Farhan discusses Scharaw et al.’s study about how the COPII machinery is used to replenish EGFR at the cell surface.
Abstract
Cell surface levels of epidermal growth factor receptors (EGFRs) are thought to be controlled mainly by endocytic trafficking, with biosynthetic EGFR trafficking presumed to be a constitutive and unregulated process. However, Scharaw et al. (2016. J. Cell Biol. http://dx.doi.org/10.1083/jcb.201601090) demonstrate a role for inducible COPII trafficking in controlling EGFR surface levels.
Receptors at the plasma membrane enable cells to sense their environment and to mount appropriate responses to variations in tonicity, ion composition, nutrients, or growth factors. The EGF receptor (EGFR) is a well-characterized, prototypical receptor tyrosine kinase that has been reported to regulate cellular growth, proliferation, survival, and differentiation (Lemmon and Schlessinger, 2010). The levels of EGFR at the cell surface determine the magnitude with which cells respond to EGF; therefore, EGFR is considered an important player in tumorigenesis. Accordingly, several therapeutic strategies against cancer target the EGFR (Arteaga and Engelman, 2014). So far, surface levels of EGFR have been reported to be controlled by the rates of EGFR endocytosis, recycling, and lysosomal degradation. Typically, prolonged exposure to EGF results in degradation of EGFR, which serves to prevent excessive stimulation of the cell with mitogens. At some point, however, degraded EGFR needs to be replaced by newly synthesized protein from the ER to maintain homeostasis. Although this assumption appears intuitive, it has not been experimentally tested. In this issue, Scharaw et al. describe a mechanism by which cell surface levels of EGFR are replenished by specific components of the ER export machinery (Scharaw et al., 2016).
The ER is the place for synthesis, quality control, and export of secretory proteins that constitute about a third of the cellular proteome (Huh et al., 2003). ER export is mediated by COPII vesicles that form ribosome-free regions of the rough ER called ER exit sites (ERES). Secretory transmembrane proteins are captured by the COPII subunit Sec24, of which four isoforms exist in mammalian cells (Sec24A–D). Only a few examples exist of cargo specificity of Sec24 isoforms (Farhan et al., 2007; Wendeler et al., 2007; Mancias and Goldberg, 2008; Reiterer et al., 2008; Sucic et al., 2011). Although COPII trafficking has been considered to be constitutive for a long time, research from the last decade has shown that it is a regulated process. For example, biogenesis of ERES can be regulated by the load of cargo in the ER (Guo and Linstedt, 2006; Farhan et al., 2008) and ER export altered by environmental factors such as growth factors (Farhan et al., 2010; Simpson et al., 2012; Tillmann et al., 2015) or nutrients (Zacharogianni et al., 2011). The response to mitogenic growth factors (such as EGF) is mediated via signaling through the RAF-MEK1/2-ERK2 cascade, which phosphorylates Sec16, a protein that regulates biogenesis and formation of ERES. This rapid response of ERES occurred within a few minutes of growth factor receptor stimulation and led to the generation of more, but smaller, ERES (Tillmann et al., 2015). This was suggested to represent an adaptive response that prepares ERES to deal with a higher load of secretory cargo, which is expected to arise in the case of persistent growth factor stimulation. However, these newly formed ERES are small and likely to disassemble if growth factor stimulation is only transient. A persistent stimulus requires consolidation of these newly formed ERES by de novo synthesis of COPII components and their regulators. Indeed, growth factor stimulation was shown to increase the levels of Sec16 (Tillmann et al., 2015). However, it remained unclear whether COPII components are also induced.
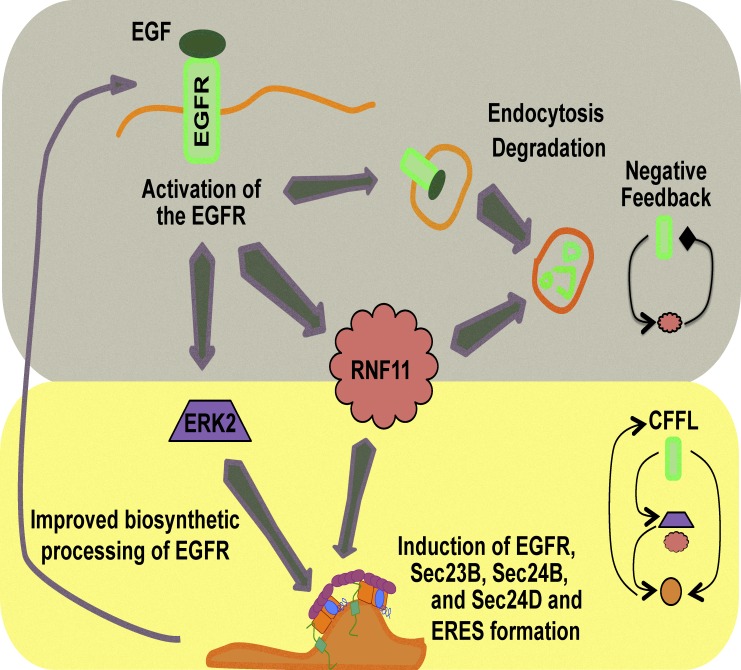
Scharaw et al. (2016) now show that prolonged (24 h) stimulation of cells with EGF increases the transcription of EGFR, as well as the COPII components Sec23B, Sec24B, and Sec24D, which are required for the ER export of EGFR. Using the “retention using selective hooks” or RUSH assay, which permits biotin-inducible release of an eGFP-EGFR fusion protein from the ER, they observed that transport of EGFR from the ER to the plasma membrane became more efficient after prolonged EGF stimulation. Of note, only the protein levels of Sec23B and Sec24D were increased, and it remains unclear why Sec24B is induced at the mRNA but not at the protein level. Nevertheless, this is the first example of a growth factor that induces the synthesis of its own receptor together with the proteins required for its ER export. Therefore, these findings by Scharaw et al. (2016) contribute toward our broader understanding of the regulation of EGFR levels at the plasma membrane. The response of the cell to prolonged stimulation with EGF represents a negative feedback loop, whereby lysosomal degradation negatively regulates the ability of cells to respond to EGF. However, earlier work (Tillmann et al., 2015) together with the work by Scharaw et al. (2016), suggest the existence of a coherent feed-forward loop (CFFL) that regulates the response of the ER export machinery to EGFR signaling and that this CFFL supports biosynthetic trafficking of the EGFR to restore its surface levels (Fig. 1). CFFLs are typically part of persistence detector systems (Lim et al., 2013) that allow cells to respond differently, depending on whether a stimulus is transient (making more, but smaller ERES) or persistent (synthesis of COPII components to support the newly formed ERES). Usually, the response to the persistent stimulus is slower, which is consistent with the observation that EGF induces the transcription of Sec23B, Sec24B, and Sec24D (Scharaw et al., 2016). The final output of the CFFL is trafficking of EGFR, and therefore acts as a positive feedback loop to EGFR signaling at the surface (Fig. 1).
Figure 1.
Schematic illustration of the mutual regulatory circuits between EGFR signaling and ER export. The top part shows that prolonged EGFR signaling will trigger lysosomal degradation of the receptor, which represents a negative feedback loop. The bottom shows that rapid (via ERK2) and slow (via RNF11) signaling leads to ERES formation at the ER (shown in orange), which conforms to a CFFL. This CFFL eventually leads to improved biosynthetic processing of the EGFR. To emphasize the two different layers of regulation, they have been drawn on differently colored backgrounds: The negative feedback circuit on gray and the CFFL on yellow.
Are these two regulatory circuits linked and, if so, how? Scharaw et al. (2016) provide a potential answer to this question. To identify potential transcriptional regulators that may mediate the increased expression of these components of the COPII machinery, the authors first searched, within an siRNA screen of genes that control secretion, for genes that are also predicted to be transcription factors or which have been annotated as being DNA binding. The initial list of 38 potential transcription factors was then prioritized in order of genes whose expression levels appear to be coregulated with Sec23B, Sec24B, and Sec24D, which suggests that they may be in the same pathway. The putative transcriptional regulator RNF11 was ranked second using this approach, and Scharaw et al. (2016) found that overexpression of RNF11 up-regulated, whereas RNF11 knockdown down-regulated, mRNA levels of these COPII components. The effect of RNF11 on the protein levels of these COPII components was not determined, but it is likely to be affected because RNF11 levels regulated the efficiency of EGFR export from the ER. Therefore, the authors propose that RNF11 is the mediator of the EGF-dependent induction of Sec23B, Sec24B, and Sec24D. It is mechanistically unclear precisely how RNF11 regulates the levels of these COPII components, but Scharaw et al. (2016) favor a direct role in gene transcription because RNF11 was shown to bind DNA (Li and Seth, 2004) and prolonged EGF treatment resulted in nuclear translocation of RNF11. In addition, Scharaw et al. (2016) demonstrate that overexpression of RNF11 is sufficient to increase expression of a luciferase reporter driven by the promoter regions of Sec24B and Sec 24D. Intriguingly, RNF11 is a RING-E3 ubiquitin ligase that was shown to play a role in the lysosomal degradation of EGFR by interacting with the ESCRT complex (Kostaras et al., 2013) or by ubiquitinating EPS15, a factor required for ligand-induced endocytosis of EGFR (Confalonieri et al., 2000; Li and Seth, 2004). Therefore RNF11 appears to be part of the negative feedback loop (EGFR degradation) as well as part of the CFFL (induction of COPII levels) that work together to control the surface levels of EGFR. It will be interesting to find out how the contribution of RNF11 to these pathways with opposing effects on EGFR levels is regulated.
A notable finding by Scharaw et al. (2016) is that the effect was very specific to EGF, because stimulation with other mitogens such as PDGF or insulin-like growth factor had no effect on the levels of EGFR or any of the COPII components. Not only was the effect specific to EGF, it also selectively augments the transport of EGFR, as no effect of EGF treatment on vesicular stomatitis virus glycoprotein (VSVG) transport was detected. This might be attributed to the fact that prolonged EGF treatment only enhanced the protein levels of Sec24D, whereas Sec24B protein levels did not change (although its mRNA levels were induced). Based on this, it could be concluded that EGFR is a client for Sec24D, although this needs to be rigorously tested in future work. In contrast, VSVG was shown to selectively bind Sec24A and Sec24B (Mancias and Goldberg, 2008), the two isoforms that are not induced by EGF treatment. More work is needed to carefully assess whether EGF treatment positively modulates the ER-to-Golgi trafficking of other cargos, in particular of Sec24D-dependent cargo. It appears safe to assume that EGF treatment will induce the secretion of other proteins because prolonged treatment with EGF is expected to induce synthesis of a wide range of proteins, of which a significant fraction are secretory proteins.
So far, there is no evidence that cargo-specific ERES exist in mammalian cells. Likewise, no evidence exists for ERES that are specific to certain Sec24 isoforms. In light of this, it is very interesting to note that the depletion of RNF11 reduced the number of Sec24B-positive ERES (Scharaw et al., 2016). This is likely a result of the reduction in the protein levels of Sec24B, because Sec31-positive ERES were unaffected, indicating that ERES, per se, are not affected by RNF11 depletion, most likely because they contain other Sec24 isoforms. However, transport of the Sec24B client VSVG was unaffected, which might be because Sec24A is still present and unaffected.
Targeting EGFR using monoclonal antibodies or small molecular inhibitors has proven to be a useful strategy against different types of malignancies such as lung and colorectal cancer (Arteaga and Engelman, 2014). Given the important role of RNF11 in the regulation of EGFR levels, it is tempting to propose RNF11 as a potential therapeutic target to control surface levels of EGFR. Of note, RNF11 was found to be overexpressed in breast cancer (Subramaniam et al., 2003). Inhibition of RNF11, on one hand, will disrupt the negative feedback loop and, on the other hand, negatively regulate biosynthetic trafficking of EGFR. Whether RNF11 inhibition is a useful therapeutic strategy will depend on the relative contribution of its effect on EGFR endocytosis versus biosynthetic trafficking on the overall levels of EGFR at the cell surface. Furthermore, it would be interesting to determine whether RNF11-overexpressing tumors harbor alterations in the levels of the COPII components described above. Therefore, like many interesting studies, the work by Scharaw et al. (2016) answers an open question in the field but at the same time raises new ones. Future efforts are needed to test how selective the effect of EGF treatment is on different types of ERES. If it is selective only to ERES that control export of EGFR, then what is the basis for this selectivity and what is its biological significance? Why is it different from previously described mechanisms for the regulation of the secretory pathway, such as the unfolded protein response? In the unfolded protein response, overexpression of a single misfolded protein can induce the expression of a large set of genes that not only handle the misfolded protein but also affect secretion of other cargo. Highly selective effects are attractive because they offer the possibility of being targeted with “surgical” precision. Whether the effect of EGF is indeed selective to the ER export of EGFR and whether other examples exist for such highly selective modes of regulation represents an important task for future research efforts.
Acknowledgments
The author declares no competing financial interests.
References
- Arteaga C.L., and Engelman J.A.. 2014. ERBB receptors: From oncogene discovery to basic science to mechanism-based cancer therapeutics. Cancer Cell. 25:282–303. 10.1016/j.ccr.2014.02.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Confalonieri S., Salcini A.E., Puri C., Tacchetti C., and Di Fiore P.P.. 2000. Tyrosine phosphorylation of Eps15 is required for ligand-regulated, but not constitutive, endocytosis. J. Cell Biol. 150:905–912. 10.1083/jcb.150.4.905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farhan H., Reiterer V., Korkhov V.M., Schmid J.A., Freissmuth M., and Sitte H.H.. 2007. Concentrative export from the endoplasmic reticulum of the γ-aminobutyric acid transporter 1 requires binding to SEC24D. J. Biol. Chem. 282:7679–7689. 10.1074/jbc.M609720200 [DOI] [PubMed] [Google Scholar]
- Farhan H., Weiss M., Tani K., Kaufman R.J., and Hauri H.P.. 2008. Adaptation of endoplasmic reticulum exit sites to acute and chronic increases in cargo load. EMBO J. 27:2043–2054. 10.1038/emboj.2008.136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farhan H., Wendeler M.W., Mitrovic S., Fava E., Silberberg Y., Sharan R., Zerial M., and Hauri H.P.. 2010. MAPK signaling to the early secretory pathway revealed by kinase/phosphatase functional screening. J. Cell Biol. 189:997–1011. 10.1083/jcb.200912082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y., and Linstedt A.D.. 2006. COPII–Golgi protein interactions regulate COPII coat assembly and Golgi size. J. Cell Biol. 174:53–63. 10.1083/jcb.200604058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh W.K., Falvo J.V., Gerke L.C., Carroll A.S., Howson R.W., Weissman J.S., and O’Shea E.K.. 2003. Global analysis of protein localization in budding yeast. Nature. 425:686–691. 10.1038/nature02026 [DOI] [PubMed] [Google Scholar]
- Kostaras E., Sflomos G., Pedersen N.M., Stenmark H., Fotsis T., and Murphy C.. 2013. SARA and RNF11 interact with each other and ESCRT-0 core proteins and regulate degradative EGFR trafficking. Oncogene. 32:5220–5232. 10.1038/onc.2012.554 [DOI] [PubMed] [Google Scholar]
- Lemmon M.A., and Schlessinger J.. 2010. Cell signaling by receptor tyrosine kinases. Cell. 141:1117–1134. 10.1016/j.cell.2010.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., and Seth A.. 2004. An RNF11: Smurf2 complex mediates ubiquitination of the AMSH protein. Oncogene. 23:1801–1808. 10.1038/sj.onc.1207319 [DOI] [PubMed] [Google Scholar]
- Lim W.A., Lee C.M., and Tang C.. 2013. Design principles of regulatory networks: searching for the molecular algorithms of the cell. Mol. Cell. 49:202–212. 10.1016/j.molcel.2012.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancias J.D., and Goldberg J.. 2008. Structural basis of cargo membrane protein discrimination by the human COPII coat machinery. EMBO J. 27:2918–2928. 10.1038/emboj.2008.208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiterer V., Maier S., Sitte H.H., Kriz A., Rüegg M.A., Hauri H.P., Freissmuth M., and Farhan H.. 2008. Sec24- and ARFGAP1-dependent trafficking of GABA transporter-1 is a prerequisite for correct axonal targeting. J. Neurosci. 28:12453–12464. 10.1523/JNEUROSCI.3451-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharaw S., Iskar M., Ori A., Boncompain G., Laketa V., Poser I., Lundberg E., Perez F., Beck M., Bork P., and Pepperkok R.. 2016. The endosomal transcriptional regulator RNF11 integrates degradation and transport of EGFR. J. Cell Biol. 10.1083/jcb.201601090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson J.C., Joggerst B., Laketa V., Verissimo F., Cetin C., Erfle H., Bexiga M.G., Singan V.R., Hériché J.K., Neumann B., et al. . 2012. Genome-wide RNAi screening identifies human proteins with a regulatory function in the early secretory pathway. Nat. Cell Biol. 14:764–774. 10.1038/ncb2510 [DOI] [PubMed] [Google Scholar]
- Subramaniam V., Li H., Wong M., Kitching R., Attisano L., Wrana J., Zubovits J., Burger A.M., and Seth A.. 2003. The RING-H2 protein RNF11 is overexpressed in breast cancer and is a target of Smurf2 E3 ligase. Br. J. Cancer. 89:1538–1544. 10.1038/sj.bjc.6601301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sucic S., El-Kasaby A., Kudlacek O., Sarker S., Sitte H.H., Marin P., and Freissmuth M.. 2011. The serotonin transporter is an exclusive client of the coat protein complex II (COPII) component SEC24C. J. Biol. Chem. 286:16482–16490. 10.1074/jbc.M111.230037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillmann K.D., Reiterer V., Baschieri F., Hoffmann J., Millarte V., Hauser M.A., Mazza A., Atias N., Legler D.F., Sharan R., et al. . 2015. Regulation of Sec16 levels and dynamics links proliferation and secretion. J. Cell Sci. 128:670–682. 10.1242/jcs.157115 [DOI] [PubMed] [Google Scholar]
- Wendeler M.W., Paccaud J.P., and Hauri H.P.. 2007. Role of Sec24 isoforms in selective export of membrane proteins from the endoplasmic reticulum. EMBO Rep. 8:258–264. 10.1038/sj.embor.7400893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharogianni M., Kondylis V., Tang Y., Farhan H., Xanthakis D., Fuchs F., Boutros M., and Rabouille C.. 2011. ERK7 is a negative regulator of protein secretion in response to amino-acid starvation by modulating Sec16 membrane association. EMBO J. 30:3684–3700. 10.1038/emboj.2011.253 [DOI] [PMC free article] [PubMed] [Google Scholar]