Summary
Tumor-infiltrating regulatory T lymphocytes (Treg) can suppress effector T cells specific for tumor antigens. Deeper molecular definitions of tumor-infiltrating-lymphocytes could thus offer therapeutic opportunities. Transcriptomes of T helper 1 (Th1), Th17, and Treg cells infiltrating colorectal or non-small-cell lung cancers were compared to transcriptomes of the same subsets from normal tissues and validated at the single-cell level. We found that tumor-infiltrating Treg cells were highly suppressive, upregulated several immune-checkpoints, and expressed on the cell surfaces specific signature molecules such as interleukin-1 receptor 2 (IL1R2), programmed death (PD)-1 Ligand1, PD-1 Ligand2, and CCR8 chemokine, which were not previously described on Treg cells. Remarkably, high expression in whole-tumor samples of Treg cell signature genes, such as LAYN, MAGEH1, or CCR8, correlated with poor prognosis. Our findings provide insights into the molecular identity and functions of human tumor-infiltrating Treg cells and define potential targets for tumor immunotherapy.
Highlights
-
•
Transcriptome analysis performed on tumor-resident CD4+ Th1, Th17, and Treg cells
-
•
Tumor-infiltrating Treg cells are defined by the expression of signature genes
-
•
Treg-specific signature genes correlate with patients’ survival in both CRC and NSCLC
Tumor-infiltrating regulatory T cells can suppress effector T cells specific for tumor antigens. De Simone et al. (2016) demonstrate that tumor-infiltrating Treg cells display specific gene signatures that were also validated at the single-cell level. These data can contribute to dissect the molecular networks underlying the biology of tumor-infiltrating Treg cells. As part of the IHEC consortium, this study integrates genetic, epigenetic, and transcriptomic profiling in three immune cell types from nearly 200 people to characterize the distinct and cooperative contributions of diverse genomic inputs to transcriptional variation. Explore the Cell Press IHEC webportal at www.cell.com/consortium/IHEC.
Introduction
The combination of genetic mutations and epigenetic modifications that are peculiar to all tumors generate antigens that T and B lymphocytes can use to specifically recognize tumor cells (Jamal-Hanjani et al., 2013). It is increasingly clear that T lymphocytes recognizing tumor-derived peptides presented by major histocompatibility complex (MHC) molecules play a central role in immunotherapy and in conventional chemo-radiotherapy of cancer (Galluzzi et al., 2015). In fact, anti-tumor T cell responses arise in cancer patients but are disabled upon tumor progression by suppressive mechanisms triggered by the interplay between malignant cells and the tumor microenvironment (Munn and Bronte, 2016). The tumor-dependent immunosuppressive mechanisms depend on the integrated action of infiltrating leukocytes and lymphocytes that upregulate a range of modulatory molecules, collectively called immune checkpoints, whose function is only partially characterized (Pardoll, 2012). Therefore, the search for agonists of co-stimulatory complexes or antagonists of inhibitory molecules to potentiate antigen-specific T cell responses is a primary goal of current anti-tumor research (Sharma and Allison, 2015, Zitvogel et al., 2013). Indeed, clinical trials have unequivocally shown that the blockade of immune checkpoints unleashes the spontaneous anti-tumor immune responses in such a powerful way that it has created a paradigm shift in cancer therapy (Śledzińska et al., 2015, Topalian et al., 2015).
Among the immune checkpoints targeted by blocking strategies, CTLA-4 has been one of the first to be translated into therapeutic applications.
Anti-CTLA-4 monoclonal antibodies (mAb) show remarkable success in metastatic melanoma, and more recently in non-small-cell lung cancer, prostate cancer, renal cell carcinoma, urothelial carcinoma, and ovarian cancer (Carthon et al., 2010, Hodi et al., 2010, van den Eertwegh et al., 2012, Yang et al., 2007). However, the fraction of patients that do not respond remains high, prompting a deeper investigation of the mechanisms underpinning the modulation of immune responses by tumors. Recent experimental evidence shows that anti-CTLA-4 mAb efficacy depends on FcγR-mediated depletion of CD4+ regulatory T cells (Treg cells) within the tumor microenvironment (Peggs et al., 2009, Selby et al., 2013, Simpson et al., 2013, Twyman-Saint Victor et al., 2015).
Treg cells, which are physiologically engaged in the maintenance of immunological self-tolerance and immune homeostasis (Josefowicz et al., 2012, Sakaguchi et al., 2008), are potent suppressors of effector cells and are found at high frequencies in various types of cancers (Fridman et al., 2012, Nishikawa and Sakaguchi, 2010). Treg cells adapt their transcriptional program to the various cytokines to which they are exposed in the inflammatory milieu (Campbell and Koch, 2011). This versatility is controlled by transcription factors generally associated with the differentiation of other effector CD4+ T cell subsets, resulting in various Treg cell populations with unique features and immunomodulatory functions (Duhen et al., 2012, Geginat et al., 2014). Moreover, Treg cells infiltrating non-lymphoid tissues are reported to exhibit unique phenotypes and transcriptional signatures, because they can display functions beyond their well-established suppressive roles, such as metabolic modulation in adipose tissue (Cipolletta et al., 2012) or regulation of tissue repair in skeletal muscle (Burzyn et al., 2013) and in lung tissue (Arpaia et al., 2015).
Treg cell depletion has been reported to increase anti-tumor specific immune responses and to reduce tumor burden (Marabelle et al., 2013, Teng et al., 2010, Walter et al., 2012). Although promising clinical results have been achieved with Treg cell depleting strategies, some relevant issues are to be addressed, for a safer, more effective, and wider clinical application of these therapies. First, severe autoimmunity can occur following systemic Treg cells depletion (Nishikawa and Sakaguchi, 2010), which could be avoided if selective depletion of tumor infiltrating Treg cells were feasible. A second issue concerns the specificity of targeting. Indeed, Treg cells share with effector lymphocytes most of the molecules targeted for therapy, which can possibly deplete also the tumor-specific effector cells. Therefore, the molecular characterization of Treg cells at different tumor sites should help to better define therapeutic targets through a better description of their signature molecules and of the network that regulates Treg cell functions in the tumor microenvironment.
Non-small-cell lung cancer (NSCLC) and colorectal cancer (CRC) are the two most frequent cancers in both genders (Torre et al., 2015). NSCLC has the worst prognosis due to its high mortality rate even in early stages. Although CRC survival rate is highly dependent on the tumor stage at diagnosis, about 50% of patients will progress to metastatic cancer (Gonzalez-Pons and Cruz-Correa, 2015). Both tumors have been targeted with therapies based on monoclonal antibodies to checkpoint inhibitors, but the outcomes have been different. While remarkable clinical success has been obtained in NSCLC, evidence of durable response in CRC is scarce with the exception of mismatch repair-deficient CRC lesions (Jacobs et al., 2015, Kroemer et al., 2015, Le et al., 2015).
Here we provide a comprehensive transcriptome analysis of human CD4+ Treg cells and effector cells (Th1 and Th17) infiltrating NSCLC or CRC and their matched normal tissues. We defined molecular signatures of tumor-infiltrating Treg cells in these two cancer types and confirmed the relevance of these signatures by single-cell analyses. These data could help a better understanding of Treg functional role at tumor sites and pave the way to the identification of therapeutic targets for more specific and safer modulation of Treg cells in cancer therapy.
Results
Tumor Infiltrating Treg Cells Upregulate Immune Checkpoints and Are Highly Suppressive
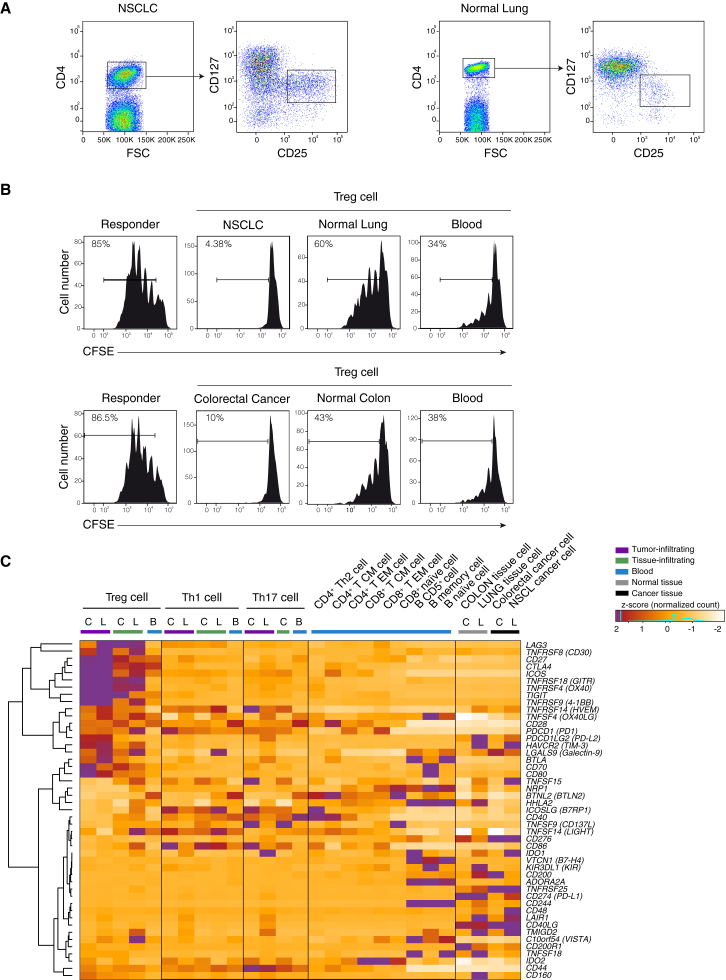
To assess the gene expression landscape of tumor infiltrating CD4+ T cells, we isolated different CD4+ lymphocytes subsets from two different tumors, NSCLC and CRC, from the adjacent normal tissues, and from peripheral blood samples. From all these tissues, we purified by flow cytometry (Figure 1A and S1A and S1B) CD4+ Treg (36 samples from 18 individuals), Th1 (30 samples from 21 individuals), and Th17 (22 samples from 14 individuals) cells (Table 1 and Table S1). To assess Treg cell function, we tested their suppressor activity and showed that Treg cells infiltrating either type of tumor tissues have a remarkably stronger suppressive activity in vitro compared to Treg cells isolated from the adjacent normal tissue and peripheral blood of the same patients (Figure 1B).
Figure 1.
Purification, Functional Characterization, and Expression of Immune Checkpoints in Tumor Infiltrating Cells
(A) Representation of the sorting strategy of Treg cells infiltrating tumor or normal tissue.
(B) Representative flow cytometry plots showing suppressive activity of Treg cells isolated from tumor (NSCLC or CRC), normal tissue and blood of the same patient. 4 × 105 carboxyfluorescein diacetate succinimidyl ester (CFSE)-labeled CD4+ naive T cells from healthy donors were cocultured with an equal number of Treg cells for 4 days with a CD3-specific mAb and CD1c+CD11c+ dendritic cells. Percentage of proliferating cells is indicated. Data are representative of three independent experiments.
(C) Z-score normalized RNA-seq expression values of immune checkpoints genes are represented as a heatmap. Cell populations are reported as a color code in the upper part of the graph, while gene names have been assigned to heatmap rows. Hierarchical clustering results are shown as a dendrogram drawn on the left side of the matrix. Colon tissues are indicated as C, lung tissues as L, and peripheral blood as B.
See also Figure S1.
Table 1.
Purification and RNA-Sequencing of Human Primary Lymphocyte Subsets
| Tissue | Subset | Sorting Phenotype | Number of Samples | Mapped Reads (M) |
|---|---|---|---|---|
| NSCLC | CD4+ Treg | CD4+ CD127− CD25+ | 8 | 587 |
| CD4+ Th1 | CD4+ CXCR3+ CCR6− | 8 | 409 | |
| CD4+ Th17 | CD4+ CCR6+ CXCR3− | 6 | 206 | |
| CRC | CD4+ Treg | CD4+ CD127− CD25+ | 7 | 488 |
| CD4+ Th1 | CD4+ CXCR3+ CCR6− | 5 | 266 | |
| CD4+ Th17 | CD4+ CCR6+ CXCR3− | 5 | 308 | |
| Lung (normal tissue) | CD4+ Treg | CD4+ CD127− CD25+ | 1 (pool of 6) | 73 |
| CD4+ Th1 | CD4+ CXCR3+ CCR6− | 1 (pool of 6) | 76 | |
| Colon (normal tissue) | CD4+ Treg | CD4+ CD127− CD25+ | 7 | 404 |
| CD4+ Th1 | CD4+ CXCR3+ CCR6− | 6 | 352 | |
| CD4+ Th17 | CD4+ CCR6+ CXCR3− | 6 | 284 | |
| PB (healthy donor) | CD4+ Treg | CD4+ CD127− CD25+ | 8 | 259 |
| CD4+ Th1 | CD4+ CXCR3+ CCR6− | 5 | 70 | |
| CD4+ Th17 | CD4+ CCR6+ CXCR3− | 5 | 77 |
For each cell subsets profiled by RNA-sequencing tissue of origin, surface marker combinations used for sorting, number of profiled samples, as well as number of mapped sequencing reads are indicated. M, million; CRC, colorectal cancer; NSCLC, non-small cell lung cancer; PB, peripheral blood.
See also Table S1.
The polyadenylated RNA fraction extracted from the sorted CD4+ Treg, Th1, and Th17 cells was then analyzed by paired-end RNA sequencing obtaining about 4 billion mapped “reads” (Table 1). First, we interrogated RNA-sequencing data of CD4+ T cells infiltrating both CRC and NSCLC and their matched normal tissues, to quantitate mRNA expression of known immune checkpoints and their ligands. Second, we analyzed RNA-seq data of CRC and NSCLC, as well as of normal colon and lung samples. We found that several immune checkpoints and their ligands transcripts were strikingly upregulated in tumor infiltrating Treg cells compared to both normal tissue and peripheral blood-derived Treg cells, as well as to T and B lymphocyte subsets purified from peripheral blood mononuclear cells (PBMCs) (Figures 1C and S1C and Table S5). Our findings highlight the specific expression patterns of immune checkpoints and their ligands in tumor infiltrating Treg and effector cells and suggest that their functional relevance should be investigated directly at tumor sites.
Tumor-Infiltrating Treg Cells Express a Specific Gene Signature
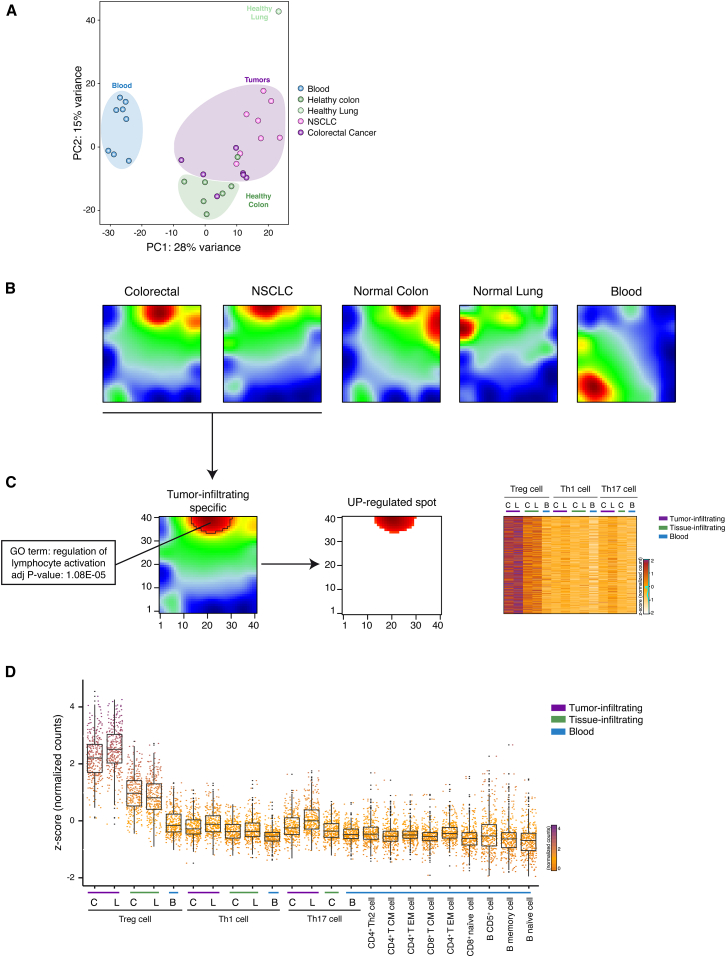
We then asked whether tumor infiltrating Treg cells could be defined by specific gene-expression patterns. First, in order to capture the overall similarity between the tumor infiltrating lymphocytes, we performed a principal components analysis (PCA) on the whole transcriptomes. Tumor-infiltrating Treg cells purified from CRC and NSCLC tissues clustered together and were clearly separated from Th1 and Th17 cells purified from CRC and NSCLC tissues (Figures S2A and S2B). PCA showed a distinct grouping of Treg cells purified from different sites; in fact, separation along the first principal component (PC1) clearly divided peripheral blood Treg cells from tissue infiltrating Treg cells (Figure 2A), whereas normal-tissue and tumor-tissue infiltrating Treg cells are mostly divided by the second component (PC2). These findings indicate that tumor-infiltrating Treg cells have specific expression patterns compared not only to other CD4+ T cell subsets but also compared to Treg cells isolated from normal tissues.
Figure 2.
SOM Analysis Identifies Co-regulated Genes in Tumor Infiltrating Treg Cells
(A) PCA has been performed on rlog-normalized (DESeq2) counts for all T regulatory cell RNA-seq samples (36 samples from 18 individuals).
(B) Self-organizing maps analysis has been performed on the RNA-seq dataset comprising Treg, Th1, and Th17 cell subsets. Bidimensional SOM profiles are reported for Treg cells.
(C) Group-centered analysis for the identification of upregulated spot (FDR < 0.1) in Treg cells infiltrating both NSCLC and CRC is described as 2D heatmap. Heatmap representing Z-score normalized expression values of genes selected from the upregulated spot is shown on the right side of the figure. Top enriched GO term (DAVID) for genes assigned to upregulated spot is reported with the corresponding significance p value. Colon tissues are indicated as C, lung tissues as L, and peripheral blood as B.
(D) Z-score normalized expression values of genes that are preferentially expressed in tumor-infiltrating Treg cells (Wilcoxon Mann Whitney test p < 2.2 × 10–16) over the listed cell subsets are represented as boxed plots. Colon tissues are indicated as C, lung tissues as L, and peripheral blood as B.
See also Figure S2.
In order to identify genes that are preferentially expressed in tumor-infiltrating lymphocytes, we performed self-organizing maps (SOM) analyses that provide a powerful way to define coordinated gene-expression patterns that are visualized in spatial proximity in a 2D mosaic grid heatmap (Wirth et al., 2012). In this way, we analyzed 7,763 genes that were differentially expressed between the different CD4+ T cell subsets purified from PBMCs and tumor tissues (DESeq2 package; FDR < 0.05). Among the different CD4+ T cell subsets (Th1, Th17, and Treg) assessed with SOM, only the tumor-infiltrating Treg cells displayed peculiar gene-expression patterns that were similar between NSCLC and CRC samples (Figures 2B and S2C), thus allowing the identification (FDR < 0.1) of transcripts upregulated in both CRC and NSCLC infiltrating Treg cells (Figure 2C and Table S2). Gene-ontology (GO) analyses of those genes upregulated in tumor infiltrating Treg cells showed significant enrichment for terms related to lymphocytes activation (Figure 2C and Table S3).
To identify signature transcripts of tumor-infiltrating Treg cells, we included in the expression pattern analyses the transcriptome datasets we previously obtained from different T and B lymphocyte subsets purified from PBMCs (Ranzani et al., 2015). In so doing, we obtained a signature of 309 transcripts whose expression is higher in tumor infiltrating Treg cells (Wilcoxon Mann Whitney test p < 2.2 × 10–16) (Figures 2D and S2D and Table S4) compared to the other lymphocyte subsets purified from non-tumoral tissues and from PBMCs of healthy or neoplastic patients.
Altogether, the data show that Treg cells display the most pronounced differences in transcripts expression among CD4+ T cell subsets infiltrating normal and tumor tissues. We defined a subset of signature genes that describe the specific gene-expression profile of tumor infiltrating Treg cells.
Gene Signature of Tumor-Infiltrating Treg Cells Is Present in Primary and Metastatic Human Tumors
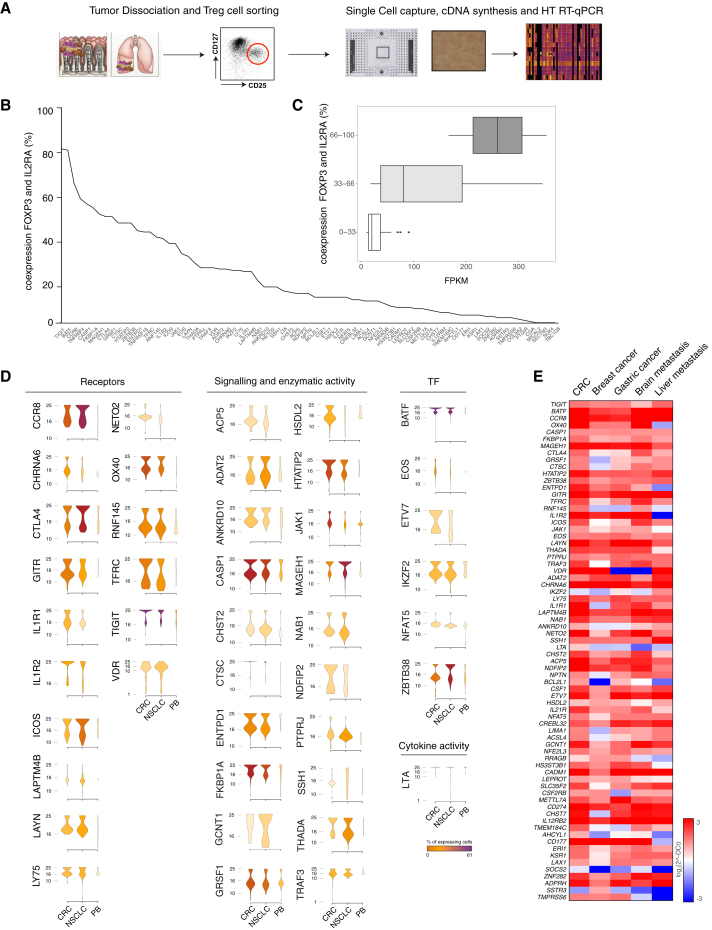
We then look at the single cell level for the differential expression profile of signature genes of tumor infiltrating Treg cells. We isolated CD4+ T cells from 5 CRC and 5 NSCLC tumor samples, as well as from 5 PBMCs of healthy individuals (Table S1), purified Treg cells, and using an automated microfluidic system (C1 Fluidigm) captured single cells (a total of 858 Treg cells: 320 from CRC and 286 from NSCLC; 252 from PBMCs of healthy individuals). We then assessed by high throughput RT-qPCR (Biomark HD, Fluidigm) the expression of 79 genes selected among the highly expressed (> 10 FKPM) tumor Treg cell signature genes (Figures 3A, S3A and S3B).
Figure 3.
Single Cell Analysis of Tumor Infiltrating Treg Cells
(A) Schematic representation of the experimental workflow. Experiments were performed on Treg cells infiltrating CRC, NSCLC, or isolated from peripheral blood of healthy donors (PB); five samples were collected for each tissue.
(B) Percentage of co-expression of signature genes with FOXP3 and IL2RA is depicted.
(C) Expression levels of the signature genes classified by the percentage of co-expression are represented as boxplot.
(D) Expression distribution (violin plots) in Treg cells infiltrating CRC, NSCLC, or PB. Plots representing the ontology classes of receptors, signaling and enzymatic activity, cytokine activity, and transcription factors are shown (Wilcoxon Mann Whitney test p < 0.05). Color gradient indicates the percentage of cells expressing each gene in Treg cells isolated from the three tissues.
(E) Gene-expression analysis of tumor Treg signature genes in different tumor types. Expression values are expressed as log2 (2ˆ-DCt).
See also Figure S3.
Notably, we found that the vast majority (75 over 79; 95%) of the tumor-infiltrating Treg cell signatures were co-expressed with bona fide Treg cell markers (i.e., FOXP3+ and IL2RA) (Figure 3B). The percentage of co-expression between these Treg cell markers and the 79 genes selected among the tumor-infiltrating-Treg-cell signature genes ranged between 81% of TIGIT and 0.59% of CGA (Figure 3B). The expression of Treg signature genes in the RNA-seq of the whole Treg cell population correlated with the percentage of single cells expressing the different genes (Figure 3C). In order to reduce the “drop-out” effect of the single cell data (i.e., events in which a transcript is detected in one cell but not in another one because the transcript is “missed” during the reverse-transcription step) (Kharchenko et al., 2014), we defined a threshold (median value t = 8.4%) based on the expression distribution for each transcript and discarded genes below this threshold (see the Supplemental Experimental Procedures). The forty-five signature transcripts of tumor infiltrating Treg cells detected above this threshold were in most cases significantly overexpressed in Treg cells from both tumors (39 over 45, 87%; Wilcoxon Mann Whitney test p < 0.05) or in one tumor type (43 over 45, 96%; Figure 3D). Homogeneity of the purified tissue infiltrating Treg cells can be affected by the carry-over of cells from other lymphocyte subsets. To quantitate this possible contamination, the single cell RT-qPCR analyses of Treg cells was performed including markers specific for other lymphocytes subsets (i.e., Th1, Th2, Th17, Tfh, CD8 T cells, B cells) (Figure S3C). Our data showed that only a very low fraction of the purified single cells displayed markers of lymphocytes subsets different from Treg cells (Figure S3C).
The overlap between the signature genes in the CRC and NSCLC infiltrating Treg cells (Figure 2D) prompted us to assess whether this signature were also enriched in Treg cells infiltrating other tumors. RNA was thus extracted from Treg cells infiltrating breast cancer, gastric cancer, brain metastasis of NSCLC, and liver metastasis of CRC. We found by RT-qPCR that tumor infiltrating Treg signatures genes were mostly upregulated also in these tumors (Figure 3E).
Overall these data show that the tumor-infiltrating Treg cell signature genes are co-expressed at single cell level with FOXP3 and IL2RA and that several primary and metastatic human tumors express the tumor-infiltrating Treg cell signature.
Gene Signature of Tumor Infiltrating Treg Cells Is Translated in a Protein Signature
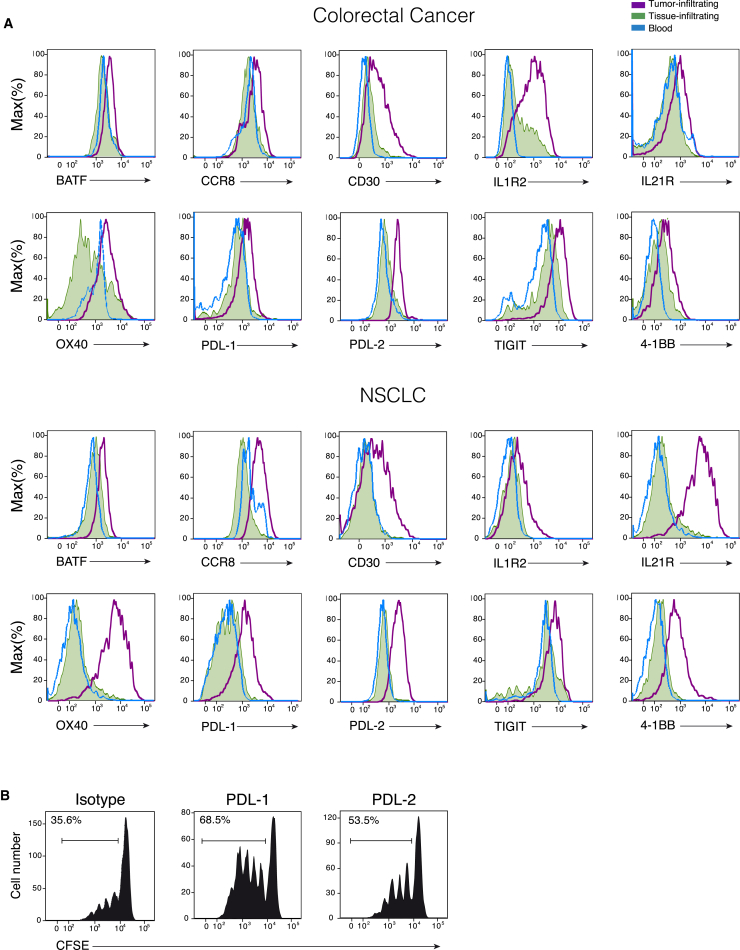
We then assessed at the single cell level by flow cytometry the protein expression of ten representative signature genes present in CRC and NSCLC infiltrating Treg cells, adjacent normal tissues, and patients PBMCs. Of the ten proteins, two were proteins (OX40 and TIGIT) whose relevance for Treg cells biology has been demonstrated (Joller et al., 2014, Voo et al., 2013), seven are proteins (BATF, CCR8, CD30, IL-1R2, IL-21R, PDL-1, and PDL-2) whose expression has never been described in tumor-infiltrating Treg cells, and one protein, 4-1BB, is a co-stimulatory receptor expressed on several hematopoietic cells, whose expression on Treg cells has been shown to mark antigen-activated cells (Schoenbrunn et al., 2012). Our findings showed that all these proteins were upregulated (Figure 4A), to different extent, in tumor infiltrating Treg cells compared to the Treg cells resident in normal tissues. Given the increasing interest in the PD1 - PDLs axis as targets for tumor immunotherapy, we assessed the effect of antibodies against PDL-1 and PDL-2 on the suppressive function of tumor-infiltrating Treg cells toward effector CD4+ T cell proliferation in vitro. We found that preincubation of tumor infiltrating Treg cells with monoclonal antibodies against PDL-1 or PDL-2 reduced their suppressive activity as demonstrated by the increased proliferation of effector CD4+ T cells (Figure 4B).
Figure 4.
Expression of Tumor-Infiltrating Treg Cells Protein Signatures in CRC and NSCLC Samples
(A) Representative flow cytometry plots for tumor (purple line) normal (green area) tissue infiltrating Treg cells and peripheral blood Treg cells (blue line) analyzed for the expression of the indicated proteins.
(B) Flow cytometry plots representative of four independent experiments showing suppressive activity of CRC infiltrating Treg cells on proliferation (shown as CFSE dilution) of CD4+ effector T cells. First panel shows the inhibitory effect of Treg cells on the effector T cell proliferation in the presence of an isotype control antibody. The other panels show the inhibitory effect of Treg cells that have been preincubated with anti PD-L1 or PD-L2 antibodies. Percentage of proliferating cells are indicated. The calculated division index is 0.26 in the presence of the control antibody; 0.57 in the presence of anti-PDL-1 and 0.39 in the presence of anti-PDL-2. Data are representative of four independent experiments.
See also Figure S4.
Altogether, our data show there is a molecular signature of tumor infiltrating Treg cells, which can be detected both at the mRNA and at the protein levels.
Expression of Tumor Treg Signature Genes Is Negatively Correlated with Patient Survival
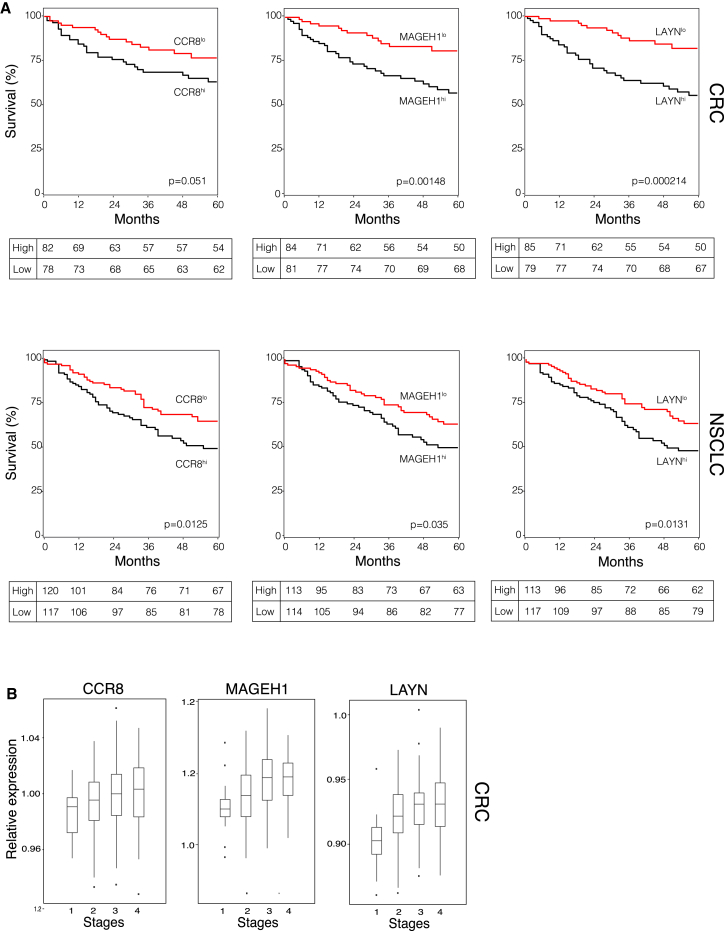
In an attempt to correlate our findings with clinical outcome, we asked whether the expression of the tumor-Treg signature transcripts correlated with disease prognosis in CRC and NSCLC patients. We therefore interrogated for expression of Treg signature genes transcriptomic datasets obtained from resected tumor tissues of a cohort of 177 CRC patients (GSE17536; Smith et al., 2010) and of a cohort of 263 NSCLC patients (GSE41271; Sato et al., 2013) and correlated high and low gene expression with the 5-year survival data. Among those genes whose expression is highly enriched in tumor-infiltrating Treg cells, we selected LAYN, MAGEH1, and CCR8 that are the three genes more selectively expressed (Figure S5A). To normalize for differences in T cell densities within the resected tumor tissues, we used the ratio between expression of the selected signature genes and CD3G. We found that high expression of the three signature genes is in all cases correlated with a significantly reduced survival (Figure 5A). We also observed that expressions of the three signature genes increased with tumor staging of CRC patients (Figure 5B).
Figure 5.
Prognostic Value of Signature Transcripts of Tumor Infiltrating Treg Cells
(A) Kaplan-Meier survival curve comparing the high and low expression of the tumor Treg signature transcripts (CCR8, MAGEH1, LAYN) normalized to the CD3G for the CRC (n = 177) and NSCLC (n = 263) studies. Univariate analysis confirmed a significant difference in overall survival curve comparing patients with high and low expression. Statistical significance was determined by the log-rank test. (CRC: p = 0.05 for CCR8, p = 1.48 × 10−3 for MAGEH1, p = 2.1 × 10−4 for LAYN; NSCLC: p = 0.0125 for CCR8, p = 0.035 for MAGEH1, p = 0.0131 for LAYN.) Each table depicts the Kaplan-Meier estimates at the specified time points.
(B) Expression distributions of CCR8, MAGEH1, and LAYN according to tumor staging at the time of surgery in the cohort of CRC patients.
See also Figure S5.
In conclusion, high expression in the whole-tumor samples of three genes (LAYN, MAGEH1, and CCR8) that are specifically and highly expressed in tumor infiltrating Treg cells correlates with a poor prognosis in both NSCLC and CRC patients.
Discussion
Diversity of tumor-infiltrating Treg cells should be fully elucidated to understand their functional relevance and prognostic significance in different types of cancer and to possibly improve the therapeutic efficacy of Treg cell modulation through the selective depletion of tumor infiltrating Treg cells. The transcriptome analysis we performed on CRC- and NSCLC-infiltrating T cells showed that tumor-infiltrating Treg cells are different from both circulating and normal tissue-infiltrating Tregs, suggesting that the tumor microenvironment influences specific gene expression in Treg cells. Our findings further support the view that Treg cells from different tissues are instructed by environmental factors to display different gene-expression profiles (Panduro et al., 2016). Indeed the list of signature genes includes a number of molecules that are consistently upregulated in tumor-infiltrating Treg cells isolated from different tumor types, and these signature genes would have not been identified if we had not profiled specifically tumor infiltrating Treg cells.
The number of genes highly expressed in tumor infiltrating cells, as defined by differential expression and SOM analyses, was significantly higher in Treg than in Th17 and Th1 cells, suggesting that Treg cells are more susceptible than other T cell subsets to external cues they are exposed to in tumor tissues. We found that tumor-infiltrating-Treg signature genes are not only largely shared between CRC- and NSCLC-infiltrating cells but are also conserved in breast and gastric cancers, as well as in CRC and NSCLC metastatic tumors (in liver and brain, respectively) suggesting that expression of these genes is a common feature of tumor infiltrating Treg cells that might correlate with Treg cell-specific function within the tumor microenvironment.
Although our knowledge on the function of immune checkpoints on lymphocytes is still incomplete, agonist or antagonist monoclonal antibodies targeting checkpoints are in clinical development. We have found that some of these checkpoints (such as GITR, OX40, TIGIT, LAG-3, and TIM-3) and some of their ligands (such as OX40LG, Galectin-9, CD70) are upregulated also in tumor-infiltrating Treg cells, and this fact should be taken into account in interpreting clinical results with checkpoint inhibitors. Indeed, it is likely that assessment of the expression of checkpoints and of their ligands on the various subsets of tumor infiltrating lymphocytes will help to elucidate conflicting results and provide the rationale for combination therapies. Therefore, expression pattern of checkpoints should be evaluated both in tumor-infiltrating lymphocytes and in tumor cells.
Single-cell analysis on selected tumor Treg signature genes confirmed the whole transcriptomic data and provided information on the expression frequency of these genes. Tumor-infiltrating Treg cells express with high frequency genes that are associated with increased suppressor activity, such as the well characterized OX40, CTLA4, and GITR. Moreover, there were a number of interesting and less expected genes the specific expression of which was validated also at the protein level. For example, IL-1R2 upregulation could be another mechanism that tumor resident Treg cells employ to dampen anti-tumor immune responses through the neutralization of IL-1β function on effector cells. PD-L1 and PD-L2 expression has been recently reported on activated T cells or APCs (Boussiotis et al., 2014, Lesterhuis et al., 2011, Messal et al., 2011) but, to the best of our knowledge, neither PD-L2 nor PD-L1 expression has ever been reported in Treg cells, and our finding that they are overexpressed in tumor infiltrating Treg cells adds an additional level of complexity to the PD1 - PD-Ls immunomodulatory axis within the tumor microenvironment. BATF is a transcription factor that has been mainly associated to Th17 development and CD8+ T cells differentiation (Murphy et al., 2013). Our findings revealed that BATF transcript is upregulated in tumor-infiltrating Treg cells more than in tumor infiltrating Th17 cells (Figure S4). Expression of BATF in CD8+ T cells is induced by IL-21 (Xin et al., 2015), and we found that IL21R is highly expressed in tumor-infiltrating Treg cells (Figure 4).
We showed that tumor-infiltrating Treg cells express high amounts of 4-1BB (CD137) a marker of TcR-mediated activation (Schoenbrunn et al., 2012) and have shown they display very high suppressor function on effector T cell proliferation. It could be that expression of the signature genes correlated with the enhanced suppressive ability and so contributed to the establishment of a strong immunosuppressive environment at tumor sites.
A corollary to our findings would have that increased number of Treg cells in the tumor environment should associate with a worst clinical outcome. In fact, when LAYN, MAGEH1, and CCR8 (which represent three of the most enriched genes in tumor-infiltrating Treg cells) are highly detected in whole-tumor samples there is a significant worsening of the 5-year survival of both CRC and NSCLC patients. Although, the functional roles in Treg cells of LAYN, a transmembrane protein with homology to c-type lectin (Borowsky and Hynes, 1998), and of MAGEH1, a member of the melanoma antigen gene family (Weon and Potts, 2015), are unknown, the high expression of the chemokine receptor CCR8 is instead intriguing. Indeed, CCL18, the ligand of CCR8 (Islam et al., 2013), is highly expressed in different tumors including NSCLC (Chen et al., 2011, Schutyser et al., 2005). The high specificity of CCR8 expression on tumor-infiltrating Treg cells suggests it could be an interesting therapeutic target to inhibit Treg cells trafficking to tumor sites, without disturbing recruitment of other effector T cells that do not express CCR8.
Considerable efforts have been recently put in the development of sophisticated bioinformatics approaches that exploit lymphocyte gene-expression data to understand the immune-modulatory networks at tumor sites, to predict clinical responses to immune-therapies, and to define therapeutic targets (Bindea et al., 2013a, Bindea et al., 2013b, Gentles et al., 2015). The data we present here represent a comprehensive RNA-sequencing analysis performed on tumor-infiltrating human CD4+ Treg, Th1, and Th17 cells. Our findings highlight the relevance of assessing gene-expression patterns of lymphocyte at tumor-sites and suggest that generation of more transcriptomic data of tumor-infiltrating lymphocyte subsets purified from different cancer types might contribute to a better understanding of the dynamics underlying immune modulation in the tumor microenvironment. Moreover, our data represent a resource to generate and validate hypotheses that will increase our knowledge on tumor-infiltrating Treg cell biology and should lead to the identification of therapeutic targets.
Experimental Procedures
Human Primary Tissues
Primary human lung or colorectal tumors and non-neoplastic counterparts were obtained from 15 and 14 patients, respectively. Patients’ records clinicopathological staging, tumor histotype, and grade are listed in Table S1. Informed consent was obtained from all patients, and the study was approved by the Institutional Review Board of the Fondazione IRCCS Ca’ Granda (approval n.30/2014). No patient received palliative surgery or neoadjuvant chemo- and/or radiotherapy. NSCLC specimens were cut into pieces and single-cell suspensions were prepared by using the Tumor Dissociation Kit, human and the gentleMACS Dissociator (Miltenyi Biotech cat. 130-095-929). Cell suspensions were than isolated by ficoll-hypaque density-gradient centrifugation (Amersham Bioscience). CRC specimens were cut into pieces, incubated in 1 mM EDTA (Sigma-Aldrich) for 50 min at 37°C, and then incubated in type D collagenase solution 0.5 mg/mL (Roche Diagnostic) for 4 hr at 37°C. T cell fractions were recovered after fractionation on a four-step gradient consisting of 100%, 60%, 40%, and 30% Percoll solutions (Pharmacia). See also Supplemental Experimental Procedures.
CD4+ T cell subsets were purified by flow cytomtery sorting using the following fluorochrome conjugated antibodies: anti-CD4 APC/Cy7 (clone OKT4), anti-CD27 Pacific Blue (clone M-T271), anti-IL7R PE (clone MB15-18C9), anti-CD25 PE/Cy7 (clone BC96), anti-CXCR3 PE/Cy5 (clone 1C6/CXCR3), anti-CCR6 APC (clone G034E3), and anti-CCR5 FITC (clone j418F1) using a FACSAria II (BD).
RNA Isolation and RNA Sequencing
RNA from tumor-infiltrating lymphocytes was isolated using mirVana Isolation Kit. Libraries for Illumina sequencing were constructed from 50 ng of total RNA with the Illumina TruSeq RNA Sample Preparation Kit v2. Paired-end sequencing (2 × 125) was then performed on an Illumina HiSeq 2500. See also Supplemental Experimental Procedures.
RNA-Seq Data Analysis, Mapping, and Quantification
Raw.fastq files were analyzed using FastQC v0.11.3, and adaptor removal was performed using cutadapt 1.8. Trimming was performed on raw reads using Trimmomatic: standard parameters for phred33 encoding were used. Reads mapping to the reference genome (GRCh38) was performed on quality-checked and trimmed reads using STAR 2.4.1c. The reference annotation is Ensembl v80. The overlap of reads with annotation features found in the reference.gtf was calculated using HT-seq v0.6.1. The output computed for each sample (raw read counts) was then used as input for DESeq2 analysis. Raw counts were normalized using DESeq2’s function “rlog,” and normalized counts were used to perform and visualize principal component analysis (PCA) results (using DESeq2’s “plotPCA” function). See also Supplemental Experimental Procedures.
Differential Expression Analysis
Differential expression analyses of tumor-infiltrating CD4+ Treg, Th1, and Th17 subsets versus CD4+ Treg, Th1, and Th17 from PBMC were performed using DESeq2. Regulated genes were selected for subsequent analyses if their expression values were found to exceed the threshold of 0.05 FDR (Benjamini-Hochberg correction).
SOM Analysis
SOM analyses were carried out using the R package oposSOM using default parameters. Expression values of genes selected in the previous differential expression step were Z-score normalized and supplied in input to the automated pipeline for SOM training and analysis. Genes from regulated spots in the bidimensional output space were selected according to FDR threshold (< 0.1) at group-level. Expression values of genes assigned to regulated spots extracted from the oposSOM output were subject to correlation analysis using model vectors to further refine the results and genes having expression profiles with p < 0.05 were discarded from further analysis and signature definition. See also Supplemental Experimental Procedures.
GO Analysis
A GO enrichment analysis was performed for biological process terms associated with genes assigned to upregulated spots in the SOM bidimensional space using DAVID. Adjusted p (< 0.05) has been used for terms ranking and selection.
Capturing of Single Cells, cDNA Preparation, and Single-Cell PCR
Treg cells from CRC and NSCLC were isolated as previously described (see also Table S1). Single cells were captured on a microfluidic chip on the C1 System (Fluidigm) and whole-transcriptome amplified cDNA was prepared on chip using the SMARTer Ultra Low RNA kit (Clontech). For qPCR experiments, harvested cDNA from single cells was pre-amplified using the same pool of TaqMan gene expression assays to be used for qPCR. Single-cell gene expression experiments were performed using the 96 × 96 quantitative PCR (qPCR) DynamicArray microfluidic chips (Fluidigm) on a BioMark real-time PCR reader following manufacturer’s instructions. A list of the 78 TaqMan assays used in this study is provided in Supplemental Experimental Procedures.
Single-Cell Data Analysis
Raw Ct data have been converted to Log2Exp. Co-expression analysis has been performed by considering both CRC and NSCLC samples and genes for which co-expression with FOXP3 and IL2RA was null were discarded for the subsequent analysis. Gene expression was depicted as violin plots after log2 scale transformation. The violin color gradient represents the percentage of cells that are expressing the gene of interest. A non-parametric test (Mann-Whitney p < 0.05) has been performed on the selected genes by comparing tumor versus peripheral blood samples (see also Supplemental Experimental Procedures).
Flow Cytometry Analysis
Surface markers were directly stained with the following fluorochrome-conjugated antibodies and analyzed by flow cytometry: anti-CD4 (OKT4), anti-PD1-LG2 (CL24F.10C12), anti-CD127 (clone RDR5), anti-CD25 (clone 4E3), anti-4-1BB (clone 4B4), anti-CCR8 (Biolegend clone L263G8), anti CD30 (eBioscience, clone Ber-H2), anti-PD-L1 (Biolegend clone 29E.2A3), anti- TIGIT (eBioscience, clone MBSA43), anti-IL1R2 (R and D clone 34141), IL21R (Biolegend clone 2G1-K12), and anti-OX40 (Biolegend clone Ber-ACT35). FOXP3 and BATF intracellular staining was performed with anti-FOXP3 antibody (clone 236A/E7), anti-BATF (clone MBM7C7), and expression analyzed by flow cytometry. See also Supplemental Experimental Procedures.
Suppression Assay
(CFSE)-labeled responders CD4+ Naive+ T cells from healthy donors were cocultured with different effector to target (E/T) ratios with unlabeled CD127−CD25lowCD4+ T cells sorted from TILs or PBMCs of patients with CRC or NSCLC, using FACS Aria II (BD Biosciences), in the presence of CD11c+CD1c+dentritic cells as antigen-presenting cells and anti-CD3 (OKT3) mAb. Proliferation of CFSE-labeled cells was assessed after 96 hr. Some suppression assays were also performed with tumor Treg cells that were preincubated with the following antibodies (at a final concentration of 20 μg/ml): anti-human PD-L1 (Biolegend clone 29E.2 A 3), anti-human PD-L2 (Biolegend clone MIH18), and anti-human Functional Grade as isotype control (eBioscience clone MBSA43).
Kaplan-Meier Analysis
The Kaplan-Meier analysis (KM) was used to compare the high and low expression of the tumor-Treg signature transcripts either CRC (GSE17536, n = 177) and NSCLC (GSE41271, n = 263) patients. See also Supplemental Experimental Procedures.
Author Contribution
M.D., A.A., G.R., and P.G. designed and performed the main experiments, analyzed the data, and contributed to the preparation of the manuscript. V.R. and R.J.P.B. set up all the bioinformatics pipelines, performed the bioinformatics analyses, and contributed to the preparation of the manuscript. S.M., M.M., M.C., E.P., C.P., M.L.S., I.P., and V.V. performed experiments and analyzed the data. S.B., V.V., N.Z., and G.B. coordinated pathology analyses. A.P., L.S., M.T., N.M., P.C.A., O.E., and L.G. coordinated clinical contributions. R.A.P., G.C., R.D.F., H.G.S., and J.G. discussed results, provided advice, and commented on the manuscript. M.D., A.A., G.R., S.A., and M.P. wrote the manuscript. S.A. and M.P. designed the study and supervised research. All authors discussed and interpreted the results.
Acknowledgments
We would like to thank S. Biffo and P. Della Bona (San Raffaele Scientific Institute, Milan, Italy) for discussions and critical revision of the manuscript. This study was supported by: the Italian Minister of Health GR2011-02351626 to V.V.; AIRC grant n° IG2015-ID17448 to J.G.; the Italian Minister of University and Research Grant CTN01_00177_817708 “DNA on Disk” to S.B.; the ERC Advanced Grant n° 269022 to S.A.; the Flagship CNR-MIUR grant “EPIGEN,” CARIPLO grant n° 2013-0955, AIRC grant n° IG2013-ID14596 and ERC Consolidator Grant n° 617978 to M.P.; and by an unrestricted grant of the “Fondazione Romeo ed Enrica Invernizzi.”
Published: November 15, 2016
Footnotes
Supplemental Information includes five figures, six tables, and Supplemental Experimental Procedures and can be found with this article online at http://dx.doi.org/10.1016/j.immuni.2016.10.021.
Contributor Information
Sergio Abrignani, Email: abrignani@ingm.org.
Massimiliano Pagani, Email: pagani@ingm.org.
Accession Numbers
The accession numbers for the data in this paper are as follows: ENA: PRJEB11844 for RNA-seq tumor and tissue infiltrating lymphocytes; ArrayExpress: E-MTAB-2319 for RNA-seq human lymphocytes datasets; ArrayExpress: E-MTAB-513 for Illumina Human BodyMap 2.0 project; GEO: GSE50760 for RNA-seq datasets CRC; GEO: GSE40419 for RNA-seq datasets NSCLC; GEO: GSE17536 for CRC expression profiling by array; and GEO: GSE41271 for NSCLC expression profiling by array.
Supplemental Information
References
- Arpaia N., Green J.A., Moltedo B., Arvey A., Hemmers S., Yuan S., Treuting P.M., Rudensky A.Y. A Distinct Function of Regulatory T Cells in Tissue Protection. Cell. 2015;162:1078–1089. doi: 10.1016/j.cell.2015.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindea G., Galon J., Mlecnik B. CluePedia Cytoscape plugin: pathway insights using integrated experimental and in silico data. Bioinformatics. 2013;29:661–663. doi: 10.1093/bioinformatics/btt019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindea G., Mlecnik B., Tosolini M., Kirilovsky A., Waldner M., Obenauf A.C., Angell H., Fredriksen T., Lafontaine L., Berger A. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity. 2013;39:782–795. doi: 10.1016/j.immuni.2013.10.003. [DOI] [PubMed] [Google Scholar]
- Borowsky M.L., Hynes R.O. Layilin, a novel talin-binding transmembrane protein homologous with C-type lectins, is localized in membrane ruffles. J. Cell Biol. 1998;143:429–442. doi: 10.1083/jcb.143.2.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boussiotis V.A., Chatterjee P., Li L. Biochemical signaling of PD-1 on T cells and its functional implications. Cancer J. 2014;20:265–271. doi: 10.1097/PPO.0000000000000059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burzyn D., Kuswanto W., Kolodin D., Shadrach J.L., Cerletti M., Jang Y., Sefik E., Tan T.G., Wagers A.J., Benoist C., Mathis D. A special population of regulatory T cells potentiates muscle repair. Cell. 2013;155:1282–1295. doi: 10.1016/j.cell.2013.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell D.J., Koch M.A. Phenotypical and functional specialization of FOXP3+ regulatory T cells. Nat. Rev. Immunol. 2011;11:119–130. doi: 10.1038/nri2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carthon B.C., Wolchok J.D., Yuan J., Kamat A., Ng Tang D.S., Sun J., Ku G., Troncoso P., Logothetis C.J., Allison J.P., Sharma P. Preoperative CTLA-4 blockade: tolerability and immune monitoring in the setting of a presurgical clinical trial. Clin. Cancer Res. 2010;16:2861–2871. doi: 10.1158/1078-0432.CCR-10-0569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Yao Y., Gong C., Yu F., Su S., Chen J., Liu B., Deng H., Wang F., Lin L. CCL18 from tumor-associated macrophages promotes breast cancer metastasis via PITPNM3. Cancer Cell. 2011;19:541–555. doi: 10.1016/j.ccr.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cipolletta D., Feuerer M., Li A., Kamei N., Lee J., Shoelson S.E., Benoist C., Mathis D. PPAR-γ is a major driver of the accumulation and phenotype of adipose tissue Treg cells. Nature. 2012;486:549–553. doi: 10.1038/nature11132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duhen T., Duhen R., Lanzavecchia A., Sallusto F., Campbell D.J. Functionally distinct subsets of human FOXP3+ Treg cells that phenotypically mirror effector Th cells. Blood. 2012;119:4430–4440. doi: 10.1182/blood-2011-11-392324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridman W.H., Pagès F., Sautès-Fridman C., Galon J. The immune contexture in human tumours: impact on clinical outcome. Nat. Rev. Cancer. 2012;12:298–306. doi: 10.1038/nrc3245. [DOI] [PubMed] [Google Scholar]
- Galluzzi L., Buqué A., Kepp O., Zitvogel L., Kroemer G. Immunological Effects of Conventional Chemotherapy and Targeted Anticancer Agents. Cancer Cell. 2015;28:690–714. doi: 10.1016/j.ccell.2015.10.012. [DOI] [PubMed] [Google Scholar]
- Geginat J., Paroni M., Maglie S., Alfen J.S., Kastirr I., Gruarin P., De Simone M., Pagani M., Abrignani S. Plasticity of human CD4 T cell subsets. Front. Immunol. 2014;5:630. doi: 10.3389/fimmu.2014.00630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentles A.J., Newman A.M., Liu C.L., Bratman S.V., Feng W., Kim D., Nair V.S., Xu Y., Khuong A., Hoang C.D. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat. Med. 2015;21:938–945. doi: 10.1038/nm.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Pons M., Cruz-Correa M. Colorectal Cancer Biomarkers: Where Are We Now? BioMed Res. Int. 2015;2015:149014. doi: 10.1155/2015/149014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodi F.S., O’Day S.J., McDermott D.F., Weber R.W., Sosman J.A., Haanen J.B., Gonzalez R., Robert C., Schadendorf D., Hassel J.C. Improved survival with ipilimumab in patients with metastatic melanoma. N. Engl. J. Med. 2010;363:711–723. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam S.A., Ling M.F., Leung J., Shreffler W.G., Luster A.D. Identification of human CCR8 as a CCL18 receptor. J. Exp. Med. 2013;210:1889–1898. doi: 10.1084/jem.20130240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs J., Smits E., Lardon F., Pauwels P., Deschoolmeester V. Immune Checkpoint Modulation in Colorectal Cancer: What’s New and What to Expect. J. Immunol. Res. 2015;2015:158038. doi: 10.1155/2015/158038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamal-Hanjani M., Thanopoulou E., Peggs K.S., Quezada S.A., Swanton C. Tumour heterogeneity and immune-modulation. Curr. Opin. Pharmacol. 2013;13:497–503. doi: 10.1016/j.coph.2013.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joller N., Lozano E., Burkett P.R., Patel B., Xiao S., Zhu C., Xia J., Tan T.G., Sefik E., Yajnik V. Treg cells expressing the coinhibitory molecule TIGIT selectively inhibit proinflammatory Th1 and Th17 cell responses. Immunity. 2014;40:569–581. doi: 10.1016/j.immuni.2014.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josefowicz S.Z., Lu L.F., Rudensky A.Y. Regulatory T cells: mechanisms of differentiation and function. Annu. Rev. Immunol. 2012;30:531–564. doi: 10.1146/annurev.immunol.25.022106.141623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharchenko P.V., Silberstein L., Scadden D.T. Bayesian approach to single-cell differential expression analysis. Nat. Methods. 2014;11:740–742. doi: 10.1038/nmeth.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroemer G., Galluzzi L., Zitvogel L., Fridman W.H. Colorectal cancer: the first neoplasia found to be under immunosurveillance and the last one to respond to immunotherapy? OncoImmunology. 2015;4:e1058597. doi: 10.1080/2162402X.2015.1058597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le D.T., Uram J.N., Wang H., Bartlett B.R., Kemberling H., Eyring A.D., Skora A.D., Luber B.S., Azad N.S., Laheru D. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N. Engl. J. Med. 2015;372:2509–2520. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesterhuis W.J., Steer H., Lake R.A. PD-L2 is predominantly expressed by Th2 cells. Mol. Immunol. 2011;49:1–3. doi: 10.1016/j.molimm.2011.09.014. [DOI] [PubMed] [Google Scholar]
- Marabelle A., Kohrt H., Sagiv-Barfi I., Ajami B., Axtell R.C., Zhou G., Rajapaksa R., Green M.R., Torchia J., Brody J. Depleting tumor-specific Tregs at a single site eradicates disseminated tumors. J. Clin. Invest. 2013;123:2447–2463. doi: 10.1172/JCI64859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messal N., Serriari N.E., Pastor S., Nunès J.A., Olive D. PD-L2 is expressed on activated human T cells and regulates their function. Mol. Immunol. 2011;48:2214–2219. doi: 10.1016/j.molimm.2011.06.436. [DOI] [PubMed] [Google Scholar]
- Munn D.H., Bronte V. Immune suppressive mechanisms in the tumor microenvironment. Curr. Opin. Immunol. 2016;39:1–6. doi: 10.1016/j.coi.2015.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy T.L., Tussiwand R., Murphy K.M. Specificity through cooperation: BATF-IRF interactions control immune-regulatory networks. Nat. Rev. Immunol. 2013;13:499–509. doi: 10.1038/nri3470. [DOI] [PubMed] [Google Scholar]
- Nishikawa H., Sakaguchi S. Regulatory T cells in tumor immunity. Int. J. Cancer. 2010;127:759–767. doi: 10.1002/ijc.25429. [DOI] [PubMed] [Google Scholar]
- Panduro M., Benoist C., Mathis D. Tissue Tregs. Annu. Rev. Immunol. 2016;34:609–633. doi: 10.1146/annurev-immunol-032712-095948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardoll D.M. The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer. 2012;12:252–264. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peggs K.S., Quezada S.A., Chambers C.A., Korman A.J., Allison J.P. Blockade of CTLA-4 on both effector and regulatory T cell compartments contributes to the antitumor activity of anti-CTLA-4 antibodies. J. Exp. Med. 2009;206:1717–1725. doi: 10.1084/jem.20082492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranzani V., Rossetti G., Panzeri I., Arrigoni A., Bonnal R.J., Curti S., Gruarin P., Provasi E., Sugliano E., Marconi M. The long intergenic noncoding RNA landscape of human lymphocytes highlights the regulation of T cell differentiation by linc-MAF-4. Nat. Immunol. 2015;16:318–325. doi: 10.1038/ni.3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi S., Yamaguchi T., Nomura T., Ono M. Regulatory T cells and immune tolerance. Cell. 2008;133:775–787. doi: 10.1016/j.cell.2008.05.009. [DOI] [PubMed] [Google Scholar]
- Sato M., Larsen J.E., Lee W., Sun H., Shames D.S., Dalvi M.P., Ramirez R.D., Tang H., DiMaio J.M., Gao B. Human lung epithelial cells progressed to malignancy through specific oncogenic manipulations. Mol. Cancer Res. 2013;11:638–650. doi: 10.1158/1541-7786.MCR-12-0634-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenbrunn A., Frentsch M., Kohler S., Keye J., Dooms H., Moewes B., Dong J., Loddenkemper C., Sieper J., Wu P. A converse 4-1BB and CD40 ligand expression pattern delineates activated regulatory T cells (Treg) and conventional T cells enabling direct isolation of alloantigen-reactive natural Foxp3+ Treg. J. Immunol. 2012;189:5985–5994. doi: 10.4049/jimmunol.1201090. [DOI] [PubMed] [Google Scholar]
- Schutyser E., Richmond A., Van Damme J. Involvement of CC chemokine ligand 18 (CCL18) in normal and pathological processes. J. Leukoc. Biol. 2005;78:14–26. doi: 10.1189/jlb.1204712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selby M.J., Engelhardt J.J., Quigley M., Henning K.A., Chen T., Srinivasan M., Korman A.J. Anti-CTLA-4 antibodies of IgG2a isotype enhance antitumor activity through reduction of intratumoral regulatory T cells. Cancer Immunol. Res. 2013;1:32–42. doi: 10.1158/2326-6066.CIR-13-0013. [DOI] [PubMed] [Google Scholar]
- Sharma P., Allison J.P. Immune checkpoint targeting in cancer therapy: toward combination strategies with curative potential. Cell. 2015;161:205–214. doi: 10.1016/j.cell.2015.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson T.R., Li F., Montalvo-Ortiz W., Sepulveda M.A., Bergerhoff K., Arce F., Roddie C., Henry J.Y., Yagita H., Wolchok J.D. Fc-dependent depletion of tumor-infiltrating regulatory T cells co-defines the efficacy of anti-CTLA-4 therapy against melanoma. J. Exp. Med. 2013;210:1695–1710. doi: 10.1084/jem.20130579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Śledzińska A., Menger L., Bergerhoff K., Peggs K.S., Quezada S.A. Negative immune checkpoints on T lymphocytes and their relevance to cancer immunotherapy. Mol. Oncol. 2015;9:1936–1965. doi: 10.1016/j.molonc.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J.J., Deane N.G., Wu F., Merchant N.B., Zhang B., Jiang A., Lu P., Johnson J.C., Schmidt C., Bailey C.E. Experimentally derived metastasis gene expression profile predicts recurrence and death in patients with colon cancer. Gastroenterology. 2010;138:958–968. doi: 10.1053/j.gastro.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng M.W., Ngiow S.F., von Scheidt B., McLaughlin N., Sparwasser T., Smyth M.J. Conditional regulatory T-cell depletion releases adaptive immunity preventing carcinogenesis and suppressing established tumor growth. Cancer Res. 2010;70:7800–7809. doi: 10.1158/0008-5472.CAN-10-1681. [DOI] [PubMed] [Google Scholar]
- Topalian S.L., Drake C.G., Pardoll D.M. Immune checkpoint blockade: a common denominator approach to cancer therapy. Cancer Cell. 2015;27:450–461. doi: 10.1016/j.ccell.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torre L.A., Bray F., Siegel R.L., Ferlay J., Lortet-Tieulent J., Jemal A. Global cancer statistics, 2012. CA Cancer J. Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- Twyman-Saint Victor C., Rech A.J., Maity A., Rengan R., Pauken K.E., Stelekati E., Benci J.L., Xu B., Dada H., Odorizzi P.M. Radiation and dual checkpoint blockade activate non-redundant immune mechanisms in cancer. Nature. 2015;520:373–377. doi: 10.1038/nature14292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Eertwegh A.J., Versluis J., van den Berg H.P., Santegoets S.J., van Moorselaar R.J., van der Sluis T.M., Gall H.E., Harding T.C., Jooss K., Lowy I. Combined immunotherapy with granulocyte-macrophage colony-stimulating factor-transduced allogeneic prostate cancer cells and ipilimumab in patients with metastatic castration-resistant prostate cancer: a phase 1 dose-escalation trial. Lancet Oncol. 2012;13:509–517. doi: 10.1016/S1470-2045(12)70007-4. [DOI] [PubMed] [Google Scholar]
- Voo K.S., Bover L., Harline M.L., Vien L.T., Facchinetti V., Arima K., Kwak L.W., Liu Y.J. Antibodies targeting human OX40 expand effector T cells and block inducible and natural regulatory T cell function. J. Immunol. 2013;191:3641–3650. doi: 10.4049/jimmunol.1202752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter S., Weinschenk T., Stenzl A., Zdrojowy R., Pluzanska A., Szczylik C., Staehler M., Brugger W., Dietrich P.Y., Mendrzyk R. Multipeptide immune response to cancer vaccine IMA901 after single-dose cyclophosphamide associates with longer patient survival. Nat. Med. 2012;18:1254–1261. doi: 10.1038/nm.2883. [DOI] [PubMed] [Google Scholar]
- Weon J.L., Potts P.R. The MAGE protein family and cancer. Curr. Opin. Cell Biol. 2015;37:1–8. doi: 10.1016/j.ceb.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth H., von Bergen M., Binder H. Mining SOM expression portraits: feature selection and integrating concepts of molecular function. BioData Min. 2012;5:18. doi: 10.1186/1756-0381-5-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin G., Schauder D.M., Lainez B., Weinstein J.S., Dai Z., Chen Y., Esplugues E., Wen R., Wang D., Parish I.A. A Critical Role of IL-21-Induced BATF in Sustaining CD8-T-Cell-Mediated Chronic Viral Control. Cell Rep. 2015;13:1118–1124. doi: 10.1016/j.celrep.2015.09.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J.C., Hughes M., Kammula U., Royal R., Sherry R.M., Topalian S.L., Suri K.B., Levy C., Allen T., Mavroukakis S. Ipilimumab (anti-CTLA4 antibody) causes regression of metastatic renal cell cancer associated with enteritis and hypophysitis. J. Immunother. 2007;30:825–830. doi: 10.1097/CJI.0b013e318156e47e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zitvogel L., Galluzzi L., Smyth M.J., Kroemer G. Mechanism of action of conventional and targeted anticancer therapies: reinstating immunosurveillance. Immunity. 2013;39:74–88. doi: 10.1016/j.immuni.2013.06.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.