Summary
Transfer RNA (tRNA) is a central component of protein synthesis and cell signaling network. One salient feature of tRNA is its heavily modified status, which can critically impact its function. Here we show that mammalian ALKBH1 is a tRNA demethylase. It mediates the demethylation of N1-methyladenosine (m1A) in tRNAs. The ALKBH1-catalyzed demethylation of the target tRNAs results in attenuated translation initiation and their decreased usage in protein synthesis. This process is dynamic and responds to glucose availability to affect translation. Our results uncover reversible methylation of tRNA as a new regulatory mechanism of post-transcriptional gene expression.
In brief
Reversible tRNA methylation helps translation respond to nutrient availability.
Introduction
In contrast to epigenetic regulations of DNA and histones, which have been intensively studied (Jenuwein and Allis, 2001; Klose and Zhang, 2007; Kohli and Zhang, 2013; Strahl and Allis, 2000; Suzuki and Bird, 2008), only recently have researchers begun to appreciate the importance of reversible RNA modifications in regulating gene expression (Fu et al., 2014; He, 2010). The discoveries of the “writer”, “reader”, and “eraser” proteins of the N6-methyladenosine (m6A) of messenger RNAs (mRNAs) and certain long noncoding RNAs (lncRNAs) have opened a new realm of dynamic regulation of gene expression at the post-transcriptional level (Dominissini et al., 2012; Jia et al., 2011; Liu et al., 2014; Ping et al., 2014; Wang et al., 2014; Wang et al., 2015; Zheng et al., 2013). Defects of the m6A methyltransferase complex result in a battery of development problems in various eukaryotic species (Yue et al., 2015). The YTH domain proteins, the m6A readers, influence the stability, translation, and splicing of the m6A-containing mRNAs (Wang et al., 2014; Wang et al., 2015; Xiao et al., 2016; Xu et al., 2014; Zhou et al., 2015).
Two RNA m6A demethylases, FTO and ALKBH5, have been identified since 2011, with both capable of removing the methyl group of m6A on poly-adenylated RNA through an iron-dependent oxidative demethylation mechanism (Fu et al., 2013; Jia et al., 2011; Zheng et al., 2013). These two demethylases have been shown to impact human body weight (Dina et al., 2007; Do et al., 2008; Frayling et al., 2007) and mouse fertility (Zheng et al., 2013), respectively. Thus far, m6A is the only documented reversible RNA modification that impacts the metabolism and fate of RNA (Dominissini et al., 2012; Liu et al., 2015; Meyer et al., 2012; Wang et al., 2014; Wang et al., 2015), although new dynamic marks have recently been identified on mRNA (Dominissini et al., 2016; Li et al., 2016).
All the aforementioned studies reveal the prominent importance of the dynamic mRNA modifications in affecting gene expression. tRNAs are known to be heavily modified (Phizicky and Hopper, 2010; Suzuki and Suzuki, 2014; Yacoubi et al., 2012). Many tRNA modifications affect cell viability, fitness, development, and a range of other cellular functions (Anderson et al., 1998; Begley et al., 2007; Chan et al., 2012; Kaiser et al., 2014; Saikia et al., 2010; Songe-Møller et al., 2010). However, a reversible tRNA modification process that could noticeably affect gene expression has yet to be shown.
FTO and ALKBH5 are RNA demethylases that belong to the human AlkB family dioxygenases that utilize a nonheme iron(II) to catalyze biological oxidation (Falnes et al., 2002; Gerken et al., 2007; Loenarz and Schofield, 2008; Trewick et al., 2002). Almost all of the AlkB family members with known biological functions work on nucleic acid substrates (Fedeles et al., 2015). Human ALKBH1 is another member of the AlkB family proteins. The deletion of Alkbh1 in mice could lead to embryonic lethality with the survived mice exhibiting neural development defects and sex-ratio distortion (Nordstrand et al., 2010; Ougland et al., 2012; Pan et al., 2008). The exact molecular function of ALKBH1 remains elusive although DNA has been suggested as a potential substrate (Muller et al., 2010; Westbye et al., 2008).
Here, we present ALKBH1 as a RNA demethylase that mediates removal of the methyl group from N1-methyladenosine (m1A) in tRNA. We further show that ALKBH1 can tune translation initiation and elongation by regulating the cellular levels of tRNAiMet and the association of other ALKBH1 target tRNAs to polysomes, thereby affecting protein synthesis. This discovery reveals a new pathway and mechanism of translation control through reversible tRNA methylation.
RESULT
ALKBH1 Binds tRNA Inside Cells
We utilized crosslinking and immunoprecipitation (CLIP) (Ule et al., 2005) followed by gel analysis to probe and identify potential RNA species bound by ALKBH1 in HeLa cells. To our surprise, CLIP identified mature tRNAs as main RNA species bound by ALKBH1 (Figure 1A). We subsequently performed high-throughput sequencing of the RNA products obtained from CLIP. Although reverse transcription and amplification of tRNAs are known to be significantly inhibited because of their heavily modified nature as compared to other RNA species (Cozen et al., 2015; Mohr et al., 2013), replicate experiments consistently showed that tRNAs are enriched in the ALKBH1-based CLIP products compared to the total RNA control using small RNA library preparation (Figure S1A and S1B). The composition of these tRNA species bound by ALKBH1 is quite different from that of cellular tRNAs in HeLa cells (Figure 1B, 1C). Further analysis of all tRNAs enriched in the CLIP products of ALKBH1 indicates that all of them contain the m1A58 modification (Machnicka et al., 2013).
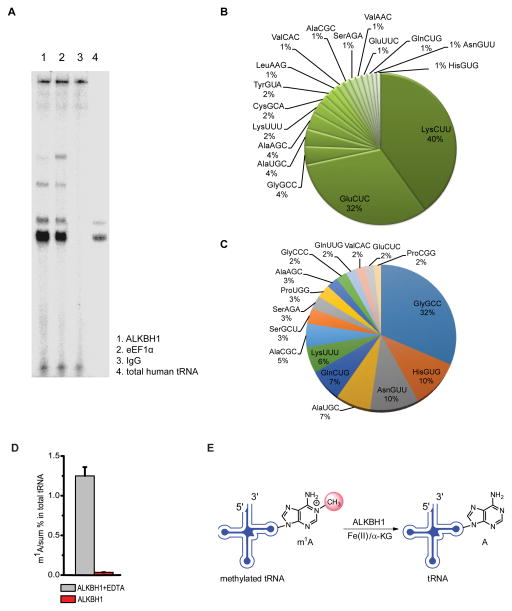
Figure 1. Human ALKBH1 Binds tRNA and Catalyzes Oxidative Demethylation of m1A in tRNA.
(A) TBE-Urea gel of ALKBH1-bound RNA species. The image shows RNA species immunoprecipitated by ALKBH1 (lane 1), eEF1α (lane 2), and IgG (lane 3) with purified total human tRNA (lane 4) used as a size marker for mature tRNAs; eEF1α was used as a known tRNA-binding protein control and IgG as a negative control. The majority of RNAs crosslinked to ALKBH1 are intact mature tRNAs.
(B) Pie chart showing ALKBH1-bound tRNAs identified by CLIP-seq.
(C) Pie chart showing tRNAs identified in the total RNA control.
(D) Demethylation of m1A in total tRNA isolated from HeLa cells by recombinant ALKBH1 in vitro. Ratios of m1A versus the sum of all four bases are shown. Error bars represent mean ± s.d., n = 8 (four biological replicates × two technical replicates).
(E) Proposed oxidative demethylation of m1A at the 58 position in tRNA by ALKBH1. See also Figure S1.
Intrigued by this observation, we performed protein sequence alignments of ALKBH1 with known tRNA-binding motifs. Indeed, we found that ALKBH1 possesses a tRNA-binding domain that is used by eukaryotic tRNA ligases to recognize tRNAs (Figure S1C) (Ghosh and Vishveshwara, 2007; Nakanishi et al., 2005).
The tRNA m1A Demethylation by ALKBH1
The CLIP results suggested that tRNAs are potential substrates of ALKBH1. We thus tested whether ALKBH1 exhibits biochemical activity on tRNA by treating the purified total tRNA from HeLa cells with recombinant ALKBH1 protein. The levels of known tRNA methylations including m1A, m7G, m5C, and m3C, were measured with ALKBH1 treatment in the presence or absence of EDTA (EDTA chelates iron and inhibits the demethylation activity) using liquid chromatography-tandem mass spectrometry (LC-MS/MS, Figure S1D) (Jia et al., 2011; Zheng et al., 2013). Incubation of total tRNA with ALKBH1 led to a dramatic decrease of the m1A level, but not levels of m7G, m5C, or m3C, in comparison to the control samples (Figure 1D and Figure S2A), suggesting that m1A58 in tRNAs is a major substrate of ALKBH1 (Figure 1E); m1A58 has been shown to be the predominant m1A modifications in tRNAs as revealed by recent m1A sequencing experiments (Cozen et al., 2015; Tserovski et al., 2016).
To analyze the observed tRNA demethylation, ALKBH1 was transiently knocked down or overexpressed in HeLa cells, and the level of m1A was quantified using LC-MS/MS. ~6% increase and ~16% decrease of the m1A level in total tRNA were consistently observed in the knockdown and overexpression samples, respectively (Figure 2A). Moreover, an ALKBH1 stable overexpression HeLa cell line showed ~28% decrease of the m1A level in tRNA compared to control cells. We also studied the Alkbh1 homozygous knockout (Alkbh1−/−) mouse embryonic fibroblast (MEF) cells that we obtained previously (Nordstrand et al., 2010). We observed ~42% increase of the m1A level in tRNA from the Alkbh1−/− MEF cells in comparison to the wild-type MEF cells (Figure 2B). A dot-blot assay using an m1A-specific antibody was also applied to the same samples, and results were consistent with LC-MS/MS measurements (Figure S2B and S2C). As expected, neither knockdown nor overexpression of ALKBH1 caused any changes of the levels of m7G, m5C, or m3C in total tRNA (Figure S2D).
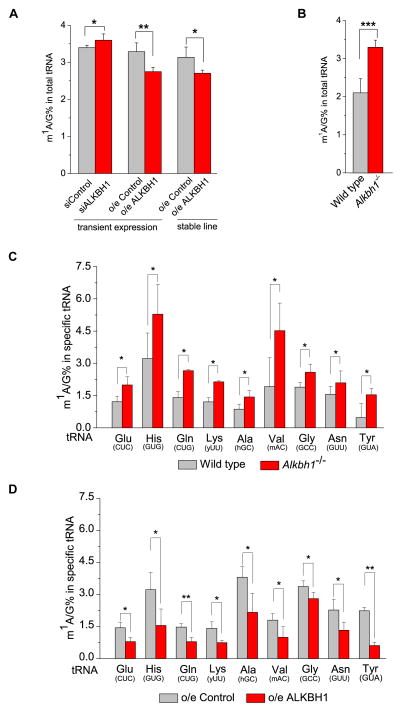
Figure 2. ALKBH1-Mediated m1A Demethylation of tRNAs inside Cells.
(A) Quantification of m1A/G ratio in total tRNA purified from HeLa cells by LC-MS/MS. The transient knockdown and overexpression of ALKBH1 in HeLa cells led to ~6% increase and ~16% decrease of the m1A/G ratio in total tRNA, respectively. Stable overexpression of ALKBH1 in HeLa cells resulted in ~21% decrease of the m1A/G ratio compared to control cells.
(B) In the Alkbh1−/− MEF cell line, the m1A/G ratio is ~42% higher compared to control cells. Control samples are shown in light grey. G is the most abundant base in tRNAs and is used to more accurately quantify the m1A level in tRNA.
(C) Quantification of m1A/G ratio in tRNAGlu(CUC), tRNAHis(GUG), tRNAGln(CUG), tRNALys(yUU), tRNAAla(hGC), tRNAVal(mAC), tRNAGly(GCC), tRNAAsn(GUU), and tRNATyr(GUA) by LC-MS/MS. Knockout of Alkbh1 in MEF cells led to a noticeable increase of m1A/G ratio in these tRNAs compared to controls. y = U,C; h = A,C,U; m = A,C.
(D) Stable overexpression of ALKBH1 in HeLa cells led to a noticeable decrease of the m1A/G ratio in these tRNAs compared to controls. p values were determined using two-tailed Student’s t-test for paired samples. Error bars represent mean ± s.d., n = 8 (four biological replicates × two technical replicates). *p < 0.05, **p < 0.01.
Because the m1A level of the total tRNA is significantly affected by ALKBH1 (Figure 2A and 2B) and all tRNAs identified by CLIP contain m1A58 (Figure 1B), we further validated the m1A demethylation activity of ALKBH1 on selected target tRNAs. Biotin-labeled DNA probes complementary to tRNA species were used to specifically isolate the corresponding tRNA from HeLa and MEF cells (Figure S2E). The m1A level of the purified specific tRNA was then measured using LC-MS/MS. The results demonstrated that the knockout of Alkbh1 led to increases of the m1A level in most tRNAs tested including tRNAGlu(CUC), tRNAHis(GUG), tRNAGln(CUG), tRNALys(yUU), tRNAAla(hGC), tRNAVal(mAC), tRNAGly(GCC), tRNAAsn(GUU), and tRNATyr(GUA) (Figure 2C, Table S1). Likewise, we measured the m1A levels of these tRNAs in the ALKBH1 stable overexpression HeLa cell line and the control cell line. Results again revealed that overexpression of ALKBH1 led to reduced m1A in these tRNAs (Figure 2D). Therefore, the majority of tRNAs identified by CLIP-seq are substrates of ALKBH1. Taking tRNAHis(GUG) as an example, which has a relatively high m1A level, ~65% increase and ~39% decrease of the m1A/G ratio were observed in the Alkbh1 knockout and ALKBH1 overexpression cells compared to corresponding controls, respectively.
m1A in tRNA as the Primary Substrate of ALKBH1
To probe whether ALKBH1 also mediates demethylation of nucleic acid substrates other than m1A in tRNA, we measured the levels of m1A and m6A in various RNA species in the Alkbh1 knockout mouse embryonic stem cells (mESCs) and MEF cells, as well as ALKBH1 knockdown HeLa cells, and compared to the corresponding control cells. No significant changes were observed for m1A or m6A in ribosome RNA (rRNA) when comparing Alkbh1 heterozygous knockout (Alkbh1+/−) mESCs to Alkbh1−/− mESCs, and between the wild-type MEF cells and Alkbh1−/− MEF cells, respectively. Similarly, transient knockdown of ALKBH1 did not alter the m1A or m6A level in rRNA in comparison to the corresponding controls (Figure S2F). Similar measurements were performed on mRNA and the results showed that ALKBH1 does not noticeably affect the mRNA m6A level (Figure S2G). Biochemical experiments further confirmed that m6A in RNA is less likely a substrate of ALKBH1 (Figure S2H).
We observed a slight increase of the total m1A in mRNA in Alkbh1−/− mESCs when compared to Alkbh1+/− mESCs, while overexpression of ALKBH1 in HeLa cells induced a slight m1A decrease in mRNA (Figure S2G). These results suggested that ALKBH1 may mediate demethylation of m1A in mRNA; however, these changes are quite small comparing to those observed for tRNA. Because ALKBH1 possesses a tRNA-binding domain and the substrate m1A58 resides in the loop region of a stem-loop motif in tRNA, we reasoned that ALKBH1 may preferentially mediate m1A demethylation in tRNA with selectivity towards a stem-loop. We synthesized four RNA probes with m1A in stem-loop sequences mimicking the T Ψ C loops in tRNAiMet and tRNAHis(GUG) as well as in two unstructured sequences as controls (Figure S2I). Both tRNAiMet and tRNAHis(GUG) contain m1A58 at the T Ψ C loop region with distinct sequences. Indeed, ALKBH1 showed high preference to the stem-loop structure as the measured maximal velocity values of m1A demethylation toward both stem-loop probes are much higher than those for the unstructured probes. This biochemical result, combined with quantifications of modification changes in various cells and CLIP-seq data, strongly support m1A in tRNA as the primary substrate of ALKBH1. We could not exclude the possibility that certain structured m1A sites in mRNA may also serve as substrates of ALKBH1.
Lastly, DNA N6-methyldeoxyadenosine (6mA) has been proposed as a substrate of ALKBH1 in a recent study (Wu et al., 2016). Our biochemical assay did not reveal observable DNA 6mA demethylation activity by ALKBH1 (Figure S2H). In addition, we have measured the genomic DNA N6-methyldeoxyadenosine (6mA) levels in Alkbh1+/− and Alkbh1−/− mESC cells without noticeable changes of 6mA observed (Figure S2J).
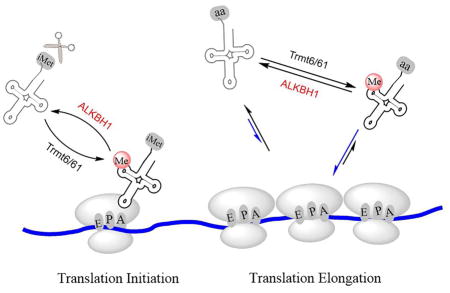
ALKBH1 Affects Cellular tRNAiMet Level and Translation Initiation
Although ALKBH1 can demethylate most m1A sites in tRNAs, there are exceptions. The m1A levels of tRNAPhe(GAA) and tRNASec(UCA) were not appeared to be affected by ALKBH1 (Figure S3A and S3B); these tRNAs are not among CLIP targets found for ALKBH1 and tRNASec(UCA) is known to possess structural features distinct from other tRNAs (Itoh et al., 2009). Our CLIP-seq also did not reveal tRNAiMet as potential binding substrates of ALKBH1. However, previous studies have reported that m1A58 is essential for the function of tRNAiMet in yeast, and a loss of m1A leads to polyadenylation and subsequent degradation of tRNAiMet (Anderson et al., 1998). The m1A58-dependent tRNAiMet stability may also be true in human cells. The demethylation of m1A58 in tRNAiMet may lead to rapid degradation of the demethylated tRNAiMet, leaving the isolated tRNAiMet in our experiment always fully methylated.
To biochemically probe potential m1A demethylation of tRNAiMet by ALKBH1, we pulled down tRNAiMet by using its complementary DNA probe. For comparison, we also pulled out an elongator tRNA, tRNAeMet(CAU) which also was not present as a binding substrate in our CLIP-seq experiment. Biochemical results showed that ALKBH1 is capable of demethylating m1A in both tRNAiMet and tRNAeMet(CAU) (Figure S3C). Genome-wide tRNA sequencing was performed to estimate levels of every tRNA species in the Alkbh1−/− and control cells (Zheng et al., 2015). The result showed modest change for many tRNAs in the Alkbh1−/− cells compared to the wild-type control; however, the level of tRNAiMet was increased by ~1.9-fold (Figure S3D). Northern blot analysis was used to further probe changes in the Alkbh1−/− and control cell lines with a significantly increased tRNAiMet level (~1.6-fold) observed in the Alkbh1−/− cell line (Figure S3E). We further probed the level of tRNAiMet using transient knockdown of ALKBH1 in comparison to the control HeLa cells. The northern blot result revealed that transient knockdown of ALKBH1 in HeLa cells led to ~3-fold increase of the tRNAiMet level compared to the control (Figure 3A).
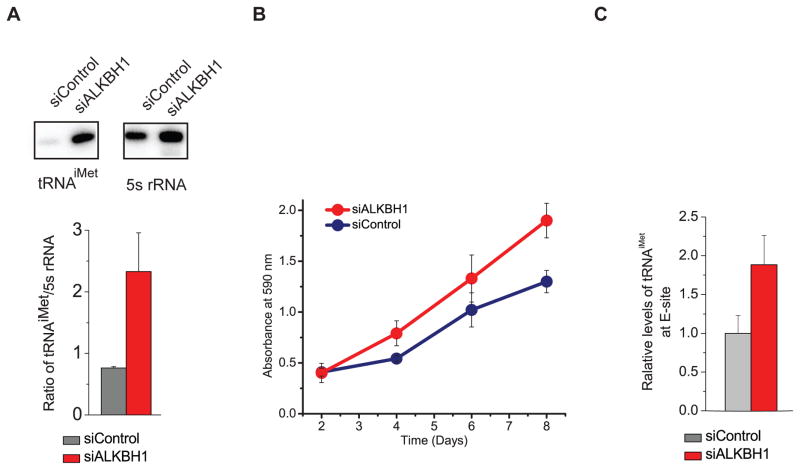
Figure 3. ALKBH1 Affects the Cellular Level of tRNAiMet and Cell Proliferation.
(A) The knockdown of ALKBH1 in HeLa cells increased the cellular level of tRNAiMet compared to the control (5s rRNA was used as the loading control). One representative result was shown in the upper panel. Four biological replicates were performed with statistic errors calculated. The relative ratios of tRNAiMet to 5s rRNA were compared in the ALKBH1 knockdown HeLa cells (red) and control samples (grey).
(B) The knockdown of ALKBH1 promoted proliferation of HeLa cells.
(C) The knockdown of ALKBH1 in HeLa cells increased the tRNAiMet level in the initiating ribosome compared to the control.
See also Figure S3.
Our observations suggested that tRNAiMet is a substrate of ALKBH1 and that the ALKBH1-mediated m1A58 demethylation may affect the stability of tRNAiMet, which could impact the global programming of tRNAs. The cellular level of tRNAiMet is known to significantly impact translation initiation and cell proliferation (Macari et al., 2016; Pavon-Eternod et al., 2013). Indeed, a noticeably increased cell proliferation was observed in the ALKBH1 knockdown cells compared to control cells (Figure 3B). To probe the impact of the elevated tRNAiMet caused by ALKBH1 knockdown on translation initiation, a key step critical to cell proliferation, we treated the ALKBH1 knockdown and control HeLa cells with ribosome E site inhibitor lactimidomycin (LTM) and separated the initiating ribosome (Lee et al., 2012). We measured the absolute levels of tRNAiMet in the initiating ribosome and in the total RNA with 5s rRNA as the loading control. We consistently observed that ALKBH1 knockdown leads to increased cellular tRNAiMet as well as increased tRNAiMet in the initiating ribosome (Figure 3C and S3F). The polysome profile results supported increased 80S monosome assembly upon ALKBH1 knockdown (Figure S3G). Taken together, the elevated tRNAiMet induced by ALKBH1 knockdown promotes translation initiation and cell proliferation as a functional consequence.
m1A Methylated tRNAs are Enriched in Polysomes
Our previous studies have shown that hypermodification at the m1A58 site can alter the association of certain tRNAs with polysomes (Saikia et al., 2010). The ALKBH1-catalyzed demethylation could function as a switch to regulate the utilization of certain tRNAs in protein synthesis by tuning their m1A58 levels. We separated tRNA in the translation-inactive non-polysome fractions (< 80S) as well as tRNA associated with translation-active polysomes (> 80S) from HeLa cells. Three selected tRNAs, tRNAVal(mAC), tRNAHis(GUG), and tRNAGly(GCC), were subsequently extracted from these fractions using biotinylated complementary DNA probes that are specific for each tRNA. LC-MS/MS measurements confirmed that the m1A-hypermodified tRNAVal(mAC), tRNAHis(GUG), and tRNAGly(GCC) preferentially associate with the translation-active polysomes (Figure 4A). We further examined HeLa cells with stable overexpression of ALKBH1, in which tRNAHis(GUG) and tRNAGly(GCC) were hypomethylated. Northern blot analysis revealed significantly reduced populations of these two tRNAs (73% reduction for tRNAGly(GCC) and 57% reduction for tRNAHis(GUG)) in the translationally active pool versus the inactive pool (Figure S4A and S4B).
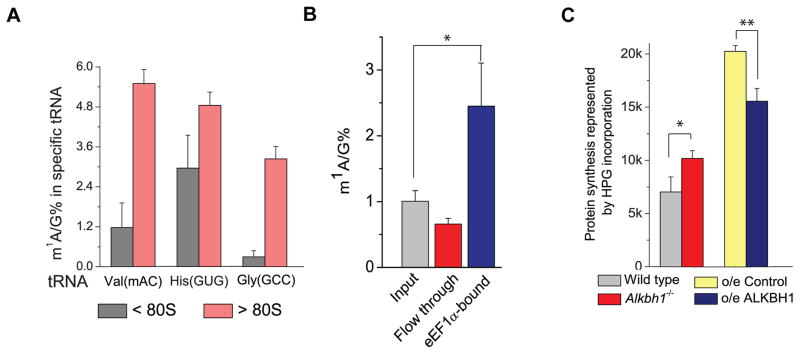
Figure 4. The Methylated tRNA Speices are Preferrentially Used to Promote Translation.
(A) Hypermethylated tRNAVal(mAC), tRNAHis(GUG), and tRNAGly(GCC) preferentially associate with the polysomes. Quantification of the m1A/G ratio in tRNAVal(mAC), tRNAHis(GUG), and tRNAGly(GCC) extracted from the translationally inactive portions (< 80S) and the translationally active portions (> 80S) in ALKBH1 stable overexpression HeLa cells and the control cells. Error bars represent mean ± s.d., n = 8 (four biological replicates × two technical replicates).
(B) The elongation factor-1 protein (eEF1α) was pulled down from HeLa cells using a specific anti-eEF1α antibody and then incubated with an excess amount of total tRNA purified from same cells. LC-MS/MS measurements showed enrichment of m1A in the eEF1α-bound portion and depletion of m1A in the flow through portion. Error bars represent mean ± s.d., n = 8 (four biological replicates × two technical replicates).
(C) Quantification of protein synthesis in MEF cells by flow cytometry. HPG incorporation into MEF control cells (grey), Alkbh1−/− MEF cells (red), HeLa control cells (yellow), and ALKBH1 stable overexpression HeLa cells (blue) were recorded 1 h after administration. Six biological replicates were performed; for ALKBH1 stable overexpression compared to the control, p = 0.012. For Alkbh1−/− MEF vs. control, p = 0.003. P values were determined using two-tailed Student’s t-test for paired samples. *p < 0.05, **p < 0.01. Error bars represent mean ± s.d., n = 9 (three biological replicates × three technical replicates).
See also Figure S4.
We next studied how m1A-methylated tRNAs might be preferentially recognized and delivered to translation-active polysomes. We pulled down eEF1α, the elongation factor protein known to deliver aminoacyl-tRNA to translation-active polysomes, from HeLa cells using an anti-eEF1α specific antibody (Figure S4C). The immunoprecipitated eEF1α was incubated with total tRNA purified from the same cells. We observed ~2.5-fold enrichment of m1A in the eEF1α-bound fraction (Figure 4B), confirming enrichment of m1A-methylated tRNAs in the translation-active pool. eEF1α may preferentially bind m1A-methylated tRNA. It is also possible that the methylated tRNAs are preferentially aminoacylated and subsequently recognized by eEF1α, although fully unmodified tRNAs can be readily charged in vitro, and many tRNA synthetases are not known to interact with the tRNA region where m1A58 resides (Giegé et al., 1998).
The ALKBH1-Mediated tRNA Demethylation Impacts Translation Elongation
In order to confirm the effect of the ALKBH1-dependent tRNA demethylation on protein synthesis, we used an alkyne-modified glycine analog, L-homopropargylglycine (HPG), to metabolically label newly synthesized proteins in Alkbh1−/−and wild-type MEF cells, as well as in ALKBH1 stable overexpression HeLa cells versus control cells. The HPG-labeled cells were then fluorescently modified, and analyzed by flow cytometry. The results showed a notable increase of protein synthesis rate in the Alkbh1−/− cells compared to the wild-type cells. In contrast, ALKBH1 overexpression led to a reduction of the protein synthesis rate compared to the control (Figure 4C and Figure S4D–S4I). These results are consistent with the promotion of the translation initiation by the knockdown of ALKBH1. The results also support the preferential recruitment of m1A-methylated tRNAs to the active translation pool which promotes translation elongation.
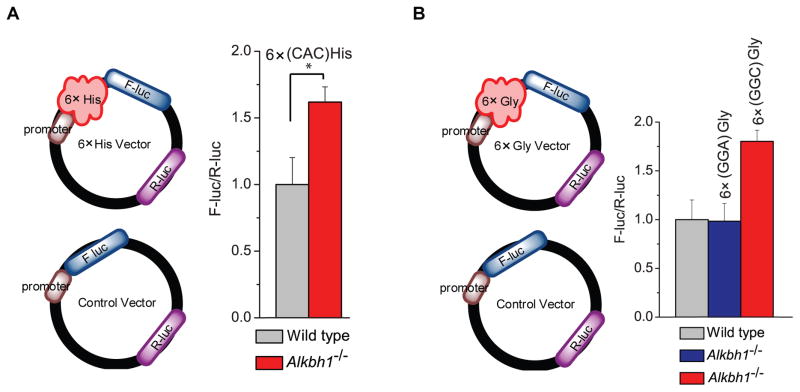
Because we observed up to 3-fold change of the total cellular tRNAiMet level and ~2-fold increase of tRNAiMet in initiating ribosome with ALKBH1 knockdown inside HeLa cells, the effect of ALKBH1 on translation initiation is significant. To investigate the translation elongation effect of ALKBH1 that is independent of translation initiation, we first designed a reporter assay with six repeated specific codon sequences (6×CAC(His) for tRNAHis(GUG) and 6×GGC(Gly) for tRNAGly(GCC)) added to the 5′ of firefly luciferase (F-luc) (Figure 5); a second Renilla luciferase (R-luc) encoded in the same plasmid was used to normalize the expression efficiency. To further normalize translation differences between Alkbh1−/− and wild-type MEF cells introduced by any other factor including initiation (i.e. the cellular levels of tRNAiMet and other tRNAs), a control reporter (F-luc plus R-luc) devoid of these 6× sequences was also transfected. The normalization factor from this control reporter was applied to the 6×CAC(His)-reporter signal. For the mRNA reporter fused with 6×CAC(His), ~1.6-fold elevated translation was observed in the Alkbh1−/− MEF cells compared to the wild-type MEF cells (Figure 5A). Similarly, 6×GGC(Gly) was inserted in the reporter plasmid. tRNAGly(GCC) cannot decode the GGA codon; therefore, we also inserted 6×GGA(Gly) into a negative control reporter. We observed elevated translation (~1.8-fold) for the 6×GGC(Gly)-reporter in the Alkbh1−/− cell compared to the wild-type MEF cells, but not the negative control reporter with 6×GGA(Gly) sequences (Figure 5B).
Figure 5. A Reporter Assay Confirming Translation Regulation by the ALKBH1-Meidated tRNA Demethylation.
(A) Scheme of the reporter assay on the left: the RNA reporter vector encodes firefly luciferase (F-luc) as the primary reporter and Renilla luciferase (R-luc) on the same plasmid as the internal transfection control. The effect of tRNAHis(GUG) in protein translation was revealed by a reporter assay. 6×CAC(His)-coding sequences (recognized by tRNAHis(GUG)) were inserted after the PLK promoter region of F-luc as the positive reporter (noted as 6×CAC(His)); this insertion in the luciferase reporter led to a noticeable increase of protein synthesis in the Alkbh1−/− MEF cells compared to the wild-type MEF cells. The control reporter without any insertion was used to normalize the translation differences between the two cells lines. Error bars represent mean ± s.d., n = 12 (three biological replicates × four technical replicates).
(B) Scheme of the reporter assay on the left. The effect of tRNAGly(GCC) in protein translation was revealed by a reporter assay. 6×GGC(Gly)-coding sequences were inserted after the promoter region of F-luc as the positive reporter (noted as 6×GGC(Gly)); 6× glycine-coding sequences of GGA(Gly) were fused to the 5′end of F-luc as the negative control (noted as 6×GGA(Gly)); another control vector is devoid of any amino acid insertion (noted as control). The insertion of 6×GGC(Gly)-coding sequences in the luciferase reporter led to a ~1.8-fold increased protein translation in the Alkbh1−/− MEF cells compared to the wild-type MEF cells; this effect was not observed with the negative control of 6×GGA(Gly) insertion. The control reporter without any insertion was used to normalize the translation differences between the two cells lines. Error bars represent mean ± s.d., n = 12 (three biological replicates × four technical replicates).
See also Figure S5.
We further constructed two mutant reporters with selected codons mutated from matching tRNA anti-codons that are substrates of ALKBH1 to codons matching isoacceptor tRNAs that are not substrates of ALKBH1 (Figure S5). Mutant reporter 1 contains mutations of three codons (Lys8, Lys9, and Gly10, see Supplemental Information) and mutant reporter 2 contains mutations of three codons (Ala64, Ala66, and Lys68, see Supplemental Information) of F-luc, respectively. We transfected the mutant reporters with the wild-type reporter control to ALKBH1 knockdown cells and control cells, respectively. We observed that the mutations led to noticeably reduced protein synthesis when normalized to the control reporter (without mutations) upon ALKBH1 knockdown (Figure S5A, B). Together, these results indicate that a deficiency of ALKBH1 leads to increased m1A levels of specific tRNAs and augments translation elongation from the corresponding codons.
An ALKBH1-dependent Translation Control in Responses to Glucose Deprivation
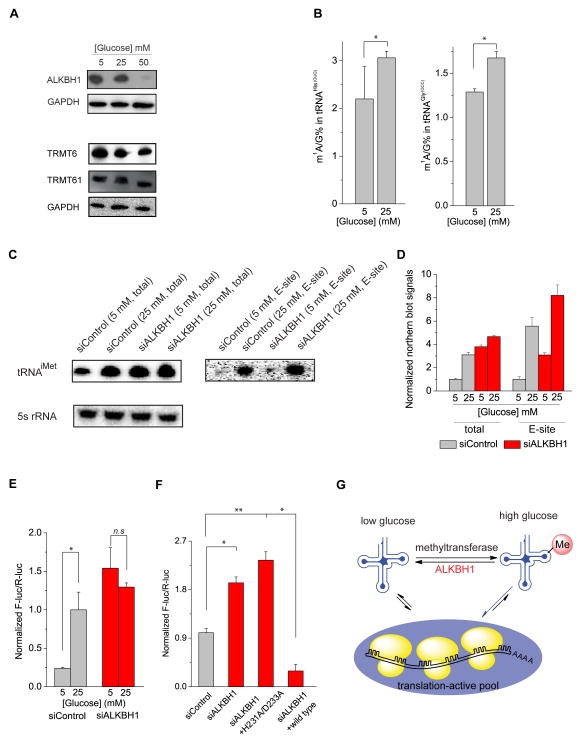
We then investigated the dynamics of ALKBH1 under different physiological conditions. We noticed an increased expression of ALKBH1 in HeLa cells under glucose deprivation conditions; while the level of the m1A58 methyltransferase heterodimer Trmt6/Trmt61 remained mostly unchanged (Figure 6A, Figure S6). The elevated ALKBH1, induced by the reduced glucose concentration in the growth medium, led to progressively decreased m1A methylation in tRNAHis(GUG) and tRNAGly(GCC) as expected (Figure 6B); the expression level of tRNAiMet also decreased under glucose deprivation, which can be reversed by the knockdown of ALKBH1 (Figure 6C). After treating cells with ribosome E site inhibitor LTM and separating the initiating ribosome, we measured the levels of tRNAiMet in the initiating ribosome. The knockdown of ALKBH1 induced a noticeable increase of tRNAiMet in the initiating ribosome in comparison to the knockdown control, indicating a translation repressing role of ALKBH1 (Figure 6C and 6D). The inhibition of translation initiation induced by the elevated ALKBH1 is likely a mechanism cells employ to globally repress translation upon glucose deprivation.
Figure 6. The ALKBH1-Mediated tRNA Demethylation in Response to Glucose Availability.
(A) Western blot showing increased concentrations of glucose in the growth medium led to decreased ALKBH1 expression in HeLa cells; protein levels of the m1A58-transferase Trmt6/Trmt61 heterodimer remained mostly unchanged.
(B) LC-MS/MS quantification of the m1A/G ratio in tRNAHis(GUG) and tRNAGly(GCC) in HeLa cells cultured with 5 and 25 mM of glucose. The increased concentration of glucose led to increased m1A methylation in tRNAHis(GUG) and tRNAGly(GCC). Error bars represent mean ± s.d., n = 8 (four biological replicates × two technical replicates).
(C) Glucose deprivation decreased the cellular level of tRNAiMet, which could be reversed by ALKBH1 knockdown. (5s rRNA was used as the loading control). Glucose deprivation also noticeably decreased the level of tRNAiMet in the initiating ribosome. The knockdown of ALKBH1 led to elevated tRNAiMet in the initiating ribosome in comparison to the control in HeLa cells cultured with 5 mM and 25 mM glucose, respectively. 500 ng of total RNA extracted from the initiating ribosomes were applied in the northern blot analysis. One representative result was shown.
(D) The levels of tRNAiMet as shown in (C) were quantified by using Quantity One.
(E) The 6×CAC(His)-reporter assay showing increased translation in HeLa cells with elevated glucose levels in the culture medium. This effect could be reversed by transient knockdown of ALKBH1. Error bars represent mean ± s.d., n = 12 (three biological replicates × four technical replicates).
(F) Knockdown of ALKBH1 in HeLa cells growing at 25 mM glucose led to an increased translation of the 6×CAC(His)-reporter. The effect could be reversed by transient overexpression of the wild-type ALKBH1 but not a catalytically inactive ALKBH1 H228A/D231A mutant. P values were determined using two-sided Student’s t-test for paired samples. *p < 0.05, **p < 0.01. Error bars represent mean ± s.d., n = 12 (three biological replicates × four technical replicates). n.s. represents not significant.
(G) A proposed model of ALKBH1-dependent regulation of translation. The m1A58 methylated tRNAs are preferentially used in the translation-active pool; the ALKBH1-mediated tRNA m1A demethylation, which responds to the glucose deprivation, tunes the association of specific tRNAs to polysomes and thus represses protein synthesis.
See also Figure S6.
To examine effects on translation elongation with the elevated ALKBH1 upon glucose deprivation, we again employed the 6×CAC(His)-reporter normalized to the control (Figure 5A) in HeLa cells growing with different concentrations of glucose. Significantly decreased protein synthesis from the reporter mRNA was observed in cells growing in the medium with reduced glucose; this effect could again be reversed by the transient knockdown of ALKBH1 (Figure 6E). In addition, the knockdown of ALKBH1 in HeLa cells (growing at 25 mM glucose) resulted in an increased translation of the 6×CAC(His)-reporter (Figure 6F). This effect could be reversed by transient overexpression of the wild-type ALKBH1 but not a catalytically inactive ALKBH1 H228A/D231A mutant, further confirming that the ALKBH1-catalyzed demethylation affects protein synthesis in response to the availability of glucose (Figure 6G). With mutant reporters (Figure S5) we also observed that the mutations led to noticeable increases in the normalized protein synthesis over the control reporter upon glucose starvation (Figure S5C), consistent with the elevated cellular ALKBH1 in glucose starving cells. Thus, ALKBH1 can affect both translation initiation and translation elongation upon glucose deprivation.
DISCUSSION
Here we report ALKBH1 as a tRNA demethylase which catalyzes demethylation of m1A in tRNAs. A tRNA-binding motif exists in ALKBH1 and it preferentially demethylates m1A in a stem-loop structure. CLIP sequencing data showed that ALKBH1 binds m1A58-containing tRNAs. Further biochemistry characterizations revealed that ALKBH1 possesses effective tRNA m1A demethylation activity in vitro and inside cells. We showed that ALKBH1 controls cellular levels of tRNAiMet to affect translation initiation, and that the m1A-methylated tRNAs are preferentially recruited to polysomes to promote translation elongation. The ALKBH1-mediated tRNA demethylation controls the utility of the target tRNAs in translation, thereby directly influencing protein synthesis. This process is dynamic and is used by human cells to respond to glucose deprivation. This discovery opens a potential new paradigm of reversible tRNA methylation in affecting gene expression.
Nucleic Acid Substrate(s) of ALKBH1
ALKBH1 has been suggested to work on nucleic acid substrates in the past (Fedeles et al., 2015). A recent publication describes ALKBH1 as a 6mA demethylase (Wu et al., 2016). We have tested the activity of ALKBH1 towards 6mA in DNA in vitro and inside cells. The reported demethylase activity towards 6mA DNA was too low to be observed in our experiments (Figure S2H). Although we could not completely exclude 6mA in DNA as a substrate of ALKBH1, all of our biochemical studies of the same mESC cell lines as well as other mammalian cell lines do not currently support 6mA in DNA as a substrate of ALKBH1.
ALKBH1 possesses a tRNA-binding motif. Our CLIP sequencing data and demethylation activity analysis of ALKBH1 indicate that ALKBH1 binds and acts preferentially on tRNA over mRNA in cell lines we studied. If ALKBH1 can gain access to mRNA it is possible that ALKBH1 may also mediate mRNA m1A demethylation to affect specific developmental or cellular processes in mammalian systems, which should be investigated in the future.
tRNA m1A Demethylation by ALKBH1
The m1A methylation endows tRNA with a positive charge which can significantly affect its structure and interaction with potential partner proteins. This methylation is known to be critical to the stability of tRNAiMet (Anderson et al., 1998). m1A has recently been discovered as another mRNA modification, in addition to m6A, pseudouridine, and m5C. The presence of m1A at the 5′ UTR region in mRNA has been suggested to promote translation (Dominissini et al., 2016; Li et al., 2016). ALKBH3, a DNA repair enzyme that demethylates N1-methyldeoxyadenosine in single-stranded DNA (Aas et al., 2003; Duncan et al., 2002) has been suggested as a potential demethylase of m1A in mRNA (Li et al., 2016), the functional relevance of which still requires further investigations. ALKBH3 is also capable of repairing tRNA alkylation damages (Aas et al., 2003), again the biological relevance of which was not clear. We show that m1A in tRNA is the main substrate of ALKBH1. Many tRNA species contain this methylation (Machnicka et al., 2013), with a substantial fraction of them subjected to ALKBH1-mediated demethylation.
The cellular level of tRNAiMet can be significantly affected by ALKBH1, suggesting an ALKBH1-mediated demethylation of m1A58 of tRNAiMet. m1A58 is essential for the stability and function of tRNAiMet in yeast (Anderson et al., 1998); this is also likely to be the case in humans. In human cells, it is possible that ALKBH1 encounters tRNAiMet and catalyzes its m1A58 demethylation which then leads to tRNAiMet degradation. Subsequent dissociation of ALKBH1 from tRNAiMet may explain the lack of tRNAiMet as a binding substrate of ALKBH1 in the CLIP result. Importantly, we observed significant effects of ALKBH1 on the total cellular tRNAiMet level and subsequent translational initiation.
Translation Regulation through Reversible tRNA Methylation
Translational control is an essential means in the regulation of gene expression in a myriad of cellular events. The pathways of translation regulation are complex in mammals, reflecting the complexity of mammalian development, signaling, and response to environmental cues. Global regulation of translation is often through influencing the activity of the translation initiation factors and the availability of the ribosomes (Gebauer and Hentze, 2004; Hershey et al., 2012). The rate-limiting step in translational control is typically at the initiation step. Phosphorylation of eIF2α, 4E-BPS, and other initiation factors negatively impacts translation (Pavitt and Ron, 2012). In addition to the regulation of translation initiation, mRNA can be sequestered into stress granules or P-bodies upon translation inactivation in response to stresses (Decker and Parker, 2012). Nonsense mediated decay ensures translation fidelity by screening mRNAs for premature stop codons and mediating their decay (Hentze and Kulozik, 1999; Maquat, 2004). In contrast to the global translational regulation, microRNA can attenuate translation by interacting directly to the target mRNAs (Bartel, 2009; Ha and Kim, 2014). Transacting proteins can affect translation rate by specifically recognizing a particular mRNA site, such as the 3′ UTR (Gebauer et al., 2012).
Dynamic RNA modifications also emerged as a new type of regulatory mechanism of translation. For instance, the m6A reader protein YTHDF1 specifically associates with the m6A sites and promotes translation by interacting with the translation initiation complex (Wang et al., 2015). The recently identified mRNA modification m1A in mRNA is suggested to be related to active translation as well (Dominissini et al., 2016; Li et al., 2016), although the m1A-binding protein and the underlying mechanism have yet to be unveiled.
tRNA modifications have long been thought to affect translation by fine-tuning tRNA structures and their interactions with the corresponding mRNA codons (Agris, 2008; Agris et al., 2007). Most of the known cases involve tRNA modifications that impact specific tRNAs. For example, m1A58 is indispensable for the proper folding of tRNAiMet; under stressed conditions, such as heat shock, the loss of m1A58 results in tRNAiMet degradation (Wang et al., 2008). We show here that a group of tRNAs can be subjected to a more global regulation through the ALKBH1-mediated demethylation of a common m1A modification. tRNAs are abundant in all cells; however, the m1A-modified tRNAs, at least those that are ALKBH1 substrates, preferentially associate with polysomes to support translation. The current work represents the discovery of the first tRNA demethylase and the first reversible tRNA modification that could broadly affect protein synthesis.
Cytosine-5 RNA methylation has very recently been shown to protect tRNA from cleavage and modulate global protein synthesis and cell fate (Blanco et al., 2016). The ALKBH1-mediated tRNA demethylation directly affects translation initiation through controlling the cellular levels of tRNAiMet, and impacts translation elongation by tuning the methylation status and availability of target tRNAs. These effects enable rapid responses to cellular signaling cues, such as the glucose deprivation shown in this work (Figure 6G). Severe embryonic development defects had been reported in Alkbh1 knockout mice (Nordstrand et al., 2010). The survived Alkbh1 knockout mice show defects in tissues originating from the ectodermal lineage (Pan et al., 2008) and impaired neuronal development (Ougland et al., 2012). These phenotypes all indicate functional significance of ALKBH1 in mammals. Our previous discoveries of FTO and ALKBH5 as mRNA m6A demethylases sparked the emergence of the epitranscriptomics field (Jia et al., 2011; Zheng et al., 2013). The discovery presented here on tRNA m1A demethylation by ALKBH1 should stimulate future studies on reversible tRNA methylation in translation control and its impacts on mammalian developments.
STAR METHODS
Detailed methods are provided in the online version of this paper and include the following:
CONTACT FOR REAGENT AND RESOURCE SHARING
-
METHOD DETAILS
Luciferase Reporter Assay
CLIP
Analysis of RNA Sequencing Results
Cloning, Expression, and Purification of ALKBH1
Mammalian Cell Culture, siRNA Knockdown (KD), and Plasmid Transfection
ALKBH1 Stable Overexpression HeLa Cell Line Construction
Glucose Deprivation
tRNA Isolation
Biotinylated Single-stranded DNA Probes
Biochemistry Assay of ALKBH1 Activity in vitro
Quantitative Analysis of the m1A Level Using LC-MS/MS
Dot Blot Assay
Western Blot
Cell Viability Assay
Northern Blot
Quantification of tRNAiMet in the Initiating Ribosome
eEF1α Immunoprecipitation
Measurement of the Protein Synthesis Rate
QUANTIFICATION AND STATISTICAL ANALYSIS
-
DATA AND SOFTWARE AVAILABILITY
Data Resources
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests may be directed to, and will be fulfilled by the lead contact corresponding author Chuan He (chuanhe@uchicago.edu).
METHOD DETAILS
Luciferase Reporter Assay
pmirGlo luciferase expression vector (Promega) was used to construct the reporter plasmid, which contained both a firefly luciferase (F-luc), and a Renilla luciferase (R-luc). F-luc-6× His(CAC) reporter plasmid was obtained by inserting CACCACCACCACCACCAC before the F-luc coding region; F-luc-6×Gly(GGC) reporter plasmid was obtained by inserting GGCGGCGGCGGCGGCGGC before the F-luc coding region; F-luc-6×Gly(GGA) reporter plasmid was obtained by inserting GGAGGAGGAGGAGGAGGA before the F-luc coding region. Two F-luc mutant reporters were made by using site-directed mutagenesis. For mutant reporter 1, the sequences of Lys8, Lys9, and Gly10 were mutated from the tRNA anti-codons being the substrates of ALKBH1 to the ones not. For mutant reporter 2, the sequences of Ala64, Ala66, and Lys68 were mutated from the tRNA anti-codons being the substrates of ALKBH1 to the ones not. Basic setting: 500 ng of reporter plasmids (pmirGlo empty vector or pmirGlo-specific tRNA anti-codon inserted vector) were transfected into Alkbh1−/− and wild-type MEF cells in a six-well plate. After 6 hours, each well was re-seeded into a 96-well plate (1:20). After 24 h, the cells in 96-well plate were assayed by Dual-Glo Luciferase Assay system (Promega). Renilla Luciferase (R-luc) was used to normalize firefly luciferase (F-luc) activity to evaluate the translation efficiency of the reporter. Glucose deprivation was applied 4 h after the reporter plasmids were transfected and kept for another 12 h prior to the luciferase assay.
CLIP
10 plates of HeLa cells in 10 cm petri dish were grown until confluency of 80% (~8 million cells). The medium was aspirated and 5 ml of ice-cold PBS was subsequently added to the culture dish. Cells were irradiated once with 400 mJ/cm2 at 254 nm in Stratalinker on ice. After irradiation, cells were harvested by centrifugation at 4000 rpm for 3 min at 4 °C. The cell pellets were then lysed in lysis buffer (1× PBS supplemented with 0.1% SDS, 1% Nonidet P-40, 0.5% Sodium Deoxycholate, Protease Inhibitor Cocktail (1 tablet/50 ml) and RNase inhibitor) and kept on ice. After 4 hours of incubation, the cell pellets were subjected to centrifugation at 17,000 g at 4 °C for 30 min; the supernatant was carefully collected. 50 μl of protein A beads were added into the supernatant to preclear the supernatant at 4 °C for 1 h which was then subjected to centrifugation at 17,000 g at 4 °C for 10 min. The supernatant was collected in a fresh tube. 50 μl of antibody-conjugated protein A beads were subsequently added to the supernatant and kept at 4 °C for overnight with rotation. On the next day, the supernatant was discarded. The beads were then washed for 3 times with 1 ml of high salt buffer (5× PBS supplemented with 0.1% SDS, 1% Nonidet P-40 and 0.5% Sodium Deoxycholate) and another 3 times with 1 ml of wash buffer (20 mM Tris-HCl, pH 7.4, 10 mM MgCl2 and 0.2% Tween-20). The RNAs were then isolated by 200 μl of elution buffer (100 mM Tris-HCl, pH 7.4, 10 mM EDTA, and 1% SDS) containing 2 mg/ml proteinase K under incubation at 50 °C for 30 min. Subsequently, the sample was subjected to centrifugation at 17,000 g at 4 °C for 15 min. The beads were removed and RNAs were recovered by Phenol/chloroform extraction. The RNA species were then 3′ 32P-labeled and subjected to TBE-Urea-PAGE separation. eEF1α and IgG were employed as positive and negative controls in this experiment while human total tRNA was utilized as a size marker for intact tRNA. The TBE-Urea gel was finally exposed to phosphoimager.
Analysis of RNA Sequencing Results
(1) CLIP
Raw reads from CLIP samples were first trimmed according to recommended settings using Trimmomatic with a minimum length of 25 nucleotides (Bolger et al., 2014). Processed reads were then mapped to the hg19 genome using bowtie with the following settings (-v 2 -m 10 -best -strata -p 12), and the subsequent SAM outputs were analyzed with default PARalyzer parameters. From genomic coordinates, refseq IDs and region ID (genomic, tRNA, intergenic, rRNA, etc.) were assigned using custom scripts in order to classify PARalyzer clusters. Conversion rate and cluster assignment and enrichment were determined using custom Python scripts from PARalyzer output. Ratios of each class of small RNAs in the total RNA dataset were calculated and compared to the RNA classes in the CLIP dataset.
(2) tRNA Seq
Data analysis method was adapted from our previous work (Zheng et al., 2015). Briefly, tRNA-seq raw reads were trimmed and processed using Trimmomatic (Bolger et al., 2014) with a minimum length of 16 nucleotides. Processed reads were then aligned with bowtie2 to culled tRNA fasta libraries from the Genomic tRNA database for mm10 and hg19 with CCA appended to every sequence. Bowtie2 parameters were sensitive settings, which allowed for up to 2 mismatches. Reads mapping to multiple isodecoders were discarded. Downstream isodecoder abundance was performed using custom Python scripts.
Cloning, Expression, and Purification of ALKBH1
The human ALKBH1 gene (GenBank Accession NM_006020.2) with deletion of the N-terminal 36 amino acids was subcloned into a pMCSG19 vector by ligation-independent cloning (LIC) to generate the plasmid pMCSG19-His-ALKBH1 (Donnelly et al., 2006). Human ALKBH1 was then expressed in a BL21 (DE3) E. coli strain containing the plasmid pRK1037. Cells were induced with 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) for 24 h at 16 °C. The soluble fraction was purified using a Ni-NTA column (QIAGEN), a gel-filtration column (Superdex-200, Pharmacia), and finally an ion-exchange column (PE 4.6/100, Mono Q). FLAG-tagged ALKBH1 was cloned into vector pcDNA 3.0 (between EcoRI and XhoI restriction enzyme cutting site). High-purity plasmids used for mammalian cell transfection were prepared using HiSpeed Plasmid Maxi Kit (QIAGEN).
The plasmid with site-directed mutagenesis was constructed using QuikChange II site-directed mutagenesis kit (QIAGEN).
The following pair of primers were used:
-
H231A/D233A_forward: GGAATCGCCGTAGCCAGATCTGAG
H231A/D233A_reverse: CTCAGATCTGGCTACGGCGATTCC
-
Reporter mutant 1: Lys8(CUU to UUU) Lys9(CUU to UUU) Gly10(GCC to CCC)
Mutant1_forward: GAAAAACATTAAAAAAGGGCCAGCGCCATTCTACCCACTCGAAG
Mutant1_reverse: GCATCTTCCATGGTGGCTTTACCAACAGTACCGGATTG
Reporter mutant 2: Ala64(UGC to CGC) Ala66(AGC to CGC) Lys68(CUU to UUU)
Mutant2_forward: TGGCGGAAGCGATGAAACGCTATGGGCTGAATACAAACCATC
Mutant2_reverse: GCCGAACGCTCATCTCGAAGTACTCGGCGTAGGTAATG
Mammalian Cell Culture, siRNA Knockdown (KD), and Plasmid Transfection
HeLa cell line was purchased from ATCC (CCL-2) and grown in DMEM (Gibco, 11965) media supplemented with 10% FBS (Gibco, 10438-026) and 1× Pen/Strep (Gibco, 15140). Transfections were performed using Lipofectamine RNAiMAX (Invitrogen) for siRNA knockdown, and Lipofectamine 2000 (Invitrogen) for plasmid transfection, respectively, following the manufacturer’s procedure. Two synthesized duplex RNAi oligos targeting human ALKBH1 mRNA sequences were used: 5′-ACAAGUACUUCUUCGGCGA-3′ (392–410 bp); 5′-GCGCCGUCAUCAACGACUA-3′ (566–584 bp); a scrambled duplex RNAi oligo (5′-UUCUCCGAACGUGUCACGU) was used as a negative control.
The Alkbh1 knockout MEF cell line and the control cell line as well as Alkbh1 knockout MES cell line and the control cell line were previously reported (Ougland et al., 2012).
ALKBH1 Stable Overexpression HeLa Cell Line Construction
The plasmid for pMSCV-FLAG-HA-ALKBH1 expression was transfected into HeLa cells. The cells were selected under 1 mg/ml puromycin for two weeks. During the selection period, cells were resuspended every two days with fresh DMEM medium supplemented with 10% FBS and puromycin. After 7 days, the survived cells were separated as single cell in 96-well plate and subjected to puromycin selection for another 7 days. The survived cells from each of the 96-well plate were collected as monoclonal ALKBH1 stable overexpression HeLa cell lines. The expression of ALKBH1 was confirmed by using both anti-FLAG and anti-HA antibodies.
Glucose Deprivation
Cells were subjected to glucose deprivation (glucose-depleted medium supplemented with 2% FBS, 1× Pen/Strep, and 5 mM glucose) for 8 h. The cells were subsequently harvested for western blot analysis; the tRNAs were extracted from the treated cells for LC-MS/MS analysis.
In the reporter assay, the experiments were carried out right after 8 h of glucose starvation in ALKBH1 knockdown versus control cells, overexpression of ALKBH1 wild-type or inactive mutant ALKBH1 (H231A/D233A) versus control cells.
tRNA Isolation
RNA species smaller than 200 nucleotides were extracted using mirVana™ miRNA Isolation Kit (Life Technologies). The small RNAs were further loaded onto a 15% TBE-Urea gel and the specific tRNA bands were sliced and recovered from the gel. Streptavidin-conjugated M-280 magnetic Dynabeads (Invitrogen) were used for specific tRNA isolation. 20 μl of RNase-free beads were generated according to the manufacturer’s instructions, washed once with buffer A (10 mM Tris-HCl, pH 7.5, 2 mM EDTA, 2 M NaCl), and finally resuspended in 20 μl of buffer A. Subsequently, 200 μM of biotinylated oligonucleotides in 10 μl of water were mixed with an equal volume of Dynabeads in buffer A and incubated at room temperature for 30 min with gentle mixing. After the incubation, the oligonucleotide-coated Dynabeads were then washed for four times in buffer B (5 mM Tris-HCl, pH 7.5, 1 mM EDTA, 1 M NaCl) and equilibrated in 6× SSC solution (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate, pH 7.0). The oligonucleotide-coated Dynabeads and total tRNA in 6× SSC solutions were heated for 10 min at 75°C before they were pooled and incubated together at 75 °C for another 10 min. Thereafter, the suspension was placed at room temperature for 3 hours to allow binding of the tRNAs to the complementary DNA strands on the dynabeads. The oligonucleotide-coated Dynabeads were washed, in succession, three times with 3× SSC, twice with 1× SSC, and several times with 0.1× SSC until the absorbance of the wash solution at 260 nm was close to zero. tRNA retained on the beads was eluted three times using RNase-free water.
Biotinylated Single-Stranded DNA Probes
DNA probes were designed to complement with the 3′ gene sequences of tRNAVal(mAC), tRNAGly(GCC), tRNAHis(GUG), tRNAGlu(CUC), tRNAGln(CUG), tRNALys(yUU), tRNATyr(GUA), tRNAAla(hGC), tRNAiMet(CAU), tRNAeMet(CAU), tRNAPhe(GAA), tRNASec(UCA), and tRNAAsn(GUU). Probe for tRNATyr(GUA) is located in the middle of the mature tRNA. Because human and mouse share identical sequences in these regions of the selected tRNAs, the same DNA probes were utilized for both human and mouse tRNA selection. The probes used for tRNA selection are listed below:
-
For tRNAVal(mAC) selection: (m = A,C)
5′biotin-TGTTTCCGCCCGGTTTCGAACCGGGGACCTTTCGCGT
-
For tRNAGly(GCC) selection:
5′biotin-TGCATTGGCCGGGAATCGAACCCGGGGCCTC
-
For tRNAHis(GUG) selection:
5′-biotin-TGCCGTCACTCGGATTCGAACCGAGGTTGCTG
-
For tRNAGlu(CUC) selection:
5′biotin-TTCCCTGACCGGGAATCGAACCCGGGCCG
-
For tRNAGln(CUG) selection:
5′biotin-AGGTCCCACCGAGATTTGAACTCGGATCGCTGG
-
For tRNALys(yUU) selection: (y = C,U)
5′biotin-CCAACGTGGGGCTCGAACCCACGACCCT
-
For tRNATyr(GUA) selection:
5′biotin-CTAAGGATCTACAGTCCTCCGCTCTACCAGCT
-
For tRNAAla(hGC) selection: (h = A,C,U)
5′biotin-TGGAGGATGCGGGCATCGATCCCGCTACC
-
For tRNAAsn(GUU) selection:
5′biotin-CGTCCCTGGGTGGGCTCGATCCACCAACC
-
For tRNAiMet(CAU) (tRNAiMet) selection:
5′biotin-TAGCAGAGGATGGTTTCGATCCATCA
-
For tRNASec(UCA) selection:
5′biotin-CGCCCGAAATGGAATTGAACCACTCTGTCG
-
For tRNAeMet(CAU) selection:
5′biotin-TGCCCCGTGTGAGGATCGAACTCACGACCT
-
For tRNAPhe(GAA) selection:
5′biotin-TGCCGAAACCCGGGATCGAACCAGGGAC
Biochemistry Assay of ALKBH1 Activity in vitro
The demethylation activity assay was performed in standard 100 μl of reaction mixture containing ALKBH1 (2 nmol), TBE-Urea gel purified tRNA (400 ng), KCl (100 mM), MgCl2 (2 mM), SUPERNase In (0.2 U/μl, life technology), L-ascorbic acid (2 mM), α-ketoglutarate (300 μM), (NH4)2Fe(SO4)2·6H2O (150 μM), and 50 mM of MOPS buffer (pH 6.5). The reaction was incubated overnight at 16 °C, and quenched by addition of 5 mM of EDTA. 60 μl of phenol-chloroform was added to the reaction tube and mixed well (an equal volume of phenol/CHCl3 must be always used). The water phase, which contained the RNA, was collected and subjected to ethanol precipitation. The pellet was finally dissolved into the desired amount of water and analyzed.
Biochemical demethylation assays of ALKBH1 towards m1A and m6A in single-stranded RNA (ssRNA) and 1mA and 6mA in double-stranded DNA (dsDNA) were conducted in 50 μl of reaction mixture containing 1 nmol ALKBH1, 100 ng probes, KCl (100 mM), MgCl2 (2 mM), SUPERNase In (0.2 U/μl, life technology), L-ascorbic acid (2 mM), α-ketoglutarate (300 μM), (NH4)2Fe(SO4)2·6H2O (150 μM), and 50 mM of MOPS buffer (pH 6.5). The reaction was incubated at 37 °C for 1 h and quenched by addition of 5 mM of EDTA. The samples were then centrifuged at 13000 g for 30 min. The supernatant was collected for RNA or DNA digestion prior to LC-MS/MS analysis.
For the steady-state kinetics of the ALKBH1-catalyzed demethylation of m1A in the stem-loop structured RNA and in unstructured RNA probes, all reactions were performed at 37 °C with 50 mol% of ALKBH1 used.
For the annealing of dsDNA, the complementary strands were kept at 10 μM and heated at 95 °C for 3 min before cooling down to room temperature over 2 h.
The sequences of the probes are listed below:
-
6mA-containing dsDNA oligo (#1):
5′-CATGATACCTTATGGAA(6mA)AGCATGCTTGTATTT-3′
3′-AAATACAAGCATGCTTTTCCATAAGGTATCATG-5′
-
1mA-containing dsDNA oligo (#2):
5′-CATGATACCTTATGGAA(1mA)AGCATGCTTGATTT-3′
3′-AAATACAAGCATGCTTTTCCATAAGGTATCATG-5′
-
m6A-containing unstructured single-stranded RNA oligo (#3):
5′-GAUGGAUCG(m6A)AACCAUCG
-
m1A-containing unstructured single-stranded RNA oligo (#4):
5′-AGUUCCGCG (m1A)AACCAUG
-
m1A-containing unstructured single-stranded RNA oligo (#5):
5′-CACGGUUCG (m1A)UUCAAAG
-
m1A-containing stem-loop RNA oligo 1 (TΨC loop in tRNAiMet, #6):
5′-GAUGGAUCG(m1A)AACCAUCG
-
m1A-containing stem-loop RNA oligo 2 (TΨC loop in tRNAHis(GUG), #7):
5′-CCCGGUUCG(m1A)UUCCCGG
Quantitative Analysis of the m1A Level Using LC-MS/MS
400 ng of tRNA, purified by using TBE-Urea gel, was digested by nuclease P1 (2 U) in 30 μl of buffer containing 25 mM of NaCl, and 2.5 mM of ZnCl2 at 37 °C for 1 h, followed by the addition of NH4HCO3 (100 mM) and alkaline phosphatase (0.5 U). After an additional incubation at 37 °C for 1 h, the solution was diluted to 100 μl, filtered through PVDF filter, and 10 μl of the solution was injected into HPLC-QQQ-MS/MS. Nucleosides were separated by reverse-phase ultra-performance liquid chromatography on a C18 column with online mass spectrometry detection using Agilent 6410 QQQ triple-quadruple LC mass spectrometer in positive electrospray ionization mode. The nucleosides were quantified using the nucleoside-to-base ion mass transitions of 282.1 to 150.0 (m1A), 268.0 to 136.0 (A), 298.1 to166.1 (m7G, extension time 0.8 min.), 258.2 to 126.1 (m3C), 258.0 to 126.0 (m5C), 245.0 to 113.1 (U), 244.0 to 112.0 (C) and 244.0 to112.0 (G). Quantification was performed by comparison with the standard curve obtained from pure nucleoside standards running at the same batch. The ratio of m1A to the sum of A, U, G, and C was determined based on the calculated concentrations.
Dot Blot Assay
In the dot-blot analysis, a m1A monoclonal antibody was used to measure the m1A levels in total tRNA of HeLa cells with transient overexpression of ALKBH1 versus control (empty vector pcDNA 3.0 transfection), in total tRNA of Alkbh1−/− versus wild-type MEF cells, and in total tRNA of HeLa cells of ALKBH1 stable overexpression versus the control. tRNA was firstly isolated and denatured by heating at 95 °C for 1 min, followed by chilling on ice directly. Twofold serial dilutions were spotted on an Amersham Hybond-N+ membrane optimized for nucleic acid binding (GE Healthcare). After UV crosslinking in a Stratagene Stratalinker 2400 UV Crosslinker, the membrane was washed by 1× PBST buffer, blocked with 5% of milk in 1× PBST, and incubated with anti-m1A monoclonal antibody (1: 1,000; MEDICAL&BIOLOGICAL LABORATORIES CO., LTD) overnight at 4 °C. After incubating with horseradish peroxidase (HRP)-conjugated anti-mouse IgG secondary antibody, the membrane was visualized by ECL Western Blotting Detection Kit (Thermo). To ensure an equal amount of mRNA was spotted on the membrane, the same blot was stained with 0.02% methylene blue in 0.3 M sodium acetate (pH 5.2).
To test the specificity of m1A antibody (1: 1,000; MEDICAL&BIOLOGICAL LABORATORIES CO., LTD) and m6A antibody (1: 2,000), we applied these antibodies to a synthesized oligonucleotide encoding an ACAUG sequence; the second A was in the form of unmodified A, m6A, and m1A, respectively.
Western Blot
The expression levels of ALKBH1 and the heterodimer TRMT6/TRMT61 in HeLa cells growing at different glucose concentrations were analyzed by western blot. HeLa cells were cultured at different glucose concentration for 8 h before they were lysed and subjected to western blot analysis. Monoclonal rabbit anti-ALKBH1 antibody (Abcam, ab128895) was applied with a dilution of 1: 1,000 for western blot. Polyclonal rabbit anti-TRMT6 antibody (Sigma, SAB2107213) and polyclonal rabbit anti-TRMT61 antibody (Sigma SAB2700607) were both used with 1: 400 dilution. Monoclonal rabbit anti-eEF1α antibody (Abcam, ab157455) was used with 1: 1,000 dilution.
Cell Viability Assay
Cell viability was measured by using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay. Cells with ALKBH1 knockdown and control cells were seeded into 96-well plate at equal quantity of 3,000 cells per well. MTT assay was conducted on every other day till the eighth day; 20 μl of MTT solution (1 mM) was added to each well and the mixture was incubated at 37 °C for 4 hours. The solution in each well was aspirated. The resulted purple crystals were dissolved in 100 μl DMSO and subsequently transferred into a new 96-well plate for UV-Vis measurement. The absorbance at 620 nm was recorded and plotted.
Northern Blot
In the northern blot analysis, 1 μg of total RNA was mixed with Novex® TBE-Urea Sample Buffer (2×) and heated at 95 °C for 3 min. Samples were chilled on ice before being subjected to a 15% TEB-Urea gel for electrophoresis separation. After electrophoresis, the RNA was transferred to positively charged Amersham Hybond-N+ membrane and subsequently immobilized by UV-crosslinking by using UV stratalinker 2400. The membrane was further subjected to prehybridization with buffer containing 5× SSC, 5× Denhardt’s-solution, 0.1% SDS; 100 μg/ml salmon sperm DNA for 2 h prior to the incubation with 5′-32P labeled tRNA detecting probes. After incubation, the membrane was washed several times with washing buffer containing 2× SSC and 0.5% SDS. The washed membrane was finally subjected to phosphoimager exposure.
In order to quantify the levels of tRNAiMet, His(GUG), Gly(GCC), Val(mAC) in ALKBH1 knockdown cells versus knockdown control cells, Alkbh1 knockout cells versus knockout control cells, total RNAs were extracted from cultured cells. 1μg of total RNAs were used for each sample in the northern blot analysis. 5s rRNA was used as a loading control.
In order to quantify the levels of tRNAHis(GUG) and tRNAGly(GCC) in the translational active (> 80S) and inactive component (< 80S), the translational active and inactive components were separated using sucrose gradient. The total RNAs were extracted and then followed the aforementioned procedure. 5s rRNA was used as a loading control.
The sequences of the northern probes are listed below:
For tRNAGly(GCC): 5′-GCATTGGCCAGGAATCGAAGCCCGG
For tRNAHis(GUG): 5′-TGCCGTGACTCGGATTCGAACCGA
-
For tRNAiMet:
5′TGGTAGCAGAGGATGGTTTCGATCCATCGACCTCTGGGTTATGGGCCCAGCACGCTTCCGCTGCGCCACTCTGCT
For tRNAVal(mAC): 5′-AGGCGAACGTGATAACCACTACACTACGGAAAC
For 5S rRNA: 5′-GGCCATACCACCCTGAACGCGCCCGATC
Quantification of tRNAiMet in the Initiating Ribosome
Cells were treated with 50 μM Lactimidomycin (LTM) for 30 min. And then cells were harvested and lysed prior to size exclusion chromatography using MicroSpin™ S-400 HR Columns. 100 μl of cell lysate were saved as input. The collected flow-through from the size exclusion column contains the initiating ribosomes. The total RNAs from the whole cell lysate and the initiating ribosomes were extracted for northern blot analysis. 5s rRNA was used as the loading control.
eEF1α Immunoprecipitation
Three plates of HeLa cells were lysed and the supernatant was saved for immunoprecipitation. 50 μl of eEF1α antibody-conjugated protein A beads were subsequently added to the supernatant and kept at 4 °C overnight with rotation. On the next day, one portion of the immunoprecipitated protein was subjected to western blot analysis to test eEF1α pull-down specificity. ~300 ng eEF1α was incubated with 1 μg of tRNA purified from HeLa cells in 200 μl IPP buffer (150 mM NaCl, 0.1% NP-40, 10 mM Tris, pH 7.4, 40 U/ml RNase inhibitor, 0.5 mM DTT) for 2 hours. The unbound tRNA was also collected from the flow-through fraction. After three times of wash with IPP buffer, proteinase K was supplemented to the incubation pool to release the tRNAs that were bound to eEF1α. LC-MS/MS was used to measure the level of m1A in the tRNAs in the input, flow-through, and eEF1α–bound fractions.
Measurement of the Protein Synthesis Rate
HPG (Life Technologies; 50 nM final concentration) was added to the culture medium, and incubated for 1 h. Cells were then removed from plates and washed twice in Ca2+- and Mg2+-free phosphate buffered saline (PBS). Cells were fixed in 0.5 ml of 1% paraformaldehyde (Affymetrix) in PBS for 15 min on ice. Cells were washed in PBS, and permeabilized in 200 μl PBS supplemented with 3% fetal bovine serum (Sigma) and 0.1% saponin (Sigma) for 5 min at room temperature. The azide-alkyne reaction was performed with the Click-iT Cell Reaction Buffer Kit (Life Technologies) and azide-modified Alexa Fluor 488 (Life Technologies) at 5 mM final concentration. After a 30-minute reaction, cells were washed twice in 3% BSA. All cells were filtered through a 40-μm cell strainer to obtain single cell suspensions. For flow cytometric analysis, all sorted fractions were double sorted to ensure high purity. Data were analyzed by FlowJo (Tree Star) software.
QUANTIFICATION AND STATISTICAL ANALYSIS
P values were determined using two-tailed Student’s t-test for paired samples. *p < 0.05, **p < 0.01. Error bars represent mean ± s.d.. n.s. means not significant.
DATA AND SOFTWARE AVAILABILITY
Data Resources
Sequencing data have been deposited into the Gene Expression Omnibus (GEO) under the accession number GSE65299 (See also Table S2).
Supplementary Material
(A) Pie chart classification of the identified RNA species in the total RNA controls and
(B) ALKBH1-CLIP samples from Figure 1A.
(C) Sequence alignment of human ALKBH1 and human methionine tRNA ligase MSE1. Highly conserved residues were shown in red text and boxed in blue; strictly conserved residues were shown in red background. The tRNA-binding domain was depicted in black box. This figure was prepared by using ESPript (Robert and Gouet, 2014).
(D) The amount of each nucleoside was quantified by its integrated area in the corresponding LC-MS/MS chromatogram. The m1A standard calibration curve was obtained using five standard samples with different concentrations of pure m1A.
(A) The in vitro demethylation by ALKBH1 in total RNA extracted from HeLa cells. Demethylation reactions were performed in the presence (depicted in grey) or absence of EDTA (depicted in red); EDTA chelates cofactor iron and inactivates ALKBH1. The levels of m3C/G, m7G/G, and m5C/G in total tRNA were measured. n.s. represents not significant. Error bars represent mean ± s.d., n = 8 (four biological replicates × two technical replicates).
(B) From left to right: dot-blot analysis of m1A levels in total tRNA of HeLa cells with transient knockdown of ALKBH1 by siALKBH1 and control siRNA; the m1A levels in total tRNA of HeLa cells with transient overexpression of ALKBH1 versus the control (empty vector pcDNA 3.0 transfection); the m1A levels in total tRNA of Alkbh1−/− versus wild-type MEF cells; the m1A levels in total tRNA of HeLa cells with ALKBH1 stable overexpression versus the control (empty vector).
(C) The specificities of m1A and m6A antibodies were tested against ACAUG RNA oligonucleotides; the underlined A was in the form of unmethylated A, m6A, or m1A, respectively.
(D) The levels of m3C/G, m7G/G, and m5C/G in total tRNA were measured in the samples of ALKBH1 knockdown, ALKBH1 overexpression, and relevant controls. The expression changes of ALKBH1 do not appear to induce any noticeable changes on the ratios of m3C/G, m7G/G, and m5C/G in HeLa cells.
(E) The gel image of 32P-labeled total tRNA and tRNA using the tRNAHis(GUG)-specific biotinylated complementary DNA oligo after selection. tRNA array image showing signals in the application of total tRNA (left), tRNAHis(GUG) after selection using the biotin-labeled DNA complementary oligo (middle), and the array layout with the probes for tRNAHis(GUG) indicated in black squares. The result showed that the biotin-labeled DNA complementary oligo to tRNAHis(GUG) was able to specifically isolate only tRNAHis(GUG).
(F) Neither m6A nor m1A level in rRNA was significantly perturbed by the ALKBH1 knockdown or Alkbh1 knockout.
(G) The m6A level in mRNA was not significantly perturbed by changes of ALKBH1. The m1A level in mRNA was slightly altered by changing ALKBH1.
(H) Biochemical demethylation assays of dsDNA probes containing 6mA and 1mA (#1 and #2, sequences are listed in the Supplementary Information), m6A and m1A in linear ssRNA probes (#3, and #4), and m1A in a stem-loop structured probe (#6) using recombinant ALKBH1 in vitro. The reactions were run with 2 nmol ALKBH1 and 100 ng of each probe at 37 °C for 1 h. The demethylation percentage mediated by ALKBH1 on the individual probe was shown. Error bars represent mean ± s.d., n = 4 (four biological replicates).
(I) The plot of the ALKBH1-catalyzed demethylation (%) versus time (min) towards m1A in stem-loop structured RNA probes that mimic the TΨC loops of tRNAiMet and tRNAHis(GUG) and in unstructured RNA probes at pH 7.0 at 37 °C. Error bars represent mean ± s.d., n = 3 (three biological replicates).
(J) The basal level of 6mA in genomic DNA from mouse embryonic stem cells (mESCs) is low. We did not observe noticeable changes of the 6mA level in the Alkbh1 knockout mESCs compared to the wild-type control.
(A) Specific tRNAs were isolated from the Alkbh1−/− and wild-type MEF cells; the extracted tRNAs were then subjected to LC-MS/MS analysis. No significant changes of the m1A level were observed for these tRNA species in cells with Alkbh1−/− versus the wild-type and
(B) ALKBH1 stable overexpression versus control. n.s. represents not significant. Error bars represent mean ± s.d., n = 8 (four biological replicates × two technical replicates).
(C) Demethylation of m1A in tRNAiMet and tRNAeMet(CAU) extracted from HeLa cells by recombinant ALKBH1 in the presence or absence of EDTA. Error bars represent mean ± s.d., n = 4 (four biological replicates).
(D) tRNA sequencing (DM-tRNA-seq) was performed to compare the levels of tRNAs in the ALKBH1 knockdown cells and the control cells. The levels of tRNAs in the ALKBH1 knockdown cells were normalized to the knockdown controls; the ratios were shown in the heat map where tRNAs were grouped according to the anticodon nucleotides.
(E) The total RNA were extracted from wild-type and Alkbh1−/− MEF cells. The levels of tRNAiMet, tRNAGly(GCC), tRNAVal(mAC), and tRNAHis(GUG) were quantified by northern blots using the radioactively labeled complementary oligos. Slight increases of tRNAGly(GCC), tRNAHis(GUG), and tRNAVal(mAC) were observed accompanied by a ~1.6-fold increase of tRNAiMet in the Alkbh1−/− MEF cells in comparison to the wild-type MEF cells.
(F) The knockdown of ALKBH1 increased tRNAiMet level in the initiating ribosome as well as the cellular level of tRNAiMet compared to the knockdown control (5s rRNA was used as the loading control). The representative result was shown in the left panel. The level of tRNAiMet in the initiating ribosome and the relative ratios of tRNAiMet to 5s rRNA were compared in the ALKBH1 knockdown HeLa cells (red) and control samples (grey).
(G) Polysome profiling suggests increased assembly of 80S monosomes upon ALKBH1 knockdown.
(A) The translation-active pool (> 80S) and translationally inactive pool (< 80S) were separated and then subjected to TBE-Urea gel separation and northern blot. The result revealed that ALKBH1 overexpression reduced the level of tRNAHis(GUG) and tRNAGly(GCC) in the translationally active pool. Error bars represent mean ± s.d., n = 2 (two biological replicates)
(B) The representative images of the northern blot analysis of the translationally active pool (> 80S) and translation-inactive pool (< 80S).
(C) The anti-eEF1α antibody can specifically pull down eEF1α from HeLa cells. Silver staining of a SDS-PAGE gel showing the purity of eEF1α in the IP fraction.
(D) Translation rates in wild-type MEF cells were measured by using flow-cytometry (FACS) analysis, after incorporation of HPG and labeling with Alexa 488 fluorophore. The same gating criteria were used to study other cells in E-G.
(E) The gating strategy is applied to Alkbh1−/− MEF cells
(F) The gating strategy is applied to control HeLa cells
(G) The gating strategy is applied to ALKBH1 stable overexpression HeLa cells. FSC denotes forward scatter, SSC denotes side scatter.
(H) HPG incorporation into wild-type (dark blue) and Alkbh1−/− (red) MEF cells
(I) HPG incorporation into ALKBH1 stable overexpression (magenta) and control (grey) HeLa cells.
(A) Scheme of the reporter assay: the RNA reporter vector encodes firefly luciferase (F-luc) as the primary reporter and Renilla luciferase (R-luc) on the same plasmid as the internal transfection control. The codon usage effect resulted by the ALKBH1-mediated tRNA demethylation was revealed by recording the mutant reporter protein synthesis normalized over the control reporter without mutations.
(B) Mutant reporter 1 contains three mutations, Lys8(CUU to UUU), Lys9(CUU to UUU), Gly10(GCC to CCC), and mutant reporter 2 contains three mutations, Ala64(UGC to CGC), Ala66(AGC to CGC), Lys68(CUU to UUU), at the F-luc region (noted as mut-F-luc); both F-luc mutants led to noticeable decreases of normalized protein synthesis (over the control reporter without mutations) in ALKBH1 knockdown cells versus control cells.
(C) Both mutant reporters also showed increases of normalized protein synthesis (over the control reporter without mutations) under glucose starvation conditions versus glucose rich conditions. The control reporter without mutations was used to normalize the translation differences between the two cells lines. Error bars represent mean ± s.d., n = 8 (four biological replicates × two technical replicates).
(A) Western blot analysis of the levels of 0.3, 0.75, 1.5, and 3 μg ALKBH1.
(B) Western blot analysis of the levels of 0.5, 1, 3, and 10 μg of TRMT6.
(C) Western blot analysis of the levels of 0.5, 1, 3, and 10 μg of TRMT61. The standard curves were obtained by fitting the intensities of the individual proteins as the function of the protein concentrations. Using these standard curves, we determined the levels of ALKBH1, TRMT6, and TRMT61 at 5, 25, and 50 mM of glucose growth conditions. The results revealed the decreased cellular levels of ALKBH1 (~3.8, ~3.2, and ~1.1 μg for the amount of protein applied) with the increase of the concentrations of glucose in the growth medium; however, the levels of TRMT6 and TRMT61 remained almost unchanged at 5, 25, and 50 mM of glucose growth conditions (TRMT6, ~9.2, ~8.8, and ~8.3 μg for the amount of protein applied; and TRMT61, ~6.8, ~6.5, and ~6.6 μg for the amount of protein applied).
m1A methylated human tRNAs are listed. The m1A sites are known to be present in the Modomics database (Machnicka et al., 2013) for human or mammalian tRNAs or detected in (Cozen et al., 2015).
Sample identifications and high-throughput sequencing reads are listed. There are two replicates for CLIP (Control), three replicates for CLIP (ALKBH1), two replicates for tRNA seq of siControl, and two replicates for tRNA seq of siALKBH1.
Highlights.
ALKBH1 catalyzes the demethylation of m1A in tRNA
The m1A demethylation affects the tRNAiMet level and translation initiation
The ALKBH1-mediated tRNA demethylation attenuates translation elongation
Reversible tRNA methylation dynamically regulates translation
Acknowledgments
This work is supported by the National Institute of Health (GM071440 to C.H, and GM113194 to C.H. and T.P.). The Mass Spectrometry Facility of the University of Chicago is funded by National Science Foundation (CHE-1048528). The University of Chicago Cancer Center is supported by National Institute of Health CA014599. C.H. is an investigator of the Howard Hughes Medical Institute. We thank S.F. Reichard for editing the manuscript.
Footnotes
Supplemental Information includes Extended Experimental Procedures, six figures can be found with this article online.
AUTHOR CONTRIBUTION
F. L. and C.H. designed experiments with input from T.P. F. L., Y.F. X. W. and J.W. performed experiments and analyzed data. G. Luo and W.C. processed the high-throughput RNA sequencing and proteomic data with help from D.H. X.Y. W. helped with the CLIP experiment. H.M. helped with the stable line construction and protein mass spectroscopy experiment. A.K. provided the Alkbh1−/− MEF cell line and the control cell line. G.Z. Z.H. Q.D. and M.E. assisted experiments and data analysis. F. L. and C.H. wrote the manuscript with comments from T. P.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aas PA, Otterlei M, Falnes PO, Vagbo CB, Skorpen F, Akbari M, Sundheim O, Bjoras M, Slupphaug G, Seeberg E, et al. Human and bacterial oxidative demethylases repair alkylation damage in both RNA and DNA. Nature. 2003;421:859–863. doi: 10.1038/nature01363. [DOI] [PubMed] [Google Scholar]
- Agris PF. Bringing order to translation: the contributions of transfer RNA anticodon-domain modifications. EMBO reports. 2008;9:629–635. doi: 10.1038/embor.2008.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agris PF, Vendeix FAP, Graham WD. tRNA’s Wobble Decoding of the Genome: 40 Years of Modification. J Mol Biol. 2007;366:1–13. doi: 10.1016/j.jmb.2006.11.046. [DOI] [PubMed] [Google Scholar]
- Anderson J, Phan L, Cuesta R, Carlson BA, Pak M, Asano K, Bjork GR, Tamame M, Hinnebusch AG. The essential Gcd10p-Gcd14p nuclear complex is required for 1-methyladenosine modification and maturation of initiator methionyl-tRNA. Genes Dev. 1998;12:3650–3662. doi: 10.1101/gad.12.23.3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: Target Recognition and Regulatory Functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begley U, Dyavaiah M, Patil A, Rooney JP, DiRenzo D, Young CM, Conklin DS, Zitomer RS, Begley TJ. Trm9-Catalyzed tRNA Modifications Link Translation to the DNA Damage Response. Mol Cell. 2007;28:860–870. doi: 10.1016/j.molcel.2007.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco S, Bandiera R, Popis M, Hussain S, Lombard P, Aleksic J, Sajini A, Tanna H, Cortés-Garrido R, Gkatza N, et al. Stem cell function and stress response are controlled by protein synthesis. Nature. 2016;534:335–340. doi: 10.1038/nature18282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CT, Pang YL, Deng W, Babu IR, Dyavaiah M, Begley TJ, Dedon PC. Reprogramming of tRNA modifications controls the oxidative stress response by codon-biased translation of proteins. Nat Commun. 2012;3:937. doi: 10.1038/ncomms1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cozen AE, Quartley E, Holmes AD, Hrabeta-Robinson E, Phizicky EM, Lowe TM. ARM-seq: AlkB-facilitated RNA methylation sequencing reveals a complex landscape of modified tRNA fragments. Nat Meth. 2015;12:879–884. doi: 10.1038/nmeth.3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker CJ, Parker R. P-Bodies and Stress Granules: Possible Roles in the Control of Translation and mRNA Degradation. CSH Perspect Biol. 2012;4:a012286. doi: 10.1101/cshperspect.a012286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dina C, Meyre D, Gallina S, Durand E, Korner A, Jacobson P, Carlsson LMS, Kiess W, Vatin V, Lecoeur C, et al. Variation in FTO contributes to childhood obesity and severe adult obesity. Nat Genet. 2007;39:724–726. doi: 10.1038/ng2048. [DOI] [PubMed] [Google Scholar]
- Do R, Bailey SD, Desbiens K, Belisle A, Montpetit A, Bouchard C, Pérusse L, Vohl MC, Engert JC. Genetic Variants of FTO Influence Adiposity, Insulin Sensitivity, Leptin Levels, and Resting Metabolic Rate in the Quebec Family Study. Diabetes. 2008;57:1147–1150. doi: 10.2337/db07-1267. [DOI] [PubMed] [Google Scholar]
- Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- Dominissini D, Nachtergaele S, Moshitch-Moshkovitz S, Peer E, Kol N, Ben-Haim MS, Dai Q, Di Segni A, Salmon-Divon M, Clark WC, et al. The dynamic N1-methyladenosine methylome in eukaryotic messenger RNA. Nature. 2016;530:441–446. doi: 10.1038/nature16998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly MI, Zhou M, Millard CS, Clancy S, Stols L, Eschenfeldt WH, Collart FR, Joachimiak A. An expression vector tailored for large-scale, high-throughput purification of recombinant proteins. PERP. 2006;47:446–454. doi: 10.1016/j.pep.2005.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan T, Trewick SC, Koivisto P, Bates PA, Lindahl T, Sedgwick B. Reversal of DNA alkylation damage by two human dioxygenases. Proc Natl Acad Sci USA. 2002;99:16660–16665. doi: 10.1073/pnas.262589799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falnes PO, Johansen RF, Seeberg E. AlkB-mediated oxidative demethylation reverses DNA damage in Escherichia coli. Nature. 2002;419:178–182. doi: 10.1038/nature01048. [DOI] [PubMed] [Google Scholar]
- Fedeles BI, Singh V, Delaney JC, Li D, Essigmann JM. The AlkB Family of Fe(II)/α-Ketoglutarate-dependent Dioxygenases: Repairing Nucleic Acid Alkylation Damage and Beyond. J Biol Chem. 2015;290:20734–20742. doi: 10.1074/jbc.R115.656462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, Lindgren CM, Perry JRB, Elliott KS, Lango H, Rayner NW, et al. A Common Variant in the FTO Gene Is Associated with Body Mass Index and Predisposes to Childhood and Adult Obesity. Science. 2007;316:889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Dominissini D, Rechavi G, He C. Gene expression regulation mediated through reversible m6A RNA methylation. Nat Rev Genet. 2014;15:293–306. doi: 10.1038/nrg3724. [DOI] [PubMed] [Google Scholar]
- Fu Y, Jia G, Pang X, Wang RN, Wang X, Li CJ, Smemo S, Dai Q, Bailey KA, Nobrega MA, et al. FTO-mediated formation of N6-hydroxymethyladenosine and N6-formyladenosine in mammalian RNA. Nat Commun. 2013;4:1798–1805. doi: 10.1038/ncomms2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebauer F, Hentze MW. Molecular mechanisms of translational control. Nat Rev Mol Cell Biol. 2004;5:827–835. doi: 10.1038/nrm1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebauer F, Preiss T, Hentze MW. From Cis-Regulatory Elements to Complex RNPs and Back. CSH Perspect Biol. 2012;4:a012245. doi: 10.1101/cshperspect.a012245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerken T, Girard CA, Tung YCL, Webby CJ, Saudek V, Hewitson KS, Yeo GSH, McDonough MA, Cunliffe S, McNeill LA, et al. The Obesity-Associated FTO Gene Encodes a 2-Oxoglutarate-Dependent Nucleic Acid Demethylase. Science. 2007;318:1469–1472. doi: 10.1126/science.1151710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh A, Vishveshwara S. A study of communication pathways in methionyl-tRNA synthetase by molecular dynamics simulations and structure network analysis. Proc Natl Acad Sci USA. 2007;104:15711–15716. doi: 10.1073/pnas.0704459104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giegé R, Sissler M, Florentz C. Universal rules and idiosyncratic features in tRNA identity. Nucleic Acids Res. 1998;26:5017–5035. doi: 10.1093/nar/26.22.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- He C. Grand Challenge Commentary: RNA epigenetics? Nat Chem Biol. 2010;6:863–865. doi: 10.1038/nchembio.482. [DOI] [PubMed] [Google Scholar]
- Hentze MW, Kulozik AE. A Perfect Message: RNA Surveillance and Nonsense-Mediated Decay. Cell. 1999;96:307–310. doi: 10.1016/s0092-8674(00)80542-5. [DOI] [PubMed] [Google Scholar]
- Hershey JWB, Sonenberg N, Mathews MB. Principles of Translational Control: An Overview. CSH Perspect Biol. 2012;4:a011528. doi: 10.1101/cshperspect.a011528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh Y, Chiba S, Sekine S-i, Yokoyama S. Crystal structure of human selenocysteine tRNA. Nucleic Acids Res. 2009;37:6259–6268. doi: 10.1093/nar/gkp648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the Histone Code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, Yi C, Lindahl T, Pan T, Yang YG, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011;7:885–887. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser S, Rimbach K, Eigenbrod T, Dalpke AH, Helm M. A modified dinucleotide motif specifies tRNA recognition by TLR7. RNA. 2014;20:1351–1355. doi: 10.1261/rna.044024.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose RJ, Zhang Y. Regulation of histone methylation by demethylimination and demethylation. Nat Rev Mol Cell Biol. 2007;8:307–318. doi: 10.1038/nrm2143. [DOI] [PubMed] [Google Scholar]
- Kohli RM, Zhang Y. TET enzymes, TDG and the dynamics of DNA demethylation. Nature. 2013;502:472–479. doi: 10.1038/nature12750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Liu B, Lee S, Huang SX, Shen B, Qian SB. Global mapping of translation initiation sites in mammalian cells at single-nucleotide resolution. Proc Natl Acad Sci USA. 2012;109:E2424–E2432. doi: 10.1073/pnas.1207846109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Xiong X, Wang K, Wang L, Shu X, Ma S, Yi C. Transcriptome-wide mapping reveals reversible and dynamic N1-methyladenosine methylome. Nat Chem Biol. 2016;12:311–316. doi: 10.1038/nchembio.2040. [DOI] [PubMed] [Google Scholar]
- Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, Jia G, Yu M, Lu Z, Deng X, et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol. 2014;10:93–95. doi: 10.1038/nchembio.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Dai Q, Zheng G, He C, Parisien M, Pan T. N6-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature. 2015;518:560–564. doi: 10.1038/nature14234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loenarz C, Schofield CJ. Expanding chemical biology of 2-oxoglutarate oxygenases. Nat Chem Biol. 2008;4:152–156. doi: 10.1038/nchembio0308-152. [DOI] [PubMed] [Google Scholar]
- Macari F, El-houfi Y, Boldina G, Xu H, Khoury-Hanna S, Ollier J, Yazdani L, Zheng G, Bieche I, Legrand N, et al. TRM6/61 connects PKC[alpha] with translational control through tRNAiMet stabilization: impact on tumorigenesis. Oncogene. 2016;35:1785–1796. doi: 10.1038/onc.2015.244. [DOI] [PubMed] [Google Scholar]
- Machnicka MA, Milanowska K, Osman Oglou O, Purta E, Kurkowska M, Olchowik A, Januszewski W, Kalinowski S, Dunin-Horkawicz S, Rother KM, et al. MODOMICS: a database of RNA modification pathways--2013 update. Nucleic Acids Res. 2013;41:D262–D267. doi: 10.1093/nar/gks1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maquat LE. Nonsense-mediated mRNA decay: splicing, translation and mRNP dynamics. Nat Rev Mol Cell Biol. 2004;5:89–99. doi: 10.1038/nrm1310. [DOI] [PubMed] [Google Scholar]
- Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012;149:1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr S, Ghanem E, Smith W, Sheeter D, Qin Y, King O, Polioudakis D, Iyer VR, Hunicke-Smith S, Swamy S, et al. Thermostable group II intron reverse transcriptase fusion proteins and their use in cDNA synthesis and next-generation RNA sequencing. RNA. 2013;19:958–970. doi: 10.1261/rna.039743.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller TA, Meek K, Hausinger RP. Human AlkB homologue 1 (ABH1) exhibits DNA lyase activity at abasic sites. DNA Repair (Amst) 2010;9:58–65. doi: 10.1016/j.dnarep.2009.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi K, Ogiso Y, Nakama T, Fukai S, Nureki O. Structural basis for anticodon recognition by methionyl-tRNA synthetase. Nat Struct Mol Biol. 2005;12:931–932. doi: 10.1038/nsmb988. [DOI] [PubMed] [Google Scholar]
- Nordstrand LM, Svard J, Larsen E, Nilsen A, Ougland R, Furu K, Lien GF, Rognes T, Namekawa SH, Lee JT, et al. Mice lacking Alkbh1 display sex-ratio distortion and unilateral eye defects. PLoS One. 2010;5:e13827. doi: 10.1371/journal.pone.0013827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ougland R, Lando D, Jonson I, Dahl JA, Moen MN, Nordstrand LM, Rognes T, Lee JT, Klungland A, Kouzarides T, et al. ALKBH1 is a histone H2A dioxygenase involved in neural differentiation. Stem Cells. 2012;30:2672–2682. doi: 10.1002/stem.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Z, Sikandar S, Witherspoon M, Dizon D, Nguyen T, Benirschke K, Wiley C, Vrana P, Lipkin SM. Impaired placental trophoblast lineage differentiation in Alkbh1(-/-) mice. Dev Dyn. 2008;237:316–327. doi: 10.1002/dvdy.21418. [DOI] [PubMed] [Google Scholar]
- Pavitt GD, Ron D. New Insights into Translational Regulation in the Endoplasmic Reticulum Unfolded Protein Response. CSH Perspect Biol. 2012;4:a012278. doi: 10.1101/cshperspect.a012278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavon-Eternod M, Gomes S, Rosner MR, Pan T. Overexpression of initiator methionine tRNA leads to global reprogramming of tRNA expression and increased proliferation in human epithelial cells. RNA. 2013;19:461–466. doi: 10.1261/rna.037507.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phizicky EM, Hopper AK. tRNA biology charges to the front. Genes Dev. 2010;24:1832–1860. doi: 10.1101/gad.1956510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ping XL, Sun BF, Wang L, Xiao W, Yang X, Wang WJ, Adhikari S, Shi Y, Lv Y, Chen YS, et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014;24:177–189. doi: 10.1038/cr.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert X, Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42:W320–W324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saikia M, Fu Y, Pavon-Eternod M, He C, Pan T. Genome-wide analysis of N1-methyladenosine modification in human tRNAs. RNA. 2010;16:1317–1327. doi: 10.1261/rna.2057810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Songe-Møller L, van den Born E, Leihne V, Vågbø CB, Kristoffersen T, Krokan HE, Kirpekar F, Falnes PØ, Klungland A. Mammalian ALKBH8 Possesses tRNA Methyltransferase Activity Required for the Biogenesis of Multiple Wobble Uridine Modifications Implicated in Translational Decoding. Mol Cell Biol. 2010;30:1814–1827. doi: 10.1128/MCB.01602-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Suzuki T. A complete landscape of post-transcriptional modifications in mammalian mitochondrial tRNAs. Nucleic Acids Res. 2014;42:7346–7357. doi: 10.1093/nar/gku390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trewick SC, Henshaw TF, Hausinger RP, Lindahl T, Sedgwick B. Oxidative demethylation by Escherichia coli AlkB directly reverts DNA base damage. Nature. 2002;419:174–178. doi: 10.1038/nature00908. [DOI] [PubMed] [Google Scholar]
- Tserovski L, Marchand V, Hauenschild R, Blanloeil-Oillo F, Helm M, Motorin Y. Highthroughput sequencing for 1-methyladenosine (m(1)A) mapping in RNA. Methods. 2016;107:110–121. doi: 10.1016/j.ymeth.2016.02.012. [DOI] [PubMed] [Google Scholar]
- Ule J, Jensen K, Mele A, Darnell RB. CLIP: A method for identifying protein–RNA interaction sites in living cells. Methods. 2005;37:376–386. doi: 10.1016/j.ymeth.2005.07.018. [DOI] [PubMed] [Google Scholar]
- Wang X, Jia H, Jankowsky E, Anderson JT. Degradation of hypomodified tRNAiMet in vivo involves RNA-dependent ATPase activity of the DExH helicase Mtr4p. RNA. 2008;14:107–116. doi: 10.1261/rna.808608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, Fu Y, Parisien M, Dai Q, Jia G, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505:117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zhao Boxuan S, Roundtree Ian A, Lu Z, Han D, Ma H, Weng X, Chen K, Shi H, He C. N6-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell. 2015;161:1388–1399. doi: 10.1016/j.cell.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westbye MP, Feyzi E, Aas PA, Vagbo CB, Talstad VA, Kavli B, Hagen L, Sundheim O, Akbari M, Liabakk NB, et al. Human AlkB homolog 1 is a mitochondrial protein that demethylates 3-methylcytosine in DNA and RNA. J Biol Chem. 2008;283:25046–25056. doi: 10.1074/jbc.M803776200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TP, Wang T, Seetin MG, Lai Y, Zhu S, Lin K, Liu Y, Byrum SD, Mackintosh SG, Zhong M, et al. DNA methylation on N6-adenine in mammalian embryonic stem cells. Nature. 2016;532:329–333. doi: 10.1038/nature17640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao W, Adhikari S, Dahal U, Chen YS, Hao YJ, Sun BF, Sun HY, Li A, Ping XL, Lai WY, et al. Nuclear m6A Reader YTHDC1 Regulates mRNA Splicing. Mol Cell. 2016;61:507–519. doi: 10.1016/j.molcel.2016.01.012. [DOI] [PubMed] [Google Scholar]
- Xu C, Wang X, Liu K, Roundtree IA, Tempel W, Li Y, Lu Z, He C, Min J. Structural basis for selective binding of m6A RNA by the YTHDC1 YTH domain. Nat Chem Biol. 2014;10:927–929. doi: 10.1038/nchembio.1654. [DOI] [PubMed] [Google Scholar]
- Yacoubi BE, Bailly M, Crécy-Lagard Vd. Biosynthesis and Function of Posttranscriptional Modifications of Transfer RNAs. Annu Rev Genet. 2012;46:69–95. doi: 10.1146/annurev-genet-110711-155641. [DOI] [PubMed] [Google Scholar]
- Yue Y, Liu J, He C. RNA N6-methyladenosine methylation in post-transcriptional gene expression regulation. Genes Dev. 2015;29:1343–1355. doi: 10.1101/gad.262766.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G, Dahl JA, Niu YM, Fedorcsak P, Huang CM, Li CJ, Vagbo CB, Shi Y, Wang WL, Song SH, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49:18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G, Qin Y, Clark WC, Dai Q, Yi C, He C, Lambowitz AM, Pan T. Efficient and quantitative high-throughput tRNA sequencing. Nat Meth. 2015;12:835–837. doi: 10.1038/nmeth.3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Wan J, Gao X, Zhang X, Jaffrey SR, Qian SB. Dynamic m6A mRNA methylation directs translational control of heat shock response. Nature. 2015;526:591–594. doi: 10.1038/nature15377. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Pie chart classification of the identified RNA species in the total RNA controls and
(B) ALKBH1-CLIP samples from Figure 1A.
(C) Sequence alignment of human ALKBH1 and human methionine tRNA ligase MSE1. Highly conserved residues were shown in red text and boxed in blue; strictly conserved residues were shown in red background. The tRNA-binding domain was depicted in black box. This figure was prepared by using ESPript (Robert and Gouet, 2014).
(D) The amount of each nucleoside was quantified by its integrated area in the corresponding LC-MS/MS chromatogram. The m1A standard calibration curve was obtained using five standard samples with different concentrations of pure m1A.
(A) The in vitro demethylation by ALKBH1 in total RNA extracted from HeLa cells. Demethylation reactions were performed in the presence (depicted in grey) or absence of EDTA (depicted in red); EDTA chelates cofactor iron and inactivates ALKBH1. The levels of m3C/G, m7G/G, and m5C/G in total tRNA were measured. n.s. represents not significant. Error bars represent mean ± s.d., n = 8 (four biological replicates × two technical replicates).
(B) From left to right: dot-blot analysis of m1A levels in total tRNA of HeLa cells with transient knockdown of ALKBH1 by siALKBH1 and control siRNA; the m1A levels in total tRNA of HeLa cells with transient overexpression of ALKBH1 versus the control (empty vector pcDNA 3.0 transfection); the m1A levels in total tRNA of Alkbh1−/− versus wild-type MEF cells; the m1A levels in total tRNA of HeLa cells with ALKBH1 stable overexpression versus the control (empty vector).
(C) The specificities of m1A and m6A antibodies were tested against ACAUG RNA oligonucleotides; the underlined A was in the form of unmethylated A, m6A, or m1A, respectively.
(D) The levels of m3C/G, m7G/G, and m5C/G in total tRNA were measured in the samples of ALKBH1 knockdown, ALKBH1 overexpression, and relevant controls. The expression changes of ALKBH1 do not appear to induce any noticeable changes on the ratios of m3C/G, m7G/G, and m5C/G in HeLa cells.
(E) The gel image of 32P-labeled total tRNA and tRNA using the tRNAHis(GUG)-specific biotinylated complementary DNA oligo after selection. tRNA array image showing signals in the application of total tRNA (left), tRNAHis(GUG) after selection using the biotin-labeled DNA complementary oligo (middle), and the array layout with the probes for tRNAHis(GUG) indicated in black squares. The result showed that the biotin-labeled DNA complementary oligo to tRNAHis(GUG) was able to specifically isolate only tRNAHis(GUG).
(F) Neither m6A nor m1A level in rRNA was significantly perturbed by the ALKBH1 knockdown or Alkbh1 knockout.
(G) The m6A level in mRNA was not significantly perturbed by changes of ALKBH1. The m1A level in mRNA was slightly altered by changing ALKBH1.
(H) Biochemical demethylation assays of dsDNA probes containing 6mA and 1mA (#1 and #2, sequences are listed in the Supplementary Information), m6A and m1A in linear ssRNA probes (#3, and #4), and m1A in a stem-loop structured probe (#6) using recombinant ALKBH1 in vitro. The reactions were run with 2 nmol ALKBH1 and 100 ng of each probe at 37 °C for 1 h. The demethylation percentage mediated by ALKBH1 on the individual probe was shown. Error bars represent mean ± s.d., n = 4 (four biological replicates).
(I) The plot of the ALKBH1-catalyzed demethylation (%) versus time (min) towards m1A in stem-loop structured RNA probes that mimic the TΨC loops of tRNAiMet and tRNAHis(GUG) and in unstructured RNA probes at pH 7.0 at 37 °C. Error bars represent mean ± s.d., n = 3 (three biological replicates).
(J) The basal level of 6mA in genomic DNA from mouse embryonic stem cells (mESCs) is low. We did not observe noticeable changes of the 6mA level in the Alkbh1 knockout mESCs compared to the wild-type control.
(A) Specific tRNAs were isolated from the Alkbh1−/− and wild-type MEF cells; the extracted tRNAs were then subjected to LC-MS/MS analysis. No significant changes of the m1A level were observed for these tRNA species in cells with Alkbh1−/− versus the wild-type and
(B) ALKBH1 stable overexpression versus control. n.s. represents not significant. Error bars represent mean ± s.d., n = 8 (four biological replicates × two technical replicates).
(C) Demethylation of m1A in tRNAiMet and tRNAeMet(CAU) extracted from HeLa cells by recombinant ALKBH1 in the presence or absence of EDTA. Error bars represent mean ± s.d., n = 4 (four biological replicates).
(D) tRNA sequencing (DM-tRNA-seq) was performed to compare the levels of tRNAs in the ALKBH1 knockdown cells and the control cells. The levels of tRNAs in the ALKBH1 knockdown cells were normalized to the knockdown controls; the ratios were shown in the heat map where tRNAs were grouped according to the anticodon nucleotides.
(E) The total RNA were extracted from wild-type and Alkbh1−/− MEF cells. The levels of tRNAiMet, tRNAGly(GCC), tRNAVal(mAC), and tRNAHis(GUG) were quantified by northern blots using the radioactively labeled complementary oligos. Slight increases of tRNAGly(GCC), tRNAHis(GUG), and tRNAVal(mAC) were observed accompanied by a ~1.6-fold increase of tRNAiMet in the Alkbh1−/− MEF cells in comparison to the wild-type MEF cells.
(F) The knockdown of ALKBH1 increased tRNAiMet level in the initiating ribosome as well as the cellular level of tRNAiMet compared to the knockdown control (5s rRNA was used as the loading control). The representative result was shown in the left panel. The level of tRNAiMet in the initiating ribosome and the relative ratios of tRNAiMet to 5s rRNA were compared in the ALKBH1 knockdown HeLa cells (red) and control samples (grey).
(G) Polysome profiling suggests increased assembly of 80S monosomes upon ALKBH1 knockdown.
(A) The translation-active pool (> 80S) and translationally inactive pool (< 80S) were separated and then subjected to TBE-Urea gel separation and northern blot. The result revealed that ALKBH1 overexpression reduced the level of tRNAHis(GUG) and tRNAGly(GCC) in the translationally active pool. Error bars represent mean ± s.d., n = 2 (two biological replicates)
(B) The representative images of the northern blot analysis of the translationally active pool (> 80S) and translation-inactive pool (< 80S).
(C) The anti-eEF1α antibody can specifically pull down eEF1α from HeLa cells. Silver staining of a SDS-PAGE gel showing the purity of eEF1α in the IP fraction.
(D) Translation rates in wild-type MEF cells were measured by using flow-cytometry (FACS) analysis, after incorporation of HPG and labeling with Alexa 488 fluorophore. The same gating criteria were used to study other cells in E-G.
(E) The gating strategy is applied to Alkbh1−/− MEF cells
(F) The gating strategy is applied to control HeLa cells
(G) The gating strategy is applied to ALKBH1 stable overexpression HeLa cells. FSC denotes forward scatter, SSC denotes side scatter.
(H) HPG incorporation into wild-type (dark blue) and Alkbh1−/− (red) MEF cells
(I) HPG incorporation into ALKBH1 stable overexpression (magenta) and control (grey) HeLa cells.
(A) Scheme of the reporter assay: the RNA reporter vector encodes firefly luciferase (F-luc) as the primary reporter and Renilla luciferase (R-luc) on the same plasmid as the internal transfection control. The codon usage effect resulted by the ALKBH1-mediated tRNA demethylation was revealed by recording the mutant reporter protein synthesis normalized over the control reporter without mutations.
(B) Mutant reporter 1 contains three mutations, Lys8(CUU to UUU), Lys9(CUU to UUU), Gly10(GCC to CCC), and mutant reporter 2 contains three mutations, Ala64(UGC to CGC), Ala66(AGC to CGC), Lys68(CUU to UUU), at the F-luc region (noted as mut-F-luc); both F-luc mutants led to noticeable decreases of normalized protein synthesis (over the control reporter without mutations) in ALKBH1 knockdown cells versus control cells.
(C) Both mutant reporters also showed increases of normalized protein synthesis (over the control reporter without mutations) under glucose starvation conditions versus glucose rich conditions. The control reporter without mutations was used to normalize the translation differences between the two cells lines. Error bars represent mean ± s.d., n = 8 (four biological replicates × two technical replicates).
(A) Western blot analysis of the levels of 0.3, 0.75, 1.5, and 3 μg ALKBH1.
(B) Western blot analysis of the levels of 0.5, 1, 3, and 10 μg of TRMT6.
(C) Western blot analysis of the levels of 0.5, 1, 3, and 10 μg of TRMT61. The standard curves were obtained by fitting the intensities of the individual proteins as the function of the protein concentrations. Using these standard curves, we determined the levels of ALKBH1, TRMT6, and TRMT61 at 5, 25, and 50 mM of glucose growth conditions. The results revealed the decreased cellular levels of ALKBH1 (~3.8, ~3.2, and ~1.1 μg for the amount of protein applied) with the increase of the concentrations of glucose in the growth medium; however, the levels of TRMT6 and TRMT61 remained almost unchanged at 5, 25, and 50 mM of glucose growth conditions (TRMT6, ~9.2, ~8.8, and ~8.3 μg for the amount of protein applied; and TRMT61, ~6.8, ~6.5, and ~6.6 μg for the amount of protein applied).
m1A methylated human tRNAs are listed. The m1A sites are known to be present in the Modomics database (Machnicka et al., 2013) for human or mammalian tRNAs or detected in (Cozen et al., 2015).
Sample identifications and high-throughput sequencing reads are listed. There are two replicates for CLIP (Control), three replicates for CLIP (ALKBH1), two replicates for tRNA seq of siControl, and two replicates for tRNA seq of siALKBH1.