Few tools exist to examine the inner nuclear membrane (INM) in living cells, and the INM-specific proteome is poorly characterized. Smoyer et al. combine a split-GFP screen and fluorescence correlation spectroscopy analysis to identify membrane proteins that access the INM and study INM-specific interactions.
Abstract
Understanding the protein composition of the inner nuclear membrane (INM) is fundamental to elucidating its role in normal nuclear function and in disease; however, few tools exist to examine the INM in living cells, and the INM-specific proteome remains poorly characterized. Here, we adapted split green fluorescent protein (split-GFP) to systematically localize known and predicted integral membrane proteins in Saccharomyces cerevisiae to the INM as opposed to the outer nuclear membrane. Our data suggest that components of the endoplasmic reticulum (ER) as well as other organelles are able to access the INM, particularly if they contain a small extraluminal domain. By pairing split-GFP with fluorescence correlation spectroscopy, we compared the composition of complexes at the INM and ER, finding that at least one is unique: Sbh2, but not Sbh1, has access to the INM. Collectively, our work provides a comprehensive analysis of transmembrane protein localization to the INM and paves the way for further research into INM composition and function.
Introduction
Within the double lipid bilayer of the nuclear envelope (NE), the genomic material of the cell is copied, read, and stored as the instruction manual for the cell and its progeny. The inner and outer nuclear membranes (INM and ONM) are joined at numerous sites where nuclear pore complexes (NPCs) reside. The ONM is contiguous with the ER, whereas the INM is thought to be molecularly distinct, containing integral and peripheral membrane proteins such as lamins, lamin B receptor, LEM domain–containing proteins, and SUN domain–containing proteins, which play vital roles in nuclear organization and function (Mekhail and Moazed, 2010; Starr and Fridolfsson, 2010; Rothballer and Kutay, 2013). The distinct composition of the INM is thought to arise by one of two mechanisms: NPCs function as a barrier to restrict the passage of integral membrane proteins with large extraluminal domains from the ONM to the INM, or NPCs control the nucleocytoplasmic transport of soluble proteins that function as tethers for INM proteins with small extraluminal domains (Katta et al., 2014; Ungricht and Kutay, 2015). In addition, a recently discovered INM-associated destruction (INMAD) pathway serves as a quality control pathway in yeast, targeting misfolded and damaged INM proteins for ubiquitin-mediated proteolysis (Foresti et al., 2014; Khmelinskii et al., 2014). This system, involving the INM E3 ligases Asi1 and Asi3, may also recognize aberrant proteins that reach the INM because of leakage through NPCs, ensuring that INM integrity and function are preserved.
Understanding protein dynamics at the INM is important given that proteins embedded in the INM function in chromosome organization within the nucleus, distribution of NPCs, and maintenance of NE structure. Mutations in known components of the INM or their binding partners result in a spectrum of human diseases, collectively known as the laminopathies (Dauer and Worman, 2009; Burke and Stewart, 2014; Davidson and Lammerding, 2014). However, our experimental toolkit to study the INM is currently extremely limited. Because the INM and ONM are separated by only 10–50 nm, EM is the only unequivocal method for determining INM versus ONM localization. Furthermore, because biochemical methods for studying INM composition depend on in silico subtraction or comparative analysis of nuclear and microsomal membrane samples (an ER-derived fraction formed in vitro), proteins that have dual functions in the INM and ONM/ER are overlooked (e.g., Strambio-de-Castillia et al., 1995; Schirmer et al., 2003; Korfali et al., 2010, 2012; Wilkie et al., 2011). EM and biochemical fractionation also provide only a snapshot of cell populations, so analysis of protein dynamics is difficult or impossible.
Our objective was to design a fluorescence-based assay to study INM localization on a system-wide level in living cells. Using split-GFP (Cabantous et al., 2005; Cabantous and Waldo, 2006), we tested all known and predicted transmembrane proteins in budding yeast in vivo for localization to the INM. We found known NE proteins enriched at the INM, as well as components of other organelles such as the ER. SBH1 and SBH2 are nonessential genes, and it is thought that Sbh1 and Sbh2 play partially overlapping roles in stabilizing ER complexes containing Sec61 and Ssh1, respectively (Finke et al., 1996; Jan et al., 2014). Interestingly, Sbh2 but not Sbh1 localized to the INM using our approach, lending evidence to the idea that ER components have differential access to the INM. To further explore this idea, we combined split-GFP with fluorescence cross-correlation spectroscopy to study the composition of the Sec61 complex at the ER and INM. Our work illustrates how split-GFP can be used to identify proteins that access the INM and study INM-specific interactions, tools that are important to understand INM function.
Results
INM localization and topology can be visualized using split-GFP
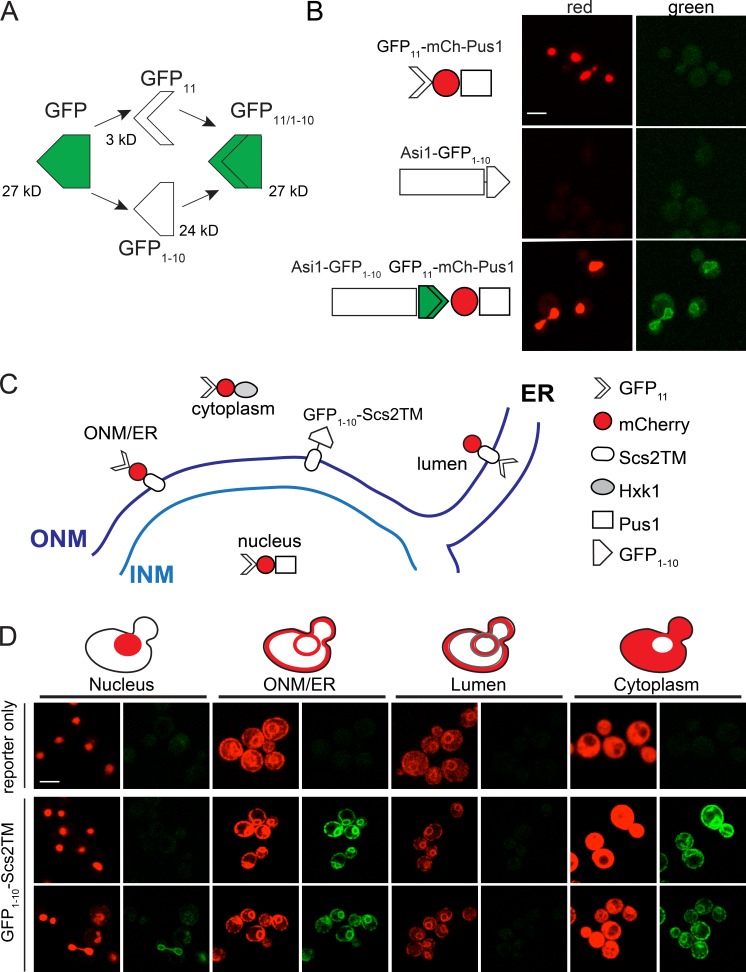
Superfolder GFP can be split asymmetrically into two parts, GFP11 (3 kD) and GFP1–10 (24 kD), that do not fluoresce individually but can reconstitute a working GFP when expressed in the same cellular compartment (Fig. 1 A; Cabantous et al., 2005; Cabantous and Waldo, 2006). We adapted this system for use in Saccharomyces cerevisiae by yeast codon-optimizing GFP11 and fusing it to the coding sequence of the soluble nuclear protein Pus1 expressed under the NOP1 promoter; in most cases, this was also fused to mCherry. The high expression levels ensured that complementation by GFP11 was not limiting. When NOP1pr-GFP11-mCherry-PUS1 was expressed alone, no fluorescence in the green channel was observed, but fluorescence in the red channel showed strong localization to all regions of the nucleus, similar to previously described versions of this marker (Fig. 1 B, top; Hellmuth et al., 1998; Friederichs et al., 2011; Witkin et al., 2012). We developed additional reporters to serve as controls, including a marker to detect proteins on the ONM or cytoplasmic face of the ER (GFP11-mCherry-Scs2TM), a luminal marker for proteins located in the ER or the space between the INM and ONM (mCherry-Scs2TM-GFP11), and a soluble, cytoplasmic marker (GFP11-mCherry-Hxk1). A schematic showing the reporters and example images of each are in Fig. 1 (C and D).
Figure 1.
Split-GFP to study protein localization. (A) Schematic of the GFP complementation system. Protein molecular masses in kilodaltons based on amino acid composition. (B) Schematics show the GFP11-mCherry-Pus1 reporter and INM protein Asi1-GFP1–10, which were expressed alone (top and middle) or together in yeast. Fluorescence in the red (561-nm) and green (488-nm) channels is shown. Residual background is autofluorescence, which was shown by multispectral imaging (not depicted). (C) Schematic illustrating the localization of GFP11-mCherry reporters to detect signal inside the nucleus (GFP11-mCherry-Pus1), ONM/ER surface (GFP11-mCherry-Scs2TM), ER lumen (mCherry-Scs2TM-GFP11), and cytoplasm (GFP11-mCherry-Hxk1). An ONM/ER protein containing the Scs2 transmembrane domain fused to GFP1–10 on its cytoplasmic side is also shown. (D) Example images of reporters alone (top) or with GFP1–10-Scs2TM (bottom). Bars, 2 µm.
We also yeast codon-optimized GFP1–10 and cloned it into tagging cassettes identical to those used in creating the yeast GFP collection. This allowed us to generate GFP1–10–tagged copies of genes expressed under the endogenous promoter as the sole copy. Asi1-GFP1–10 showed no fluorescence in red or green channels (Fig. 1 B, middle). When GFP11-mCherry-Pus1 was added, fluorescence at 488 nm was observed at the nuclear periphery (Fig. 1 B, bottom). This result is consistent with previous work showing that Asi1 localizes to the INM with its C terminus inside the nucleus (Boban et al., 2006). A second well-characterized INM protein, Heh2, fused with GFP1–10 at either its C or N terminus (denoted Heh2-GFP1–10 or GFP1–10-Heh2, respectively), also exhibited fluorescence at 488 nm with the GFP11-mCherry-Pus1 reporter (Fig. 2). Having both N and C termini facing the nucleoplasm is consistent with current topological predictions for Heh2 and other LEM domain–containing proteins (King et al., 2006).
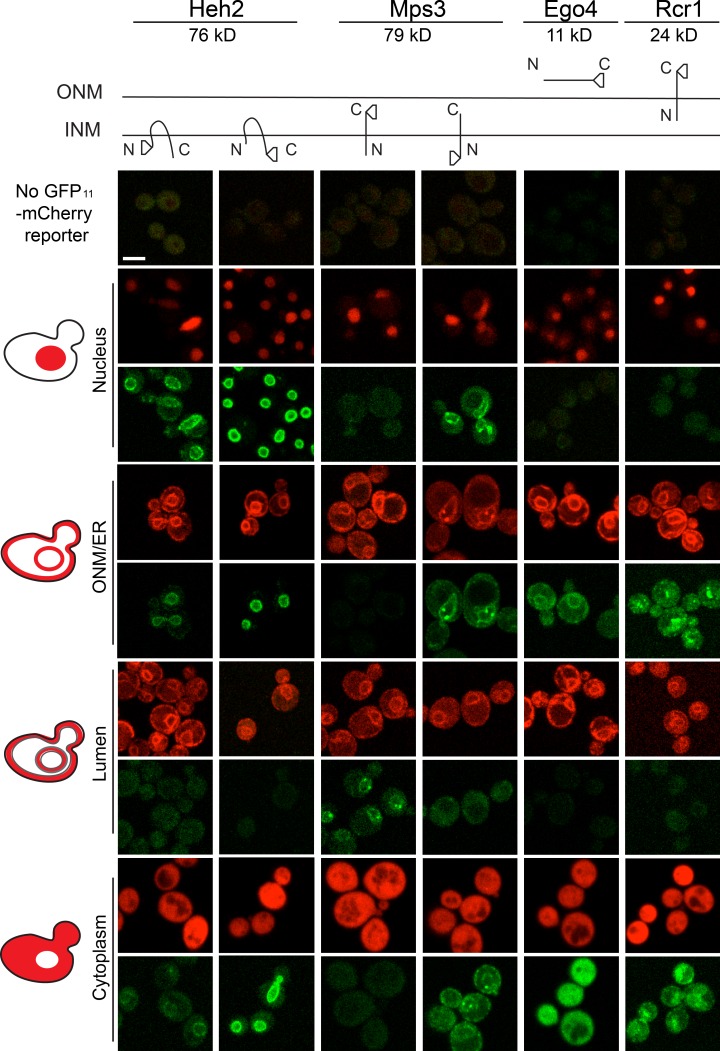
Figure 2.
Protein topology determination using split-GFP. Schematics of Heh2, Mps3, Rcr1, and Ego4 are shown along with the location of the GFP1–10 fusion and estimated size in kD. Images of strains containing the tagged proteins and the nucleus (GFP11-mCherry-Pus1), ONM/ER surface (GFP11-mCherry-Scs2TM), ER lumen (mCherry-Scs2TM-GFP11), and cytoplasm (GFP11-mCherry-Hxk1) reporters. Bar, 2 µm.
The C terminus of Mps3 is in the lumen, and its N terminus is in the nucleoplasm (Jaspersen et al., 2002; Nishikawa et al., 2003). The nuclear reporter showed a strong signal in the green channel with GFP1–10-Mps3 but not with Mps3-GFP1–10, whereas the luminal marker exhibited fluorescence with Mps3-GFP1–10 but not GFP1–10-Mps3 (Fig. 2). GFP1–10-Mps3 also interacted with the cytoplasmic and ONM/ER markers (Fig. 2; unpublished data). At least three explanations could account for detection of GFP1–10-Mps3 on the ONM/ER. First, differences in the levels or functionality of N- and C-terminally GFP1–10–tagged Mps3 might result in protein accumulation in the ONM/ER. Second, a pool of Mps3 incorporated into the ER may have yet to be transported to the INM. Last, a bona fide population of Mps3 could function outside of the INM. We observed a similar distribution for other INM proteins such as Heh2 that was proportional to its expression level (Fig. 2), lending support to the second model. Detection of GFP1–10-Mps3 at the spindle pole body (SPB) with the ONM/ER marker is perhaps not surprising because Mps3 localizes to the membrane region of the SPB where INM and ONM are contiguous, as shown previously by immuno-EM (Jaspersen et al., 2002).
The strong binding affinity of GFP11 for GFP1–10 may cause ectopic localization to the INM (or nucleus) by GFP11-mCherry-Pus1 binding to proteins outside of the nucleus before nuclear translocation. Analysis of a series of overexpressed GFP1–10 reporters fused to blue fluorescent protein and binucleated cells formed using the kar1Δ15 mutant suggested that this “dragging” force is weak, particularly for membrane-associated proteins (Fig. S1 and not depicted). Further evidence to suggest that dragging does not contribute significantly to detection of proteins at the INM came from analysis of small soluble proteins or ER membrane proteins such as Ego4 (10.8 kD) and Rcr1 (23.9 kD; cytoplasmic domain 16.9 kD), which showed no signal with GFP11-mCherry-Pus1. However, both Ego4-GFP1–10 and Rcr1-GFP1–10 showed fluorescence with other reporters, including GFP11-mCherry-Hxk1 in the cytoplasm and GFP11-mCherry-Scs2TM on the cytoplasmic face of the ONM/ER (Fig. 2). Similar results were observed when GFP1–10-Scs2TM distribution was examined with all reporters (Fig. 1 D). Collectively, these data strongly suggest that split-GFP methodology is specific and can be used to visualize proteins at the INM, as well as determine their topology.
A genome-wide screen for INM proteins
The budding yeast proteome has been extensively studied, including localization analysis of 4,156 C-terminal GFP fusions (Huh et al., 2003; Breker et al., 2013; Chong et al., 2015) and C- and N-terminal fusions to 1,759 components of the endomembrane system (Yofe et al., 2016). In these system-wide studies, genes that localize to the NE were identified. However, because the INM and ONM cannot be resolved using conventional methods, only a small number of genes (12) are annotated as components of the yeast INM in the Saccharomyces Genome Database (SGD): Asi1 and Asi3 ubiquitin ligases and their binding partner Asi2; the LEM-domain proteins, Heh1/Src1 and Heh2; and the Doa10 ubiquitin ligase (Deng and Hochstrasser, 2006; King et al., 2006; Zargari et al., 2007). Other INM proteins such as the SUN protein Mps3, the NPC and SPB component Ndc1, and the ER protein Sec61 are not annotated despite experimental evidence demonstrating both localization and function (Chial et al., 1998; Jaspersen et al., 2002; Deng and Hochstrasser, 2006). Even with these additions, the number of yeast INM components is considerably smaller than the 100–1,000 estimated NE transmembrane proteins in higher eukaryotes (Schirmer et al., 2003; Korfali et al., 2010, 2012). It is unclear in many cases whether these NE transmembrane proteins function at the INM, simply diffuse in and out of the nucleus, or are retained in the nucleus after membrane reformation at the end of mitosis. Because budding yeast undergo a closed mitosis, NPC-mediated transport is the sole route in or out of the INM, with the exception of nuclear degradation.
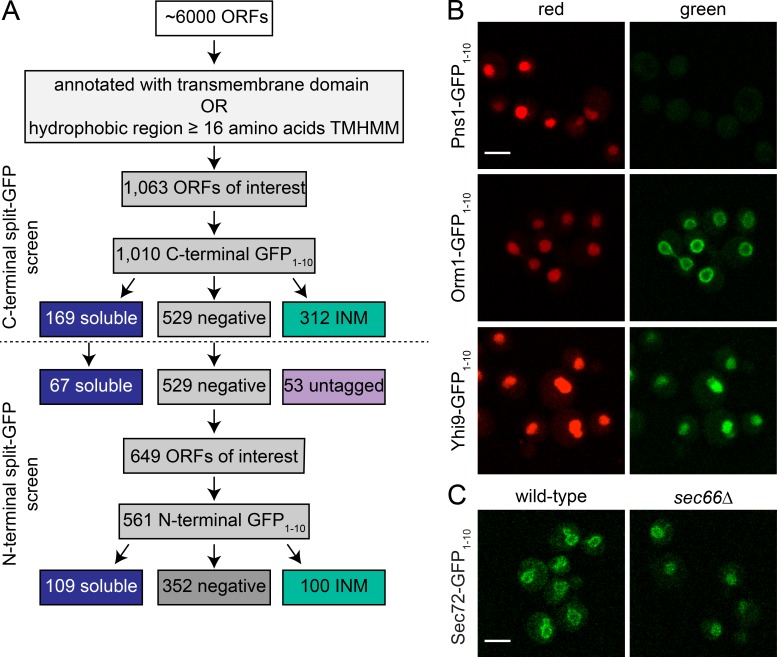
To systematically address what proteins in yeast access the INM, we constructed and verified a library of genes fused to GFP1–10 in a derivative of the haploid yeast strain BY4742 containing NOP1pr-GFP11-mCherry-PUS1 on a centromeric plasmid. Because we were specifically interested in integral membrane proteins, a list of 1,063 ORFs that encode proteins with known or putative transmembrane helices (see Materials and methods) was compiled. Although it is possible that tagging certain classes of membrane proteins may affect protein localization and function, we did not remove genes from analysis based on predicted motifs, so our screen was as unbiased as possible. Our workflow is shown in Fig. 3 A and is described as follows.
Figure 3.
A screen to identify INM composition. (A) A library of 1,063 possible transmembrane proteins was defined as described in the text, and a C-terminal GFP1–10 collection of strains was imaged. Because the C-terminal epitope may affect localization or function, negatives were subjected to N-terminal tagging and screening along with 67 randomly chosen soluble hits from the C-terminal screen. (B) Representative images of negative (Pns1-GFP1–10), INM (Orm1-GFP1–10), and soluble (Yhi9-GFP1–10) localization. Representative images from subclasses of INM and nuclear localization shown in Fig. S2. (C) Sec72-GFP1–10 localization in wild-type and sec66Δ cells containing GFP11-mCherry-Pus1. Bars, 2 µm.
In yeast, C-terminal fusions of genes to fluorescent proteins are relatively straightforward to create and, in most cases, result in functional fusion proteins expressed at endogenous levels (Ghaemmaghami et al., 2003; Huh et al., 2003; Yofe et al., 2016). Therefore, we began analysis with 1,010 C-terminally tagged GFP1–10 strains that were grown to mid-log phase and screened for INM localization by scanning for green signal at four different 488-nm laser settings, with increasing power (Table S3). If a signal was observed, at least three sets of images were acquired. Images were manually inspected and assigned into one of three major categories of phenotypes: no signal (529), localization to the NE that is consistent with INM signal (312), and soluble nuclear localization (169). Example images of each phenotype are shown in Fig. 3 B and Fig. S2; a summary of results is listed in Table S3. Most proteins that localized to the nuclear periphery showed a uniform distribution across the nuclear surface, similar to images of Orm1 (Fig. 3 B), but some showed a punctate pattern or localized to discrete foci (Fig. S2 A and Table S3). The nature of these puncta is largely unknown. However, we presume based on gene function that foci formed by Pom34, Pom33, Per33, Ndc1, and Pom152 are NPCs, whereas the bright localization of Nbp1, Ndc1, and Mps3 to one or two spots represents the SPB. In most cases, split-GFP results were consistent with predicted or known topologies; a comparison of the 195 C-terminal hits that overlap with a large-scale topology screen resulted in 177 having the same inside prediction (Table S4; Kim et al., 2006).
Proteins that were soluble or not localized to the INM were further analyzed, because protein topology or C-terminal sorting motifs could result in these false-negative categories. Most (125) of the 169 C-terminal tagged proteins that showed soluble nuclear signal did not contain a bona fide membrane domain predicted using transmembrane helices hidden Markov model (TMHMM) or Phobius (Table S3), so we chose a randomly selected set of 67 genes for further analysis by tagging with GFP1–10 at the N terminus along with testing negatives from the C-terminal screen and genes we were unable to C-terminally tag (presumably the tag resulted in a nonviable gene product). For genes that lacked a cleaved signal sequence, the constitutive CDC42 promoter followed by GFP1–10 was placed at the 5′ end of the ORF using PCR; for genes with a signal sequence, a similar construct containing the Kar2 signal sequence before GFP1–10 was used. Although this places genes under the control of a nonnative promoter, the moderate expression levels of CDC42 are similar to those of genes in this study, many of which are thought to be controlled post-transcriptionally (Cho et al., 1998; Yofe et al., 2016), and we found that its use did not result in mislocalization of Heh2 or Mps3 (Fig. 2). We successfully screened an additional 560 fusion proteins, including 32 that were not in our C-terminal library (Fig. 3 A). Of these, 100 localized to the nuclear periphery, 109 showed soluble nuclear signal, and 352 were negative (Table S3). We were unable to make C- or N-terminal GFP1–10 constructs for 21 genes.
Of the 67 soluble hits retested using N-terminal GFP1–10, 50 remained soluble, 12 were negative for split-GFP fluorescence, and five (Asg7, Opt1, Tna1, Vma21, and YBR220c) predicted to have a transmembrane domain exhibited signal at the INM (Table S3). This confirms that the soluble signal we observed is primarily caused by the lack of a bona fide transmembrane domain rather than the tag disrupting protein function and/or localization. Interestingly, some proteins such as Sec72 localize to the nuclear periphery despite the lack of a membrane-spanning or membrane-associated domain, suggesting that an interaction with a membrane protein tethers them at the NE. Consistent with this idea, we found that Sec72-GFP1–10 shows peripheral localization in wild-type cells but soluble localization in cells lacking Sec66, its membrane receptor (Fig. 3 C; Feldheim and Schekman, 1994).
Additional INM assays for a subset of split-GFP hits
Of the 1,063 proteins in our library, 412 showed positive signal in our split-GFP assay at the nuclear periphery, including known INM components such as Heh1, Heh2, Asi1, Asi2, Asi3, Trm1, Ste14, and Ste24 and all membrane components of the NPC and SPB with the exception of Mps2 (Fig. S2 and Table S3). This clearly demonstrates the effectiveness of the split-GFP assay in identification of known INM components. However, the large number of positives was somewhat surprising, so we validated several hits using independent assays.
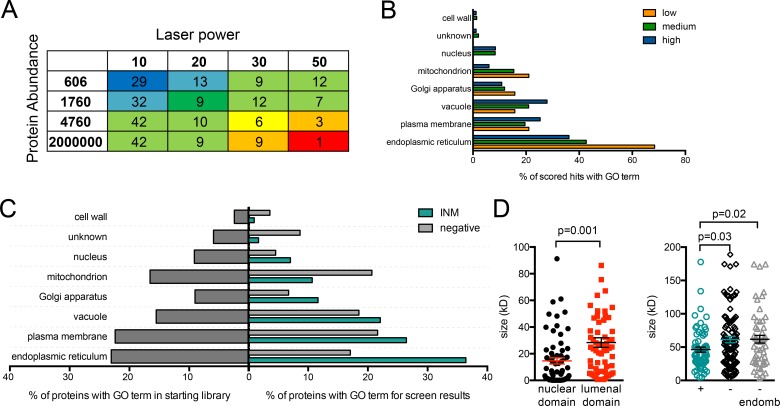
GFP1–10 and GFP11 show linear binding over at least four orders of magnitude (Cabantous et al., 2005; Cabantous and Waldo, 2006; Kamiyama et al., 2016). Thus, detection of a protein at the INM using split-GFP is related to both protein abundance and the fraction of protein present at the INM. To prioritize hits for follow-up analysis, we combined available whole-cell estimates of protein levels (Ghaemmaghami et al., 2003) with our INM detection information to assign 245 of 412 INM hits a confidence score (Fig. 4 A and Table S4). Nonabundant proteins that gave an INM signal with a lower laser power had high confidence scores, whereas highly abundant proteins that were only observed using higher laser power settings had low scores. 83 and 143 of our hits had high and medium confidence scores, respectively. Known nuclear proteins that were enriched in our screen had high and medium confidence scores, strongly suggesting that the confidence score was correlated with bona fide INM components (Fig. 4, B and C).
Figure 4.
Analysis of INM hits shows overlap with the ER. (A) A scoring matrix for INM hits based on detection at the INM (binned into four categories based on laser power) and whole-cell protein abundance data from Ghaemmaghami et al. (2003) (binned into four categories). In this way, 242 of 412 hits were ranked: blue and dark green, high-confidence hits; light green, medium confidence; yellow, orange, or red, low confidence. Laser power, abundance, and confidence score for each INM hit listed in Table S4. No abundance data were available for 170 hits, so they were not scored, although most are likely to be high-confidence hits because protein detection fell below detection limits in previous work. Because of the use of the CDC42pr, some genes in this category may represent products not normally expressed under standard vegetative growth conditions. (B) GO slim component analysis of the proteins in each confidence category of low, medium, and high. (C) Enrichment for cellular components based on GO slim annotations in SGD was performed for the library (left) and for genes in each category (right). (D) For single-pass integral membrane proteins localized to the INM (n = 65), sizes of cytoplasmic/nucleoplasmic and luminal domains were determined based on amino acid composition. Total molecular masses for INM hits as well as single-pass proteins that were negative (n = 110)/negative with GO component term: endomembrane system (n = 54) were plotted. The mean and SEM are shown on each plot. P-values were calculated by Student’s t test. One split-GFP negative was removed by the Grubs test: Csf1, molecular mass = 338 kD.
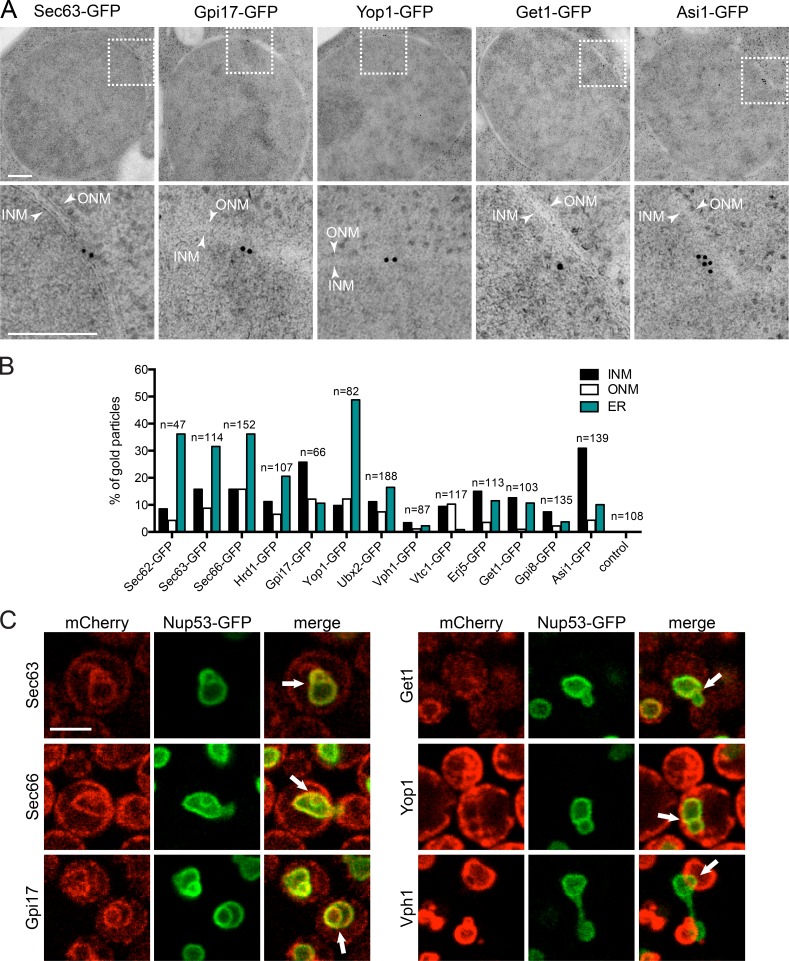
Proteins that have low (Sec62, Sec63, Sec66, Sec72, Hrd1, Erg6, Aur1, Tpo4, and Ubx2), medium (Get1, Gpi17, Vph1, and Vtc1), and high (Yop1 and Gpi8) confidence scores and nonscoring hits (Erj5) were further characterized to determine whether the low scoring hits showed evidence of INM access in the absence of split-GFP. We used two independent assays to verify localization, neither of which was tied to split-GFP. In the first approach, 12 endogenously expressed GFP-tagged proteins were analyzed by immuno-EM with an anti-GFP primary antibody and a secondary antibody conjugated to colloidal gold. In our experience, the GFP antibody results in high signal to noise; however, a few gold particles were typically observed in the cytoplasm and nucleoplasm in untagged control samples. Because we were interested in INM localization, we quantitated the number of gold particles within 50 nm of the NE—a distance that is less than the 60-nm size of the primary and secondary antibodies. Most showed localization to both sides of the NE but not to the luminal space (Fig. 5, A and B). Enrichment at the ONM and ER is consistent with the known function of many proteins in the ER; however, our data also indicate that a previously overlooked minority of GFP-tagged protein is present at the INM, consistent with our split-GFP results.
Figure 5.
INM localization of split-GFP hits using two independent assays. (A) Immuno-EM of nuclei from cells expressing Sec63-GFP (low score), Gpi17-GFP (medium score), Yop1-GFP (high score), Get1-GFP (medium score), and Asi1-GFP (no score). Below is a magnified region of the NE (box) with at least one gold particle located near the INM. The INM and ONM are marked with arrows. Bars, 200 nm. (B) Gold particles were counted in at least 10 cells for 12 hits. The number and percentage of gold particles at the INM, ONM, and ER is shown. (C) Single-plane images shown of theta nuclei induced by overexpression of Nup53-GFP (green) in 2% galactose for 5 h at 23°C. Arrows in merge images point to extra inner nuclear lamellae signal, where colocalization of the indicated mCherry-tagged protein was assayed. Vph1-mCherry showed overlap with Nup53-GFP signal at some points of the NE. Bars, 2 µm. Additional images are in Fig. S3.
In the second approach, we took advantage of the fact that overexpression of the NPC subunit Nup53 results in overproliferation of the INM, resulting in intranuclear lamellae. These INM sheets can sometimes be observed crossing through the nucleus, resulting in a distinctive nucleus that resembles the Greek letter theta (θ), known as a theta nucleus (Marelli et al., 2001; Deng and Hochstrasser, 2006). We created C-terminal mCherry fusions to 10 of 12 genes listed earlier, as well as four additional low-scoring INM hits, in a strain containing GAL1-NUP53-GFP. After 5 h of induction in galactose, cells containing high levels of Nup53-GFP were examined. We could detect all proteins except Tpo4-mCherry and Vph1-mCherry in the theta nuclei formed in these cells (Figs. 5 C and S3). Tpo4-mCherry was detected at the cell periphery and the vacuole, as previously reported (Huh et al., 2003; Yofe et al., 2016). Vph1-mCherry colocalizes with Nup53-GFP at sites where the nucleus and vacuole are in contact; it also extends into nuclei in small protrusions, but full extensions across the nucleus were not observed, preventing us from drawing conclusions about its localization using this approach (Fig. 5 C and not depicted). These data are independent evidence that hits from our split-GFP screen localize to the yeast INM; after removal of Tpo4, our final INM hit list contained 411 components.
The validation assays provide compelling evidence that the majority of the 411 proteins isolated in our screen have access to the INM in yeast. Because of the affinity of GFP1–10 and GFP11, we are able to detect proteins that partially or transiently access the INM, which would be difficult to identify using other approaches. It is important to note that our ability to visualize a protein at the INM using split-GFP does not demonstrate that it functions in this compartment. In addition, our list is likely not comprehensive, as it seems probable that the composition of the INM may be linked to growth conditions.
ER components showed enriched INM access
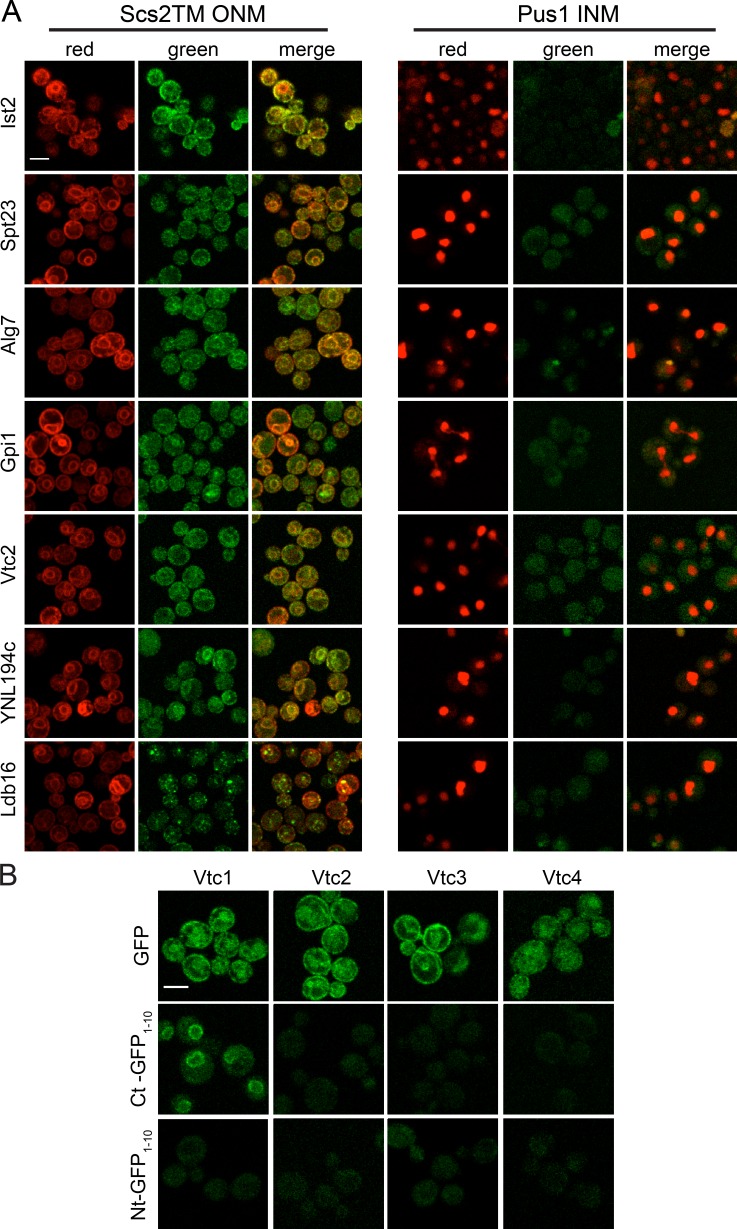
Slim gene ontology (GO) analysis showed enrichment for ER components in our 411 hits (Fig. 4 C), and 12 of the 19 low-scoring hits in our scoring matrix were abundant components of the ER (Fig. 4 B and Table S4). Because of the high degree of overlap with the ER and because the ONM/ER and INM are contiguous at NPCs, we were interested in determining whether all ER membrane proteins access the INM. Using the ONM/ER marker GFP11-mCherry-Scs2TM, we showed that Rcr1, Ist2, Mga2, Spt23, Alg7, Ire1, and Gpi1 were detected on the ER and ONM. However, none could be visualized using GFP11-Pus1-mCherry at the INM, similar to GFP1–10-Scs2TM (Figs. 1 D, 2, and 6 A; and not depicted). This result, combined with our immuno-EM and theta nuclei data, strongly supports the idea that the INM shares a specific subset of proteins with the ER (Fig. 6 A and not depicted).
Figure 6.
ER components unable to access the INM. (A) Ist2, Spt23, Alg7, and Gpi1 (ER proteins), Vtc2 (vacuole), YNL194c (plasma membrane), and Ldb16 (lipid droplet) were detected on the ONM/ER using the GFP11-mCherry-Scs2TM reporter. None gave signal with GFP11-mCherry-Pus1 at the INM. (B) Representative images of log-phase cells containing GFP (top), C-terminal GFP1–10 (middle), and N-terminal GFP1–10 (bottom) tagged versions of Vtc1, Vtc2, Vtc3, and Vtc4. Bars, 2 µm.
Some ER- and membrane-associated proteins contain sorting motifs and/or glycophosphatidylinositol (GPI)-linked or tail-anchor signals that may be masked by an epitope such as GFP1–10. Growth assays on a variety of media and growth conditions suggested that most of our C-terminal fusions are functional (Fig. S4 A and not depicted), and we also localized N-terminally tagged versions of many of these proteins. We found that 5 of 23, 8 of 40, 1 of 3, and 1 of 4 tested GPI-linked, tail-anchored, HDEL-containing, and CAAX-containing proteins, respectively, were hits in our screen (Table S5). Localization of C-terminally tagged Vtc1 and Vtc4 were recently proposed to be degraded at the INM via the INMAD pathway (Khmelinskii et al., 2014). In our screen, we detected Vtc1 but not other subunits of the vacuolar transporter chaperone complex (Vtc2, Vtc3, or Vtc4) at the INM (Fig. 6 B). Of the 20 putative INMAD pathway targets proposed (Khmelinskii et al., 2014), 8 of the 15 tested localized to the INM, similar to the fraction of targets of the ER-associated ligase, Hrd1 (10 of 20 tested).
Proteins with small extraluminal domains have greater INM access
No particular motif was enriched above background within the 411 hits, lending evidence to the idea that a simple localization sequence is unlikely to confer INM localization (Katta et al., 2014; Ungricht and Kutay, 2015). Using the transmembrane predictor programs TMHMM and Phobius, we estimated molecular mass before and after the transmembrane domain of predicted single-pass membrane proteins (Table S4). Proteins that were INM hits were more likely to have small extraluminal domains (mean molecular mass of 14 ± 2 kD, n = 65) compared with non-INM proteins (29 ± 4 kD, n = 110; Fig. 4 D; P = 0.001). Additionally, overall size appears to be a factor in accessing the INM; predicted single-pass transmembrane proteins that localized to the INM were significantly smaller (46 ± 4 kD, n = 65) than proteins that did not localize (61 ± 5 kD, n = 110 for all, P = 0.03; 62 ± 6 kD, n = 54 for endomembrane proteins, P = 0.02; Fig. 4 D). Recent work suggests that INM transport occurs primarily through diffusion, which would require that INM components be small, particularly in terms of the extraluminal domain (Boni et al., 2015; Ungricht et al., 2015). The size-dependent leak through NPCs proposed in yeast would also bias the INM toward smaller proteins (Popken et al., 2015). Our data extend these studies, implicating size as a general global feature of proteins found at the INM. However, it is important to note that some membrane proteins as large as 178 kD (Pep1) were detected at the INM. Three (Heh1, 95 kD; Heh2, 76 kD; and Mps3, 79 kD) of the 15 INM proteins >60 kD are transported by alternative pathways requiring active transport (Table S4; King et al., 2006; Gardner et al., 2011; Meinema et al., 2011).
Fluorescence correlation spectroscopy estimates that a fraction of ER protein accesses the INM
An extension of the idea that proteins reach the INM primarily by diffusion is that ER complex assembly may limit INM access because of size—only unassembled subunits would be free to diffuse in and out of the INM. Therefore, it was somewhat surprising to find that many or all subunits of ER complexes had INM access (for example, five of five protein mannosyltransferase subunits; 4/5 GPI-anchor transamidase subunits (one not tested); 4/4 Sec63 complex subunits; and 2/3 and 3/3 members of the Sec61 and Ssh1 translocons, respectively). It is possible that these proteins are unassembled, an idea that we tested using GFP and split-GFP in combination with fluorescence correlation spectroscopy (FCS) and fluorescence cross-correlation spectroscopy (FCCS) to measure the diffusion rate and binding affinity of proteins at the INM, ONM, and ER—metrics that are directly related to the type and amount of protein complex formed at each location.
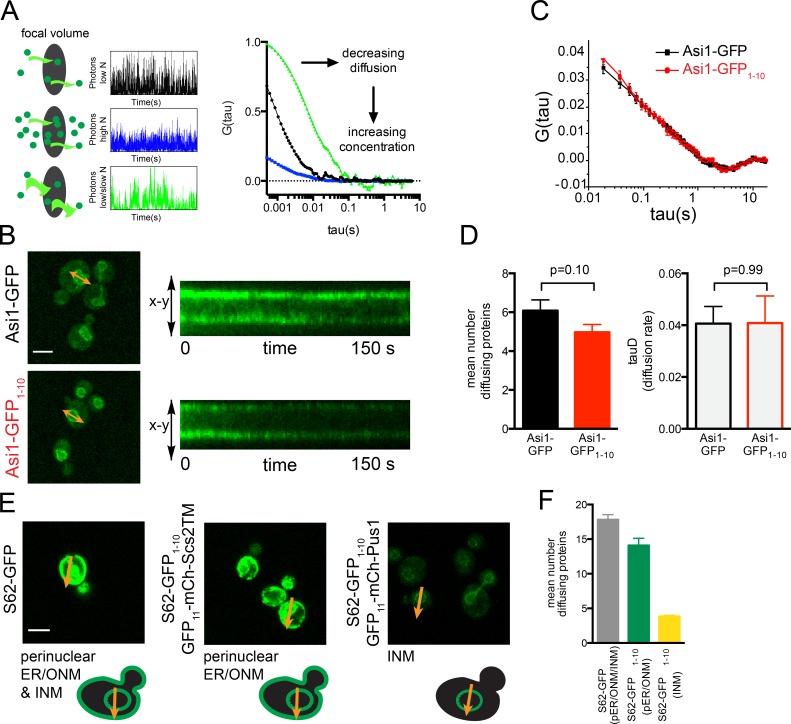
In FCS, intensity fluctuations of fluorescently labeled molecules within a defined region of the cell are recorded over a period of time (Fig. 7 A). Correlation analysis is then performed and used to extract information about the fluorescent molecules, such as the number of diffusing molecules and transit time (Slaughter et al., 2011). Although FCS does not reveal the identity of possible binding partners, it provides information about the size and heterogeneity of mobile complexes containing the fluorescently labeled protein and can be done together with confocal linescanning to measure fluctuations at the ER using GFP-tagged proteins and INM using reconstituted split-GFP.
Figure 7.
FCS can be used in combination with split-GFP. (A) Schematic demonstrating the principle of FCS. Simulated data and subsequent correlation curves for diffusing molecules through the focal volume of samples with increased concentration (middle) or decreased diffusion rate (bottom). The amplitude of the correlation is inversely proportional to the number of diffusing molecules (n), and the slope (τD) is related to the diffusion rate. (B) Localization of Asi1-GFP and Asi1-GFP1–10/GFP11-mCherry-Pus1. A line profile was generated spanning the NE, and linescanning FCS data were collected. Each scan can be visualized as a kymograph, which shows the fluctuations in molecules as they traverse the NE over time. (C) Mean autocorrelation curves of multiple Asi1-GFP (n = 25) and Asi1-GFP1–10 (n = 34) images and their fits are shown. (D) From each of these scans, the diffusion rate and number of diffusing molecules were determined, and mean values are plotted. (E) Focal volumes defined by arrows to include the perinuclear ER for Sec62-GFP, the ONM for Sec62-GFP1–10/GFP11-mCherry-Scs2TM, or the INM for Sec62-GFP1–10/GFP11-mCherry-Pus1. (F) The number of diffusing molecules based on FCS of 20 individual scans for each was calculated, and mean values are shown. Bars, 2 µm. In, C, D, and F, error bars indicate SEM. P-values were determined by Student’s t test. All values were highly statistically significant (P < 0.01) unless noted otherwise. FCS of Sec61 and Ssh1 at the NE and cortical ER depicted in Fig. S5.
One concern was that the GFP11-mCherry-Pus1 reporter would interfere with analysis, because it adds ∼60 kD to the INM protein. However, when we compared FCS of the INM protein Asi1 visualized using GFP1–10 bound to the reporter to that of Asi1-GFP, we observed virtually identical diffusion rates, and the concentrations of Asi1-GFP (n = 10.15 ± 0.92) and Asi1-GFP1–10 (n = 8.29 ± 0.67) were statistically similar despite decreased brightness of reconstituted split-GFP compared with GFP (Fig. 7, B–D). The concentration similarity is expected based on the localization of Asi1 to the INM, whereas the similarity in diffusion coefficient may reflect the slow diffusion of Asi1 in a large protein complex such that addition of the reporter does not appreciably change the diffusion properties (Boban et al., 2006; Zargari et al., 2007). Thus, it is possible to use split-GFP and GFP in combination with FCS to study the composition of protein complexes within the cell.
Unlike Asi1, which localizes exclusively to the INM, other hits are found at the cortical ER that is adjacent to the cell periphery in yeast and at the perinuclear region, which presumably is a mixture of protein at the INM and ONM/perinuclear ER. To confirm this idea, we analyzed FCS taken in the perinuclear region using Sec62-GFP and found that the mean number of diffusing molecules was the sum of the values on the INM and ONM, which were determined using Sec62-GFP1–10 with the INM or ONM reporter (Fig. 7, E and F). Based on this finding, we can estimate the fraction of GFP attributable to protein at the INM by dividing the mean number of diffusing reconstituted split-GFP molecules at the INM by the number of GFP molecules at the perinuclear region. For Sec62, 18% of the NE Sec62-GFP in the cell is at the INM. Estimates for other proteins based on FCS are listed in Table S1. These data further show that a fraction of ER proteins indeed access the INM.
FCCS shows a distinct INM variant of an ER protein complex
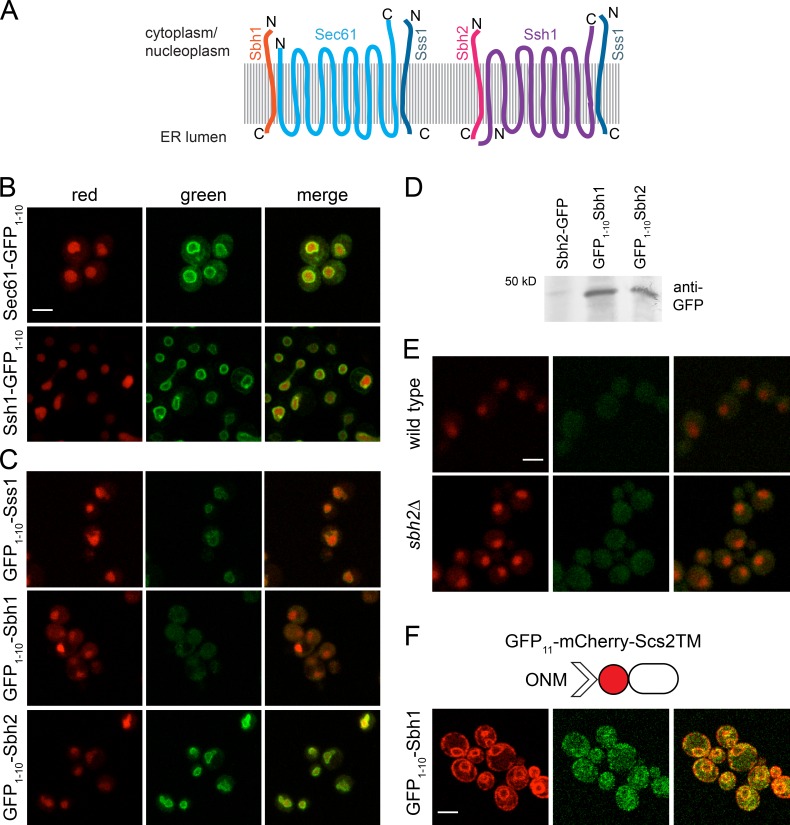
In the ER, Sec61 is part of a heterotrimeric translocon composed of Sec61/Sec61α, Sbh1/Sec61β, and Sss1/Sec61γ (Park and Rapoport, 2012). Yeast also contains a second translocon composed of Ssh1, Sbh2, and Sss1 (Panzner et al., 1995; Finke et al., 1996). Sec61, Ssh1, and Sbh2 were hits in our screen (Table S3 and Fig. 8, A and B), and we could detect GFP1–10-Sss1 at the INM using split-GFP in the presence of an untagged wild-type copy of SSS1 (Fig. 8 C). However, we were unable to demonstrate that GFP1–10-Sbh1 accesses the INM even in cells lacking SBH2 (Fig. 8 E). The inability of GFP1–10-Sbh1 to access the INM was not caused by poor growth, expression, folding, or topology, as we were able to detect the protein using the ONM/ER marker and by Western blotting (Fig. 8, C and F; and Fig. S4 B). Therefore, the observation that Sec61 and Sss1 access the INM but Sbh1 does not suggests that Sec61 may exist as a monomer on the INM. Alternatively, it could assemble into an INM-specific complex, in which Sbh2 replaces Sbh1, which is homologous over 50% of its length.
Figure 8.
Sbh2, but not Sbh1, localizes to the INM. (A) Schematic of the Sec61 and Ssh1 heterotrimers, including protein topology. (B and C) Localization of Sec61-GFP1–10 and Ssh1-GFP1–10 (B) and N-terminal GFP1–10 fusions to Sss1, Sbh1, and Sbh2 (C). (D) Western blotting with anti-GFP antibodies of the indicated strains shows that GFP1–10-Sbh1 is expressed. The 50-kD molecular mass marker is shown. (E) Localization of GFP1–10-Sbh1 expressed under its native promoter in wild-type and sbh2Δ cells. In B–E, cells contained GFP11-mCherry-Pus1 (red); INM localization based on GFP1–10 (green). (F) GFP1–10-Sbh1 localization at the ONM/ER was tested using the ONM/ER reporter, GFP11-mCherry-Scs2TM. Bars, 2 µm.
FCCS is a method that queries the diffusion of pairwise combinations of fluorescently labeled proteins to determine whether they are present in the same mobile complex (Fig. 9 A). To test whether Sec61 interacts with Sbh2 at the INM, we performed FCCS using Sec61-mCherry and GFP1–10-Sbh2, expressed under its native promoter. As controls, we also performed FCCS using Ssh1-mCherry and GFP1–10-Sbh2 because biochemical work suggests that the two interact (Finke et al., 1996). In addition, we tested FCCS between pairs of Sec61-mCherry/Ssh1-mCherry and GFP-Sbh1/GFP-Sbh2 on the cortical ER to compare this approach with coimmunoprecipitation and copurification data (Panzner et al., 1995; Finke et al., 1996; Harada et al., 2011). Two important modifications to our system were needed for FCCS analysis. First, mCherry was removed from the INM reporter so that we could study endogenously expressed SEC61 or SSH1 fused to mCherry in combination with GFP or split-GFP. Second, because the NE pool of Sec61-mCherry and Ssh1-mCherry is composed of protein present on the INM and ONM (Fig. 7, E and F), it was necessary to account for the amount of protein at the INM. For this, the fraction of mCherry bound to reconstituted split-GFP was computationally adjusted by the percentage of mCherry on the INM, which was calculated by comparison of FCS measurements for GFP and split-GFP tagged Sec61 and Ssh1 (see Materials and Methods; Fig. S5).
Figure 9.
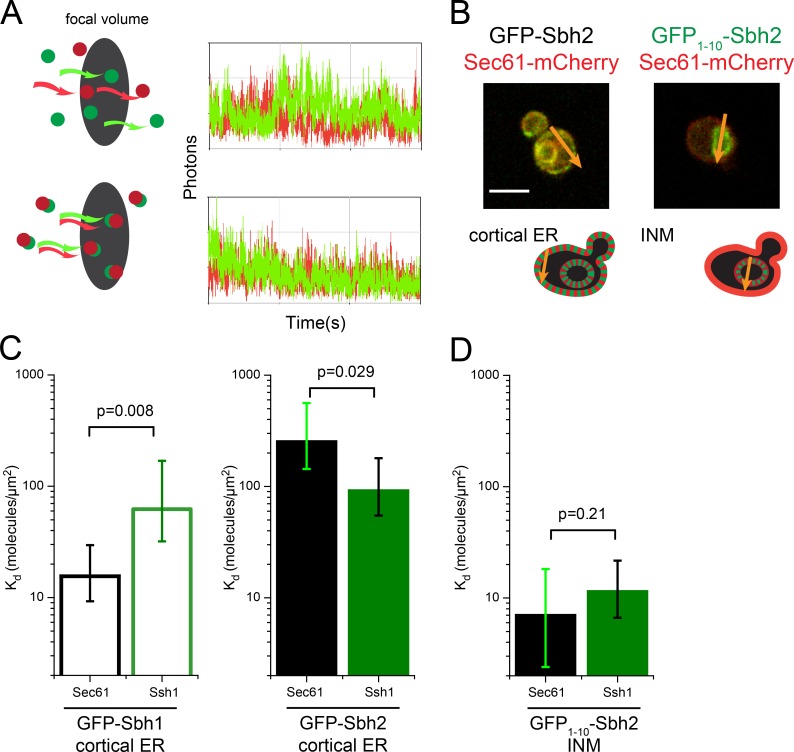
FCCS with split-GFP reveals novel INM complex. (A) Principle of FCCS is illustrated for randomly diffusing red and green particles (top) and codiffusing particles (bottom) using simulated data. (B) Focal volumes defined by arrows to include the cortical ER for Sbh2-GFP or INM for GFP1–10-Sbh2/GFP11-Pus1 (green) in strains containing Sec61-mCherry (red) show how FCCS data were acquired at specific regions within the cell. Bar, 2 µm. (C) Kd for Sbh1 and Sbh2 with Sec61 (n = 21, Sbh1; n = 23, Sbh2) and Ssh1 (n = 37, Sbh1; n = 27, Sbh2) at the cortical ER was determined by cross-correlation analysis as described in Materials and Methods from FCCS data. (D) Kd for Sbh2 with Sec61 (n = 45) and Ssh1 (n = 24) at the INM. Because Sbh1 did not localize to INM, no FCCS at this location could be acquired. Error bars in C and D indicate SEM. P-values were calculated by Monte Carlo simulation.
From FCCS analysis, we derived dissociation constants that express the relative affinity of the two molecules in question for each other: a high Kd (molecules/micrometers squared) is indicative of a weak or transient interaction, whereas a low Kd suggests that the two proteins are tightly bound (Slaughter et al., 2011). At the cortical ER, we found that GFP-Sbh1 had a lower Kd with Sec61-mCherry than with Ssh1-mCherry (15.6 vs. 62.2, P = 0.008), whereas the reverse was true for GFP-Sbh2 (Fig. 9 C); the Kd for Sbh2-Sec61 is 250.6 and for Sbh2-Ssh1 is 90.8 (P = 0.029). Interestingly, the Kd value calculated for GFP-Sbh2 and Ssh1-mCherry is similar to that of GFP-Sbh1 and Ssh1-mCherry, whereas the Kd between GFP-Sbh2 and Sec61-mCherry is >15-fold higher (Fig. 9 C). This data are consistent with previous studies showing that Sbh1 is the preferred binding partner of Sec61, whereas Ssh1 mainly associates with Sbh2 (Panzner et al., 1995; Finke et al., 1996). Our data suggest that the majority of this specificity may arise from a lack of Sec61 and Sbh2 binding at the cortical ER.
When we examined Sec61-mCherry and Ssh1-mCherry binding to INM reconstituted GFP1–10-Sbh2, we found that Sbh2 codiffuses with Sec61 and Ssh1 at the INM with a Kd of 6.9 and 11.3 molecules/µm2 (P = 0.21), respectively (Fig. 9 D). Surprisingly, binding between Sbh2 and Sec61 at the INM is greater than that of Sbh2 and Ssh1 in the same compartment (Fig. 9 D). In addition, 54% of Sec61-mCherry is associated with reconstituted GFP1–10-Sbh2 at the INM, whereas only 29% of Ssh1-mCherry is bound to INM GFP1–10-Sbh2. The diffusion characteristics of Sbh2 on the NE are more similar to Sec61 than Ssh1 (Sbh2 = 0.073, Sec61 = 0.096, Ssh1 = 0.133 µm2/s). Collectively, these data support the idea that Sbh2 has the ability to associate with Sec61 at the INM.
Discussion
Our study represents an important extension to localization studies using GFP in that we were able to detect proteins specifically at the INM using fluorescence microscopy. Our hits show minimal overlap (28) with the top 200 yeast NE proteins from Chong et al., 2015: most (155) of their NE hits were not in our screen because they lack a transmembrane domain, whereas 12 were negative, 3 were soluble, and 2 were untagged. This observation suggests that many proteins at the nuclear periphery in yeast associate with the membrane through interactions with integral membrane components, as we showed for Sec72.
Through the use of split-GFP, we found a significant overlap between proteins that are able to access the INM and proteins that function in the ER. Although this may not be entirely unexpected given that components of the ER have been shown to localize to the INM in other imaging studies (Saksena et al., 2004, 2006; Deng and Hochstrasser, 2006), our work suggests that roughly 35% of the proteins that access the INM come from the ER. By combining split-GFP with FCCS, we provided evidence that the INM is not simply an extension of the ER but rather is a distinct membrane compartment. Although our data do not show that proteins such as Sec61 and Sbh2 function within the nucleus as they do in the ER, the localization of membrane-based components from the ER and other organelles may explain aspects of nuclear membrane biology including synthesis of INM lipids, regulated control of NE breakdown, and fusion or targeting and degradation of misfolded/damaged INM proteins.
One of our most surprising results was the large number of proteins (411) that were able to access the INM. We estimate that <5–10% of total cellular Sec61 and Sec62 is at the INM, a level that falls significantly below the threshold limit used in most proteomic studies of NE composition. This enrichment would also be challenging to detect using traditional imaging. A focused screen using GFP-tagged versions of endomembrane proteins that included 386 of our 412 INM components hinted at nuclear functions for only 19, including Heh1/2, Mps3, Pom34, Pom152, Ndc1, Asi1/3, Nsg1, Vma22, Nvj1, Lac1, Hmg1, Ubc7, Ydc1, Sec72, and 3 uncharacterized ORFs (Yofe et al., 2016). The major advantage to split-GFP is that we only examine the INM pool. The strong interaction between GFP1–10 and GFP11 may also slow diffusion and turnover, providing us with an extended picture of proteins that access the INM. Although this may increase our ability to visualize proteins in the INM, it does not indicate whether the proteins (or protein complexes studied by FCCS) are transient or intermediate species in the INM.
An intriguing question raised by our data is how a subset of ER and other membrane proteins comes to reside at the INM, and whether protein complexes assemble before or after translocation through the NPC. No particular targeting motif was identified, yet most INM proteins contained small extraluminal domains. These proteins are likely to reach the INM by passive diffusion through peripheral channels in the NPC. However, size alone is insufficient to account for INM localization, as many small soluble and membrane proteins do not reach the nucleus. Although it is possible they may be tethered in larger complexes, recent data suggest that the NPC barrier is permeable to proteins with domains as large as 90–100 kD (Popken et al., 2015). We therefore propose that the INM proteome is defined based on diffusion, retention, and degradation of INM proteins. Because protein localization to the INM can be assayed in live cells using split-GFP, the methodology presented in this study opens new avenues to address the relative contributions of these pathways to defining the INM proteome under a wide variety of conditions.
More than half (230/411) of the yeast INM proteome has a clear human ortholog; therefore, understanding how these proteins localize and function at the INM under different growth conditions will help elucidate their role in human disease, aging, cell stress, and cell-cycle arrest. The steady-state abundance of our 411 INM hits does not appear to be cell-cycle regulated; however, careful quantitation of INM levels, analysis of INM transport, and use of multiple reporters will be needed to confirm this finding. Our approach is not restricted to the INM, as illustrated by the ONM/ER, cytoplasmic, and luminal reporters. These modified reporters can be introduced into strains generated in this study to test localization to other subcellular compartments to further refine our knowledge of protein localization and targeting at the single-cell level. Recent work from Rogers and Rose (2014) illustrates the utility of split-GFP in the detection of Kar5 at the INM and ONM during nuclear membrane fusion in yeast. In addition, split-GFP can be used to determine protein topology, as we showed for Mps3 and Heh2. A similar split-GFP system to detect topology, ER membrane association, and even INM access has been developed in mammalian cells; therefore, the methods we have developed here should be easily portable to study protein dynamics and interactions at the single-cell level in other eukaryotic systems (Feinberg et al., 2008; Kilpatrick et al., 2012; Hyun et al., 2015; Kamiyama et al., 2016).
Materials and methods
Yeast strains and plasmids
All GFP1–10 and GFP strains are derivatives of BY (can1Δ::STE2pr-SpHIS5 lyp1Δ his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 LYS2). Split-GFP reporters consisted of pRS315-NOP1pr-GFP11-mCherry-PUS1 (pSJ1321) or pRS315-NOP1pr-GFP11-PUS1 (pSJ1679) for the INM, pRS315-NOP1pr-GFP11-mCherry-HXK1 (pSJ1996) for the cytosol, pRS315-NOP1pr-GFP11-mCherry-SCS2TM (pSJ1568) for the ONM, and pRS315-NOP1pr-mCherry-SCS2TM-GFP11 (pSJ1602) for the ER lumen. The INM split-GFP reporter pRS315-NOP1pr-GFP11-mCherry-PUS1 (pSJ1321) was made in three steps. First, ApaI and XhoI restriction sites were introduced at the 5′ end of the GFP and an NheI site at the 3′ end of the GFP in pNOP1pr-GFP-PUS1-LEU2 (Hellmuth et al., 1998) using the QuikChange II mutagenesis kit (Agilent Technologies). A PCR fragment containing a yeast codon-optimized version of mCherry was amplified and inserted at XhoI and NheI sites (in place of the GFP gene), and a dsDNA-containing, yeast codon-optimized GFP11 was cloned at ApaI and XhoI sites. The INM reporter lacking mCherry (pRS315-NOP1pr-GFP11-PUS1 [pSJ1679]) was made by digesting pSJ1321 with ApaI and NheI to remove GFP11-mCherry and replacing it with a PCR product for GFP11 alone. To make the cytoplasmic marker, the PUS1 gene was replaced with HXK1 using the NheI and SphI sites. To create the ONM/ER split-GFP reporter pRS315-NOP1pr-GFP11-mCherry-SCS2TM (pSJ1568), the PUS1 gene was replaced in pSJ1321 by the sequence for the Scs2 transmembrane domain (residues 223–244) using in-frame NheI and SacI restriction sites engineered into the PCR primers. To create pRS316-NOP1pr-GFP1–10-SCS2TM (pSJ2039), the NOP1pr-GFP1–10-SCS2TM-ADH1tr sequence was made by gene synthesis and inserted into the SacI/NotI sites in pRS316. The luminal marker (pSJ1602) was made by moving the GFP11 to the other side of the Scs2 transmembrane domain. A yeast codon-optimized version of GFP1–10 was made by gene synthesis and inserted into the PacI site of pFA6-link-yEGFP-CaURA3MX (pKT209; Sheff and Thorn, 2004) in place of GFP to create pSJ1256 (pFA6-link-yGFP1–10-CaURA3MX) for C-terminal tagging. N-terminal tagging plasmids were created in pFA6-NATMX4 (Goldstein and McCusker, 1999). 737 bp of the CDC42 promoter immediately upstream of the start codon was PCR amplified along with GFP1–10 and cloned into the SacI–SpeI sites to create pS1643 (pFA6-NATMX-CDC42pr-yGFP1–10), using Gibson assembly (New England BioLabs, Inc.). A second version (pSJ1726 [pFA6-NATMX-CDC42pr-KAR2SS-GFP1–10]) containing the coding sequence for residues 1–87 of Kar2, which includes the signal sequence before GFP1–10, was also made by Gibson assembly. For theta nuclei experiments, strains were made in W303 (ade2-1 trp1-1 leu2-3,112 ura3-1 his3-11,15 can1-100 RAD5+); genes were tagged with mCherry using pFA6-mCherry-KANMX in a strain containing GAL-NUP53-GFP. Strains used for each experiment are listed in Table S2.
Standard techniques were used for yeast growth and manipulations. Growth of strains containing GFP1–10-tagged GPI-anchored proteins and Sec61/Ssh1 translocon subunits was tested on plates containing yeast extract peptone dextrose (YPD), YPD + 2.5 mM DTT (D9779-10G; Sigma-Aldrich), YPD + 0.5 µg/ml myriocin (M1177-5MG; Sigma-Aldrich), YPD + 0.25 µg/ml tunicamycin, and YPD + 0.5 µg/ml tunicamycin (T7765; Sigma-Aldrich).
Library construction
Genes associated with the GO annotation of integral component of membrane or transmembrane were compiled using SGD, and TMHMM (Krogh et al., 2001) was used to predict additional genes containing hydrophobic stretches of greater than 16 aa using a version of the genome downloaded on June 10, 2012.
PCR primers to tag genes at the C terminus were designed as follows: F5 primer, 60 bp of gene-specific sequence immediately before the stop codon followed by 5′-GGTGACGGTGCTGGTTTA-3′; R3 primer, 60 bp of gene-specific sequence immediately after the stop codon on the reverse strand followed by 5′-TCGATGAATTCGAGCTCG-3′. PCR primers to tag genes at the N terminus were designed as follows: forward primer, 60 bp of gene-specific sequence up to the start codon followed by 5′-GGTCGACGGATCCCCGGGTT-3′; reverse primer, 60 bp of gene-specific sequence immediately after the start codon on the reverse strand followed by 5′-AGAACCACCACCAGAACCAC-3′. PCR products amplified from DNA templates using a high-fidelity polymerase were transformed into SLJ7859 (MATα can1Δ::STE2pr-Sp-HIS5 lyp1Δ his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 LYS2 pCEN/ARS-LEU2-NOP1pr-GFP11-mCherry-PUS1 [pSJ1321]). Integration of GFP1–10 at N or C terminus of the target gene and confirmation that the tagged gene was the sole copy in the cell were verified by PCR as described (Gardner and Jaspersen, 2014). Two independent isolates of each product were frozen as glycerol stocks. Replicates of the library were created to facilitate distribution to the community upon request.
INM screen
Cells were grown overnight at 23°C in SC-Leu to mid-log phase. Samples were immobilized between a slide and a no. 1.5 coverslip before imaging with a 40×, 1.2-NA, Plan Apochromatic objective on a Confocor 3 (ZEISS) using the avalanche photodiode imaging module. The following parameters were used: fluorophores were excited using a 488-nm argon laser line, GFP emission was collected through a BP 505- to 540-nm filter, and mCherry was collected through a BP 615- to 680-nm filter. Using an acousto-optic tunable filter dampener, a 488-nm laser power range between 2 and 20 µW was scanned to detect green fluorescence. If signal was observed, at least three sets of images were acquired using AIM v.4.2 software (ZEISS). Images were collected with six to eight image stacks with a 0.5-µm step size through the cells at RT. Images were processed using ImageJ software (National Institutes of Health). Localization to the INM, nucleus, or other compartment was determined by manual inspection of the images and bioinformatics analysis of hits as performed using Yeast Mine tools at SGD. For data in Fig. 4 D, molecular mass analysis was performed on single-pass transmembrane proteins using Phobius and TMHMM. A summary of all genes and their localization patterns is listed in Table S3. A complete repository of all images is available at http://research.stowers.org/jaspersenlab/542807/. Maximum-intensity projections over two to five z-slices were created (except single-plane images shown for theta nuclei experiments: Figs. 5 C and S3), and images were then binned 2 × 2 and smoothed with a Gaussian blur of 0.05 for figures.
Linescanning FCS and FCCS
Linescanning FCS and FCCS were performed as described (Slaughter et al., 2011; Chen et al., 2014). For FCCS, the following parameters were used: fluorophores were excited using a 488- and 561-nm argon laser line, GFP emission was collected through a BP 505- to 540-nm filter, and mCherry was collected through an LP 580-nm filter, with the pinhole set to 1.78 airy units. Lines through the nucleus and cortical ER were selected to cross a central focal plane perpendicular to the nuclear and cell periphery, as depicted. Linescanning time series were collected with a line size of 512 pixels and an effective line time for both channels of 15.3 ms. The pixel size was 22 nm, resulting in a total line size of 11.3 µm. The pixel dwell time was 6.4 µs. The number of particles per focal volume was calculated from the amplitude of the correlation curve using N = γ/G(0), with a γ value of 0.45 for 2D diffusion on a membrane (Slaughter et al., 2013). The diffusion coefficient was calculated from the transit time (τD) of the correlation decay based on D = ro2/4 τD, using an ro value of 0.17 µm (Slaughter et al., 2013). Linescanning kymographs were analyzed using custom software in ImageJ, available at http://research.stowers.org/imagejplugins.
Derivation of binding values using GFP and split-GFP
Fusions to GFP and mCherry were made by PCR tagging as previously described (Gardner and Jaspersen, 2014). Because GFP and mCherry allow for visualization of NE protein at both the INM and ONM, it was necessary for us to account for the fraction of mCherry protein at the INM because this is the only pool of protein able to diffuse with split-GFP. To do this, we used the relative particle number from split-GFP versus GFP-tagged strains at the NE to estimate the proportion of particles at the INM versus the whole NE in FCCS experiments: fINM = NE GFP/INM split-GFP. We then made the assumption that mCherry-tagged particles were present at the same proportion in these compartments. This proportion was used to calculate the number of bound mCherry particles at the INM: Nbound,INM = Nbound × fINM.
Once the number of free and bound particles was obtained, it was possible to estimate the dissociation constant as follows (relative Kd in units of number of particles per focal volume): Kd = Ng × fg,bound × Nr × fr,bound/(0.5 × Ng,bound × Nr,bound), with a volume of 0.6 µm2, as measured with a fluorescent bead (Slaughter et al., 2013). This assumes that our equilibrium can be approximated as a first-order reaction. Note that we have averaged the Nbound value from the red and green signals. These are not identical because of noise and deviations from first-order equilibrium. Particle numbers and raw fractions bound were determined for each individual scanning FCS run, and SEMs were propagated through subsequent calculations by Monte Carlo simulations.
Immuno-EM
GFP-tagged strains used in this analysis were taken from the yeast GFP collection (Huh et al., 2003). Log-phase cells cultured overnight in YPD at 30°C were harvested and frozen on the EM-Pact (Leica Biosystems) at ∼2,050 bar, transferred under liquid nitrogen into 0.2% uranyl acetate and 1% water in acetone, and transferred to the AFS (Leica Biosystems). The freeze substitution protocol was as follows: −90° to −80° over 60 h, −80° to −60° over 4 h, −60° for 4 h, −60° to −50° over 6 h, and −50° to −20°C over 6 h. Samples were then removed from the AFS and processed for immuno-EM as previously described (Giddings et al., 2001). In brief, 60-nm serial thin sections were cut on a UC6 (Leica Biosystems); blocked for 1 h with 0.8% BSA/0.1% gelatin in PBST (10 mM sodium phosphate, pH 7.4, 150 mM sodium chloride, and 0.1% Tween); and immunostained with anti-GFP antibodies (a gift from M. Rout, Rockefeller University, New York, NY) for 2 h at RT, washed in a stream of PBST, and floated on 12 nm colloidal gold–conjugated anti–rabbit secondary antibodies (Jackson ImmunoResearch Laboratories, Inc.) diluted in PBST for 1 h. After washes in PBST and then water, samples were stained with uranyl acetate and Sato’s lead. Images were acquired on a FEI Technai Spirit.
Online supplemental material
Fig. S1 shows that karyogamy-defective cells exhibit two distinct nuclei. In Fig. S2, split-GFP illustrates other localization phenotypes. In Fig. S3, theta nuclei verify several low scoring-INM hits. Fig. S4 shows growth of selected split-GFP–tagged ER–INM hits. In Fig. S5, FCS of Sec61 and Ssh1 reveals different characteristics in the cortical and perinuclear ER. Table S1 shows FCS data for ER translocon proteins. Table S2 shows yeast strains used in this study. Table S3 shows results of split-GFP1–10 screen. Table S4 shows proteins that access the yeast INM based on split-GFP1–10 screen. Table S5 shows the contribution of proteins with known C-terminal motifs to our library.
Supplementary Material
Acknowledgments
We are grateful to Ann Sizemore, Ariel Paulson, and Madelaine Gogol for help during this project and to Jaspersen laboratory members for comments on the manuscript. Original data underlying this manuscript can be downloaded from the Stowers Original Data Repository at http://www.stowers.org/pubs/LIBPB-1117.
S.L. Jaspersen is supported by the Stowers Institute for Medical Research and the American Cancer Society (RSG-11-030-01-CSM). C.J. Smoyer is a predoctoral researcher in the Graduate School of the Stowers Institute.
The authors declare no competing financial interests.
Author contributions: C.J. Smoyer, J.R. Unruh, B.D. Slaughter, and S.L. Jaspersen conceived of using split-GFP for INM studies; C.J. Smoyer and S.S. Katta developed tools for its use in yeast and performed pilot experiments; S. McCroskey and W.D. Bradford made the yeast libraries that were screened by C.J. Smoyer with assistance from L. Stoltz, S.E. Smith, J.M. Gardner, and M. McClain; FCS and FCCS experiments were performed and analyzed by C.J. Smoyer using image analysis tools developed by J.R. Unruh, with assistance from B.D. Slaughter; and C.J. Smoyer and S.L. Jaspersen wrote the paper with input from all the authors.
Footnotes
Abbreviations used:
- FCCS
- fluorescence cross-correlation spectroscopy
- FCS
- fluorescence correlation spectroscopy
- GPI
- glycophosphatidylinositol
- INM
- inner nuclear membrane
- INMAD
- INM-associated destruction
- NE
- nuclear envelope
- NPC
- nuclear pore complex
- ONM
- outer nuclear membrane
- SGD
- Saccharomyces genome database
- SPB
- spindle pole body
- TMHMM
- transmembrane helices hidden Markov model
- YPD
- yeast extract peptone dextrose
References
- Boban M., Zargari A., Andréasson C., Heessen S., Thyberg J., and Ljungdahl P.O.. 2006. Asi1 is an inner nuclear membrane protein that restricts promoter access of two latent transcription factors. J. Cell Biol. 173:695–707. 10.1083/jcb.200601011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boni A., Politi A.Z., Strnad P., Xiang W., Hossain M.J., and Ellenberg J.. 2015. Live imaging and modeling of inner nuclear membrane targeting reveals its molecular requirements in mammalian cells. J. Cell Biol. 209:705–720. 10.1083/jcb.201409133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breker M., Gymrek M., and Schuldiner M.. 2013. A novel single-cell screening platform reveals proteome plasticity during yeast stress responses. J. Cell Biol. 200:839–850. 10.1083/jcb.201301120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke B., and Stewart C.L.. 2014. Functional architecture of the cell’s nucleus in development, aging, and disease. Curr. Top. Dev. Biol. 109:1–52. 10.1016/B978-0-12-397920-9.00006-8 [DOI] [PubMed] [Google Scholar]
- Cabantous S., and Waldo G.S.. 2006. In vivo and in vitro protein solubility assays using split GFP. Nat. Methods. 3:845–854. 10.1038/nmeth932 [DOI] [PubMed] [Google Scholar]
- Cabantous S., Terwilliger T.C., and Waldo G.S.. 2005. Protein tagging and detection with engineered self-assembling fragments of green fluorescent protein. Nat. Biotechnol. 23:102–107. 10.1038/nbt1044 [DOI] [PubMed] [Google Scholar]
- Chen J., Smoyer C.J., Slaughter B.D., Unruh J.R., and Jaspersen S.L.. 2014. The SUN protein Mps3 controls Ndc1 distribution and function on the nuclear membrane. J. Cell Biol. 204:523–539. 10.1083/jcb.201307043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chial H.J., Rout M.P., Giddings T.H., and Winey M.. 1998. Saccharomyces cerevisiae Ndc1p is a shared component of nuclear pore complexes and spindle pole bodies. J. Cell Biol. 143:1789–1800. 10.1083/jcb.143.7.1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho R.J., Campbell M.J., Winzeler E.A., Steinmetz L., Conway A., Wodicka L., Wolfsberg T.G., Gabrielian A.E., Landsman D., Lockhart D.J., and Davis R.W.. 1998. A genome-wide transcriptional analysis of the mitotic cell cycle. Mol. Cell. 2:65–73. 10.1016/S1097-2765(00)80114-8 [DOI] [PubMed] [Google Scholar]
- Chong Y.T., Koh J.L., Friesen H., Duffy S.K., Cox M.J., Moses A., Moffat J., Boone C., and Andrews B.J.. 2015. Yeast proteome dynamics from single cell imaging and automated analysis. Cell. 161:1413–1424. 10.1016/j.cell.2015.04.051 [DOI] [PubMed] [Google Scholar]
- Dauer W.T., and Worman H.J.. 2009. The nuclear envelope as a signaling node in development and disease. Dev. Cell. 17:626–638. 10.1016/j.devcel.2009.10.016 [DOI] [PubMed] [Google Scholar]
- Davidson P.M., and Lammerding J.. 2014. Broken nuclei—lamins, nuclear mechanics, and disease. Trends Cell Biol. 24:247–256. 10.1016/j.tcb.2013.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng M., and Hochstrasser M.. 2006. Spatially regulated ubiquitin ligation by an ER/nuclear membrane ligase. Nature. 443:827–831. 10.1038/nature05170 [DOI] [PubMed] [Google Scholar]
- Feinberg E.H., Vanhoven M.K., Bendesky A., Wang G., Fetter R.D., Shen K., and Bargmann C.I.. 2008. GFP reconstitution across synaptic partners (GRASP) defines cell contacts and synapses in living nervous systems. Neuron. 57:353–363. 10.1016/j.neuron.2007.11.030 [DOI] [PubMed] [Google Scholar]
- Feldheim D., and Schekman R.. 1994. Sec72p contributes to the selective recognition of signal peptides by the secretory polypeptide translocation complex. J. Cell Biol. 126:935–943. 10.1083/jcb.126.4.935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finke K., Plath K., Panzner S., Prehn S., Rapoport T.A., Hartmann E., and Sommer T.. 1996. A second trimeric complex containing homologs of the Sec61p complex functions in protein transport across the ER membrane of S. cerevisiae. EMBO J. 15:1482–1494. [PMC free article] [PubMed] [Google Scholar]
- Foresti O., Rodriguez-Vaello V., Funaya C., and Carvalho P.. 2014. Quality control of inner nuclear membrane proteins by the Asi complex. Science. 346:751–755. 10.1126/science.1255638 [DOI] [PubMed] [Google Scholar]
- Friederichs J.M., Ghosh S., Smoyer C.J., McCroskey S., Miller B.D., Weaver K.J., Delventhal K.M., Unruh J., Slaughter B.D., and Jaspersen S.L.. 2011. The SUN protein Mps3 is required for spindle pole body insertion into the nuclear membrane and nuclear envelope homeostasis. PLoS Genet. 7:e1002365 10.1371/journal.pgen.1002365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner J.M., and Jaspersen S.L.. 2014. Manipulating the yeast genome: Deletion, mutation, and tagging by PCR. Methods Mol. Biol. 1205:45–78. 10.1007/978-1-4939-1363-3_5 [DOI] [PubMed] [Google Scholar]
- Gardner J.M., Smoyer C.J., Stensrud E.S., Alexander R., Gogol M., Wiegraebe W., and Jaspersen S.L.. 2011. Targeting of the SUN protein Mps3 to the inner nuclear membrane by the histone variant H2A.Z. J. Cell Biol. 193:489–507. 10.1083/jcb.201011017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami S., Huh W.K., Bower K., Howson R.W., Belle A., Dephoure N., O’Shea E.K., and Weissman J.S.. 2003. Global analysis of protein expression in yeast. Nature. 425:737–741. 10.1038/nature02046 [DOI] [PubMed] [Google Scholar]
- Giddings T.H. Jr., O’Toole E.T., Morphew M., Mastronarde D.N., McIntosh J.R., and Winey M.. 2001. Using rapid freeze and freeze-substitution for the preparation of yeast cells for electron microscopy and three-dimensional analysis. Methods Cell Biol. 67:27–42. 10.1016/S0091-679X(01)67003-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein A.L., and McCusker J.H.. 1999. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast. 15:1541–1553. [DOI] [PubMed] [Google Scholar]
- Harada Y., Li H., Wall J.S., Li H., and Lennarz W.J.. 2011. Structural studies and the assembly of the heptameric post-translational translocon complex. J. Biol. Chem. 286:2956–2965. 10.1074/jbc.M110.159517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellmuth K., Lau D.M., Bischoff F.R., Künzler M., Hurt E., and Simos G.. 1998. Yeast Los1p has properties of an exportin-like nucleocytoplasmic transport factor for tRNA. Mol. Cell. Biol. 18:6374–6386. 10.1128/MCB.18.11.6374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh W.K., Falvo J.V., Gerke L.C., Carroll A.S., Howson R.W., Weissman J.S., and O’Shea E.K.. 2003. Global analysis of protein localization in budding yeast. Nature. 425:686–691. 10.1038/nature02026 [DOI] [PubMed] [Google Scholar]
- Hyun S.I., Maruri-Avidal L., and Moss B.. 2015. Topology of endoplasmic reticulum-associated cellular and viral proteins determined with split-GFP. Traffic. 16:787–795. 10.1111/tra.12281 [DOI] [PubMed] [Google Scholar]
- Jan C.H., Williams C.C., and Weissman J.S.. 2014. Principles of ER cotranslational translocation revealed by proximity-specific ribosome profiling. Science. 346:1257521 10.1126/science.1257521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspersen S.L., Giddings T.H. Jr., and Winey M.. 2002. Mps3p is a novel component of the yeast spindle pole body that interacts with the yeast centrin homologue Cdc31p. J. Cell Biol. 159:945–956. 10.1083/jcb.200208169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiyama D., Sekine S., Barsi-Rhyne B., Hu J., Chen B., Gilbert L.A., Ishikawa H., Leonetti M.D., Marshall W.F., Weissman J.S., and Huang B.. 2016. Versatile protein tagging in cells with split fluorescent protein. Nat. Commun. 7:11046 10.1038/ncomms11046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katta S.S., Smoyer C.J., and Jaspersen S.L.. 2014. Destination: Inner nuclear membrane. Trends Cell Biol. 24:221–229. 10.1016/j.tcb.2013.10.006 [DOI] [PubMed] [Google Scholar]
- Khmelinskii A., Blaszczak E., Pantazopoulou M., Fischer B., Omnus D.J., Le Dez G., Brossard A., Gunnarsson A., Barry J.D., Meurer M., et al. . 2014. Protein quality control at the inner nuclear membrane. Nature. 516:410–413. 10.1038/nature14096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilpatrick L.E., Briddon S.J., and Holliday N.D.. 2012. Fluorescence correlation spectroscopy, combined with bimolecular fluorescence complementation, reveals the effects of β-arrestin complexes and endocytic targeting on the membrane mobility of neuropeptide Y receptors. Biochim. Biophys. Acta. 1823:1068–1081. 10.1016/j.bbamcr.2012.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Melén K., Osterberg M., and von Heijne G.. 2006. A global topology map of the Saccharomyces cerevisiae membrane proteome. Proc. Natl. Acad. Sci. USA. 103:11142–11147. 10.1073/pnas.0604075103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King M.C., Lusk C.P., and Blobel G.. 2006. Karyopherin-mediated import of integral inner nuclear membrane proteins. Nature. 442:1003–1007. 10.1038/nature05075 [DOI] [PubMed] [Google Scholar]
- Korfali N., Wilkie G.S., Swanson S.K., Srsen V., Batrakou D.G., Fairley E.A., Malik P., Zuleger N., Goncharevich A., de Las Heras J., et al. . 2010. The leukocyte nuclear envelope proteome varies with cell activation and contains novel transmembrane proteins that affect genome architecture. Mol. Cell. Proteomics. 9:2571–2585. 10.1074/mcp.M110.002915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korfali N., Wilkie G.S., Swanson S.K., Srsen V., de Las Heras J., Batrakou D.G., Malik P., Zuleger N., Kerr A.R., Florens L., and Schirmer E.C.. 2012. The nuclear envelope proteome differs notably between tissues. Nucleus. 3:552–564. 10.4161/nucl.22257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A., Larsson B., von Heijne G., and Sonnhammer E.L.. 2001. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 305:567–580. 10.1006/jmbi.2000.4315 [DOI] [PubMed] [Google Scholar]
- Marelli M., Lusk C.P., Chan H., Aitchison J.D., and Wozniak R.W.. 2001. A link between the synthesis of nucleoporins and the biogenesis of the nuclear envelope. J. Cell Biol. 153:709–724. 10.1083/jcb.153.4.709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinema A.C., Laba J.K., Hapsari R.A., Otten R., Mulder F.A., Kralt A., van den Bogaart G., Lusk C.P., Poolman B., and Veenhoff L.M.. 2011. Long unfolded linkers facilitate membrane protein import through the nuclear pore complex. Science. 333:90–93. 10.1126/science.1205741 [DOI] [PubMed] [Google Scholar]
- Mekhail K., and Moazed D.. 2010. The nuclear envelope in genome organization, expression and stability. Nat. Rev. Mol. Cell Biol. 11:317–328. 10.1038/nrm2894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa S., Terazawa Y., Nakayama T., Hirata A., Makio T., and Endo T.. 2003. Nep98p is a component of the yeast spindle pole body and essential for nuclear division and fusion. J. Biol. Chem. 278:9938–9943. 10.1074/jbc.M210934200 [DOI] [PubMed] [Google Scholar]
- Panzner S., Dreier L., Hartmann E., Kostka S., and Rapoport T.A.. 1995. Posttranslational protein transport in yeast reconstituted with a purified complex of Sec proteins and Kar2p. Cell. 81:561–570. 10.1016/0092-8674(95)90077-2 [DOI] [PubMed] [Google Scholar]
- Park E., and Rapoport T.A.. 2012. Mechanisms of Sec61/SecY-mediated protein translocation across membranes. Annu. Rev. Biophys. 41:21–40. 10.1146/annurev-biophys-050511-102312 [DOI] [PubMed] [Google Scholar]
- Popken P., Ghavami A., Onck P.R., Poolman B., and Veenhoff L.M.. 2015. Size-dependent leak of soluble and membrane proteins through the yeast nuclear pore complex. Mol. Biol. Cell. 26:1386–1394. 10.1091/mbc.E14-07-1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers J.V., and Rose M.D.. 2014. Kar5p is required for multiple functions in both inner and outer nuclear envelope fusion in Saccharomyces cerevisiae. G3 (Bethesda). 5:111–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothballer A., and Kutay U.. 2013. The diverse functional LINCs of the nuclear envelope to the cytoskeleton and chromatin. Chromosoma. 122:415–429. 10.1007/s00412-013-0417-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saksena S., Shao Y., Braunagel S.C., Summers M.D., and Johnson A.E.. 2004. Cotranslational integration and initial sorting at the endoplasmic reticulum translocon of proteins destined for the inner nuclear membrane. Proc. Natl. Acad. Sci. USA. 101:12537–12542. 10.1073/pnas.0404934101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saksena S., Summers M.D., Burks J.K., Johnson A.E., and Braunagel S.C.. 2006. Importin-alpha-16 is a translocon-associated protein involved in sorting membrane proteins to the nuclear envelope. Nat. Struct. Mol. Biol. 13:500–508. 10.1038/nsmb1098 [DOI] [PubMed] [Google Scholar]
- Schirmer E.C., Florens L., Guan T., Yates J.R.I. III, and Gerace L.. 2003. Nuclear membrane proteins with potential disease links found by subtractive proteomics. Science. 301:1380–1382. 10.1126/science.1088176 [DOI] [PubMed] [Google Scholar]
- Sheff M.A., and Thorn K.S.. 2004. Optimized cassettes for fluorescent protein tagging in Saccharomyces cerevisiae. Yeast. 21:661–670. 10.1002/yea.1130 [DOI] [PubMed] [Google Scholar]
- Slaughter B.D., Unruh J.R., and Li R.. 2011. Fluorescence fluctuation spectroscopy and imaging methods for examination of dynamic protein interactions in yeast. Methods Mol. Biol. 759:283–306. 10.1007/978-1-61779-173-4_17 [DOI] [PubMed] [Google Scholar]
- Slaughter B.D., Unruh J.R., Das A., Smith S.E., Rubinstein B., and Li R.. 2013. Non-uniform membrane diffusion enables steady-state cell polarization via vesicular trafficking. Nat. Commun. 4:1380 10.1038/ncomms2370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr D.A., and Fridolfsson H.N.. 2010. Interactions between nuclei and the cytoskeleton are mediated by SUN-KASH nuclear-envelope bridges. Annu. Rev. Cell Dev. Biol. 26:421–444. 10.1146/annurev-cellbio-100109-104037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strambio-de-Castillia C., Blobel G., and Rout M.P.. 1995. Isolation and characterization of nuclear envelopes from the yeast Saccharomyces. J. Cell Biol. 131:19–31. 10.1083/jcb.131.1.19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungricht R., and Kutay U.. 2015. Establishment of NE asymmetry—targeting of membrane proteins to the inner nuclear membrane. Curr. Opin. Cell Biol. 34:135–141. 10.1016/j.ceb.2015.04.005 [DOI] [PubMed] [Google Scholar]
- Ungricht R., Klann M., Horvath P., and Kutay U.. 2015. Diffusion and retention are major determinants of protein targeting to the inner nuclear membrane. J. Cell Biol. 209:687–703. 10.1083/jcb.201409127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkie G.S., Korfali N., Swanson S.K., Malik P., Srsen V., Batrakou D.G., de las Heras J., Zuleger N., Kerr A.R., Florens L., and Schirmer E.C.. 2011. Several novel nuclear envelope transmembrane proteins identified in skeletal muscle have cytoskeletal associations. Mol. Cell. Proteomics. 10:003129 10.1074/mcp.M110.003129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witkin K.L., Chong Y., Shao S., Webster M.T., Lahiri S., Walters A.D., Lee B., Koh J.L., Prinz W.A., Andrews B.J., and Cohen-Fix O.. 2012. The budding yeast nuclear envelope adjacent to the nucleolus serves as a membrane sink during mitotic delay. Curr. Biol. 22:1128–1133. 10.1016/j.cub.2012.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yofe I., Weill U., Meurer M., Chuartzman S., Zalckvar E., Goldman O., Ben-Dor S., Schütze C., Wiedemann N., Knop M., et al. . 2016. One library to make them all: Streamlining the creation of yeast libraries via a SWAp-Tag strategy. Nat. Methods. 13:371–378. 10.1038/nmeth.3795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zargari A., Boban M., Heessen S., Andréasson C., Thyberg J., and Ljungdahl P.O.. 2007. Inner nuclear membrane proteins Asi1, Asi2, and Asi3 function in concert to maintain the latent properties of transcription factors Stp1 and Stp2. J. Biol. Chem. 282:594–605. 10.1074/jbc.M609201200 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.