0.19 | 1.26 | 0.34 | 2.24 | 0.031 | 1.62 | 0.06 |
HR = Hazard Ratio* a hazard ratio of 1 indicates no difference between the two subgroups such that a HR < 1 indicates an increased survival for the first variables listed.
p-value from Chi-square test using the Cox proportional hazards model.
Figure 1.
Overall survival for DCK protein expression, multivariate analysis. A. All patients. B Gemcitabine Treatment Arm. C 5-FU treatment arm
RRM1 Protein Expression
Of the 229 patient tumor samples evaluated, 196 had analyzable RRM1 immunostaining. There were no positive statistical associations between baseline characteristics and missing and determined RRM1 protein expression (Supplementary Table 4 and Supplementary Table 5). For nucleus staining maximum of the triplicate tissues from a single patient, the median of the average of all TMA was 971.38 (min-max: 27.14–3370.34), which is used as the cut-off to divide patients into “Low RRM1” and “High RRM1”. “High RRM1” staining was seen in 44 of 92 patients in the gemcitabine treatment arm, and 54 of 104 patients randomized to the 5-FU treatment arm.
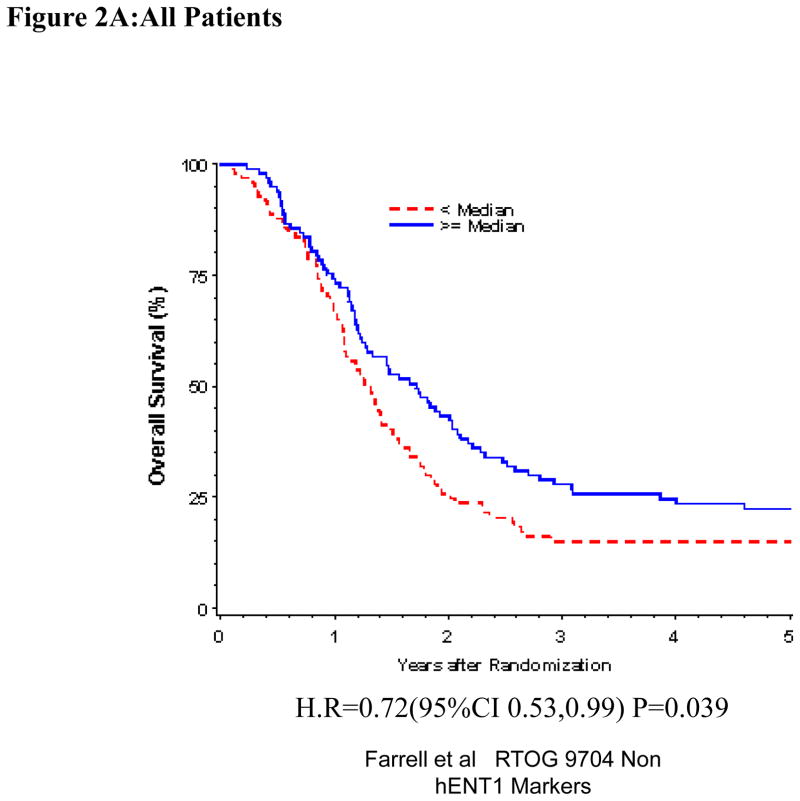
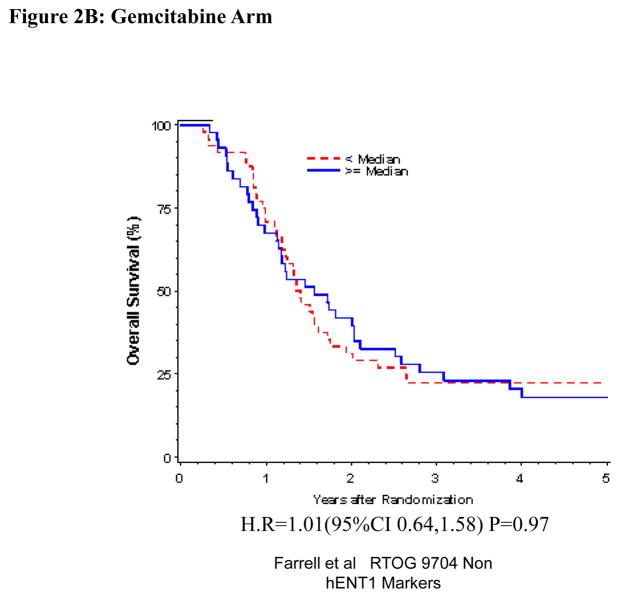
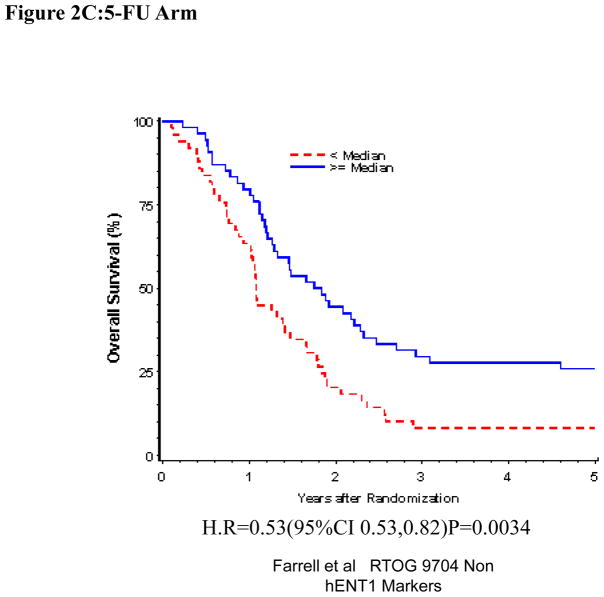
Univariate and Multivariate analyses for dichotomized RRM1: “Low RRM1” vs. “High RRM1” was performed for all patients (Table 1). RRM1 expression was independently and significantly associated with OS (H.R.= 0.71, p=0.031), but not DFS (H.R.=0.84, p=0.26) despite adjusting for baseline characteristics in these multivariate models for the overall treatment group (Table 1, Figure 2A). We observed statistically significant prolonged OS (H.R. = 0.51 p=0.0021) and DFS (H.R. = 0.65 p=0.036) for patients with “High RRM1” as compared to patients with “Low RRM1” for all patients treated with 5-FU (Table 1, Figure 2C). No association between RRM1 protein expression and OS and DFS was seen in the gemcitabine treated group (Table 1, Figure 2B).
Figure 2.
Overall survival for RRM1 protein expression, multivariate analysis. A. All patients. B Gemcitabine Treatment Arm. C 5-FU treatment arm
RRM2 Protein Expression
Of the 229 patient tumor samples evaluated, 189 had analyzable RRM2 immunostaining. There were no positive statistical associations between baseline characteristics and missing and determined RRM2 protein expression (Supplementary Table 6 and Supplementary Table 7). “High RRM2” staining was seen in 9 of 88 patients in the gemcitabine treatment arm, and 12 of 101 patients randomized to the 5-FU treatment arm.
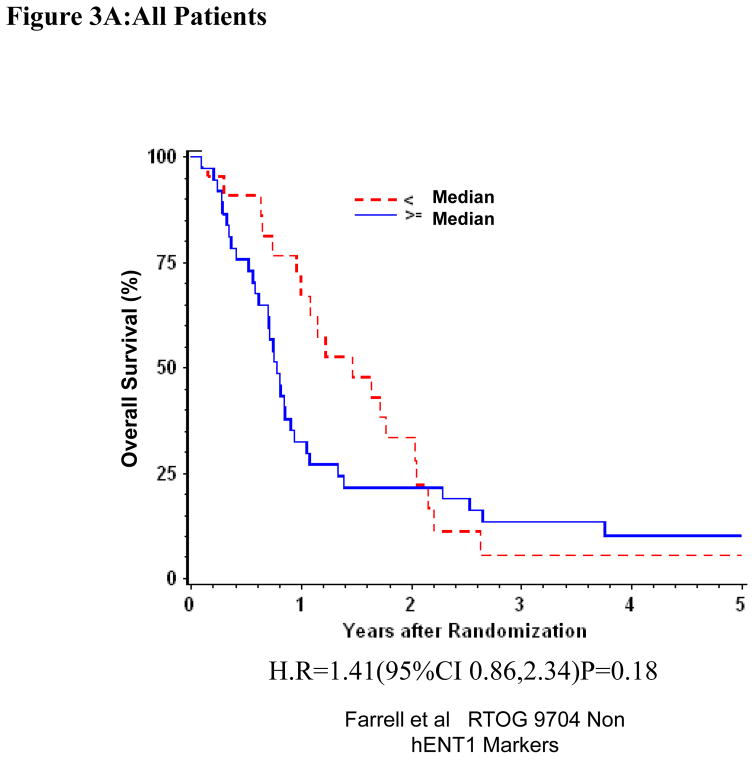
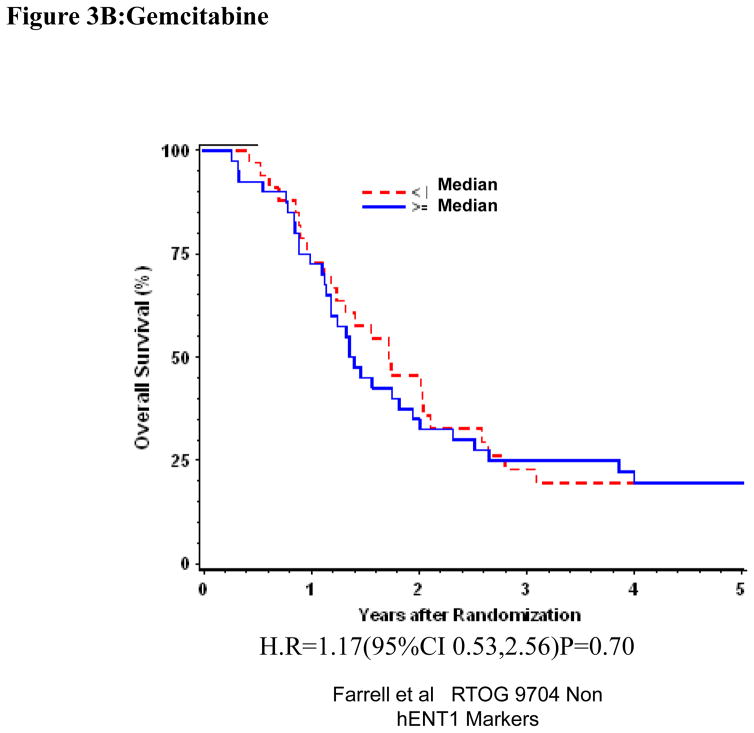
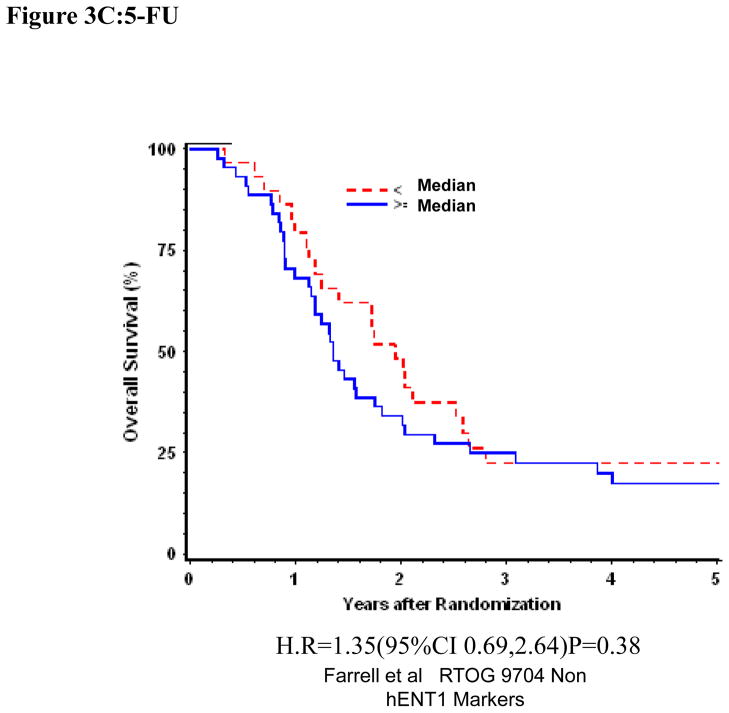
Univariate and multivariate analyses for dichotomized RRM2: “Low RRM2” vs. “High RRM2” was performed for all patients (Table 1). RRM2 expression was independently and significantly associated with DFS despite adjusting for baseline characteristics in these multivariate models for the overall treatment group (H.R. = 1.62 (95% CI: 1.01, 2.60) p=0.045)(Table 1, Figure 3A)). We observed prolonged disease-free survival which approached statistical significance for patients with “Low RRM2” as compared to patients with “High RRM2” for all patients treated with gemcitabine (H.R. = 2.24 (95% CI: 1.08,4.68) p=0.031)(Table 1, Figure 3B). No association between RRM2 protein expression and OS was seen in the gemcitabine treated group. In the 5-fluorouracil treatment arm, RRM2 expression was not significantly associated with OS or DFS after adjusting for baseline characteristics in these multivariate models. (Table 1, Figure 3C)
Figure 3.
Overall survival for RRM2 protein expression, multivariate analysis. A. All patients. B Gemcitabine Treatment Arm. C 5-FU treatment arm
P53R2 Protein Expression
Of the 229 patient tumor samples evaluated, 163 had analyzable p53R2 immunostaining. There were no positive statistical associations between baseline characteristics and missing and determined p53R2 protein expression (Supplementary Table 8 and Supplementary Table 9). For cytoplasm mean, the median is 1.3 (min-max: 0.00–2.00) to dichotomize the results in to “Low p53R2” and “High p53R2”. “High p53R2” expression was seen in 44 of 74 patients in the gemcitabine treatment arm, and 55 of 89 patients randomized to the 5-FU treatment arm.
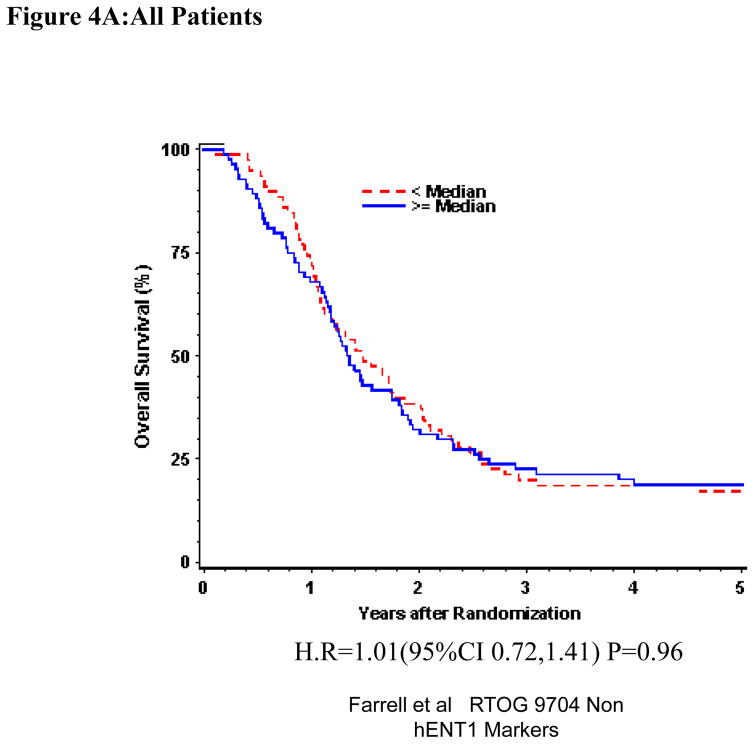
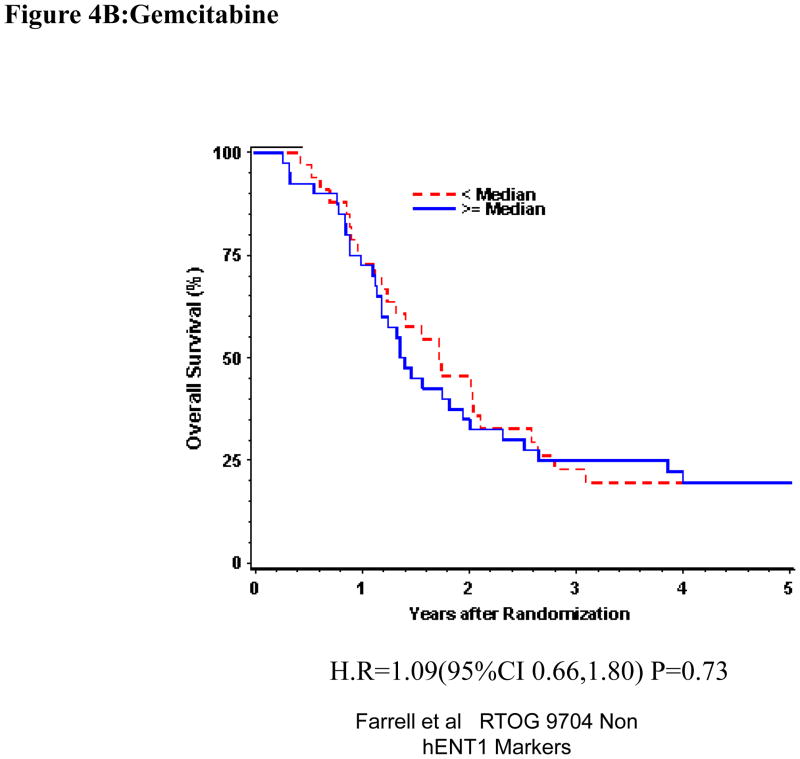
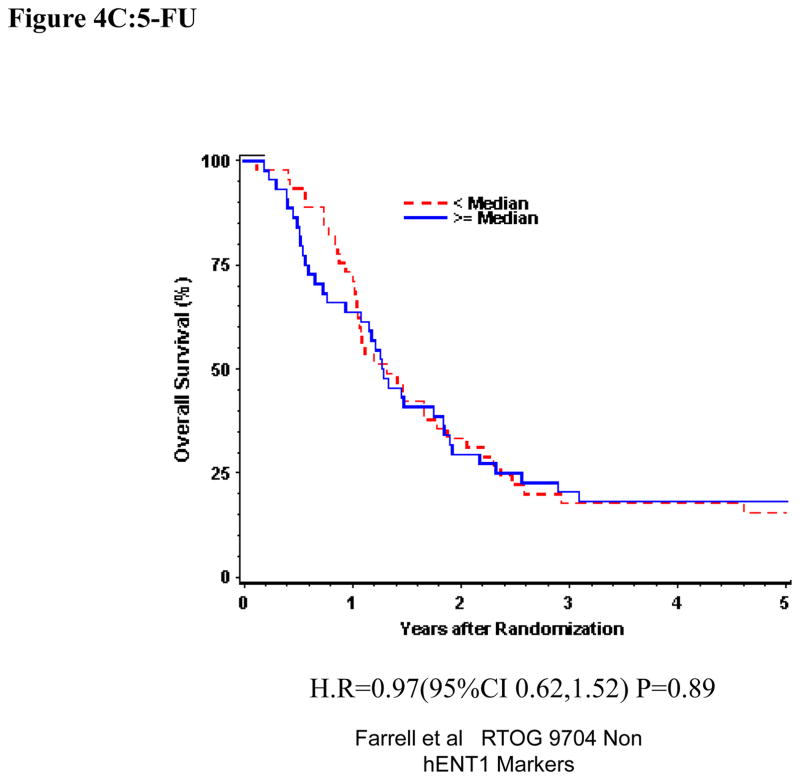
Univariate and multivariate analyses for dichotomized p53R2: “Low p53R2” vs. “High p53R2” was performed for all patients. P53R2 (Table 1) expression was not significantly associated with OS or DFS despite adjusting for baseline characteristics in these multivariate models for the overall treatment group (OS: H.R. 1.07, p =0.71, DFS (H.R. 1.01, p=0.97)) (Table 1, Figure 4A). Again, p53R2 proteins expression was not significantly associated with OS or DFS among those patients randomized to either the gemcitabine treatment arm (OS: H.R.=1.36, p=0.25, DFS: H.R.=1.62, p=0.06) or the 5-FU treatment arm (OS: H.R.=0.98, p=0.92, DFS: H.R.= 0.76 p=0.23 on multivariate analysis (Table 1 Figure 4B,C).
Figure 4.
Overall survival for p53R2 protein expression, multivariate analysis. A. All patients. B Gemcitabine Treatment Arm. C 5-FU treatment arm
DISCUSSION
Several lessons were learned from the initial RTOG 9704 study of hENT1 which supported its role as a promising predictive marker of treatment response for precision medicine in pancreatic cancer, and its subsequent validation in other studies.(5) Firstly, the importance of stage of disease and treatment setting is important when discussing the predictive value of a possible marker. The studies supporting hENT1 are all in the adjuvant setting, (6–13) with no strong evidence to support its role in the advanced or metastatic stage,(12, 29) and unclear significance in the neoadjuvant setting.(30, 31). In addition, the methodology for quantitating the marker is important. It is important to know whether protein or RNA is being measured; what type of antibody is being used; whether a tissue micro-array to increase tumor representation is employed, and the marker scoring system used including marker location (e.g. cytoplasm vs. nuclear location) and appropriate cut-offs. For example, if mRNA is being measured, then microdissection of tumor is important.(32, 33) Finally the availability of well characterized prospectively treated patients with both gemcitabine and non-gemcitabine treatment arms is important to formally assess the predictive value of a marker, rather than just the prognostic value.(5, 11, 12) Hence this study aimed to validate other non-hENT1 gemcitabine related markers using the RTOG 9704 tissue in the adjuvant setting and using previously standardized IHC methodology.
DCK is the enzyme responsible for the rate limiting step which converts the prodrug gemcitabine to its active mono phosphorylated form.(34) One study correlated high DCK mRNA levels with prolonged disease free survival in patients with pancreas cancer receiving palliative gemcitabine(33), and another showing no correlation at all.(32) Our data does not support the predictive role for tumoral cytoplasmic DCK protein to determine outcome response to gemcitabine in the adjuvant setting in pancreatic cancer. This is in contradistinction to other IHC studies which have suggested DCK as a gemcitabine predictive marker (Table 2). (7, 14, 15). While these studies used a similar antibody and DCK cytoplasmic scoring system, two are small retrospective studies which did not use TMA analysis or included heterogeneous treatment groups.(14, 15) The more recent large study in the adjuvant setting is complicated by its retrospective tissue and data collection methodology, and the absence of a randomized non-gemcitabine treatment arm.(7) Interestingly, our current study did appear to support an association between high levels of DCK and overall survival for the entire study group (both treatment arms), likely due to the effect seen in the 5-FU treatment arm. There is some preclinical cell data supporting the association between high DCK levels and 5-FU sensitivity but very little is known about the precise mechanism.(35) Similarly cell data has shown that RRM1 significantly contributes to the induction of DNA damage by 5-FU possibly related to RAD51 focus formation.(36) It is possible that it reflects an increased sensitivity of proliferating cells to 5-FU with dCK and possibly RRM1 being key enzymes of DNA synthesis
Table 2.
DCK, RRM1 and RRM2 Expression by Immunohistochemistry in Pancreatic Cancer
| DCK Protein Expression | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Author | Year | Gemcitabine | Non-Gemcitabine | Antibody | IHC | Scores Used | TMA | Treatment Design | O.S. association with DCK |
| Farrell | Current | 88 | 98 | Rabbit Polyclonal | Semiquant | Cyto only | Yes | Prospective Randomized | No |
| Sebastiani | 2006 | 44 | 0 | Rabbit Polyclonal | Semiquant | No | Retrospective Non Randomized | Yes/p=0.04 (UniVar) | |
| Marechal | 2010 | 45 | 0 | Rabbit Polyclonal | Semiquant | Cyto only | Yes | Prospective Non Randomized | Yes/p=0.002 |
| Marechal | 2012 | 243 | 49 | Rabbit Polyclonal | Semiquant | Cyto only | Yes | Retrospective Non Randomized | Yes/p=0.012 |
| RRM1 Protein Expression | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Author | Year | Gemcitabine | Non-Gemcitabine | Antibody | IHC | Scores Used | TMA | Treatment Design | O.S. association with RRM1 |
| Farrell | Current | 92 | 104 | Rabbit Polyclonal | Quantitative | Nuclear | Yes | Prospective Randomized | No |
| Fisher | 2012 | 74(33)* | 0 | Rabbit Polyclonal | Semiquant | Cyto and Nuclear | No | Retrospective Non Randomized | No |
| Marechal | 2012 | 243 | 49 | Rabbit Polyclonal | Semiquant | Nuclear | Yes | Retrospective Non Randomized | No |
| Valsecchi | 2012 | 82 | 0 | Rabbit Polyclonal | Semiquant | Cytoplasmic | No | Retrospective Non Randomized | No |
| Nakagawa | 2012 | 109 | 0 | Rabbit Polyclonal | Semiquant | Nuclear | Yes | Retrospective Non Randomized | Yes/p=0.019 |
| Akita | 2009 | 28 | 40 | Rabbit Polyclonal | Quantitative | Nuclear | Yes | Retrospective Non Randomized | Yes/p=0.0010 |
| RRM2 Protein Expression | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Author | Year | Gemcitabine | Non-Gemcitabine | Antibody | IHC | Scores Used | TMA | Treatment Design | O.S. association with RRM1 |
| Farrell | Current | 88 | 101 | Rabbit Polyclonal | Semiquant | Cytoplasm | Yes | Prospective Randomized | No |
| Xie | 2012 | 44 | 0 | Goat polyclonal | Semiquant | Nuclear | Yes | Retrospective Non Randomized | No |
| Fisher | 2012 | 74(33)* | 0 | Mouse monoclonal | Semiquant | Cyto and Nuclear | No | Retrospective Non-Randomized | Yes/p=0.02 |
33 of the reported 74 patients definitely received gemcitabine
IHC, immunohistochemistry
Semiquant, semiquantitative
Cyto, cytoplasm
TMA, tissue microarray
O.S., overall survival
RRM1, a multimeric enzyme that converts ribonucleotides to deoxyribonucleosides has been reported as a predictive marker of gemcitabine treatment response in pancreatic cancer in several preclinical studies..(37, 38). Several very small clinical studies in pancreatic cancer have demonstrated an association between low RRM1 expression (as determined by quantitative reverse transcription PCR,(37, 39) or immunohistochemistry.(16)) In the study showing an association with survival in a gemcitabine treated cohort and low RRM1 protein expression, only nuclear expression was measured and the association was not seen for patients with high expression of RRM1. Similar to our larger study, there are also several other large RNA based and immunohistochemistry studies which do not demonstrate the predictive value of RRM1 (Table 2) (7, 17, 18, 32, 40). Two studies which have demonstrated the predictive value of combining low RRM1 and high hENT1 to predict improved survival with or without the expression of other markers such as RRM2 and dCK. (9, 33).
We demonstrated an association between high tumor RRM2 protein expression and decreased disease free survival in patients with pancreatic cancer receiving adjuvant therapy in a prospective randomized trial. This effect was not seen in patients treated with 5-FU alone, suggesting that tumor RRM2 protein levels may function as a predictive marker of response to treatment with gemcitabine. This is in keeping with previous non-pancreas cancer data which associated high levels of RRM2 expression (either by protein or gene expression) with gemcitabine chemoresistance and worse survival.(27, 41) The importance the ribonucleotidase reductase RRM2 enzyme in cancer prognosis and as a potential predictive marker for response to gemcitabine treatment, has been reported in several preclinical cancer models including lung cancer (25, 42–44) and pancreatic cancer.(32, 34, 37, 45–47) However, the other clinical data supporting the role for RRM2 as a predictive marker of gemcitabine response is conflicting. Lower pre-treatment RRM2 mRNA expression of endoscopic ultrasound-fine needle aspiration biopsy specimens from patients with pancreatic cancer was associated with improved median survival and overall response rate after gemcitabine treatment..(45) However, in a larger study of both RRM1 and RRM2 gene expression in laser microdissected pancreatic surgical or biopsy specimens, there was no association between either gene expression and pancreatic cancer disease survival after treatment with gemcitabine.(32)A recent non-TMA retrospective study RRM2 protein expression study in the adjuvant setting with pancreatic cancer, supports the concept of high RRM2 expression corresponding with worse overall and disease free survival in the overall patient group as well as the 74 patients receiving adjuvant therapy (Table 2).
The p53-inducible p53R2 gene also plays an important role in DNA repair and synthesis after DNA damage.(48).(49, 50) Expression of p53R2 has been variably correlated with overall prognosis in patients with esophageal squamous cell carcinoma, early stage non-small cell lung cancer and colon cancer.(27, 41) To date there has been no study of p53R2 in pancreatic cancer, either as a prognostic or predictive biomarkers. This current study does not favor p53R2 to be either prognostic or predictive of treatment response in the adjuvant pancreatic cancer setting.
In conclusion, our current study does not support the predictive roles of DCK, RRM1, or p53R2 in the precision medicine guided treatment of pancreatic cancer with gemcitabine in the adjuvant setting, but does support the possible predictive value of RRM2. While our study did not combine multiple markers as has been done with other studies, it is unlikely to have proven of benefit in this particular study. For now, efforts should focus on hENT1 as a valid predictive marker of treatment response of gemcitabine in pancreas cancer.
Supplementary Material
Footnotes
Conflict of Interest: The authors have no conflict of interest to declare.
Contribution of Authors
James J. Farrell, MD, Conception, Data Collection, Data Analysis, Writing, Approval
Jennifer Moughan, MS, Conception, Data Collection, Data Analysis, Writing, Approval
Jonathan L. Wong, BS, Conception, Data Collection, Data Analysis, Writing, Approval
William F. Regine, MD, FACR, FACRO, Conception, Data Collection, Approval
Paul Schaefer MD, Data Collection, Approval
Al B. Benson III, MD FACP FASCO, Data Collection, Approval
John S. Macdonald, MD, Data Collection, Approval
Xiyong Liu, MD, PhD, Data Collection, Approval
Yun Yen, MD, PhD, Data Collection, Approval
Raymond Lai, MD, PhD, Data Collection, Approval
Zhong Zheng, MD, PhD, Data Collection, Approval
Gerold Bepler MD, PhD, Data Collection, Approval
Chandan Guha, MD, PhD, Data Collection, Approval
Hany Elsaleh, MD, PhD, Conception, Data Collection, Data Analysis, Writing Approval
Contributor Information
James J. Farrell, Yale Center for Pancreatic Disease, Yale School of Medicine, New Haven, CT.
Jennifer Moughan, Statistics and Data Management Center, NRG Oncology, Philadelphia, PA.
Jonathan L. Wong, Department of Medicine, John A Burns School of Medicine, University of Hawaii, Honolulu, HI.
William F. Regine, Department of Radiation Oncology, University of Maryland, Baltimore, MD.
Paul Schaefer, Oncology Program, Toledo Clinic, Toledo, OH.
Al B. Benson, III, Department of Medicine, Northwestern University Feinberg School of Medicine, Chicago, Il.
John S. Macdonald, Saint Vincent Comprehensive Cancer Center, New York, NY.
Xiyong Liu, California Cancer Institute, Temple City, CA.
Yun Yen, Graduate Institute of Medical Sciences, College of Medicine, Taipei Medical University, Taipei, Taiwan. Program for Cancer Biology and Drug Discovery, College of Medical Science and Technology, Taipei Medical University, Taipei, Taiwan.
Raymond Lai, Department of Pathology, Cross Cancer Center, University of Alberta, Edmonton, Canada.
Zhong Zheng, Department of Pathology, H. Lee Moffitt Cancer Center, Tampa, FL.
Gerold Bepler, Karmanos Cancer Institute, Detroit, MI. Department of Oncology and Cancer Biology Graduate Program, Wayne State University, Detroit, MI.
Chandan Guha, Department of Radiation Oncology, Montifiore Medical Center, Bronx, NY.
Hany Elsaleh, Department of Radiation Oncology, The Canberra Hospital, Australian National University, Canberra, Australia.
References
- 1.Conroy T, Desseigne F, Ychou M, et al. FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N Engl J Med. 2011;364:1817–1825. doi: 10.1056/NEJMoa1011923. [DOI] [PubMed] [Google Scholar]
- 2.Von Hoff DD, Ervin T, Arena FP, et al. Increased survival in pancreatic cancer with nab-paclitaxel plus gemcitabine. N Engl J Med. 2013;369:1691–1703. doi: 10.1056/NEJMoa1304369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moore MJ, Goldstein D, Hamm J, et al. Erlotinib plus gemcitabine compared with gemcitabine alone in patients with advanced pancreatic cancer: a phase III trial of the National Cancer Institute of Canada Clinical Trials Group. J Clin Oncol. 2007;25:1960–1966. doi: 10.1200/JCO.2006.07.9525. [DOI] [PubMed] [Google Scholar]
- 4.Regine WF, Winter KA, Abrams RA, et al. Fluorouracil vs gemcitabine chemotherapy before and after fluorouracil-based chemoradiation following resection of pancreatic adenocarcinoma: a randomized controlled trial. JAMA. 2008;299:1019–1026. doi: 10.1001/jama.299.9.1019. [DOI] [PubMed] [Google Scholar]
- 5.Farrell JJ, Elsaleh H, Garcia M, et al. Human equilibrative nucleoside transporter 1 levels predict response to gemcitabine in patients with pancreatic cancer. Gastroenterology. 2009;136:187–195. doi: 10.1053/j.gastro.2008.09.067. [DOI] [PubMed] [Google Scholar]
- 6.Spratlin J, Sangha R, Glubrecht D, et al. The absence of human equilibrative nucleoside transporter 1 is associated with reduced survival in patients with gemcitabine-treated pancreas adenocarcinoma. Clin Cancer Res. 2004;10:6956–6961. doi: 10.1158/1078-0432.CCR-04-0224. [DOI] [PubMed] [Google Scholar]
- 7.Marechal R, Bachet JB, Mackey JR, et al. Levels of gemcitabine transport and metabolism proteins predict survival times of patients treated with gemcitabine for pancreatic adenocarcinoma. Gastroenterology. 2012;143:664–674. e1–6. doi: 10.1053/j.gastro.2012.06.006. [DOI] [PubMed] [Google Scholar]
- 8.Morinaga S, Nakamura Y, Watanabe T, et al. Immunohistochemical analysis of human equilibrative nucleoside transporter-1 (hENT1) predicts survival in resected pancreatic cancer patients treated with adjuvant gemcitabine monotherapy. Ann Surg Oncol. 2012;19:S558–564. doi: 10.1245/s10434-011-2054-z. [DOI] [PubMed] [Google Scholar]
- 9.Nakagawa N, Murakami Y, Uemura K, et al. Combined analysis of intratumoral human equilibrative nucleoside transporter 1 (hENT1) and ribonucleotide reductase regulatory subunit M1 (RRM1) expression is a powerful predictor of survival in patients with pancreatic carcinoma treated with adjuvant gemcitabine-based chemotherapy after operative resection. Surgery. 2013;153:565–575. doi: 10.1016/j.surg.2012.10.010. [DOI] [PubMed] [Google Scholar]
- 10.Kondo N, Murakami Y, Uemura K, et al. Combined analysis of dihydropyrimidine dehydrogenase and human equilibrative nucleoside transporter 1 expression predicts survival of pancreatic carcinoma patients treated with adjuvant gemcitabine plus S-1 chemotherapy after surgical resection. Ann Surg Oncol. 2012;19:S646–55. doi: 10.1245/s10434-011-2140-2. [DOI] [PubMed] [Google Scholar]
- 11.Greenhalf W, Ghaneh P, Neoptolemos JP, et al. Pancreatic cancer hENT1 expression and survival from gemcitabine in patients from the ESPAC-3 trial. J Natl Cancer Inst. 2014;106:djt347. doi: 10.1093/jnci/djt347. [DOI] [PubMed] [Google Scholar]
- 12.Poplin E, Wasan H, Rolfe L, et al. Randomized, multicenter, phase II study of CO-101 versus gemcitabine in patients with metastatic pancreatic ductal adenocarcinoma: including a prospective evaluation of the role of hENT1 in gemcitabine or CO-101 sensitivity. J Clin Oncol. 2013;31:4453–4461. doi: 10.1200/JCO.2013.51.0826. [DOI] [PubMed] [Google Scholar]
- 13.Xiao JC, Zhang TP, Zhao YP. Human equilibrative nucleoside transporter 1 (hENT1) predicts the Asian patient response to gemcitabine-based chemotherapy in pancreatic cancer. Hepatogastroenterology. 2013;60:258–262. doi: 10.5754/hge12687. [DOI] [PubMed] [Google Scholar]
- 14.Sebastiani V, Ricci F, Rubio-Viqueira B, et al. Immunohistochemical and genetic evaluation of deoxycytidine kinase in pancreatic cancer: relationship to molecular mechanisms of gemcitabine resistance and survival. Clin Cancer Res. 2006;12:2492–2497. doi: 10.1158/1078-0432.CCR-05-2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marechal R, Mackey JR, Lai R, et al. Deoxycitidine kinase is associated with prolonged survival after adjuvant gemcitabine for resected pancreatic adenocarcinoma. Cancer. 2010;116:5200–5206. doi: 10.1002/cncr.25303. [DOI] [PubMed] [Google Scholar]
- 16.Akita H, Zheng Z, Takeda Y, et al. Significance of RRM1 and ERCC1 expression in resectable pancreatic adenocarcinoma. Oncogene. 2009;28:2903–2909. doi: 10.1038/onc.2009.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fisher SB, Patel SH, Bagci P, et al. An analysis of human equilibrative nucleoside transporter-1, ribonucleoside reductase subunit M1, ribonucleoside reductase subunit M2, and excision repair cross-complementing gene-1 expression in patients with resected pancreas adenocarcinoma: implications for adjuvant treatment. Cancer. 2013;119:445–453. doi: 10.1002/cncr.27619. [DOI] [PubMed] [Google Scholar]
- 18.Valsecchi ME, Holdbrook T, Leiby BE, et al. Is there a role for the quantification of RRM1 and ERCC1 expression in pancreatic ductal adenocarcinoma? BMC Cancer. 2012;12:104. doi: 10.1186/1471-2407-12-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xie H, Lin J, Thomas DG, et al. Ribonucleotide reductase M2 does not predict survival in patients with resectable pancreatic adenocarcinoma. Int J Clin Exp Pathol. 2012;5:347–355. [PMC free article] [PubMed] [Google Scholar]
- 20.Ko AH, Tempero MA. Personalized medicine for pancreatic cancer: a step in the right direction. Gastroenterology. 2009;136:43–45. doi: 10.1053/j.gastro.2008.11.027. [DOI] [PubMed] [Google Scholar]
- 21.Plunkett W, Huang P, Searcy CE, et al. Gemcitabine: preclinical pharmacology and mechanisms of action. Semin Oncol. 1996;23:3–15. [PubMed] [Google Scholar]
- 22.Lin ZP, Belcourt MF, Cory JG, et al. Stable suppression of the R2 subunit of ribonucleotide reductase by R2-targeted short interference RNA sensitizes p53(−/−) HCT-116 colon cancer cells to DNA-damaging agents and ribonucleotide reductase inhibitors. J Biol Chem. 2004;279:27030–27038. doi: 10.1074/jbc.M402056200. [DOI] [PubMed] [Google Scholar]
- 23.Regine WF, Abrams RA. Adjuvant therapy for pancreatic cancer: current status, future directions. Semin Oncol. 2006;33:S10–13. doi: 10.1053/j.seminoncol.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 24.Hatzis P, Al-Madhoon AS, Jullig M, et al. The intracellular localization of deoxycytidine kinase. J Biol Chem. 1998;273:30239–30243. doi: 10.1074/jbc.273.46.30239. [DOI] [PubMed] [Google Scholar]
- 25.Zheng Z, Chen T, Li X, et al. DNA synthesis and repair genes RRM1 and ERCC1 in lung cancer. N Engl J Med. 2007;356:800–808. doi: 10.1056/NEJMoa065411. [DOI] [PubMed] [Google Scholar]
- 26.Liu X, Zhang H, Lai L, et al. Ribonucleotide reductase small subunit M2 serves as a prognostic biomarker and predicts poor survival of colorectal cancers. Clin Sci (Lond) 2013;124:567–578. doi: 10.1042/CS20120240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu X, Lai L, Wang X, et al. Ribonucleotide reductase small subunit M2B prognoses better survival in colorectal cancer. Cancer Res. 2011;71:3202–3213. doi: 10.1158/0008-5472.CAN-11-0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu X, Zhou B, Xue L, et al. Metastasis-suppressing potential of ribonucleotide reductase small subunit p53R2 in human cancer cells. Clin Cancer Res. 2006;12:6337–6344. doi: 10.1158/1078-0432.CCR-06-0799. [DOI] [PubMed] [Google Scholar]
- 29.Eto K, Kawakami H, Kuwatani M, et al. Human equilibrative nucleoside transporter 1 and Notch3 can predict gemcitabine effects in patients with unresectable pancreatic cancer. Br J Cancer. 2013;108:1488–1494. doi: 10.1038/bjc.2013.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murata Y, Hamada T, Kishiwada M, et al. Human equilibrative nucleoside transporter 1 expression is a strong independent prognostic factor in UICC T3-T4 pancreatic cancer patients treated with preoperative gemcitabine-based chemoradiotherapy. J Hepatobiliary Pancreat Sci. 2012;19:413–425. doi: 10.1007/s00534-011-0440-3. [DOI] [PubMed] [Google Scholar]
- 31.Kawada N, Uehara H, Katayama K, et al. Human equilibrative nucleoside transporter 1 level does not predict prognosis in pancreatic cancer patients treated with neoadjuvant chemoradiation including gemcitabine. J Hepatobiliary Pancreat Sci. 2012;19:717–722. doi: 10.1007/s00534-012-0514-x. [DOI] [PubMed] [Google Scholar]
- 32.Giovannetti E, Del Tacca M, Mey V, et al. Transcription analysis of human equilibrative nucleoside transporter-1 predicts survival in pancreas cancer patients treated with gemcitabine. Cancer Res. 2006;66:3928–3935. doi: 10.1158/0008-5472.CAN-05-4203. [DOI] [PubMed] [Google Scholar]
- 33.Fujita H, Ohuchida K, Mizumoto K, et al. Gene expression levels as predictive markers of outcome in pancreatic cancer after gemcitabine-based adjuvant chemotherapy. Neoplasia. 2010;12:807–817. doi: 10.1593/neo.10458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakano Y, Tanno S, Koizumi K, et al. Gemcitabine chemoresistance and molecular markers associated with gemcitabine transport and metabolism in human pancreatic cancer cells. Br J Cancer. 2007;96:457–463. doi: 10.1038/sj.bjc.6603559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bergman AM, Giaccone G, van Moorsel CJ, et al. Cross-resistance in the 2′,2′-difluorodeoxycytidine (gemcitabine)-resistant human ovarian cancer cell line AG6000 to standard and investigational drugs. Eur J Cancer. 2000;36:1974–1983. doi: 10.1016/s0959-8049(00)00246-x. [DOI] [PubMed] [Google Scholar]
- 36.Aoki Y, Sakogawa K, Hihara J, et al. Involvement of ribonucleotide reductase-M1 in 5-fluorouracilinduced DNA damage in esophageal cancer cell lines. Int J Oncol. 2013;42:1951–1960. doi: 10.3892/ijo.2013.1899. [DOI] [PubMed] [Google Scholar]
- 37.Nakahira S, Nakamori S, Tsujie M, et al. Involvement of ribonucleotide reductase M1 subunit overexpression in gemcitabine resistance of human pancreatic cancer. Int J Cancer. 2007;120:1355–1363. doi: 10.1002/ijc.22390. [DOI] [PubMed] [Google Scholar]
- 38.Kurata N, Fujita H, Ohuchida K, et al. Predicting the chemosensitivity of pancreatic cancer cells by quantifying the expression levels of genes associated with the metabolism of gemcitabine and 5-fluorouracil. Int J Oncol. 2011;39:473–482. doi: 10.3892/ijo.2011.1058. [DOI] [PubMed] [Google Scholar]
- 39.Xie H, Jiang W, Jiang J, et al. Predictive and prognostic roles of ribonucleotide reductase M1 in resectable pancreatic adenocarcinoma. Cancer. 2013;119:173–181. doi: 10.1002/cncr.27715. [DOI] [PubMed] [Google Scholar]
- 40.Kim R, Tan A, Lai KK, et al. Prognostic roles of human equilibrative transporter 1 (hENT-1) and ribonucleoside reductase subunit M1 (RRM1) in resected pancreatic cancer. Cancer. 2011;117:3126–3134. doi: 10.1002/cncr.25883. [DOI] [PubMed] [Google Scholar]
- 41.Hsu NY, Wu JY, Liu X, et al. Expression status of ribonucleotide reductase small subunits hRRM2/p53R2 as prognostic biomarkers in stage I and II non-small cell lung cancer. Anticancer Res. 2011;31:3475–3481. [PubMed] [Google Scholar]
- 42.Ceppi P, Volante M, Novello S, et al. ERCC1 and RRM1 gene expressions but not EGFR are predictive of shorter survival in advanced non-small-cell lung cancer treated with cisplatin and gemcitabine. Ann Oncol. 2006;17:1818–1825. doi: 10.1093/annonc/mdl300. [DOI] [PubMed] [Google Scholar]
- 43.Shibata S, Chow W, Frankel P, et al. A phase I study of oxaliplatin in combination with gemcitabine: correlation of clinical outcome with gene expression. Cancer Chemother Pharmacol. 2007;59:549–557. doi: 10.1007/s00280-006-0297-3. [DOI] [PubMed] [Google Scholar]
- 44.Bepler G, Kusmartseva I, Sharma S, et al. RRM1 modulated in vitro and in vivo efficacy of gemcitabine and platinum in non-small-cell lung cancer. J Clin Oncol. 2006;24:4731–4737. doi: 10.1200/JCO.2006.06.1101. [DOI] [PubMed] [Google Scholar]
- 45.Itoi T, Sofuni A, Fukushima N, et al. Ribonucleotide reductase subunit M2 mRNA expression in pretreatment biopsies obtained from unresectable pancreatic carcinomas. J Gastroenterol. 2007;42:389–394. doi: 10.1007/s00535-007-2017-0. [DOI] [PubMed] [Google Scholar]
- 46.Duxbury MS, Ito H, Zinner MJ, et al. Inhibition of SRC tyrosine kinase impairs inherent and acquired gemcitabine resistance in human pancreatic adenocarcinoma cells. Clin Cancer Res. 2004;10:2307–2318. doi: 10.1158/1078-0432.ccr-1183-3. [DOI] [PubMed] [Google Scholar]
- 47.Duxbury MS, Whang EE. RRM2 induces NF-kappaB-dependent MMP-9 activation and enhances cellular invasiveness. Biochem Biophys Res Commun. 2007;354:190–196. doi: 10.1016/j.bbrc.2006.12.177. [DOI] [PubMed] [Google Scholar]
- 48.Tanaka H, Arakawa H, Yamaguchi T, et al. A ribonucleotide reductase gene involved in a p53-dependent cell-cycle checkpoint for DNA damage. Nature. 2000;404:42–49. doi: 10.1038/35003506. [DOI] [PubMed] [Google Scholar]
- 49.Xue L, Zhou B, Liu X, et al. Wild-type p53 regulates human ribonucleotide reductase by protein-protein interaction with p53R2 as well as hRRM2 subunits. Cancer Res. 2003;63:980–986. [PubMed] [Google Scholar]
- 50.Okumura H, Natsugoe S, Yokomakura N, et al. Expression of p53R2 is related to prognosis in patients with esophageal squamous cell carcinoma. Clin Cancer Res. 2006;12:3740–3745. doi: 10.1158/1078-0432.CCR-05-2416. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.