Abstract
Nutrient restriction, also known as caloric restriction, has been extensively examined for its positive impact on lifespan, immune system boost, and aging. In addition, nutrient restriction is implicated in decreasing cancer initiation and progression. Given the phenotypic changes associated with nutrient restriction, we hypothesized significant protein expression alterations must be associated with caloric restriction. To compare the molecular and phenotypic changes caused by glucose restriction and fetal bovine serum (FBS) restriction there is need for an efficient model system. We establish three dimensional cell culture models, known as spheroids, in the HCT 116 colorectal cancer cell line as a high throughput model for studying the proteomic changes associated with nutrient restriction. Flow cytometry was used to assess apoptosis and autophagy levels in the spheroids under nutrient restriction. Isobaric tags for relative and absolute quantification (iTRAQ) and liquid chromatography tandem mass spectrometry were used to determine differential protein abundances between the nutrient restriction conditions. We identified specific proteins that have implications in cancer progression and metastasis that are differentially regulated by restriction of either glucose or serum. These proteins include the up regulation of sirtuin 1 (SIRT1) and protein inhibitor of activated STAT 1 (PIAS1) and down regulation of multi-drug resistance protein (MRP1) and Zinc finger and BTB domain-containing protein 7A (ZBTB7). The results indicate nutrient restriction causes lower apoptotic and higher autophagy rates in HCT 116 spheroids. In addition, proteins shown to be differentially regulated by both glucose and serum restriction were similarly regulated.
Keywords: Nutrient restriction, three dimensional cell culture, autophagy, apoptosis, quantitative proteomics, colorectal cancer
Graphical Abstract
1. Introduction
Dietary intervention has long been studied for health benefits [1]. Nutrient restriction (NR) is a form of dietary intervention defined as providing essential nutrients but restricting total caloric intake. There are several different forms of NR including limiting glucose, amino acids, or growth factors. NR has been shown to lengthen lifespan in mice, chimpanzees and humans [2]. NR can also act as a potent protector against cancer [1–3]. Data in recent years has shown that different forms of NR in cancer patients in conjunction with traditional chemotherapy has the ability to increase malignant cell death and decrease the common side effects of chemotherapy treatment [4]. It has been shown that there is a decrease in the incidence of cancer associated with NR, but the molecular mechanisms by which NR decreases tumorigenesis and metastasis in pronounced tumors is not fully understood [5].
NR causes metabolic stress which activates both cell death and cell survival pathways in cancer cells. Metabolic stress can directly lead to cell death via apoptosis and necrosis, but the same stress signals can also lead to autophagy which can paradoxically cause cell death or permit cell survival [6]. In autophagy, a double-membraned organelle, called an autophagosome, forms around nonspecific bulk proteins that are then degraded through subsequent fusion to the lysosome [7],[8].The amino acids that are produced from autophagy can be utilized by cells during an energy crisis to produce necessary proteins for survival [9]. This process allows nutrient scavenging during periods of metabolic stress and can be a means by which cancer cells survive in low nutrient conditions, thus promoting tumorigenesis [10–12]. Conversely, persistent autophagy can eventually lead to caspase-independent cell death, known as type II programmed cell death, and cancer cells can utilize autophagy in this fashion to inhibit tumorigenesis [9],,[13],[14]. The role of autophagy in cancer is context dependent and may vary depending on the stage of a cancer [14]. With a better understanding of the global protein changes that are involved with metabolic stress, specifically NR, we may be able to unravel the complexity of the role of autophagy in cancer.
In order to study the global protein changes occurring as a result of NR, we needed to establish a model system. Previous research using rhesus monkeys and rodent models have provided insight into the molecular effects of NR, however replicates in these model systems are limited [15]. In addition, the effects of NR may be more profound in short-lived species, like mice, where major nutrient loss is catastrophic as compared to longer-lived humans who can adapt to major food loss. Thus, the response of NR in mice may differ from humans. Cell cultures offer many positive attributes, including rapid growth time and larger numbers of replicates than animal studies. For these reasons, human cell lines are an attractive model system to study the proteomic changes associated with NR.
Human cell lines can be grown as two dimensional (2D) cultures or, in some cases, in more complicated three dimensional (3D) assemblies. 2D cell cultures, which are typically grown in monolayers, are simple to cultivate. However, 2D cell cultures lacks complex cell organization, morphology, and the molecular signaling that is present in vivo [16]. To study the protein abundance changes of NR in cancer, an alternative model system that contains complexities absent in 2D cell cultures is advantageous. Three dimensional (3D) cell cultures, known as spheroids, are an attractive alternative. Spheroids are tumor mimics that are more complex than 2D cell culture, while also more feasible with higher throughput than animal models [17],[18],[16]. We have previously demonstrated that spheroids have a pathophysiological gradient of cellular viability resulting from chemical gradients of nutrients and oxygen. Both the physiological and chemical gradients observed in spheroids mimic ones observed in solid tumors in vivo [19]. The colon carcinoma cell line HCT 116 grown in spheroids form a tumor-like pathophysiological gradient after 13 days of in vitro growth. 3D cell culture allows for multiple variables to be tested in a high throughput manner.
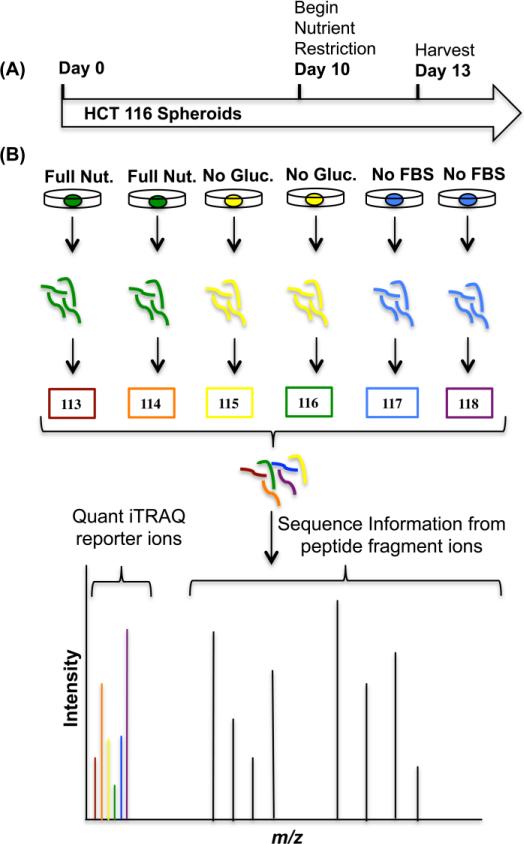
In order to establish 3D cell culture as an efficient model system to study NR, we compared two types of NR, glucose or fetal bovine serum (FBS) restriction, for 72 hours prior to harvesting proteins (Figure 1A). In case studies of NR as an adjuvant cancer treatment in humans, fasting was implemented for 48-140 hours prior to or following chemotherapy treatment. All types of fasting regimes show a decrease in chemotherapy side effects as reported by patients [20]. In addition, fasting has also been shown to increase the efficacy of chemotherapy drugs in 2D cell culture, after NR of 24 hours [21].
Figure 1.
A) Treatment plan for the HCT 116 spheroids to include ten days of normal growth, followed by 72 hours of nutrient restriction prior to harvesting protein at day 13. B) iTRAQ experimental design. Spheroids are grown, in biological duplicate (25 spheroids for each sample), subject to either no glucose or no FBS, homogenized, and harvested for protein. Subsequent proteins are digested into tryptic peptides. The primary amines of the peptides are labeled with six unique isobaric tags with specific reporter ion m/z values. Labeled digests are mixed into one sample. The combined sample is then analyzed using LC-MS/MS to quantitate and identify proteins in different biological samples.
There are different types of NR. For example, reducing glucose levels will directly affect cancer cells, who rely on glucose for survival, and has been a proposed adjuvant cancer treatment [21]. Most cancer cells rely on the breakdown of glucose to generate energy, a process that is known as the Warburg effect, a hallmark of cancer [22],[23]. Alternatively, the reduction of growth factors will directly affect the mTOR (mammalian Target of Rapamycin) cascade, which is the major regulator of coordinating nutrient availability and energy response to growth factors [24], [25]. In the scientific community, there is a lack of consensus on which type of diet is best when NR is used for cancer prevention [4]. Therefore, elucidating the proteomic changes resulting from glucose or serum restriction will help determine which form of NR may benefit cancer treatment.
In this study, we evaluated the phenotypic changes of NR with flow cytometry based apoptotic and autophagic assays. Autophagy is known to occur in the center of solid tumors and will be prevalent in HCT 116 spheroids where the center of the spheroid has decreased oxygen and nutrient availability [6]. Our hypothesis is that NR will increase the amount of autophagy in HCT 116 spheroids due to metabolic stress and change the amount of apoptotic cells.
Proteomics holds promise for new discoveries related to NR as an adjuvant cancer treatment [26]. The molecular mechanisms that underlie the anti-cancer effect of NR are poorly understood. Understanding global proteomic changes due to NR can help unravel these molecular mechanisms and be used to identify potential drug targets. Previous studies have examined the mRNA expression levels but not the protein levels of NR [27]. Mass spectrometry is a powerful tool for quantifying proteomic changes in different biological conditions [27],[28]. To evaluate the proteome, we used a bottom-up approach to isolate tryptic peptides, then separated the peptides by liquid chromatography and detected the peptides with electrospray-ionization (ESI) tandem mass spectrometry [29], [30]. For quantification, we used isobaric tags for relative and absolute quantification (iTRAQ) which allows identification of peptide fragments at the same time as protein quantification with reporter ions in the low mass range of the mass spectrum (113-119 m/z) (Figure 1B) [31]. We hypothesize that protein abundance will be differentially expressed when HCT 116 spheroids are subject to NR. With this work, we identify the proteomic changes that accompany NR in 3D cell culture.
2. Methods and materials
2.1. Cell Culture Spheroid Growth and Treatment
The human colon carcinoma cell line HCT 116 was obtained from American Type Culture Collection (ATCC)a and grown in RPMI 1640 cell culture medium (Life Technologies)b supplemented with 10% fetal bovine serum (FBS) (Thermo Scientific)c. The provider assured authentication of the cell line by cytogenetic analysis. Spheroid cell culture was carried out using agarose-coated 96- well plates by seeding 7000 cells into the middle of each well as previously described.[32] Half the volume of the culture medium was changed every 48 h after 4 days in culture. For spheroids starved of glucose or FBS, on day 10 of culture, media was changed to RPMI 1640 containing either no glucose (Life Technologies)b or no FBS (Thermo Scientific)c respectively as shown in Figure 1A. The spheroids were deprived of glucose or FBS for 72 hours, and harvested at day 13. Full nutrient spheroids continued to have media changed every 48 h until harvested also at day 13 with all samples. For flow cytometry controls, spheroids were treated with 2 μM rapamycin (CYTO-ID® Autophagy Detection Kit - ENZO)d and 25 μM chloroquine (CYTO-ID® Autophagy Detection Kit - ENZO) d for 24 hours prior to harvest.
2.2. Flow Cytometry Sample Preparation and Analysis
For the apoptotic assay, the FITC Annexin V Apoptosis Detection Kit with 7-AAD (BioLegend)e was used. Twelve spheroids grown for 13 days were collected and washed with 2 mL of warmed PBS (Gibco)f, and then transferred to a well of a 12 well microtiter plates (Thermo)c containing 2 mL 0.05% Trypsin (HyClone Laboratories)g. The plate was placed into the 37°C incubator for a total of 15 min, pipetting the mixture every three minutes. The trypsin was then neutralized with 3 mL of complete RPMI 1640 medium and solution was filtered through a 40 μm cell strainer (Corning Incorporated)h to a 15 mL conical vial. The well was rinsed with 1 mL of complete RPMI 1640 medium and the contents transferred to the 15 mL conical vial through the cell strainer. The vial was centrifuged at 1000 rpm for 5 min to collect single cells. Supernantant was aspirated and the cells were resuspended in 2 mL of PBS and centrifuged at 1000 rpm for 5 min. Supernantant was aspirated and cells were resuspended in 100 μL of PBS and transferred to a flow cytometry tube (VWR)i, 5 μL of Annexin V was added and left standing in the dark for 15 min at RT, 5 μL of 7-AAD was added to the tube along with 400 μL of binding buffer. Samples were kept on ice and transported to the flow cytometer for analysis on a BD LSRFortessa X-20j in the red (APC) and green (FITC) channel. Forward scatter (FS) and side scatter (SS) were used to identify single cells. The blank (unstained) sample, and single-stained samples, that is those labeled with one fluorochrome, were analyzed for adjustment of the parameters in each channel, for generating the compensation matrix, and for setting up the quadrant marker.
For the autophagy assay, the CYTO-ID® Autophagy Detection Kit (ENZO)d was used. The previous flow cytometry protocol was used, and cells were resuspended in 250 μL 1X Assay Buffer and 250 μL of the diluted CYTO-ID Green stain solution was added. The samples were incubated at RT for 45 min then centrifuged at 1000 rpm for 5 min. The supernatant was aspirated and cells were resuspended in 500 μL 1X Assay Buffer and analyzed on a BD LSRFortessa X-20 flow cytometerj in the green (FITC) channel. Two positive controls were used, Rapamycin, an inducer of autophagy, and chloroquine, an inhibitor of late phase autophagy that causes an accumulation of autophagolysosomes. HCT 116 ATG16L1 −/− spheroids were used as a negative control. ATG16L1 is a protein necessary for autophagy and the knockout cell line is incapable of autophagy. ATGL1 −/− spheroids have less autophagic vesicles than the full nutrient control samples. The blank (unstained) sample was used to gate for negative cell population in this analysis.
2.3. Sample Preparation for Proteomic Analysis
Whole spheroids (25 per biological replicate, 50 per biological condition) were homogenized and lysed in 300 μL lysis solution (8 M urea with 75 mM NaCl, 50 mM Tris-HCl (pH 8.2), 10 mM sodium pyrophosphate, 1 mM phenylmethane sulfonyl fluoride protease inhibition (PMSF), 1 mM Na3VO4, 1 mM NaF, 1 mM β-glycerophosphate, and 1 EDTA-free protease inhibitor cocktail). The solution was mechanically pipetted up and down several times. The solution was then sonicated on ice for three rounds of 30 seconds on, 30 seconds off at 15% amplification. Sample was spun down for 10 min at 10,000 × g and supernatant was removed. Protein amount was quantified using Pierce BCA Protein Assay Kit (Thermo)c. 100 μg of protein was denatured at 37 °C for 1 h, reduced with 5 mM DTT at 37 °C for 1 h, and alkylated with 14 mM iodoacetamide for 30 min at RT. The reaction was then quenched with 5 mM DTT at RT for 25 min. Proteins were diluted with 25 mM Tris-HCl (pH 8.2) and digested with trypsin in the presence of 1 mM CaCl2 overnight at 37 °C. The resulting peptides were desalted with 100 mg C18 Sep-Paks (Oasis)k. The tryptic peptides from the 6 different samples were then labeled with isobaric tags for relative and absolute quantification (iTRAQ) 8-plex reagents according to the manufacturer’s protocols (AB Sciex)l. The labeled samples were then mixed and dried down and desalted with 50 mg C18 Sep-Paks. The sample was then resuspended in 120 μL Buffer A (10 mM KH2PO4 (aq), 20% ACN, pH 2.85) for strong cation exchange (SCX) liquid chromatography fractionation.
2.4. SCX Fractionation
The labeled sample was fractionated using a Waters Alliance HPLC System SCX (Waters)k via SCX liquid chromatography. The sample (100 μL) was loaded onto an SCX guard column (2.1 mm i.d. X 50 mm length, 5 μM particles, Agilent Technologies)m. The mobile phase gradient was generated using Buffer A (10 mM KH2PO4 (aq), 20% ACN, pH 2.85) and Buffer B (1 M KCl in Buffer A, pH 2.85) at a flow rate 0.25 mL/min. The method was 60 minutes in total. The first 10 min of the method consisted of washing with 100% Buffer A to remove excess iTRAQ reagent. Following the wash, peptides were fractionated by a linear gradient from 100% Buffer A and 100% Buffer B collecting fractions every min for a total of 31 fractions collected. The column was then washed with 100% Buffer B and equilibrated with 100% Buffer A. Fractions from min 10-15 were combined as one fraction, due to low concentration of sample eluting from the column in the first five minutes of the gradient.
2.5. Peptide Desalting
The collected SCX fractions were dried down and resuspended in 30 μL of 0.1% Formic Acid (FA) solution and 20 μL were taken and desalted with a 5 μg C18 ZipTip (ZTC18S096, Millipore)n. The desalted peptides were resuspended in 5 μL of 0.1% FA, followed by analysis by ultra-performance liquid chromatography (UPLC)-ESI-MS/MS analysis.
2.6. UPLC-ESI-MS/MS analysis
Peptide separation was performed by a nanoACQUITY UltraPerformance LC® (UPLC®) system (Waters)k with mobile phases Buffer A (0.1% FA in water) and Buffer B (0.1% FA in ACN). For each sample, 2 μL of peptides were automatically loaded onto a commercial C18 reversed phase column (Waters, 100 μm × 100 mm, 1.7 μm particle, part No. 186003546, column temperature 40°C) k The LC method was 90 minutes, with the following gradient: 2% buffer B for 8 min at a flow rate of 1 μL/min, followed by 3-step gradient separation, 60 min to 30%, 75 min to 50% B, 1 min to 85% B, and maintained at 85% B for 5 min. The column was equilibrated for 10 min with 2% B.
The eluted peptides were then analyzed via electrospray ionization (ESI) by a Q-Exactive mass spectrometer (Thermo Fisher Scientific)c. The following parameters were used for the analysis: electrospray voltage was set to 2.1 kV and the ion transfer tube temperature was 280°C. The data acquisition was programmed in data dependent acquisition (DDA) mode with the S-Lens RF level was 50.00. A top 12 method was used and Full MS scans were acquired in Orbitrap mass analyzer 350–1800 m/z range with resolution of 70,000 and the number of microscans set to 1. The target value was 1.00E + 06, and maximum injection time was 250 ms. the isolation window was set as 1.0 m/z. The normalized collision energy was 31.0%, and tandem mass spectra were acquired in the Orbitrap mass analyzer with resolution 17,500 with the fixed first mass m/z 100.0. The target value was 1.00E + 06 and maximum injection time was 120 ms. The number of microscans was set to 1 and the ion selection threshold was 5.0E + 04 counts. Peptide match and exclude isotopes were turned on. Dynamic exclusion was set as 40s.
2.7. Data Analysis
Raw data files were searched with MaxQuant version 1.3.05 using the Andromeda search engine [33]. Peptides were searched against reverse sequence data to determine false discovery rate (FDR) with the FDR set to 0.01 for peptide and protein identifications. iTRAQ 8plex as the sample type, with Iodacetamide as a fixed modification, trypsin as the digestion enzyme, urea denaturation, and instrument Orbitrap MS and MS/MS were set as the parameters. Reactome was used for GO molecular pathway analysis.
2.8. Quantitative Real Time PCR
RNA was harvested from 12 HCT 116 spheroids at day 13 staying consistent with all previous experiments in biological duplicate with varying conditions: full nutrients, 72 hours of glucose deprivation, and 72 hours of FBS deprivation (Figure 1A) using the RNeasy Kit (Qiagen)o using the manufactures instructions. The RNA (40 μg) was reverse transcribed into cDNA using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems)p in a thermocycler for 30 min at 16 °C, 30 min at 42 °C, followed by 5 min at 85 °C. The ΔΔCt method was used for quantification; cDNA was prepared in triplicate using the YWHAZ gene for gene normalization and PerfeCta SYBRGreen FastMix (Quanta Biosciences)q. The quantitative Real Time PCR was performed on a StepOne-Plus (Applied Biosystems) p using the following program: samples were held at 95 °C for 2 min, 40 cycles of 95 °C for 15s and 60 °C for 60s. (Primer sequences 5’-3’: PIAS1_1: ccacaacaagtgcagcaaat, PIAS1_2:ttgtggacaactggtttctga, ARAP3_1: gagacgctttgtgcagttca, ARAP3_2:ggtcatctcaatggcagtca, ZBTB3_1: tgggagcgaaaaagatctctac, ZBTB3_2:ccatagtaccccacgaagga, SIRT1_1: acccatggaggatgaaagtg, SIRT1_2:ctccatcagtcccaaatcc, MRP1_1: cctggacaaagccagagaag, MRP1_2:tctgccattctgaaacacca, LC3b_1: gagagcagcatccaaccaa, LC3b_2:tgacatggtcaggtacaagga)
2.9 Statistical Analyses
All experiments were performed in biological and technical replicate. Data were analyzed using analysis of variance, followed by Student-Newman-Keuls test and unpaired Students t test for comparison of means to detect differences compared with control. For RT-PCR data, the expression of genes were normalized against YWHAZ and compared as a fold change relative to the basal values. Values are reported as means ± SD and differences were considered to be statistically significant at */#P < .05 and **P < .01.
3. Results
This study aims to better understand the molecular mechanisms of NR in 3D cell culture tumor mimics. This goal was achieved by evaluating the changes associated with glucose or serum (FBS) restriction in HCT 116 spheroids by quantifying the apoptotic cell population, autophagic vesicle formation, and global proteome changes.
3.1. Apoptotic and Autophagy Assays
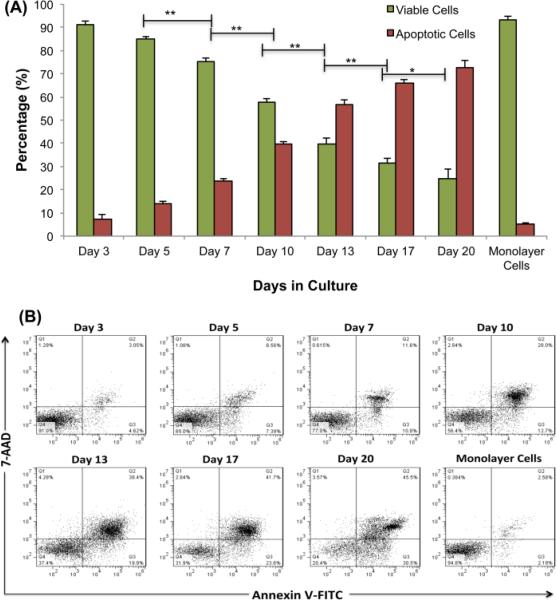
We previously reported that after 13 days of growth, spheroids have developed the pathophysiological gradients seen in solid tumors in vivo [34]. In order to establish a baseline of cell populations in full nutrient HCT 116 spheroids, we examined the apoptotic rate over 20 days of culture using flow cytometry (Figure 2). To be able to evaluate the different cell populations present, the spheroids were trypsinized to single cells before analysis. Spheroids form after three days of culture and are composed of 90% viable cells. The percentage of viable cells significantly decreases, and appropriately, the percentage of apoptotic cells significantly increases along the time course. By day 20, the spheroids consist of only 25% viable cells (Figure 2A). This increase in the number of apoptotic cells over time agrees with the previous results [19]. These results establish a baseline of apoptotic cell percentage as well as the use of 3D cell culture as an appropriate model system to study the effects of NR in cancer progression.
Figure 2.
A) and B) Time course apoptotic assay using flow cytometry. Twelve spheroids were trypsinized to single cells and stained with annexin V to identify early apoptotic cells and 7-aad to determine late apoptotic cells and quantified using the fluorescence in the red and green channel respectively. Values are reported as means ± SD of duplicates, each repeated 4 times (n = 4). Differences are judged to be significant at *P < .05, **P < .01 (Student unpaired t test) compared with respective control groups. We have labeled only statistical differences in viable cells, but when the viable cells are statistically different, the apoptotic cells are also statistically different.
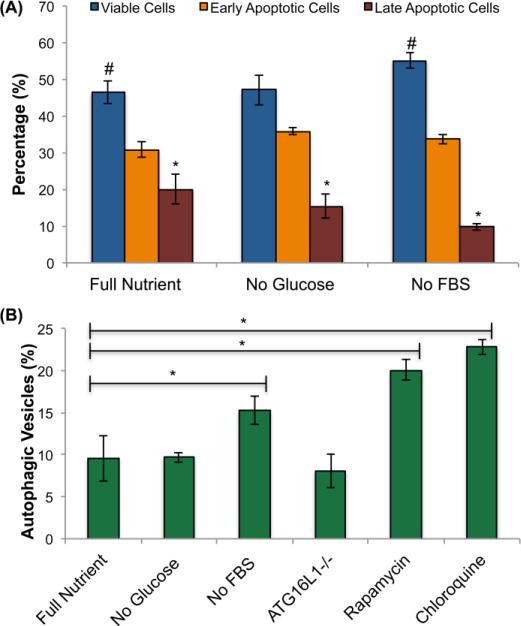
After establishing a baseline apoptotic rate in spheroids, we determined the apoptotic cell percentage as well as the rate of autophagic vesicle formation in HCT 116 spheroids subjected to NR. The spheroids were deprived of either glucose or serum (FBS) for 72 hours prior to harvest at day 13 (Figure 1A) and evaluated using flow cytometry assays for apoptosis and autophagy rates (Figure 3). Glucose deprivation causes no statistically significant change in the number of viable cells, but a statistically significant decrease in late apoptotic/necrotic cells was observed (Figure 3A). When spheroids are deprived of serum, we observe a statistically significant increase in viable cells as well as a statistically significant decrease in the number of late apoptotic/necrotic cells (Figure 3A). We find it intriguing that NR is causing a decrease in late apoptotic cells with glucose or serum restriction. The metabolic stress initiated by NR is likely activating cell survival pathways. One pathway that could be activated in response to NR is autophagy, which is why we chose to quantify autophagic vesicle formation. The deprivation of FBS shows a statistically significant increase in autophagic vesicles, while deprivation of glucose shows no significant change in autophagic vesicles (Figure 3B). From this data, we conclude that serum restriction causes a more pronounced change in apoptosis and autophagy in HCT 116 spheroids than glucose restriction.
Figure 3.
A) Apoptotic assay and B) autophagy assay using flow cytometry. A) Twelve spheroids were trypsinized to single cells and stained with annexin V to identify early apoptotic cells and 7-aad to determine late apoptotic cells and quantified using the florescence in the red and green channel respectively. Values are reported as means ± SD of duplicates, each repeated 4 times (n = 4). Differences are judged to be significant at */#P < .05, (Student unpaired t test) compared with respective control groups. B) Twelve spheroids were trypsinized to single cells and stained with CYTO-ID autophagic vesicle marker and quantified using the florescence in the green channel. Values are reported as means ± SD of duplicates, each repeated 4 times (n = 4). Differences are judged to be significant at */#P < .05, (Student unpaired t test) compared with respective control groups.
3.2. Quantitative Proteomics
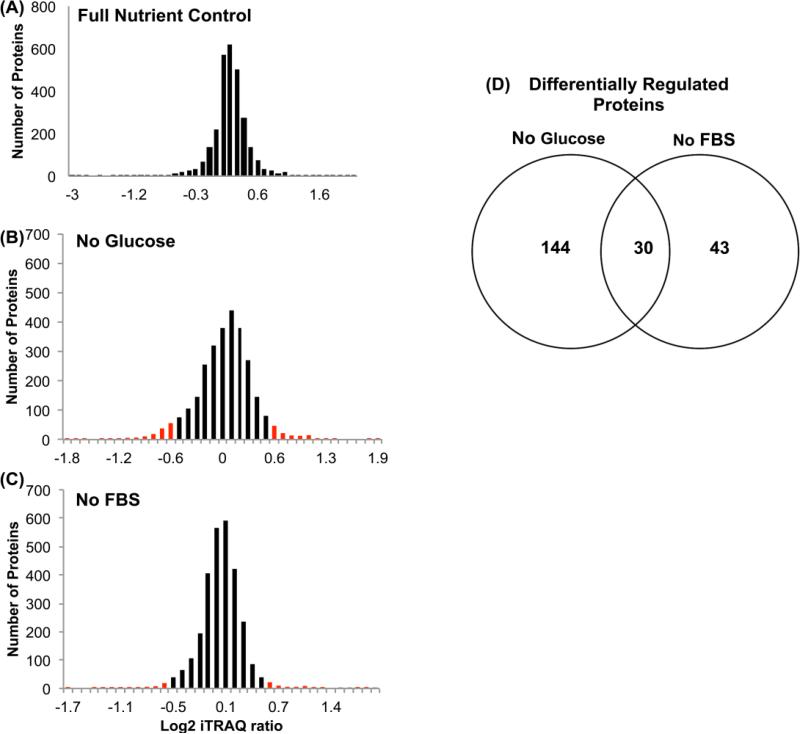
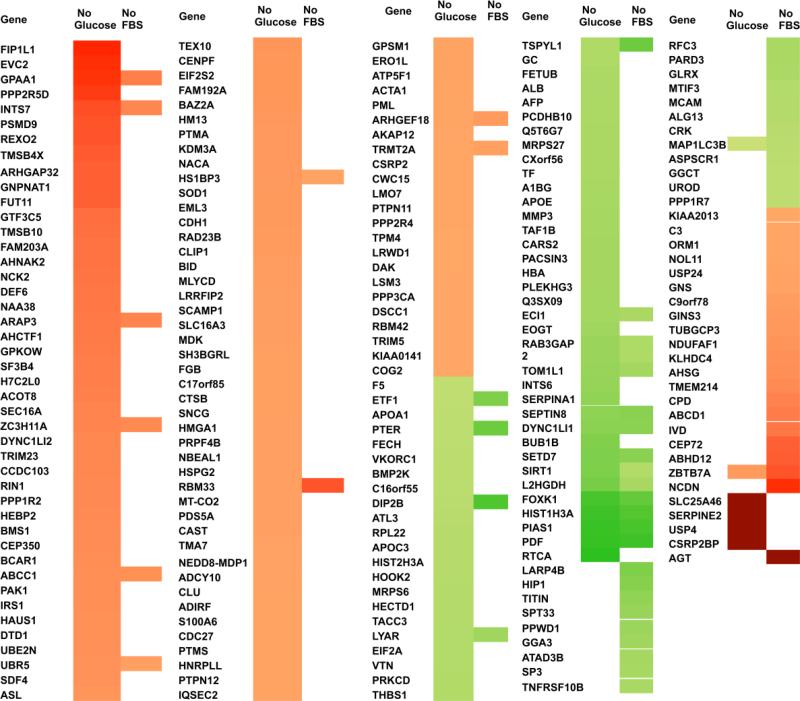
To identify and quantify the protein abundance changes due to NR, we performed iTRAQ using UPLC-MS/MS. For the quantitative proteomics study, the experimental design is depicted in Figure 2. Full nutrient, glucose deprived, and serum deprived HCT 116 spheroids were examined in biological and technical duplicate. The data was analyzed using MaxQuant software (details found in the experimental section) for a total of 2875 protein identifications (supplemental data includes a spreadsheet showing protein identification using iTRAQ UPLC-ESI-MS/MS). The full-nutrient reporter ion score was plotted on a log 2 scale histogram (Figure 4A) centered around 0. Glucose and serum deprivation reporter ions were compared to their full nutrient counterparts and plotted on a log 2 scale histogram (Figure 4B-C). Proteins found in the glucose or serum deprived data set that exceeded two standard deviations (+/− 0.678) of the full nutrient control were considered to be significantly up or down regulated. Of those proteins significantly altered, 30 proteins were found to be differentially regulated by both conditions (Figure 4D). Based on these data, glucose deprivation produces a more profound impact on protein expression, specifically causing more differentially regulated proteins while serum starvation seems to have less of an impact on global protein expression. Differentially regulated proteins are displayed in a heat map based on their log2 iTRAQ ratio value (Figure 5). From this data we concluded that the majority of proteins altered by glucose or serum deprivation are different. It is important to note that every protein altered by both conditions is altered in the same direction. Of the 30 proteins found to be up or down regulated in both NR conditions, five are closely related to pathways that are functional during cancer progression and metastasis. These five proteins include the up regulated SIRT1 and PIAS1 and the down regulated MRP1, ARAP3, and ZBTB7.
Figure 4.
(A) Distribution of iTRAQ ratios centered around 0 on a log2 scale. Differentially expressed proteins regulated (+/− 2 SD of full nutrient control mean) for no glucose and no FBS are represented in red in (B) and (C) respectively. Venn diagrams of regulated proteins in no glucose and no FBS in (D). For glucose deprivation, 175 proteins were significantly altered. For serum deprivation, 73 proteins were significantly altered, and 30 proteins were found to be differentially regulated by both conditions
Figure 5.
Log2 iTRAQ ratio of all proteins found to be differentially regulated (+/− 2 SD of full nutrient control mean) by glucose or FBS restriction represented in a heat map. NQ = Not Quantifiable, meaning that reporter ions were present in both biological replicates of full nutrient control, but not present in both biological replicates of NR. These proteins are then considered substantially down regulated due to NR but are not quantifiable.
3.3. Quantitative Real-Time PCR
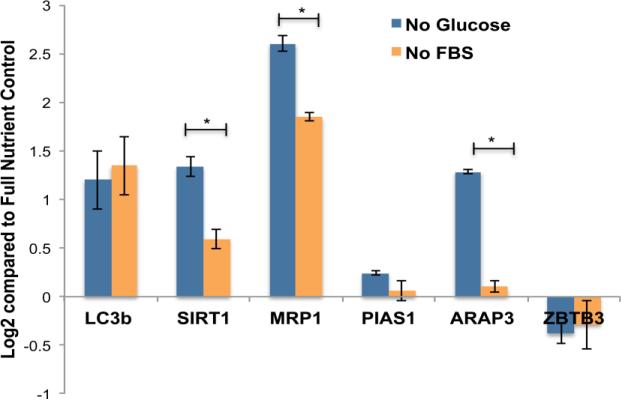
We further investigated six genes using RT-PCR to look at the changes in gene expression level due to NR (Figure 6). Five of these genes (SIRT1, PIAS1, MRP1, ARAP3, and ZBTB7) were chosen from the proteomic data because of their altered protein expression during NR. The sixth gene, LC3b, is an essential autophagy gene, whose protein product accumulates on the surface of autophagosomes. LC3b is used to indicate the levels of autophagy. As expected, the expression of LC3b was up regulated with both glucose and FBS deprivation. This data agrees with the autophagy assay and the iTRAQ data that also show an increase in LC3b expression (Figure 3b, Figure 5). In addition, the up regulation of SIRT1 and PIAS1 gene expression is in agreement with the quantitative protein expression. The down regulation of ZBTB7 gene expression also matches the protein expression data. However, MRP1 and ARAP3 show opposite gene and protein expression. Though this fact is interesting, gene expression and protein abundance often do not correlate due to regulation control at various different levels and the biochemical diversity of proteins [35].
Figure 6.
Quantitative Real Time PCR was preformed on the five genes of interest from the proteomic data (SIRT1, MRP1, PIAS1, ARAP3, ZBTB3) as well as LC3b to quantify gene expression due to nutrient restriction including glucose and FBS deprivation. Values are reported as means ± SD of duplicates, each repeated 4 times (n = 3). Differences are judged to be significant at *P < .05, **P < .01 (Student unpaired t test) compared with respective control groups.
3.4 Pathway Analysis
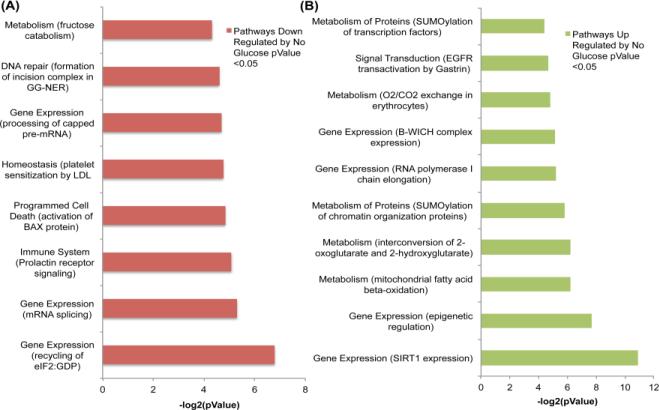
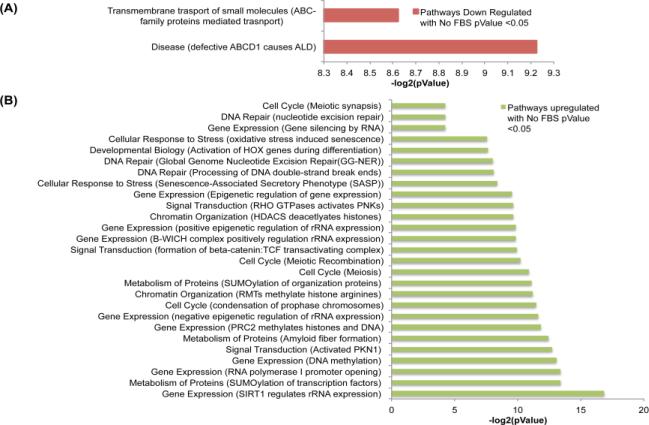
Proteins that were found to be up or down regulated following NR were analyzed using Reactome. Identified pathways with pValues < 0.05 can be found in Figure 7 and Figure 8. Of the pathways to be identified most were related to gene expression and metabolism being regulated by NR. The pathways that are regulated in common with either glucose or serum deprivation should be noted. The gene expression pathways of B-WICH complex expression, epigenetic regulation, and SIRT1 expression as well as the metabolism of SUMOylation proteins were up regulated in both glucose and serum restriction. No pathways were found to be commonly down regulated between glucose and serum restriction. This data shows that NR in HCT 116 spheroids causes protein abundance changes that result in regulation of metabolic pathways. Knowing if a certain pathway is targeted by glucose or serum deprivation in spheroids will be valuable information for future studies that will interrogate individual proteins and their role in NR for cancer treatment.
Figure 7.
Molecular Pathways up or down regulated due to treatment of no glucose for 72 hours represented in (A) and (B) respectively. Pathways were determined using Reactome.
Figure 8.
Molecular Pathways up or down regulated due to treatment of no serum for 72 hours represented in (A) and (B) respectively. Pathways were determined using Reactome.
4. Discussion
In this study, we found that NR has many effects on HCT 116 spheroids, including a decrease in late apoptotic cells, an increase in viable cells, and an increase in autophagic vesicles. In addition, either glucose or serum deprivation causes significant protein abundance changes to HCT 116 spheroids, confirming our hypothesis. We have shown that 3D cell culture is a valuable model system in which we can identify proteins that are differentially regulated as a result of NR. Proteins found to be differentially regulated by both treatments were always affected in the same way i.e. upregulated by both or down regulated by both. There are no instances of proteins being upregulated by one treatment but down regulated by the other treatment. We find this result to be very intriguing. From the data collected, we chose the following five proteins of interest. These proteins were found to be differentially regulated by both glucose and serum deprivation and have previously published implications in cancer progression and metastasis. The other 25 proteins that were found to be differentially regulated by both conditions are listed in the Figure 5 but not discussed in the manuscript due to space limitations.
SIRT1
SIRT1 is a widely studied NAD+-dependent protein deacetylase. SIRT1 has been implicated in the regulation of many physiological processes including heart disease, obesity, neurodegeneration, and cancer [36]. It has also been shown that there are benefits to the activation of SIRT1 in metabolic disease such as diabetes, heart disease, and obesity[31],[40]. SIRT1 is a member of the sirtuin family of proteins. The sirtuins are key regulators of metabolism known to be affected by NR. It has been shown that mammalian HEK293 and PC12 cells, when subjected to glucose or serum deprivation, produce an increase of SIRT1 [38]. Similarly, in our data we observe up regulation of SIRT1 when HCT 116 spheroids were subjected to either glucose or serum restriction in both the proteomic and gene expression data. This data is in agreement with other studies that do not use 3D cell culture [38], showing that using 3D cell culture to examine proteomic changes associated with NR is a reliable model system.
PIAS1
SUMOylation regulates many cellular processes, one of which is the response to cellular stress [39]. A protein is SUMOylated by covalent conjugation of small ubiquitin-like modifier (SUMO) protein, through the use of E1, E2, and E3 ligase enzymes, to lysine residues of the target protein [42],[43]. The function of PIAS1 is as an E3-type SUMO ligase. PIAS1 is up regulated when HCT116 spheroids are subject to either serum or glucose restriction. PIAS1 is also thought to be a critical regulator in the epithelial mesenchymal transition (EMT) [41]. It has been reported that the expression of PIAS1 regulates the TGFβ-induced metastasis of breast cancer with PIAS1 as a possible biomarker for metastasis in breast cancer [42]. Our data shows that either form of NR causes an up regulation of PIAS1 in the proteomic and gene expression data and SUMOylation in the gene ontology report. The up regulation of PIAS1 from NR could potentially decrease the rate of metastasis. Knockdown experiments of PIAS1 in our spheroid model will be completed in the future to further investigate the role of PIAS1.
MRP1
One of the main obstacles in cancer treatment is multidrug resistance, where cancer cells do not respond to many different cytotoxic chemotherapeutics. MRP1 (Multidrug resistance–associated protein 1) is a member of the ATP-binding cassette transmembrane transporters and causes drug resistance through its ability to pump or transport chemotherapeutic drugs from the interior of the cell outwards [43]. MRP1 is expressed in healthy tissue, but has been shown to be overexpressed in solid tumors suggesting its direct role in chemotherapy resistance by tumors in many different cancers [44]. MRP1 has been shown to be over expressed in a lung cancer cell line that was resistant to chemotherapeutics [45]. MRP1 is also suggested to be a biomarker for poor prognosis in breast cancer [46],[47]. We found that MRP1 is down regulated in the proteomic data, but up regulated in the gene expression data when HCT 116 spheroids are subject to NR. NR can cause a decrease in one of the major forms of drug resistance, making the HCT 116 spheroids more susceptible to chemotherapy. Due to the discrepancy in protein and gene expression, we plan to further investigate the relationship of NR and MRP1 expression in future studies.
ARAP3
Arf-GAP with Rho-GAP domain, ANK repeat and PH domain-containing protein 3 (ARAP3) is involved in phosphoinositide 3-kinase (PI3K) signaling pathways as a GTPase activating protein [47]. ARAP3 expression has been reported to slow cell migration, specifically, slowing peritoneal dissemination of cirrhous gastric carcinoma cells [48]. Our results show that glucose and serum deprivation causes a decrease in ARAP3 protein abundance, but an increase in gene expression was also observed. Down regulation of ARAP3 may lead to cell migration, which could promote metastasis. However as there is a difference in the gene and protein expression data, we cannot say with certainty that this would be a negative effect of NR. Since the phosphorylation state of ARAP3 is implicated in the invasiveness of cancer [48], further phosphoproteomic profiling experiments will be conducted to determine if the phosphorylation state of ARAP3 is altered during NR.
ZBTB7
ZBTB7 (zinc finger and BTB domain-containing protein 7A) is a transcriptional repressor that is linked to oncogenesis [49]. ZBTB7 has been shown to not only bind and repress the tumor suppressor protein ARF but is also associated with oncogenic transformation when overexpressed [35] [49]. In both glucose and serum restriction conditions, HCT 116 spheroids show a significant down regulation of ZBTB7 in both protein and gene expression. We conclude that NR of serum or glucose can repress the expression of a protein that has an impact on oncogenesis, and has potential as a drug target.
By employing the global proteomic methods, we were able to identify proteins regulated due to NR involved in the progression and metastasis of cancer from several metabolic pathways. Restriction of either glucose or serum causes significant abundance changes to the five proteins of interest. To determine if these five proteins have known or predicted protein-protein interactions, we performed an analysis with String (www.string-db.org). Based on the String analysis, these five proteins have no direct interaction with one another. However, all five proteins are downstream of the PI3K signaling pathway. The PI3K pathway has been extensively reviewed and is thought to be the most activated signaling pathway found in cancer, so it is not surprising that the proteins of interest are downstream of PI3K [51]. In future studies, these proteins will be further interrogated to elucidate their role with the use of NR as an adjuvant cancer treatment. The most significant finding of the quantitative proteomics portion of the study is that a protein differentially regulated by one type of NR, is never differentially regulated in the opposite direction by the other form of NR. This suggests that the type of NR is not necessarily key; rather, it is the use of NR at all in conjunction with other forms of chemotherapy treatment. It is likely that any form of NR for 72 hours will cause sufficient metabolic stress to activate stress relief pathways in HCT 116 spheroids. This metabolic stress will directly affect the mTOR cascade, which is activated by the PI3K pathway, causing differential protein expression of metabolic stress contributors as well as result in changes in apoptosis and autophagy rates.
From the data, there is evidence that serum restriction has a greater effect on apoptosis and autophagy rates, but is also accompanied by less changes in protein abundance than is observed in glucose restriction. Since we were only able to quantify a few proteins related to apoptosis and autophagy, we cannot draw broad conclusions as to whether serum restriction has greater effects on the apoptotic and autophagy pathway proteins than glucose does. Which form of NR is better for cancer treatment? The answer is likely context dependent. Based on the results of this study, one can determine if proteins in the pathways of interest were differentially regulated by glucose or FBS, and can determine if employing NR will be useful.
In conclusion, NR of serum or glucose results in significant changes of global protein expression, apoptotic cell populations, and autophagic vesicle formation in HCT 116 spheroids. One of the limitations of our study arises from our model system. While the 3D cell cultures are amenable to high-throughput research studies, they do not have the full complexity of a human tumor. Thus, they provide insight into some of the molecular pathways and processes that are altered with NR, but they cannot mimic the surrounding environment, blood supply, etc. We now have a baseline for understanding the proteomic changes resulting from NR in HCT 116 spheroids, and have shown that 3D cell culture is an efficient model system for the study of specific protein alterations associated with NR.
In the literature, there is a lack of proteomic data on the effect of food components and diet as related to cancer. It has been suggested the factors to be hindering nutritional proteomics are limited accessibility to proteomic technologies and insufficient preanalytical, sample handling, and sample processing training [52]. We believe we can make progress to the limitations by establishing 3D cell culture to study the global proteomic changes of food component restrictions. Spheroids provide the platform to identify changes in proteins that have implications in cancer, and can be used as a high throughput approach to further interrogate the molecular changes caused by NR that can decrease cancer progression.
Supplementary Material
Acknowledgment
We thank the Notre Dame Mass Spectrometry and Proteomics Facility for their assistance. In addition, we thank Katelyn Ludwig for her helpfulness in procedures and data analysis and William Andrews for artwork assistance. This work was funded by the National Institutes of Health (R01GM110406), and the National Science Foundation (CAREER Award, CHE-1351595). The Walther Cancer Foundation provided salary support for ABH.
Abbreviations
- NR
nutrient restriction
- FBS
fetal bovine serum
- iTRAQ
isobaric tags for relative and absolute quantification
- SIRT1
sirtuin 1
- PIAS1
protein inhibitor of activated STAT 1
- MRP1
multi-drug resistance protein
- ZBTB7
zinc finger and BTB domain-containing protein 7A
- 2D
two dimensional
- 3D
three dimensional
- mTOR
mammalian Target of Rapamycin
- ESI
electrospray-ionization
- PI3K
phosphoinositide 3-kinase
- SD
standard deviation
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
This material has not been presented or published elsewhere.
The authors declare no conflict of interest.
American Type Culture Collection (ATCC), P.O. Box 1549, Manassas, VA 20108 USA
Life Technologies, Thermo Fisher Scientific, 81 Wyman Street, Waltham, MA USA 02451
Thermo Scientific, 81 Wyman Street, Waltham, MA USA 02451
Enzo Life Sciences, Inc. 10 Executive Blvd, Farmingdale, NY 11735 USA
BioLegend, 9727 Pacific Heights Blvd, San Diego, CA 92121
Gibco, Thermo Fisher Scientific, 81 Wyman Street, Waltham, MA USA 02451
GE Healthcare Life Sciences, HyClone Laboratories, 925 West 1800 South, Logan, Utah 84321
Corning Incorporated, One Riverfront Plaza, Corning, NY 14831, USA
VWR International, Radnor Corporate Center, Building One, Suite 200, 100 Matsonford Road, P.O. Box 6660, Radnor, PA 19087
BD LSRFortessa X-20, BD Biosciences, 2350 Qume Drive, San Jose, CA 95131
Oasis, Waters Corporation, 34 Maple Street, Milford Massachusetts, 01757 USA
AB Sciex, 500 Old Connecticut Path, Framingham, MA 01701
Agilent Technologies, 5301 Stevens Creek Blvd, Santa Clara, CA 95051, United States
EMD Millipore, 290 Concord Road, Billerica, Massachusetts 01821 United States of America
Qiagen, 27220 Turnberry Lane, Suite 200, Valencia, CA 91355
Applied Biosystems, Thermo Fisher Scientific, 81 Wyman Street, Waltham, MA USA 02451
Quanta Biosciences, 100 Cummings Center, Suite 407J, Beverly, MA 01915
References
- 1.Tannenbaum A. The genesis and growth of tumors. II. Effects of caloric restriction per se. Cancer Res. 1942;2:460–7. [Google Scholar]
- 2.Hursting SD, Lavigne J a, Berrigan D, Perkins SN, Barrett JC. Calorie restriction, aging, and cancer prevention: mechanisms of action and applicability to humans. Annu Rev Med. 2003;54:131–52. doi: 10.1146/annurev.med.54.101601.152156. doi:10.1146/annurev.med.54.101601.152156. [DOI] [PubMed] [Google Scholar]
- 3.Longo VD, Fontana L. Calorie restriction and cancer prevention: metabolic and molecular mechanisms. Trends Pharmacol Sci. 2010;31:89–98. doi: 10.1016/j.tips.2009.11.004. doi:10.1016/j.tips.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee C, Longo VD. Fasting vs dietary restriction in cellular protection and cancer treatment: from model organisms to patients. Oncogene. 2011;30:3305–16. doi: 10.1038/onc.2011.91. doi:10.1038/onc.2011.91. [DOI] [PubMed] [Google Scholar]
- 5.Meynet O, Ricci J-E. Caloric restriction and cancer: molecular mechanisms and clinical implications. Trends Mol Med. 2014;20:419–27. doi: 10.1016/j.molmed.2014.05.001. doi:10.1016/j.molmed.2014.05.001. [DOI] [PubMed] [Google Scholar]
- 6.Amaravadi RK, Thompson CB. The Roles of Therapy-Induced Autophagy and Necrosis in Cancer T reatment. 2007;13:7271–80. doi: 10.1158/1078-0432.CCR-07-1595. doi:10.1158/1078-0432.CCR-07-1595. [DOI] [PubMed] [Google Scholar]
- 7.Florey O, Overholtzer M. Autophagy proteins in macroendocytic engulfment. Trends Cell Biol. 2012;22:374–80. doi: 10.1016/j.tcb.2012.04.005. doi:10.1016/j.tcb.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Y, Levine B. Autosis and autophagic cell death : the dark side of autophagy. 2014;22:367–76. doi: 10.1038/cdd.2014.143. doi:10.1038/cdd.2014.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lum JJ, DeBerardinis RJ, Thompson CB. Autophagy in metazoans: cell survival in the land of plenty. Nat Rev Mol Cell Biol. 2005;6:439–48. doi: 10.1038/nrm1660. doi:10.1038/nrm1660. [DOI] [PubMed] [Google Scholar]
- 10.Nixon RA, Wegiel J, Kumar A, Yu WH, Peterhoff C, Cataldo A, et al. Extensive Involvement of Autophagy in Alzheimer Disease: An Immuno-Electron Microscopy Study. J Neuropathol Exp Neurol. 2005;64:113–22. doi: 10.1093/jnen/64.2.113. doi:10.1093/jnen/64.2.113. [DOI] [PubMed] [Google Scholar]
- 11.Mizushima N, Yoshimori T, Ohsumi Y. The Role of Atg Proteins in Autophagosome Formation. 2011 doi: 10.1146/annurev-cellbio-092910-154005. doi:10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- 12.Rosenfeldt MT, Ryan KM. The role of autophagy in tumour development and cancer therapy. Expert Rev Mol Med. 2009;11:e36. doi: 10.1017/S1462399409001306. doi:10.1017/S1462399409001306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu L, Alva A, Su H, Dutt P, Freundt E, Welsh S, et al. Regulation of an ATG7-beclin 1 program of autophagic cell death by caspase-8. Science (80− ) 2004;304:1500–2. doi: 10.1126/science.1096645. doi:10.1126/science.1096645\r1096645 [pii] [DOI] [PubMed] [Google Scholar]
- 14.White E. the role for autophagy in cancer (White, 2015).pdf. 2015;125:42–6. doi: 10.1172/JCI73941. doi:10.1172/JCI73941.Autophagy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ingram DK, Zhu M, Mamczarz J, Zou S, Lane MA, Roth GS, et al. Calorie restriction mimetics: An emerging research field. Aging Cell. 2006;5:97–108. doi: 10.1111/j.1474-9726.2006.00202.x. doi:10.1111/j.1474-9726.2006.00202.x. [DOI] [PubMed] [Google Scholar]
- 16.Yamada KM, Cukierman E. Modeling Tissue Morphogenesis and Cancer in 3D. Cell. 2007;130:601–10. doi: 10.1016/j.cell.2007.08.006. doi:10.1016/j.cell.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 17.Sutherland RM. Microregions : The Spheroid Model. Science (80− ) 1988;240:177–240. doi: 10.1126/science.2451290. [DOI] [PubMed] [Google Scholar]
- 18.Pampaloni F, Reynaud EG, Stelzer EHK. The third dimension bridges the gap between cell culture and live tissue. Nat Rev Mol Cell Biol. 2007;8:839–45. doi: 10.1038/nrm2236. doi:10.1038/nrm2236. [DOI] [PubMed] [Google Scholar]
- 19.Liu X, Weaver EM, Hummon AB, Dame N, Hall NS, Dame N, et al. Evaluation of Therapeutics in Three-Dimensional Cell Culture Systems by MALDI Imaging Mass Spectrometry. 2013 doi: 10.1021/ac400519c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Safdie FM, Dorff T, Quinn D, Fontana L, Wei M, Lee C, et al. Fasting and cancer treatment in humans: A case series report. Aging (Albany NY) 2009;1:988–1007. doi: 10.18632/aging.100114. doi:.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raffaghello L, Lee C, Safdie FM, Wei M, Madia F, Bianchi G, et al. Starvation-dependent differential stress resistance protects normal but not cancer cells against high-dose chemotherapy. Proc Natl Acad Sci. 2008;105:8215–20. doi: 10.1073/pnas.0708100105. doi:10.1073/pnas.0708100105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heiden MG, Vander Cantley LC, Thompson CB, Mammalian P, Exhibit C, Metabolism A. Understanding the Warburg Effect : Cell Proliferation. Science (80-) 2009;324:1029–34. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klement RJ, Kämmerer U. Is there a role for carbohydrate restriction in the treatment and prevention of cancer? Nutr Metab (Lond) 2011;8:75. doi: 10.1186/1743-7075-8-75. doi:10.1186/1743-7075-8-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhuang Y, Chan DK, Haugrud AB, Miskimins WK. Mechanisms by which low glucose enhances the cytotoxicity of metformin to cancer cells both in vitro and in vivo. PLoS One. 2014:9. doi: 10.1371/journal.pone.0108444. doi:10.1371/journal.pone.0108444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Panieri E, Toietta G, Mele M, Labate V, Ranieri SC, Fusco S, et al. Nutrient withdrawal rescues growth factor-deprived cells from mTOR-dependent damage. Aging (Albany NY) 2010;2:487–503. doi: 10.18632/aging.100183. doi:100183 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang J, Li D, Dangott LJ, Wu G. Recent Advances in Nutritional Sciences Proteomics and Its Role in. J Chromatogr. 2006:1759–62. [Google Scholar]
- 27.Olivo-Marston SE, Hursting SD, Perkins SN, Schetter A, Khan M, Croce C, et al. Effects of calorie restriction and diet-induced obesity on murine colon carcinogenesis, growth and inflammatory factors, and MicroRNA expression. PLoS One. 2014;9:1–11. doi: 10.1371/journal.pone.0094765. doi:10.1371/journal.pone.0094765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ong S-E, Mann M. Mass spectrometry-based proteomics turns quantitative. Nat Chem Biol. 2005;1:252–62. doi: 10.1038/nchembio736. doi:10.1038/nchembio736. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Y, Fonslow BR, Shan B, Baek MC, Yates JR. Protein analysis by shotgun/bottom-up proteomics. Chem Rev. 2013;113:2343–94. doi: 10.1021/cr3003533. doi:10.1021/cr3003533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun L, Bertke MM, Champion MM, Zhu G, Huber PW, Dovichi NJ. Quantitative proteomics of Xenopus laevis embryos: expression kinetics of nearly 4000 proteins during early development. Sci Rep. 2014;4:4365. doi: 10.1038/srep04365. doi:10.1038/srep04365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Philip L, Ross‡ YNH. Multiplexed Protein Quantitation in\nSaccharomyces cerevisiae Using\nAmine-reactive Isobaric Tagging Reagents*. Mol Cell Proteomics. 2004;12:3, 1154–69. doi: 10.1074/mcp.M400129-MCP200. doi:10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- 32.Li H, Hummon AB. Imaging Mass Spectrometry of Three-Dimensional Cell Culture Systems. 2011:8794–801. doi: 10.1021/ac202356g. [DOI] [PubMed] [Google Scholar]
- 33.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–72. doi: 10.1038/nbt.1511. doi:10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 34.Liu X, Hummon AB. Mass Spectrometry Imaging of Therapeutics from Animal Models to Three-Dimensional Cell Cultures. Anal Chem. 2015;87:9508–19. doi: 10.1021/acs.analchem.5b00419. doi:10.1021/acs.analchem.5b00419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gygi SP, Rochon Y, Franza BR, Aebersold R. Correlation between protein and mRNA abundance in yeast. Mol Cell Biol. 1999;19:1720–30. doi: 10.1128/mcb.19.3.1720. doi:10.1128/MCB.19.3.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin Z, Fang D. The Roles of SIRT1 in Cancer. Genes Cancer. 2013;4:97–104. doi: 10.1177/1947601912475079. doi:10.1177/1947601912475079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brooks CL, Gu W. How does SIRT1 affect metabolism, senescence and cancer? Nat Rev Cancer. 2009;9:123–8. doi: 10.1038/nrc2562. doi:10.1038/nrc2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kanfi Y, Peshti V, Gozlan YM, Rathaus M, Gil R, Cohen HY. Regulation of SIRT1 protein levels by nutrient availability. FEBS Lett. 2008;582:2417–23. doi: 10.1016/j.febslet.2008.06.005. doi:10.1016/j.febslet.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 39.Rabellino A, Carter B, Konstantinidou G, Wu SY, Rimessi A, Byers LA, et al. The SUMO E3-ligase PIAS1 regulates the tumor suppressor PML and its oncogenic counterpart PML-RARA. Cancer Res. 2012;72:2275–84. doi: 10.1158/0008-5472.CAN-11-3159. doi:10.1158/0008-5472.CAN-11-3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Toropainen S, Malinen M, Kaikkonen S, Rytinki M, Jääskeläinen T, Sahu B, et al. SUMO ligase PIAS1 functions as a target gene selective androgen receptor coregulator on prostate cancer cell chromatin. Nucleic Acids Res. 2015;43:848–61. doi: 10.1093/nar/gku1375. doi:10.1093/nar/gku1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Netherton SJ, Bonni S. Suppression of TGFβ-induced epithelial-mesenchymal transition like phenotype by a PIAS1 regulated sumoylation pathway in NMuMG epithelial cells. PLoS One. 2010:5. doi: 10.1371/journal.pone.0013971. doi:10.1371/journal.pone.0013971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dadakhujaev S, Salazar-arcila C, Netherton SJ, Chandhoke AS, Singla AK, Jirik FR, et al. A novel role for the SUMO E3 ligase PIAS1 in cancer metastasis ABSTRACT : Oncoscience. 2014;1:229–40. doi: 10.18632/oncoscience.27. doi:10.18632/oncoscience.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Persidis A. Cancer multidrug resistance. Nat Biotechnol. 1999;17:94–5. doi: 10.1038/5289. doi:10.1038/80051. [DOI] [PubMed] [Google Scholar]
- 44.Munoz M, Henderson M, Haber M, Norris M. Role of the MRP1/ABCC1 multidrug transporter protein in cancer. IUBMB Life. 2007;59:752–7. doi: 10.1080/15216540701736285. doi:10.1080/15216540701736285. [DOI] [PubMed] [Google Scholar]
- 45.Cole S, Bhardwaj G, Gerlach J. Overexpression of a transporter gene in a multidrug-resistant human lung cancer cell line. Science (80− ) 1992;258:1650–4. doi: 10.1126/science.1360704. [DOI] [PubMed] [Google Scholar]
- 46.Nooter K, Brutel de la Riviere G, Look MP, van Wingerden KE, Henzen-Logmans SC, Scheper RJ, et al. The prognostic significance of expression of the multidrug resistance-associated protein (MRP) in primary breast cancer. Br J Cancer. 1997;76:486–93. doi: 10.1038/bjc.1997.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stacey TTI, Nie Z, Stewart A, Najdovska M, Hall NE, He H, et al. ARAP3 is transiently tyrosine phosphorylated in cells attaching to fibronectin and inhibits cell spreading in a RhoGAP-dependent manner. J Cell Sci. 2004;117:6071–84. doi: 10.1242/jcs.01526. doi:10.1242/jcs.01526. [DOI] [PubMed] [Google Scholar]
- 48.Yagi R, Tanaka M, Sasaki K, Kamata R, Nakanishi Y, Kanai Y, et al. ARAP3 inhibits peritoneal dissemination of scirrhous gastric carcinoma cells by regulating cell adhesion and invasion. Oncogene. 2011;30:1413–21. doi: 10.1038/onc.2010.522. doi:10.1038/onc.2010.522. [DOI] [PubMed] [Google Scholar]
- 49.Maeda T, Hobbs RM, Merghoub T, Guernah I, Zelent A, Cordon-Cardo C, et al. Role of the proto-oncogene Pokemon in cellular transformation and ARF repression. Nature. 2005;433:278–85. doi: 10.1038/nature03203. doi:10.1038/nature03203. [DOI] [PubMed] [Google Scholar]
- 50.Lim J-H. Zinc finger and BTB domain-containing protein 3 is essential for the growth of cancer cells. BMB Rep. 2014;47:405–10. doi: 10.5483/BMBRep.2014.47.7.075. doi:10.5483/BMBRep.2014.47.7.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu P, Cheng H, Roberts TM, Zhao JJ. Targeting the phosphoinositide 3-kinase (PI3K) pathway in cancer. Nat Rev Drug Discov. 2009;8:627–44. doi: 10.1038/nrd2926. doi:10.1038/nrd2926.Targeting. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Romagnolo DF, Milner JA. Opportunities and Challenges for. Jounal Nutr. 2012;142:225–9. doi: 10.3945/jn.111.151803. doi:10.3945/jn.111.151803.1360S. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.