Abstract
The comparison between microbial sequencing data is critical to understand the dynamics of microbial communities. The alignment-based tools analyzing metagenomic datasets require reference sequences and read alignments. The available alignment-free dissimilarity approaches model the background sequences with Fixed Order Markov Chain (FOMC) yielding promising results for the comparison of microbial communities. However, in FOMC, the number of parameters grows exponentially with the increase of the order of Markov Chain (MC). Under a fixed high order of MC, the parameters might not be accurately estimated owing to the limitation of sequencing depth. In our study, we investigate an alternative to FOMC to model background sequences with the data-driven Variable Length Markov Chain (VLMC) in metatranscriptomic data. The VLMC originally designed for long sequences was extended to apply to high-throughput sequencing reads and the strategies to estimate the corresponding parameters were developed. The flexible number of parameters in VLMC avoids estimating the vast number of parameters of high-order MC under limited sequencing depth. Different from the manual selection in FOMC, VLMC determines the MC order adaptively. Several beta diversity measures based on VLMC were applied to compare the bacterial RNA-Seq and metatranscriptomic datasets. Experiments show that VLMC outperforms FOMC to model the background sequences in transcriptomic and metatranscriptomic samples. A software pipeline is available at https://d2vlmc.codeplex.com.
Understanding the factors affecting microbe composition and the relationship between microbes and hosts depends on accurate comparison of microbial communities1. The high-throughput sequencing data of microbial communities harbor the whole DNA/RNA information for elaborate and comprehensive comparison. Generally, alignment-based sequencing comparison methods, such as the Smith-Waterman algorithm2 and BLAST3, have been extensively used to compare microbial communities based on short read data. The reads are usually mapped to known genome or pathway databases, followed by estimation of the abundance levels of genomes and/or gene families. Microbial communities are then compared based on the abundance levels. Recently, several computational tools including Kraken4, Clark5 and Kaiju6, have been developed for fast taxonomic classification of sequencing reads using hash-based k-mer indices built from reference sequences. These methods achieved comparable accuracy as that of the traditional BLAST programs, yet they are up to ~900 times4 faster than Megablast and ~10 times4 faster than MetaPhlan7. In addition, MetaPhlan7 uses only known marker genes. If communities do not share any marker genes included in MetaPhlan, the program will not be able to report the relationships among the communities. On the other hand, Kraken4, Clark5 and Kaiju6 do not have such limitations. However, the reference-based comparison approaches have several limitations: (1) Dependency on sequences of reference genomes or genes. However, a large amount of microbial genomes and gene families are unknown or incomplete, which affects the accuracy and completeness of the analysis. According to current publications, for metatranscriptomic data, there were about 19–42% unassigned reads in marine water samples8, about 10–20% unassigned reads in human small intestine microbiota9, and up-to 50% reads that cannot be assigned to reference databases in oceans with large phytoplankton10. Therefore, alignment-based methods are not applicable for microbial communities with a large amount of dark matters. (2) Current tools analyzing the microbial communities were mostly designed for metagenomics based on mark genes, such as 16S rRNA. However, for the metatranscriptomic dataset, ribosomal RNA (rRNA) transcripts are often required to be depleted in order to maximize mRNA recovery8,9. Therefore, the metagenomic tools based on 16S rRNA marker genes are not suitable to analyze metatranscriptomic data. Among the limited metatranscriptomic analytic tools, some were designed for Illumina paired-end10 or single/paired-end data11, or only used to evaluate the gene expression level12. A previous study11 compared four taxonomical classification tools based on a common metatranscriptomic data and obvious differences among the taxonomical analytic results were observed, which was the second figure in original paper11. (3) Sequence assembly is time-consuming and challenging especially for metagenome/metatranscriptome when organisms share a high volume of homologous sequences. Different assembled contigs were obtained for the same reads when using different assembly tools. Therefore, alignment-free methods provide a promising alternative for microbial community comparison, eliminating the requirements of reference sequences and assembly.
One type of alignment-free methods is based on the frequencies of k-tuples (k-words, k-mers or k-grams)13. A k-tuple is a contiguous sequence of length k. Previous studies indicate that relative k-tuple frequencies are similar across different regions of the same genome, but differ between genomes14. One of the earliest similarity measures between two sequences is D2 which measures the total number of matched k-tuples between two long sequences13. However, theoretical studies have shown that the distribution of D2 is dominated by the variance in the number of occurrences of k-tuples along individual sequences and less by the relationship between sequences15. Consequently, other similarity measures have been developed with different normalization, centralization and background models in an attempt to modify D2, including 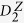 16,
16, 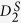 15,
15, 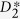 17,
17, 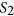 18,19 and CVTree measures. Subsequently, normalized dissimilarity measures20 based on D2,
18,19 and CVTree measures. Subsequently, normalized dissimilarity measures20 based on D2,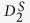 and
and 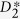 , including
, including 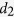 ,
, 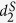 and
and 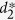 1,21 with range between 0 and 1, were developed for high-throughput sequencing data. Indeed, previous studies22 showed that k-tuple-based dissimilarity measures are effective in revealing group relationships and gradient relationships among metagenomic and metatranscriptomic samples and that
1,21 with range between 0 and 1, were developed for high-throughput sequencing data. Indeed, previous studies22 showed that k-tuple-based dissimilarity measures are effective in revealing group relationships and gradient relationships among metagenomic and metatranscriptomic samples and that 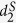 and
and 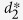 achieved the best performances in most comparisons of microbial communities.
achieved the best performances in most comparisons of microbial communities.
However, the utility of 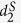 and
and 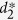 depends on a proper probability model for background genomes. To address this gap, Fixed Order Markov Chains (FOMC) were used to model the background genome sequences, as reported in previous studies22,23. There are several limitations during the applications of FOMC: (1) The order of Markov Chain (MC) needs to be set manually. However, for most microbial communities, there is no prior knowledge available for setting the MC order. (2) Furthermore, it is hard to model probabilities of different tuples using a single fixed order MC, and FOMC is not structurally rich. There are nr × (n − 1) independent parameters for an r-th order MC, where n is the number of states, that is, n = 4 for DNA or RNA sequences. When the order r equals 2 or 3, the number of parameters for the model is 48 or 192, respectively. There are no FOMCs with number of parameters between 48 and 192. (3) Thus, the number of parameters grows exponentially with the increase of order r. When sequencing depth is relatively low, the parameters, with their number growing exponentially with the increase of MC order in FOMC models, cannot be accurately estimated.
depends on a proper probability model for background genomes. To address this gap, Fixed Order Markov Chains (FOMC) were used to model the background genome sequences, as reported in previous studies22,23. There are several limitations during the applications of FOMC: (1) The order of Markov Chain (MC) needs to be set manually. However, for most microbial communities, there is no prior knowledge available for setting the MC order. (2) Furthermore, it is hard to model probabilities of different tuples using a single fixed order MC, and FOMC is not structurally rich. There are nr × (n − 1) independent parameters for an r-th order MC, where n is the number of states, that is, n = 4 for DNA or RNA sequences. When the order r equals 2 or 3, the number of parameters for the model is 48 or 192, respectively. There are no FOMCs with number of parameters between 48 and 192. (3) Thus, the number of parameters grows exponentially with the increase of order r. When sequencing depth is relatively low, the parameters, with their number growing exponentially with the increase of MC order in FOMC models, cannot be accurately estimated.
With this in mind, we introduced Variable Length Markov Chains24 (VLMC) as an alternative for FOMC to model the background genomes of microbial community in this study. VLMC adaptively determines the order of MC based on the sequence data, thus eliminating manual selection. Additionally, the number of variables in VLMC is flexible. VLMC was originally designed for modeling one long sequence and was represented as a context tree structure24,25. For high-throughput sequencing of short reads, the likelihood of underlying, or unobserved sequences cannot be calculated. As a result, the rules for pruning the tree are not clearly defined. Therefore, we first developed strategies to determine the parameters for building a context tree and then extended VLMC for high-throughput sequencing of short reads. Thus, the complete context tree is constructed from these short reads, which typically overfits the data. The number of independent parameters is Num(nodes) × 3, where the Num(nodes) is the total number of nodes in the context tree except the root-node. The tree is then pruned according to a local decision rule. Using VLMC for background modeling, 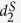 and
and 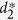 measures were then applied to compare transcriptomic or metatranscriptomic datasets. From the obtained dissimilarities among samples, the clustering trees were evaluated based on the triples distance26 between the reference and resulting trees. Experimental results show that VLMC models the position dependency in the nucleotide sequences better than FOMC, and since it is free from order selection required by FOMC, VLMC is easier to apply. Our studies also show that VLMC probability models combined with
measures were then applied to compare transcriptomic or metatranscriptomic datasets. From the obtained dissimilarities among samples, the clustering trees were evaluated based on the triples distance26 between the reference and resulting trees. Experimental results show that VLMC models the position dependency in the nucleotide sequences better than FOMC, and since it is free from order selection required by FOMC, VLMC is easier to apply. Our studies also show that VLMC probability models combined with 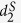 and
and 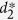 measures exhibit superior performance in clustering metatranscriptomic samples when compared to previous approaches.
measures exhibit superior performance in clustering metatranscriptomic samples when compared to previous approaches.
Results
Design of experiments
In order to explore the performance of 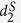 and
and 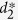 with VLMC, we designed experiments with one simulated dataset and four real datasets. The simulated metatranscriptomic dataset is composed of 90 samples belonging to 3 different groups with 5,000 genes from 5 microbes. Real dataset 1 consists of 18 and 22 RNA-Seq datasets from marine microbial eukaryotes. For 18 RNA-Seq datasets, the molecular phylogeny27 was reconstructed based on the 18S rRNA genes with maximum likelihood (ML) method. The ML phylogenetic tree was then used to evaluate the ability of VLMC as background model, combined with
with VLMC, we designed experiments with one simulated dataset and four real datasets. The simulated metatranscriptomic dataset is composed of 90 samples belonging to 3 different groups with 5,000 genes from 5 microbes. Real dataset 1 consists of 18 and 22 RNA-Seq datasets from marine microbial eukaryotes. For 18 RNA-Seq datasets, the molecular phylogeny27 was reconstructed based on the 18S rRNA genes with maximum likelihood (ML) method. The ML phylogenetic tree was then used to evaluate the ability of VLMC as background model, combined with 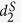 and
and 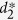 measures to compare their relationships based on the high-throughput sequencing data of individual species. For 22 RNA-Seq datasets, phylogenetic tree was built with Bayesian inference using MrBayes28 program. Dataset 2 contains 88 metatranscriptomic samples collected from the Global Ocean Sampling Expedition (GOSE), and they were used to study the effect of VLMC-based measures in identifying group relationships. Dataset 3 consists of 8 metagenomic and 8 metatranscriptomic samples from ocean depths of 25 m, 75 m, 125 m and 500 m. Dataset 4 consists of 14 metatranscriptomic samples from depths of 0.03 m and 0.08 m within a typical iron-rich microbial mat. Datasets 3 and 4 were used to study the performance of VLMC-based measures in revealing environmental gradient relationships. The triples distances were applied to evaluate the consistency between the reference and clustering trees from alignment-free measures.
measures to compare their relationships based on the high-throughput sequencing data of individual species. For 22 RNA-Seq datasets, phylogenetic tree was built with Bayesian inference using MrBayes28 program. Dataset 2 contains 88 metatranscriptomic samples collected from the Global Ocean Sampling Expedition (GOSE), and they were used to study the effect of VLMC-based measures in identifying group relationships. Dataset 3 consists of 8 metagenomic and 8 metatranscriptomic samples from ocean depths of 25 m, 75 m, 125 m and 500 m. Dataset 4 consists of 14 metatranscriptomic samples from depths of 0.03 m and 0.08 m within a typical iron-rich microbial mat. Datasets 3 and 4 were used to study the performance of VLMC-based measures in revealing environmental gradient relationships. The triples distances were applied to evaluate the consistency between the reference and clustering trees from alignment-free measures.
There is no rigorous criterion to decide the optimal length k for k-tuples. However, according to our previous experiments, generally the optimal k is 6–9. For comparison, 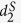 and
and 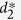 with 0–4th order FOMC, three Lp-norm measures and d2 were also applied.
with 0–4th order FOMC, three Lp-norm measures and d2 were also applied.
Experiment 1: Detecting group relationships among simulated metatranscriptomic datasets
Using a similar simulation strategy as developed in Martinez et al.11, we simulated three groups of synthetic mock communities with different expression levels using Polyester12, an RNA-Seq simulation tool. Five most abundant microbial genomes in human gut were selected based on Qin et al.29: Bacteroides vulgatus ATCC 8482, Ruminococcus torques L2−14, Faecalibacterium prausnitzii SL3/3, Bacteroides thetaiotaomicron VPI-5482 and Parabacteroides distasonis ATCC 8503. For each bacterium, a subsample of 1000 genes was randomly selected without replacement. Based on the mock community consisting of 5,000 genes from the five bacteria, we set three group centers with different gene expression levels as follows:
Among the 5000 genes, 20% showed 4-fold overexpression, 20% showed 4-fold under-expression, and 60% were normally-expressed. The simulation tool Polyester12 uses a fold change vector to specify the different expression levels among transcripts. Polyester generates the baseline read numbers from a negative binomial distribution with a preset mean value (default mean = 300), and then multiply the baseline numbers by the fold changes to simulate the transcripts with different expression levels. As shown in equation (1), A is the basic fold change vector, and 20% of the elements equal
 , 20% equal 4, and the others equal 1.
, 20% equal 4, and the others equal 1.
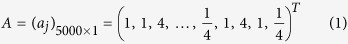 |
We then generated 90 samples belonging to 3 groups each containing 30 samples using the simulation strategy as in Jiang et al.22, shown in steps (2) and (3).
(2)The three group centers A1 A2 and A3 were generated as equation (2). Norm(μ,σ2) indicates the normal distribution with mean μ and variance σ2.
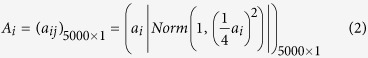 |
(3) For the qth sample 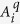 within group Ai, the expression level vector
within group Ai, the expression level vector 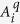 were generated using equation (3).
were generated using equation (3).
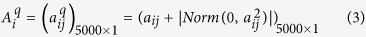 |
Based on the generated 90 expression level vectors, 90 metatranscriptomic sequencing data were simulated and the read length was 76 bp.
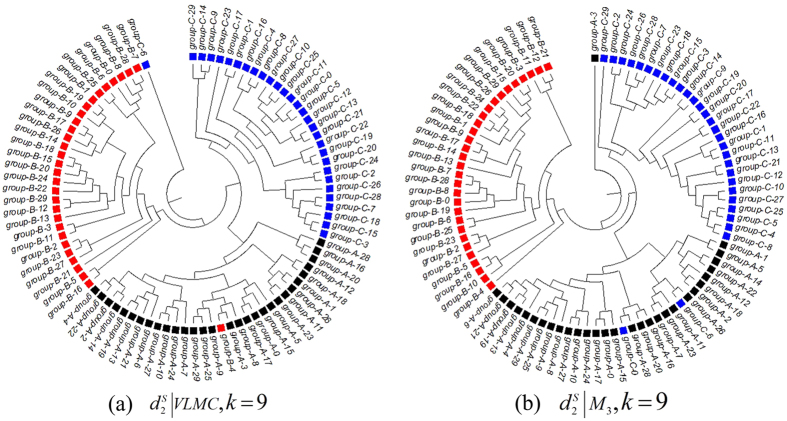
The best hierarchical clustering trees with VLMC and FOMC are shown in Fig. 1, and the corresponding triples distances are shown in Table 1. Clear groups of three simulated datasets among samples can be observed for both VLMC and FOMC. The best clustering trees with the smallest triples distance for VLMC and FOMC are both obtained in k = 9 and using 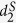 dissimilarity measure. From the clustering tree in Fig. 1, it is clear that the tree built based on VLMC is more similar to the true tree than that build based on FOMC. Quantitatively, the smallest triples distance for VLMC and FOMC are 42,973 and 43,043, respectively, where VLMC outperforms FOMC with less misclassification.
dissimilarity measure. From the clustering tree in Fig. 1, it is clear that the tree built based on VLMC is more similar to the true tree than that build based on FOMC. Quantitatively, the smallest triples distance for VLMC and FOMC are 42,973 and 43,043, respectively, where VLMC outperforms FOMC with less misclassification.
Figure 1. The clustering trees based on different models for the 90 simulation samples in Experiment 1.
(a) The best clustering tree on VLMC. (b) The best clustering tree when using FOMC and lp-norm measures. *Samples are divided into three groups A–C. Each group has 30 samples numbered from 0 to 29.
Table 1. The triples distance between the reference and the clustering trees using various background models with k = 2–9 for the simulation dataset of Experiment 1.
| k | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | |
|---|---|---|---|---|---|---|---|---|---|
| VLMC | 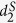 |
91845 | 90796 | 91209 | 90583 | 89748 | 74759 | 54841 | 42973 |
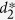 |
92398 | 91368 | 91967 | 91557 | 90858 | 91088 | 92198 | 91266 | |
| FOMC | 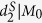 |
91737 | 91317 | 90938 | 89990 | 89176 | 73161 | 64770 | 58339 |
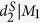 |
91845 | 90825 | 89631 | 89432 | 86202 | 74615 | 66449 | 63143 | |
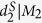 |
NA | 91215 | 90772 | 89205 | 88783 | 77420 | 61034 | 46369 | |
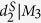 |
NA | NA | 91244 | 90672 | 89128 | 67519 | 62031 | 43047 | |
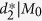 |
91799 | 91887 | 90788 | 90149 | 89170 | 83466 | 64932 | 56770 | |
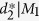 |
92398 | 90617 | 88314 | 90011 | 86425 | 73498 | 63022 | 59554 | |
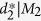 |
NA | 90684 | 90589 | 87693 | 86522 | 75092 | 58091 | 50969 | |
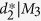 |
NA | NA | 91002 | 89840 | 89773 | 62601 | 55456 | 50293 | |
| Lp -Norm and d2 measures | d2 | 90569 | 89957 | 88967 | 88062 | 84338 | 72034 | 65577 | 63569 |
| Ch | 89553 | 89271 | 90366 | 89440 | 91424 | 90956 | 90456 | 90913 | |
| Eu | 90800 | 90930 | 89777 | 88271 | 86178 | 74766 | 67672 | 66107 | |
| Ma | 90338 | 90835 | 88814 | 89461 | 85528 | 71241 | 50671 | 47623 |
“Mi” indicates that the expected counts are calculated based on an i-th order Markov model for the background sequences. The p-values are estimated by comparing the observed triples distance to the triples distance in 3000 randomly-joined trees: for triples distance < 8000: p < 0.001.
Experiment 2: Comparison based on RNA-Seq data of Marine Microbial Eukaryotes
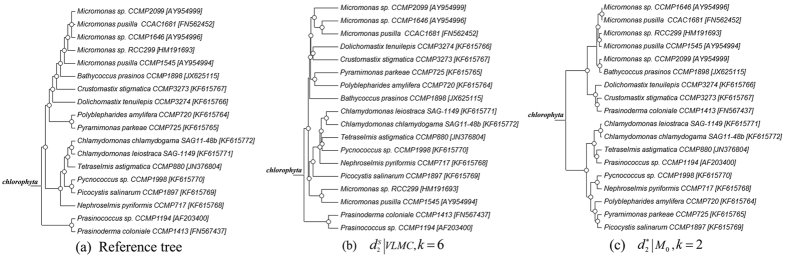
RNA-Seq data of 18 marine eukaryotes were downloaded from “The Marine Microbial Eukaryote Transcriptome Sequencing Project”30. The 18 eukaryotes are from the Phylum Chlorophyta, and the sample information is listed in Table S1 in Supplementary Section 1.1. The reference tree of the eukaryotes was extracted from the molecular phylogenetic tree built from a previous study27 that reconstructed the tree by maximum likelihood (ML) based on the 18S rRNA gene from a genome sequence or RNA-seq-based transcriptome assembly, shown in Supplementary Figure 1 of their paper27. Figure 2a shows the resulting ML of the 18 eukaryotes and it is used as a reference tree in our study. The bootstrap supports for the nodes in the phylogenetic reference tree are higher than 65%. The bootstrap support values of the nodes were calculated based on 1,000 replicates of the data with the same substitution model. The Bayesian posterior probabilities of the nodes in the tree were higher than 90%. The Bayesian analyses were performed with two independent runs with 1,000,000 generations per run. After a burn-in of 350,000 trees (that were discarded) per run, the remaining trees were used to reconstruct a consensus tree and to obtain posterior probabilities for node supports27.
Figure 2. The reference tree and the best clustering trees based on VLMC and FOMC models for 18 RNA-Seq data in Experiment 2.
(a) The molecular phylogenic tree of the 18 RNA-seq built with Maximum likelihood method on the 18S rRNA genes. (b) The best clustering tree with VLMC. (c) The best clustering tree when using FOMC and Lp-norm measures. *Samples are labeled as the Organisms-Strain-18S rRNA. For example, Micromonas pusilla CCAC1681 [FN562452] represents the organism Micromonas pusilla from strain CCAC1681 and the 18S rRNA used to construct this ML tree is FN562452. Details about sample labels can be found in Supplementary Table S4 in section 2.
Table 2 shows the triples distance between the reference and clustering trees using various dissimilarity measures and tuple length. The best clustering result with the smallest triples distance of 177 is obtained by VLMC using the dissimilarity measure 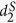 and tuple length k = 6, as shown in Fig. 2b. The topological structure is similar to that of the reference phylogenetic tree which basically includes three groups. The smallest triples distance for FOMC is 318, which was achieved by using
and tuple length k = 6, as shown in Fig. 2b. The topological structure is similar to that of the reference phylogenetic tree which basically includes three groups. The smallest triples distance for FOMC is 318, which was achieved by using 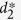 with 0-order MC and k = 2, as shown in Fig. 2c. Its overall topological structure of the clustering results is different with the phylogenetic tree in Fig. 2a. The clustering result based on VLMC is obviously better than the result based on the FOMC model.
with 0-order MC and k = 2, as shown in Fig. 2c. Its overall topological structure of the clustering results is different with the phylogenetic tree in Fig. 2a. The clustering result based on VLMC is obviously better than the result based on the FOMC model.
Table 2. The triples distance between the reference and the clustering trees using various background models with k = 2–9 for the 18 RNA-Seq data in Experiment 2.
| k | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | |
|---|---|---|---|---|---|---|---|---|---|
| VLMC | 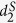 |
354 | 375 | 415 | 193 | 177 | 241 | 227 | 219 |
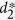 |
359 | 421 | 236 | 187 | 240 | 293 | 291 | 279 | |
| FOMC | 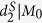 |
336 | 327 | 407 | 407 | 403 | 428 | 428 | 428 |
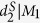 |
550 | 582 | 504 | 518 | 518 | 518 | 456 | 487 | |
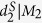 |
NA | 561 | 515 | 533 | 518 | 506 | 489 | 476 | |
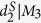 |
NA | NA | 534 | 581 | 577 | 509 | 500 | 440 | |
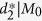 |
318 | 326 | 410 | 415 | 456 | 526 | 521 | 548 | |
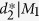 |
537 | 557 | 565 | 568 | 592 | 592 | 585 | 541 | |
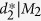 |
NA | 561 | 533 | 535 | 566 | 570 | 535 | 500 | |
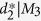 |
NA | NA | 554 | 573 | 564 | 545 | 526 | 506 | |
| Lp -Norm and d2 measures | d2 | 466 | 475 | 470 | 480 | 480 | 504 | 492 | 553 |
| Ch | 491 | 490 | 499 | 523 | 546 | 579 | 556 | 567 | |
| Eu | 470 | 474 | 474 | 479 | 494 | 564 | 564 | 530 | |
| Ma | 495 | 473 | 473 | 480 | 472 | 474 | 478 | 480 |
“Mi” indicates that the expected counts are calculated based on an i-th order Markov model for the background sequences. The p-values are estimated by comparing observed the triples distance to the triples distance in 3000 randomly-joined trees: for triples distance in 300~400: p < 0.006; for the triples distance < 300: p < 0.001.
We also analyzed RNA-Seq data from another set consisting of 22 Marine Microbial Eukaryotes from the Phylum Bacillariophyta, Chlorophyta, and Cryptophyta. The phylogenetic tree was built using MrBayes28 based on multiple alignments of 18S rRNA sequences using the default settings, and it was used as a reference tree for evaluations. The score for each branch is the Bayesian posterior probability of each partition or clade in the tree. It is the fraction of times that the partition or clade appears in the set of sampled posterior trees. The total number of samples generated from the posterior probability distribution is 1,000,000, and the beginning 25% of the samples were treated as burn-in and were discarded. The three groups Ch, Cr and Ba were clearly clustered to different groups with 100% posterior probabilities. Two internal branches in group Ba have Bayesian posterior probabilities less than 100%. The corresponding results for clustering the 22 species based on transcriptome data using FOMC and VLMC were shown in Supplementary Section 1.2. The experiment also shows the superior performance of VLMC over FOMC.
Experiment 3: Comparison based on 88 global ocean metatranscriptomic samples
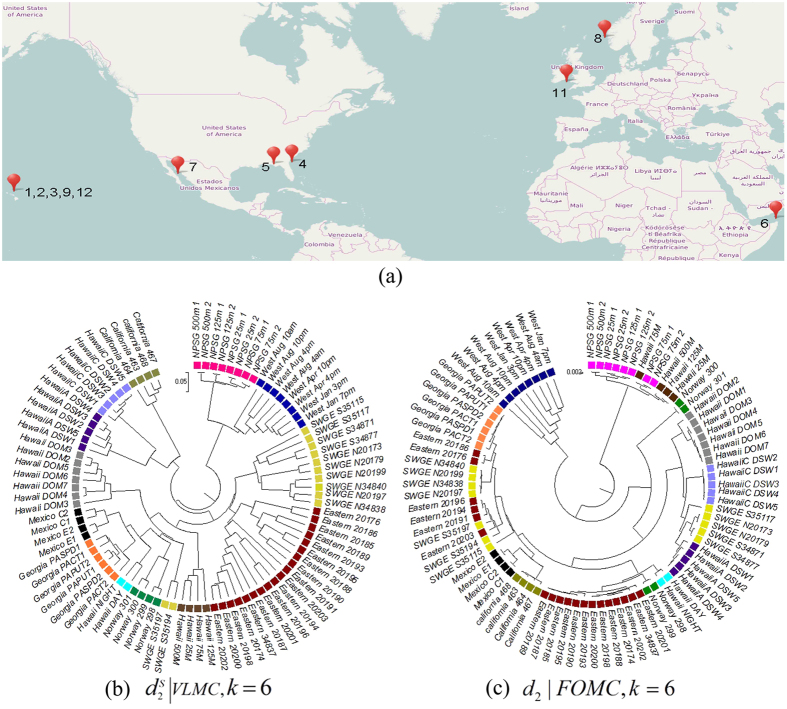
In this experiment, 88 metatranscriptomic samples collected from different global ocean locations were analyzed. These samples were downloaded from 12 different projects from Microbe (http://data.imicrobe.us/, originally belonging to CAMERA) and NCBI with 454 pyrosequencing. The descriptions and dataset IDs are given in Table S4 in Supplementary Section 2. Figure 3a shows the locations of these 88 samples. Twenty-three samples are from the subtropical north Pacific (Hawaiian), 4 from the Mexican Gulf, 4 from the California Gulf, 4 from the Norwegian Fjord, 6 from Sapelo Island (Georgia), 8 from the North Atlantic Ocean (West English Channel), 8 from North Pacific Subtropical Gyre (NPSG), and 19 from Eastern Equatorial Atlantic Ocean mixed with Amazon River plume. In addition, 12 samples were collected from different locations of Equatorial North Atlantic Ocean and South Pacific Subtropical Gyre. The map for the distribution of collecting locations was based on OpenStreetMap, and the cartography in the OpenStreetMap map tiles is licensed under CCBY-SA (www.openstreetmap.org/copyright). The license terms can be found on the link: http://creativecommons.org/licenses/by-sa/2.0/.
Figure 3. Locations of the 88 metatranscriptomic samples from global ocean, the reference tree, and the clustering trees based on different dissimilarity measures and background sequence models in Experiment 3.
(a) The distribution of the collecting locations. The map is based on OpenStreetMap and the cartography in the OpenStreetMap map tiles is licensed under CCBY-SA (www.openstreetmap.org/copyright). The license terms can be found on the link: http://creativecommons.org/licenses/by-sa/2.0/. The location labels are marked with the coordinates of sample-collecting locations in Supplementary Table S4 in section 2. (b) The clustering tree with VLMC using 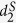 and k = 6. (c) The clustering tree with FOMC using
and k = 6. (c) The clustering tree with FOMC using 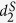 and k = 6. *‘SWGE’ (Dataset 10 in Supplemental Table S4 in section 2) samples were collected from different locations with two research cruises in the Equatorial North Atlantic Ocean and South Pacific Subtropical gyre.
and k = 6. *‘SWGE’ (Dataset 10 in Supplemental Table S4 in section 2) samples were collected from different locations with two research cruises in the Equatorial North Atlantic Ocean and South Pacific Subtropical gyre.
The clustering trees with 6-tuples based on 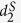 using VLMC and d2 using FOMC are shown in Fig. 3c. In VLMC, clear groups of different locations among samples can be seen. Except for the two samples from “SWGE”, all other samples are consistently grouped with the marine locations. The communities with proximate latitudes, including Eastern Equa, Atlan_Amazon and SWGE, are clustered first, which is consistent with our understanding that these communities should have greater similarity of gene expression profiles. For FOMC, samples from SWGE and the Amazon River are both scattered into several parts of the clustering. VLMC-based measures reveal location relationships of these 88 global ocean metatranscriptomic samples.
using VLMC and d2 using FOMC are shown in Fig. 3c. In VLMC, clear groups of different locations among samples can be seen. Except for the two samples from “SWGE”, all other samples are consistently grouped with the marine locations. The communities with proximate latitudes, including Eastern Equa, Atlan_Amazon and SWGE, are clustered first, which is consistent with our understanding that these communities should have greater similarity of gene expression profiles. For FOMC, samples from SWGE and the Amazon River are both scattered into several parts of the clustering. VLMC-based measures reveal location relationships of these 88 global ocean metatranscriptomic samples.
Experiment 4: Comparison of gradient relationship based on metatranscriptomic samples from different ocean depths
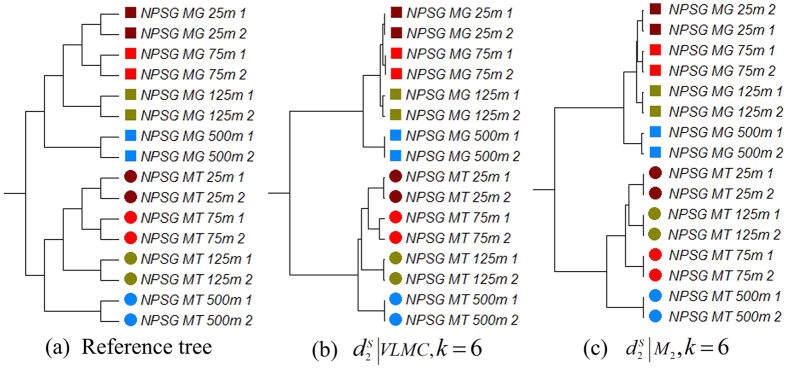
The gene expression profile of microbes can be affected by environmental factors, such as ocean depth, temperature, or pH. To evaluate the performance of the different dissimilarity measures and background sequence models in recovering the gradient relationships of microbial communities, we studied 8 metagenomic and 8 metatranscriptomic samples from depths of 25 m, 75 m, 125 m and 500 m (two replicate samples for each depth) of North Pacific Subtropical Gyre (NPSG) in ALOHA stations31 (dataset 12 in Table S4 in Supplementary Section 2).
Table 3 shows the triples distance between the reference tree and the derived clustering trees using different dissimilarity measures and background sequence models. Using the VLMC background sequence model, both 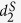 and
and 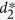 can recover the reference tree. The best results from both VLMC and FOMC background sequence models show clear separations between metagenomic and metatranscriptomic groups, as shown in Fig. 4b,c, respectively. For both background sequence models, samples from the same depth are clustered first, then the samples belonging to the photic zone (25 m, 75 m and 125 m) are merged, and, finally, samples belonging to the mesopelagic zone (500 m). However, for the FOMC background sequence model, the metatranscriptomic samples from 25 m and 125 m are clustered first, which is inconsistent with gradient relationships. In contrast, VLMC background sequence model produces clustering of metagenomic and metatranscriptomic samples as expected with 25 and 50 m first and then 125 m.
can recover the reference tree. The best results from both VLMC and FOMC background sequence models show clear separations between metagenomic and metatranscriptomic groups, as shown in Fig. 4b,c, respectively. For both background sequence models, samples from the same depth are clustered first, then the samples belonging to the photic zone (25 m, 75 m and 125 m) are merged, and, finally, samples belonging to the mesopelagic zone (500 m). However, for the FOMC background sequence model, the metatranscriptomic samples from 25 m and 125 m are clustered first, which is inconsistent with gradient relationships. In contrast, VLMC background sequence model produces clustering of metagenomic and metatranscriptomic samples as expected with 25 and 50 m first and then 125 m.
Table 3. The triples distance between the reference and the clustering trees using various background models with k = 2–9 to identify the gradient relationships of metagenomic and metatranscriptomic samples at different ocean depths in Experiment 4.
| k | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | |
|---|---|---|---|---|---|---|---|---|---|
| VLMC | 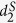 |
131 | 8 | 16 | 16 | 0 | 4 | 30 | 131 |
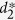 |
123 | 24 | 16 | 0 | 0 | 12 | 156 | 123 | |
| FOMC | 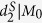 |
32 | 24 | 32 | 16 | 16 | 16 | 16 | 32 |
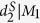 |
131 | 40 | 32 | 16 | 16 | 16 | 8 | 131 | |
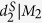 |
NA | 40 | 16 | 8 | 8 | 8 | 8 | 40 | |
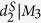 |
NA | NA | 34 | 8 | 8 | 8 | 8 | 34 | |
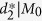 |
40 | 32 | 32 | 32 | 24 | 24 | 24 | 40 | |
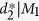 |
123 | 16 | 16 | 16 | 8 | 8 | 8 | 123 | |
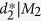 |
NA | 24 | 8 | 8 | 8 | 8 | 8 | 24 | |
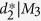 |
NA | NA | 16 | 8 | 8 | 8 | 8 | 16 | |
| Lp –Norm and d2 measures | d2 | 112 | 32 | 32 | 32 | 32 | 32 | 32 | 112 |
| Ch | 32 | 128 | 128 | 128 | 136 | 136 | 128 | 32 | |
| Eu | 112 | 32 | 32 | 32 | 32 | 32 | 40 | 112 | |
| Ma | 112 | 112 | 16 | 16 | 16 | 16 | 16 | 112 |
“Mi” indicates that the expected counts are calculated based on an i-th order Markov model for the background sequences. The p-values are estimated by comparing observed the triples distance to the triples distance in 3000 randomly-joined trees: for all triples distance in table, p < 0.001.
Figure 4. The reference and clustering trees based on various models for different depths of metatranscriptomic marine samples in Experiment 4.
(a) Reference tree of different depths of metatranscriptomic samples from the ocean. (b) The best clustering tree with VLMC background sequence model. (c) The best clustering tree when using FOMC and lp-norm measures.
Experiment 5: Comparison of gradient relationships based on metatranscriptomic samples from different iron-rich microbial mats
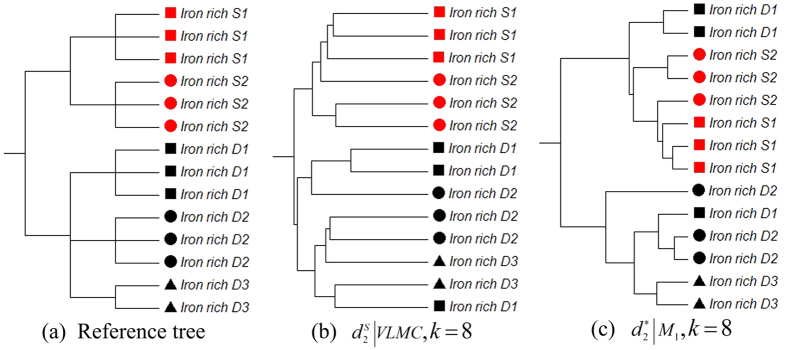
A microbial mat is a multilayered sheet of microorganisms, mainly bacteria and archaea. Previous studies32 found clear phylogenetic stratification between the surface and the deeper regions of the microbial mat where iron-oxidizing bacteria dominated the community in the upper layers, and methanothrophs contributed to the majority of sequences in the deeper layers. Therefore, in this experiment, we used our methods to study 14 metatranscriptomic samples32 to evaluate gradient relationships at different depths of the microbial mat. As shown in Figure S2 in Supplementary Section 3, the sampling site is a slow-flowing stream where two collection sites (S1, S2) are placed at 1 cm in depth (surface water), and three collection sites (D1, D2.D3) are placed in deeper regions of 7–9 cm. Three samples were collected at every collection site, except D3, where only two samples were harvested. The descriptions and dataset IDs of these samples can be found in Table S5 in Supplementary Section 3. Figure 5a shows the reference tree of the 14 microbial mat samples. Samples from S1 and S2 are marked in red, and samples from D1, D2 and D3 are marked in black. Samples at three different locations were respectively represented as squares, triangles and circles.
Figure 5. Reference and clustering trees based on different background sequence models for the metatranscriptomic mat samples in Experiment 5.
(a) Reference tree of the microbial mat data in experiment 5. (b) The best clustering tree with the VLMC background sequence model. (c) The best clustering tree with the FOMC background sequence model and lp-norm measures.
Table 4 shows the triples distance between the reference and the clustering trees. The best clustering tree was achieved by VLMC with 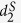 when k = 8 as shown in Fig. 5b, and the smallest triples distance is 76. Samples from surface water (S1, S2) and from deeper regions (D1, D2 and D3) are clearly separated. In contrast, the best result based on the FOMC background sequence model showed that surface samples were merged with deeper samples successively, as shown in Fig. 5c.
when k = 8 as shown in Fig. 5b, and the smallest triples distance is 76. Samples from surface water (S1, S2) and from deeper regions (D1, D2 and D3) are clearly separated. In contrast, the best result based on the FOMC background sequence model showed that surface samples were merged with deeper samples successively, as shown in Fig. 5c.
Table 4. The triples distance between the reference and the clustering trees using various background models with k = 2–9 to identify the gradient relationships of metatranscriptomic samples of microbial mats in Experiment 5.
| k | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | |
|---|---|---|---|---|---|---|---|---|---|
| VLMC | 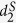 |
251 | 251 | 283 | 274 | 245 | 195 | 76 | 76 |
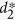 |
251 | 274 | 273 | 263 | 228 | 210 | 76 | 262 | |
| FOMC | 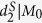 |
218 | 189 | 189 | 202 | 202 | 178 | 201 | 189 |
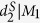 |
251 | 248 | 159 | 202 | 201 | 178 | 201 | 189 | |
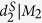 |
NA | 276 | 154 | 197 | 201 | 178 | 201 | 189 | |
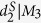 |
NA | NA | 272 | 201 | 118 | 164 | 170 | 189 | |
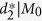 |
251 | 222 | 221 | 159 | 202 | 178 | 178 | 148 | |
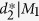 |
251 | 218 | 159 | 202 | 195 | 178 | 148 | 148 | |
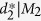 |
NA | 274 | 154 | 154 | 195 | 178 | 148 | 148 | |
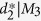 |
NA | NA | 268 | 201 | 195 | 148 | 148 | 148 | |
| Lp -Norm and d2 measures | d2 | 264 | 218 | 213 | 159 | 198 | 178 | 178 | 148 |
| Ch | 264 | 247 | 256 | 260 | 241 | 207 | 232 | 238 | |
| Eu | 264 | 218 | 213 | 159 | 198 | 165 | 162 | 162 | |
| Ma | 264 | 218 | 213 | 159 | 198 | 206 | 206 | 202 |
“Mi” indicates that the expected counts are calculated based on an i-th order Markov model for the background sequences. The p-values are estimated by comparing the observed triples distance to the triples distance in 3000 randomly-joined trees: for the triples distance < 155:p < 0.001.
The two-dimension Principal Component Analysis (PCA) plots based on the optimal results from FOMC and VLMC when k = 8 are shown in Fig. 6a,b, respectively. The PCA plot based on VLMC reflects the gradient information for collection depths and sites as the first and second principal component. We also plotted the PCA figures for k = 7 and 9, shown in Figure S3 in Supplementary Section 4. Although k = 7 and 9 are not the optimal value for VLMC, they still can separate the different depths and collecting sites. In comparison, the PCA ordinates based on FOMC did not show clear separations, and some points are shown as outliers.
Figure 6. PCA ordinates of samples in Experiment 5.
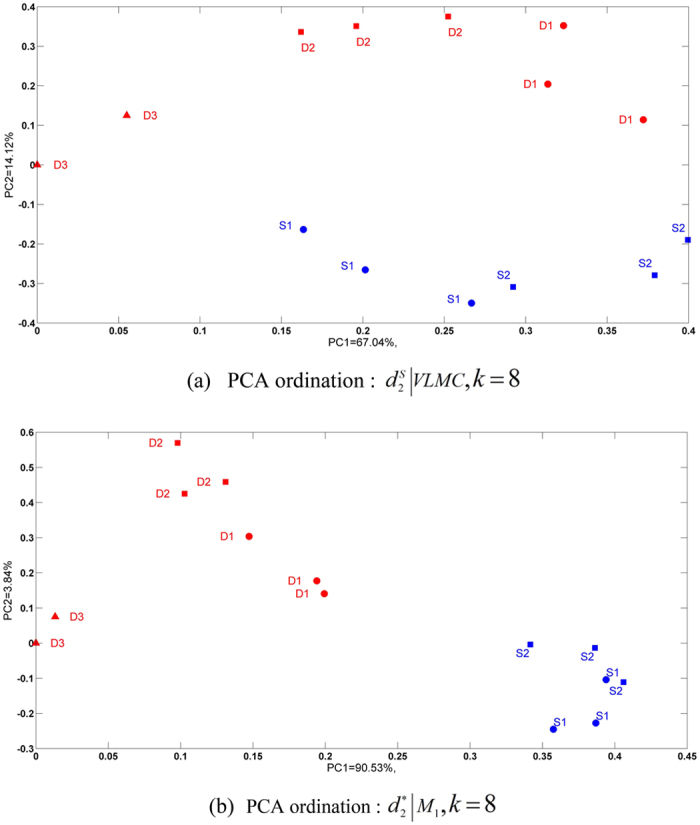
(a) Two-dimensional PCA plot based on FOMC. (b) Two-dimensional PCA plot based on VLMC.
Discussion and Conclusions
In this study, we developed theoretical and computational approaches to model background sequences using VLMCs based on short reads from high throughput sequencing. We compared the performances of VLMC and FOMC with 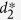 and
and 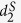 , as well as d2 and three Lp-norm measures, to model the background sequence with one simulated dataset, three real metatranscriptomic datasets, and one real RNA-seq dataset. VLMC outperformed FOMC in all experiments; and
, as well as d2 and three Lp-norm measures, to model the background sequence with one simulated dataset, three real metatranscriptomic datasets, and one real RNA-seq dataset. VLMC outperformed FOMC in all experiments; and 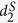 together with VLMC, as background sequence model, outperformed FOMC in all experiments. Experiments show that VLMC builds the model with adaptive and variable MC according to the metatranscriptomic data, exempting from manual selection of a fix MC order. Compared with FOMC, VLMC are more structural rich and easy to use. Based on the experimental results, we show that
together with VLMC, as background sequence model, outperformed FOMC in all experiments. Experiments show that VLMC builds the model with adaptive and variable MC according to the metatranscriptomic data, exempting from manual selection of a fix MC order. Compared with FOMC, VLMC are more structural rich and easy to use. Based on the experimental results, we show that 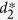 and
and 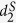 dissimilarity measures combined with VLMC background model can identify the underlying relationships among samples from different microbial communities. They can also reveal the gradient relationship among the samples. Therefore, such dissimilarity measures should be adopted in comparative transcriptomic and metatranscriptomic studies.
dissimilarity measures combined with VLMC background model can identify the underlying relationships among samples from different microbial communities. They can also reveal the gradient relationship among the samples. Therefore, such dissimilarity measures should be adopted in comparative transcriptomic and metatranscriptomic studies.
In this study, we only applied VLMC to RNA-Seq or metatranscriptomic datasets. We also attempted to apply VLMC to metagenomic datasets, but here, VLMC does not achieve obvious improvements compared with the results of FOMC. For instance, we applied VLMC to analyze a real mammalian gut metagenomic dataset33. It includes 21 samples from mammalian species of herbivores and 7 samples from species of carnivores. As shown in Figure S4 in Supplementary section 5, results indicate that VLMC is less effective than FOMC in distinguishing between the two mammalian sample types. This could be attributed to the inclusion of both expressed and non-expressed regions in the whole genomes, making them heterogeneous. One model cannot fit the data well resulting in a simple independent identically distributed yielding the most meaningful results in most cases. Since the transcriptome only includes expressed regions, they will most likely be homogeneous, and a Markov model may fit better. Thus, while VLMC can improve performance for metatranscriptomic datasets, it does not show obvious improved performance for metagenomic datasets.
Alignment-free method avoids the complications of alignment-based approach, and is able to process the microbial community with a large amount of dark matters. However, it does not provide detail insights of microbial communities and further biological interpretation. To answer such questions, alignment-based methods are still needed.
Methods
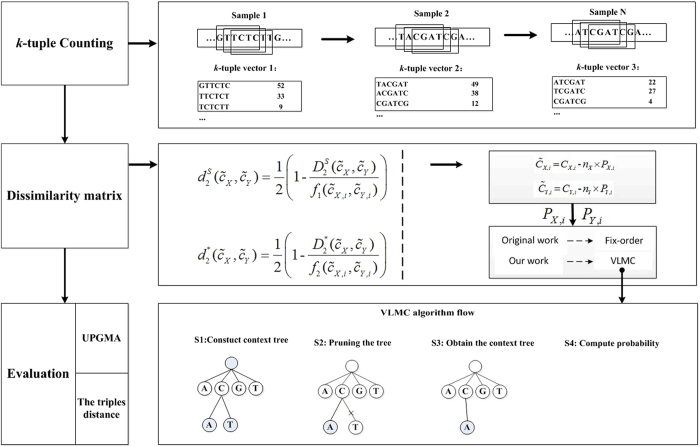
Processing flow chart
The processing procedure consists of three main steps: (1) calculating k-tuple frequency; (2) calculating the probability of each tuple based on VLMC and applying various dissimilarity measures to k-tuple frequencies; and (3) evaluating different dissimilarity measures and models for background sequences. We used UPGMA34 for hierarchical clustering based on dissimilarity matrix and applied the triples distance26 to evaluate consistency between the reference tree and the clustering tree. We extended the VLMC algorithm to make it suitable for high-throughput sequencing data and then applied VLMC to model the underlying background genomes in 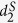 and
and 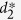 dissimilarity measures. Figure 7 shows the flow chart, and the details of these steps are given below.
dissimilarity measures. Figure 7 shows the flow chart, and the details of these steps are given below.
Figure 7. Flow chart of our approach:
(1) The frequency vector of 1–10 tuples is generated from the sequencing data. (2) Markov transition probability of each tuple is calculated based on VLMC, and different dissimilarity measures are applied to k-tuple sequence signature. (3) Measured dissimilarities are evaluated. We used UPGMA for hierarchical clustering based on dissimilarity matrix and applied the triples distance to evaluate the consistency between the reference and the clustering trees.
Calculating k-tuple frequency
Alignment-free methods use k-tuple frequencies as sequence signatures to represent each metatranscriptomic datum. In our study, k-tuple frequencies from k = 1 to a maximum k value are calculated with our developed pipeline, taking complementary strands into consideration. The maximum k value is d + 1, where d is the depth of the full prefix tree constructed in step (1). In our study, the depth of the prefix tree is 10. The k-tuple frequencies are used in constructing prefix tree, calculating the transition probabilities and compute dissimilarity measures.
Dissimilarity measures based on k-tuple frequency
The dissimilarity between two samples is calculated based on the frequency vectors using various measures, including measures with background model normalization such as 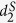 and
and 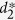 with VLMC/FOMC background sequence models, and measures without background model normalization such as d2, Ma, Ch and Eu in our study. The calculation of
with VLMC/FOMC background sequence models, and measures without background model normalization such as d2, Ma, Ch and Eu in our study. The calculation of 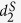 and
and 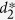 is described briefly as follows21:
is described briefly as follows21:
Let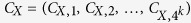 and
and 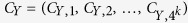 represent the k-tuple frequency vectors of sequencing data X and Y, Let
represent the k-tuple frequency vectors of sequencing data X and Y, Let 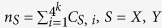 be the sum of the counts of all k-tuples. The
be the sum of the counts of all k-tuples. The 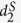 and
and 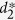 dissimilarity measures are defined in equations (1) and (2), where
dissimilarity measures are defined in equations (1) and (2), where 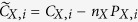 and
and 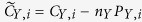 . The ranges of
. The ranges of 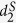 and
and 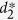 are between 0 and 1.
are between 0 and 1.
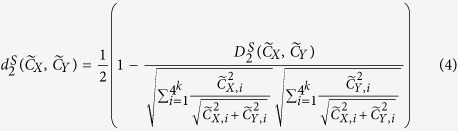 |
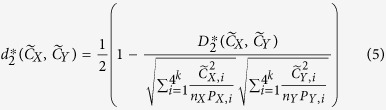 |
where PX,i and PY,i are the probability of the ith k-tuple based on X and Y, respectively. The probabilities are calculated based on a specific probabilistic model. For example, consider a 5-tuple “GCTAC”. Then P(GCTAC) can be calculated as:
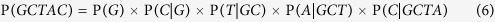 |
In previous studies1, FOMC was used to compute transition probability with fixed order r. For example, when r = 2,
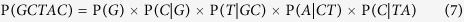 |
In application, the order of MC needs to be set manually. But for most microbial communities, there is no prior knowledge available for MC order. Furthermore, it is hard to model probabilities of different tuples using a single fixed order MC. Variable Length Markov Chains24 (VLMC) model the background genomes selecting the MC order adaptively in a data-driven way. For example, the probability (3) might be represented as formula (8) after determining the order in VLMC:
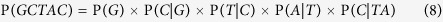 |
Thus VLMC is more structurally rich and the number of variables is flexible. VLMC was originally designed to model long sequences24,35 and was represented as a context tree structure25. In our study, VLMC was extended to model the background genomes based on short reads from high throughput sequencing.
VLMC for modeling background genomes with high-throughput sequencing data
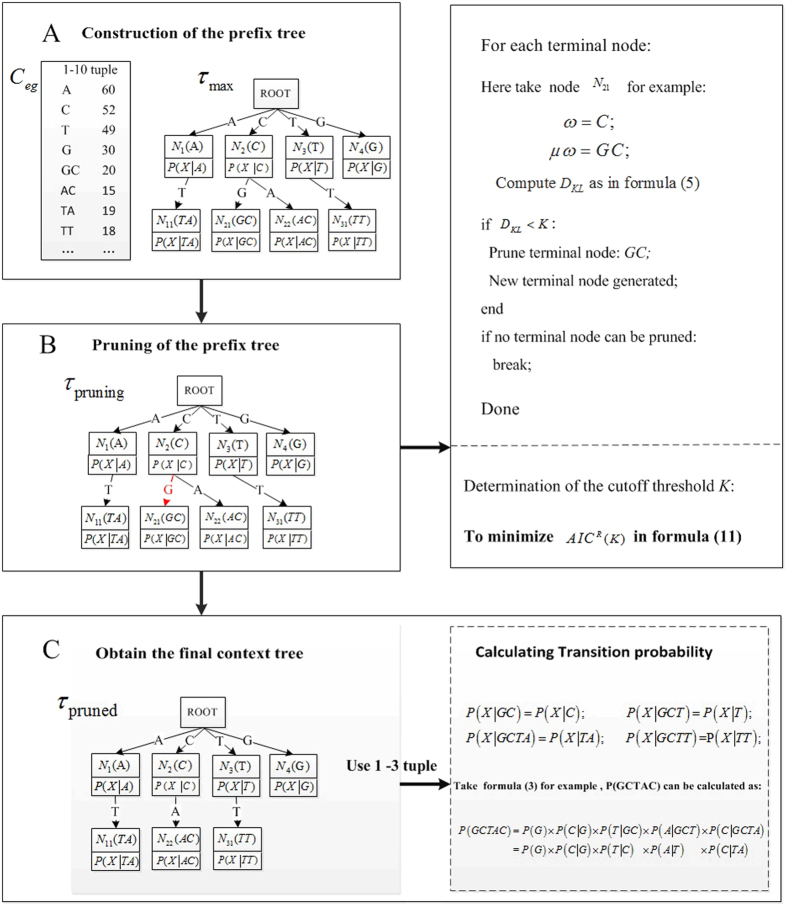
The VLMC for high-throughput sequencing data is implemented with the following three steps: (1) A full prefix tree is built based on 1, 2, …, 10-tuple frequency vectors, but the tree usually overfits the data. (2) The tree is subsequently pruned to remove redundant branches based on Kullback-Leibler divergence36, and the pruned tree is also called a context tree25. (3) Transition probabilities are calculated with respect to the MC orders from the context tree, and the probabilities of k-tuples are then computed accordingly. A specific example is given in Fig. 8. The three steps were inspired by the original VLMC method on a single genomic sequence proposed by Bühlmann P. and Wyner, A. J. in 199924.
Figure 8. Flow chart showing the construction of VLMC based on high-throughput sequencing data.
(A) Construction of the prefix tree. (B) Pruning of the prefix tree. (C) Calculation of probability based on the pruned (or context) tree.
Step 1: Generating a prefix tree τ max based on tuple frequency
We first generate a tree τmax to store tuples in the frequency vector. The tree τmax is actually a prefix tree growing downwards, where each node in the tree represents a tuple. The lth level nodes represent tuples of length l. In our study, the maximum depth of the tree τmax is up to 10. The following logic determines the relationships connecting nodes. If a node represents the l-tuple ω ∈{A,C,G,T}l, l = 1, 2, …, 9, then its offspring represents the (l + 1)-tuple word μω (μ is a character in front of ω, μ ∈{A,C,G,T}. For a node representing ω, the transition probability is calculated as PX(X|ω) = CX,ωX/CX,ω and saved at the node. In practice, the construction of τmax based on 1, 2, …, k-tuple frequency vectors is fast.
In Fig. 8A, τmax is generated based on frequency vector Cgg. Node N2(C) represents tuple C, and its offspring N21(GC) represents tuple GC. Additionally, each node is associated with the transition probability from corresponding tuple to X (X∈{A,C,G,T}). Node N2(C) is associated with P(X|C), and node N21(GC) is associated with P(X|GC).
Step 2: Pruning the tree τ max
The next step involves pruning the tree τmax to remove redundant branches. If the probability P(X|μω) for a terminal node μω is the same as its parent node’s transition probability P(X|ω), meaning that the transition probability of μω can be replaced by that of ω, then the terminal node μω can be pruned from the branch. In our study, Kullback-Leibler divergence is a measure of the distance between two probability distributions P(X|μω) and P(X|ω). Accordingly, Kullback-Leibler divergence36 is applied to compare P(X|μω) and P(X|ω), which is denoted as DKL(P(X|μω)||P(X|ω)). A value of DKL(P(X|μω)||P(X|ω)) less than a threshold value K indicates that no information is lost when P(X|ω) is used to approximate P(X|μω), thereby allowing μω to be pruned. DKL(P(X|μω)||P(X|ω)) is given by formula (9), and N(*) is the frequency.
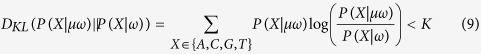 |
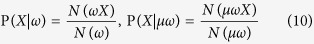 |
Taking Fig. 8B as an example, the Kullback-Leibler divergence between N21(GC) and N2(C) is calculated to determine whether nodeN21(GC) should be pruned:
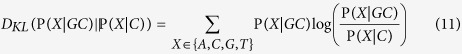 |
Suppose that threshold K is set to 5, then node N21(GC) should be pruned if
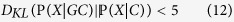 |
The pruning is implemented for each terminal node until no branches can be pruned. K is the threshold that determines the degree of pruning. A larger K means greater conditional latitude in branch pruning, in turn producing a smaller tree.
Similar to the study of Mächler and Bühlmann35, the determination of K is implemented through the optimization of Akaike Information Criterion (AIC)37 designed for high-throughput sequencing data. AIC measures the relative quality of statistical models for a given set of data. AIC is originally defined as
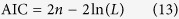 |
where L is the maximum value of the likelihood function for a statistical model, indicating the goodness of fit of the model, and n is the number of parameters in the model, indicating the complexity of the model.
Here we develop the AIC calculation algorithm for high-throughput sequencing data. Given high-throughput sequencing data with M reads of length β,
 |
where Sj is the jth read, Sji is the ith nucleotide of jth read, and Sji ∈ {A,C,G,T}. Then, AIC with pruning threshold K is defined as:
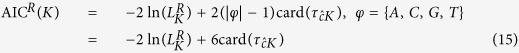 |
where card (τĉK) denotes the number of nodes in the context tree τĉK, and 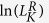 is the log-pseudo-likelihood under a fitted VLMC model with threshold K. The superscript R denotes the short read data. The log-pseudo-likelihood of the sequencing data is
is the log-pseudo-likelihood under a fitted VLMC model with threshold K. The superscript R denotes the short read data. The log-pseudo-likelihood of the sequencing data is
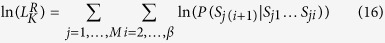 |
where P(Sj(i+ 1)|Sj1…Sji) is the estimated transition probability from the high-throughput sequencing data. The optimal K is determined by minimizing the formula AICR(K).
The two steps of tree building and pruning for high throughput sequencing data is extended from the original algorithm for long sequences from Bühlmann et al.24. The pruning step starts from the terminal nodes and the procedure is repeated until no more pruning is possible. The algorithm is greedy, so it is possible that the final pruned context tree is not the global optimal one. The R-package for long sequences35 developed in 2012 follows the same greedy algorithm.
Step 3: Calculating probabilities of tuples based on the context tree
The corresponding probabilities of tuples are calculated based on the context tree. The number of independent parameters is Num(nodes) × 3, where the Num(nodes) is the total number of nodes in the context tree except the root-node. Taking the context tree in Fig. 8C as an example, Node N21(GC) was pruned away; therefore, in tuple GCX, G has no effect on the transition probability from GC to state X. Thus, P(X|GC) can be replaced by P(X|C) and stored in node N2(C) of the context tree in Fig. 8C.
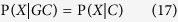 |
The tuples in the node of context tree can be of variable length, allowing the VLMC model to estimate the transition probability. The corresponding probabilities of tuples used in 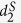 and
and 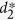 are then computed based on the transition probabilities. For example in Fig. 8C, the probability of 5-tuple word “GCTAC”,
are then computed based on the transition probabilities. For example in Fig. 8C, the probability of 5-tuple word “GCTAC”,
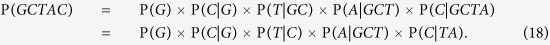 |
In the real data from marine metatranscriptome, there are ~103 nodes for the pruned context tree with 8 levels and ~102 nodes for the tree with 7 levels, which means that the number of parameters reduced from 48 × 3∼2 × 105 to ∼103–4 for r = 8; and from 47 × 3∼5 × 104 to ∼102–3 for r = 7, at least 10-fold decrease in the number of parameters.
Using a heuristic approach to search for optimal K
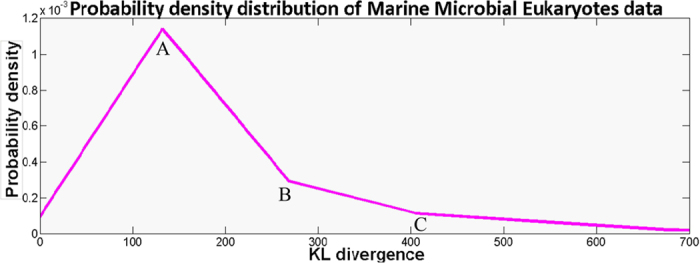
The value of K is determined by minimizing AICR(K). However, no simple analytical formula exists between K and AICR(K), making it a challenge to find the optimal K for all sequencing data. To solve this problem, we developed the following heuristic approach to determine the value of K. In our study, one branch is pruned when its Kullback-Leibler divergence is less than the threshold K. Therefore, K is meaningful only when it is within the value range of Kullback-Leibler divergence. In Experiment on 22 Marine Microbial Eukaryotes, the probability density distribution of the Kullback-Leibler divergence is shown as Fig. 9. The values of Kullback-Leibler divergence in most tuples are between 100 and 500. Optimal results are generally obtained with K setting around the peak points and the right two inflexions (point A, B and C in Fig. 9). Hence, we only implement local search around these three points for the optimal K that minimizes loss functionAICR(K).
Figure 9. Probability density distributions of 22 Marine Microbial Eukaryotes RNA-Seq data.
Sample clustering with UPGMA34 (Unweighted Pair Group Method with Arithmetic Mean) is a hierarchical clustering method initially designed for classification problems. UPGMA is now widely used for hierarchical clustering in bioinformatics based on dissimilarity matrices. The nearest two clusters are combined into a higher-level cluster. The distance between two clusters A and B is defined as the average of all distances between pairs of samples x in A and y in B. The calculation is presented in equation (14), where d (x,y) refers to the dissimilarity between sample x and sample y. This is repeated for each step. UPGMA is implemented with the function ‘upgma’ from the ‘phangorn’ toolbox of R.
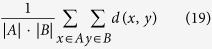 |
The selection of proper evaluation metrics: Based on the dissimilarity matrix from different background models, the hierarchical clustering trees are produced. The consistency between the reference and the clustering trees offers the metrics to evaluate the performance of the various background models. There are several metrics to measure the difference of topological structures between two trees.
Parsimony score38,39 is the most common one to compare the topological structures of two trees. The parsimony score for a tree is the sum of the smallest number of substitutions needed comparing with the reference tree, which was implemented with toolbox Mothur in our study. When one tree is binary and one tree is not binary, the parsimony score is not suitable for comparison of the trees.
Symmetric difference40 was originally defined to compare two node sets. It has been used as a criterion to evaluate the consistency between two trees38. Two trees A and B have the same leaves, and their node sets are 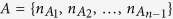 and
and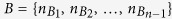 . The symmetric difference between A and B is defined as
. The symmetric difference between A and B is defined as
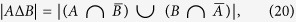 |
i.e., the set of nodes present in one tree, but not in the other tree, where |*| is the number of elements, and 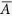 and
and 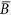 are the complements of set A and set B, respectively. Compared with the parsimony score, symmetric difference does not use branch length information, only tree topologies. Moreover, symmetric difference has taken the order of hierarchical clustering into consideration, making the comparison more sensitive. Symmetric difference is calculated with Treedist from Phylip.
are the complements of set A and set B, respectively. Compared with the parsimony score, symmetric difference does not use branch length information, only tree topologies. Moreover, symmetric difference has taken the order of hierarchical clustering into consideration, making the comparison more sensitive. Symmetric difference is calculated with Treedist from Phylip.
The triples distance26, another tree comparison metric to measure the distance between binary26 or non-binary trees41, is also used. In our study, some reference trees are rooted non-binary trees. The measures are based on the topologies of the input trees induce on triplets; that is, on three-element subsets of the set of species. Triplet based distances provide a robust and fine-grained measure of the similarities between trees41, which was developed as toolbox TreeCmp42.
The above three metrics have different characteristics and application scopes of their own. In Supplementary Section 7, we constructed example trees and measure their distances with the three metrics. Table S6 shows the three metrics for experiment 5, and the three metrics reflect general consistent tendency of tree distance. These two experiments show that the triples distance is most suitable and has high accuracy to evaluate the consistence of topologies of two trees.
Principal component analysis (PCA)43 is an important tool to analyze a multivariate data table in which observations are described by several inter-correlated quantitative dependent variables. Its goal is to extract the important information from the table and to represent the information as a set of new orthogonal variables called principal components. In R ‘ape’ toolbox, the functions princomp and prcomp can be used for principal component analysis.
Additional Information
Accession codes: https://d2vlmc.codeplex.com.
How to cite this article: Liao, W. et al. Alignment-free Transcriptomic and Metatranscriptomic Comparison Using Sequencing Signatures with Variable Length Markov Chains. Sci. Rep. 6, 37243; doi: 10.1038/srep37243 (2016).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
This research is supported by the National Natural Science Foundation of China (61203282, 61202144, 61503314, 61673324), U.S. National Science Foundation grants (DMS-1518001, OCE-1136818), China Scholarship Council (201606315011) and Natural Science Foundation of Fujian (2016J01316).
Footnotes
Author Contributions Y.W. and F.S. planned the project; W.L. and J.R. developed the model and designed the experiments; W.L., K.W. and S.W. realized the models and implemented the experiments; F.Z. analyzed the results; Y.W., W.L. and F.S. wrote the main manuscript. All authors read and approved the final manuscript.
References
- Wang Y., Liu L., Chen L., Chen T. & Sun F. Comparison of metatranscriptomic samples based on k-tuple frequencies. PloS One 9, e84348 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith T. F. & Waterman M. S. Identification of common molecular subsequences. Journal of Molecular Biology 147, 195–197 (1981). [DOI] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W. & Lipman D. J. Basic local alignment search tool. Journal of Molecular Biology 215, 403–410 (1990). [DOI] [PubMed] [Google Scholar]
- Wood D. E. & Salzberg S. L. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biology 15 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ounit R., Wanamaker S., Close T. J. & Lonardi S. CLARK: fast and accurate classification of metagenomic and genomic sequences using discriminative k-mers. BMC Genomics 16 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzel P., Ng K. L. & Krogh A. Fast and sensitive taxonomic classification for metagenomics with Kaiju. Nature Communications 7, 11257 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segata N. et al. Metagenomic microbial community profiling using unique clade-specific marker genes. Nature Methods 9, 811–814 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Tyson G. W. & DeLong E. F. Metatranscriptomics reveals unique microbial small RNAs in the ocean’s water column. Nature 459, 266–226 (2009). [DOI] [PubMed] [Google Scholar]
- Leimena M. M., Ramiro-Garcia J. & Davids M. A comprehensive metatranscriptome analysis pipeline and its validation using human small intestine microbiota datasets. BMC Genomics 14, 530 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adria M., David M. S. & Colleen A. D. Comparative metatranscriptomics identifies molecular bases for the physiological responses of phytoplankton to varying iron availability[J]. Proceedings of the National Academy of Sciences 109, 317–325 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez X. et al. MetaTrans: an open-source pipeline for metatranscriptomics. Scientific Reports 6, 26447 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazee A. C., Jaffe A. E., Langmead B. & Leek J. T. Polyester: simulating RNA-seq datasets with differential transcript expression. Bioinformatics 31, 2778–2784 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippert R. A., Huang H. & Waterman M. S. Distributional regimes for the number of k-word matches between two random sequences. Proceedings of the National Academy of Sciences 99, 13980–13989 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlin S., Mrazek J. & Campbell A. M. Compositional biases of bacterial genomes and evolutionary implications. Journal of Bacteriology 179, 3899–3913 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinert G., Chew D., Sun F. & Waterman M. S. Alignment-free sequence comparison (I): statistics and power. Journal of Computational Biology 16, 1615–1634 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantorovitz M. R., Robinson G. E. & Sinha S. A statistical method for alignment-free comparison of regulatory sequences. Bioinformatics 23, i249–i255 (2007). [DOI] [PubMed] [Google Scholar]
- Wan L., Reinert G., Sun F. & Waterman M. S. Alignment-free sequence comparison (II): theoretical power of comparison statistics. Journal of Computational Biology 17, 1467–1490 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Q. & Wang T. Comparison study on k-word statistical measures for protein: From sequence to ‘sequence space’. BMC Bioinformatics 9, 394 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Q., Yang Y. & Wang T. Markov model plus k-word distributions: a synergy that produces novel statistical measures for sequence comparison. Bioinformatics 24, 2296–2302 (2008). [DOI] [PubMed] [Google Scholar]
- Qi J., Wang B. & Hao B. L. Whole proteome prokaryote phylogeny without sequence alignment: a K-string composition approach. Journal of Molecular Evolution 58, 1–11 (2004). [DOI] [PubMed] [Google Scholar]
- Song K. et al. Alignment-free sequence comparison based on next-generation sequencing reads. Journal of Computational Biology 20, 64–79 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang B. et al. Comparison of metagenomic samples using sequence signatures. BMC Genomics 13, 730 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J., Song K., Deng M. & Reinert G. Inference of Markovian properties of molecular sequences from NGS data and applications to comparative genomics. Bioinformatics 32, 993–1000 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bühlmann P. & Wyner A. J. Variable length Markov chains. The Annals of Statistics 27, 480–513 (1999). [Google Scholar]
- Rissanen J. A universal data compression system. IEEE Transactions On Information Theory 29, 656–664 (1983). [Google Scholar]
- Critchlow D. E., Pearl D. K. & Qian C. The triples distance for rooted bifurcating phylogenetic trees. Systematic Biology 45, 323–334 (1996). [Google Scholar]
- Duanmu D. et al. Marine algae and land plants share conserved phytochrome signaling systems. Proceedings of the National Academy of Sciences 111, 15827–15832 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck J. P. & Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17, 754–755 (2001). [DOI] [PubMed] [Google Scholar]
- Qin J. et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling P. J. et al. The Marine Microbial Eukaryote Transcriptome Sequencing Project (MMETSP): illuminating the functional diversity of eukaryotic life in the oceans through transcriptome sequencing. PLoS Biol 12(6), e1001889 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karl D., Bidigare R. & Letelier R. Long-term changes in plankton community structure and productivity in the North Pacific Subtropical Gyre: the domain shift hypothesis. Deep Sea Research Part II: Topical Studies in Oceanography 48, 1449–1470 (2001). [Google Scholar]
- Quaiser A. et al. Unraveling the stratification of an iron-oxidizing microbial mat by metatranscriptomics. PLoS One 9(7) e102561 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muegge B. D., Kuczynski J. & Knights D. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science 332, 970–974 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murtagh F. Complexities of hierarchic clustering algorithms: State of the art. Computational Statistics Quarterly 1, 101–113 (1984). [Google Scholar]
- Mächler M. & Bühlmann P. Variable length Markov chains: methodology, computing, and software. Journal of Computational and Graphical Statistics 13(2), 435–455 (2012). [Google Scholar]
- Kullback S. & Leibler R. A. On Information and Sufficiency. Annals of Mathematical Statistics 22, 79–86 (1951). [Google Scholar]
- Akaike H. Factor analysis and AIC. Psychometrika 52, 317–332 (1987). [Google Scholar]
- Robinson D. & Foulds L. R. Comparison of phylogenetic trees. Mathematical Biosciences 53, 131–147 (1981). [Google Scholar]
- Schloss P. D. & Handelsman J. Introducing TreeClimber, a test to compare microbial community structures. Applied and Environmental Microbiology 72, 2379–2384 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penny D. & Hendy M. The use of tree comparison metrics. Systematic Zoology 34, 75–82 (1985). [Google Scholar]
- Bansal M. S., Dong J. & Fernández-Baca D. Comparing and aggregating partially resolved trees. Theoretical Computer Science 412, 6634–6652 (2011). [Google Scholar]
- Bogdanowicz D., Giaro K. & Wróbel B. TreeCmp: Comparison of Trees in Polynomial Time. Evolutionary Bioinformatics Online 8, 475–487 (2012). [Google Scholar]
- Wold S., Esbensen K. & Geladi P. Principal component analysis. Chemometrics and Intelligent Laboratory Systems 2, 37–52 (1987). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.