Maintenance of EGFR plasma membrane levels is critical for cell functioning. Scharaw et al. demonstrate that endosomal RNF11 is required for transcriptional up-regulation of COPII components, specifically facilitating EGFR transport in response to its lysosomal degradation after EGF stimulation.
Abstract
Stimulation of cells with epidermal growth factor (EGF) induces internalization and partial degradation of the EGF receptor (EGFR) by the endo-lysosomal pathway. For continuous cell functioning, EGFR plasma membrane levels are maintained by transporting newly synthesized EGFRs to the cell surface. The regulation of this process is largely unknown. In this study, we find that EGF stimulation specifically increases the transport efficiency of newly synthesized EGFRs from the endoplasmic reticulum to the plasma membrane. This coincides with an up-regulation of the inner coat protein complex II (COPII) components SEC23B, SEC24B, and SEC24D, which we show to be specifically required for EGFR transport. Up-regulation of these COPII components requires the transcriptional regulator RNF11, which localizes to early endosomes and appears additionally in the cell nucleus upon continuous EGF stimulation. Collectively, our work identifies a new regulatory mechanism that integrates the degradation and transport of EGFR in order to maintain its physiological levels at the plasma membrane.
Introduction
EGF receptor (EGFR) is a type I receptor tyrosine kinase, which can be bound at its extracellular domain by growth factors such as the EGF. EGF binding triggers dimerization and auto-phosphorylation of the receptor, driving the recruitment of effector proteins and the activation of multiple signaling cascades important for cell proliferation, differentiation, migration, and survival (Schlessinger, 2000; Lemmon and Schlessinger, 2010). EGF binding also causes EGFR internalization into endosomes, from where the receptor is either recycled back to the cell surface or degraded by the endo-lysosomal pathway. Degradation diminishes EGFR signaling until the amount of receptor at the cell surface has been restored by signaling-induced synthesis and subsequent transport (Earp et al., 1986; Roepstorff et al., 2009). Accordingly, EGFR signaling and subsequent cellular responses depend on the amount of EGFR at the cell surface, which is controlled by the rates of EGFR internalization, recycling, degradation, new synthesis, and transport to the cell surface. Deregulations of these control pathways have been implicated in a variety of human carcinomas (Yarden and Sliwkowski, 2001). The mechanism underlying the transport of newly synthesized EGFR to the cell surface after degradation has not yet been determined. In eukaryotic cells, receptor proteins are transported from their site of synthesis in the ER to their site of action by passing multiple steps within the secretory pathway. Receptor recruitment and sorting decisions are already made at discrete ER subdomains called ER exit sites, where the protein complex SEC23/24 of the inner coat protein complex II (COPII) recognizes sorting signals in receptor cytoplasmic domains (Aridor et al., 1998; Kuehn et al., 1998). The proteins of the inner COPII coexist in mammalian cells as multiple paralogues with potentially divergent functions (Jensen and Schekman, 2011; Zanetti et al., 2011). SEC23 has two different paralogues, termed SEC23A and SEC23B, whereas SEC24 has four different paralogues, termed SEC24A–D. Although their precise function is largely unknown, paralogues of SEC24 have been implicated in the selective recruitment of transmembrane proteins into COPII transport vesicles through their interaction with the ER export motifs of different proteins (Farhan et al., 2007; Wendeler et al., 2007; Merte et al., 2010; Sucic et al., 2011). We and others have previously shown a link between EGF ligand stimulation and an adaptation of COPII transport vesicle organization and protein transport through the secretory pathway (Farhan et al., 2010; Simpson et al., 2012; Tillmann et al., 2015). These findings raise the intriguing question of whether EGF ligand stimulation induces the transport of newly synthesized EGFR to the cell surface through changes in the secretory pathway components. Here, we demonstrate that EGF stimulation leads to an up-regulation of the inner COPII paralogues SEC23B, SEC24B, and SEC24D and that this up-regulation requires the transcriptional regulator (TR) RNF11. We further show that these SEC23/SEC24 paralogues are necessary for the specific transport of newly synthesized EGFR from the ER to the cell surface. We propose that this regulatory mechanism is critical for the maintenance of physiological EGFR levels at the plasma membrane after EGF-induced degradation and that abnormal regulation of this process may contribute to uncontrolled proliferation in human carcinomas.
Results
EGF stimulation increases EGFR transport efficiency
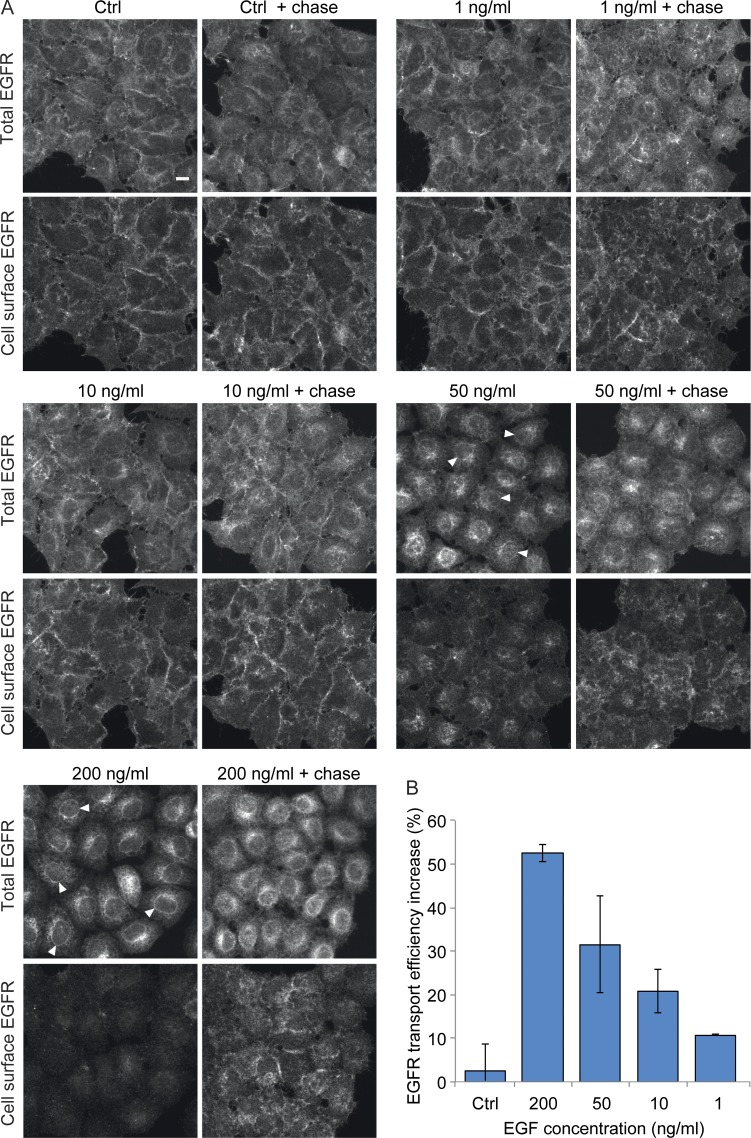
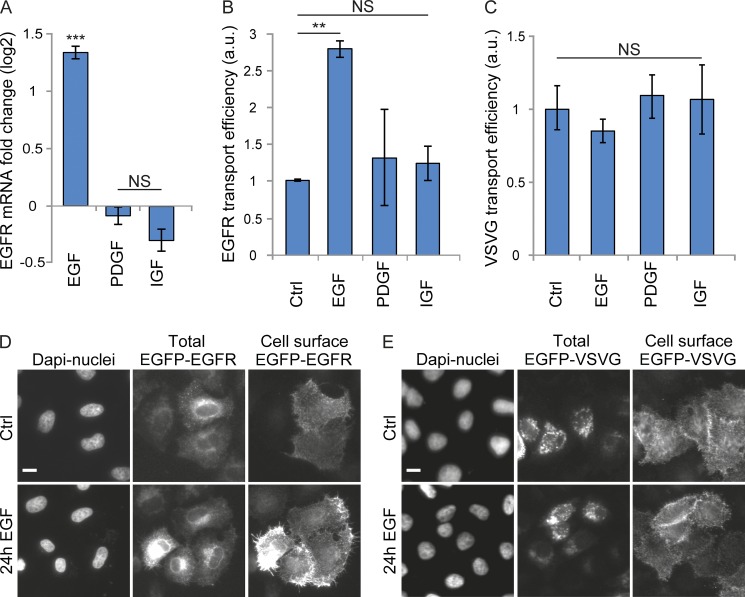
In a previous study, we have shown that stimulation of HeLa cells with high concentrations of EGF can result in up to 80% EGFR degradation within 2 h after stimulation (Laketa et al., 2014), raising the question of how plasma membrane EGFR levels can be maintained under these conditions. We hypothesized that EGFR synthesis and transport efficiency along the secretory pathway would increase upon EGF stimulation in order to restore EGFR levels at the plasma membrane. To test this hypothesis, we first monitored endogenous EGFR localization by immunostaining after continuous stimulation at different EGF concentrations. We found that at low EGF concentrations (1–10 ng/ml), endogenous EGFR localized mostly to the plasma membrane and, to a much lesser extent, to internal structures (Fig. 1 A). In contrast, at higher EGF concentrations (50 ng/ml and 200 ng/ml), the amount of endogenous EGFR at the plasma membrane was strongly reduced, and the majority of EGFR localized to internal structures most likely resembling the ER, as diagnosed by the labeling of reticular structures and of the nuclear envelope (Fig. 1 A and Fig. S1). Time course analysis showed that stimulation with 200 ng/ml EGF for 1 h already results in a significant depletion of EGFR at the plasma membrane (Fig. S1). At this time point, EGFR localized predominantly to internal structures resembling endo-lysosomal structures and, at 6 h or later, to those resembling the ER and the Golgi complex (Fig. S1). Interestingly, when EGF was washed out and cells chased in EGF-free medium for a further 2 h, EGFR plasma membrane levels increased strongly (Fig. 1 A). Quantification of the respective fluorescent signals showed that the ratio of EGFR at the cell surface to total EGFR increased during the chase, consistent with an efficient transport of the EGFR from the ER to the plasma membrane (Fig. 1 B). This increase was EGF concentration dependent and occurred already at 1 ng/ml, although it was most apparent at EGF concentrations of 50 ng/ml (32%) and 200 ng/ml (52%; Fig. 1 B). We next tested whether EGF stimulation–induced plasma membrane EGFR depletion leads to new synthesis of EGFR, accounting for the EGFR observed at the ER. We measured the mRNA level of EGFR after 24 h of continuous EGF stimulation. This revealed a 2.5-fold increase in the EGFR mRNA level compared with nontreated cells (Fig. 2 A). This increase was specific for EGF stimulation, as EGFR expression was unaffected by PDGF or insulin-like growth factor (IGF) stimulation (Fig. 2 A). We sought for a way to investigate EGF stimulation–dependent EGFR transport efficiency toward the plasma membrane after synchronized release from the ER. To this end, we used a retention using selective hooks (RUSH) expression construct encoding for EGFP-tagged EGFR, which allows for the synchronized release of EGFP-EGFR from the ER upon biotin treatment (Boncompain et al., 2012). Continuous 200-ng/ml EGF stimulation for 24 h increased EGFP-EGFR transport efficiency from the ER to the plasma membrane in cells on average by 2.8-fold, whereas PDGF- and IGF-stimulated cells did not show any changes (Fig. 2, B and D). Already, 10 ng/ml EGF stimulation was sufficient to increase EGFP-EGFR transport efficiency by 1.4-fold (Fig. S2 A). In contrast, none of the growth factor stimulations significantly affected vesicular stomatitis virus glycoprotein (VSVG) transport efficiency (Fig. 2, C and E). When we analyzed a possible dependence of EGFP-EGFR transport efficiency on its expression levels, an inhibition in transport efficiency was observed in cells expressing high EGFP-EGFR levels compared with those expressing lower levels (Fig. S2, B and C). However, the transport efficiency increase induced by EGF stimulation in comparison to nontreatment was comparable at different expression level intervals (see example experiment shown in Fig. S2, C–E).
Figure 1.
EGF stimulation increases endogenous EGFR transport efficiency. (A and B) Endogenous EGFR transport efficiency was determined in HeLa cells stimulated for 18 h with 1, 10, 50, or 200 ng/ml EGF. This was followed by a wash out and 2-h chase in starvation medium. (A) Endogenous EGFR protein was localized by confocal microscopy either both intracellularly and at the cell surface (total EGFR) or at the cell surface only. Arrowheads point to the nuclear envelope in example cells. Images are sum projections of z stacks covering the entire cell volume acquired on a confocal laser-scanning microscope. Bar, 10 µm. (B) Quantification of the endogenous EGFR transport efficiency increase percentage in nontreated and stimulated cells calculated as described in Materials and methods. Data are means of two independent biological experiments with a minimum of 10,000 analyzed cells per experiment ± SEM.
Figure 2.
EGF stimulation increases EGFP-EGFR transport efficiency to the plasma membrane. HeLa cells were stimulated for 24 h with either 200 ng/ml EGF, PDGF, or IGF. This was followed by Q-RT-PCR experiments to study the stimulation effect on EGFR mRNA levels and the RUSH transport assay to study the effect on cargo transport efficiency. (A) EGFR log2 mRNA fold change of stimulated cells normalized to nontreated cells and GAPDH mRNA levels. (B and C) Quantification of the EGFP-EGFR (B) and EGFP-VSVG (C) transport efficiency of stimulated cells normalized to nontreated cells. a.u., arbitrary units. (D and E) Representative cells of EGFP-EGFR (D) and EGFP-VSVG (E) transport from the ER to the plasma membrane in nontreated or EGF-stimulated cells. All data are means ± SEM (n = 3; t test: **, P < 0.01; ***, P < 0.001). Images in D and E were acquired on an automated ScanˆR microscope. Bars, 10 µm.
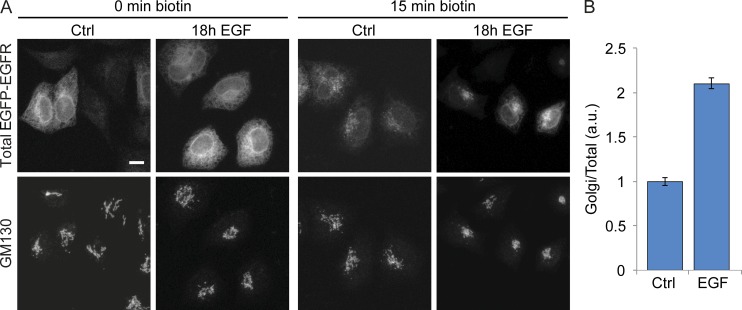
Next, we asked where the EGFP-EGFR transport change between the ER and plasma membrane occurs upon EGF stimulation. To address this, we quantified the arrival of EGFP-EGFR at the Golgi complex (Fig. 3). At 15 min after EGFP-EGFR release from the ER, a significant amount of EGFP-EGFR had already arrived in the Golgi complex, revealed by its colocalization with the Golgi marker GM130 (Fig. 3 A). Quantification showed that EGFP-EGFR localized to the Golgi complex was 2.1-fold higher in EGF-stimulated cells when compared with nonstimulated cells (Fig. 3 B), suggesting that EGF stimulation enhances ER to Golgi transport efficiency. Finally, we compared the EGFP-EGFR transport efficiency of cells exposed to continuous EGF stimulation with those exposed to EGF 10-min pulse stimulation over different times. Continuous EGF stimulation increased EGFP-EGFR transport efficiency with a maximum of 3.7-fold at 18 h, whereas pulse stimulation reached a maximum of 2.2-fold at 12 h (Fig. S2 F).
Figure 3.
EGF stimulation increases EGFP-EGFR transport efficiency to the Golgi complex. HeLa cells were stimulated for 18 h with 200 ng/ml EGF, and the RUSH transport assay was performed to study the effect on EGFP-EGFR transport efficiency from the ER to the Golgi complex. (A) Representative cells of EGFP-EGFR ER to Golgi transport before (left) and 15 min after biotin-induced EGFP-EGFR release from the ER (right). EGFP-EGFR fluorescence intensity at the Golgi complex was investigated using GM130 as a cis-Golgi marker. Images are sum projections of z stacks covering the entire cell volume acquired on a confocal laser-scanning microscope. Bar, 10 µm. (B) Quantification of the EGFP-EGFR transport efficiency to the Golgi complex of stimulated cells normalized to nontreated cells. Data are means of at least 30 cells per condition ± SEM. a.u., arbitrary units.
Altogether, these results show that EGF stimulation specifically increases transport of newly synthesized EGFR to maintain its plasma membrane levels.
EGF stimulation up-regulates the inner COPII paralogues SEC23B, SEC24B, and SEC24D
We next searched for changes in the early secretory pathway that could potentially lead to this specific increase in EGFR transport efficiency upon EGF stimulation. Earlier work suggests that the transcription of COPII genes is regulated in response to increased secretory demands (Shaffer et al., 2004; Abrams and Andrew, 2005; Saito et al., 2009; Fox et al., 2010; Iyer et al., 2013). We therefore tested whether the level of transcription of any COPII components was altered by continuous EGF stimulation in parallel with the increase in EGFR synthesis. We found that the mRNA levels of the inner COPII paralogues SEC23B, SEC24B, and SEC24D were increased upon 200 ng/ml EGF stimulation, with a maximum reached 12 h after stimulation (Fig. 4 A and Fig. S3 A). An increase in SEC23B and SEC24D mRNA levels was also seen at 10 ng/ml EGF (Fig. S3 B). Stimulation of cells with concentrations of EGF <10 ng/ml had no significant effect on the expression of inner COPII paralogues (Fig. S3 B). EGF stimulation also increased the mRNA levels of the COPI components COPD and ARF1 (Fig. 4 A). Stimulation of cells with the growth factors PDGF or IGF had no detectable effect on the mRNA levels of SEC23B, SEC24B, SEC24D, or COPD, whereas the mRNA levels of ARF1 were increased by both EGF and IGF stimulation (Fig. 4 B; unpublished data). Further experiments focused on inner COPII paralogues, as they showed the largest increase in mRNA levels in response to EGF stimulation. Increases in SEC23B and SEC24D mRNA levels upon EGF stimulation were validated in A431 cells (Fig. 4 B). Moreover, A431 cells, which have high EGFR expression levels (Lin et al., 1984), revealed elevated basal expression levels of specifically SEC23B and SEC24D in comparison to HeLa cells (Fig. 4 C). Altogether, these data show that increased EGFR transport efficiency after an extended period of continuous EGF stimulation coincides with elevated mRNA levels of the inner COPII paralogues SEC23B, SEC24B, and SEC24D in parallel to increased EGFR synthesis.
Figure 4.
EGF stimulation up-regulates COPII paralogues in HeLa and A431 cells. HeLa cells and A431 cells were stimulated for 12 or 24 h with 200 ng/ml EGF, PDGF, or IGF. The effect on COP paralogue expression levels was investigated by Q-RT-PCR and targeted proteomics. Q-RT-PCR results were normalized to nontreated cells and GAPDH mRNA levels. Targeted proteomics results were normalized to nontreated cells and prelamin A/C protein levels. (A) Log2 mRNA fold changes of COP paralogues and EGFR in HeLa cells treated for 12 h with EGF (n = 4). (B) Log2 mRNA fold changes of COPII paralogues in A431 cells treated for 24 h with EGF, PDGF, or IGF (n = 6). (C) Basal log2 mRNA fold change expression of COPII paralogues in nontreated A431 cells compared with nontreated HeLa cells (n = 3). Data in A–C are means ± SEM (t test; *, P < 0.05; **, P < 0.01; ***, P < 0.001). (D) Log2 protein abundances of COPII paralogues in HeLa cells treated for 24 h with EGF (n = 3). Data are medians ± median absolute deviation.
If the increase in transport efficiency of newly synthesized EGFR is mediated by SEC23B, SEC24B, and SEC24D, they must be up-regulated not only at the RNA level, but also at the protein level. To provide evidence for this, we used targeted proteomics to quantify the cellular levels of COPII paralogues in response to EGF stimulation (Table S3). Our analysis showed that 24 h of continuous EGF stimulation increased SEC23B and SEC24D protein levels (Fig. 4 D). Furthermore, SEC24A levels were also increased (Fig. 4 D), although they were previously unaffected at the mRNA level by EGF stimulation. The increase in the protein levels of SEC24A could therefore be caused by a posttranslational protein regulation mechanism, leading to, e.g., reduced protein degradation. Unfortunately, we were unable to reliably evaluate SEC24B protein changes by targeted proteomics because of technical limitations.
SEC23B, SEC24B, and SEC24D are required for EGFR transport
Selective cargo protein recognition was previously suggested to depend on specific inner COPII paralogues and their interactions with cargo-specific ER export motifs (Farhan et al., 2007; Wendeler et al., 2007; Bonnon et al., 2010; Merte et al., 2010; Sucic et al., 2011). We therefore tested whether the inner COPII paralogues up-regulated by EGF stimulation could specifically mediate EGFR transport to the plasma membrane. Knockdown of the inner COPII paralogues SEC23B, SEC24B, or SEC24D strongly inhibited EGFP-EGFR transport to levels comparable to brefeldin A (BFA) treatment (Fig. 5 A; Lippincott-Schwartz et al., 1989; Miller et al., 1992). In contrast, knockdown of SEC23A, SEC24A, or SEC24C had no apparent effect on EGFP-EGFR transport. Transport of the EGFP-VSVG was not significantly affected by any knockdown of inner COPII paralogues; only double knockdown of SEC23A and SEC23B resulted in an efficient transport inhibition (Fig. 5 A). In SEC23B or SEC23B and SEC23A double knockdown cells, EGFP-EGFR remained arrested in the ER, as indicated by the nuclear envelope and reticular network localization of the marker transport protein (Fig. 5 B). In contrast, EGFP-EGFR was found at the Golgi complex and the plasma membrane in SEC23A knockdown cells (Fig. 5 B). EGFP-tagged VSVG localized at the Golgi complex and the plasma membrane upon release after both SEC23A and SEC23B knockdowns (Fig. 5 B). Only double knockdown of SEC23A and SEC23B caused EGFP-VSVG to be retained mostly in the ER (Fig. 5 B). Collectively, these data demonstrate that the inner COPII paralogues SEC23B, SEC24B, and SEC24D are required for the transport of EGFR from the ER to the plasma membrane.
Figure 5.
SEC23B, SEC24B, and SEC24D are required for EGFP-EGFR transport. Specificity of inner COPII paralogues for EGFP-EGFR transport efficiency was investigated by performing the RUSH transport assay after 48 h of siRNA-mediated COPII gene knockdown. Results were normalized to nonsilencing negative control siRNA. In control experiments, the RUSH transport assay was performed after 30 min of BFA treatment or after 48 h of simultaneous knockdown of SEC23A and SEC23B. (A) Quantification of the EGFP-EGFR and EGFP-VSVG transport efficiency. Data are means ± SEM (n = 4; t test: *, P < 0.05; **, P < 0.01; ***, P < 0.001). a.u., arbitrary units. (B) Representative cells for EGFP-EGFR and EGFP-VSVG transport from the ER to the plasma membrane after SEC23A, SEC23B, or simultaneous knockdown (KD) of SEC23A and SEC23B. Arrows point to the plasma membrane, arrowheads to the Golgi complex, and dashed arrows to the nuclear envelope. Images have been exposed such that ER staining (on the nuclear envelope) could also be seen. Crosses mark cells without EGFP-EGFR/VSVG expression. Images were acquired on an automated ScanˆR microscope. Bar, 10 µm.
Identification of putative TRs of SEC23B, SEC24B, and SEC24D
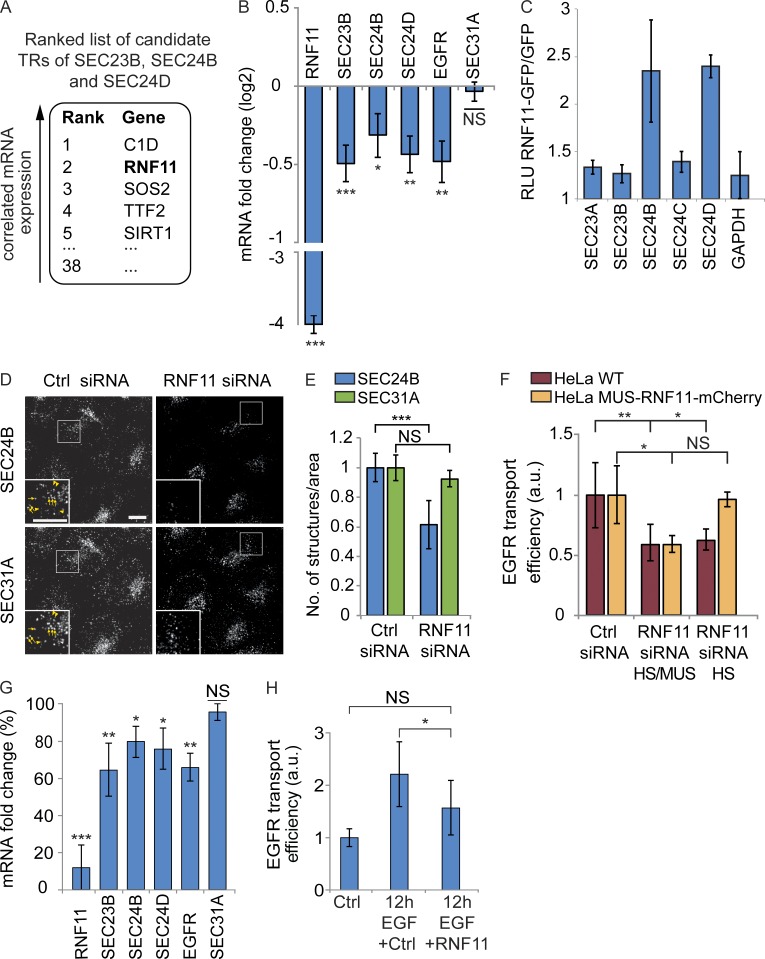
As we observed that EGF stimulation specifically increased SEC23B, SEC24B, and SEC24D mRNA levels, we set out to identify possible TRs mediating the mRNA changes of these COPII paralogues. A genome-wide RNAi screen has previously identified 554 “hit genes” with a role in secretion (Simpson et al., 2012). Using the DNA-binding domain transcription factor prediction database and the Gene Ontology annotation term “DNA binding,” we identified 38 putative TRs among the hit genes of this study (Table S4). These putative TRs could potentially regulate secretion through transcriptional control of secretory pathway genes. To prioritize the list of TR candidates, we performed coexpression analysis and searched for TRs that show similar expression patterns with SEC23B, SEC24B, and SEC24D under various cellular conditions. For this analysis, we used one of the largest publicly available gene expression datasets derived from human cell lines subjected to a wide range of perturbations (Lamb et al., 2006). We hypothesized that functional related TRs that regulate COPII-mediated EGFR sorting and transport are likely to be coexpressed with SEC23B, SEC24B, and SEC24D under different cellular conditions (Stuart et al., 2003; van Noort et al., 2003). Finally, the 38 TRs were ranked based on their coexpression with SEC23B, SEC24B, and SEC24D (see Materials and methods for details; Table S4), which revealed ranked candidate TRs that might participate in the regulation of the mRNA levels of SEC23B, SEC24B, and SEC24D (Fig. 6 A).
Figure 6.
RNF11 is required for the EGF stimulation–induced up-regulations. Coexpression analysis was performed to predict candidate TRs of SEC23B, SEC24B, and SEC24D. To experimentally validate the prediction, Q-RT-PCR, luciferase assay, confocal microscopy, and the RUSH transport assay were performed in HeLa cells. (A) Top five–ranked candidate TRs coexpressed with SEC23B, SEC24B, and SEC24D. (B) Log2 mRNA fold changes of COPII paralogues and EGFR after siRNA-mediated RNF11 knockdown. Results were normalized to nonsilencing negative control siRNA and to GAPDH mRNA levels (n = 8). (C) Luciferase assay was performed with luciferase reporter containing inner COPII or GAPDH gene promotor. The luciferase signal was analyzed as the ratio of relative luminescence units of RNF11-GFP–transfected cells to GFP-transfected cells in at least two independent biological experiments. (D and E) Representative cells (D) and quantification (E) of the RNF11 knockdown effect on the number of SEC24B- and SEC31A-labeled ER exit sites. Arrows in insets point to ER exit sites with SEC24B and SEC31A labeling, and arrowheads point to SEC31A sites with reduced SEC24B labeling. Images are max projections of z stacks covering the entire cell volume acquired on a confocal laser-scanning microscope. Bar, 10 µm. Data are means with a minimum of 100 analyzed cells ± standard deviation. (F) Quantification of the EGFP-EGFR transport efficiency in HeLa WT cells and in HeLa cells stably expressing mouse RNF11-mCherry (MUS-RNF11-mCherry) after knockdown of the human and mouse RNF11 gene (RNF11 siRNA HS/MUS) or of the human RNF11 gene (RNF11 siRNA HS; n = 3). Images for the quantification were acquired on an automated ScanˆR microscope. (G) Log2 mRNA fold changes of COPII paralogues and EGFR after RNF11 knockdown, followed by 12-h 200-ng/ml EGF stimulation (n = 6). Results are represented as the percentage of fold change reductions normalized to the fold changes induced with nonsilencing negative control siRNA, followed by 12-h 200-ng/ml EGF stimulation (100%). (H) Quantification of the EGFR transport efficiency of cells after RNF11 knockdown, followed by 12-h 200-ng/ml EGF stimulation (n = 3). All data, except when stated otherwise, are means ± SEM (t test: *, P < 0.05; **, P < 0.01; ***, P < 0.001). a.u., arbitrary units.
RNF11 regulates SEC23B, SEC24B, and SEC24D expression and EGFR transport efficiency
As the second ranked TR RNF11 (ring finger protein 11; Fig. 6 A) has been implicated in EGFR signaling and degradation (Burger et al., 2006; Chen et al., 2008; Kostaras et al., 2013), we experimentally tested the first two ranked TRs for a possible role in the transcriptional regulation of SEC23B, SEC24B, and SEC24D. Knockdown of the highest ranked TR C1D (C1D nuclear receptor corepressor; Fig. 6 A) had no significant effect on SEC23B, SEC24B, and SEC24D mRNA levels (Fig. S4 A). On the contrary, RNF11 knockdown specifically reduced the mRNA levels of SEC23B, SEC24B, SEC24D, and also of EGFR (Fig. 6 B). Hence, we analyzed the role of RNF11 as a TR of SEC23B, SEC24B, and SEC24D in the context of EGFR signaling in more detail. We found that RNF11-GFP overexpression specifically increased the mRNA levels of SEC23B, SEC24D, and EGFR (Fig. S4 B), although on average only 30% of the cells proved to be transfected. We aimed to independently confirm these results and overexpressed RNF11 together with luciferase reporter constructs monitoring the transcriptional activity of the promotor regions of the respective inner COPII paralogues. This revealed an enhanced transcriptional activity of SEC24B and SEC24D by 2.3- and 2.4-fold, respectively, induced by RNF11 coexpression (Fig. 6 C).
Consistent with the observed RNF11-mediated expression changes of the COPII paralogues, RNF11 knockdown reduced the number of ER exit sites, which were significantly labeled for SEC24B (39% reduction compared with control treated cells; Fig. 6, D and E). RNF11 overexpression instead had the opposite effect and increased the number of ER exit sites significantly labeled for SEC24B (27% increase compared with control cells; Fig. S4 C). In contrast, the total number of SEC31A-positive ER exit sites remained unchanged under RNF11 knockdown or overexpression conditions (Fig. 6, D and E; and Fig. S4 C).
Next, we down-regulated RNF11 and investigated the effect on EGFP-EGFR transport efficiency using the RUSH system. To confirm the specificity of the RNF11 knockdown phenotype, we used both HeLa wild-type (WT) cells and HeLa cells stably expressing mouse mCherry-tagged RNF11 (MUS-RNF11-mCherry). siRNA targeting of both human and mouse RNF11 (RNF11 HS/MUS) caused a 41% reduction of EGFP-EGFR transport efficiency in both HeLa WT and MUS-RNF11-mCherry cells (Fig. 6 F). siRNA targeting of just human RNF11 (RNF11 HS) caused a 38% reduction of EGFP-EGFR transport efficiency in HeLa WT cells. This decrease in transport efficiency was efficiently rescued in HeLa MUS-RNF11-mCherry cells (Fig. 6 F). RNF11 was specifically required for EGFP-EGFR transport, as EGFP-VSVG transport in HeLa WT cells was not affected by RNF11 knockdown under the conditions used here (Fig. S4 D).
Finally, we determined whether RNF11 mediates the increased expression of SEC23B, SEC24B, SEC24D, and EGFR and hence the increase in EGFR transport upon EGF stimulation. We found that RNF11 knockdown led to a significantly smaller mRNA increase after 12-h EGF stimulation compared with control treated cells (Fig. 6 G). Moreover, using the RUSH system, we observed that RNF11 knockdown led to a significant reduction of the EGFP-EGFR transport efficiency increase after 12-h EGF stimulation (Fig. 6 H).
Collectively, these data support the hypothesis that the predicted candidate TR RNF11 is involved in the up-regulation of the inner COPII paralogues SEC23B, SEC24B, and SEC24D in response to EGF stimulation and is required for EGFR transport to the plasma membrane.
RNF11 appears in the nucleus upon EGF stimulation in a PI3K/Akt-dependent manner
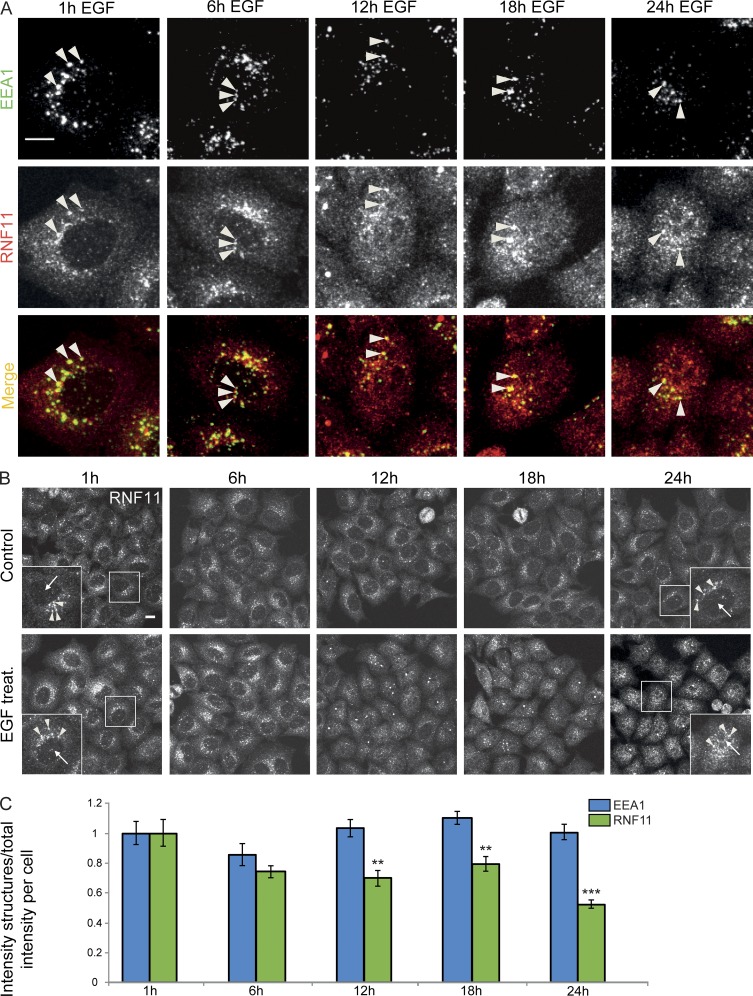
Overexpressed RNF11 localizes on early and late endosomes, where it participates in EGFR trafficking to lysosomes for degradation (Santonico et al., 2010; Kostaras et al., 2013, 2014). In addition, constitutively active serine/threonine-protein kinase Akt was suggested to induce RNF11 localization changes to the nucleus (Connor et al., 2005). As RNF11 had previously also been proposed to bind DNA (Li and Seth, 2004; Azmi and Seth, 2005), we speculated whether RNF11 could directly participate in the transcriptional changes of SEC23B, SEC24B, and SEC24D in the nucleus. Immunostaining showed that in nontreated cells, endogenous RNF11 localized to the cytoplasm and punctate structures, which colocalized with the early endosome marker EEA1 (Fig. 7 A). We found a partial relocalization of RNF11 to the nucleus, starting at 12 h of continuous EGF stimulation (Fig. 7, A and B; and Fig. 8 A). Nuclear RNF11 appearance was EGF concentration dependent and was already observed at 10 ng/ml EGF in 38% of the cells and increased with higher EGF concentrations, with a maximum at EGF concentrations of 200 ng/ml (78% of cells with nuclear RNF11; Fig. 8, A and B). We confirmed the specificity of the endogenous RNF11 antibody staining in the nucleus by siRNA-mediated knockdown of RNF11, which resulted in a strong reduction of the nuclear and cytoplasmic punctate antibody labeling (Fig. S5, A and B). RNF11 intensity in EEA1-positive early endosomes decreased over the time course of EGF stimulation, in parallel with its increase in the nucleus, whereas the intensity of EEA1-positive early endosomes remained unchanged (Fig. 7 C). This finding is consistent with a translocation of the RNF11 protein from early endosomes to the nucleus under continuous EGF stimulation.
Figure 7.
RNF11 appears in the nucleus upon EGF stimulation. Endogenous RNF11 protein localization was investigated by confocal microscopy in HeLa cells treated with 200 ng/ml EGF for various times and compared with nontreated cells. (A) Coimmunostaining of endogenous EEA1 and RNF11 after a time course of EGF stimulation. Arrowheads mark colocalized structures. (B) Endogenous RNF11 localization after a time course of EGF stimulation. Arrowheads mark RNF11 structures on early endosomes, and arrows point to the nucleus area. Images in A and B are max projections of z stacks covering the entire cell volume acquired on a confocal laser-scanning microscope. Bars, 10 µm. (C) Quantification of RNF11/EEA1 punctate structure intensity compared with total RNF11/EEA1 intensity per cell. Data are means ± SEM (n = 3; t test: **, P < 0.01; ***, P < 0.001).
Figure 8.
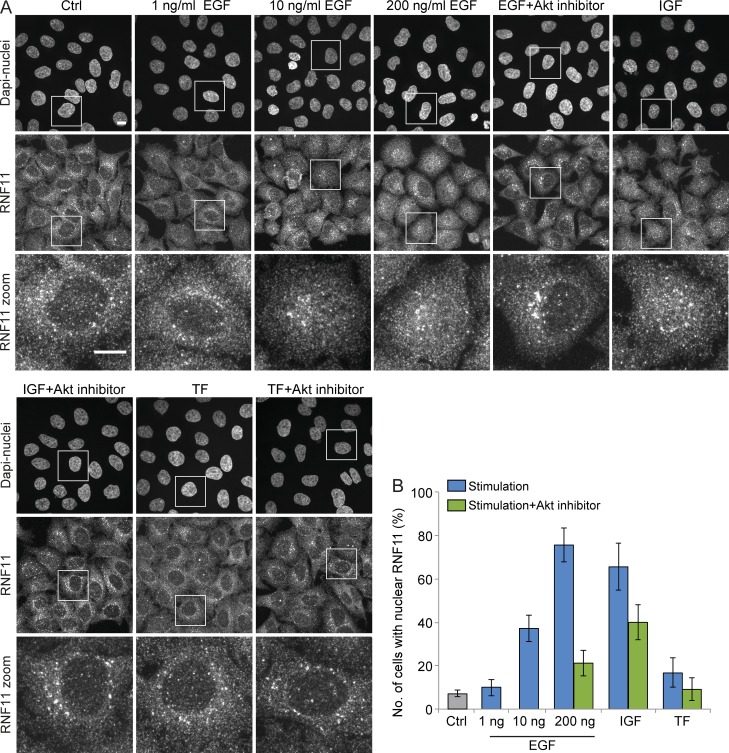
RNF11 relocation is PI3K/Akt dependent. Endogenous RNF11 protein localization was investigated by confocal microscopy in HeLa cells nontreated or stimulated for 12 h with 1, 10, or 200 ng/ml EGF, 200 ng/ml IGF, or 300 µg/ml transferrin (TF). Stimulations were additionally combined with 1 µM PI3K/Akt inhibitor treatment. (A) Staining of the nucleus and immunostaining of endogenous RNF11. Images are max projections of z stacks covering the entire cell volume acquired on a confocal laser-scanning microscope. Bar, 10 µm. (B) Quantification of the percentage of cells with nuclear RNF11 staining. Data are means with at least 200 analyzed cells per condition ± SEM of different fields of view.
To start to gain mechanistic insight into how RNF11 might translocate to the nucleus upon EGF stimulation, we followed quantitatively RNF11 localization in cells stimulated with EGF, IGF, and transferrin and tested the dependence of RNF11 relocation on Akt signaling. We found that IGF treatment, which is known to result in Akt activation, led to a nuclear relocation of RNF11 in a comparable number of cells as EGF stimulation (Fig. 8, A and B). Additional treatment of cells with a PI3K/Akt inhibitor strongly reduced the number of cells with nuclear RNF11 from 76% to 21% after EGF stimulation and from 66% to 40% after IGF stimulation, in agreement with the hypothesis that RNF11 relocation to the nucleus induced by EGF or IGF stimulation requires PI3K/Akt signaling. Stimulation of cells with transferrin had no apparent effect on RNF11 localization, and nuclear RNF11 was observed in <10% of the cells, similar to nontreated cells (Fig. 8 B).
Discussion
EGF stimulation of cells results in EGFR internalization, followed by its significant degradation via the endo-lysosomal pathway. This raises the question of how EGFR levels at the cell surface are maintained under situations of continuous EGF stimulation. In this study, we show that continuous EGF treatment of cells results in a depletion of the EGFR at the cell surface in an EGF concentration–dependent manner and accumulation of newly synthesized EGFR in internal membranes such as the ER and Golgi complex. Under these conditions, EGFR biosynthesis is increased two- to threefold coincident with an enhancement of EGFR transport from the ER to the cell surface. This is accompanied by RNF11-dependent up-regulation of the inner COPII paralogues SEC23B, SEC24B, and SEC24D, which themselves are specifically required for EGFR plasma membrane transport. Altogether, our work identifies a new mechanism to maintain cell surface EGFR homeostasis after receptor degradation.
Our observation that EGF stimulation induces new synthesis of EGFR in a concentration-dependent manner is in agreement with earlier work (Earp et al., 1986; Roepstorff et al., 2009). The identified increase in EGFR transport to the cell surface, as found for the first time in this study, was also EGF concentration dependent and was stronger after continuous EGF stimulation in comparison to a short stimulation pulse. Our localization experiments monitoring endogenous EGFR provide evidence that this enhanced EGFR transport is most likely caused by persistent EGFR internalization from the plasma membrane, followed by its degradation and an increased requirement to transport newly synthesized EGFR to the plasma membrane, situations that we on purpose achieved using high EGF concentrations over a long time period of continuous stimulation. We were able to measure increased EGFR transport efficiency from the ER to the plasma membrane upon EGF stimulation either by monitoring endogenous EGFR during a starvation-medium chase after EGF stimulation or alternatively by quantifying the transport of an EGFP-EGFR RUSH transport marker that can be retained in the ER and released from it by biotin addition, thereby allowing its synchronized transport through the secretory pathway. This synchronized transport of EGFP-EGFR RUSH revealed that transport after EGF stimulation occurs at the ER/Golgi boundary, as the marker arrived more efficiently in the Golgi complex compared with control cells after a short period of release from the ER. The increased EGFR transport efficiency was observed only at extended periods of time after EGF stimulation, indicating that the transport efficiency increase observed here is caused by long-term cellular adaptations, e.g., by transcriptional changes of transport machinery, rather than by posttranslational modifications of transport machinery by rapid signaling as previously described (Palmer et al., 2005; Farhan et al., 2010; Sharpe et al., 2011). In line with this notion, we found that mRNA levels of SEC23B, SEC24B, and SEC24D, components of the inner COPII complex that mediates ER exit, increased in parallel to new EGFR synthesis upon long-term EGF stimulation. The observed EGF stimulation–induced up-regulations of COPII components were subtle but appear to be sufficient to help to maintain EGFR levels at the plasma membrane under continuous EGF stimulation. In line with this view, other fundamental biological processes have also been shown to be regulated by subtle transcript alterations (Hughes et al., 2000; Mootha et al., 2003; Ori et al., 2015).
Based on our data, at least two different explanations for the dependence of EGFR transport on RNF11 function exist. First, upon EGF-induced EGFR degradation, an RNF11-independent mechanism leads to the up-regulation of SEC23B, SEC24B, and SEC24D, and hence to adapted EGFR transport. The RNF11 requirement in this case might arise from its role on early endosomes (Kostaras et al., 2013, 2014), where its levels are critical to balance between EGFR degradation and recycling (Kostaras et al., 2013). Alternatively, a more intriguing possibility is that RNF11 itself transmits the signal of EGFR degradation from early endosomes into the nucleus, where it directly or indirectly participates in the transcriptional regulation of SEC23B, SEC24B, and SEC24D. Although we cannot exclude the first scenario, the latter is supported by several of our observations. First, after EGF stimulation, the fraction of RNF11 protein on early endosomes is reduced, and RNF11 appears in the nucleus. Second, the first appearance of RNF11 in the nucleus at 12 h after EGF stimulation is in line with the time point of increased SEC23B, SEC24B, and SEC24D mRNA levels and enhanced EGFR transport, consistent with the hypothesis that nuclear RNF11 is required for the transcriptional changes of these COPII paralogues. In further support of this scenario, RNF11 has been shown to bind DNA (Li and Seth, 2004; Azmi and Seth, 2005), and we show that its overexpression results in an increase in the efficiency of luciferase-based SEC24B and SEC24D reporters measuring the promotor activity of the respective genes. Interestingly, the up-regulation of SEC23B by EGF stimulation appears to follow a different mechanism. Although we find that RNF11 is sufficient to mediate mRNA changes of SEC23B, it was not sufficient to increase the efficiency of the SEC23B-specific luciferase reporter. A possible mechanism for SEC23B up-regulation by EGF stimulation could be by mRNA stabilization mechanisms involving RNF11. For a transcriptional up-regulation of SEC23B, additional factors to RNF11 would be required.
In our experiments, we have started to explore how RNF11 relocates to the nucleus mechanistically, and we found that RNF11 nuclear appearance was achieved not only by EGF but also by IGF stimulation. In both cases, RNF11 nuclear appearance was inhibited by a PI3K/Akt inhibitor, consistent with earlier findings showing that EGF or IGF is able to trigger PI3K/Akt signaling efficiently (Vivanco and Sawyers, 2002; Ma et al., 2015). These results were surprising because IGF stimulation had no effect on EGFR transport or COPII component expression levels, both shown here to require RNF11. One explanation for these results could be that IGF-induced nuclear RNF11 is not sufficient to mediate the up-regulation of SEC23/SEC24 components on its own, and factors additional to RNF11 are necessary to mediate the EGF-stimulated up-regulation of SEC23/24 and EGFR transport. The existence of such factors might also explain the significant but rather moderate effects of RNF11 knockdown or overexpression on SEC23/24 mRNA levels.
We show that EGF stimulation up-regulated, in an RNF11-dependent manner, specifically SEC23B, SEC24B, and SEC24D, but not other COPII paralogues, excluding a general increase in expression of secretory transport machinery upon EGF stimulation. We find that SEC23B, SEC24B, and SEC24D are specifically required for the transport of EGFR and show with the example of SEC24B that RNF11 knockdown reduces the number of ER exit sites containing significant amounts of SEC24B, whereas the number of ER exit sites containing the outer COPII component SEC31 remains unaffected. This suggests that RNF11 perturbation does not affect the formation of functional COPII vesicles as such, but rather reduces the amounts of specific COPII components required for EGFR transport. The identification of paralogue-specific EGFR transport strongly supports the proposed role of inner COPII paralogues in cargo-specific transport signal recognition and subsequent cargo-specific ER export (Wendeler et al., 2007; Bonnon et al., 2010; Jensen and Schekman, 2011; Zanetti et al., 2011). Whereas it is tempting to speculate that this is a general mechanism for the transport of different cargo proteins, we show that inner COPII paralogues exhibit redundant functions for the transport of VSVG, as well as other cargoes such as TNF (unpublished data). This suggests the coexistence of cargo proteins that require specific paralogues for their transport and cargo proteins that are transported by one or another paralogue.
EGF concentrations as low as 10 ng/ml also induced RNF11 translocation to the nucleus, increased mRNA levels of SEC23/24 genes, and increased EGFR transport efficiency, suggesting that these effects are EGF specific and are not induced by, e.g., cellular stress or unfolded protein response, as they may occur under high EGF concentrations (e.g., 200 ng/ml). EGF concentration in the serum has been mostly reported to occur between 0.2 and 2 ng/ml (Joh et al., 1986; Nexø et al., 1992; Marquèze-Pouey et al., 2014). At these low EGF concentrations, the effects described here could not be detected in our experimental setups. However, in this context, it is worth noting that growth factor concentrations in tumors can become excessively high locally compared with plasma concentrations (Stockhammer et al., 2000; Salmaggi et al., 2003). This has not been shown for EGF. However we believe that it is not unreasonable to speculate that EGF concentrations also could be excessively high locally in tumors. In this case, the mechanism of continuous EGFR degradation from the plasma membrane described here, subsequent increased EGFR synthesis, and EGFR transport will be extremely interesting and of high pathophysiological relevance. Along this line, RNF11 was found to be overexpressed in breast and prostate cancer (Kitching et al., 2003; Subramaniam et al., 2003). We show that RNF11 overexpression is sufficient to increase the expression of EGFR and of those COPII paralogues required for EGFR transport to the plasma membrane. Whether abnormal EGFR cell surface levels linked to RNF11 overexpression could play a critical role in cancer will be an interesting question for future research.
Materials and methods
Cell lines and reagents
HeLa Kyoto cells, A431 cells, and HeLa Kyoto cells stably expressing mouse RNF11-mCherry-LAP (MCB_7996) in a bacterial artificial chromosome were cultured in DMEM (Thermo Fisher Scientific) supplemented with 10% FCS (PAA Laboratories GmbH) and 1% l-glutamine (Sigma-Aldrich) at 37°C in 5% CO2. HeLa Kyoto cells stably expressing mouse RNF11-mCherry-LAP were generated as previously described (Poser et al., 2008). Experiments were performed 24 h after seeding the cells. Antibodies were used as follows: rabbit RNF11 (HPA045781; used at a 1:43 dilution; Atlas Antibodies), mouse GFP (11814460001; clones 7.1 and 13.1, used at a 1:100 dilution; Roche), mouse SEC31A (612351; clone 32, used at a 1:50 dilution; BD), mouse EEA1 (610457; clone 14, used at a 1:100 dilution; BD), rabbit SEC24B (12042; used at a 1:100 dilution; Cell Signaling Technology), mouse anti-VG (used at a 1:100 dilution; gift from T. Kreis, University of Geneva, Geneva, Switzerland), mouse EGFR detecting an extracellular EGFR domain (sc-101; clone R-1, used at a 1:250 dilution; Santa Cruz Biotechnology, Inc.), and rabbit EGFR (sc-03; clone 1005, used at a 1:500 dilution; Santa Cruz Biotechnology, Inc.). All goat Alexa Fluor–conjugated secondary antibodies were purchased from Thermo Fisher Scientific or Molecular Probes (A21236 and A11001 were used at a 1:200 dilution and A11011 and A11008 at a 1:400 dilution). The following chemicals were purchased from Sigma-Aldrich: DAPI, d-biotin, EGF, PDGF-BB, IGF-I, apo-transferrin, and BFA. PI3K/Akt inhibitor (Fan et al., 2006) was obtained from Selleckchem.
Growth factor experiments
HeLa Kyoto cells were grown in 6-well culture dishes (Thermo Fisher Scientific) for 24 h in complete medium at 37°C in 5% CO2. Thereafter, the medium was exchanged to medium without FCS for 24 h. HeLa cells were then treated with 1, 10, 50, 100, or 200 ng/ml EGF, 200 ng/ml PDGF-BB or IGF-I, or 300 µg/ml apo-transferrin for times as stated (control cells remained nontreated). After this, cells were used for RUSH transport assay, endogenous EGFR transport assay, quantitative real-time-PCR (Q-RT-PCR), targeted proteomics, or immunostaining experiments.
RNAi experiments
For RNAi experiments, HeLa Kyoto cells were grown in 6-well culture dishes for 24 h in complete medium at 37°C in 5% CO2. Cells were transfected with the SilencerSelect siRNAs (Thermo Fisher Scientific) listed in Table S1 using Lipofectamine 2000 transfection reagent (Invitrogen) according to the manufacturer’s instructions. Final siRNA concentrations were 15 nM for all siRNAs. For Q-RT-PCR or immunostaining experiments, cells were lysed or fixed after 48-h siRNA transfection. For siRNA experiments followed by the RUSH transport assay (see RUSH transport assay section), cells were transfected with RUSH constructs 24 h after siRNA transfection and then incubated for another 24 h. For siRNA experiments followed by growth factor stimulation (see the previous section), after 24 h of siRNA transfection, cells were treated with medium without FCS for 24 h and then growth factor stimulated for times as stated. If the RUSH assay (see RUSH transport assay section) was additionally performed, cells were transfected with RUSH constructs 24 h before growth factor treatment was over and incubated for 24 h.
GFP-cDNA overexpression
HeLa Kyoto cells were grown in 6-well culture dishes for 24 h in complete medium at 37°C in 5% CO2. Afterward, cells were transfected with a human RNF11 cDNA clone carboxy-terminally linked to turboGFP (pCMV6-AC-GFP; OriGene) using FuGENE6 (Roche) according to the manufacturer’s instructions. 24 h after transfection, cells were lysed and used for Q-RT-PCR experiments or fixed and immunostained.
Q-RT-PCR
For the analysis of mRNA levels, cells were grown in 6-well culture dishes for 24 h in complete medium at 37°C in 5% CO2. Cells were lysed, and total RNA was extracted using the InviTrap Spin Universal RNA Mini kit (Stratec Molecular). cDNA was obtained using the SuperScript III First-Strand Synthesis SuperMix (Invitrogen), all according to the manufacturer’s instructions. The Q-RT-PCR was performed in internal triplicates with the specific and efficient (efficiency between 90 and 110%) primer pairs (Sigma-Aldrich) listed in Table S2. The Q-RT-PCR reaction was performed using the SYBR green detection reagent (Applied Biosystems) in StepOne Real-Time PCR System machines (Applied Biosystems) using the StepOne software v2.3, all according to the manufacturer’s instructions with standard Q-RT-PCR cycling conditions. For each biological condition, the fold change (2−ΔΔCT) of target mRNA was normalized to that of GAPDH.
Endogenous EGFR transport assay
HeLa Kyoto cells were grown on 15-mm glass coverslips in 6-well culture dishes for 24 h in complete medium at 37°C in 5% CO2. The medium was exchanged to medium without FCS for another 24 h. Cells were stimulated with various concentrations of EGF for 18 h. Thereafter, cells were fixed and stained or washed and incubated for a further 2 h in starvation medium before fixation and staining. Cell surface EGFR was detected using a primary mouse anti-EGFR antibody recognizing an extracellular EGFR domain and a secondary Alexa Fluor 647 anti–mouse antibody. This was followed by cell permeabilization and total EGFR detection using a primary rabbit anti-EGFR antibody and a secondary Alexa Fluor 488 anti–rabbit antibody. DAPI staining (0.2-µg/ml final concentration) was performed to highlight the nucleus. Images of the nucleus (DAPI), total EGFR (A488), and plasma membrane EGFR (A647) were acquired on an automated ScanˆR microscope (Olympus) with an Orca R2 camera (Hamamatsu Photonics) and a UPlanAPO 20×/0.7 NA air objective using the software ScanR 2.1.0.15. 100 positions per coverslip were imaged with roughly 100 cells per position, which led to at least 10,000 imaged cells per condition. The transport efficiency to the plasma membrane was determined as the ratio of cell surface EGFR fluorescence (A647 signal intensity) to total EGFR fluorescence (A488 signal intensity) as previously described (Simpson et al., 2007, 2012). The mean transport efficiency was calculated from individual cells for each condition, and the EGFR transport efficiency increase was calculated by the following formula: (Ratio (2h) − Ratio (0h))/Ratio (0h).
RUSH transport assay
HeLa Kyoto cells were grown on 15-mm glass coverslips in 6-well culture dishes for 24 h in complete medium at 37°C in 5% CO2. Thereafter, cells were transfected with RUSH cDNA expression constructs encoding for EGFP-tagged EGFR or VSVG using Lipofectamine LTX (Thermo Fisher Scientific), according to the manufacturer’s instructions. The VSVG RUSH construct was cloned as previously described (Boncompain et al., 2012). The RUSH expression construct for EGFR contained the ER hook streptavidin-KDEL. Human EGFR gene excluding its endogenous signal peptide was purchased as a synthetic gene. To generate the RUSH plasmid Str-KDEL_SBP-EGFP-EGFR, human EGFR fragment was inserted downstream of the IL2 signal peptide-SBP-EGFP cassette using the FseI and PacI restriction sites. Endocytosis of SBP-EGFP-EGFR after exposure to 50 ng/ml human EGF of HeLa cells expressing RUSH EGFR in the presence of biotin was observed, confirming that the RUSH EGFR construct is functional. RUSH constructs were expressed for 24 h. Thereafter, release of the RUSH constructs was induced by the addition of biotin for 15 min (for ER to Golgi transport) or 45 min (for ER to plasma membrane transport) at a final concentration of 40 µM in the culture medium as previously described (Boncompain et al., 2012). The release was additionally performed in the presence of 100 µg/ml cycloheximide to stop new protein synthesis. In parallel, a control sample to monitor leakiness of the RUSH construct–encoded cargo protein before the release was left for each condition without biotin (no release). After the release, cells were fixed and immunostained as previously described (Boncompain and Perez, 2014). In brief, primary antibody staining was performed using mouse anti-GFP antibody for EGFP-EGFR or mouse anti-VG antibody for EGFP-VSVG and secondary antibody staining using Alexa Fluor 647 anti–mouse antibody and DAPI staining to highlight the nucleus. When the RUSH assay was performed after siRNA-mediated gene knockdown, cell lysates were additionally taken from the 6-well culture dishes, and Q-RT-PCR experiments were performed to confirm the siRNA-mediated mRNA reduction of the investigated genes. Only samples with at least 80% mRNA reduction were used for immunostaining. Images of the nucleus (DAPI), total EGFP-EGFR/EGFP-VSVG (A488), and plasma membrane EGFP-EGFR/EGFP-VSVG (A647) were acquired on an automated ScanˆR microscope with an Orca R2 camera and UPlanAPO 20×/0.7 NA air objective using the software ScanR 2.1.0.15. 100 positions per coverslip were imaged with roughly 100 cells per position, which led to at least 10,000 imaged cells per condition. Images were analyzed automatically using CellProfiler version 2.0.11047. In brief, based on predefined size and shape parameters, stained nuclei were detected. Subsequently, the borders of single cells were estimated based on digital dilation of the nuclei masks. A488 and A647 fluorescence signal intensities were measured within the single cell areas. The transport efficiency to the plasma membrane was determined as the ratio of cell surface EGFP-EGFR/EGFP-VSVG fluorescence (A647 signal intensity) to total EGFP-EGFR/EGFP-VSVG fluorescence (A488 signal intensity) as previously described (Simpson et al., 2007, 2012). For each condition, the mean transport ratio was calculated from individual cells and corrected to the mean transport ratio of the cells at no release. Only cells expressing EGFP-EGFR/EGFP-VSVG (transfected) were analyzed, which was defined by using the total values (A488 signal intensity) of cells without EGFP-EGFR/EGFP-VSVG expression (untransfected) as a threshold.
Targeted proteomics
Targeted proteomics assays for COPII components were developed as previously described (Ori et al., 2014). In brief, proteotypic peptides for the COPII components SEC23A, SEC23B, SEC24A, SEC24B, SEC24C, SEC24D, and SEC31A were selected from an in-house build spectral library generated from HeLa Kyoto total cell lysate. Isotopically labeled versions of the selected peptides were synthetized (JPT Technologies) and used for assay development and as spike-in internal standards. Two proteotypic peptides were used for relative protein quantification across samples, with the exception of SEC31A, for which four proteotypic peptides were used. Two additional peptides derived from prelamin A/C were included and used to normalize peptide ratios across samples (i.e., to account for variations in the amount of internal standard peptides that were spiked in). Control (nontreated) and 24-h EGF-stimulated cells were lysed and digested into peptides as described previously (Ori et al., 2014). After protein digestion, desalted peptides were spiked in with a pool of synthetic peptides for COPII components and iRT-kit (Biognosys AG) for retention time calibration (Escher et al., 2012). Assays for both the endogenous (light) and reference (heavy) peptides were recorded in schedule mode using a triple quadrupole mass spectrometer (TSQ Vantage) connected to a nanoAcquity ultra performance liquid chromatography (UPLC) system (Waters). Digested peptides were separated on a BEH300 C18 (75 µm × 250 mm, 1.7 µm) nanoAcquity UPLC column with a 75-min linear gradient between 3 and 35% (vol/vol) acetonitrile in 0.1% (vol/vol) formic acid at a flow rate of 300 nl/min. Data were recorded using scheduled acquisition with a fixed cycle time of 2.5 s, and the number of coeluting transitions was limited to 100 per cycle. Data acquisition was performed using 4-min-long time windows centered around the approximate retention time of targets relative to the iRT-kit. Peptide ratios were estimated using the summed intensity of all transitions and normalized to the mean ratio of the prelamin A/C peptides across samples. Protein fold changes were estimated using linear mixed-effect models (as implemented in R package ‘nlme;’ version 3.1–103) as described previously by Ori et al. (2013). Assay refinement, recalibrations, and data analysis were performed using SpectroDive (Biognosys AG).
Coexpression analysis
For coexpression analysis, the Connectivity Map resource was used, which contains 6,100 gene expression profiles from four human cancer cell lines treated with more than a thousand drug-like molecules (Lamb et al., 2006). To minimize batch effect, normalization and filtering steps were performed as described previously (Iskar et al., 2010). 990 drug-induced gene expression profiles from the human cancer cell lines HL60, MCF7, and PC3 were analyzed (see supplemental data in Iskar et al., 2013). In each cell line, probe sets were filtered if not called “present” in >50% of the experiments. For genes represented by multiple probe sets, the probe set with the highest variance (ranked highest in mean across three cell lines) was selected as the representative of the gene. Large-scale coexpression analysis was performed on all 554 secretion screen “hit genes” (Simpson et al., 2012) that are known or predicted to be transcription regulators, as determined by their presence in the DNA-binding domain: Transcription factor prediction database (Wilson et al., 2007) or their annotation with the GO term “DNA-binding activity” (GO:0003677). The list of TRs was limited to available probe sets that were measured in the Connectivity Map resource by the HT_HG-U133A microarray platform. In total, from the 554 hit genes, 38 TRs detected as “present” in at least two out of three cell lines were retained for further analysis. Drug-induced gene expression profiles from three cell lines were used to calculate all the pairwise Pearson’s correlations between TRs and the SEC23B, SEC24B, and SEC24D COPII components. For each TR, an average coexpression similarity was calculated in each cell line by averaging their Pearson correlation. Next, we used the nonparametric rank product (RankProd) method to assess the significance of coexpression similarity (Hong et al., 2006). RankProd combines the ranked lists of coexpression values from multiple cell lines to identify TRs significantly coexpressed with the COPII components SEC23B, SEC24B, and SEC24D. Finally, all the TRs were ranked based on their corrected p-value (estimated percentage of false prediction) in the order of coexpression from high to low.
Confocal microscopy and segmentation analysis
Confocal microscopy experiments were performed with fixed and immunostained cell samples on a microscope (LSM780; ZEISS) with a Plan Apochromat 63×/1.4 NA oil differential interference contrast objective and the ZEN (black edition) 2012 SP1 8.1 software (ZEISS). DAPI was detected by using a 405-nm laser with a 407–489-nm emission filter; a 488/561-nm laser with a 497–558-nm emission filter was used for Alexa Fluor 488–labeled proteins; a 488/561-nm laser with a 559–655-nm emission filter was used for Alexa Fluor 568–labeled proteins; and a 633-nm laser with a 640–715-nm emission filter was used for Alexa Fluor 647–labeled proteins. Z stacks of images covering the entire cell thickness were acquired. Using ImageJ version 1.46r (National Institutes of Health), the stacks were projected (maximum or sum intensity), the background was subtracted, and the images were smoothed. To determine the number of SEC24B/SEC31A structures per cell area, cells were manually segmented, and SEC24B/SEC31A structures were determined by thresholding. Only the peripheral structures were considered, where the space between the punctae allowed the segmentation of individual structures. The total number of detected structures per cell was then divided by the total cell area. For RNF11/EEA1 intensity measurements, cells were manually segmented, and RNF11/EEA1 structures were determined by thresholding. The fluorescence intensity of detected RNF11/EEA1 punctate structures per cell was divided by the total RNF11/EEA1 intensity per segmented cell.
Luciferase assay
HeLa Kyoto cells were grown in 96-well plates for 24 h in complete medium at 37°C in 5% CO2. Luciferase assays were performed with the following promoter luminescent reporter gene constructs: SEC23A, SEC23B, SEC24B, SEC24C, SEC24D, and GAPDH (Active Motif). All promotor constructs were obtained from the Active Motif LightSwitch Promoter Reporter GoClone Collection (Trinklein et al., 2003). Cells were cotransfected with 50 ng GoClone plasmid DNA per well and with 10 ng of either turboGFP-tagged RNF11 or GFP empty vector according to the manufacturer’s instructions (Active Motif). Plasmid DNA expression time was 24 h for all experiments. Luciferase reporter signal was measured with the LightSwitch Luciferase Assay System (Active Motif) on a SpectraMax L luminometer according to the manufacturer’s instructions. Relative luminescence units of GFP-expressing cells were normalized to RNF11-GFP–expressing cells. To achieve this, in parallel to the 96-well plate for the luciferase assay, a 96-well glass-bottom plate was assessed on a standard wide-field fluorescence microscope for the transfection efficiency of cells with GFP or RNF11-GFP in each well. Relative luminescence units measured in the wells of GFP-expressing cells were then corrected for the higher expression efficiency (on average 1.5-fold higher) compared with RNF11-GFP–expressing cells.
Statistical analysis
All data were analyzed for significance using a Welch’s t test. Effects were compared with nontreated samples or samples treated with nonsilencing negative control siRNA and were considered as significant if the p-values were *, P < 0.05; **, P < 0.01; or ***, P < 0.001.
Online supplemental material
Fig. S1 illustrates endogenous EGFR protein localizations induced by EGF stimulation for various times. Fig. S2 illustrates EGF stimulation–induced EGFP-EGFR transport efficiency changes. Fig. S3 demonstrates the mRNA level changes of inner COPII components and EGFR caused by EGF stimulation. Fig. S4 shows the effect of C1D knockdown and RNF11 overexpression on inner COPII gene expression and the effect of RNF11 knockdown on EGFP-VSVG transport. Fig. S5 illustrates the validation of the RNF11 siRNA knockdown efficiency. Table S1 is a list of the used siRNAs. Table S2 is a list of the used primer pairs. Table S3 contains the designed assay parameters for the targeted proteomics analysis. Table S4 is the ranked list of TRs coexpressed with SEC23B, SEC24B, and SEC24D. Additional data are available in the JCB DataViewer at https://doi.org/10.1083/JCB.201601090.dv.
Supplementary Material
Acknowledgments
We are grateful to P. Ronchi and C. Godlee for critical reading of the manuscript. We thank the Advanced Light Microscopy Facility at the European Molecular Biology Laboratory (EMBL) for support. We also gratefully acknowledge support from EMBL’s genomics and proteomics facilities. We thank M. Reiss for support with sample preparation, C. Tischer and A. Halavatyi for their help with image analysis, J. Krijgsveld, C. Hughes, and W. Elings for support in the design of targeted proteomic assays, and Biognosys AG for access to the SpectroDive software.
S. Scharaw was supported by an EMBL International PhD Program fellowship.
The authors declare no competing financial interests.
Author contributions: S. Scharaw and R. Pepperkok designed and coordinated the study. S. Scharaw performed and analyzed all experiments, with the exception of targeted proteomics experiments performed and analyzed by A. Ori under the supervision of M. Beck. M. Iskar performed the coexpression analysis under the supervision of P. Bork. G. Boncompain and F. Perez contributed material support and helped to optimize the RUSH transport assay. V. Laketa provided unpublished experimental information. E. Lundberg provided unpublished information about antibodies and the staining protocols. I. Poser generated the mouse mCherry-tagged RNF11 stable cell line. S. Scharaw and R. Pepperkok interpreted the data and wrote the manuscript with input from all authors.
Footnotes
Abbreviations used:
- BFA
- brefeldin A
- COP
- coat protein complex
- EGFR
- EGF receptor
- IGF
- insulin-like growth factor
- Q-RT-PCR
- quantitative real-time PCR
- RUSH
- retention using selective hooks
- TR
- transcriptional regulator
- VSVG
- vesicular stomatitis virus glycoprotein
- WT
- wild type
References
- Abrams E.W., and Andrew D.J.. 2005. CrebA regulates secretory activity in the Drosophila salivary gland and epidermis. Development. 132:2743–2758. 10.1242/dev.01863 [DOI] [PubMed] [Google Scholar]
- Aridor M., Weissman J., Bannykh S., Nuoffer C., and Balch W.E.. 1998. Cargo selection by the COPII budding machinery during export from the ER. J. Cell Biol. 141:61–70. 10.1083/jcb.141.1.61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azmi P., and Seth A.. 2005. RNF11 is a multifunctional modulator of growth factor receptor signalling and transcriptional regulation. Eur. J. Cancer. 41:2549–2560. 10.1016/j.ejca.2005.08.020 [DOI] [PubMed] [Google Scholar]
- Boncompain G., and Perez F.. 2014. Synchronization of secretory cargos trafficking in populations of cells. Methods Mol. Biol. 1174:211–223. 10.1007/978-1-4939-0944-5_14 [DOI] [PubMed] [Google Scholar]
- Boncompain G., Divoux S., Gareil N., de Forges H., Lescure A., Latreche L., Mercanti V., Jollivet F., Raposo G., and Perez F.. 2012. Synchronization of secretory protein traffic in populations of cells. Nat. Methods. 9:493–498. 10.1038/nmeth.1928 [DOI] [PubMed] [Google Scholar]
- Bonnon C., Wendeler M.W., Paccaud J.P., and Hauri H.P.. 2010. Selective export of human GPI-anchored proteins from the endoplasmic reticulum. J. Cell Sci. 123:1705–1715. 10.1242/jcs.062950 [DOI] [PubMed] [Google Scholar]
- Burger A., Amemiya Y., Kitching R., and Seth A.K.. 2006. Novel RING E3 ubiquitin ligases in breast cancer. Neoplasia. 8:689–695. 10.1593/neo.06469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Zhou Z., Liu R., Li Y., Azmi P.B., and Seth A.K.. 2008. The WW domain containing E3 ubiquitin protein ligase 1 upregulates ErbB2 and EGFR through RING finger protein 11. Oncogene. 27:6845–6855. 10.1038/onc.2008.288 [DOI] [PubMed] [Google Scholar]
- Connor M.K., Azmi P.B., Subramaniam V., Li H., and Seth A.. 2005. Molecular characterization of ring finger protein 11. Mol. Cancer Res. 3:453–461. 10.1158/1541-7786.MCR-04-0166 [DOI] [PubMed] [Google Scholar]
- Earp H.S., Austin K.S., Blaisdell J., Rubin R.A., Nelson K.G., Lee L.W., and Grisham J.W.. 1986. Epidermal growth factor (EGF) stimulates EGF receptor synthesis. J. Biol. Chem. 261:4777–4780. [PubMed] [Google Scholar]
- Escher C., Reiter L., MacLean B., Ossola R., Herzog F., Chilton J., MacCoss M.J., and Rinner O.. 2012. Using iRT, a normalized retention time for more targeted measurement of peptides. Proteomics. 12:1111–1121. 10.1002/pmic.201100463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Q.W., Knight Z.A., Goldenberg D.D., Yu W., Mostov K.E., Stokoe D., Shokat K.M., and Weiss W.A.. 2006. A dual PI3 kinase/mTOR inhibitor reveals emergent efficacy in glioma. Cancer Cell. 9:341–349. 10.1016/j.ccr.2006.03.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farhan H., Reiterer V., Korkhov V.M., Schmid J.A., Freissmuth M., and Sitte H.H.. 2007. Concentrative export from the endoplasmic reticulum of the γ-aminobutyric acid transporter 1 requires binding to SEC24D. J. Biol. Chem. 282:7679–7689. 10.1074/jbc.M609720200 [DOI] [PubMed] [Google Scholar]
- Farhan H., Wendeler M.W., Mitrovic S., Fava E., Silberberg Y., Sharan R., Zerial M., and Hauri H.P.. 2010. MAPK signaling to the early secretory pathway revealed by kinase/phosphatase functional screening. J. Cell Biol. 189:997–1011. 10.1083/jcb.200912082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox R.M., Hanlon C.D., and Andrew D.J.. 2010. The CrebA/Creb3-like transcription factors are major and direct regulators of secretory capacity. J. Cell Biol. 191:479–492. 10.1083/jcb.201004062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong F., Breitling R., McEntee C.W., Wittner B.S., Nemhauser J.L., and Chory J.. 2006. RankProd: a bioconductor package for detecting differentially expressed genes in meta-analysis. Bioinformatics. 22:2825–2827. 10.1093/bioinformatics/btl476 [DOI] [PubMed] [Google Scholar]
- Hughes T.R., Marton M.J., Jones A.R., Roberts C.J., Stoughton R., Armour C.D., Bennett H.A., Coffey E., Dai H., He Y.D., et al. 2000. Functional discovery via a compendium of expression profiles. Cell. 102:109–126. 10.1016/S0092-8674(00)00015-5 [DOI] [PubMed] [Google Scholar]
- Iskar M., Campillos M., Kuhn M., Jensen L.J., van Noort V., and Bork P.. 2010. Drug-induced regulation of target expression. PLOS Comput. Biol. 6:e1000925 10.1371/journal.pcbi.1000925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iskar M., Zeller G., Blattmann P., Campillos M., Kuhn M., Kaminska K.H., Runz H., Gavin A.-C., Pepperkok R., van Noort V., and Bork P.. 2013. Characterization of drug-induced transcriptional modules: towards drug repositioning and functional understanding. Mol. Syst. Biol. 9:662. 10.1038/msb.2013.20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer S.C., Ramachandran Iyer E.P., Meduri R., Rubaharan M., Kuntimaddi A., Karamsetty M., and Cox D.N.. 2013. Cut, via CrebA, transcriptionally regulates the COPII secretory pathway to direct dendrite development in Drosophila. J. Cell Sci. 126:4732–4745. 10.1242/jcs.131144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen D., and Schekman R.. 2011. COPII-mediated vesicle formation at a glance. J. Cell Sci. 124:1–4. 10.1242/jcs.069773 [DOI] [PubMed] [Google Scholar]
- Joh T., Itoh M., Katsumi K., Yokoyama Y., Takeuchi T., Kato T., Wada Y., and Tanaka R.. 1986. Physiological concentrations of human epidermal growth factor in biological fluids: use of a sensitive enzyme immunoassay. Clin. Chim. Acta. 158:81–90. 10.1016/0009-8981(86)90118-X [DOI] [PubMed] [Google Scholar]
- Kitching R., Wong M.J., Koehler D., Burger A.M., Landberg G., Gish G., and Seth A.. 2003. The RING-H2 protein RNF11 is differentially expressed in breast tumours and interacts with HECT-type E3 ligases. Biochim. Biophys. Acta. 1639:104–112. 10.1016/j.bbadis.2003.07.001 [DOI] [PubMed] [Google Scholar]
- Kostaras E., Sflomos G., Pedersen N.M., Stenmark H., Fotsis T., and Murphy C.. 2013. SARA and RNF11 interact with each other and ESCRT-0 core proteins and regulate degradative EGFR trafficking. Oncogene. 32:5220–5232. 10.1038/onc.2012.554 [DOI] [PubMed] [Google Scholar]
- Kostaras E., Pedersen N.M., Stenmark H., Fotsis T., and Murphy C.. 2014. SARA and RNF11 at the crossroads of EGFR signaling and trafficking. Methods Enzymol. 535:225–247. 10.1016/B978-0-12-397925-4.00014-6 [DOI] [PubMed] [Google Scholar]
- Kuehn M.J., Herrmann J.M., and Schekman R.. 1998. COPII-cargo interactions direct protein sorting into ER-derived transport vesicles. Nature. 391:187–190. 10.1038/34438 [DOI] [PubMed] [Google Scholar]
- Laketa V., Zarbakhsh S., Traynor-Kaplan A., Macnamara A., Subramanian D., Putyrski M., Mueller R., Nadler A., Mentel M., Saez-Rodriguez J., et al. 2014. PIP3 induces the recycling of receptor tyrosine kinases. Sci. Signal. 7 10.1126/scisignal.2004532 [DOI] [PubMed] [Google Scholar]
- Lamb J., Crawford E.D., Peck D., Modell J.W., Blat I.C., Wrobel M.J., Lerner J., Brunet J.P., Subramanian A., Ross K.N., et al. 2006. The connectivity map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 313:1929–1935. 10.1126/science.1132939 [DOI] [PubMed] [Google Scholar]
- Lemmon M.A., and Schlessinger J.. 2010. Cell signaling by receptor tyrosine kinases. Cell. 141:1117–1134. 10.1016/j.cell.2010.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., and Seth A.. 2004. An RNF11: Smurf2 complex mediates ubiquitination of the AMSH protein. Oncogene. 23:1801–1808. 10.1038/sj.onc.1207319 [DOI] [PubMed] [Google Scholar]
- Lin C.R., Chen W.S., Kruiger W., Stolarsky L.S., Weber W., Evans R.M., Verma I.M., Gill G.N., and Rosenfeld M.G.. 1984. Expression cloning of human EGF receptor complementary DNA: gene amplification and three related messenger RNA products in A431 cells. Science. 224:843–848. 10.1126/science.6326261 [DOI] [PubMed] [Google Scholar]
- Lippincott-Schwartz J., Yuan L.C., Bonifacino J.S., and Klausner R.D.. 1989. Rapid redistribution of Golgi proteins into the ER in cells treated with brefeldin A: evidence for membrane cycling from Golgi to ER. Cell. 56:801–813. 10.1016/0092-8674(89)90685-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C.X., Reinert T., Chmielewska I., and Ellis M.J.. 2015. Mechanisms of aromatase inhibitor resistance. Nat. Rev. Cancer. 15:261–275. 10.1038/nrc3920 [DOI] [PubMed] [Google Scholar]
- Marquèze-Pouey B., Mailfert S., Rouger V., Goaillard J.M., and Marguet D.. 2014. Physiological epidermal growth factor concentrations activate high affinity receptors to elicit calcium oscillations. PLoS One. 9:e106803 10.1371/journal.pone.0106803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merte J., Jensen D., Wright K., Sarsfield S., Wang Y., Schekman R., and Ginty D.D.. 2010. Sec24b selectively sorts Vangl2 to regulate planar cell polarity during neural tube closure. Nat. Cell Biol. 12:41–46: 1–8. 10.1038/ncb2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller S.G., Carnell L., and Moore H.H.. 1992. Post-Golgi membrane traffic: brefeldin A inhibits export from distal Golgi compartments to the cell surface but not recycling. J. Cell Biol. 118:267–283. 10.1083/jcb.118.2.267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootha V.K., Lindgren C.M., Eriksson K.F., Subramanian A., Sihag S., Lehar J., Puigserver P., Carlsson E., Ridderstråle M., Laurila E., et al. 2003. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34:267–273. 10.1038/ng1180 [DOI] [PubMed] [Google Scholar]
- Nexø E., Jørgensen E., and Hansen M.R.. 1992. Human epidermal growth factor—on molecular forms present in urine and blood. Regul. Pept. 42:75–84. 10.1016/0167-0115(92)90025-P [DOI] [PubMed] [Google Scholar]
- Ori A., Banterle N., Iskar M., Andrés-Pons A., Escher C., Khanh Bui H., Sparks L., Solis-Mezarino V., Rinner O., Bork P., et al. 2013. Cell type-specific nuclear pores: a case in point for context-dependent stoichiometry of molecular machines. Mol. Syst. Biol. 9:648 10.1038/msb.2013.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ori A., Andrés-Pons A., and Beck M.. 2014. The use of targeted proteomics to determine the stoichiometry of large macromolecular assemblies. Methods Cell Biol. 122:117–146. 10.1016/B978-0-12-417160-2.00006-0 [DOI] [PubMed] [Google Scholar]
- Ori A., Toyama B.H., Harris M.S., Bock T., Iskar M., Bork P., Ingolia N.T., Hetzer M.W., and Beck M.. 2015. Integrated transcriptome and proteome analyses reveal organ-specific proteome deterioration in old rats. Cell Syst. 1:224–237. 10.1016/j.cels.2015.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer K.J., Konkel J.E., and Stephens D.J.. 2005. PCTAIRE protein kinases interact directly with the COPII complex and modulate secretory cargo transport. J. Cell Sci. 118:3839–3847. 10.1242/jcs.02496 [DOI] [PubMed] [Google Scholar]
- Poser I., Sarov M., Hutchins J.R., Hériché J.K., Toyoda Y., Pozniakovsky A., Weigl D., Nitzsche A., Hegemann B., Bird A.W., et al. 2008. BAC TransgeneOmics: a high-throughput method for exploration of protein function in mammals. Nat. Methods. 5:409–415. 10.1038/nmeth.1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roepstorff K., Grandal M.V., Henriksen L., Knudsen S.L., Lerdrup M., Grøvdal L., Willumsen B.M., and van Deurs B.. 2009. Differential effects of EGFR ligands on endocytic sorting of the receptor. Traffic. 10:1115–1127. 10.1111/j.1600-0854.2009.00943.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito A., Hino S., Murakami T., Kanemoto S., Kondo S., Saitoh M., Nishimura R., Yoneda T., Furuichi T., Ikegawa S., et al. 2009. Regulation of endoplasmic reticulum stress response by a BBF2H7-mediated Sec23a pathway is essential for chondrogenesis. Nat. Cell Biol. 11:1197–1204. 10.1038/ncb1962 [DOI] [PubMed] [Google Scholar]
- Salmaggi A., Eoli M., Frigerio S., Silvani A., Gelati M., Corsini E., Broggi G., and Boiardi A.. 2003. Intracavitary VEGF, bFGF, IL-8, IL-12 levels in primary and recurrent malignant glioma. J. Neurooncol. 62:297–303. 10.1023/A:1023367223575 [DOI] [PubMed] [Google Scholar]
- Santonico E., Belleudi F., Panni S., Torrisi M.R., Cesareni G., and Castagnoli L.. 2010. Multiple modification and protein interaction signals drive the Ring finger protein 11 (RNF11) E3 ligase to the endosomal compartment. Oncogene. 29:5604–5618. 10.1038/onc.2010.294 [DOI] [PubMed] [Google Scholar]
- Schlessinger J. 2000. Cell signaling by receptor tyrosine kinases. Cell. 103:211–225. 10.1016/S0092-8674(00)00114-8 [DOI] [PubMed] [Google Scholar]
- Shaffer A.L., Shapiro-Shelef M., Iwakoshi N.N., Lee A.H., Qian S.B., Zhao H., Yu X., Yang L., Tan B.K., Rosenwald A., et al. 2004. XBP1, downstream of Blimp-1, expands the secretory apparatus and other organelles, and increases protein synthesis in plasma cell differentiation. Immunity. 21:81–93. 10.1016/j.immuni.2004.06.010 [DOI] [PubMed] [Google Scholar]
- Sharpe L.J., Luu W., and Brown A.J.. 2011. Akt phosphorylates Sec24: new clues into the regulation of ER-to-Golgi trafficking. Traffic. 12:19–27. 10.1111/j.1600-0854.2010.01133.x [DOI] [PubMed] [Google Scholar]
- Simpson J.C., Cetin C., Erfle H., Joggerst B., Liebel U., Ellenberg J., and Pepperkok R.. 2007. An RNAi screening platform to identify secretion machinery in mammalian cells. J. Biotechnol. 129:352–365. 10.1016/j.jbiotec.2006.12.027 [DOI] [PubMed] [Google Scholar]
- Simpson J.C., Joggerst B., Laketa V., Verissimo F., Cetin C., Erfle H., Bexiga M.G., Singan V.R., Hériché J.K., Neumann B., et al. 2012. Genome-wide RNAi screening identifies human proteins with a regulatory function in the early secretory pathway. Nat. Cell Biol. 14:764–774. 10.1038/ncb2510 [DOI] [PubMed] [Google Scholar]
- Stockhammer G., Obwegeser A., Kostron H., Schumacher P., Muigg A., Felber S., Maier H., Slavc I., Gunsilius E., and Gastl G.. 2000. Vascular endothelial growth factor (VEGF) is elevated in brain tumor cysts and correlates with tumor progression. Acta Neuropathol. 100:101–105. 10.1007/s004010051199 [DOI] [PubMed] [Google Scholar]
- Stuart J.M., Segal E., Koller D., and Kim S.K.. 2003. A gene-coexpression network for global discovery of conserved genetic modules. Science. 302:249–255. 10.1126/science.1087447 [DOI] [PubMed] [Google Scholar]
- Subramaniam V., Li H., Wong M., Kitching R., Attisano L., Wrana J., Zubovits J., Burger A.M., and Seth A.. 2003. The RING-H2 protein RNF11 is overexpressed in breast cancer and is a target of Smurf2 E3 ligase. Br. J. Cancer. 89:1538–1544. 10.1038/sj.bjc.6601301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sucic S., El-Kasaby A., Kudlacek O., Sarker S., Sitte H.H., Marin P., and Freissmuth M.. 2011. The serotonin transporter is an exclusive client of the coat protein complex II (COPII) component SEC24C. J. Biol. Chem. 286:16482–16490. 10.1074/jbc.M111.230037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillmann K.D., Reiterer V., Baschieri F., Hoffmann J., Millarte V., Hauser M.A., Mazza A., Atias N., Legler D.F., Sharan R., et al. 2015. Regulation of Sec16 levels and dynamics links proliferation and secretion. J. Cell Sci. 128:670–682. 10.1242/jcs.157115 [DOI] [PubMed] [Google Scholar]
- Trinklein N.D., Aldred S.J., Saldanha A.J., and Myers R.M.. 2003. Identification and functional analysis of human transcriptional promoters. Genome Res. 13:308–312. 10.1101/gr.794803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Noort V., Snel B., and Huynen M.A.. 2003. Predicting gene function by conserved co-expression. Trends Genet. 19:238–242. 10.1016/S0168-9525(03)00056-8 [DOI] [PubMed] [Google Scholar]
- Vivanco I., and Sawyers C.L.. 2002. The phosphatidylinositol 3-kinase AKT pathway in human cancer. Nat. Rev. Cancer. 2:489–501. 10.1038/nrc839 [DOI] [PubMed] [Google Scholar]
- Wendeler M.W., Paccaud J.P., and Hauri H.P.. 2007. Role of Sec24 isoforms in selective export of membrane proteins from the endoplasmic reticulum. EMBO Rep. 8:258–264. 10.1038/sj.embor.7400893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson D., Charoensawan V., Kummerfeld S.K., and Teichmann S.A.. 2007. DBD--taxonomically broad transcription factor predictions: new content and functionality. Nucleic Acids Res. 36:D88–D92. 10.1093/nar/gkm964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarden Y., and Sliwkowski M.X.. 2001. Untangling the ErbB signalling network. Nat. Rev. Mol. Cell Biol. 2:127–137. 10.1038/35052073 [DOI] [PubMed] [Google Scholar]
- Zanetti G., Pahuja K.B., Studer S., Shim S., and Schekman R.. 2011. COPII and the regulation of protein sorting in mammals. Nat. Cell Biol. 14:20–28. 10.1038/ncb2390 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.