Abstract
Sesame is one of the oldest oilseed crops grown mainly in Africa and Asia. Although genetic and genomic studies on sesame have started late, the past 5 years have witnessed extensive progresses in these areas on this crop. Important genomic sequence resources such as functional markers, genes and QTLs linked to agronomically important traits, have been generated through linkage mapping and association analysis to assist sesame improvement programs. However, most of these data are scattered in different maps making them hard to be exploited efficiently in breeding programs. In this study, we report a comprehensive physical map gathering 151 published genomic sequence resources which highlighted some hotspot functional regions in the sesame genome. Moreover, 83,135 non-redundant SSRs have been supplied along with their physical position and motif composition. This will assist future research in fine mapping or pinpointing more functional genes based on the already published QTLs and functional markers. This physical map represents a good landmark for further non-overlapping genetic and genomic studies working towards sesame improvement.
Electronic supplementary material
The online version of this article (doi:10.1007/s12298-016-0385-8) contains supplementary material, which is available to authorized users.
Keywords: Sesamum indicum, QTL, Functional genes, SSR, Physical map
Sesame (Sesamum indicum L., 2n = 26) is one of the oldest cultivated plants mainly grown in tropical and subtropical regions of Asia, Africa and South America (Bedigian 2003). Its seeds are a source of excellent vegetable oil and are a reservoir of nutritional components with numerous beneficial effects along with health promotion in humans (Pathak et al. 2014). Sesame is relatively drought resistant (Langham 2007); its cultivation is much cheaper than other crops and requires little care. Hence, it represents a significant crop for small-holders in marginal areas. Although it is an ancient crop with high economic value, sesame is listed as a neglected and underutilized crop species by the International Plant Genetic Resources Institute (IPGRI) due to its limited cultivation and expansion in the world. Consequently, its improvement by modern technology such as genetic and genomic tools has been lagging behind other model plants (Dossa et al. 2016a).
Genetic research in sesame was initiated lately and the first linkage map was set in 2009 using 220 EST-SSR (Expressed Sequence Tags- Simple Sequence Repeats), AFLP (Amplified Fragment Length Polymorphism) and RSAMPL (Random Selective Amplification of Microsatellite Polymorphic Loci) markers (Wei et al. 2009). However, the past 5 years have witnessed extensive progresses in genetic and genomic studies in this crop. In 2013, 02 dense genetic maps have been published using different types of markers including SSR, AFLP, SLAFseq (Specific-Locus Amplified Fragment sequencing) and RSAMPL (Zhang et al. 2013a, b. These studies enabled the identification of QTLs linked to the seed coat color which is an important agronomic trait in sesame, as it is associated with seed biochemical properties, antioxidant content and activity and even, disease resistance of sesame (Zhang et al. 2013a, b). Later on, using the RADseq technology (Restriction Site Associated DNA sequencing), Wu et al. (2014) constructed a high-density linkage map with nearly 1230 markers and identified several QTLs linked to grain yield-related traits. In addition, Zhang et al. (2014) identified some QTLs linked to waterlogging tolerance that could help to enhance sesame which is known to be naturally susceptible to waterlogging. Further significant works in QTLs identification have also been published based on different type of markers, populations and traits (Zhao et al. 2013; Rao et al. 2014; Liu et al. 2015; Wang et al. 2016). Meanwhile, association mapping studies have also been conducted in sesame and led to the identification of several functional markers linked to important traits mainly the oil content and quality (Wei et al. 2013; Li et al. 2014). During the same period, the first genome sequence of sesame was published and has since brought sesame research into the functional genomics era (Wang et al. 2014a, b). Thus, the functional characterization of the 27,148 genes of the sesame genome has become a hot topic. A comprehensive genome-wide association study (GWAS) recently published by Wei et al. (2015) shed more light on the functions of several genes in the sesame genome.
The aforementioned works generated important amounts of genomic sequence information that could be exploited to speed-up sesame improvement strategies in the future. However, most of these genetic data are scattered throughout different linkage maps built on various populations, making them very hard to be exploited for further experiments. With limited research teams working on sesame, it is crucial to avoid duplication of efforts and redundancy in research but better opting for complementary studies. Moreover, until now only few SSR markers are available for sesame research, failing to represent adequately the sesame genome although their importance for gene mapping as well as marker-assisted selection (MAS) is now well established.
Here we present a comprehensive physical map gathering some public available functional markers, genes, QTLs and SSRs identified from whole genome scan, which will provide better options for further non-overlapping genetic and genomic studies towards sesame improvement.
To build this physical map, a literature search of published articles on QTLs, functional markers and genes related to different traits in sesame, has been conducted using NCBI and Google scholar. However it is worth mentioning that all scientific papers published in sesame research could not be found in these two databases. Articles related to these contents have been downloaded and screened for the consistency of their results, the quality of the journal and mainly the primers used (This is discussed in results section). In total, we retained 10 articles including the works of Wei et al. (2013), Zhao et al. (2013), Li et al. (2014), Wu et al. (2014), Zhang et al. (2013a, b), Liu et al. (2015), Wei et al. (2015), Wang et al. (2014a, b, 2016) and took advantage of their data in this study (Table 1).
Table 1.
Summary of the published data used in this study
| Authors | Gene/QTL/marker | Numbers |
|---|---|---|
| Wei et al. (2013) | Marker | 10 |
| Zhao et al. (2013) | Marker | 1 |
| Li et al. (2014) | Marker | 18 |
| Wang et al. (2014a, b) | Gene | 10 |
| Wu et al. (2014) | QTL | 15 |
| Zhang et al. (2014a) | Marker | 2 |
| Zhang et al. (2014b) | QTL | 3 |
| Liu et al. (2015) | Marker | 1 |
| Wei et al. (2015) | Gene | 46 |
| Wang et al. (2016) | QTL | 45 |
| Total | 151 |
A local bank with the retrieved sequences of the functional markers, genes and markers flanking the QTLs was generated in order to find their physical positions in the sesame genome. The online database on sesame: Sinbase (http://ocri-genomics.org/Sinbase/) (Wang et al. 2014a, b), was employed and the sequences were BLASTn searched against the sesame genome version 1 (Wang et al. 2014a, b). A cut off E-value ≥ 1e-40 were set for gene sequences where as an E-value = 10 was set for sequences of the functional markers and markers flanking the QTLs. Particularly for the sequences of the functional markers and markers flanking QTLs, to improve the sequence similarity and alignment with the genome sequence, we concatenated the two primers sequences (reverse and forward) into one sequence separated by ~20 nucleotides prior submitting to BLASTn search, with a default value of 10 words and the low complexity filter turned off. The orientation of the two primer sequences is not significant as BLAST is able to search both strands for matches.
The linkage group (LG) and the position (bp) in the genome for each sequence were recorded and confirmed using the BLASTn algorithm (Altschul et al. 1990) in NCBI. All sequences that hit on different sesame LGs in both databases were filtered out.
For SSR identification, the whole genome was scanned for SSR 1oci with the software SciRoKo 3.4 (Kofler et al. 2007). The parameters were set for detection of mono, di, tri, tetra, penta, and hexa -nucleotide (nt) motifs with a minimum of 15, 7, 5, 3, 3 and 3 repeats, respectively (under the mismatched and fixed penalty search mode). Based on these data, the Mapchart version 2.3 (Voorrips 2002) was used to successfully map the QTLs, functional genes and markers onto the 16 LGs of the sesame genome. It is noteworthy that a new version of the sesame genome with 13 chromosomes has been updated recently, but it is not yet available either in Sinbase or NCBI (Wang et al. 2016).
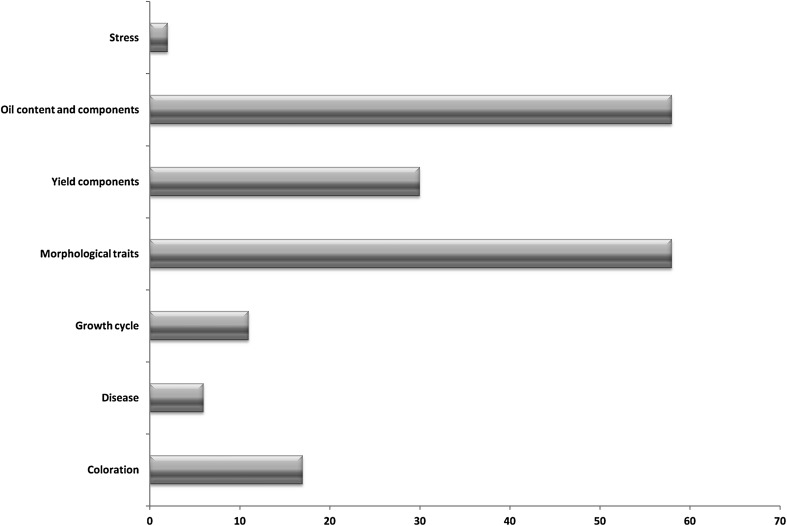
Sesame, with its diploid character and small genome size (~370 Mb), constitutes the model plant species for genetic studies among the major oilseed crops. In total, 151 genomic sequences including 56 functional genes, 63 QTLs and 32 functional markers were retrieved and used in this study (Table 1). Functional markers and markers flanking QTLs which were retained in this study were mostly SSR and SRAP, as these markers are more reproducible and easy to locate in the genome. For instance, Rao et al. (2014) reported several interesting QTLs, but their works were based on RAPD universal primers which are hard to be physically localized in the genome. Overall, the genomic sequences mapped could be classified into 7 functional categories (Fig. 1). Results showed that till date, functional studies in sesame mainly focused on three main traits including oil yield and quality, morphological and yield component traits. Sesame is mainly cultivated for its seeds which are highly valued because of its excellent oil quality and contains of the highest oil contents (~55 %) among major oil crops (Wei et al. 2015). The seed is also rich in protein, vitamins, including niacin, minerals and lignans, such as sesamolin and sesamin (Nakimi 1995; Moazzami and Kamal-Eldin 2006). It is thus comprehensible that many studies attempted to decipher the genetic basis of the oil yield and quality in sesame as well as high seed yield trait. On the other hand, very few studies have been undertaken on stress tolerance and disease resistance in sesame which is known as a very hardy plant able to sustain harsh environments (Langham 2007). However, many diseases have been reported in sesame as well as susceptibility to some environmental conditions such as waterlogging, severe drought, heat and salinity (Langham et al. 2008; Langham 2007; Bahrami and Razmjoo 2012; Wei et al. 2013; Wang et al. 2016, Dossa et al. 2016b, c). Thus, future researches need to address these issues as they are seriously affecting sesame productivity.
Fig. 1.
Proportion of the genomic sequences regrouped into seven functional categories
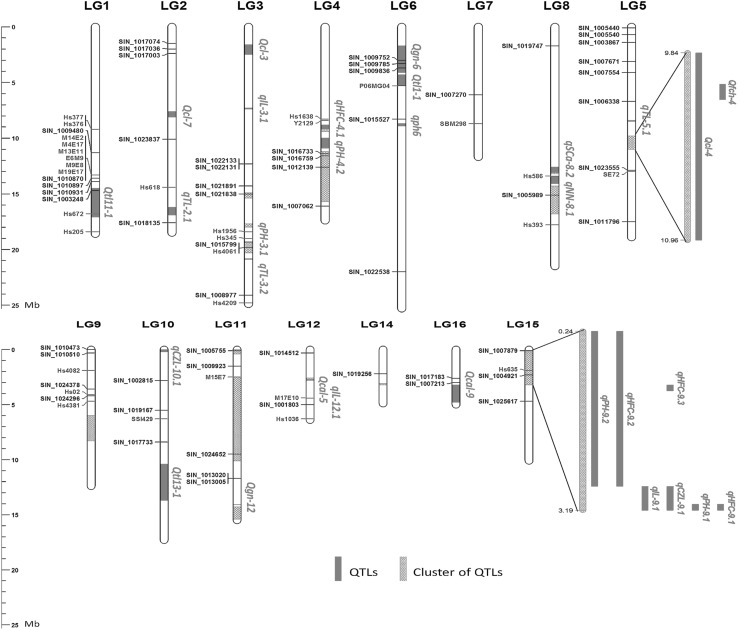
The genomic sequences retrieved have been successfully mapped onto 15 of the 16 LGs (Except LG13) in the sesame genome (Fig. 2). The physical position details of each sequence could be found in the Online Resource Table S1. Interestingly, results highlighted some hotspot genomic regions involved in the control of various traits. For instance, we found 15 clusters of different QTLs controlling various traits. Furthermore, many clusters of functional genes, markers and QTLs could also be observed. These regions need to be more excavated using bioinformatics, map-based cloning and transgenic studies to pinpoint the functional genes. Also, we found that some genomic sequences reported in different studies to be correlated to the same traits and clustered together in this physical map or are closely located. Good illustration for this are the genes SIN_1010870, SIN_1010931, SIN_1003248 (Wei et al. 2015) and the markers M14E2, M4E17, M13E11, M9E8, M19E17, E6M9 (Wei et al. 2013), Hs377, Hs376 (Li et al. 2014) which were described as linked to oil yield and components were all found closely located on the LG1. Therefore, this map provides an opportunity to cross-validate findings of different studies and perform comparative genomic analysis. Results compiled in this study represent a good reference for future studies either for better deciphering the hotspot genomic regions and find out the best candidate genes or to embrace new important traits and avoiding duplication of efforts in sesame improvement perspectives.
Fig. 2.
Distribution of important functional genes, markers and QTLs on the linkage groups LG1–LG16 of the sesame genome. Bars represent the LGs and locus names in green and blue indicate functional markers and genes respectively. Red hatched regions of the LGs represent clusters of QTLs while segments colored in red represent single QTLs (color figure online)
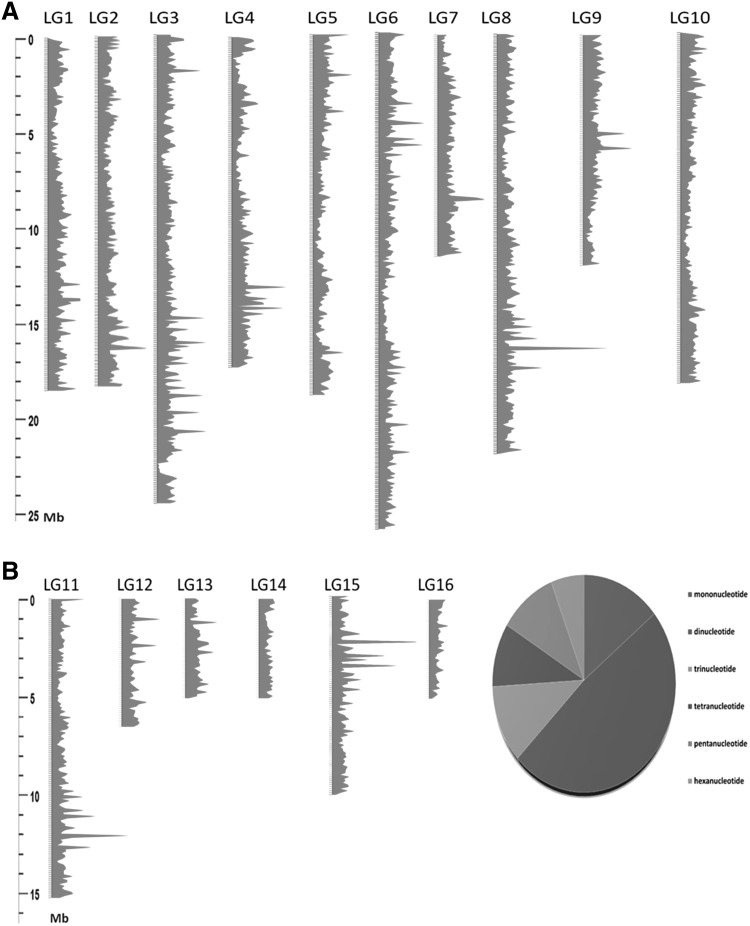
Together with these genomic resources, we successfully identified 83,135 non-redundant SSRs in the whole genome of sesame including 74,672 (89.33 % of all SSRs) which were mapped onto the 16 LGs (Table 2; Fig. 3a). Dinucleotide repeats were the most abundant SSR type identified (48.25 % of all SSRs), followed by mononucleotide SSRs (14.30 %) as shown in Fig. 3b. The LG3 displayed the highest number of SSRs (11.4 % of all SSRs) while and the LG16 harbored the lowest number (1.76 %) and the average density was 302 SSRs/Mb. The number of SSRs identified in our study and the SSR density were higher than previous reports of Wei et al. (2014) who only focused on 19 % of the scaffolds present in the sesame genome. The released microsatellite resources and their genome locations are expected to assist germplasm management, genetic studies, gene fine mapping and molecular breeding applications leading to the increase of sesame productivity.
Table 2.
Abundance and distribution of SSR types in the LGs of the sesame genome
| Number of SSRs | Frequency % | |
|---|---|---|
| SSR motifs | ||
| AAAAG/CTTTT | 972 | 1.2 |
| AAG/CTT | 1056 | 1.3 |
| AAATT/AATTT | 1059 | 1.3 |
| AATT/AATT | 1070 | 1.3 |
| AAAG/CTTT | 1117 | 1.3 |
| ATC/GCT | 1141 | 1.4 |
| C/G | 2184 | 2.6 |
| AAAAT/ATTTT | 3005 | 3.6 |
| AAAT/ATTT | 3754 | 4.5 |
| AAT/ATT | 5174 | 6.2 |
| AG/CT | 8932 | 10.7 |
| A/T | 9707 | 11.7 |
| AC/GT | 9937 | 12.0 |
| AT | 21,228 | 25.5 |
| LGs | ||
| LG15 | 3948 | 5.3 |
| LG9 | 3957 | 5.3 |
| LG10 | 5000 | 6.7 |
| LG5 | 5255 | 7.0 |
| LG11 | 5285 | 7.1 |
| LG4 | 5476 | 7.3 |
| LG1 | 5858 | 7.8 |
| LG2 | 6070 | 8.1 |
| LG8 | 7085 | 9.5 |
| LG6 | 7651 | 10.2 |
| LG3 | 8527 | 11.4 |
Only SSR motifs with a frequency >1 % and LGs with SSRs >5 % are listed
Fig. 3.
a Genome-wide densities of microsatellites in the assembled LGs of sesame. On the figure, the red curves represent the frequencies of microsatellites in a window of 100 Kb. b Percentage of different repeat motifs in the sesame genome (color figure online)
The physical map constructed in the present study provides not only the genomic distribution of 151 genomic sequences including QTLs, functional genes and markers linked to some important traits in sesame but also sheds light on some functional genomic regions which need future thorough researches to effectively pinpoint the functional genes. In addition, complete SSR resources present in the sesame genome have been supplied along with their physical positions and characteristics. The results of this study will undoubtedly assist and guide breeders and geneticists to accelerate sesame improvement studies. Finally, this map needs to be updated regularly as research progresses and transformed into an online interactive database for more convenience in its exploitation.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
The author wishes to thank all scientists and research teams devoted to sesame crop research. Author is sincerely grateful to Uche Arinze and Allen James Bohr for their help in language editing of the manuscript.
Compliance with ethical standards
Conflict of interest
The author declares there is no conflict of interest.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bahrami H, Razmjoo J. Effect of salinity stress (NaCl) on germination and early seedling growth of ten sesame cultivars (Sesamum indicum L.) Int J Agric Sci. 2012;2:529–537. [Google Scholar]
- Bedigian D. Evolution of sesame revisited: domestication, diversity and prospects. Genet Resour Crop Evol. 2003;50:779–787. doi: 10.1023/A:1025029903549. [DOI] [Google Scholar]
- Dossa K, Wei X, Zhang Y, Fonceka D, Wenjuan Y, Diouf D, Boshou L, Cissé N, Zhang X. Analysis of genetic diversity and population structure of sesame accessions from Africa and Asia as major centers of its cultivation. Genes. 2016;7:14. doi: 10.3390/genes7040014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dossa K, Wei X, Li D, Zhang Y, Wang L, Fonceka D, Yu J, Diouf D, Boshou L, Cissé N, Zhang X. Insight into the AP2/ERF transcription factor superfamily in sesame (Sesamum indicum) and expression profiling of the DREB subfamily under drought stress. BMC Plant Biol. 2016;16:171. doi: 10.1186/s12870-016-0859-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dossa K, Niang M, Assogbadjo AE, Cissé N, Diouf D. Whole genome homology-based identification of candidate genes for drought resistance in (Sesamum indicum L.) Afr J Biotechnol. 2016;15:1464–1475. doi: 10.5897/AJB2016.15420. [DOI] [Google Scholar]
- Kofler R, Schlotterer C, Lelley T. SciRoKo: a new tool for whole genome microsatellite search and investigation. Bioinformatics. 2007;23:1683–1685. doi: 10.1093/bioinformatics/btm157. [DOI] [PubMed] [Google Scholar]
- Langham DR. Phenology of sesame. In: Janick J, Whipkey A, editors. Issues in new crops and new uses. Alexandria: ASHS Press; 2007. pp. 144–182. [Google Scholar]
- Langham DR, Riney J, Smith G, Wiemers T (2008) Sesame harvest guide. www.sesaco.net
- Li C, Miao H, Wei L, Zhang T, Han X, Zhang H. Association mapping of seed oil and protein content in Sesamum indicum L. PLoS One. 2014;9:e105757. doi: 10.1371/journal.pone.0105757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Zhou X, Wu K, Yang M, Zhao Y. Inheritance and molecular mapping of a novel dominant genic male-sterile gene in Sesamum indicum L. Mol Breed. 2015;35:9. doi: 10.1007/s11032-015-0189-5. [DOI] [Google Scholar]
- Moazzami AA, Kamal-Eldin A. Sesame seed is a rich source of dietary lignans. J Am Oil Chem Soc. 2006;8:719–723. doi: 10.1007/s11746-006-5029-7. [DOI] [Google Scholar]
- Nakimi M. The chemistry and physiological functions of sesame. Food Rev Int. 1995;11:281–329. doi: 10.1080/87559129509541043. [DOI] [Google Scholar]
- Pathak N, Rai AK, Kumari R, Bhat KV. Value addition in sesame: a perspective on bioactive components for enhancing utility and profitability. Pharmacogn Rev. 2014;8:147–155. doi: 10.4103/0973-7847.134249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao VRP, Prasuna K, Anuradha G, Srividya A, Vemireddy LR, Shankar VG, Sridhar S, Jayaprada M, Reddy RK, Reddy ENP, Siddiq EA. Molecular mapping of important agro-botanic traits in sesame. Electron J Plant Breed. 2014;5:475–488. [Google Scholar]
- Voorrips RE. MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered. 2002;93:77–78. doi: 10.1093/jhered/93.1.77. [DOI] [PubMed] [Google Scholar]
- Wang L, Yu J, Li D, Zhang X. Sinbase: an integrated database to study genomics, genetics and comparative genomics in Sesamum indicum. Plant Cell Physiol. 2014;0:1–7. doi: 10.1093/pcp/pcu175. [DOI] [PubMed] [Google Scholar]
- Wang L, Yu S, Tong C, Zhao Y, Liu Y, Song C, Zhang Y, Zhang X, Wang Y, Hua W, Li D, Li D, Li F, Yu J, Xu C, Han X, Huang S, Tai S, Wang J, Xu X, Li Y, Liu S, Varshney R, Wang J, Zhang X. Genome sequencing of the high oil crop sesame provides insight into oil biosynthesis. Genome Biol. 2014;15:R39. doi: 10.1186/gb-2014-15-2-r39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Xia Q, Zhang Y, Zhu X, Zhu X, Li D, Ni X, Gao Y, Xiang H, Wei X, Yu J, Quan Z, Zhang X. Updated sesame genome assembly and fine mapping of plant height and seed coat color QTLs using a new high-density genetic map. BMC Genom. 2016;17:31. doi: 10.1186/s12864-015-2316-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei LB, Zhang HY, Zheng YZ, Miao HM, Zhang TZ, Guo WZ. A genetic linkage map construction for sesame (Sesamum indicum L.) Genes Genom. 2009;31:199–208. doi: 10.1007/BF03191152. [DOI] [Google Scholar]
- Wei W, Zhang Y, Lü H, Li D, Wang L, Zhang X. Association analysis for quality traits in a diverse panel of Chinese sesame (Sesamum indicum L.) germplasm. J Integr Plant Biol. 2013;55:745–758. doi: 10.1111/jipb.12049. [DOI] [PubMed] [Google Scholar]
- Wei X, Wang L, Zhang Y, Qi X, Wang X, Ding X, Zhang J, Zhang X. Development of simple sequence repeat (SSR) markers of sesame (Sesamum indicum) from a genome survey. Molecules. 2014;19:5150–5162. doi: 10.3390/molecules19045150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Liu K, Zhang Y, Feng Q, Wang L, Zhao Y, Li D, Zhao Q, Zhu X, Zhu X, Li W, Fan D, Gao Y, Lu Y, Zhang X, Tang X, Zhou C, Zhu C, Liu L, Zhong R, Tian Q, Wen Z, Weng Q, Han B, Huang X, Zhang X. Genetic discovery for oil production and quality in sesame. Nat Commun. 2015;6:8609. doi: 10.1038/ncomms9609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K, Liu H, Yang M, Tao Y, Ma H, Wu W, Zuo Y, Zhao Y. High-density genetic map construction and QTLs analysis of grain yield-related traits in sesame (Sesamum indicum L.) based on RAD-Seq technology. BMC Plant Biol. 2014;14:274. doi: 10.1186/s12870-014-0274-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Miao H, Wei L, Li C, Zhao R, Wang C. Genetic analysis and QTL mapping of seed coat color in sesame (Sesamum indicum L.) PLoS One. 2013;8:e63898. doi: 10.1371/journal.pone.0063898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wang L, Xin H, Li D, Ma C, Ding X, Hong W, Zhang X. Construction of a high-density genetic map for sesame based on large scale marker development by specific length amplified fragment (SLAF) sequencing. BMC Plant Biol. 2013;13:141. doi: 10.1186/1471-2229-13-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wang L, Li D, Gao Y, Lü H, Zhang X. Mapping of sesame waterlogging tolerance QTL and identification of excellent waterlogging tolerant germplasm. Sci Agric Sin. 2014;47:422–430. [Google Scholar]
- Zhao Y, Yang M, Wu K, Liu H, Wu J, Liu K. Characterization and genetic mapping of a novel recessive genic male sterile gene in sesame (Sesamum indicum L.) Mol Breed. 2013;32:901–908. doi: 10.1007/s11032-013-9919-8. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.