ABSTRACT
Chromatin structure and function are determined by a plethora of proteins whose genome-wide distribution is typically assessed by immunoprecipitation (ChIP). Here, we developed a novel tool to investigate the local chromatin environment at specific DNA sequences. We combined the programmable DNA binding of dCas9 with the promiscuous biotin ligase BirA* (CasID) to biotinylate proteins in the direct vicinity of specific loci. Subsequent streptavidin-mediated precipitation and mass spectrometry identified both known and previously unknown chromatin factors associated with repetitive telomeric, major satellite and minor satellite DNA. With super-resolution microscopy, we confirmed the localization of the putative transcription factor ZNF512 at chromocenters. The versatility of CasID facilitates the systematic elucidation of functional protein complexes and locus-specific chromatin composition.
KEYWORDS: biotinylation, CRISPR/Cas, CasID, chromatin composition, repetitive elements
Introduction
Regulation of gene expression involves a yet undetermined number of nuclear proteins ranging from tightly bound histones to loosely attached or transiently interacting factors that directly and indirectly bind DNA sequences along the genome. Establishment, maintenance and alteration of functional DNA states during development and disease requires dynamic changes in local enrichment and posttranslational modification of chromatin proteins. The genome-wide distribution of a given protein is traditionally determined by chromatin immunoprecipitation (ChIP) and subsequent sequencing of co-precipitated DNA fragments. However, ChIP experiments rely on the availability of suitable antibodies and provide data on global antigen distribution rather than local chromatin composition.
Previously described strategies to directly analyze chromatin complexes such as HyCCaPP (Hybridization Capture of Chromatin Associated Proteins for Proteomics)1 and PICh (Proteomic Isolation of Chromatin fragments)2 were based on chemical crosslinking and precipitation with complementary DNA probes. Alternatively, DNA binding proteins were used for chromatin precipitation and subsequent analysis by mass spectrometry.3-5
For visualization and manipulation, specific genomic loci can be targeted by different recombinant DNA binding proteins such as engineered polydactyl zinc finger proteins (PZFs),6 designer transcription activator-like effectors (dTALEs)7,8 or an enzymatically dead Cas9 (dCas9).9-11 Whereas target specificity of PZFs and dTALEs is determined by their amino acid sequence, DNA binding of dCas9 is programmed by an easily exchangeable single guide RNA (sgRNA).12
Here, we exploited the RNA-programmable DNA binding of dCas9 to direct a biotin ligase to specific genomic sites and mark adjacent chromatin proteins for subsequent identification by mass spectrometry. Proximity-dependent biotin identification (BioID) employs a promiscuous biotin ligase (BirA*) fused to a target protein for biotinylation of proteins within a 10 nm range.13,14 Biotinylated proteins can then be identified by robust streptavidin-mediated capture and subsequent mass spectrometry. Based on BirA* and dCas9 we developed a hybrid approach (CasID) to elucidate chromatin composition at specific DNA sequences.
Results and discussion
Immunofluorescence microscopy reveals protein biotinylation at targeted loci
To evaluate whether the CasID approach is suited to biotinylate proteins at specific genomic loci we constructed a BirA*-dCas9-eGFP fusion (Fig. 1). We co-transfected C2C12 myoblasts with this BirA*-dCas9-eGFP construct and a sgRNA plasmid, targeting dCas9 to either telomeres, major or minor satellite sequences. We previously showed that all sgRNAs used in this study successfully target dCas9-eGFP to the desired loci.10 Although here dCas9 is tagged on both N- (BirA*) and C-terminus (eGFP), we observed specific recruitment to the designated sequences (Supplementary Fig. 1). In control cells without sgRNA expression, BirA*-dCas9-eGFP shows a diffuse localization throughout the cell and a nucleolar enrichment (Supplementary Fig. 1). Importantly, in the presence of functional sgRNAs, BirA*-dCas9-eGFP was targeted to the respective loci and co-localized with a strong biotin signal, when the growth medium was supplemented with exogenous biotin (Fig. 2). These results demonstrate that the promiscuous biotin ligase BirA* can be directed to endogenous loci via dCas9.
Figure 1.
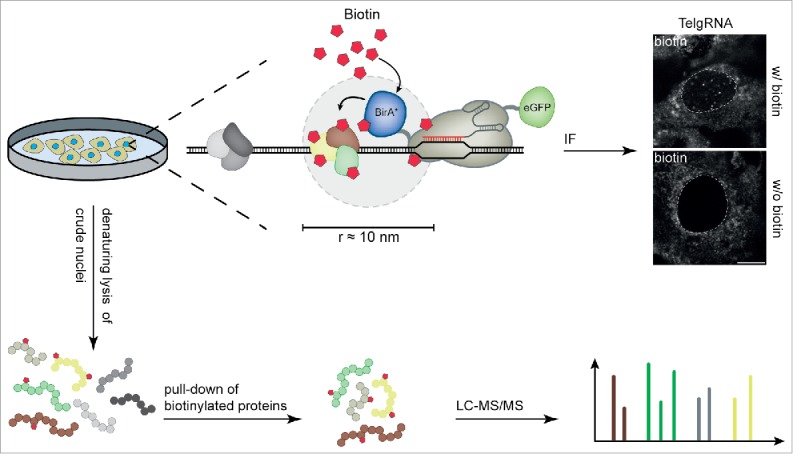
Workflow for CasID. BirA*-dCas9-eGFP/sgRNA expressing cells are cultured in growth medium without exogenous biotin. The BirA*-dCas9-eGFP fusion is directed to the desired target by sequence complementarity between sgRNA and the genomic locus. Upon addition of biotin to the medium, BirA* ligates biotin to lysine residues of proteins in close proximity. Successful biotinylation of locus-associated proteins can directly be visualized via immunofluorescence microscopy. For mass-spectrometric analysis, cells are harvested, followed by isolation of crude nuclei. After a denaturing lysis, biotinylated proteins can be pulled from the lysate with streptavidin and subjected to mass spectrometry. White dashed lines indicate the border between nucleus and cytoplasm. Scale bar: 10 µm.
Figure 2.
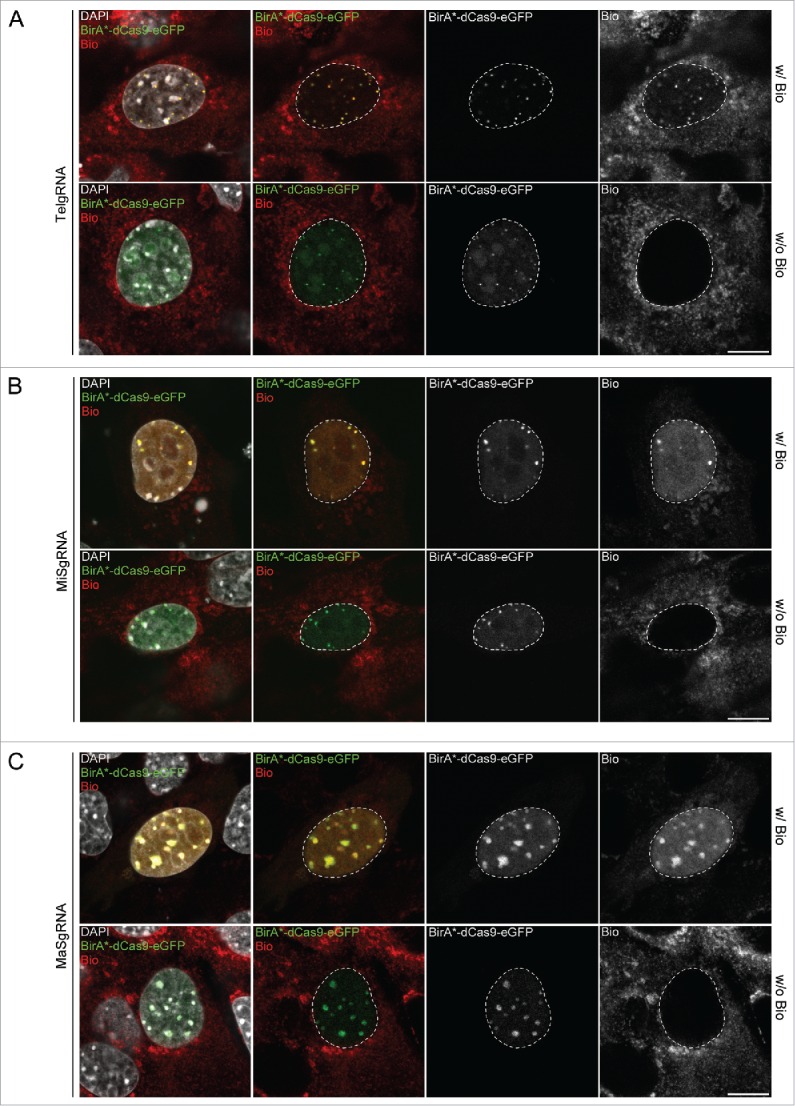
Targeted biotinylation of telomeres, major and minor satellites. Representative confocal images of C2C12 cells, co-transfected with CAG-BirA*-dCas9-eGFP and a plasmid encoding either telomere- (A, TelgRNA), minor satellite- (B, MiSgRNA) or major satellite-specific sgRNA (C, MaSgRNA). Nuclear enrichment of biotin at targeted sequences is only detectable after addition of exogenous biotin. White dashed lines indicate the border between nucleus and cytoplasm. Scale bar: 10 µm.
Determination of local chromatin composition at distinct genomic loci by mass spectrometry
To identify proteins associated with distinct genomic regions, cells stably expressing BirA*-dCas9-eGFP targeted to either telomeric regions, minor satellite repeats or major satellite repeats were supplemented with 50 µM biotin for 24 h, representing standard BioID conditions.13 We enriched for biotinylated proteins from crude nuclear extract with streptavidin-coated magnetic beads and analyzed them via tandem mass spectrometry (LC-MS/MS, Fig. 1). With label free quantification, we compared protein levels in pulldowns from cells expressing both BirA*-dCas9-eGFP and a sgRNA with control samples of cells stably expressing untargeted BirA*-dCas9-eGFP (without any sgRNA). Common BioID contaminants,15 like endogenously biotinylated mitochondrial carboxylases were found in all pulldowns including the negative control (Supplementary Table S1). Besides proteins predicted to associate with DNA, we also detected numerous unexpected proteins in our dataset (Supplementary Table S1) providing a basis for the identification of new chromatin factors and their future comprehensive characterization. For statistical analysis in a two-sided Student's T-test, only proteins present in at least 3 out of 4 biological replicates were included.
First, we targeted telomeric regions and observed a strong enrichment of several proteins when compared to pulldowns from control cells (Fig. 3A). Most prominent among these significantly enriched proteins were TERF2, TINF2 and ACD which are components of the shelterin complex known to directly bind telomeric DNA.16 We did not identify additional shelterin components which could be explained by sterical hindrances leading to an selective labeling of complex subunits. Altogether, these data show that CasID is suitable to investigate the native protein environment at specific genomic loci in mammalian cells.
Figure 3.

Chromatin composition of distinct genomic loci determined by mass spectrometry. Volcano plots of proteins enriched at telomeric regions (A), major satellites (B) and minor satellites (C), respectively. Black: significantly enriched/de-enriched proteins relative to BirA*-dCas9-eGFP control cells without sgRNA. FDR = 0.01, S0 = 0.1, n = 4. (See Table S1.) (D) Overlap between proteins identified at major satellites by CasID and candidates from PICh analysis.17 (E) Overlap between proteins significantly enriched at minor and major satellite repeats. (F) Localization of ZNF512-eGFP at major satellite repeats in transiently transfected C2C12 cells. Blow-ups depict DAPI and eGFP signal of boxed regions. Conventional confocal microscopy (upper panel) shows a homogeneous and strong association of ZNF512 at heterochromatin and high-resolution microscopy (3D-SIM, lower panel) reveals a network-like structure. Scale bars: 10 µm (confocal) and 5 µm (3D-SIM). Scale bars in blow-ups: 2 µm (confocal) and 1 µm (3D-SIM).
Second, we investigated the local protein environment at major satellite repeats. Here, we find not only known heterochromatic proteins such as MECP2, SMCHD1 and HP1BP3 but also previously uncharacterized proteins like ZNF512 (Fig. 3B). We validated the localization of ZNF512 by recombinantly expressing a GFP fusion (ZNF512-eGFP) which showed a distinct signal at heterochromatic loci in C2C12 cells (Fig. 3F). ZNF512 strongly associates with the major satellites also during mitosis (Supplementary Fig. 2), hinting at a structural or regulatory role for this protein throughout the cell cycle. One third of the proteins significantly enriched at major satellite repeats were also found in a data set obtained by PICh in mouse embryonic stem cells17 (Fig. 3D). Proteins found in both studies as well as those exclusively detected by CasID are categorized as DNA and RNA binding proteins or repressors (Supplementary Fig. 3A). In contrast to PICh, CasID requires BirA*-dCas9 to be introduced in target cells, yet it can be performed with considerably smaller sample sizes (∼4 × 107 vs. ∼8 × 108 cells per sample17) rendering CasID feasible and cost-effective. In total, fewer proteins were considered significant with CasID, which may be caused by a stringent statistical cutoff (FDR = 0.01) as well as the proximity-dependent nature of the CasID strategy. Collectively, these results validate CasID as a novel method to study local chromatin composition.
Third, we explored proteins in close proximity to minor satellite repeats and obtained both enriched and de-enriched proteins (Fig. 3C). To our knowledge, this is the first data set describing the protein environment of this genomic element. Among the significantly enriched proteins 12 annotated repressors or chromatin regulators and 25 DNA binding- or zinc finger motif containing-proteins were identified (Supplementary Fig. 3B). Furthermore, we find the known centromere-associated proteins CENPC18 and PCM1,19 which may reflect the close proximity of minor satellite repeats and centromeric regions or functions of these factors outside centromeres. Notably, the overlap between minor satellites and major satellite-associated proteins comprises only 9 out of 96 proteins (Fig. 3E), suggesting a distinct protein landscape of these two heterochromatic regions.
In summary, with CasID we developed a simple and robust workflow for in vivo labeling and systematic elucidation of locus specific chromatin composition that does not require prior cell fixation or protein cross-linking. We validated CasID for repetitive sequences where multiple Cas9 molecules are recruited to one target site. This approach could be extended to single copy loci by either using multiple sgRNAs, larger sample sizes and/or adapted pulldown conditions. In general, CasID experiments could be further fine-tuned by varying concentration and duration of biotin pulses and the use of a smaller biotin ligase (BioID2)20 with various linker lengths. While traditional ChIP techniques produce data on genome-wide distribution of specific antigens, CasID allows to study local chromatin composition including the identification of new factors. Therefore, ChIP and CasID are complementary approaches that bring together global and local views of dynamic and functional chromatin complexes and thus help to reveal how these complexes control structure and function of the genome and how they change during development and disease.
Material and methods
Cell culture and transfection
C2C12 cells21 were cultured at 37°C and 5 % CO2 in Dulbecco's modified Eagle's medium (DMEM, Sigma), supplemented with 20 % fetal bovine serum (FBS, Biochrom), 2 mM L-glutamine (Sigma), 100 U/ml penicillin and 100 µg/ml streptomycin (Sigma). For the CasID assay the culture medium was additionally supplemented with 50 µM biotin (Sigma) one day prior to analysis. For transfections, ∼5 × 105 cells were seeded in a p35 plate one day prior of transfection and transfections were performed with Lipofectamine® 3000 (Thermo Fisher Scientific) according to the manufacturer's instructions.
Plasmid generation
All plasmid and primer sequences can be found in Supplementary Tables S2 and S3, respectively. To generate the BirA*-dCas9-eGFP construct, BirA* was amplified from pcDNA3.1-mycBioID13 (Addgene plasmid #35700) with primers BirA*-F and BirA*-R. The resulting PCR product was ligated into the XbaI site of pCAG-dCas9-eGFP10 via Gibson Assembly (New England Biolabs). To generate the pEX-A-U6-sgRNA-PuroR plasmid, the PGK-PuroR cassette was amplified from pPthc-Oct3/422 and ligated into the SacI site of pEX-A-sgRNA10 via Gibson Assembly. sgRNA protospacer sequences were introduced into pEX-A-U6-sgRNA-PuroR by circular amplification as described previously.10 The Znf512-sequence was amplified from wt E14 cDNA with gene specific primers and cloned between the AsiSI/NotI sites of pCAG-eGFP23 via Gibson Assembly. The H2B-mRFP expression plasmid was described previously.24
Generation of stable cell lines
C2C12 cells were transfected with pCAG-BirA*-dCas9-eGFP using Lipofectamine® 3000 according to the manufacturer's instructions. Twenty-four h after transfection, the culture medium was supplemented with 10 µg/ml blasticidin S (Thermo Fisher Scientific). After two weeks of selection, eGFP-positive cells were single-cell sorted with a FACS Aria II (Becton Dickinson). A clonal cell line, stably expressing BirA*-dCas9-eGFP was used as entry cell line for transfections with sgRNA plasmids. Twenty-four h after transfection, the medium was supplemented with 2 µg/ml puromycin (Applichem). Two weeks after the start of selection, puromycin resistant cells were single-cell sorted. Individual clones (C2C12BirA*-dCas9-eGFP/sgRNA) were checked for correct BirA*-dCas9-eGFP localization by epifluorescence microscopy.
Immunofluorescence staining and image acquisition
Immunofluorescence staining was performed as described previously.25 Briefly, C2C12 cells transfected with pCAG-BirA*-dCas9-eGFP and the respective sgRNA were grown on coverslips (thickness 1.5H, 170 µm ± 5 µm; Marienfeld Superior), washed with phosphate buffered saline (PBS) 24 h after addition of 50 µM biotin and fixed with 3.7 % formaldehyde for 10 min. After permeabilization with 0.5 % Triton X-100 in PBS, cells were transferred into blocking buffer (0.02 % Tween, 2 % bovine serum albumin and 0.5 % fish skin gelatin in PBS) and incubated for 1 h. Antibodies were diluted in blocking buffer and cells were incubated with antibodies in a dark, humidified chamber for 1 h at room temperature (RT). Nuclei were counterstained with DAPI (200 ng/ml in PBS, 1 µg/ml in PBS for 3D-SIM). Coverslips were mounted with antifade medium (Vectashield, Vector Laboratories) and sealed with nail polish. Immuno-fluorescence in situ hybridization (FISH) detection of telomeres was performed as described previously.10 Primary antibodies used in this study were: anti-GFP (1:400, Roche), anti-H3K9me3 (1:500, Active Motif), anti-CENP-B (1:500, Abcam), Streptavidin conjugated to Alexa 594 (1:800, Dianova) and GFP-booster conjugated to Atto 488 (1:200, Chromotek). Secondary antibodies used in this study were: anti-rabbit IgG conjugated to Alexa 594 (1:400, Thermo Fisher Scientific) and anti-mouse IgG conjugated to Alexa 488 (1:300, Invitrogen).
Single optical sections or stacks of optical sections were acquired with a Leica TCS SP5 confocal microscope using a Plan Apo 63x/1.4 NA oil immersion objective. Super-resolution images were acquired with a DeltaVision OMX V3 3D-SIM microscope (Applied Precision Imaging, GE Healthcare), equipped with a 100x/1.4 Plan Apo oil immersion objective and Cascade II EMCCD cameras (Photometrics). Optical sections were acquired with a z-step size of 125 nm using 405 and 488 nm laser lines and SI raw data were reconstructed using the SoftWorX 4.0 software (Applied Precision). For long-term imaging experiments, C2C12 cells were seeded on 8-well chamber slides (ibidi) and transfected with ZNF512-eGFP and H2B-mRFP. Images were obtained with an UltraVIEW VoX spinning disc microscope (PerkinElmer), equipped with a 63x/1.4 NA Plan-Apochromat oil immersion objective and a heated environmental chamber set to 37°C and 5 % CO2. Confocal z-stacks of 12 µm with a step size of 2 µm were recorded every 30 min for ∼20 h. Image processing and assembly of the figures was performed with FIJI26 and Photoshop CS5.1 (Adobe), respectively.
Denaturing pulldown of biotinylated proteins and sample preparation for mass spectrometry
Four × 107 C2C12BirA*-dCas9-eGFP/sgRNA cells incubated for 24 h with 50 µM biotin were processed as described previously.27-29 In brief, cells were washed once in buffer A (10 mM HEPES/KOH pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 0.15 % NP-40, 1× protease inhibitor (SERVA)), then lysed in buffer A and homogenized using a pellet pestle. After centrifugation (15 min, 3200 rcf, 4°C), the pellet was washed once with PBS. Crude nuclei were resuspended in BioID lysis buffer (0.2 % SDS, 50 mM Tris/HCl pH 7.4, 500 mM NaCl, 1 mM DTT, 1× protease inhibitor), 0.2 % Triton-X100 was added and proteins were solubilized via sonication (Diagenode Bioruptor®, 200 W, 15 min, 30 s “on,” 1 min “off”). Lysates were 2-fold diluted with 50 mM Tris/HCl pH 7.4, centrifuged (10 min, 16000 rcf, 4°C) and the supernatant was incubated with 50 µl M-280 Streptavidin Dynabeads (Life Technologies) overnight at 4°C with rotation. A total of 5 washing steps were performed: once with wash buffer 1 (2 % SDS), wash buffer 2 (0.1 % desoxycholic acid, 1 % Triton X-100, 1 mM EDTA, 500 mM NaCl, 50 mM HEPES/KOH pH 7.5), wash buffer 3 (0.5 % desoxycholic acid, 0.5 % NP-40, 1 mM EDTA, 500 mM NaCl, 10 mM Tris/HCl pH 7.4) and twice with 50 mM Tris/HCl pH 7.4. Proteins bound to the streptavidin beads were digested as previously described.29 Beads were resuspended in digestion buffer (2 M Urea in Tris/HCl pH 7.5), reduced with 10 mM DTT and subsequently alkylated with 50 mM chloroacetamide. A total of 0.35 µg trypsin (Pierce, Thermo Scientific) was used for overnight digestion at RT. Desalting of peptides prior to LC-MS/MS analysis was performed via StageTips.30
LC-MS/MS analysis
Tandem mass spectrometry analysis was performed as described previously.27 In brief, reconstituted peptides (20 µl mobile phase A: 2% v/v acetonitrile, 0.1% v/v formic acid) were analyzed using a EASY-nLC 1000 nano-HPLC system connected to a LTQ Orbitrap Elite mass spectrometer (Thermo Fisher Scientific). For peptide separation, a PepMap RSLC column (75 μm ID, 150 mm length, C18 stationary phase with 2 µm particle size and 100 Å pore size, Thermo Fisher Scientific) was used, running a gradient from 5% to 35% mobile phase B (98% v/v acetonitrile, 0.1% v/v formic acid) at a flow rate of 300 nl/min. For data-dependent acquisition, up to 10 precursors from a MS1 scan (resolution = 60,000) in the range of m/z 250-1800 were selected for collision-induced dissociation (CID: 10 ms, 35% normalized collision energy, activation q of 0.25).
Computational analysis
Raw data files were searched against the UniprotKB mouse proteome database (Swissprot)31 using MaxQuant (Version 1.5.2.8)32 with the MaxLFQ label free quantification algorithm.33 Additionally to common contaminants specified in the MaxQuant “contaminants.fasta” file, a custom-made file containing sequences of BirA*-dCas9 and fluorescence proteins was included in the database search. Trypsin/P derived peptides with a maximum of 3 missed cleavages and a protein false discovery rate of 1 % were set as analysis parameters. Carbamidomethylation of cysteine residues was considered a fixed modification, while oxidation of methionine, protein N-terminal acetylation and biotinylation were defined as variable modifications.
For evaluation of the identified protein groups, Perseus (Version 1.5.2.6) was used.32 The data set was filtered for common contaminants classified by the MaxQuant algorithm and only proteins quantified in at least 3 out of 4 replicates per cell line were subjected to statistical analysis. For minor satellite repeats, the dataset was further filtered to exclude proteins only detected in the control sample. Missing values were replaced by a constant value of 17 for significance testing with a two-sided Student's T-test and a permutation based FDR calculation. Venn diagrams were obtained using the Webtool of the University of Gent (http://bioinformatics.psb.ugent.be/webtools/Venn/).
Supplementary Material
Abbreviations
- BioID
proximity dependent biotin identification
- ChIP
chromatin immuno precipitation
- dCas9
enzymatically dead Cas9
- eGFP
enhanced green fluorescent protein
- FDR
false discovery rate
- LC-MS/MS
liquid chromatography coupled to tandem mass spectrometry
- MaS
major satellite repeats
- MiS
minor satellite repeats
- PICh
Proteomic Isolation of Chromatin fragments
- sgRNA
single guide RNA
- Tel
telomere
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors would like to thank Joel Ryan (LMU Munich) and Susanne Leidescher (LMU Munich) for help with live cell and super-resolution imaging, suggestions on the manuscript and FISH staining. T.A., E.S. and P.R. are members of the DFG Graduiertenkolleg GRK1721. E.S. gratefully acknowledges the International Max Planck Research School for Molecular and Cellular Life Sciences.
Funding
This work was supported by the Deutsche Forschungsgemeinschaft [SFB1064 and SFB646 to H.L.] and the European Research Council [MolStruKT StG no. 638218 to F.H.]. Funding for open access charge: Deutsche Forschungsgemeinschaft.
References
- [1].Kennedy-Darling J, Guillen-Ahlers H, Shortreed MR, Scalf M, Frey BL, Kendziorski C, Olivier M, Gasch AP, Smith LM. Discovery of Chromatin-Associated proteins via sequence-specific capture and mass spectrometric protein identification in saccharomyces cerevisiae. J Proteome Res 2014; 13:3810-25; PMID:24999558; http://dx.doi.org/ 10.1021/pr5004938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Dejardin J, Kingston RE. Purification of proteins associated with specific genomic Loci. Cell 2009; 136:175-86; PMID:19135898; http://dx.doi.org/ 10.1016/j.cell.2008.11.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Fujita T, Asano Y, Ohtsuka J, Takada Y, Saito K, Ohki R, Fujii H. Identification of telomere-associated molecules by engineered DNA-binding molecule-mediated chromatin immunoprecipitation (enChIP). Scientific Reports 2013; 3:3171; PMID:24201379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Waldrip ZJ, Byrum SD, Storey AJ, Gao J, Byrd AK, Mackintosh SG, Wahls WP, Taverna SD, Raney KD, Tackett AJ. A CRISPR-based approach for proteomic analysis of a single genomic locus. Epigenetics 2014; 9:1207-11; PMID:25147920; http://dx.doi.org/ 10.4161/epi.29919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Grolimund L, Aeby E, Hamelin R, Armand F, Chiappe D, Moniatte M, Lingner J. A quantitative telomeric chromatin isolation protocol identifies different telomeric states. Nat Commun 2013; 4:2848; PMID:24270157; http://dx.doi.org/ 10.1038/ncomms3848 [DOI] [PubMed] [Google Scholar]
- [6].Klug A. The discovery of zinc fingers and their development for practical applications in gene regulation and genome manipulation. Quarterly Rev Biophys 2010; 43:1-21; PMID:20478078; http://dx.doi.org/ 10.1017/S0033583510000089 [DOI] [PubMed] [Google Scholar]
- [7].Miyanari Y, Ziegler-Birling C, Torres-Padilla ME. Live visualization of chromatin dynamics with fluorescent TALEs. Nat Structural Mol Biol 2013; 20:1321-4; PMID:24096363; http://dx.doi.org/ 10.1038/nsmb.2680 [DOI] [PubMed] [Google Scholar]
- [8].Thanisch K, Schneider K, Morbitzer R, Solovei I, Lahaye T, Bultmann S, Leonhardt H. Targeting and tracing of specific DNA sequences with dTALEs in living cells. Nucleic Acids Res 2013; PMID:24371265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Chen B, Gilbert LA, Cimini BA, Schnitzbauer J, Zhang W, Li GW, Park J, Blackburn EH, Weissman JS, Qi LS, et al.. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell 2013; 155:1479-91; PMID:24360272; http://dx.doi.org/ 10.1016/j.cell.2013.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Anton T, Bultmann S, Leonhardt H, Markaki Y. Visualization of specific DNA sequences in living mouse embryonic stem cells with a programmable fluorescent CRISPR/Cas system. Nucleus (Austin, Tex) 2014; 5:163-72; PMID:24637835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 2013; 152:1173-83; PMID:23452860; http://dx.doi.org/ 10.1016/j.cell.2013.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science 2013; 339:823-6; PMID:23287722; http://dx.doi.org/ 10.1126/science.1232033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Roux KJ, Kim DI, Raida M, Burke B. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J Cell Biol 2012; 196:801-10; PMID:22412018; http://dx.doi.org/ 10.1083/jcb.201112098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Kim DI, Birendra KC, Zhu W, Motamedchaboki K, Doye V, Roux KJ. Probing nuclear pore complex architecture with proximity-dependent biotinylation. Proc Natl Acad Sci U S A 2014; 111:E2453-61; PMID:24927568; http://dx.doi.org/ 10.1073/pnas.1406459111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Lambert JP, Tucholska M, Go C, Knight JD, Gingras AC. Proximity biotinylation and affinity purification are complementary approaches for the interactome mapping of chromatin-associated protein complexes. J Proteomics 2015; 118:81-94; PMID:25281560; http://dx.doi.org/ 10.1016/j.jprot.2014.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Liu D, O'Connor MS, Qin J, Songyang Z. Telosome, a Mammalian Telomere-associated complex formed by multiple telomeric proteins. J Biol Chem 2004; 279:51338-42; PMID:15383534; http://dx.doi.org/ 10.1074/jbc.M409293200 [DOI] [PubMed] [Google Scholar]
- [17].Saksouk N, Barth TK, Ziegler-Birling C, Olova N, Nowak A, Rey E, Mateos-Langerak J, Urbach S, Reik W, Torres-Padilla ME, et al.. Redundant mechanisms to form silent chromatin at pericentromeric regions rely on BEND3 and DNA methylation. Mol Cell 2014; 56:580-94; PMID:25457167; http://dx.doi.org/ 10.1016/j.molcel.2014.10.001 [DOI] [PubMed] [Google Scholar]
- [18].Guenatri M, Bailly D, Maison C, Almouzni G. Mouse centric and pericentric satellite repeats form distinct functional heterochromatin. J Cell Biol 2004; 166:493-505; PMID:15302854; http://dx.doi.org/ 10.1083/jcb.200403109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Balczon R, Bao L, Zimmer W. PCM-1, A 228-kD centrosome autoantigen with a distinct cell cycle distribution. J Cell Biol 1994; 124:783-93; PMID:8120099; http://dx.doi.org/ 10.1083/jcb.124.5.783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Kim DI, Jensen SC, Noble KA, Kc B, Roux KH, Motamedchaboki K, Roux KJ. An improved smaller biotin ligase for BioID proximity labeling. Mol Biol Cell 2016; 27(8):1188-96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Yaffe D, Saxel O. Serial passaging and differentiation of myogenic cells isolated from dystrophic mouse muscle. Nature 1977; 270:725-7; PMID:563524; http://dx.doi.org/ 10.1038/270725a0 [DOI] [PubMed] [Google Scholar]
- [22].Masui S, Shimosato D, Toyooka Y, Yagi R, Takahashi K, Niwa H. An efficient system to establish multiple embryonic stem cell lines carrying an inducible expression unit. Nucleic Acids Res 2005; 33:e43; PMID:15741176; http://dx.doi.org/ 10.1093/nar/gni043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Meilinger D, Fellinger K, Bultmann S, Rothbauer U, Bonapace IM, Klinkert WE, Spada F, Leonhardt H. Np95 interacts with de novo DNA methyltransferases, Dnmt3a and Dnmt3b, and mediates epigenetic silencing of the viral CMV promoter in embryonic stem cells. EMBO Rep 2009; 10:1259-64; PMID:19798101; http://dx.doi.org/ 10.1038/embor.2009.201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Martin RM, Cardoso MC. Chromatin condensation modulates access and binding of nuclear proteins. FASEB J 2010; 24:1066-72; PMID:19897663; http://dx.doi.org/ 10.1096/fj.08-128959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Solovei I, Cremer M. 3D-FISH on cultured cells combined with immunostaining. Methods Mol Biol 2010; 659:117-26; http://dx.doi.org/ 10.1007/978-1-60761-789-1_8 [DOI] [PubMed] [Google Scholar]
- [26].Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al.. Fiji: an open-source platform for biological-image analysis. Nat Methods 2012; 9:676-82; PMID:22743772; http://dx.doi.org/ 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Mulholland CB, Smets M, Schmidtmann E, Leidescher S, Markaki Y, Hofweber M, Qin W, Manzo M, Kremmer E, Thanisch K, et al.. A modular open platform for systematic functional studies under physiological conditions. Nucleic Acids Res 2015; PMID:26007658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Roux KJ, Kim DI, Burke B. BioID: A screen for protein-protein interactions. Curr Protoc Protein Sci 2013; 19(23):1-19.23.14; PMID:24510646 [DOI] [PubMed] [Google Scholar]
- [29].Baymaz HI, Spruijt CG, Vermeulen M. Identifying nuclear protein-protein interactions using GFP affinity purification and SILAC-based quantitative mass spectrometry. Meth Mol Biol 2014; 1188:207-26; http://dx.doi.org/ 10.1007/978-1-4939-1142-4_15 [DOI] [PubMed] [Google Scholar]
- [30].Rappsilber J, Mann M, Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat Protocols 2007; 2:1896-906; PMID:17703201; http://dx.doi.org/ 10.1038/nprot.2007.261 [DOI] [PubMed] [Google Scholar]
- [31].Consortium TU. UniProt: a hub for protein information. Nucleic Acids Res 2015; 43:D204-D12; PMID:25348405; http://dx.doi.org/ 10.1093/nar/gku989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotech 2008; 26:1367-72; http://dx.doi.org/ 10.1038/nbt.1511 [DOI] [PubMed] [Google Scholar]
- [33].Cox J, Hein MY, Luber CA, Paron I, Nagaraj N, Mann M. Accurate Proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol Cell Proteomics 2014; 13:2513-26; http://dx.doi.org/ 10.1074/mcp.M113.031591 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


