Abstract
The recognition and pairing of homologous chromosomes during meiosis is a complex physical and molecular process involving a combination of polymer dynamics and molecular recognition events. Two highly conserved features of meiotic chromosome behavior are the attachment of telomeres to the nuclear envelope and the active random motion of telomeres driven by their interaction with cytoskeletal motor proteins. Both of these features have been proposed to facilitate the process of homolog pairing, but exactly what role these features play in meiosis remains poorly understood. Here we investigate the roles of active motion and nuclear envelope tethering using a Brownian dynamics simulation in which meiotic chromosomes are represented by a Rouse polymer model subjected to tethering and active forces at the telomeres. We find that tethering telomeres to the nuclear envelope slows down pairing relative to the rates achieved by un-attached chromosomes, but that randomly-directed active forces applied to the telomeres speeds up pairing dramatically in a manner that depends on the statistical properties of the telomere force fluctuations. The increased rate of initial pairing cannot be explained by stretching out of the chromosome conformation but instead seems to correlate with anomalous diffusion of sub-telomeric regions.
Introduction
The pairing of homologous chromosomes is a fascinating physical process that poses unique challenges from a polymer dynamics perspective. Homolog pairing is a key biological phenomenon that underlies Mendelian inheritance during meiosis but also occurs outside of meiosis in diverse contexts including DNA repair [1–3], transvection (reviewed in [4]) and X-chromosome inactivation [5–7]. However homologous pairing is most studied in meiosis since homologous association is critical to the proper segregation of chromosome during the first meiotic division.
Several steps are involved in associating homologous chromosomes together during meiosis (Figure 1A). First homologous chromosomes become aligned and physically near each other, a process termed pairing (reviewed in [8]). For most organisms with notable exceptions in C. elegans and Drosophila, stable pairing is thought to be mediated by double-strand breaks (DSBs) which produce single stranded DNA regions that are then compared to the other chromosome to assess homology. Recombination of DNA initiated at DSBs leads to crossovers and gene conversions that exchange genetic information. The resulting crossovers are a crucial part of the tension-sensing mechanism that aligns homologs properly along the meiotic spindle to permit correct separation during meiosis I. After pairing, a protein structure called the synaptonemal complex assembles between the two homologs gluing them together, a process termed synapsis that is unique to meiosis.
Figure 1.
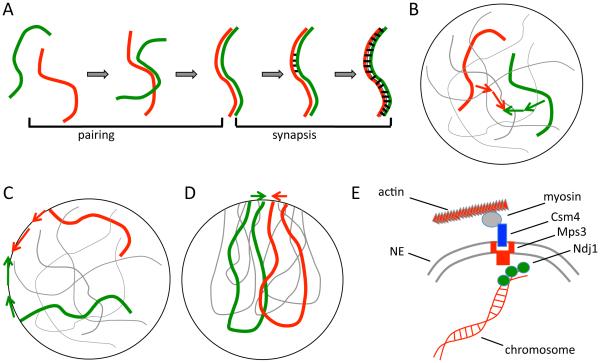
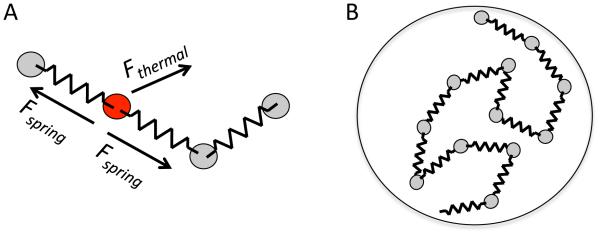
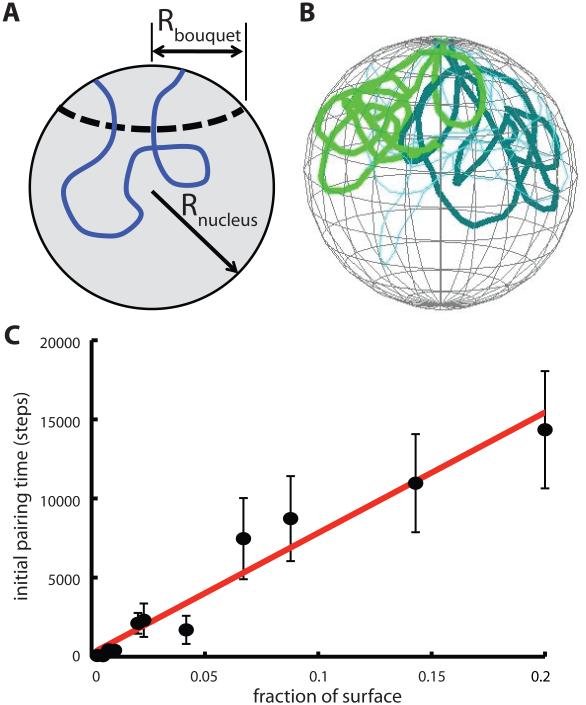
Meiotic chromosome dynamics. (A) Meiotic recombination is preceded by two distinct physical processes. First, homologous chromosomes, which initially are spatially separated in the nucleus, come together and align in a process known as pairing. Then, after pairing, a protein-based synaptonemal complex is assembled that links the two homologs together, a process known as synapsis. (B) Homolog pairing is a challenging physical problem if chromosomes must find each other by diffusion within the volume of the nucleus. (C) Telomeres of meiotic chromosomes are usually attached to the nuclear envelope, which has been proposed to speed up pairing by reducing the dimensionality of the search space from 3D to 2D. (D) Telomeres often cluster in a sub-region of the nuclear envelope, creating a chromosome configuration known as the bouquet. (E) Telomeres of meiotic chromosomes are subject to active forces generated by the cytoskeleton, to which they are coupled by NE-spanning protein complexes. This panel illustrates the telomere motion machinery of budding yeast, in which the telomere associated protein NDJ1p attaches to the NE spanning complex composed of MPS3p and CSM4p, which is then pulled along actin filaments by a myosin motor.
Whether pairing occurs in somatic vs. meiotic cells, this proposal seeks to understand the dynamics of pairing as a physical process, with a particular focus on the role of mechanical forces and constraints applied to the telomeres. Homolog pairing during meiosis is characterized by the need of homologous loci to find each other and become physically associated, despite potentially being separated at random locations on the order of microns apart (Figure 1B). One highly conserved feature of meiotic chromosomes is the attachment of telomeres to the nuclear envelope (NE) (reviewed in [9]). There have been several proposals for how telomere-NE interaction might help to promote pairing via physical mechanisms. One interesting suggestion is that by constraining telomeres to the surface of an approximately spherical nucleus, the pairing process for telomeres would reduce the dimensionality of the search from a 3D search to a 2D search (Figure 1C) [10]. This proposal was inspired by the fact that a particle undergoing a random walk will eventually visit every point in a two dimensional space but not a three dimensional space.
In addition to being attached to the nuclear envelope, in some cases telomeres cluster non-randomly to a sub-region or patch on the nuclear envelope, which has been proposed to increase the efficiency of pairing by restricting the surface area within which the telomeres could execute their 2D search process. Such clustering would lead to a chromosome conformation called a “bouquet” in which telomeres cluster in a limited region of the nuclear surface like the stems of flowers in a bouquet, with the remainder of the chromosome arms spilling out like flowers (Figure 1D).
Both proposals, that meiotic pairing is enhanced by restricted dimensionality and clustering respectively, are fundamentally based on the same idea: that NE interactions of telomeres enhance pairing by restricting the space in which the search takes place. However, an alternative hypothesis concerning the role of telomere-NE interactions stems from the observation that meiotic chromosomes undergo active motion [11]. The molecular basis for this motion has emerged during the past decade, based on discoveries that telomeres are subjected to forces generated by actin-myosin or dynein-microtubules, depending on the species [12–16]. Meiotic telomeres are coupled to the cytoskeleton through the nuclear envelope via SUN and KASH domain proteins such as Mps3 [17] (Figure 1E). Because these active forces are applied to telomeres through the NE, this suggests that perhaps telomeres interact with the NE primarily in order to allow them to harness force generation by the cytoskeleton. Mutants that affect this motion seem to affect the overall outcome of meiosis, but the nature of the effects has been difficult to interpret. Mutants with reduced motion often show delays in the completion of meiosis [18] and reduced rates of collision between homologous loci [19,20], suggesting an impairment in the process of homology searching. But other studies have found that the overall level of crossing over is increased in motion-impairing mutants [21,22], rather than decreased as one might expect if motion facilitates recombination. The question thus remains, what are these movements for? The most naive way we could imagine for actin-driven motion to promote pairing would be for the motion to be directed so as to drive one homolog directly towards the other. This does not appear to be the case, however, as the motion appears to be random, with no directional bias of a chromosome towards its homolog. To what extent can active but randomly directed motions facilitate the pairing process? Our goal in this study is to use computational modeling to assess the potential impact of active random motion of telomeres on the NE compared to the impact of passive NE tethering, in order to better understand the functional significance of telomere-NE interactions in meiosis.
Materials and Methods
Brownian Dynamics Model of Meiotic Chromosome Dynamics
Brownian dynamics modeling has been used to model interphase chromosome dynamics [23], and here we adapt this type of approach to model meiotic chromosome dynamics. Each chromosome is represented as a list of nodes whose coordinates are stored in three dimensions. All positions are represented as real numbers with a length scale in which 1 distance unit in the simulation corresponds to 100 nm in an actual cell. The nodes represent beads connected by springs, with each node subjected to forces whose direction changes randomly at each time step with no correlation between successive time-steps, thus representing the Langevin random force. In addition to this force, each node is also subjected to forces generated by the springs linking it to its two neighboring beads. To enforce confinement inside the nuclear envelope, a repulsive force is applied to any node whose distance from the center of the nucleus exceeds the radius of the nucleus. Finally, each node is subjected to a frictional force that depends on its velocity. Together these forces yield the following equation of motion, which is used to update the velocity and position of each node at each time step:
| (1) |
where xi is the position vector for node i, vi is the velocity of node i, ζ is the friction coefficient, ks is the spring constant of the links between nodes, Leq is the equilibrium length of the links, σ is the magnitude of the Langevin random force, φ is the magnitude of the telomere random force, nr and nt are randomly directed unit vectors representing the direction of the Langevin and telomere-specific random forces, and ui represents a unit vector directed from node i-1 to node i.
This model represent the Rouse model of polymer dynamics [24], which has been shown to be consistent with experimental measurements of interphase chromatin motion [25]. The Rouse model is a “phantom polymer” model that lacks topological constraints, an assumption we make based on the successful use of phantom polymer models to represent chromosome dynamics [26] and which is supported by the fact that the activity of topoisomerases II, which passes one strand through another, is active during meiosis and that defects in topoisomerases II activity lead to meiotic arrest prior to the stage of chromosome segregation and alteration in meiotic crossover distributions [27,28]. Experimental studies have shown that sufficiently high activity of topoisomerase II can cause an entangled DNA melt to behave as a viscous fluid [29], supporting the idea that topological constraints can be neglected to a first approximation when topoisomerase II activity is high, as it is during meiosis.
This model also allows the orientation of each link between nodes to rotate freely, again in accordance with the assumptions of the Rouse model. In the simulations of Figure 9, we explored the influence of constraining link bending by implementing a worm-like chain (WLC) model, in which we add a bending force term that applies a rotation to each link depending on the angle that this link makes with its preceding link.
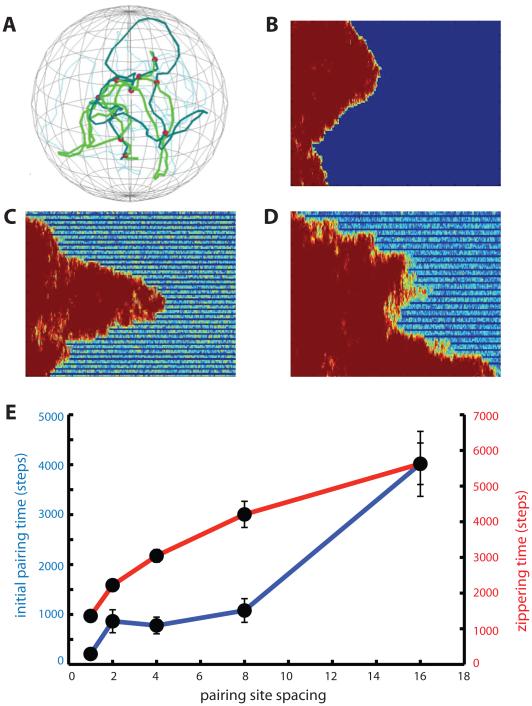
Figure 9.
DSB density determines processivity and speed of zippering. (A) snapshot of pairing simulation with 3-fold reduced density of pairing sites. (B) zippering at maximum pairing site density. (C) pairing site density reduced 3-fold. Zippering is processive but slower. (D) pairing site density reduced 10-fold, showing erratic, stepwise zippering. (E) time for initial pairing (blue) and subsequent zippering (red) versus spacing between pairing sites. A spacing of 1 means that every node in the chain is a pairing site, and was the spacing used for all preceding simulations. Error bars indicate standard error of the mean pairing times.
The simulation proceeds in two phases. In the first phase, each chromosome is initialized by picking a random point inside the nucleus as the first node, and then adding additional nodes according to a random walk, with the choice of node positions limited to points inside the nucleus. Once this initial configuration is generated, the simulation is run for 10000 iterations in order to allow the chromosome to relax into a configuration that is consistent with the simulated forces. During the second phase, the simulation continues to run but now the distance between each node and the corresponding node on its homologous chromosome is monitored, and loci that come within a defined capture radius of each other become paired. The model stores the pairing state of each node, and once a node (and its corresponding homologous node) are switched into the “paired” state, their position and velocity are constrained to be equal to each other for the remainder of the simulation. The spring force term applied at a paired node now takes into account not only the distance to the two adjacent nodes on the chromosome that contains the node in question, but also the two adjacent nodes on the homologous chromosome. For simulations of reversible pairing (Figure 10), nodes classified as paired are switched to the unpaired state with a probability given by parameter Punpair. Their position is not altered by this switch, just their paired state. This switch is performed for all loci at the beginning of each time step. Whether or not a node that has become unpaired will remain unpaired or will re-establish pairing thus depends on how much the two loci move away from each other during the ensuing timestep.
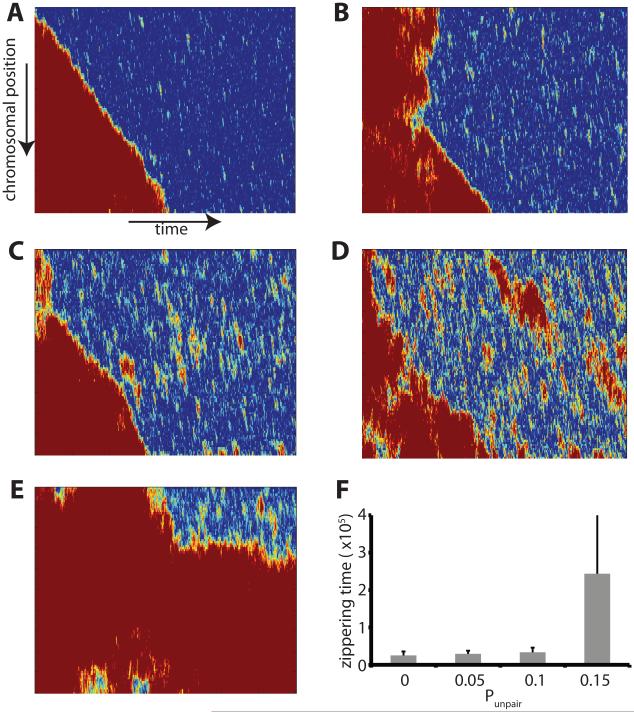
Figure 10.
Influence of reversible pairing. Simulations were performed in which pairing is no longer reversible, with each node having a fixed probability of becoming unpaired at each simulation timestep. (A-E) pairing kymographs showing progress of pairing in simulations with progressively increasing probability of unpairing. Unpairing probabilities: (A) At unpairing probability 0.1 pairing occurs by processive zippering as had been observed for irreversible pairing. (B) Processive zippering is still seen with an unpairing probability of 0.15. (C) At unpairing probability of 0.2, unpairing of extended regions becomes apparent as red zones in the kymograph. (D) At unpairing probability 0.25, pairing is approximately balanced by unpairing to yield an apparent steady state. (E) Unpairing probability 0.3 showing loss of processive pairing. All simulations in this figure were for chromosomes that were not attached to the nuclear envelope and subject only to thermal forces, without active forces applied at telomeres. (F) Graph shows the average time (in units of 10,000 timesteps) required after initial pairing until completion of pairing, which represents the time for zippering, as a function of the unpairing probability. Error bars are standard deviations.
Parameter Choice
Our model is not intended to precisely represent the actual chromosomes of any particular species, but rather is designed to be an abstract model that captures essential features of meiotic chromosome pairing. For this reason, our primary concern in choosing parameters is only to ensure that they are of the correct relative orders of magnitude. The following parameter estimates indicate the values used in our simulation.
Nucleus Radius
Since much of what is known about meiosis from a molecular perspective has been found in the model organism Saccharomyces cerevisiae (budding yeast), we have selected the yeast nucleus as a representative size scale. The radius of a meiotic yeast nucleus is approximately 1 micron. Taking our fundamental unit of length in the model to be 100 nm, the nuclear radius is then equal to 10 of these units. This same unit conversion was then applied to estimate other distance scales.
Polymer link lengths
The persistence length of yeast interphase chromatin has been measured to be approximately 100 nm [30]. While other studies have found a range of different persistence lengths including both shorter and longer values [25,31], we make the simplifying assumption that the segment length is 100 nm, corresponding to 1 distance unit in our model framework.
Chromosome Lengths
The lengths of chromosomes can vary greatly between species. In yeast, chromosome lengths can vary from hundreds of kb to 1-2 Mb. We make the approximation that all chromosomes are 1 Mb long. The linear packing density of chromatin has been estimated at 110-150 bp/nm [31], so the length of a segment (100 nm) would correspond to approximately 10kb. We thus make the assumption that a 1Mb long chromosome contains 100 segments, with the total length of the chromosome being 100 length units.
Chromosome number
Because the Rouse model is a phantom chain model, and because chromosomes only pair with their homologs, the behavior of a given polymer is not affected by the other polymers in the model. We therefore modeled just a single pair of homologous chromosomes.
Capture distance
In our model we implement the process of homology recognition by defining a capture distance, such that if two homologous loci are closer to each other than this radius, they become paired for the remainder of the simulation. We used a capture distance of 0.5 length units, corresponding to 50 nm. This defines a capture distance as half the length of a statistical segment. Based on the physical model of a polymer as a chain of random globules each corresponding to a statistical segment with diameter equal to the link length, this choice of capture distance corresponds to pairing taking place when the distance between the centers of the random globules of two homologous loci are separated by their radii, thus indicating a substantial degree of overlap between the two globules.
Frictional coefficient
Because we have not, up to this point, defined either a time or a force scale, we can choose the frictional coefficient arbitrarily. For simplicity in the simulations reported here, we defined the frictional coefficient to be 1.0
Langevin random force
After setting the frictional coefficient to 1, any given choice of time scale will then induce a choice in force scale based on the viscosity of the system, and vice versa. We arbitrarily assigned the magnitude of the Langevin force term to be 0.15. This choice then sets the force scale for the simulation, with all forces to be considered relative to the Langevin random force.
Time step
Based on the above choices for the friction coefficient and Langevin random force, the duration of a single time step can be constrained by experimental measurement of chromatin diffusion. Considering a single step of the simulation, the length of one random displacement δ is given by the product of the time step, the magnitude of the Langevin force acting during the displacement, and the friction coefficient. The product of the Langevin force and the frictional coefficient, using our assumed values, is 15 nm per second, taking into account the fact that our base length unit is 100 nm. We have previously measured the diffusion constant of yeast chromatin to be 5×10−12 cm2/s [32], which is equal to 500 nm2/s. Using the relation D=δ2/2Δt, we find that the time step Δt = 4.4 seconds. This would of course be considered a very large time step for simulating small molecules like single proteins, but given the massive size and slow diffusion of chromatin, along with the fact that meiotic pairing takes place on a 10 hour time scale, this represents a relatively small time step.
Telomere force
We model the forces applied to the telomere as a randomly directed force with a magnitude Tel_force that is an adjustable parameter of the model. To our knowledge, no direct measurements of the magnitude of the active forces at telomeres have been reported in the literature. We therefore choose a value of Tel_force equal to 1.5 in our model units, which is ten times the Langevin force, in order to represent a case in which the active force is substantially larger than the random thermal forces. This value of Tel_force is used for all active telomere motion simulations. To represent the fact that telomere forces are generated by motion along cytoskeletal filaments such as microtubules or actin filaments, we implement a persistent random walk such that the telomere force is applied in a uniform direction at successive time-steps, with a constant probability per time step Pswitch of switching to a different, randomly chosen direction. The value Pswitch is an adjustable parameter of the model and is systematically varied in the Results section to explore how the persistence of the telomere random walk affects pairing kinetics. In all cases the direction of the telomere active force is tangential to the surface of the nuclear sphere, thus representing the fact that telomeres move in the plane of the nuclear envelope.
Telomere clustering
We model the clustering of telomeres to form a bouquet by defining a spherical cap on the surface of the NE, bounded by a circle of radius Rbouquet. During the initial relaxation phase of the simulation, the location of each telomere is measured relative to the edge of this circle and if a telomere lies outside of this circle, the telomere experiences a force proportional to the distance from the edge directed towards the center of the cap. The proportionality constant was taken to be the same as that used to restrict all nodes to the interior of the NE. The widely disparate reports of bouquet clustering size scales in different organisms meant that it was not possible to choose a single radius for the circular cap. Instead, we explored a range of values for the radius. When plotting the effect of clustering on pairing times, we report the size of the cap in terms of the fraction of the nuclear surface covered by the cap. Surface area for the cap is determined by first calculating the height of the spherical cap h=R-sqrt(R2 – a2) where R is the radius of the nucleus and a is the radius of the bounding circle, and then calculating the cap surface S=2πRh.
Pairing site density
For most simulations, every node in each chain was allowed to pair with the corresponding node on the homologous chain. However for the simulations in Figure 9, the pairing site density was reduced as shown. For these simulations. pairing was only assessed at every nth node, and only those nodes were held together during subsequent joint motion of the two homologs. All simulations of reduced pairing density used the Rouse model with a nuclear radius of 10 and with telomeres attached to the NE undergoing active motion with tel_force equal to 1.5 and Pswitch equal to 0.03.
WLC Kratky-Porod bending force term
In some simulations we tested whether deviations from a Rouse polymer model would affect our predictions. We represent a worm like chain using the Kraty-Porod model in which a force is generated proportional to the bending angle between two successive links with a proportionality constant kbend. Implementing a non-Rouse worm-like chain model required us to define a biologically reasonable bending modulus for the chain. We empirically chose a bending parameter that increased the radius of gyration for the chain by a factor of 2, thus representing a moderate increase in stiffness compared to the Rouse chain. In order to calculate the radius of gyration for an unconstrained polymer, we implemented the model by removing the nuclear repulsion and NE-anchoring terms so that the chromosomes were free to move about in space without confinement, and ran the simulation for 100000 iterations, after which we calculated the radii of gyration. For the Rouse model, we obtained a value of 3.72 length units (n=48 simulations). This is close to the expected value of 4.1 for a freely jointed chain containing 100 links of length 1, which can be derived by noting that the mean squared end-to end distance of a Gaussian polymer is <R2> = Na2 where N is the number of links and a is the link length, and then using the relation rg= sqrt(<R2>/6) (Grosberg and Khoklov, 1994) [24]. Having thus confirmed that our model recapitulates the expected value for the radius of gyration of a freely jointed Rouse chain, we then tested a range of different values for kbend and found that kbend = 0.15 (which happens to be equal to the Langevin force term) caused the radius of gyration to double, to an average value of 7.93 (n=42 simulations). We used this value of kbend for all WLC model simulations in Figure 11.
Figure 11.
Pairing kinetics in worm-like chain model give similar predictions for effect of telomere force persistence as the Rouse chain model. (A) Mean time for initial pairing (blue) and zippering (red), plotted as a function of Pswitch in the persistent random walk model for telomere active forces. (B) Radius of gyration versus Pswitch.
Results
Brownian dynamics model predicts biphasic homology pairing
In order to explore the role of active forces and NE attachment in meiotic pairing, we first establish a computational model for meiotic chromosome dynamics. This model, described in details in Materials and Methods, represents each chromosome as a series of beads joined by springs, with each bead subject to randomly directed thermal forces (Figure 2A). The links between beads can rotate freely and there are no topological constrains between chains, thus yielding an implementation of the Rouse model for polymer dynamics [24]. The entire chain is confined inside a spherical nucleus (Figure 2B). Homologs are represented by pairs of bead-spring chains, and homology searching is carried out by tracking the distance between beads at corresponding positions on the two homologs. When beads come within a pre-defined capture radius, they pair irreversibly, and they are forced to remain together for the rest of the simulation.
Figure 2.
Brownian dynamics model of meiotic chromosomes. (A) Chromosomes are represented by freely-jointed bead-spring chains, with each bead subject to randomly directed thermal forces as well as spring forces oriented towards adjacent beads in the chain. (B) The bead-spring chains are confined in a spherical region that represents the interior of the nucleus. Additional constraints such as attachment to nuclear envelope and active forces applied at telomeres are described in the text.
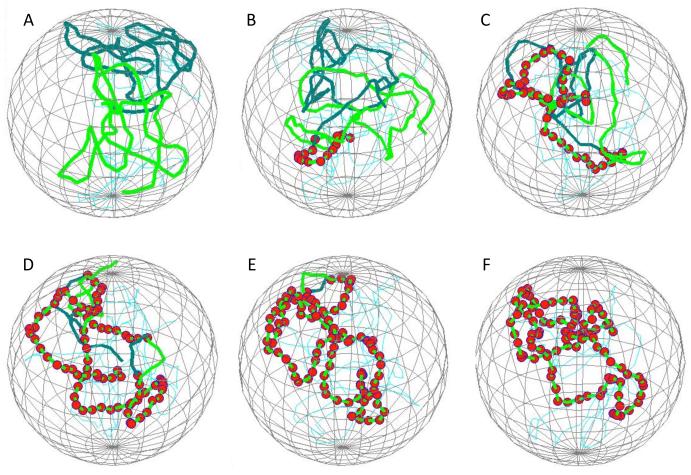
Figure 3 shows snapshots of a representative simulation run, indicating the relative position of two homologous chromosomes within a confining nuclear envelope, with individual paired loci indicated by red spheres. This particular simulation shows the zippering phenomenon previously described by [33], in which the pairing of one locus greatly facilitates the subsequent pairing of neighboring loci by constraining the neighboring loci to remain tethered near their homologs.
Figure 3.
Simulation of homolog pairing. (A) initial state following relaxation phase of the simulation, showing the configuration of chromosomes prior to enabling capture. The two homologs are shown in two different colors. Spherical mesh indicates the position of the nuclear envelope. (B) Early stage of pairing showing a short region of fourteen consecutive sites that have become paired. Red spheres indicate paired nodes. (C-E) representative snapshots at increasing times during the simulation, illustrating the elongation of the initial paired region via zippering. (F) Final state of complete pairing.
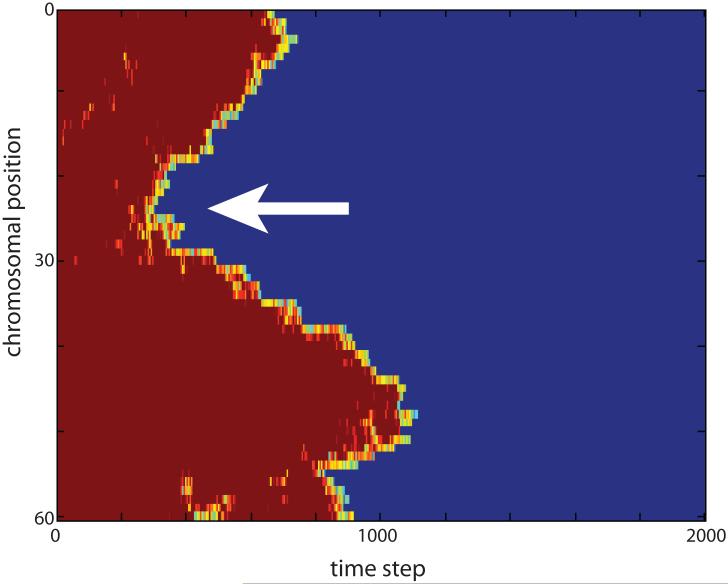
To visualize the process of pairing and to distinguish initial pairing events from subsequent zippering, we have developed a novel visual representation scheme that we refer to as the pairing kymograph (Figure 4). In a pairing kymograph, the distance between homologous loci is color-coded with dark blue representing zero distance for completely paired loci, and red representing the average distance between completely unpaired loci. Shades of yellow indicate regions that are closer than unpaired loci but not yet fully paired. Using this color scheme, the inter-homolog distance map at any point in time is represented as a vertical color bar, with the top and bottom of the bar representing the two ends of the chromosomes and all other nodes of the chromosome chain represented as intermediate positions along the bar, in the same order that they occur in the chromosome chain. These bars are then stacked left to right to indicate the distance maps at successive time-points. Pairing at a given locus is indicated by a switch from red to blue as one traverses the plot from left to right. Processive zippering is indicated by diagonal edges. Examination of such plots shows that the majority of loci pair via zippering. Pairing thus proceeds in a biphasic manner, with an initial phase in which the first paired site is established, followed by a zippering phase during which pairing extends processively and bidirectionally away from this initial pairing site. In the remainder of this study, we seek to ask how passive and active interactions of telomeres with the nuclear envelope might affect the kinetics of these two distinct phases of pairing.
Figure 4.
Pairing kymograph from a representative simulation. Vertical axis indicates position along the chromosome, horizontal axis represents time. Color encodes distance between homologous loci, with blue indicating zero distance and red indicating maximum distance. Arrow indicates an initial pairing event.
Effect of nuclear envelope attachment on homolog pairing kinetics
With our dynamic simulation of chromosome pairing, we can investigate what contributions nuclear envelope attachment makes to the pairing process. A previous computational model based on cellular automata [10] found that constraining chromosomes to the surface of a sphere allowed much more rapid pairing than was seen when chromosomes were unconstrained and had to explore the entire three dimensional volume of the simulated nucleus. Such a result seems intuitively reasonable since search in a 2D surface should be more efficient than in a 3D volume. To test whether this prediction would hold in our polymer dynamics model, we simulated meiotic pairing using the model framework described above, in which the ends of each chromosome polymer were constrained to coincide with the NE but were otherwise freely mobile, subject to a Langevin random force acting in the plane of the NE surface. We performed simulations at a range of different nuclear sizes since theoretical models of 2D and 3D diffusion to capture make different predictions for the scaling of search time to compartment size.
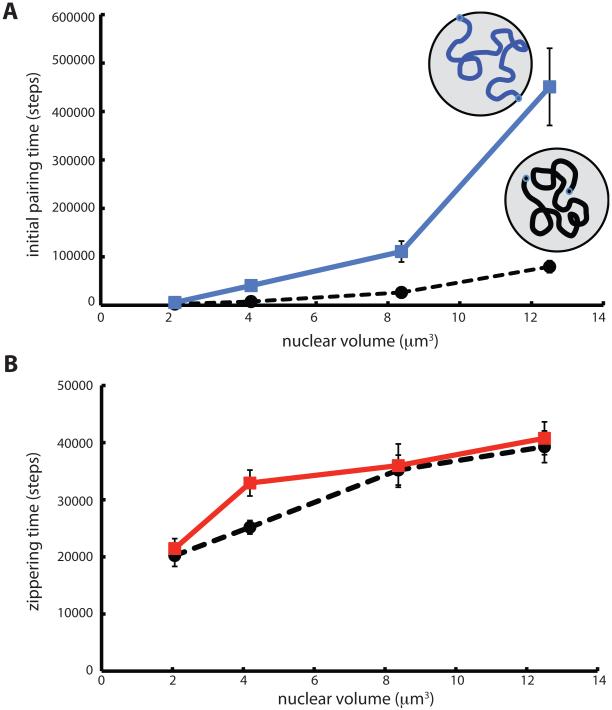
Results of the simulations of NE attachment are shown in Figure 5. We found that the initial pairing time was affected by telomere attachment to the NE, but to our surprise, attachment to the NE appears to slow down pairing, rather than speed it up (Figure 5A). At all nuclear volumes tested, the initial pairing time was longer for chromosomes whose telomeres were attached to the NE, and shorter for chromosomes that were unattached. However as the radius of the nucleus approached the radius of gyration of the chromosomes (i.e. as the nucleus became so small that each chromosome filled the entire nuclear volume) the rates for the attached and unattached cases became statistically indistinguishable. This result is intuitively reasonable since the effect of NE interactions will be small if both homologs are already extensive interdigitated. Overall, our data suggest that attachment of telomeres is a hindrance to initial pairing when the nucleus is large enough to require chromosome motion over large spatial scales.
Figure 5.
Influence of NE tethering on chromosome pairing kinetics. (A) Time for initial pairing event versus nuclear volume, plotted for chromosomes attached to the NE (blue solid line) and for chromosomes unattached to the NE (black dashed line). (B) Time for complete zippering following the initial pairing event versus nuclear volume. (red solid line) Chromosomes tethered to the NE. (black dashed line) unattached chromosomes.
In contrast to the strong effect of NE attachment on the initial pairing time, attachment made almost no difference for the zippering time (Figure 5B). The disparity in effects of attachment on initial pairing versus zippering time predicts that biological assays for a role of telomere attachment to the NE may show dramatically different results depending on whether the assay monitors initial collision or completion of pairing.
In addition to a role for telomere-NE interactions based on confining telomeres to a two-dimensional surface, it is often observed that telomeres cluster in a small sub-region of the nuclear surface, thus potentially facilitating pairing by having the telomeres start out non-randomly close together. We tested the influence of this effect by starting the simulations with telomeres assigned to random positions within a spherical cap on the NE surface. As shown in Figure 6, reducing the radius of this spherical cap does indeed increase the rate of initial pairing, while increasing the area of the patch decreases the rate of initial pairing, as expected. As indicated by the best fit line, the time to initial pairing showed an approximately linear dependence on the fraction of surface area covered by the telomere cluster patch. Linear fit of the log of pairing time to log of fraction of surface covered yields a scaling exponent of 1.3, confirming a close to linear relation.
Figure 6.
Effect of telomere clustering on pairing kinetics. (A) Diagram showing geometry of nuclear patch representation for telomere clustering. (B) Snapshot from simulation showing initial configuration of chromosomes with telomeres clustered in a patch on the NE surface. (C) Graph depicts time for initial pairing versus the size of the clustering patch, given as a fraction of the total NE surface area. Error bars are standard error of the mean pairing time. Line depicts best fit line to pairing time versus fraction of surface contained in patch.
Given our prior result that attachment to the nuclear envelope leads to slower pairing compared to chromosomes that are unconstrained by nuclear envelope attachment, we can now ask whether confinement of telomeres to a surface patch can overcome the decreasing pairing kinetics due to attachment on the surface. Is there a patch size small enough that confining telomeres on the NE becomes worthwhile in terms of facilitating pairing? In the simulations of Figure 5, we found that the average time for initial pairing for chromosomes not attached to the NE in a nucleus of radius 1 micron (volume 4 cubic microns), was 7700 time units. Using the best fit line in Figure 6, which was based on a nucleus of the same radius as Figure 5, we find that the time for initial pairing equals 7700 when the fraction of the NE surface covered by the telomere patch is 0.09. Thus, in these particular simulations, initial pairing time is less than that for unconstrained chromosomes provided the clustering patch size is smaller than 10% of the nuclear envelope surface. These results thus show that NE attachment can increase pairing, and not just impede it as shown in Figure 5, but only if the telomeres are confined to a sufficiently small region on the surface.
Active random motion of telomeres facilitates initial pairing
Telomere confinement to a 2D surface and clustering in a small spherical cap are well known features of meiosis and, as shown above, our simulation can recapitulate these effects. A less well understood aspect of the meiotic telomere-NE interaction is the fact that telomeres are subject to mechanical forces generated by motor proteins moving along cytoskeletal filaments. The telomeres are coupled to these motors by membrane spanning complexes.
What is the role of these mechanical forces? The motion is apparently not directed, in that homologous telomeres are just as likely to move away from each other as towards each other, and thus the motion is best characterized as active random motion. Given that our simulation framework could recapitulate the effects of NE confinement above, we next asked what predictions the simulation would make about the effect of active random forces applied to the telomeres. As shown in Figure 7, we found that application of force to the telomeres did indeed reduce the time required for initial pairing compared to the case with no active forces (see Figure 5 above; for comparison all simulations in Figure 7 used a nuclear volume of 4 cubic microns). Comparing Figure 7 with Figure 6, we find that in these simulations, active motion of the telomeres leads to faster pairing than does telomere clustering for most telomere cluster sizes. Only when the size of the telomere patch drops well below 1% of the nuclear surface does telomere clustering facilitate pairing more effectively than active telomere forces.
Figure 7.
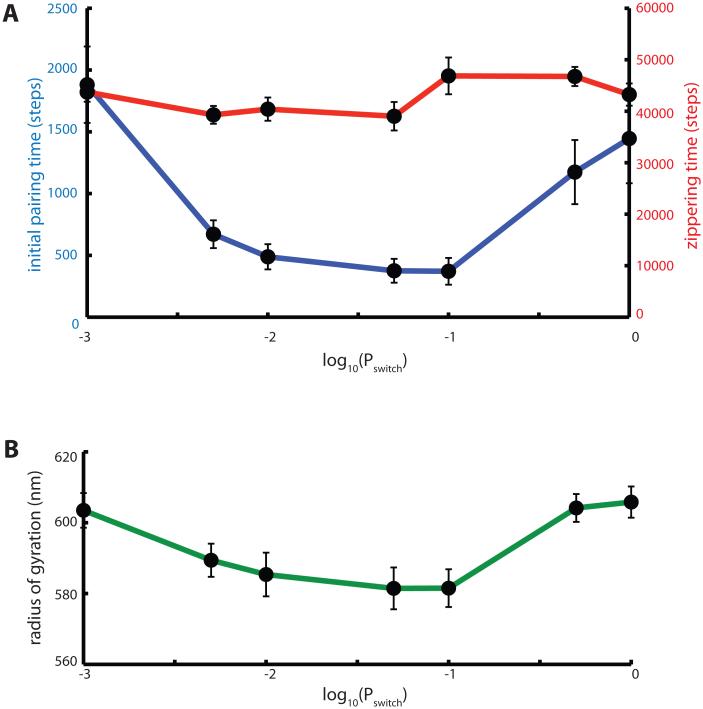
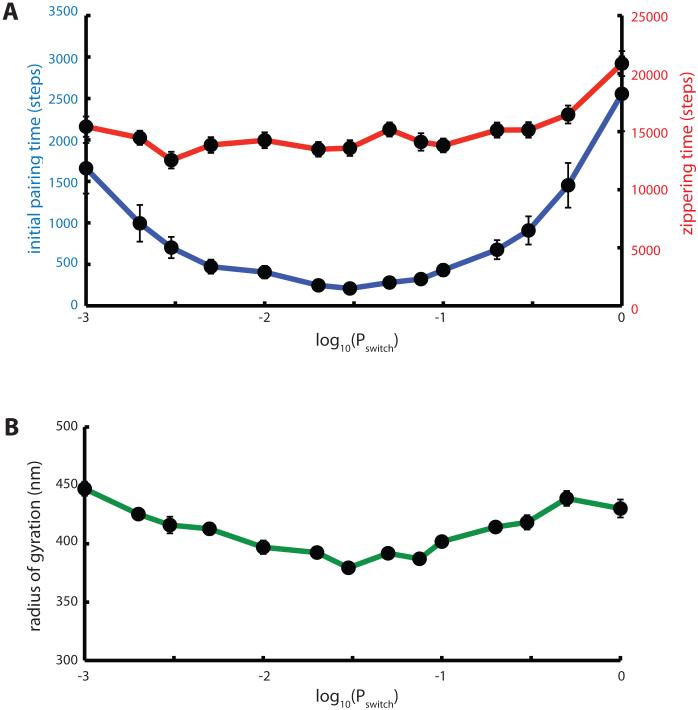
Effect of active telomere motion on pairing kinetics. (A) Influence of active motion persistence on initial pairing (blue curve) and zippering (red curve) times. Simulations were carried out with a telomere force of 1.5 units (ten times the Langevin random force applied at non-telomeric nodes) in an arbitrary direction, with the probability of the telomere force switching to a new direction given by Pswitch. The probability of switching direction is plotted on a log scale ranging from 0.001 to 1. Each datapoint is the average of 50 simulation runs. Error bars indicate standard error of the mean time. (B) Radius of gyration of simulated chromosomes plotted versus Pswitch.
Interestingly, the time to initial pairing depended strongly on the details of the telomere force fluctuations. In our model, telomere force changes direction at random intervals, with the probability of changing direction given by the parameter Pswitch. As shown by the blue curve in Figure 7A, the active forces applied to the telomere are least effective when the direction changes randomly at every time step. This is the situation in which the active force mimics a Langevin random force, with the direction of the forces uncorrelated between successive times. This would be considered “active Brownian motion” (Brangwynne 2009) [34]. Faster initial pairing is achieved when the telomere forces are correlated between successive time steps, with an optimum initial pairing time obtained when Pswitch = 0.03. Since we know that telomeres are moving on cytoskeletal elements, we expect that telomeres will be pulled in a specific direction, according to the orientation of the attached filament, for some period of time until either the filament re-orients or the telomere jumps to a different filament. We thus expect that telomere motion should resemble a persistent random walk. However, the statistical details of telomere motion have not, to our knowledge, been quantitatively measured. Our results suggest that such details could be quite important for understanding the effect that such motion has on the pairing process.
Why does active motion help increase the initial pairing? One hypothesis is that pulling forces at the telomere might help stretch out the chromosome, making it less compacted and thus increasing the overlap between chromosomes during homology searching. We tested the idea that telomere forces might contribute to pairing by de-compacting the chromosomes by calculating the radius of gyration of chromosomes, prior to initial pairing. As shown in Figure 7B, application of active forces at the telomeres did indeed alter the radius of gyration, however the radius of gyration actually was smallest for the value of Pswitch that gave the fastest initial pairing. This is the opposite relation between initial pairing time and radius of gyration to that predicted by the decompaction model. The fact that the fastest initial pairing correlates with the value of Pswitch giving the smallest radius of gyration does suggest a possible link between the two, but the nature of this relation remains to be determined.
Distribution of initial pairing sites as a function of chromosome position
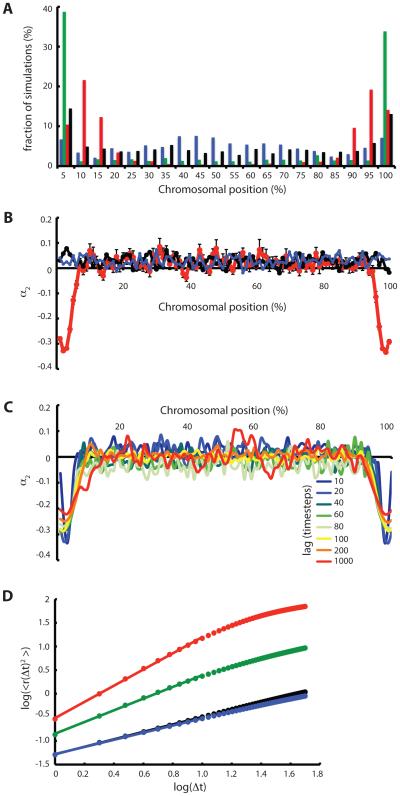
Since telomeres seem to play a critical role in meiosis, either by anchoring the chromosomes to the NE surface or by coupling chromosomes to cytoskeletal force generation systems, we asked whether telomeres would be favored over other chromosomal regions in terms of the rate of pairing. As shown in Figure 8A, we calculated the position of initial pairing and found that for unattached chromosomes, there was a slightly increased tendency for initial pairing to occur near the telomeres, possibly due to increased mobility of the polymer ends, but that this tendency was erased for chromosomes anchored to the NE. When active random motion was applied at telomeres, pairing frequencies became strongly position dependent, with initial pairing most likely to occur near the telomeres. However, the persistence of the telomere random walk had an unexpected influence on the distribution of initial pairing sites. When the active motion was of purely diffusive type, i.e. the direction of the random telomere force changed to a new direction at every time step, then initial pairing occurred almost entirely at the very terminal nodes representing the telomeres. In contrast, when telomeres were simulated to undergo a persistent random walk such as was found above to be more effective in pairing, then the position of first pairing tends not to be at the telomere itself but rather at approximately 10 % of the chromosome length away from the telomere (Figure 8A, red plot). The result of the simulation is thus that active motion of telomeres drives optimal pairing in sub-telomeric regions.
Figure 8.
Chromosomal position dependent pairing frequency and anomalous diffusion. (A) Spatial distribution of initial pairing sites along chromosome is influenced by forces applied to telomeres. (Black) Telomeres not attached to nuclear envelope. (Blue) Telomeres attached to nuclear envelope but without any active telomere forces, so that telomeres move along the surface of the NE by Brownian motion. (Green) Active telomere forces that randomly change direction at every time step. (Red) Telomere forces that change direction with probability Pswitch of 0.05, reflecting a persistent random walk. Bins represent 5% intervals of the chromosome length. Y axis indicates percentage of simulations for which the first site to pair was located at a given position. All datasets are based on 1000 simulations for each set of conditions. (B) Forces applied to telomeres drives anomalous diffusion in a chromosomal position-dependent manner. Plot show the normalized excess kurtosis α2 , a measure of anomalous diffusion, versus position on chromosome. Values near zero correspond to diffusive motion. (Red) Active telomere forces. (Black) Telomeres detached from NE and no active forces applied. (Blue) Telomeres attached to NE but without any advice telomere forces. (C) Anomalous behavior of sub-telomeric regions is observed over a range of time windows indicated by different colors. (D) Mean squared displacement versus time lag. (black) Telomeres not attached to the NE. (blue) telomeres attached but not experiencing active forces, only thermal forces equal in magnitude to the rest of the polymer nodes. (green) telomeres attached to the NE and experiencing active forces with Pswitch 1.0. (red) telomeres attached to the NE and experiencing active forces with Pswitch 0.05. Markers indicate simulation results, lines are best fit lines whose slope indicates the scaling exponent as given in the text.
Active random motion at telomeres drives anomalous diffusion of sub-telomeric chromatin
As discussed above, decompaction of the folded chromosome cannot explain the facilitated pairing seen when active telomere forces are present. What alternative mechanism might explain this effect? We note that the persistent random walk model used to represent cytoskeletal forces acting on the telomere represents a form of anomalous super-diffusion. A large body of literature has suggested that such anomalous diffusion with a long-tailed distribution of displacement probabilities can facilitate search and capture processes, for example in predator-prey interactions [35]. We therefore hypothesized that the active forces pulling telomeres along cytoskeletal filaments might contribute to pairing via such anomalous diffusion effects.
We asked whether forces applied just at the telomere could induce anomalous diffusion at positions elsewhere on the chromosome. We calculated the distribution of displacements of each locus over a time interval of 20 time steps. For standard diffusion in one dimension, such a distribution of displacements should fit a Gaussian distribution. In three dimensions, the displacement that occurs in a given time period is the square root of the sum of the squares of the individual displacements along the three Euclidean axes, hence is described by a Chi distribution with three degrees of freedom. To quantify anomalous diffusive behavior versus chromosomal position, we calculated the parameter a2 [36,37], given by the following equation:
where r is the displacement over the given time interval and brackets indicate ensemble and time averages. This parameter is simply the excess kurtosis divided by three, which is done because the expected value of the kurtosis for a Gaussian distribution is exactly 3. Kurtosis, and hence α2 , reflects both the peakedness of the distribution around its mean value as well as the weight of the tails of the distribution. For a Gaussian distribution, the kurtosis should be exactly 3, the excess kurtosis (which is the kurtosis minus three) is zero, hence α2 will have a value of zero for a Gaussian distribution, and for this reason α2 is used to quantify anomalous diffusive behavior [36,37]. For a Chi distribution with three degrees of freedom, also known as the Maxwell distribution, the normalized excess kurtosis α2 is expected to be
which is approximately 0.036. Therefore, values of that are significantly greater or smaller than 0.036 would indicate that the distribution of three-dimensional distances for a given time window is not Chi distributed, which would indicate that the underlying displacements in the three Euclidean axes are non-Gaussian distributed. Thus for the three-dimensional case, as for one-dimensional displacements, large positive or negative values of α2 would indicate anomalous diffusive motion.
As seen in Figure 8B, in the absence of active telomere forces, α2 is close to zero at all positions on the chromosome, regardless of whether or not the telomeres are anchored on the NE. In fact the mean value over all nodes is 0.027 which is closer to the expected value for a Maxwell distribution as we anticipated. In contrast, when active forces are applied to the telomeres, α2 becomes strongly negative, both at the telomeres themselves and also at sub-telomeric regions, indicating anomalous diffusion in these regions. In fact the diffusion is most strongly anomalous not at the telomeres themselves but at sub-telomeric regions, potentially consistent with the results of Figure 8A showing that initial pairing rates are highest in sub-telomeric sites. As shown in Figure 8C, the anomalous behavior depends on the time-scale over which the jump size distribution is calculated, with a decrease in α2 as the time window increases. However for all values of time window over a range of 10-1000 simulation time steps, anomalous behavior is still seen in the sub-telomeric regions compared with the rest of the chromosome.
Normalized excess kurtosis α2 provides a convenient way to identify anomalous diffusive behavior but it relies on interpretation of the kurtosis of the jump distribution. An alternative way to identify anomalous diffusion is from the scaling exponent of the mean squared displacement versus time lag. For a diffusive process, the mean squared displacement should be proportional to the time elapsed, and hence the scaling exponent should be 1. Deviations from this value indicate anomalous diffusion. In Figure 8D, we plot the mean-squared displacement versus time evaluated at the second node on the chain, chosen to represent the sub-telomeric region. For the cases of telomeres not anchored to the NE, or telomeres anchored but not experiencing any active forces, corresponding to the black and blue curves in Figure 8B, the best fit power law has an exponent of 0.79 and 0.76 respectively. For telomeres attached to the NE and experiencing active motion that switches direction randomly at each timestep (active Brownian motion), the exponent increases to 1.24, while for the case of a persistent random walk (corresponding to the red curve in Figure 8B), the exponent is 1.7. This is a substantial deviation from the predicted value of 1.0, and indicates that the active motion of telomeres can drive anomalous super-diffusion of the subtelomeric region.
Reduced pairing site density alters processivity of zippering
The effects analyzed above, namely NE tethering, active telomere motion, and telomere clustering, all influenced the first phase of pairing, but none had a significant influence on the second phase, namely the time required for chromosomes to complete pairing via zippering. We can intuitively understand this result because zippering is driven by short-range searches between loci that are tethered to each other, so that they are carrying out a search in a confined volume. For a random walk in a sufficiently small region of confinement, the mean squared displacement versus time becomes a function only of the confinement volume and not of the diffusion constant. We therefore expect that even large changes to the chromosome mobility might have little effect on the distribution of actual displacements observed during relevant time scales. If we view processive zippering as a series of short-range highly constrained searches, we would expect the zippering speed to be relatively independent of chromosome motion per se, as we have seen.
This conceptual picture of zippering as a series of constrained searches predicts that zippering speed and processivity should be highly sensitive to the tethering radius between loci adjacent to the most recently paired sites. This radius depends on the length along the chromosome polymer between pairing sites, in other words, it depends on the pairing site density. If pairing site density is reduced, then once a given site is paired, the adjacent pairing sites will be tethered by a longer tether radius, and thus their probability of pairing per unit time is reduced. These effects are recapitulated in our simulations. As shown in Figure 9A, we modified the simulation so that instead of letting every node in the chain be a potential pairing site, we take every n-th node and define it as a pairing site. Comparing the pairing kymographs for simulations in which every node is a pairing site (Figure 9B) with kymographs in which pairing site density is reduced (Figure 9C and D), we find that reduction in pairing site density leads first to a slower but still processive zippering process (based on the slope of the edges in the kymograph) and eventually leads to zippering that is no longer smooth but instead becomes erratic. These effects are manifest at the level of zippering times. As shown in Figure 9E, both initial pairing time and zippering time are increased when pairing site density is decreased. The zippering time showed a sub-linear dependence on pairing site spacing. These results, taken together, suggest that mutations reducing pairing site density should have a much stronger effect on zippering rates than would mutations affecting telomere-NE interactions or motility.
Reversible pairing alters processivity of zippering
Simulations thus far have assumed that once a locus pairs, it remains paired forever. This assumption of irreversible pairing may not be biologically realistic since most biochemical interactions are characterized by a finite binding energy and thus a non-zero off-rate. In order to ask how reversible binding and the affinity between paired loci may affect zippering, we conducted simulations in which every node has a fixed and equal probability of becoming unpaired at each time step. Results are plotted as pairing kymographs in Figure 10 A-E. Unpairing probability up to 0.2 still allows processive zippering. For unpairing probability of 0.25 (Figure 10D), stretches of processive zippering still occur but extensive unpairing occurs at a rate comparable to pairing, so that the system reaches a steady state of pairing and unpairing and never completely zippers. For unpairing probability of 0.3 (Figure 10E), processive zippering is not seen, and instead paired regions increase and decrease in length.
Not only does reversible pairing still allow for processive zippering, it has very little effect on the zippering rate at least for small enough rates of unpairing. Figure 10F indicates that the effect on the time required for zippering is negligible for unpairing probabilities in the range 0 – 0.1. When the unpairing probability gets larger, our criterion for evaluating the zippering time (based on the time elapsed between initial pairing and complete pairing of the whole chromosome) becomes inapplicable since complete pairing never occurs due to the continual presence of one or more unpaired sites.
We conclude that even substantial rates of spontaneous unpairing still allow processive zippering, likely because even if two loci become unpaired during the simulation, they will be located near each other and are likely to re-pair, particularly if neighboring loci are still paired and thus acting as tethers. However, the face that stretches of paired loci can become unpaired, and processive zippering cease, when unpairing rates are high enough suggests that experimental measurement of unpairing kinetics will provide important information for understanding the overall process of meiotic pairing.
Wormlike chain model simulations
All of the above simulations employed the Rouse chain model, a highly simplified version of a real polymer chain. In order to ask how sensitive our results are to the specific form of the polymer model, we repeated the key analyses using a wormlike chain (WLC) representation of the polymer, which we implement using the Kratky-Porod model. In this model, the equation of motion is augmented with a bending force term that depends on the angle between successive link vectors. Our choice of bending force is discussed in Materials and Methods.
As shown in Figure 11A, we find that the pairing times are increased compared to the Rouse model (Figure 7), but we still see the general trend that initial pairing is a stronger function of the telomere random walk persistence than is the subsequent zippering time. Compared to the Rouse model, the radius of gyration is larger for the WLC model (Figure 11B), which is the expected result since the chromosome polymer is now less flexible. But we continue to observe the strong negative correlation between initial pairing time and radius of gyration that was seen in Figure 7 for the Rouse model.
Parameter sensitivity analysis
The model employed here involves a number of parameters that describe the mechanical properties of the polymer models used to represent chromosome dynamics. As discussed in Materials and Methods, we have attempted to provide reasonable estimated values based on known literature, but these are in general order of magnitude estimates, raising the question of whether our results are robust to variation in the parameter values. To address this point, we have varied six key model parameters – the length of links in the bead-spring chain, the frictional coefficient, the spring constant for extension of links, the spring constant for repulsion by the nuclear envelope, the capture distance for pairing, and the magnitude of the Langevin random force (Table 1). We increased and decreased each of these parameters by a factor of two above and below the default value used in our simulations (except for friction, which made the model unstable if increased by a factor of two, in which case we only increased it by a factor of 1.5), and used these altered parameters sets to run simulations under three cases – unattached telomeres subject only to Brownian motion, telomeres anchored to the nuclear envelope, and telomeres subjected to active random forces at the nuclear envelope. As shown in Table 1, we find that for all three conditions, two-fold increases or decreases in model parameters generally produce less than a two fold change in initial pairing time. The key result concerning pairing kinetics, namely that attaching telomeres to the nuclear envelope impedes pairing compared to unattached chromosomes, but active random motion speeds up pairing, is still seen for all parameter variations tested. We therefore conclude that our results are robust at least with respect to small variations in parameter values.
Table 1.
Sensitivity of initial pairing times to model parameter variation.
| Parameter | Unattached | Attached Passive | Attached Active |
|---|---|---|---|
| Default | 9500 + 5700 | 240000 + 100000 | 210 + 150 |
| link length/2 | 8400 + 4800 | 190000 + 100000 | 120 + 70 |
| link length*2 | 340+ 83 | 2200 + 1700 | 60 + 26 |
| friction/2 | 5100 + 4000 | 7200 + 1900 | 180 + 76 |
| friction*1.5 | 14000 + 4500 | 61000 + 23000 | 430 + 72 |
| k_link/2 | 370 + 29 | 32000 + 22000 | 290 + 120 |
| k_link*2 | 15000 + 2400 | 200000 + 160000 | 200 + 100 |
| k_nuc/2 | 8900 + 4500 | 410000 + 150000 | 270 + 100 |
| k_nuc*2 | 8500 + 3300 | 32000 + 24000 | 380 + 250 |
| d_capture/2 | 12000 + 3000 | 210000 + 150000 | 320 + 140 |
| d_capture*2 | 2600 + 2000 | 200000 + 160000 | 120 + |
| F_Langevin/2 | 7300 + 1600 | 220000 + 150000 | 500 + 100 |
| F_Langevin*2 | 1700 + 710 | 8000 + 6400 | 260 + 100 |
Time for first pairing are given for the default parameters used throughout the paper and for simulations in which single parameters were varied according to the factor specified. Simulations were performed for three cases: telomeres unattached to the nuclear envelope (Unattached column), telomeres attached to the nuclear envelope but subject only to thermal motion (Attached Passive), and telomeres attached and subject to active force according to a persistent random walk with Pswitch of 0.03 (Attached Active). For each parameter set three separate simulations were run. Errors are reported as standard error of the mean. All values are rounded to two significant digits. Definitions and default values of parameters are given in Materials and Methods. Briefly, “link length” describes the distance between adjacent nodes in each chain, “friction” is the friction coefficient, “k_link” is a spring constant for the bead-spring chain, “k_nuc” is a spring constant that implements confinement by the nuclear envelope, “d_capture” is the capture distance for pairing nodes, and “F_Langevin” is the magnitude of the random thermal forces per time step acting at each node.
Discussion and Conclusions
Comparison with previous studies
Our simulations gave an entirely different result concerning the role of NE attachment than that predicted by [10], in that we see pairing is FASTER when the chromosomes are internal, not attached. We speculate this is because the telomeres do not diffuse freely in 2D on the surface because they are constrained through their attachment to the rest of the chromosome, which then acts like an anchor due to viscous drag. In order for the telomeres to undergo extensive motion over the nuclear surface, the rest of the chromosome would have to be dragged along in a parallel direction. This may explain why the proposed acceleration of search by confinement to a 2D search was not observed. As for the reduction in search efficiency caused by attachment to the NE, we speculate that the loss of mobility of the telomeres caused by reducing their range of motion from three dimensions to two may propagate to the rest of the chromosome causing slower search. Such an interpretation is supported by computational simulations of a two-dimensional bead-spring polymer model similar to our three dimensional model [38], which reported that immobilization of a chromosome polymer at the ends results in confinement of chromatin motion along the entire length of the chromosome. In the context of meiosis, the results of Verdaasdonk et al. [38] predict that all loci on a chromosome whose telomeres are immobilized on the nuclear envelope should be less effective at searching for their homolog. In our model results of Figure 5, we do not immobilize the telomeres on the nuclear envelope – they are still able to diffuse within the plane of the nuclear surface. Nevertheless, this two-dimensional diffusion does represent a type of tethering, less extreme than complete immobilization, and hence our results can be viewed as a generalization of the rigid anchoring results of Verdaasdonk et al [38].
Penfold and co-workers (2012) [39] implemented a computational model of chromosome conformation during meiosis. This model did not represent or track chromosome dynamics, but rather explored how tethering of telomeres on the nuclear envelope might influence the spatial distribution of the chromosomes within the nucleus. One interesting result of that study was the finding that tethering telomeres on the nuclear envelope, in the absence of telomere clustering, increased the average distance between homologous loci along the chromosomes, but that clustering was able to overcome this effect depending on how close together the telomeres were in the cluster. These results are consistent with our results (Figures 5 and 6), and suggest that the delayed pairing that we observed when telomeres were anchored but not clustered might at least in part be due to larger average distances between the chromosomes at the onset of homology searching.
Predictions of phenotypes based on simulation results
Our model makes testable predictions concerning the role of telomere attachment and active motion in homolog pairing, which in turn lead to predictions about mutant phenotypes. Figures 5 indicates that attaching chromosomes to the NE, in the absence of strong telomere clustering or active telomere forces, will lead to a reduction in pairing efficiency. In yeast, the extent of telomere clustering is subtle, and based on the results of Figure 6C, unlikely to have a major impact on pairing rates. But telomere attachment and forces are predicted to have strong effects. Since telomere attachment in the absence of active forces reduces pairing rates below those attainable when chromosomes are detached from the NE, our results predict mutations that stop telomere motion but leave the telomeres anchored should lead to slower pairing than mutations that cause the telomeres to detach completely. Genetic experiments are consistent with this prediction. In csm4 mutants, telomeres are attached to the NE but do not undergo active motion because they are not linked to the cytoskeleton [21,22]. In ndj1 mutants, telomeres detach entirely from the NE, so that they lack both attachment and active motion [22]. Consistent with the predictions of our model, csm4 mutants have a much stronger effect on meiotic outcomes than ndj1 mutants, showing longer delays and greater loss of gamete viability [14].
A second set of predictions concern the effect of altering DSB density. The results of Figure 9 indicate that reduction in pairing site density will slow down zippering and make the process less processive. In the cell, homolog searching is thought to be carried out in parallel by a finite number of sites, which correspond to the sites of DSBs. DSBs are resected to form 3’ single-stranded tails that are used to sense homology via base-pairing interactions. On the other hand, it has also been proposed to homolog pairing might be carried out independently of base pairing interactions via chromosome associated proteins such as cohesins that form a bar code on the surface of different chromosomes [40]. In this latter model, altering DSB density would not affect density of pairing sites. Our results predict that mutations which increase or decrease the total number of DSBs in the genome should have a corresponding effect on the pairing site density, and thus alter the initial pairing time and the zippering processivity. Measurement of zippering processivity in mutants that reduce the DSB density [41] may thus be able to test whether pairing site spacing is in fact dictated by DSBs.
Active random motion and the distribution of pairing centers
Living cells are seething with random movement, including both thermally-driven Brownian movement and active motility driven by motor proteins. Although motors often drive long range processive directional movement, it is becoming appreciated that motors acting over short distances in random directions can produce random movement that resembles Brownian motion but with a much higher diffusion constant, a phenomenon known as active diffusion [34]. The idea of active random motion has been discussed mainly in terms of cytoplasmic transport, where the phenomenon can be clearly described but where the functional relevance remains poorly understood. There is evidence for active diffusion of interphase chromatin [42] but its function is also unclear. Active telomere-led meiotic chromosome motion represents a unique opportunity to study active diffusion in a context where its biological importance has already been documented.
It has long been hypothesized that active motion of chromatin helps to speed up the random search for homologous sequences in the crowded environment of the nucleus [14,21]. Passive diffusion of chromatin is highly constrained [32,43] and also shows evidence for sub-diffusive effects [25,44,45], which would tend to make the processes of random homology search exceedingly slow. Active movement could speed up the search process by producing a higher effective diffusion constant. Active motions could also cause the motion to become super-diffusive by changing the distribution of random displacements towards a longer-tailed distribution. It is well established that random movements with long tail distributions, such as Lévy flights, are more effective as random search processes [35] and are for this reason often employed as a foraging or hunting strategy by animals [46], although the efficacy can depend on confinement and other details of the situation. Superdiffusive, long-tail random motion is documented for interphase chromatin [42,47] but has not been studied in meiosis. Our results suggest that cytoskeletal forces acting on telomeres can drive anomalous diffusion of sub-telomeric chromatin to speed up collisions. The restriction of the anomalous diffusion to sub-telomeric positions (Figure 8B), and the result that initial pairing is most likely to occur in sub-telomeric regions when telomeres undergo persistent random walks (Figure 8A) may help to explain why in some species, in which homolog pairing is mediated by specific “pairing centers”, these pairing centers tend to be located asymmetrically on the chromosome with a strong bias towards the chromosome ends [48–51].
Acknowledgments
This work was supported by NIH grant R01 GM097213 (JCF).
References
- [1].Brandt VL, Hewitt SL, Skok J a. It takes two: communicatation between homologous alleles preserves genomic stability during V(D)J recombination. Nucleus. 2010;1:23–9. doi: 10.4161/nucl.1.1.10595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Doetschman T, Gregg RG, Maeda N, Hooper ML, Melton DW, Thompson S, Smithies O. Targetted correction of a mutant HPRT gene in mouse embryonic stem cells. Nature. 1987;330:576–8. doi: 10.1038/330576a0. [DOI] [PubMed] [Google Scholar]
- [3].Tichy ED, Pillai R, Deng L, Liang L, Tischfield J, Schwemberger SJ, Babcock GF, Stambrook PJ. Mouse embryonic stem cells, but not somatic cells, predominantly use homologous recombination to repair double-strand DNA breaks. Stem Cells Dev. 2010;19:1699–711. doi: 10.1089/scd.2010.0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Duncan IW. Transvection effects in Drosophila. Annu. Rev. Genet. 2002;36:521–56. doi: 10.1146/annurev.genet.36.060402.100441. [DOI] [PubMed] [Google Scholar]
- [5].Bacher CP, Guggiari M, Brors B, Augui S, Clerc P, Avner P, Eils R, Heard E. Transient colocalization of X-inactivation centres accompanies the initiation of X inactivation. Nat. Cell Biol. 2006;8:293–9. doi: 10.1038/ncb1365. [DOI] [PubMed] [Google Scholar]
- [6].Xu N, Tsai C-L, Lee JT. Transient homologous chromosome pairing marks the onset of X inactivation. Science. 2006;311:1149–52. doi: 10.1126/science.1122984. [DOI] [PubMed] [Google Scholar]
- [7].Masui O, Bonnet I, Le Baccon P, Brito I, Pollex T, Murphy N, Hupé P, Barillot E, Belmont AS, Heard E. Live-cell chromosome dynamics and outcome of X chromosome pairing events during ES cell differentiation. Cell. 2011;145:447–58. doi: 10.1016/j.cell.2011.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Zickler D, Kleckner N. Recombination, Pairing, and Synapsisof Homologs during Meiosis. Cold Spring Harb Perspect Biol. 2015;7:a016626. doi: 10.1101/cshperspect.a016626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Scherthan H. Telomere attachment and clustering during meiosis. Cell. Mol. Life Sci. 2007;64:117–24. doi: 10.1007/s00018-006-6463-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Dorninger D, Karigl G, Loidl J. Simulation of chromosomal homology searching in meiotic pairing. J. Theor. Biol. 1995;176:247–60. doi: 10.1006/jtbi.1995.0195. [DOI] [PubMed] [Google Scholar]
- [11].Parvinenen M, Soderstrom K. Chromosome rotation and the formation of synapsis. Nature. 1976;260:534–5. doi: 10.1038/260534a0. [DOI] [PubMed] [Google Scholar]
- [12].Chikashige Y, Ding DQ, Funabiki H, Haraguchi T, Mashiko S, Yanagida M, Hiraoka Y. Telomere-led premeiotic chromosome movement in fission yeast. Science. 1994;264:270–3. doi: 10.1126/science.8146661. [DOI] [PubMed] [Google Scholar]
- [13].Scherthan H, Wang H, Adelfalk C, White EJ, Cowan C, Cande WZ, Kaback DB. Chromosome mobility during meiotic prophase in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 2007;104:16934–9. doi: 10.1073/pnas.0704860104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Conrad MN, Lee CY, Chao G, Shinohara M, Kosaka H, Shinohara A, Conchello JA, Dresser ME. Rapid Telomere Movement in Meiotic Prophase Is Promoted By NDJ1, MPS3, and CSM4 and Is Modulated by Recombination. Cell. 2008;133:1175–87. doi: 10.1016/j.cell.2008.04.047. [DOI] [PubMed] [Google Scholar]
- [15].Koszul R, Kleckner N. Dynamic chromosome movements during meiosis: a way to eliminate unwanted connections? Trends Cell Biol. 2009;19:716–24. doi: 10.1016/j.tcb.2009.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Sato A, Isaac B, Phillips CM, Rillo R, Carlton PM, Wynne DJ, Kasad RA, Dernburg AF. Cytoskeletal Forces Span the Nuclear Envelope to Coordinate Meiotic Chromosome Pairing and Synapsis. Cell. 2009;139:907–19. doi: 10.1016/j.cell.2009.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Conrad MN, Lee C-Y, Wilkerson JL, Dresser ME. MPS3 mediates meiotic bouquet formation in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 2007;104:8863–8. doi: 10.1073/pnas.0606165104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Prasada Rao HBD, Shinohara M, Shinohara A. Mps3 SUN domain is important for chromosome motion and juxtaposition of homologous chromosomes during meiosis. Genes to Cells. 2011;16:1081–96. doi: 10.1111/j.1365-2443.2011.01554.x. [DOI] [PubMed] [Google Scholar]
- [19].Lee CY, Conrad MN, Dresser ME. Meiotic chromosome pairing is promoted by telomere-led chromosome movements independent of bouquet formation. PLoS Genet. 2012:8. doi: 10.1371/journal.pgen.1002730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Lui DY, Cahoon CK, Burgess SM. Multiple Opposing Constraints Govern Chromosome Interactions during Meiosis. PLoS Genet. 2013:9. doi: 10.1371/journal.pgen.1003197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Kosaka H, Shinohara M, Shinohara A. Csm4-dependent telomere movement on nuclear envelope promotes meiotic recombination. PLoS Genet. 2008:4. doi: 10.1371/journal.pgen.1000196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Wanat JJ, Kim KP, Koszul R, Zanders S, Weiner B, Kleckner N, Alani E. Csm4, in collaboration with Ndj1, mediates telomere-led chromosome dynamics and recombination during yeast meiosis. PLoS Genet. 2008:4. doi: 10.1371/journal.pgen.1000188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Ehrlich L, Münkel C, Chirico G, Langowski J. A Brownian dynamics model for the chromatin fiber. Comput. Appl. Biosci. 1997;13:271–9. doi: 10.1093/bioinformatics/13.3.271. [DOI] [PubMed] [Google Scholar]
- [24].Grosberg AY, Khokhlov AR. Statistical Physics of Macromolecules. 1994 American Institute of Physics. [Google Scholar]
- [25].Hajjoul H, Mathon J, Ranchon H, Goiffon I, Mozziconacci J, Albert B, Carrivain P, Victor JM, Gadal O, Bystricky K, Bancaud A. High-throughput chromatin motion tracking in living yeast reveals the flexibility of the fiber throughout the genome. Genome Res. 2013;23:1829–38. doi: 10.1101/gr.157008.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Sikorav JL, Jannink G. Kinetics of chromosome condensation in the presence of topoisomerases: a phantom chain model. Biophys. J. 1994;66:827–37. doi: 10.1016/s0006-3495(94)80859-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Rose D, Holm C. Meiosis-specific arrest revealed in DNA topoisomerase II mutants. Mol. Cell. Biol. 1993;13:3445–55. doi: 10.1128/mcb.13.6.3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Zhang L, Wang S, Yin S, Hong S, Kim KP, Kleckner N. Topoisomerase II mediates meiotic crossover interference. Nature. 2014;511:551–6. doi: 10.1038/nature13442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Kundukad B, Van Der Maarel JRC. Control of the flow properties of DNA by topoisomerase II and its targeting inhibitor. Biophys. J. 2010;99:1906–15. doi: 10.1016/j.bpj.2010.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Dekker J. Mapping in vivo chromatin interactions in yeast suggests an extended chromatin fiber with regional variation in compaction. J. Biol. Chem. 2008;283:34532–40. doi: 10.1074/jbc.M806479200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Bystricky K, Heun P, Gehlen L, Langowski J, Gasser SM. Long-range compaction and flexibility of interphase chromatin in budding yeast analyzed by high-resolution imaging techniques. Proc. Natl. Acad. Sci. U. S. A. 2004;101:16495–500. doi: 10.1073/pnas.0402766101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Marshall WF, Straight A, Marko JF, Swedlow J, Dernburg A, Belmont A, Murray AW, Agard DA, Sedat JW. Interphase chromosomes undergo constrained diffusional motion in living cells. Curr. Biol. 1997;7:930–9. doi: 10.1016/s0960-9822(06)00412-x. [DOI] [PubMed] [Google Scholar]
- [33].Zickler D. Development of the synaptonemal complex and the “recombination nodules” during meiotic prophase in the seven bivalents of the fungus Sordaria macrospora Auersw. Chromosoma. 1977;61:289–316. doi: 10.1007/BF00288615. [DOI] [PubMed] [Google Scholar]
- [34].Brangwynne CP, Koenderink GH, MacKintosh FC, Weitz DA. Intracellular transport by active diffusion. Trends Cell Biol. 2009;19:423–7. doi: 10.1016/j.tcb.2009.04.004. [DOI] [PubMed] [Google Scholar]
- [35].Viswanathan GM, Buldyrev SV, Havlin S, da Luz MG, Raposo EP, Stanley HE. Optimizing the success of random searches. Nature. 1999;401:911–4. doi: 10.1038/44831. [DOI] [PubMed] [Google Scholar]
- [36].Marcus a H, Schofield J, Rice S a. Experimental observations of non-Gaussian behavior and stringlike cooperative dynamics in concentrated quasi-two-dimensional colloidal liquids. Phys. Rev. E. Stat. Phys. Plasmas. Fluids. Relat. Interdiscip. Topics. 1999;60:5725–36. doi: 10.1103/physreve.60.5725. [DOI] [PubMed] [Google Scholar]
- [37].Parry BR, Surovtsev IV, Cabeen MT, O’Hern CS, Dufresne ER, Jacobs-Wagner C. The bacterial cytoplasm has glass-like properties and is fluidized by metabolic activity. Cell. 2014;156:183–94. doi: 10.1016/j.cell.2013.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Verdaasdonk JS, Vasquez PA, Barry RM, Barry T, Goodwin S, Forest MG, Bloom K. Centromere tethering confines chromosome domains. Mol. Cell. 2013;52:819–31. doi: 10.1016/j.molcel.2013.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Penfold CA, Brown PE, Lawrence ND, Goldman ASH. Modeling meiotic chromosomes indicates a size dependent contribution of telomere clustering and chromosome rigidity to homologue juxtaposition. PLoS Comput. Biol. 2012:8. doi: 10.1371/journal.pcbi.1002496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Ishiguro KI, Kim J, Shibuya H, Hernández-Hernández A, Suzuki A, Fukagawa T, Shioi G, Kiyonari H, Li XC, Schimenti J, Höög C, Watanabe Y. Meiosis-specific cohesin mediates homolog recognition in mouse spermatocytes. Genes Dev. 2014;28:594–607. doi: 10.1101/gad.237313.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Rockmill B, Lefrançois P, Voelkel-Meiman K, Oke A, Roeder GS, Fung JC. High Throughput Sequencing Reveals Alterations in the Recombination Signatures with Diminishing Spo11 Activity. PLoS Genet. 2013:9. doi: 10.1371/journal.pgen.1003932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Weber SC, Spakowitz a. J, Theriot J a. Nonthermal ATP-dependent fluctuations contribute to the in vivo motion of chromosomal loci. Proc. Natl. Acad. Sci. 2012;109:7338–43. doi: 10.1073/pnas.1119505109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Vazquez J, Belmont AS, Sedat JW. Multiple regimes of constrained chromosome motion are regulated in the interphase Drosophila nucleus. Curr. Biol. 2001;11:1227–39. doi: 10.1016/s0960-9822(01)00390-6. [DOI] [PubMed] [Google Scholar]
- [44].Fritsch CC, Langowski J. Anomalous diffusion in the interphase cell nucleus: The effect of spatial correlations of chromatin. J. Chem. Phys. 2010:133. doi: 10.1063/1.3435345. [DOI] [PubMed] [Google Scholar]
- [45].Burnecki K, Kepten E, Janczura J, Bronshtein I, Garini Y, Weron A. Universal algorithm for identification of fractional Brownian motion. A case of telomere subdiffusion. Biophys. J. 2012;103:1839–47. doi: 10.1016/j.bpj.2012.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Sims DW, Humphries NE, Bradford RW, Bruce BD. Levy flight and Brownian search patterns of a free-ranging predator reflect different prey field characteristics. J. Anim. Ecol. 2012;81:432–42. doi: 10.1111/j.1365-2656.2011.01914.x. [DOI] [PubMed] [Google Scholar]
- [47].Levi V, Ruan Q, Plutz M, Belmont AS, Gratton E. Chromatin dynamics in interphase cells revealed by tracking in a two-photon excitation microscope. Biophys. J. 2005;89:4275–85. doi: 10.1529/biophysj.105.066670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].McKim KS, Howell AM, Rose AM. The effects of translocations on recombination frequency in Caenorhabditis elegans. Genetics. 1988;120:987–1001. doi: 10.1093/genetics/120.4.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Villeneuve AM. A cis-acting locus that promotes crossing over between X chromosomes in Caenorhabditis elegans. Genetics. 1994;136:887–902. doi: 10.1093/genetics/136.3.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].MacQueen AJ, Phillips CM, Bhalla N, Weiser P, Villeneuve AM, Dernburg AF. Chromosome sites play dual roles to establish homologous synapsis during meiosis in C. elegans. Cell. 2005;123:1037–50. doi: 10.1016/j.cell.2005.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Phillips CM, Meng X, Zhang L, Chretien JH, Urnov FD, Dernburg AF. Identification of chromosome sequence motifs that mediate meiotic pairing and synapsis in C. elegans. Nat. Cell Biol. 2009;11:934–42. doi: 10.1038/ncb1904. [DOI] [PMC free article] [PubMed] [Google Scholar]