Abstract
Expansions of a hexanucleotide repeat (GGGGCC) in the noncoding region of the C9orf72 gene are the most common genetic cause of amyotrophic lateral sclerosis (ALS) and frontotemporal dementia. Decreased expression of C9orf72 is seen in expansion carriers, suggesting loss of function may play a role in disease. We find that two independent mouse lines lacking the C9orf72 ortholog (3110043O21Rik) in all tissues developed normally and aged without motor neuron disease. Instead, C9orf72 null mice developed progressive splenomegaly and lymphadenopathy with accumulation of engorged macrophage-like cells. C9orf72 expression was highest in myeloid cells, and loss of C9orf72 led to lysosomal accumulation and altered immune responses in macrophages and microglia, with age-related neuroinflammation similar to C9orf72 ALS but not sporadic ALS patient tissue. Thus, C9orf72 is required for normal function of myeloid cells, and altered microglial function may contribute to neurodegeneration in C9orf72 expansion carriers.
One Sentence Summary
Loss of C9orf72 disrupts microglial function and may contribute to neurodegeneration in C9orf72 expansion patients.
Amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD) are neurodegenerative disorders with overlapping clinical presentations, pathology, and genetic origins (1, 2). Expansions of a GGGGCC hexanucleotide repeat in the first intron/promoter of the C9orf72 gene are the most commonly identified genetic cause of ALS/FTD (3, 4), and are found in other neurodegenerative diseases (5). Microglial dysfunction is strongly tied to ALS/FTD pathogenesis (6) with mutations in progranulin causing FTD (7, 8), and variants in the microglial expressed genes TREM2 and TBK1 implicated in ALS (9–11). However, no connection has been made between microglial function and C9orf72, where focus instead has been on its role in neurons (12, 13). While the repeat expansion leads to decreased C9orf72 expression in patient tissues, most research has focused on gain of function toxicity as the primary mechanism in disease rather than loss of function (14–18).
To investigate the function of the mouse orthologue of C9orf72 (3110043O21Rik, referred to as C9orf72 below), we analyzed two independent loss of function alleles in mice (Fig. S1, S2). C9orf72+/− and C9orf72−/− mice showed normal weight gain and lifespan, had normal sensorimotor coordination, limb strength, femoral motor and sensory axon counts, muscle electrophysiology, and no evidence of neurodegeneration on histology through advanced age (17 months) (Fig. S1–S3). The only histologic abnormalities in the nervous system were rare chromatolytic structures on H&E staining, found in gray and white matter of the spinal cord that did not increase with age or show reactive gliosis (Fig. S3). All studies were performed using the Knockout Mouse Project line except where specified.
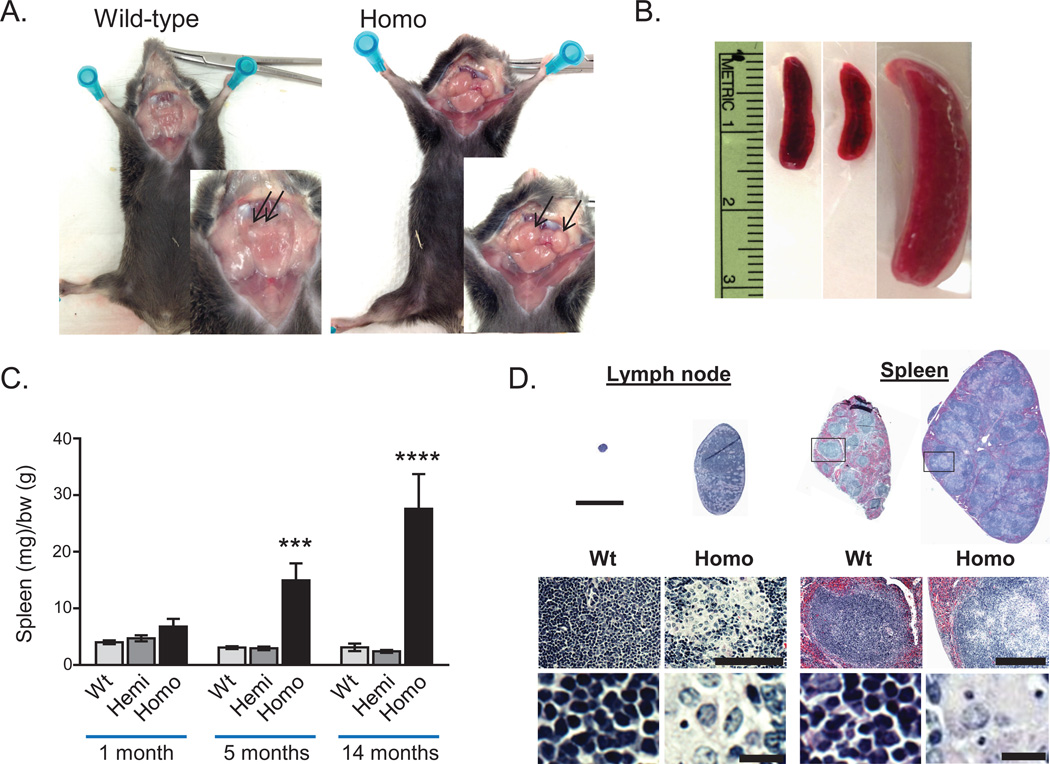
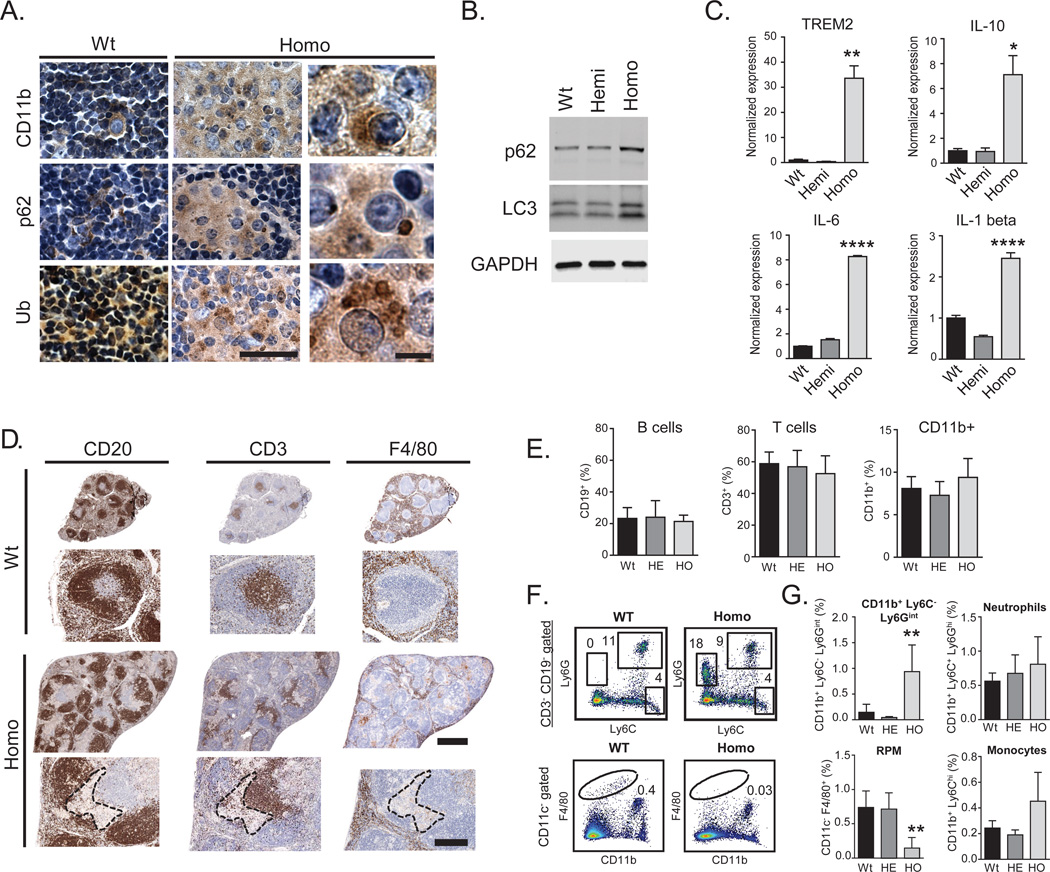
C9orf72−/− mice from both lines developed visibly enlarged cervical lymph nodes and spleens (Fig. 1A,B), detectable as early as 1 month, that slowly enlarged with age (Fig. 1C; Fig. S2). No gross or histological defects were observed in other organs at 5 months. Histology of lymph nodes and the white pulp of the spleen showed disruption of the normal follicular structure by enlarged debris-filled cells (Fig. 1D) that expressed CD11b and contained ubiquitin and p62 positive vacuoles consistent with macrophages (Fig. 2A; Fig. S4). Immunoblotting confirmed increased p62 and LC3, indicating an increase in components of the autophagy machinery in homozygote spleens (Fig. 2B). Massive upregulation of Trem2 expression was observed in C9orf72−/− spleens, a cell surface receptor expressed by macrophages/monocytes, as were inflammatory cytokines including IL-1 beta, IL-6, and IL-10 (Fig. 2C). Despite the altered follicular architecture, there were no differences in the proportion of B cells, T cells or CD11b+ myeloid cells (Fig. 2D; Fig. S2J). However, flow cytometry revealed changes in myeloid subsets, including emergence of a CD11b+Ly6C−Ly6Gint population unique to C9orf72−/− mice, and a decrease in F4/80+ red pulp macrophages, supporting a selective effect on myeloid populations in the spleen (Fig. 2E–G). Complete blood counts and flow cytometry of bone marrow were normal in C9orf72−/− mice at 5 months, (Fig. S5), supporting that splenic enlargement was not related to deficient hematopoiesis in bone marrow.
Fig. 1. Generation of C9orf72 (3110043O21Rik) null mice.
(A) Gross images of cervical lymphadenopathy (arrows) in C9orf72−/− mice (9 months of age). (B) Gross images of splenomegaly (12 months of age). (C) Spleen weights (mg) normalized to body weight (g) at indicated ages (***p=0.0008, ****p<0.0001, two-way ANOVA). (D) H&E staining of wild-type and homozygote lymph nodes and spleens at 5 months (top; scale bar = 3 mm) showing disruption of follicular architecture in null mice by large cells with swollen cytoplasm (below). Scale bars = 100µm, 10µm (lymph node) and 300µm and 10µm (spleen).
Fig. 2. C9orf72 null mice develop progressive splenomegaly with engorged macrophages, altered monocyte populations and inflammation.
(A) Enlarged cells in homozygote spleens (5 months) stained for CD11b, and contained p62 and ubiquitin (Ub) accumulations. Scale bar = 100 µm and 20 µm. (B) Western blot of spleen lysates showed an increase in p62 and LC3 in C9orf72−/− mice (n=3; 14 months). (C) qRT-PCR analysis of spleens (14 months) showed an increase in macrophage marker Trem2 (**p=0.008), and cytokines IL-10 (*p = 0.035), IL-6 (****p<0.0001), and IL-1 beta (****p<0.0001; one-way ANOVA). (D) Immunostains of wild-type and C9orf72−/− spleens (5 months) for CD20 (B cells), CD3 (T cells), and F4/80 (red pulp macrophages). Dotted outline highlights region of abnormal CD11b+ cells in the C9orf72−/− spleens. Scale bar = 1mm and 300 µm. (E) FACS analysis of spleens (5 months). (F) Dot plots and (G) bar graphs showed a unique population of CD11b+ Ly6C−Ly6Gint cells in C9orf72−/− spleens, and a decrease in F4/80+ red pulp macrophages compared to wild-type or hemizygotes (n=4; 5 months) (**p=0.01, one-way ANOVA).
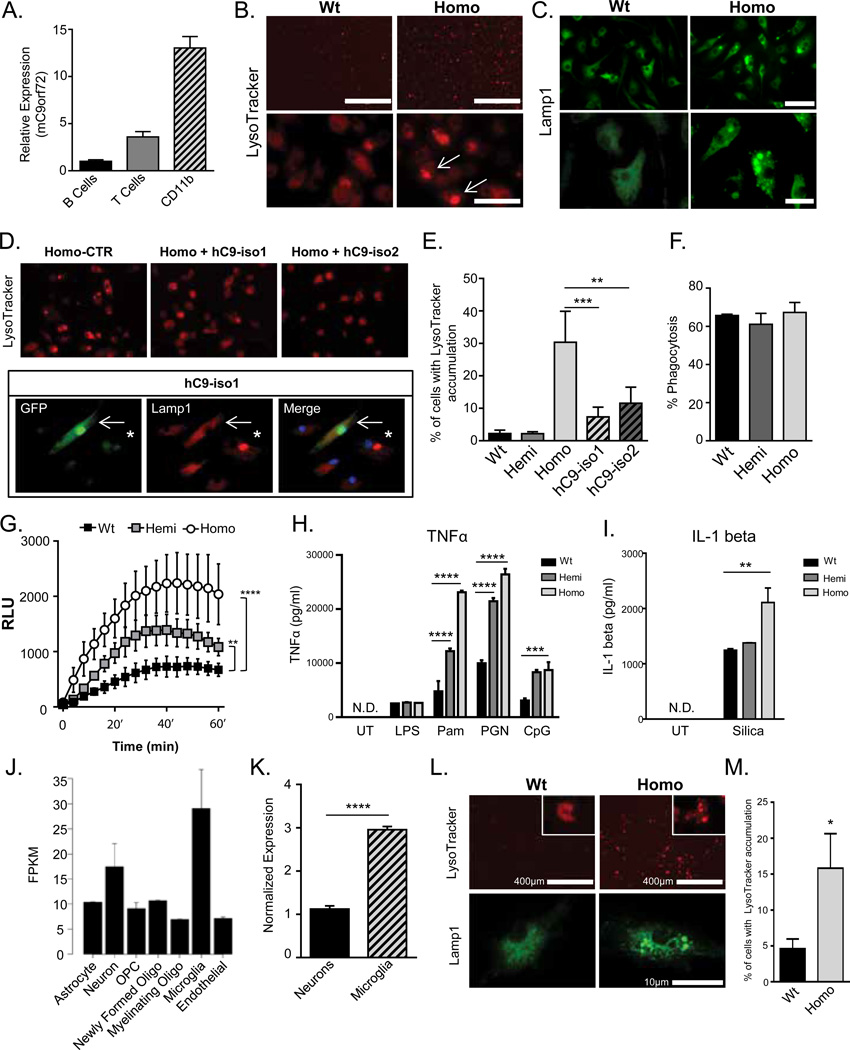
Given the progressive splenomegaly with altered myeloid cells, and the buildup of engorged macrophages with accumulations of LC3 and p62 in the spleens of C9orf72−/− mice, we hypothesized that C9orf72 protein is important for endosomal trafficking in macrophages. We first examined expression of C9orf72 by fluorescence-activated cell sorting (FACs) different populations from wild-type mouse spleens and found that C9orf72 was expressed at high levels in CD11b+ (myeloid cells), compared to CD3+ (T-cell) and CD19+ (B-cell) populations (Fig. 3A). Query of the immunological genome project (www.immgen.org) confirmed that expression of C9orf72 was highest in macrophages and dendritic cells compared to other immune cells (Fig. S6A,B). Pathway analysis (19) of the 35 genes in the C9orf72 constellation was significant for only one pathway, lysosomal function (Bonferroni p=2.32−6) (Fig. S6C). To examine whether C9orf72 is necessary for macrophage function, we isolated bone marrow derived macrophages (BMDMs) from C9orf72−/− mice and stained them for endosomal markers. BMDMs from C9orf72−/− mice showed marked accumulation of lysotracker and Lamp1 positive vesicles, supporting a defect in late endosome/lysosomal trafficking (Fig. 3B,C). No changes in the early or late endosomal markers Rab5 or Rab7 were observed (Fig. S7, S8). Accumulation of lysotracker and Lamp1 positive vesicles was rescued by viral expression of human C9orf72, indicating this defect is due to loss of C9orf72 (Fig. 3D,E). C9orf72−/− BMDMs showed normal initial phagocytosis of zymosan particles (Fig. 3F); however BMDMs from both C9orf72−/− and to a lesser extent C9orf72+/− mice showed enhanced production of phagocyte oxidase-derived reactive oxygen species (ROS) after feeding with zymosan particles (Fig. 3G), which has been reported in cells with defective fusion of phagosomes to lysosomes (20). BMDMs from C9orf72−/− and C9orf72+/− mice also showed enhanced cytokine production in response to several immune stimuli including those sensed in endosomal/lysosomal compartments such as peptidoglycan, CpG and silica (Fig. 3H,I). Thus, C9orf72 is critical for proper function of macrophages, and loss of C9orf72 leads to a pro-inflammatory state that likely drives the splenic and lymph node hyperplasia. While hemizygous mice did not have a phenotype at the tissue level, haploinsufficiency of C9orf72 led to altered inflammatory responses in macrophages at the cellular level, which could lead to a physiological phenotype when the system is stressed.
Fig. 3. Analysis of macrophages and microglia from C9orf72 deficient mice.
(A) qRT-PCR analysis from B cells, T cells, and CD11b+ cells FAC sorted from wild-type mouse spleen (n=2). (B,C) Bone marrow derived macrophages (BMDMs) from C9orf72−/− mice showed accumulation of LysoTracker and Lamp1 stained vesicles compared to wild-type (Wt). Scale bar = 50 µm and 20 µm. (D) C9orf72−/− BMDMs treated with lentivirus encoding either human C9orf72 isoform 1-IRES-GFP (hC9-iso1) or isoform 2-IRES-GFP (hC9-iso2). LysoTracker (top panel) or Lamp1(bottom panel) accumulation was rescued by either hC9-iso1 or hC9-iso2 (top panel). Arrow: hC9-iso1 infected cell; asterisk: uninfected cell. (E) Quantitation of LysoTracker accumulation in BMDMs of the indicated genotype, or homozygotes treated with hC9-iso1 and hC9-iso2 lentivirus. (***p=0.0002, **p=0.0018, one-way ANOVA). (F) BMDMs fed with fluorescent zymosan particles for 15 minutes and then analyzed by FACS analysis. (G) ROS production by BMDMs after zymosan ingestion in indicated genotypes (****p=<0.0001, two way ANOVA). (H) C9orf72+/− and C9orf72−/− BMDMs showed increased TNFα production after stimulation with Pam3CSK4 (Pam), peptidoglycan (PGN) and CpG, but not lipopolysaccharide (LPS) (****p<0.0001, ***p=0.0002, two way ANOVA. N.D. – not detected). (I) IL-1 beta production after stimulation with silica (*p<0.05, two-way ANOVA). (J) RNA-seq of C9orf72 in indicated cell types from the cerebral cortex (21). (K) qRT-PCR of C9orf72 from neurons and microglia isolated from adult mouse brain. (L) Microglia purified from C9orf72−/− mice showed accumulation of LysoTracker and Lamp1 positive enlarged vesicles. (M) Quantification of percentage of microglia with enlarged LysoTracker positive vesicles (*p=0.027, one-tailed t-test).
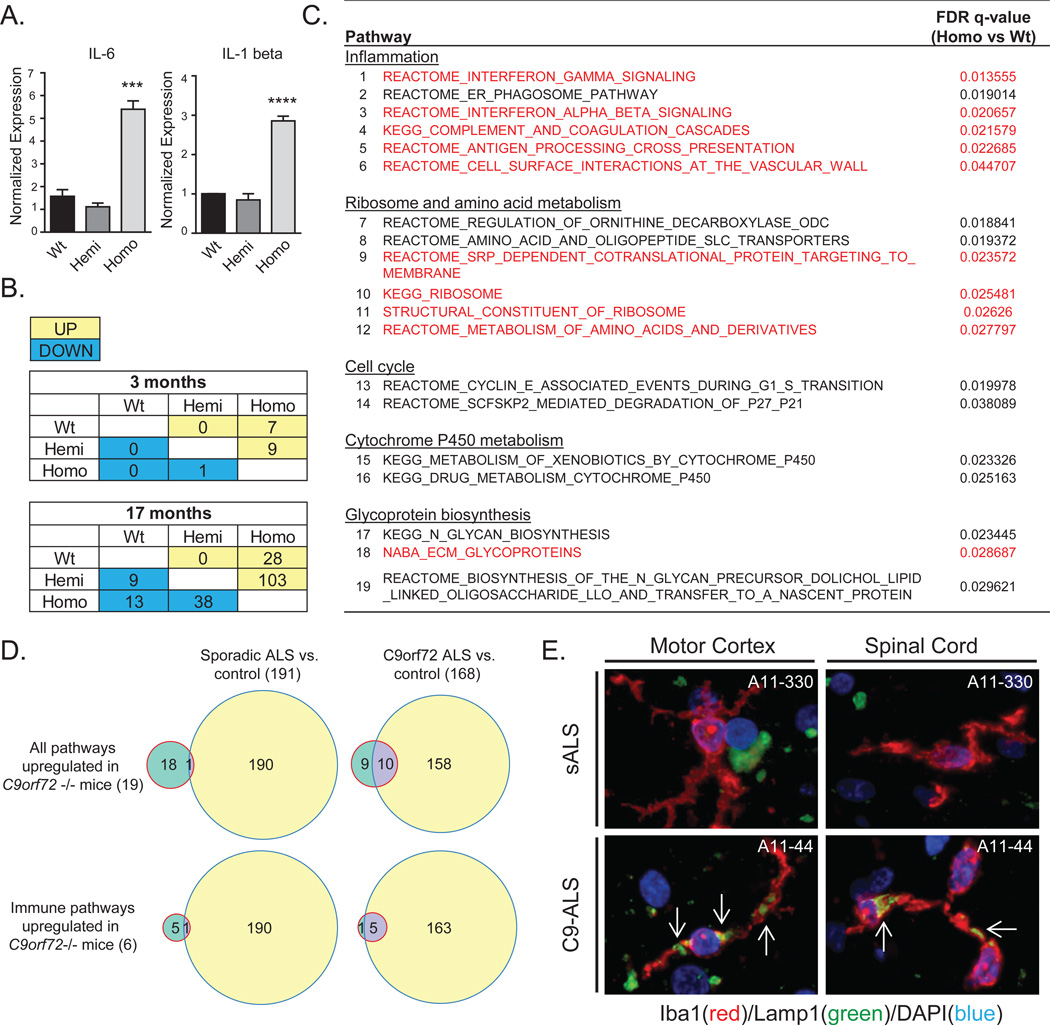
The defects in C9orf72−/− BMDMs raised the possibility that other myeloid cells, including resident microglia in the brain, also require C9orf72 for normal function. Although an earlier report suggested that microglia express low levels of C9orf72 (12), we observed that microglia showed the highest levels of C9orf72 expression of any cell type in the brain in published datasets (21–23) (Fig. 3J), and on qRT-PCR of cells isolated from adult mouse brain (Fig. 3K). Microglia from C9orf72−/− mice showed accumulation of lysotracker and Lamp1 positive structures, similar to BMDMs (Fig. 3L,M), while primary cortical neurons did not (Fig. S9). To probe the functional state of microglia lacking C9orf72 we performed qRT-PCR on spinal cord microglia isolated from C9orf72−/− mice, and found increased levels of cytokines IL-6 and IL-1b, supporting that the altered lysosomal function leads to a pro-inflammatory state (Fig. 4A) similar to that observed in BMDMs.
Fig. 4. Neuroinflammation in C9orf72−/− mice and C9orf72 expansion patient tissue.
(A) qRT-PCR of inflammatory cytokines (IL-6 and IL-1 beta) in microglia isolated from C9orf72−/− mice. (***p=0.0007; ****p=<0.0001, one-way ANOVA). (B) Tables showing the number of up and down-regulated pathways on GSEA (FDR<0.05) of RNA-seq from 3 and 17 month old lumbar spinal cords. (C) Table of up-regulated pathways in C9orf72−/− vs. C9orf72+/− and wild-type mouse spinal cords (FDR<0.05) at 17 months. Pathways up-regulated in both C9orf72−/− mice and human C9-ALS brain tissue are highlighted in red. (D) Top: Venn diagrams showing overlap between the 19 up-regulated pathways in C9orf72−/− mice from (C), and those up-regulated in cortex or cerebellum of sporadic ALS (left), or C9orf72 ALS (right). Bottom: Venn diagrams for the immune pathways from (C). (E) Human motor cortex and spinal cord from C9-ALS and sALS cases double-labelled with Iba1 (red) to identify microglia, and Lamp1 (green). Large accumulations of Lamp1 immunoreactivity (white arrows) were detected in activated microglia of C9-ALS but not sALS tissue.
Although we did not see overt neurodegeneration in C9orf72−/− mice, given the pro-inflammatory phenotype in isolated microglia, we used transcriptional profiling to investigate C9orf72 deficient nervous tissue in greater detail. Gene set enrichment analysis (GSEA) on RNA-seq of spinal cords from young animals (3 months) showed little difference between genotypes. By contrast in aged animals (17 months) a large number of pathways were altered in C9orf72−/− vs. C9orf72+/− or wild-type animals (FDR<0.05) (Fig. 4B). We focused on the 19 pathways upregulated in C9orf72−/− vs. C9orf72+/− and control animals for further analysis (Fig. S10). Of these 19 pathways, almost a third (6/19) were related to inflammation (Fig. 4C). To determine if similar changes are observed in C9orf72 ALS (C9-ALS) tissue, we analyzed a recent RNA-seq dataset that includes normal controls, sporadic ALS (sALS), and C9-ALS cases (24). Of the 19 upregulated pathways in C9orf72−/− mice, there was little overlap (1/19) with pathways upregulated in sporadic ALS brain tissue (frontal cortex or cerebellum; Fig 4D). By contrast, the majority (10/19) of pathways upregulated in C9orf72−/− mice were also upregulated in C9-ALS patient brains, including nearly all of the immune pathways (5/6), and a direct comparison showed a significant increase in inflammatory pathways in C9-ALS vs. sALS cases (Fig. S11). Finally, we performed immunostaining for Iba1 and Lamp1 on motor cortex and spinal cord tissue from C9-ALS (n=3) and sALS (n=3) cases. While frequent reactive microglia were present in all ALS cases, microglia containing large accumulations of Lamp1 positive material were only observed in the C9-ALS cases (Fig. 4E; Fig S11). Thus both transcriptome and histologic analysis of C9-ALS patient tissue are consistent with the idea that the decreased C9orf72 expression in C9-ALS leads to altered microglial function and neuroinflammation.
In summary, the loss of C9orf72 in mice led to age-related inflammation in the spleen and nervous system, with defects in lysosomal trafficking and immune responses in macrophages and microglia. The disruption of lysosomal function in macrophages is consistent with the idea that C9orf72 is a member of the DENN family of Rab-GEFs involved in late endosomal trafficking and autophagy (25–27). Our data support a model where C9orf72 regulates maturation of phagosomes to lysosomes in macrophages, as we observed both altered responses to immune stimuli including those sensed in endosomal/lysosomal compartments (PGN, CpG and silica) in BMDMs lacking C9orf72. Our findings also support that loss of C9orf72 function could impact neurodegeneration in C9-ALS and FTD, by diminishing the ability of microglia to clear aggregated proteins, and/or altering their immune responses. Of note our findings of altered immune responses in haploinsufficient macrophages support that even this partial decrease in C9orf72 levels could affect microglial function (3, 28–30). Furthermore, these data raise the possibility of a “dual-effect” mechanism for the pathogenesis of a single gene defect – that gain of function manifestations of C9orf72 expansion (RNA foci and RAN dipeptides) in neurons are coupled with “primed” and dysfunctional microglia, which ultimately results in neurodegeneration (31). Given that many ALS genes are involved in late endosomal trafficking and lysosome function (TBK1, TMEM106B, OPTN, SQSTM1, UBQLN2, VCP, CHMP2B, PGRN) (32), and are expressed in both neurons and microglia, the concept of a dual-effect mechanism may generalize to other forms of inherited ALS.
Finally, our findings raise important considerations about therapeutic knockdown of C9orf72 in the nervous system. While these approaches effectively target gain of function manifestations in neurons, they could exacerbate microglial dysfunction by further suppressing C9orf72, unless they specifically target repeat containing transcripts (33). Indeed an initial report of C9orf72 knockdown in mice using ASOs revealed up-regulation of immune markers in the nervous system including Trem2 and Tyrobp (34), suggesting that innate immune function should be monitored when performing C9orf72 knockdown strategies in humans.
Supplementary Material
Acknowledgments
We thank V. Funari for assistance with RNA-sequencing; A. Cammack for assisting with patient tissue, and A. Koehne for assisting with pathology evaluation. This work was supported by NIH Grants NS069669 (R.H.B), NS087351 (C.M.L), GM085796 (D.M.U.), NS078398 (T.M.M.), UL1TR000124, the Robert and Louise Schwab family, the Cedars-Sinai ALS Research Fund (R.H.B.), the Cedars-Sinai Board of Governors Regenerative Medicine Institute. Dr. Miller has served on medical advisory boards for Ionis Pharmaceuticals and Biogen Idec. Mouse line F12 is available through the Jackson Repository #27068, C57BL/6J-3110043O21Rik<em5Lutzy>/J. RNA-seq data are located in the Gene Expression Omnibus accession number GSE77681.
Footnotes
Supplementary Materials:
Materials and Methods
Figures S1–S11
External Databases: Gene Expression Omnibus accession GSE77681
Excel files: GSEA_17m_C9orf72_null ; GSEA_human_C9ALS_sALS
References and Notes
- 1.Lomen-Hoerth C, Anderson T, Miller B. The overlap of amyotrophic lateral sclerosis and frontotemporal dementia. Neurology. 2002 Oct 8;59:1077. doi: 10.1212/wnl.59.7.1077. [DOI] [PubMed] [Google Scholar]
- 2.Chen-Plotkin AS, Lee VM, Trojanowski JQ. TAR DNA-binding protein 43 in neurodegenerative disease. Nat Rev Neurol. 2010 Apr;6:211. doi: 10.1038/nrneurol.2010.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DeJesus-Hernandez M, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011 Oct 20;72:245. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Renton AE, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011 Oct 20;72:257. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cooper-Knock J, Shaw PJ, Kirby J. The widening spectrum of C9ORF72-related disease; genotype/phenotype correlations and potential modifiers of clinical phenotype. Acta Neuropathol. 2014 Mar;127:333. doi: 10.1007/s00401-014-1251-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hooten KG, Beers DR, Zhao W, Appel SH. Protective and Toxic Neuroinflammation in Amyotrophic Lateral Sclerosis. Neurotherapeutics. 2015 Jan 8; doi: 10.1007/s13311-014-0329-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baker M, et al. Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature. 2006 Aug 24;442:916. doi: 10.1038/nature05016. [DOI] [PubMed] [Google Scholar]
- 8.Cruts M, et al. Null mutations in progranulin cause ubiquitin-positive frontotemporal dementia linked to chromosome 17q21. Nature. 2006 Aug 24;442:920. doi: 10.1038/nature05017. [DOI] [PubMed] [Google Scholar]
- 9.Cady J, et al. TREM2 variant p.R47H as a risk factor for sporadic amyotrophic lateral sclerosis. JAMA neurology. 2014 Apr;71:449. doi: 10.1001/jamaneurol.2013.6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cirulli ET, et al. Exome sequencing in amyotrophic lateral sclerosis identifies risk genes and pathways. Science. 2015 Mar 27;347:1436. doi: 10.1126/science.aaa3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Freischmidt A, et al. Haploinsufficiency of TBK1 causes familial ALS and fronto-temporal dementia. Nat Neurosci. 2015 Mar 24; doi: 10.1038/nn.4000. [DOI] [PubMed] [Google Scholar]
- 12.Suzuki N, et al. The mouse C9ORF72 ortholog is enriched in neurons known to degenerate in ALS and FTD. Nat Neurosci. 2013 Dec;16:1725. doi: 10.1038/nn.3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koppers M, et al. C9orf72 ablation in mice does not cause motor neuron degeneration or motor deficits. Ann Neurol. 2015 Jun 5; doi: 10.1002/ana.24453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fratta P, et al. Homozygosity for the C9orf72 GGGGCC repeat expansion in frontotemporal dementia. Acta Neuropathol. 2013 Sep;126:401. doi: 10.1007/s00401-013-1147-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mizielinska S, Isaacs AM. C9orf72 amyotrophic lateral sclerosis and frontotemporal dementia: gain or loss of function? Curr Opin Neurol. 2014 Oct;27:515. doi: 10.1097/WCO.0000000000000130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang K, et al. The C9orf72 repeat expansion disrupts nucleocytoplasmic transport. Nature. 2015 Sep 3;525:56. doi: 10.1038/nature14973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Freibaum BD, et al. GGGGCC repeat expansion in C9orf72 compromises nucleocytoplasmic transport. Nature. 2015 Sep 3;525:129. doi: 10.1038/nature14974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jovicic A, et al. Modifiers of C9orf72 dipeptide repeat toxicity connect nucleocytoplasmic transport defects to FTD/ALS. Nat Neurosci. 2015 Sep;18:1226. doi: 10.1038/nn.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 20.Ma J, Becker C, Reyes C, Underhill DM. Cutting edge: FYCO1 recruitment to dectin-1 phagosomes is accelerated by light chain 3 protein and regulates phagosome maturation and reactive oxygen production. J Immunol. 2014 Feb 15;192:1356. doi: 10.4049/jimmunol.1302835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y, et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci. 2014 Sep 3;34:11929. doi: 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sharma K, et al. Cell type- and brain region-resolved mouse brain proteome. Nat Neurosci. 2015 Dec;18:1819. doi: 10.1038/nn.4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Butovsky O, et al. Identification of a unique TGF-beta-dependent molecular and functional signature in microglia. Nat Neurosci. 2014 Jan;17:131. doi: 10.1038/nn.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prudencio M, et al. Distinct brain transcriptome profiles in C9orf72-associated and sporadic ALS. Nat Neurosci. 2015 Aug;18:1175. doi: 10.1038/nn.4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang D, Iyer LM, He F, Aravind L. Discovery of Novel DENN Proteins: Implications for the Evolution of Eukaryotic Intracellular Membrane Structures and Human Disease. Frontiers in genetics. 2012;3:283. doi: 10.3389/fgene.2012.00283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levine TP, Daniels RD, Gatta AT, Wong LH, Hayes MJ. The product of C9orf72, a gene strongly implicated in neurodegeneration, is structurally related to DENN Rab-GEFs. Bioinformatics. 2013 Feb 15;29:499. doi: 10.1093/bioinformatics/bts725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Farg MA, et al. C9ORF72, implicated in amytrophic lateral sclerosis and frontotemporal dementia, regulates endosomal trafficking. Hum Mol Genet. 2014 Jul 1;23:3579. doi: 10.1093/hmg/ddu068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xi Z, et al. Hypermethylation of the CpG Island Near the GC Repeat in ALS with a C9orf72 Expansion. Am J Hum Genet. 2013 May 22; doi: 10.1016/j.ajhg.2013.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Belzil VV, et al. Reduced C9orf72 gene expression in c9FTD/ALS is caused by histone trimethylation, an epigenetic event detectable in blood. Acta Neuropathol. 2013 Dec;126:895. doi: 10.1007/s00401-013-1199-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Russ J, et al. Hypermethylation of repeat expanded C9orf72 is a clinical and molecular disease modifier. Acta Neuropathol. 2015 Jan;129:39. doi: 10.1007/s00401-014-1365-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perry VH, Holmes C. Microglial priming in neurodegenerative disease. Nat Rev Neurol. 2014 Apr;10:217. doi: 10.1038/nrneurol.2014.38. [DOI] [PubMed] [Google Scholar]
- 32.Peters OM, Ghasemi M, Brown RH., Jr Emerging mechanisms of molecular pathology in ALS. J Clin Invest. 2015 Jun;125:2548. doi: 10.1172/JCI82693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sareen D, et al. Targeting RNA foci in iPSC-derived motor neurons from ALS patients with a C9ORF72 repeat expansion. Science translational medicine. 2013 Oct 23;5:208ra149. doi: 10.1126/scitranslmed.3007529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lagier-Tourenne C, et al. Targeted degradation of sense and antisense C9orf72 RNA foci as therapy for ALS and frontotemporal degeneration. Proc Natl Acad Sci U S A. 2013 Nov 19;110:E4530. doi: 10.1073/pnas.1318835110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.