Abstract
Oomycete pathogen, Phytophthora capsici is devastating for black pepper (Piper nigrum L.) and causes foot rot disease at all stages of plant growth. Phytophthora secretes a glucanase inhibitor protein (GIP), which is capable of inhibiting defence proteins like endoglucanases. In this particular study Quantitative PCR analysis, molecular docking studies and analysis of sequences of Glucanase inhibitor protein and beta-1,3 glucanse genes were done mainly depending on the data derived from Phytophthora capsici whole genome sequencing and Piper colubrinum RNA-sequencing (RNA-Seq). Amino acid sequence length of GIP gene from P. capsici was about 353 amino acids and that of glucanase pcEGase gene from P. colubrinum was about 312 amino acids. GIP gene from P. capsici showed high level of expression at early hours of the inoculation time period and pcEGase gene showed high level of expression at 16 hpi. High level of expression of pcEGase gene at 16 hpi is an indication that the GIP gene is successfully inhibited by the glucanase protein from the plant. Moreover insilico studies gave some hint on the importance of certain sites on the surfaces of both interacting proteins that might be having a role in binding of the two proteins and subsequent reactions thereof. Insilico analysis also conclusively proved that inhibition of glucanase inhibitor protein is mainly caused by recognition of an arginine as well as an isoleucine residue during the interaction of the two proteins.
Keywords: Piper colubrinum, Phytophthora capsici, Black pepper, Molecular docking, qRT-PCR
Introduction
Foot rot is a serious disease affecting black pepper (Piper nigrum L.) and is caused by Phytophthora capsici. Piper colubrinum, an exotic and wild species related to black pepper, from South America is widely known to be resistant to Phytophthora.
Plant apoplast is a place where numerous proteases are synthesized, especially when the plant is attacked by a pathogen. Pathogens produce certain proteins called glucanase inhibitor proteins GIPs which eventually targets plant glucanases present in the apoplast. GIPs were initially cloned in Phytophthora sojae and the GIPs showed homology to the chymotrypsin class of serine proteases (Damasceno et al. 2008).
GIPs inhibits the beta-1,3-glucanases produced by a particular host, eventually protecting the oomycete cell wall from any degradation caused by the plant glucanases (York et al. 2004). To date, there is no information regarding GIP genes of P. capsici infecting black pepper counteracting on beta-1,3-glucanase from P. colubrinum, a highly resistant species. In this study, we report the characterization of these genes and their expression by quantitative reverse transcription polymerase chain reaction (qRT-PCR), in planta after inoculating with the concerned pathogen. An effort was also made to find out domains in GIP protein that binds to the beta-1,3-glucanase through molecular modeling and docking studies and, also the sequence characterization and expression analysis of both GIP and glucanase gene were indeed studied to get a comprehensive understanding of the interaction between the two proteins.
Methods
Phytophthora isolates and plant material
Phytophthora isolate, 05–06 from ICAR-IISR was used for conducting experiments and the mycelium obtained from the same isolate was used for RNA isolation from the pathogen. P. colubrinum plants grown under controlled condition were used for carrying out the experiment. The Phytophthora isolates were grown on carrot agar at 25 °C for 3 days. The leaves of P. colubrinum were cleaned with autoclaved DEPC water. The mycelia discs were prepared aseptically by cutting with 10 mm diameter cork borer at the periphery of the culture plates. The mycelia disc (10 mm diameter) from the carrot agar plates were inoculated on the lower side of the leaves, without detaching from the plant. It was then covered by sterile cotton wetted with DEPC water. For the qRT-PCR studies, inoculated parts of the leaves were collected at 2, 4, 8, 16, 24, 48 h post inoculation (hpi).
RNA extraction and cDNA synthesis
RNA isolation from both pathogen and plant materials were done based on Trizol method strictly following the manufacturers instruction. The extracted RNA was treated with DNAse (Fermentas) to remove any traces of DNA as per the manufacturer instructions. The extracted RNA was treated with DNAse (Fermentas) to remove any traces of DNA as per standard protocol. Biophotometer plus (Eppendorf) was used to quantify the extracted RNA and this was stored at –80 ºC until further use. The cDNA was stored at −20 °C until further use.
qRT-PCR primer design
Actin genes were used as endogenous controls for conducting qRT-PCR for both plant and pathogen RNA’s. Primers for the beta-1,3-glucanase and GIP genes were designed from the sequence databases maintained at IISR by using Primer 3 software (http://primer3plus.com/). Primer pairs (Table 1) were tested to check their specificity.
Table 1.
Primer sequences used for this study by quantitative reverse transcription PCR followed by inoculation with Phytophthora capsici on Piper colubrinum
| Sl. no | Primer name | Length | Sequence | Amplicon size | |
|---|---|---|---|---|---|
| 1. | Phytophthora GIP | 20 bp | F | AAGGCGCTCAAGTCTACCAC | 198 |
| R | ACGAGCGTAAATAGCCGGAG | ||||
| 2. | Phytophthora actin | 20 bp | F | CCCATCTACGAGGGTTACGC | 121 |
| R | CCGTGGTGGTGAACGAGTAA | ||||
| 3. | Piper colubrinum actin | 20 bp | F | CACATCTGCTGGAAGGTGCT | 143 |
| R | CACTACTGCAGAGCGGGAAA | ||||
| 4. | Beta-1,3-Glucanase | 24 bp | F | CGATGGGTACGTACGTGAACAGTG | 190 |
| 20 bp | R | CCGGCCTACAAAATGACATC | |||
All the real time PCR reactions were performed using three independent biological replicates and three technical replicates for each of the individual biological replicates. qPCR analysis of genes were carried out at various time period intervals. Real time PCR analysis was done using SYBR Green PCR master mix (Qiagen) following manufacturer’s instructions. (CT) values or threshold cycle values were computed using Rotor gene–Q software version 2.0.2 (Qiagen). Melt curve analysis was also performed to assess the specificity of the PCR products.
Bioinformatics analysis
Sequence data and phylogeny
β-1,3-glucanase gene from P. colubrinum has been obtained from gene ontology annotation done with transcriptome data and the pathogenic effector domain—glucanase inhibitor protein (GIP) has been identified from P. capsici 05–06 whole genome annotation data. ORFs were identified using CLC genomic workbench 7.0. The ORF start and stop position were trimmed using BioEdit version 7.0.5.3. Using CLC genomic workbench 7.0 Coding sequences (CDS) and six frame proteins were predicted. Functionally related sequences for each gene were then obtained for phylogenetic analysis using the blast hit. Phylogenetic analysis was done using MEGA5 with top hit data of delta Blast analysis. Secondary and tertiary structural study was conducted using Phyre2, I-tasser and Modeller 9.10.
Protein structure modeling
Beta-1, 3-glucanase protein of P. colubrinum model was built based on template 2B9L using Modeller 9.10. The template 2BL9 had the highest percent (70 %) similarities to the sequence of endo beta-1,3-glucanase of H. brasiliensis among other proteins.
A modeled structure of glucanase inhibitor protein from P. capsici was built based on template 2B9L. The template 2BL9 had the highest percent (40 %) similarities to the sequence of GIP among other proteins.
PSIPRED Protein Sequence Analysis Workbench (http://bioinf.cs.ucl.ac.uk/psipred/) was used to detect or predict the secondary structures of proteins. The active sites were predicted for both proteins using LIGSITE csc http://projects.biotec.tu-dresden.de/pocket/. Structure models were validated using PROCHECK (Laskowski et al. 1993), psi and phi torsion angles were checked using the Ramchandran plot (Ramachandran et al. 1963) which provides a comprehensive check on the stereochemistry of a protein structures.
Results
Expression analysis of the genes
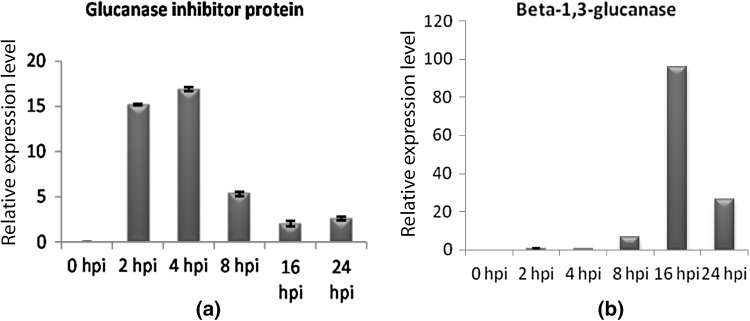
All qPCR primer pairs designed for gene expression produced specific products of expected size as revealed by the conventional PCR assay. P. capsici GIP gene was found to be expressed in both mycelia used for challenge inoculation and in planta. Significant expression of GIP following P. capsici inoculation was observed at 2 hpi, and the expression was enhanced further to maximum level at 4 hpi and thereafter it declined from 8 hpi onwards. Thus the highest level of expression of GIP gene was observed during the initial hours of infection (Fig. 1a). The beta-1,3-glucanase gene from P. colubrinum exhibited lower expression up to 4 hpi and significant increase in expression was observed by 8 hpi (Fig. 1b) and then a sharp increase in expression was shown at 16 hpi. Gradual down regulation of the gene was exhibited in the later phases particularly at 24 and 48 hpi.
Fig. 1.
Real time PCR results show relative expression levels (Y axis) of glucanase inhibitor protein (a) and beta-1,3-glucanase (b)
Sequence data and phylogeny
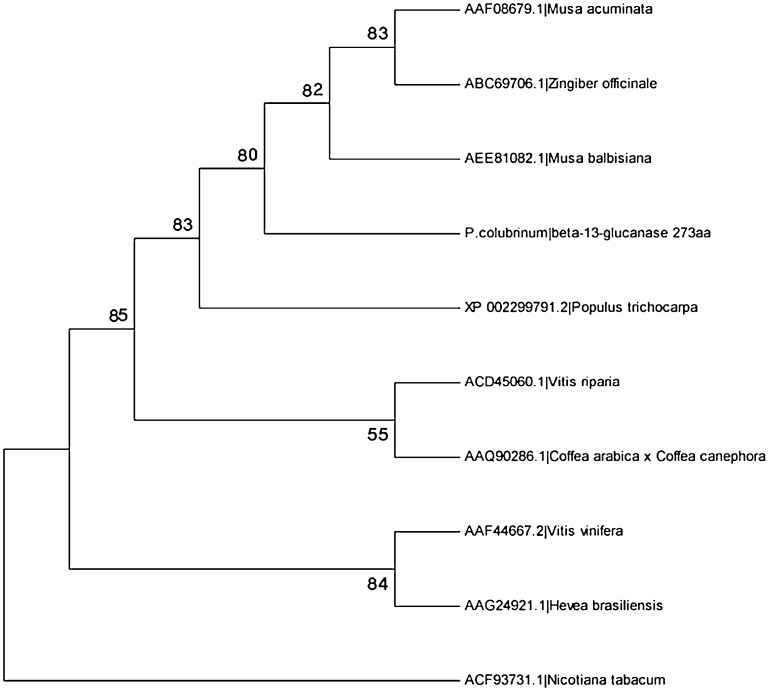
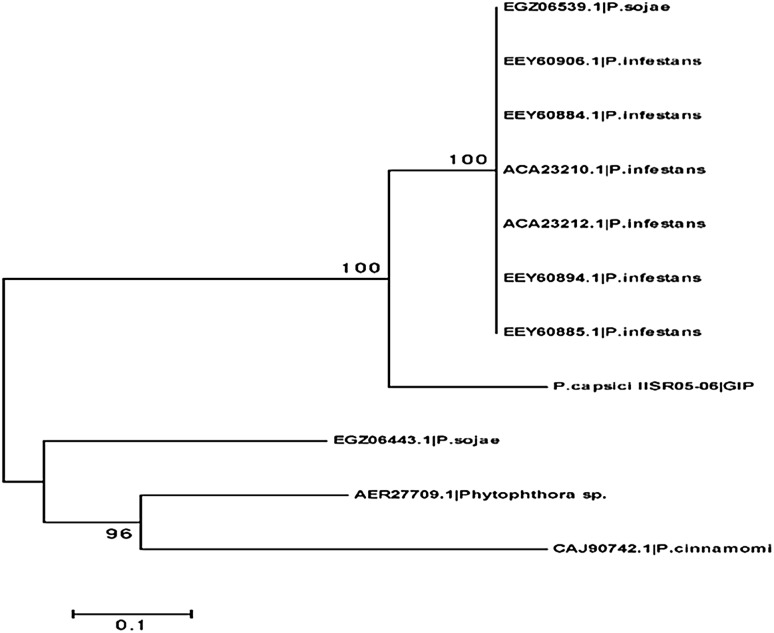
The ORF prediction gives out beta-1,3-glucanase from P. colubrinum with length, 864 and Glucanase inhibitor protein of P. capsici with length 1899. The CDS prediction gives out length of 818 for beta-1,3-glucanase from P. colubrinum and 1059 for P. capsici GIP. The translation of these CDS gives out beta-1,3-glucanase with 273 amino acid and GIP with 353 amino acid. Phylogenetic analysis was done using PSI-blast for each gene. Phylogenetic tree was drawn for each functional genes separately; beta-1,3-glucanase with other Piper species (Fig. 2) and GIP with other Phytophthora species (Fig. 3) and Phylogenetic trees shows similarity of these genes with evolutionarily related ones.
Fig. 2.
Phylogenetic tree for beta-1,3-glucanase
Fig. 3.
Phylogenetic tree for GIP gene
Protein structure prediction and evaluation
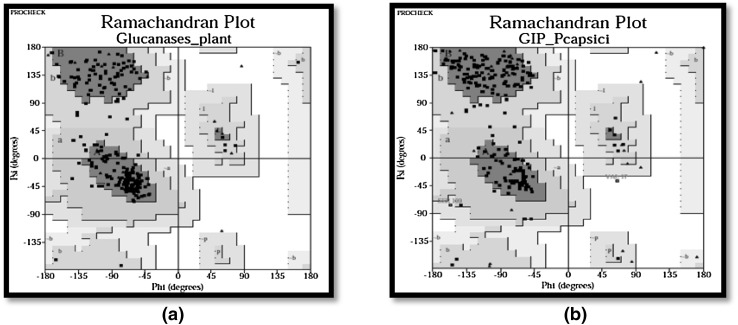
Modelled structure of beta-1,3-glucanase protein revealed that, out of 273 residues, almost 88 % were modelled with by single highest scoring template and with 100.0 % confidence. Verify_3D predicts 99.59 % residues to have an average score of >0.2 and Errat factor was 78.319. Modelled structure of glucanase inhibitor protein from P. capsici revealed that out of 353 residues, 96 % was modeled utilizing the highest scoring template. PROCHECK analysis was done for testing the modeled structures internal consistency and reliability. Ramachandran plot analysis of pcEGase indicated that 83.7 % residues belong to most favored region and 16.3 % residues in additional allowed regions. Altogether 0.0 % residues were found in both generously allowed regions and disallowed regions. Analysis by Ramachandran plot in GIP showed that, 84.3 % residues were in the most favored region, 14.7 % residues belong to additional allowed regions, 0.5 % of residues were found to be in generously allowed regions and disallowed regions had about 0.5 % residues (Fig. 4a, b).
Fig. 4.
a Ramachandran plot of beta-1,3-glucanase, b Ramachandran plot of GIP gene
Docking studies of beta-1,3-glucanase with glucanase inhibitor protein
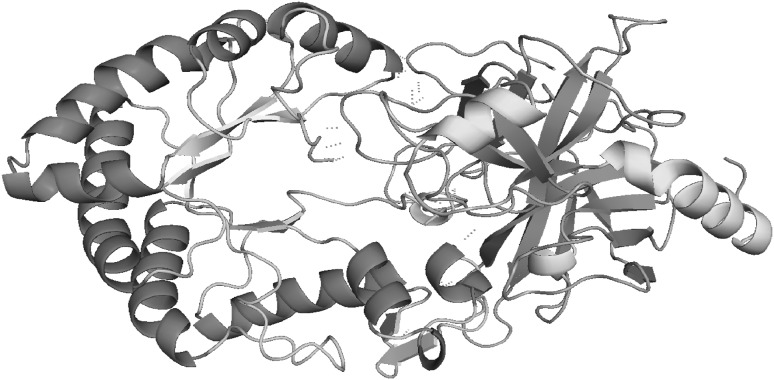
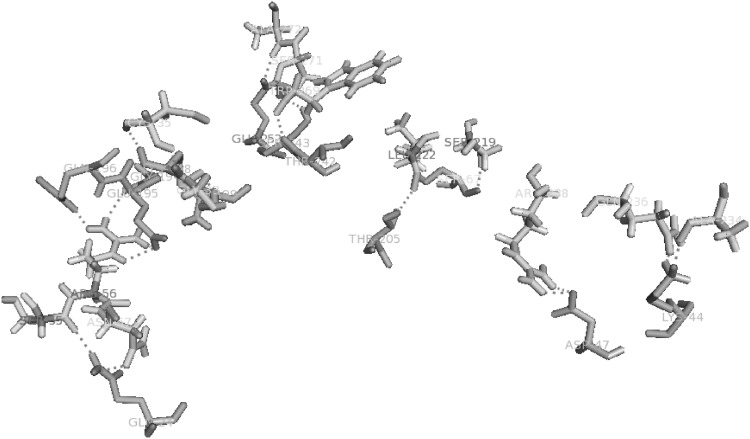
Docking of Glucanase Inhibitor Protein to sites on Endo-β-1,3-Glucanases was done using Cluspro 2.0. This particular interaction between two proteins was analyzed using Swiss PDB viewer and PyMOL v1.7.2, for having better information about what are all the key residues that are involved in the binding process. The docked complex comprising pcEGase and GIP, revealed 21 polar hydrogen bond interactions. The interaction and binding of the inhibitor to target enzyme was between following residues, as given in (Table 2). The modeling studies indicate that, GIP inhibition is mediated by an obstruction in glucanase active site (Figs. 5, 6).
Table 2.
List of interacting residues of GIP and pcEGase and the number of hydrogen bonds associated with each residue
| Serial no. | Interacting residue of GIP | Interacting residue of EGase | No. of H-bond |
|---|---|---|---|
| 1 | ASP47 | ARG238 | 2 |
| 2 | LYS44 | GLN236 | 1 |
| 3 | LYS44 | ALA234 | 1 |
| 4 | SER67 | SER219 | 1 |
| 5 | THR205 | LEU222 | 1 |
| 6 | GLU252 | ALA272 | 1 |
| 7 | GLU252 | SER271 | 1 |
| 8 | THR242 | THR269 | 1 |
| 9 | SER243 | THR269 | 1 |
| 10 | SER199 | GLY33 | 2 |
| 11 | LYS198 | CYS35 | 2 |
| 12 | GLY194 | ARG36 | 2 |
| 13 | GLU195 | ARG36 | 2 |
| 14 | ILU196 | ARG36 | 1 |
| 15 | GLN24 | ASN57 | 1 |
| 16 | GLN24 | SER55 | 1 |
Fig. 5.
Model of GIP-EGase complex interaction. Polar covalent hydrogen bond interactions are shown in blue dashed lines. GIP shown as Helix Sheet Loop and EGase shown in Helix Sheet Loop (color figure online)
Fig. 6.
Polar covalent Interaction of GIP with EGase active site residues. The interactions are shown in blue dashed lines (color figure online)
Discussion
Plants have developed an immune system which is switched on by certain microbe-associated molecules or pathogen associated molecules (PAMP/MAMP) and is therefore called PAMP-triggered immunity (Thomma et al. 2011). In PAMP-triggered immunity (PTI), hydrolases like endo glucanase and hydrolase inhibitors like glucanase inhibitor proteins are important players involved in resistance response of the plant. After pathogen recognition, plants secrete hydrolases, such as β-1,3-glucanase to degrade components of the pathogen cell wall, which is one of the PAMP-triggered defense responses. The degraded cell wall fragments are used as PAMPs to further boost PTI (Shetty et al. 2009). The recognition of a pathogen boosts levels of hydrolases in plants and the abundance in levels of these proteins can be associated with disease resistance. The quantitative or RNA transcript expression levels of glucanase inhibitor from P. capsici was studied in planta through real time PCR and the expression levels of beta-1,3-glucanase induced in P. colubrinum was studied in parallel. This study was done to understand the interaction mechanism of GIP with pcEGases in the resistant host plant and also to know about their role in pathogenesis.
In the current study, expression of P. capsici GIP was found at an increased level during early hours of infection, which can be attributed to initial success in counteracting the β-1,3-glucanase gene from P. colubrinum. The glucanase inhibitor GIP1 from P. sojae was found to inhibit endoglucanase from soybean (Misas-Villamil and van der Hoorn 2008). The temporal expression patterns of PiGIP genes in P. infestans after the pathogen inoculation were observed throughout, starting from the inoculation to different stages of the pathogen, using semi quantitative reverse transcription-polymerase chain reaction (RT-PCR) and PiGIP transcripts were found to be up-regulated in planta (Torto et al. 2002). Ham et al. (1997) found out that P. sojae GIPs inhibits soybean endoglucanase activity. Temporal expression studies on GIP family from P. infestans revealed that the genes exhibited distinct patterns of expression during an infection process (Damasceno et al. 2008). Immunoblot analysis showed the presence of PiGIPs and EGases in the tomato apoplast during infection (Damasceno et al. 2008). Studies on different black pepper cultivars revealed higher expression of β-1,3-glucanase in tolerant varieties of black pepper (Stephen et al. 2001; Nazeem et al. 2008; Vandana et al. 2014). A variety of proteins that is indeed associated with attack, defense, and counter defense are produced during Phytophthora–plant interaction (Kamoun 2006).
The maximum expression of beta-1,3-glucanase gene observed during 16 hpi indicate the preparation undertaken by the plant to counteract on the challenge caused by the pathogen through the increased expression of glucanase inhibitor gene (GIPs) during initial stages of infection. Nazeem et al. (2008) detected presence of defense related proteins including beta-1,3-glucanase in black pepper as well as in P. colubrinum in response to P. capsici infection. Being an incompatible interaction, the suppression of defense-related endo-glucanase by GIP of P. capsici was presumably not successful at later stages (after 16hpi) and hypersensitive reaction was observed in the inoculation sites by 24 hpi. Two isozymes, EGaseA and EGaseB where found in soybean which differed in their response to (GIPs) produced by P. sojae (Bishop et al. 2005). Based on above studies we may expect the possibility of the presence of more isoforms of beta-1,3-glucanase in P. colubrinum also, which have to be studied in detail in the future. The expression of both EGases and GIPs in tomato apoplast points towards a scenario of these proteins being involved in a molecular arms race in plants in general (Dawkins and Krebs 1979).
Thus the binding of GIP to pcEGase shows resemblance to a trypsin–like manner binding, which occurs by recognition of an arginine residue on the substrate (Damasceno et al. 2008), as revealed by docking studies which was based on template 2BL9. The template 2BL9 had the highest percent (70 %) similarities to the sequence of endo beta-1,3-glucanase of H. brasiliensis and it also has the highest percent (40 %) similarities to the sequence of GIP among other proteins. That was the important reason, the particular template was chosen for docking studies.
The positively selected sites in both GIPs and pcEGases are evolving rapidly and are found to be in close proximity to each other. This indeed points out to an arms race which is supposed to be taking place between the pathogen and the host (Damasceno et al. 2008). Positively selected residue on EGase protein modifies the interaction with GIP and thereby influences the glycan hydrolytic activity (Bishop et al. 2005). This might be the reason for the successful counteraction shown by pcEGase upon inhibition by GIP and thereby its expression increases subsequently at an increased level resulting in a resistance response to the pathogen.
Conclusion
The results showed the important role played by β-1,3-glucanase in P. colubrinum with respect to the response towards P. capsici and the gene could be directly linked to the high level of resistance besides other major genes involved in the interaction. Insilico studies of pcEGase and GIP, especially docking experiments carried out revealed that GIP caused inhibition at the active site of pcEGase. But studies have proved that the positively selected EGases residues are capable of overcoming the inhibition caused by GIP and these proteins are able to express successfully in order to challenge the pathogen. The same situation arises here also where EGase expression is found to be very high during the interaction with the pathogen which might be one of the reasons for the resistance response shown by P. colubrinum. Studies on more genes in P. capsici and counteracting genes in P. colubrinum will help to understand the incompatible Piper–Phytophthora interactions more thoroughly.
References
- Bishop JG, Ripoll DR, Bashir S, Damasceno CMB, Seeds JD, Rose JKC (2005) Selection on glycine β-1,3-endoglucanase genes differentially inhibited by a Phytophthora glucanase inhibitor protein. Genetics 169:1009–1019 [DOI] [PMC free article] [PubMed]
- Damasceno MB, John GB, Daniel R, Joe W, Kamoun S, Rose KC (2008) Structure of the glucanase inhibitor protein (GIP) family from Phytophthora species suggests coevolution with plant endo-β-1,3-Glucanases. Mol Plant Microbe Interact 21(6):820–830 [DOI] [PubMed]
- Dawkins R, Krebs JR. Arms races between and within species. Proc R Soc Lond Biol Sci. 1979;205:489–511. doi: 10.1098/rspb.1979.0081. [DOI] [PubMed] [Google Scholar]
- Ham KS, Wu SC, Darvill AG, Albersheim P (1997) Fungal pathogens secrete an inhibitor protein that distinguishes isoforms of plant pathogenesis-related endo-beta-1,3-glucanases. Plant J 11:169–179
- Kamoun S (2006) A catalogue of the effector secretome of plant pathogenic oomycetes. Annu Rev Phytopathol 44:41–60 [DOI] [PubMed]
- Misas-Villamil JC, van der Hoorn RA (2008) Enzyme-inhibitor interactions at the plant–pathogen interface. Curr Opin Plant Biol 11:380–388 [DOI] [PubMed]
- Nazeem PA, Achuthan CR, Babu TD, Parab GV, Girija D, Keshavachandran R, Samiyappan R (2008) Expression of pathogenesis related proteins in black pepper (Piper nigrum L.) in relation to Phytophthora foot rot disease. J Trop Agric 46(1–2):45–51
- Shetty N, Jensen JD, Kundsen A, Finnie C, Geshi N, Blennow A, Collinge DB, Jorgensen HJL (2009) Effects of β-1,3-glucan from Septoria tritici on structural defence responses in wheat. J Exp Bot 60:4287–4300 [DOI] [PubMed]
- Stephen RJ, Anandaraj M, Sarma YR (2001) Induction of PR proteins and defense related enzymes in black pepper due to inoculation with Phytophthora capsici. Ind Phytopathol 54:23–28
- Thomma BPHJ, Nürnberger T, Joosten MHAJ (2011) Of PAMPs and effectors: the blurred PTI-ETI dichotomy. Plant Cell 23:4–15 [DOI] [PMC free article] [PubMed]
- Torto TA, Rauser L, Kamoun S (2002) The pigp1 gene of the oomycete Phytophthora infestans encodes a fungal-like endopolygalacturonase. Curr Genet 40:385–390 [DOI] [PubMed]
- Vandana VV, Suseela BR, Shamina A (2014) Biochemical defense responses of black pepper pepper (Piper nigrum L.) lines to Phytophthora capsici. Physiol Mol Plant Pathol 88:18–27
- York WS, Qin Q, Rose JK (2004) Proteinaceous inhibitors of endo beta-glucanases. Biochim Biophys Acta 1696:223–233 [DOI] [PubMed]