Abstract
Recombinant Escherichia coli is a desirable platform for the production of many biological compounds including poly(3-hydroxyalkanoates), a class of naturally occurring biodegradable polyesters with promising biomedical and material applications. Although the controlled production of desirable polymers is possible with the utilization of fatty acid feedstocks, a central challenge to this biosynthetic route is the improvement of the relatively low polymer yield, a necessary factor of decreasing the production costs. In this study we sought to address this challenge by deleting arcA and ompR, two global regulators with the capacity to inhibit the uptake and activation of exogenous fatty acids. We found that polymer yields in a ΔarcA mutant increased significantly with respect to the parental strain. In the parental strain, PHV yields were very low but improved 64-fold in the ΔarcA mutant (1.92–124 mg L−1) The ΔarcA mutant also allowed for modest increases in some medium chain length polymer yields, while weight average molecular weights improved by approximately 1.5-fold to 12-fold depending on the fatty acid substrate utilized. These results were supported by an analysis of differential gene expression, which showed that the key genes (fadD, fadL, and fadE) encoding fatty acid degradation enzymes were all upregulated by 2-, 10-, and 31-fold in an ΔarcA mutant, respectively. Additionally, the short chain length fatty acid uptake genes atoA, atoE and atoD were upregulated by 103-, 119-, and 303-fold respectively, though these values are somewhat inflated due to low expression in the parental strain. Overall, this study demonstrates that arcA is an important target to improve PHA production from fatty acids.
Electronic supplementary material
The online version of this article (doi:10.1186/s13568-016-0291-z) contains supplementary material, which is available to authorized users.
Keywords: Polyhydroxyalkanoates, Biodegradable polymer, Escherichia coli, Fatty acid metabolism, arcA, Beta-oxidation
Introduction
Poly(3-hydroxyalkanoates), or PHAs, are a group of biodegradable polyesters produced by a variety of microorganisms as a form of carbon storage (Lu et al. 2009). These PHAs are typically classified as short chain-length (SCL) PHAs, which contain repeating units of 3–5 carbons, and medium chain-length (MCL) PHAs containing 6–14 carbons. The physical properties of PHAs are dependent on monomer composition; SCL PHAs are generally stiff and brittle while MCL PHAs are more elastomeric, and co-polymerization of the two groups allows for great variability in material properties (Laycock et al. 2013). Previous studies have shown that MCL PHAs can be effectively produced in recombinant E. coli lacking the fatty acid degradation gene fadB when utilizing a related carbon source such as fatty acids, although the monomer composition of the resulting polymers was heterogenous and uncontrolled (Langenbach et al. 1997; Qi et al. 1997).
Recently, it was shown that the monomer identity can be precisely controlled in both PHA homo- and co-polymers synthesized by recombinant Escherichia coli strain LSBJ (Tappel et al. 2012a, b). This was accomplished by deleting both the fadB and fadJ genes in E. coli LS5218, recombinantly co-expressing the phaJ4 gene from Pseudomonas putida KT2440 with the highly active and broad substrate utilizing phaC1(STQK) genes, and feeding in specific ratios of fatty acids for conversion to PHAs (Fig. 1). This system allowed for strict control of repeating unit composition which enables great control over the physical properties of PHA polymers produced using this system, unlocking the potential for tailoring PHA materials for click-chemistry modifications (Levine et al. 2015, 2016; Pinto et al. 2016). Although these previous studies addressed control of the monomer composition and thus physical and chemical properties of PHA polymers, overall polymer yields were still relatively low and some fatty acid substrates had poor incorporation into either PHA homo- or copolymers. The previously defined system relied heavily on the deletion of the fadR gene in E. coli LS5218 for constitutive expression of the genes encoding enzymes from the β-oxidation pathway (Spratt et al. 1981). In a previous study, researchers demonstrated that the inhibition of β-oxidation intermediates using acrylic acid was an effective strategy for improving PHA biosynthesis, particularly in combination with a fadR deletion (Qi et al. 1998). In addition to FadR, there are three other transcriptional regulators, ArcA, OmpR, and CRP-cAMP, that are known to inhibit the expression of genes involved in β-oxidation. For this study, we focused on the regulators ArcA and OmpR because CRP-cAMP is known to act as a transcriptional activator of β-oxidation in the absence of glucose, and only exhibits repression when glucose is present (Fic et al. 2009). We hypothesized that removal of transcriptional regulators that inhibit expression of β-oxidation related genes would result in improved flux through fatty acid catabolic pathways to increase PHA polymer yields in our engineered system. Therefore, in this study E. coli LSBJ was engineered to improve the biosynthesis of PHA from fatty acid substrates by removing the global regulatory genes arcA and ompR.
Fig. 1.
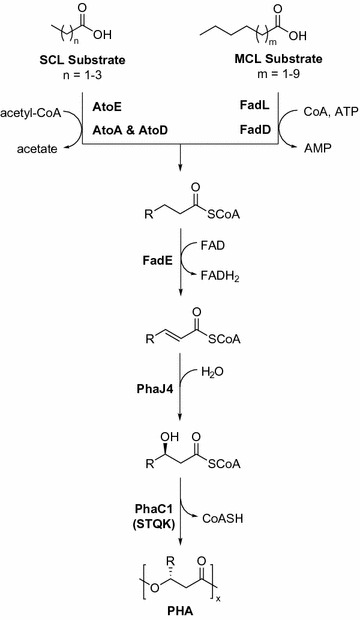
Biosynthesis of PHA in E. coli LSBJ utilizing short-chain-length (SCL) and medium-chain-length (MCL) fatty acids. The absence of fadB and fadJ in E. coli LSBJ in combination with the plasmid-borne recombinant enzymes PhaC1(STQK) and PhaJ4 establishes a linear pathway for the production of PHA polymers from free fatty acids. Extracellular fatty acids are transported across the outer membrane dependent on size; SCL and shorter MCL fatty acids can diffuse across the outer membrane, while longer MCL fatty acids can be transported by the long-chain fatty acid transporter FadL (Lepore et al. 2011). Inner membrane transport and activation is accomplished by the SCL-specific Ato system (AtoEAD) or the MCL-specific acyl-CoA synthetase FadD (Kameda and Nunn 1981; Theodorou et al. 2006). Acyl-CoA substrates are converted into enoyl-CoA by the acyl-CoA dehydrogenase enzyme FadE (Campbell and Cronan 2002), and are unable to proceed further through β-oxidation due to the absence of FadB and FadJ. The enoyl-CoA pool is then converted to (R)-3-hydroxyacyl-CoA by the R-specific enoyl-CoA hydratase PhaJ4 (Tsuge et al. 2003), and finally polymerized by the PHA synthase PhaC1(STQK) (Takase et al. 2003, 2004). This system allows for the biosynthesis of PHA polymers with tightly controlled repeating unit composition, as the number of carbons present in the fatty acid substrate is retained as the total number of carbons in each repeating unit
The transcriptional regulator OmpR functions as a response regulator of the two component regulatory EnvZ/OmpR system, which exhibits control over the expression of outer membrane porins in response to osmolarity (Mizuno and Mizushima 1990). The sensor kinase EnvZ autophosphorylates in response to extracellular osmolarity and transphosphorylates OmpR (OmpR-P), which binds to DNA and alters expression of genes within the OmpR regulon (Forst and Roberts 1994; Matsubara and Mizuno 1999). The most well-studied members of the OmpR regulon are OmpF and OmpC, outer membrane porins that control the diffusion of small hydrophilic molecules (Aiba and Mizuno 1990; Mizuno and Mizushima 1990; Silhavy and Pratt 1995). However, OmpR controls numerous other transporter genes including fadL (Table 1), as well as genes involved in amino acid metabolism and flagellar biosynthesis (Higashitani et al. 1993; Oshima et al. 2002).
Table 1.
Regulation Targets of ArcA and OmpR
| Target Gene | Description | Reference |
|---|---|---|
| arcA | ||
| fadL | Long-chain fatty acid transporter, experimental evidence | Cho et al. (2006) |
| fadD | Acyl-CoA synthetase, experimental evidence | Cho et al. (2006) |
| fadE | Acyl-CoA dehydrogenase, experimental evidence | Cho et al. (2006) |
| ompR | ||
| fadL | Long-chain fatty acid transporter, predicted | Higashitani et al. (1993) |
The global transcriptional dual regulator ArcA functions as the response regulator of the two-component regulatory ArcAB system, which regulates the expression of genes involved in aerobic and anaerobic metabolism in response to oxygen availability (Iuchi and Ec 1988; Lynch and Lin 1996). During conditions of decreasing oxygen availability, ArcA is activated through a phosphorelay mechanism and binds to the consensus sequence 5′-wGTTAATTAw-3′ (w is A or T) located in numerous genes, including several genes involved in β-oxidation shown in Table 1, and acts as either a repressor or activator (Iuchi and Lin 1992; Lynch and Lin 1996; Liu and Wulf 2004; Cho et al. 2006; Peña-Sandoval and Georgellis 2010). It has previously been shown that the redox biochemistry and transcriptional regulation of an ΔarcA mutant strain is significantly altered during microaerobic growth conditions, and to a lesser degree during aerobic growth conditions (Oshima et al. 2002; Alexeeva et al. 2003; Shalel-Levanon et al. 2005). The work presented here demonstrates for the first time an arcA deletion mutant combined with a fadR mutation to improve the biosynthesis of PHA polymers derived from fatty acid substrates.
Methods
Materials
A complete list of strains, plasmids, and primers used for this study is shown in Table 2. All strains were grown in Lennox Broth (LB; composition per liter: 10 g tryptone, 5 g yeast extract, and 5 g sodium chloride, pH 7.0) purchased from Difco, and the antibiotics kanamycin (50 mg L−1) and ampicillin (100 mg L−1) were added to media throughout the experiment as appropriate. The fatty acids sodium butyrate (Alfa Aesar), pentanoic acid (Alfa Aesar), hexanoic acid (Alfa Aesar), heptanoic acid (Alfa Aesar), sodium octanoate (Sigma Aldrich), decanoic acid (Alfa Aesar), and dodecanoic acid (Acros Organics) were used as feed supplements for PHA production (12 mM), along with the surfactant Brij-35 (Fisher Scientific, 4.0 g L−1). Sodium phosphate dodecahydrate (Acros Organics, 8 mM) was added when noted. Sodium hydroxide (5 M) was used to adjust the pH to 7.0 when necessary. Primers were ordered from Integrated DNA Technologies (IDT). ACS HPLC-grade chloroform and methanol were used for gas chromatography (GC) sample preparation and polymer purification.
Table 2.
Strains, plasmids, and primers
| Escherichia coli | Relevant characteristics | Source or reference |
|---|---|---|
| LSBJ | fadB::Cm, ΔfadJ, atoC512 (Const), fadR601 | Tappel et al. (2012a) |
| RSC02 | ΔarcA LSBJ | This study |
| RSC04 | ΔompR LSBJ | This study |
| RSC06 | ΔarcA, ΔompR LSBJ | This study |
| Plasmids | ||
| pKD46 | λ Red recombinase expression plasmid; expresses exo, β, and γ genes from λ phage; P-araB promoter; araC; AmpR; temperature sensitive replicon | Datsenko and Wanner (2000) |
| pKD13 | Neomycin phosphotransferase flanked by FLP recombinase recognition targets, AmpR, KmR | Datsenko and Wanner (2000) |
| pCP20 | FLP recombinase expression plasmid, AmpR, temperature sensitive replicon | Datsenko and Wanner ( 2000) |
| pBBR-C1J4SII | pBBR1MCS-2 derivative ΔphaAB, phaJ4, phaC1 (STQK) | Tappel et al. (2012b) |
| Primers a,b | Sequence (5′ to 3′) | |
| pKD13.F.arcA | ATGCAGACCCCGCACATTCTTATCGTTGAAGACGAGTTGGTAACACGCAAGTGTAGGCTGGAGCTGCTTC | |
| pKD13.R.arcA | TTAATCTTCCAGATCACCGCAGAAGCGATAACCTTCACCGTGAATGGTGGATTCCGTGGATCCGTCGACC | |
| pKD13.F.ompR | ATGCAAGAGAACTACAAGATTCTGGTGGTCGATGACGACATGCGCCTGCGGTGTAGGCTGGAGCTGCTTC | |
| pKD13.R.ompR | TTAGAACATTACCTTATGACCGTACTGCTCAAGAATGCCTTTCACGCGTTATTCCGTGGATCCGTCGACC | |
| arcA.check.F/R | GTTAATTTGCAGCATGCATCAGG/GACGATGAGTTACGTATCTGG | |
| ompR.check.F/R | AAATTGTTGCGAACCTTTGG/GCAATAACGTACGGGCAAAT | |
| qAtoA.F/R | GGTGCAGCCATGTTTGATAG/CGCGAGGTTTGCTTCTTC | |
| qAtoD.F/R | ACTTGGCAACCTGACCTATC/GACCAGTTCATCTGGCTCTAC | |
| qAtoE.F/R | ACTCGGTATCGCTTACCTTG/GCAGACCCGCAATCATAAAC | |
| qFadD.F/R | TCTCCAGTCTGCATCTTTCC/CCATAGCCTTCCAGCAGATAC | |
| qFadE.F/R | TTACCCGTCTGGATGAACTG/GACGGCTTTCTTCAGCTTTC | |
| qFadL.F/R | GGGCGCTTCTATTACCTCTAA/TTTCAAGGTCGGTTGTACCC | |
| qRpoD.F/R | GAGCAAGGCTATCTGACCTATG/GCCCATGTCGTTGATCATTTG | |
aUnderlined sequences are homologous to the gene to be deleted
bForward and reverse primers are denoted with an F or R, respectively, and primers used for qPCR are denoted with a q
Gene deletions
The deletion of the arcA and ompR genes was accomplished using the λ red recombinase protocol, a commonly used method for nonpolar gene deletion, as previously described (Datsenko and Wanner 2000; Tappel et al. 2012b). Briefly, knockout cassettes were generated using PCR with gene-specific primers and the kanamycin resistance marker from pKD13 (Table 2). PCR was performed using PrimeSTAR HS polymerase (Takara) following the manufacturers recommended protocol. The λ red recombinase was expressed using plasmid pKD46, and knockout cassettes introduced by electroporation (1500 V, 5 ms; BTX ECM 399). Successfully recombination was determined by antibiotic selection and loci screening using check primers (Table 2). Antibiotic resistance was removed by the expression of FLP recombinase from the pCP20 plasmid, and successful deletions were confirmed by loss of antibiotic resistance and by PCR using loci check primers (Table 2). Deletion mutants ΔarcA, ΔompR, and the double deletion ΔarcA ΔompR) were named RSC02, RSC04, and RSC06, respectively (Table 2).
PHA production
Protocols for PHA production and cell harvest were adapted from a previous study (Tappel et al. 2012b), with several modifications. For initial preliminary experiments, LSBJ, RSC02, RSC04, and RSC06 were made chemically competent and transformed with pBBR-C1J4SII following standard procedures (Sambrook and Russell 2001), to express PHA synthase and enoyl-CoA hydratase. Transformants were grown on LB-agar plates, and single colonies were used to inoculate separate 2 mL LB seed cultures, in triplicate for each strain. Seed cultures were grown for 16 h at 37 °C and 200 rpm, and used to inoculate 100 mL of growth media in 500 mL baffled shake flasks (to final concentration of 0.5%). Growth media contained LB, Brij-35, decanoic acid, and kanamycin. Cultures were grown for 48 h at 30 °C and 250 rpm on a rotary shaker, and were then harvested following previously published methods (Tappel et al. 2012b).
A more robust test of PHA homopolymer production was performed between LSBJ and RSC02 using the methods described above, with two key differences. A variety of fatty acids were tested in the growth media: sodium butyrate, pentanoic, hexanoic, heptanoic, sodium octanoate, decanoic, and dodecanoic. In addition, sodium phosphate dodecahydrate was added to the growth media for these experiments, which acts as a buffer for the shorter chain fatty acids. This addition also provides a significant source of phosphate to the media, so to keep growth conditions consistent, sodium phosphate dodecahydrate was added to every other experiment in this study.
GC analysis
The yields and repeating unit compositions of PHA polymers were determined using GC, as previously described (Braunegg et al. 1978; Tappel et al. 2012b). Briefly, dried cells (15–20 mg) were dissolved in 2 mL of sulfuric acid: methanol solution (15:85) and 2 mL of chloroform and heated at 100 °C for 140 min in a 10 mL pressure vial (Kimax). The samples were cooled to room temperature followed by the addition of 1 mL of Nanopure filtered water, after which all samples were mixed by vortex. Aqueous and organic layers were allowed to separate for 20 min. The organic layer was passed through a 0.45 μm polytetrafluoroethylene (PTFE) syringe filter (Restek). An aliquot of 500 μL of each filtered sample was mixed with 500 μL of methyl octanoate standard (1 gL−1) in chloroform in a 2 mL GC vial. Samples were injected and separated using a GC 2010 Gas Chromatograph with an AOC-20i autoinjector with a flame ionization detector. Shimadzu’s GCSolution software was used to analyze the data, and statistical significance of triplicate samples was determined using a two-tailed Student’s t test with a 95% confidence interval (α = 0.05).
Polymer purification and molecular weight determination
PHA homopolymers were extracted from residual dried cell samples from the LSBJ and RSC02 biosynthesis experiments by combining each set of triplicate samples into single 10 mL pressure vials (Kimax), adding 6 mL of chloroform, and incubating at 100 °C for 1 h. Each sample was filtered through a 0.45 μm PTFE syringe filter (Restek) into a 20 mL scintillation vial and rinsed twice with 2 mL aliquots of chloroform. Samples were concentrated to relative dryness using a rotary evaporator, and redissolved in 1 mL chloroform. Crude polymers were purified by non-solvent precipitation in cold methanol as described previously (Pinto et al. 2016), with several modifications. Briefly, dissolved samples were added dropwise to 10 mL of ice-cold methanol (4 °C) with rapid stirring. The solution was centrifuged (3452×g, 30 min, 4 °C) to pellet the PHA, decanted and washed with an additional 5 mL of methanol, and re-centrifuged. The supernatant was decanted, and the pellet dissolved in approximately 2 mL of chloroform to transfer to a scintillation vial. Samples were concentrated in a rotary evaporator, and evaporated to dryness under high vacuum for 4 h.
The weight average (Mw) and number average (Mn) molecular weights for each sample were determined by gel permeation chromatography (GPC) as described previously (Pinto et al. 2016). Briefly, PHA solutions of approximately 1.0 g L−1 were prepared by dissolution in chloroform and passed through a syringe filter (0.45 μm PTFE). Samples were injected (50 μL) into a Shimadzu LC-20AD liquid chromatograph equipped with a Shimadzu SIL-20A autosampler, a Shimadzu CTO-20A column oven, and a Shimadzu RID-10A refractive index detector. Samples were passed through an 8 × 50 mm styrenedivinylbenzene (SDV) guard column (5 μm particles; Polymer Standards Service) and an 8 × 300 mm SDV analytical column (5 μm particles; mixed bed porosity; max molecular weight 1E6 Da; Polymer Standards Service product sda0830051lim). The column oven was maintained at 40 °C with a 1 mL min−1 mobile phase of chloroform. Molecular weight standards of polystyrene with a narrow polydispersity index were used for calibration. Shimadzu’s LCsolution software was used to analyze the data. GPC chromatograms are available as supplemental material (Additional file 1: Figure S1).
Growth profiles of LSBJ and RSC02
An analysis of the growth profiles for LSBJ and RSC02 was performed under the PHA homopolymer biosynthesis conditions utilizing two separate substrates, sodium butyrate and decanoic acid. Media and growth conditions were identical to those in the PHA production section. A 1 mL aliquot was removed from each culture to measure the OD600 every hour using a spectrophotometer (Genesys 10S) until the stationary phase was observed. A 5 mL aliquot was removed from each culture at both 24 and 48 h to analyze PHA concentrations. The 5 mL samples were harvested and analyzed by GC as described above.
RNA extraction and qPCR analysis
RNA from E. coli LSBJ and RSC02 was isolated and purified as described previously (Lundgren et al. 2013; Sarwar et al. 2016). To isolate RNA for real-time quantitative PCR (qPCR), each strain was grown in duplicate in 100 ml of growth media (as described above, with sodium phosphate dodecahydrate and decanoic acid) in 500 ml baffled shake flasks at 30 °C and 250 rpm to an OD600 of ~0.6. Cultures were immediately stabilized by adding 1 ml of RNAprotect Bacteria reagent (Qiagen) to 0.5 ml of culture. Cells were then lysed with lysozyme and proteinase K as described in the manufacturer’s protocol. The total RNA was subsequently purified from the lysed cells with the RNeasy minikit (Qiagen) by using an on-column DNase digestion step. PCR and a Bioanalyzer were used to check the RNA for DNA contamination, quality, and concentration.
The iScript cDNA synthesis kit (Bio-Rad) was used to generate cDNA from 1 μg of the purified RNA samples. A tenfold dilution series of the pooled cDNA from the two duplicate RNA samples from LSBJ or RSC02 was used for the qPCR experiments. The qPCR experiments were performed in triplicate. The expression of several important genes for β-oxidation was normalized to rpoD, a housekeeping gene with stable expression during exponential growth (Jishage et al. 1996). Primers for qPCR were designed to produce ~100 bp amplicons of each of the following genes: atoA, atoD, atoE, fadD, fadE, fadL, and rpoD (Table 2). The qPCR mixtures contained 300 mM of each primer, 10 μl of the 2 × iQ SYBR green Supermix (Bio-Rad), 5 μl of diluted cDNA, and nuclease-free water to a total volume of 20 μl. qPCR was performed on the MiniOpticon system (Bio-Rad) with the following conditions: 1 cycle of 95 °C for 2 min and 40 cycles of 95 °C for 15 s, 55 °C for 30 s, and 72 °C for 30 s. Window-of-linearity R 2 values and amplification efficiency values ranged from 0.990 to 1.0 and 90.0% to >100%, respectively. The dilution series with the highest R 2 values was used to calculate relative gene expression of RSC02 compared to LSBJ using the Pfaffl method (Fleige et al. 2006). Amplification efficiencies and threshold cycle (Cq) values were calculated using the program LinRegPCR (Ruijter et al. 2009).
Results
PHA homopolymer production
Our goal was to develop a strain capable of producing PHA polymers with controlled repeating unit compositions and increased yields. To achieve this, the mutant strains RSC02, RSC04, and RSC06 were derived from E. coli LSBJ by the deletion of arcA, ompR, and a tandem arcA/ompR deletion, respectively. The amount of poly(3-hydroxydecanoate) (PHD) produced by these strains was then compared to E. coli LSBJ while expressing PhaJ4 and PhaC1(STQK) in a set of preliminary experiments. RSC02 produced significantly more PHD than other strains, with a yield of 0.353 g L−1 (Fig. 2). Strains RSC04 and RSC06 were not found to be significantly different from LSBJ (Fig. 2; Additional file 1: Table S1).
Fig. 2.
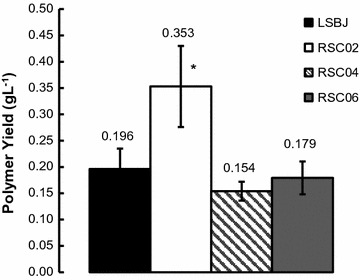
Comparison of poly(3-hydroxydecanoate) (PHD) yield between LSBJ and mutant strains RSC02, RSC04, and RSC06. The average yield achieved by RSC02 is significantly greater than that of LSBJ, while RSC04 and RSC06 are not significantly different from LSBJ. All values are averages of triplicate experiments plus or minus the standard deviation around those averages. An asterisk denotes a statistically significant difference compared to LSBJ (Student’s t-test, two-tailed, α = 0.05)
To further investigate the effect of the ΔarcA mutation on PHA polymer production, PHA homopolymer biosynthesis was characterized utilizing a variety of fatty acids. The fatty acids used for the production of PHA homopolymers were sodium butyrate, valeric acid, hexanoic acid, heptanoic acid, sodium octanoate, decanoic acid, and dodecanoic acid respectively. Analysis by GC showed significant increases in the amount of polymer produced by E. coli RSC02 when compared to E. coli LSBJ for nearly all fatty acid substrates, particularly for those with six or fewer carbons (Table 3). Overall the parental strain, LSBJ, produced very little short chain-length PHAs, with yields of only 3.04 mg L−1 of poly(3-hydroxybutyrate) (PHB) and 1.92 mg L−1 of poly(3-hydroxyvalerate) (PHV), and only 44.8 mg L−1 of poly(3-hydroxyhexanoate) (PHHx), the shortest medium chain-length polymer (Table 3). We observed significant increases in the amount of PHA synthesized by RSC02 of approximately 3750, 6360, and 485% when cells utilized sodium butyrate, pentanoic acid, or hexanoic acid as substrates, respectively (Table 3). For these shorter chain-length PHA polymers the identity was confirmed by 1H-NMR due to the large differences in production between strains (Additional file 1: Figure S2). Of those PHA polymers tested with greater than 6 carbons per repeating unit, only poly(3-hydroxyheptanoate) (PHHp) and PHD yields were significantly different between strains, with an increase of approximately 61 and 115 percent observed for RSC02, respectively (Table 3).
Table 3.
PHA yield comparison between LSBJ and RSC02
| Substrate | Strain | CDW (gL−1)a | PHAa (wt%) | PHA concentration (mg L−1)a | Percent increaseb |
|---|---|---|---|---|---|
| Sodium butyrate | LSBJ | 0.80 ± 0.03 | 0.38 ± 0.05 | 3.04 ± 0.27 | 3750 |
| RSC02 | 0.85 ± 0.03 | 13.7 ± 0.95 | 117 ± 11.1* | ||
| Pentanoic acid | LSBJ | 0.76 ± 0.05 | 0.23 ± 0.02 | 1.92 ± 0.42 | 6360 |
| RSC02 | 1.14 ± 0.04* | 11.1 ± 1.01 | 124 ± 7.25* | ||
| Hexanoic acid | LSBJ | 0.85 ± 0.01 | 5.47 ± 0.97 | 44.8 ± 6.13 | 485 |
| RSC02 | 1.06 ± 0.01* | 27.3 ± 4.28 | 262 ± 59.0* | ||
| Heptanoic acid | LSBJ | 0.93 ± 0.01 | 23.4 ± 0.48 | 198 ± 4.27 | 61 |
| RSC02 | 1.02 ± 0.06 | 30.2 ± 2.28 | 319 ± 22.8* | ||
| Sodium octanoate | LSBJ | 1.22 ± 0.05 | 44.5 ± 8.68 | 543 ± 110 | 1.10 |
| RSC02 | 1.03 ± 0.01* | 54.1 ± 1.06 | 549 ± 12.5 | ||
| Decanoic acid | LSBJ | 1.23 ± 0.05 | 29.3 ± 2.15 | 281 ± 152 | 115 |
| RSC02 | 1.49 ± 0.02* | 40.4 ± 1.30 | 603 ± 26.4* | ||
| Dodecanoic acid | LSBJ | 1.31 ± 0.17 | 23.5 ± 5.42 | 303 ± 38.4 | 7.26 |
| RSC02 | 1.11 ± 0.03* | 29.3 ± 5.21 | 325 ± 64.0 |
*Denotes statistically significant difference compared to LSBJ (Student’s t-test, two-tailed, α = 0.05)
aAll values are averages of biological triplicate experiments plus or minus the standard deviation about those averages
bPercent increase calculated as the increase in PHA concentration from RSC02 compared to LSBJ
Molecular weight comparison
To compare differences in physical properties of the polymers synthesized by E. coli LSBJ and RSC02, samples were analyzed by gel permeation chromatography (GPC) to determine the number average molecular weight (Mn), the weight average molecular weight (Mw), and the polydispersity (Mw/Mn) (Table 4). The polymers extracted from RSC02 had greater molecular weights than those from LSBJ for every polymer except poly(3-hydroxydodecanoate) (PHDD), which had a Mn of approximately 60 kDa and a Mw of approximately 172 kDa for both strains (Table 4). In addition, the polymers extracted from RSC02 had a greater degree of polydispersity, again with the exception of PHDD (Table 4).
Table 4.
PHA molecular weight data
| PHAa | Strain | Mw (kDa) | Mn (kDa) | Mw/Mn |
|---|---|---|---|---|
| PHB | LSBJ | ND | ND | ND |
| RSC02 | 390 | 117 | 3.3 | |
| PHV | LSBJ | 18 | 16 | 1.2 |
| RSC02 | 243 | 79 | 3.1 | |
| PHHx | LSBJ | 134 | 77 | 1.7 |
| RSC02 | 408 | 171 | 2.4 | |
| PHHp | LSBJ | 219 | 106 | 2.1 |
| RSC02 | 319 | 134 | 2.4 | |
| PHO | LSBJ | 157 | 73 | 2.1 |
| RSC02 | 285 | 99 | 2.9 | |
| PHD | LSBJ | 145 | 50 | 2.9 |
| RSC02 | 234 | 67 | 3.5 | |
| PHDD | LSBJ | 173 | 58 | 3.0 |
| RSC02 | 172 | 60 | 2.9 |
a PHB poly(3-hydroxybutyrate); PHV poly(3-hydroxyvalerate); PHHx poly(3-hydroxyhexanoate); PHHp poly(3-hydroxyheptanoate); PHO poly(3-hydroxyoctanoate); PHD poly(3-hydroxydecanoate); PHDD poly(3-hydroxydodecanoate); ND not detected
Growth profile of LSBJ and RSC02
We visually observed that RSC02 cultures appeared less optically dense than those of LSBJ during the first day of growth, which would be indicative of an increased lag time for this strain. To quantify this observation, we investigated the growth profiles of LSBJ and RSC02 under PHA biosynthesis conditions supplemented with either sodium butyrate or decanoic acid. There was both an increase in lag time and a decrease in the growth rate of RSC02 relative to LSBJ for both substrates (Fig. 3). Regardless of strain, when the fatty acid substrate was sodium butyrate the lag phase was considerably shorter and the stationary phase was reached more rapidly (Fig. 3). Despite the increased lag phase duration and slightly slower growth rate, RSC02 reached the same culture density as LSBJ when supplemented with sodium butyrate (OD600 ~ 3.5, 15 h), and reached a higher OD600 when supplemented with decanoic acid of ~5.0 compared to ~3.6 in LSBJ by hour 17 (Fig. 3).
Fig. 3.
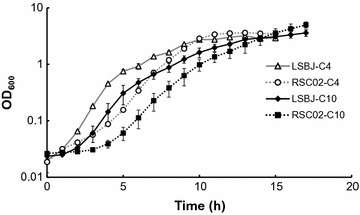
Growth profile of E. coli LSBJ and RSC02 during PHA-biosynthesis utilizing sodium butyrate (C4) or decanoic acid (C10). For both substrates, RSC02 displayed an extended lag time and slightly slower growth rate relative to LSBJ; however, RSC02 cultures reached approximately the same density as LSBJ upon reaching the stationary phase. For both strains, growth with sodium butyrate as the substrate caused a shorter lag phase than with decanoic acid substrate. All values are the averages of triplicate experiments plus or minus the standard deviation around those averages
PHA yield at 24 vs 48 h
To determine whether the increased lag phase in RSC02 had a negative effect on PHA yield earlier in the production cycle, small subsamples were removed from both LSBJ and RSC02 shake flasks after 24 h and 48 h during the growth profile experiments. Similar to the data shown in Table 3, LSBJ produced very little PHB at either 24 or 48 h, and there was very little change between the two time points (Fig. 4). However, LSBJ yields of PHD increased from 22.6 to 31.7% of cell dry weight, an increase of approximately 40% (Fig. 4). RSC02 produced significantly more polymer than LSBJ regardless of the time or fatty acid substrate; PHB yield increased from 8.2 to 12.3% from 24 to 48 h, while PHD yield increased by ~20% between time points from 37.2 to 45.0% (Fig. 4).
Fig. 4.
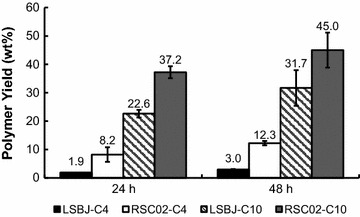
Comparison of PHA yield as a percentage of dry weight between LSBJ and RSC02 at 24 h and 48 h, utilizing either sodium butyrate (C4) or decanoic acid (C10). LSBJ saw insignificant changes in PHB production between 24 and 48 h, while RSC02 increased slightly from 8.2 to 12.3% of cell dry weight. PHD yield from LSBJ increased from 22.6 to 31.7% between 24 and 48 h, an increase of ~40%, while PHD yield from RSC02 increased from 37.2 to 45.0%, an increase of ~20%. Regardless of the time, RSC02 produced more polymer than LSBJ in all cases. All values are the averages of triplicate experiments plus or minus the standard deviation about those averages
Relative gene expression of RSC02
To analyze the relative expression of fatty acid degradation genes, RNA was isolated from mid-exponential phase cultures of LSBJ and RSC02 and reverse transcribed into cDNA for qPCR. An appropriate OD600 value (~0.6) for mid-exponential phase was determined using the previously defined growth profile (Fig. 3). All the genes analyzed were upregulated to some degree in RSC02; SCL fatty acid uptake genes atoA, atoE and atoD were all upregulated to an extremely large degree (>100-fold), while fadD, fadE, and fadL were all upregulated to a lesser degree (2-, 10-, and 31-fold) (Fig. 5). It is important to note that the expression values reported for the ato system may be exaggerated due to the low number of transcripts observed for LSBJ (as observed by a late-cycle emergence of the fluorescence during qPCR analysis).
Fig. 5.
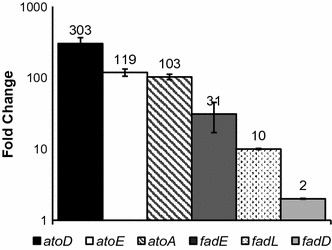
Relative gene expression of RSC02 compared to LSBJ, normalized to rpoD and measured as fold changes. The expression of genes related to fatty acid degradation was significantly increased in the RSC02 strain during mid-exponential growth phase (OD600 of ~0.6). The SCL fatty acid degradation genes atoA, atoE, and atoD were massively upregulated by 103-fold, 119-fold, and 303-fold respectively. MCL fatty acid degradation genes fadD, fadL, and fadE were upregulated by twofold, tenfold, and 31-fold respectively. Relative gene expression was calculated from qPCR fluorescence data using the LineRegPCR software (Ruijter et al. 2009) to calculate amplification efficiency and Cq values, and the Pfaffl method used to derive relative expression values with LSBJ as the calibrator (Fleige et al. 2006). All values are the averages of triplicate experiments plus or minus the standard deviation about those averages
Discussion
PHA homopolymer production
The purpose of developing the mutant strains RSC02, RSC04, and RSC06 was to improve PHA biosynthesis by removing regulatory genes known to interact with components of the PHA biosynthesis pathway. Based on the evidence seen in Fig. 2, the singular deletion of arcA conferred a significant increase in the production of PHD compared to the parental LSBJ. A reasonable explanation for this observation is that ArcA becomes active in E. coli LSBJ during the growth conditions utilized for polymer production and inhibits the transcription of fadL, fadD, and fadE, which are known targets of ArcA regulation (Table 1). In an arcA deletion mutant this inhibition cannot occur, likely resulting in higher basal transcription of these fad genes. The deletion of ompR and the double deletion of ompR/arcA provided no benefit to PHD biosynthesis, and no statistically significant differences were observed between these mutants and LSBJ (Fig. 2). One possibility for this apparent lack of effect is that OmpR regulatory target FadL is not active towards decanoic acid, which has been demonstrated previously (Black 1990). However, the similarity between the RSC06 double deletion mutant and the RSC04 mutant suggests another possibility; the loss of regulation by OmpR may yield a mildly toxic phenotype that inhibits cell growth and metabolism. This possibility is supported by both the observation that both RSC04 and RSC06 had significantly lower cell dry weights than either LSBJ and RSC02 (Additional file 1: Table S1), and by previous reporting from Oshima et al. (2002) that showed a marked growth deficiency in E. coli ΔompR mutants. These findings provided motivation for further characterization of the RSC02 mutant.
A variety of fatty acid substrates were used to characterize the differences in PHA homopolymer production between RSC02 and LSBJ. In nearly all cases, the deletion of arcA resulted in a significantly higher polymer yield (Table 3). Most surprisingly, the largest increases in polymer yield observed between RSC02 and LSBJ were for the two SCL polymers, PHB and PHV, as well as the shortest MCL polymer, PHHx (Table 3). This result is interesting considering that there are fewer steps involved in SCL fatty acid metabolism that are regulated by ArcA. Short chain fatty acids enter the β-oxidation cycle via enzymes derived from the ato operon, bypassing both FadL and FadD (Fig. 1). This operon has no known interaction with ArcA and is instead regulated by the response regulator AtoC, which is present in both LSBJ and RSC02 as a constitutively expressed mutant that confers a high level of atoDAEB transcription (Spratt et al. 1981; Jenkins and Nunn 1987; Kyriakidis and Tiligada 2009). One possible explanation for this drastic difference in SCL PHA production is that secondary metabolite pools are different between the two strains; polyamines for example have been shown to have significant effects on the regulation of AtoC (Kyriakidis and Tiligada 2009), and previous studies have shown that aspects of polyamine metabolism are affected by the deletion of arcA (Partridge et al. 2006).
The differences in MCL PHA production between LSBJ and RSC02 are more varied than those observed for SCL polymers. No significant difference was found between strains when comparing the yields of poly(3-hydroxyoctanoate) (PHO) and PHDD; however, the yields of poly(3-hydroxyheptanoate) (PHHp) and PHD were significantly increased in RSC02 (Table 3). The most reasonable explanation for this is that there are numerous differences in the substrate specificities of the enzymes involved in the PHA biosynthesis pathway. The long chain fatty acid transporter FadL is predominantly active on fatty acids containing 16 or more carbons, and no binding has been observed for decanoic acid, therefore it is unlikely that changes in fadL expression would account for the differences we observed (Black 1990). In contrast, the acyl-CoA synthetase FadD has high activity towards 12 and 10 carbon fatty acids and only low activity for 8 and 6 carbon fatty acids (Iram and Cronan 2006; Ford and Way 2015), while the recombinant enzymes PhaJ4 and PhaC1(STQK) each have their own well-documented substrate specificities that could contribute to the observed variation without any direct regulation by ArcA (Tsuge et al. 2003; Matsumoto et al. 2005; Sato et al. 2011). However, a complete understanding of fatty acid flux through this PHA biosynthesis pathway cannot be achieved with the current lack of information regarding the enzymatic activity of the acyl-CoA dehydrogenase FadE.
Comparison of the molecular weights of PHAs produced by LSBJ and RSC02
The molecular weight data showed a great degree of variability between PHAs, as well as between LSBJ and RSC02, with polydispersity values (Mw/Mn) from 1.2 to 3.3 (Table 4). The molecular weight of polymers produced by RSC02 were higher for all PHA polymers produced except for PHDD, and in general the polydispersity indices were higher for RSC02-derived polymers as well. Both the PHO and PHD molecular-number-average-molecular weights (Mn) were substantially lower for LSBJ than in previous studies, while the molecular weight and polydispersity for PHDD was found to be significantly higher than previously reported (Liu et al. 2011; Tappel et al. 2012b). A reliable comparison of the molecular weights observed in this study to other studies is difficult due to the variety of growth conditions and pathways employed. However, a possible explanation for the differences observed between LSBJ and RSC02 is that increased basal expression of β-oxidation genes improves the supply of 3-hydroxy fatty acyl-CoA monomers to the PhaC1 (STQK) polymer synthase. The evidence summarized in Table 4 suggests that the molecular machinery in E. coli plays some role in determining the molecular weight of the polymers produced.
A comparison between the polymer yield data in Table 3 and the molecular weight data in Table 4 appears to support this explanation. The PHA homopolymers with the greatest increases in yield (PHB, PHV, and PHHx) similarly showed the greatest increases in molecular weight (with the partial exception of PHB, which was not extracted in a great enough quantity to detect by GPC). The PHHp and PHD obtained from RSC02, which also saw moderate improvement in terms of yield, was observed to have a higher molecular weight than that obtained from LSBJ, although to a lesser extent than the shorter chain length PHA homopolymers. Interestingly, PHO produced by RSC02 had a higher molecular weight despite there being no significant difference in yield, while PHDD was not significantly different in either of those measurements. While it appears that improving the utilization of fatty acids also improves PHA molecular weight using this biosynthetic platform, these minor discrepancies with regards to PHO reveal that this relationship is more complex than that.
Another possible explanation for this increase in molecular weights could be that less ethanol is produced by RSC02 than LSBJ. It is typical for bacterial cultures grown into stationary phase in shake flasks to reach some level of microaerobiosis, leading to the production of fermentative byproducts such as ethanol (Gupta and Rao 2003; Losen et al. 2004). It was previously reported that the supplementation of ethanol in cultures of recombinant E. coli led to a decrease in the molecular weight of PHB due to a chain transfer reaction from PhaC to ethanol (Hiroe et al. 2013). In E. coli, the reversible enzyme AdhE is largely responsible for ethanol flux within the cell, allowing for both the biosynthesis and degradation of ethanol (Membrillo-Hernandez et al. 2000). It has been previously shown that an ΔarcA mutant has an improved ethanol tolerance compared to wild type, and it has been hypothesized that this tolerance is derived from increased expression of adhE along with enzymes involved in the tricarboxylic acid cycle (TCA), which could drive the breakdown of ethanol to acetyl-CoA (Goodarzi et al. 2010). There is also evidence in the literature showing that adhE expression is significantly higher in an E. coli ΔarcA mutant than in wild type under a range of microaerobic conditions (Shalel-Levanon et al. 2005). It is therefore possible that endogenously produced ethanol is reducing PHA molecular weights via chain termination in LSBJ, and that this effect could be mitigated by an improved flux of ethanol back to acetyl-CoA. However, further investigation of this possibility is required as there is currently no direct evidence to support this explanation.
Growth profiles of LSBJ and RSC02
We investigated our observation that RSC02 had slower growth than LSBJ by recording hourly culture densities, and found that while RSC02 had a significantly longer lag phase, the growth rate was nearly identical to LSBJ, with cultures reaching similar final cell densities regardless of which fatty acid substrate was provided (Fig. 3). These observations match the evidence found in the Keio collection of single-gene knockouts, which showed that arcA is nonessential, an ΔarcA mutant grows only slightly slower than wild type, and reaches only marginally lower culture densities (Baba et al. 2006). Despite the slightly slower growth observed with RSC02, the strain is capable of producing significantly more polymer than LSBJ even after only 24 h of growth (Fig. 4). This offers an advantage when using this strain for a large-scale continuous fermentation, and shows that the lengthened lag phase does not significantly impede PHA biosynthesis.
Relative gene expression of RSC02
Our analysis of the qPCR results shows a clear increase in the expression of each of the fatty acid degradation genes tested (Fig. 5). Although the improvements to fadD, fadL, and fadE were expected and support our hypothesis, the magnitude of the increase in the three ato genes came as a surprise. As we mentioned previously, ArcA is not known to directly regulate expression of the ato system, and both LSBJ and RSC02 harbor a constitutively expressed mutant atoC gene (Spratt et al. 1981; Jenkins and Nunn 1987; Kyriakidis and Tiligada 2009). Comparing the drastic differences seen in SCL PHA yields between LSBJ and RSC02 provides strong evidence to suggest that the similarly drastic increases in gene expression are responsible for these effects (Table 3). These results support the idea that the ΔarcA mutation indirectly effects the expression of the ato system, possibly mediated by an altered polyamine metabolism.
The differences between the relative expression of fadD, fadL and fadE also raise some interesting observations. For example, fadD was only modestly up-regulated (twofold) and its protein product has a high activity towards both 10 and 12 carbon fatty acid substrates; however, while PHD biosynthesis was significantly improved PHDD biosynthesis was not (Table 3; Fig. 5). Similarly, fadL expression was significantly increased in RSC02 (tenfold) which does have limited binding affinity for 12 carbon fatty acids (Black 1990), and yet this does not appear to improve PHDD biosynthesis either. These results suggest that the binding affinity of FadE may be an important limiting factor in PHA biosynthesis using this system, considering that fadE was the most highly up-regulated of these three genes (31-fold) and yet only the yields of PHHx, PHHp, and PHD were significantly improved (Table 3; Fig. 5). While the binding affinity of FadE is not well-known, our results suggest that it may have low binding affinity towards octanoyl-CoA and dodecanoyl-CoA, although further investigation is needed to test that hypothesis.
Conclusion
One of the largest challenges still facing the PHA industry is the relatively low yield of polymer obtained, which contributes to the overall cost (Kaur 2015). The strain E. coli RSC02 that was developed in this study offers a significant improvement to the previously reported strain, E. coli LSBJ. The most significant improvements were seen in the biosynthesis of PHB, PHV, and PHHx, with modest increases observed in PHHp and PHD. These results are supported by the increased expression of atoA, atoD, and atoE which correlates with improved PHB and PHV biosynthesis, and also the increased expression of fadD, fadL, and fadE which may contribute to the increased biosynthesis of PHHx, PHHp, and PHD.
Authors’ contributions
RS performed the gene deletion experiments, the growth and harvest of all bacterial cultures, the GC preparation and analysis, the polymer extraction and purification, the compositional verification using H-NMR, the molecular weight analysis using GPC, and the gene expression analysis. RS also drafted the manuscript in its entirety. LJ participated in the growth and harvest of bacterial cultures for the PHA homopolymer experiments using various fatty acids, assisted in the preparation and analysis of samples for GC, performed the growth profile analysis, analyzed the polymer yields at 24 and 48 h, and drafted the figures and methods corresponding to the growth profile. BL provided a significant contribution to the design of the study, and also participated in the critical revision of each draft of the manuscript. CN conceived of the study initially and participated in its design, and also assisted in the drafting and revision of the manuscript. All authors read and approved the final manuscript.
Acknowledgements
We would like to thank the SUNY ESF Upper Division Honors Program and the Center for Applied Microbiology for their assistance with this work.
Compliance with ethical standards and competing interests
This study was funded by the National Science Foundation (NSF) Grant CBET-1263905. The authors declare that they have no competing interests. This article does not contain any studies with human participants or animals performed by any of the authors.
Additional file
Additional file 1: Figure S1A. GPC chromatogram of purified poly(3-hydroxybutyrate) (C4) produced by RSC02, plotted as normalized intensity vs retention time. No data available for LSBJ. Figure S1B, C GPC chromatogram of purified poly(3-hydroxyvalerate) (C5) produced by LSBJ (B) and RSC02 (C), plotted as normalized intensity vs retention time. Figure S1D, E GPC chromatogram of purified poly(3-hydroxyhexanoate) (C6) produced by LSBJ (D) and RSC02 (E), plotted as normalized intensity vs retention time. Figure S1F, G GPC chromatogram of purified poly(3-hydroxyheptanoate) (C7) produced by LSBJ (F) and RSC02 (G), plotted as normalized intensity vs retention time. Figure S1H, I GPC chromatogram of purified poly(3-hydroxyoctanoate) (C8) produced by LSBJ (H) and RSC02 (I), plotted as normalized intensity vs retention time. Figure S1J, K GPC chromatogram of purified poly(3-hydroxydecanoate) (C10) produced by LSBJ (J) and RSC02 (K), plotted as normalized intensity vs retention time. Figure S1L, M GPC chromatogram of purified poly(3-hydroxydodecanoate) (C12) produced by LSBJ (L) and RSC02 (M), plotted as normalized intensity vs retention time. Figure S2A 1H-NMR (600 MHz, CDCl3); δ 5.28-5.23 (sext, 1H), 2.62-2.45 (m, 2H), 1.28-1.25 (d, 3H). Figure S2B 1H-NMR (600 MHz, CDCl3); δ 5.17-5.13 (p, 1H), 2.59-2.50 (m, 2H), 1.66-1.59 (m, 2H), 0.91-0.88 (t, 3H). The asterisk (*) at δ 1.53 denotes a water impurity (Fulmer et al. 2010). Figure S2C 1H-NMR (600 MHz, CDCl3); δ 5.21-5.18 (p, 1H), 2.59-2.48 (m, 2H), 1.60-1.54 (m, 2H), 1.35-1.27 (m, 2H), 0.91 (t, 3H). The asterisk (*) at δ 1.53 denotes a water impurity (Fulmer et al. 2010). Table S1: Poly(3-hydroxydecanoate) yield comparison between LSBJ, RSC02, RSC04.
Contributor Information
Ryan A. Scheel, Email: rascheel@syr.edu
Liyuan Ji, Email: jil198@potsdam.edu.
Benjamin R. Lundgren, Email: brlundgr@syr.edu
Christopher T. Nomura, Email: ctnomura@esf.edu
References
- Aiba H, Mizuno T. Phosphorylation of a bacterial activator protein, OmpR, by a protein kinase, EnvZ, stimulates the transcription of the ompF and ompC genes in Escherichia coli. FEBS Lett. 1990;261:19–22. doi: 10.1016/0014-5793(90)80626-T. [DOI] [PubMed] [Google Scholar]
- Alexeeva S, Hellingwerf KJ, de Mattos MJT. Requirement of ArcA for redox regulation in Escherichia coli under microaerobic but not anaerobic or aerobic conditions. J Bacteriol. 2003;185:204–209. doi: 10.1128/JB.185.1.204-209.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006 doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black PN. Characterization of FadL-specific fatty acid binding in Escherichia coli. Biochim Biophys Acta. 1990;1046:97–105. doi: 10.1016/0005-2760(90)90099-J. [DOI] [PubMed] [Google Scholar]
- Braunegg G, Sonnleitner B, Lafferty RM. A rapid gas chromatographic method for the determination of poly-β-hydroxybutyric acid in microbial biomass. Eur J Appl Microbiol Biotechnol. 1978;6(29):37. [Google Scholar]
- Campbell JW, Cronan JE. The enigmatic Escherichia coli fadE gene is yafH. J Bacteriol. 2002;184:3759–3764. doi: 10.1128/JB.184.13.3759-3764.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho BK, Knight EM, Palsson BO. Transcriptional regulation of the fad regulon genes of Escherichia coli by ArcA. Microbiol Read Engl. 2006;152:2207–2219. doi: 10.1099/mic.0.28912-0. [DOI] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fic E, Bonarek P, Gorecki A, Kedracka-Krok S, Mikolajczak J, Polit A, Tworzydlo M, Dziedzicka-Wasylewska M, Wasylewski Z. cAMP receptor protein from Escherichia coli as a model of signal transduction in proteins—a review. J Mol Microbiol Biotechnol. 2009;17:1–11. doi: 10.1159/000178014. [DOI] [PubMed] [Google Scholar]
- Fleige S, Walf V, Huch S, Prgomet C, Sehm J, Pfaffl MW. Comparison of relative mRNA quantification models and the impact of RNA integrity in quantitative real-time RT-PCR. Biotechnol Lett. 2006;28:1601–1613. doi: 10.1007/s10529-006-9127-2. [DOI] [PubMed] [Google Scholar]
- Ford TJ, Way JC (2015) Enhancement of E. coli acyl-CoA synthetase FadD activity on medium chain fatty acids. PeerJ 3:e1040. doi: 10.7717/peerj.1040 [DOI] [PMC free article] [PubMed]
- Forst SA, Roberts DL. Signal transduction by the EnvZ-OmpR phosphotransfer system in bacteria. Res Microbiol. 1994;145:363–373. doi: 10.1016/0923-2508(94)90083-3. [DOI] [PubMed] [Google Scholar]
- Goodarzi H, Bennett BD, Amini S, Reaves ML, Hottes AK, Rabinowitz JD, Tavazoie S (2010) Regulatory and metabolic rewiring during laboratory evolution of ethanol tolerance in E. coli. Mol Syst Biol. doi: 10.1038/msb.2010.33 [DOI] [PMC free article] [PubMed]
- Gupta A, Rao G. A study of oxygen transfer in shake flasks using a non-invasive oxygen sensor. Biotechnol Bioeng. 2003;84:351–358. doi: 10.1002/bit.10740. [DOI] [PubMed] [Google Scholar]
- Higashitani A, Nishimura Y, Hara H, Aiba H, Mizuno T, Horiuchi K. Osmoregulation of the fatty acid receptor gene fadL in Escherichia coli. Mol Gen Genet MGG. 1993;240:339–347. doi: 10.1007/BF00280384. [DOI] [PubMed] [Google Scholar]
- Hiroe A, Hyakutake M, Thomson NM, Sivaniah E, Tsuge T. Endogenous Ethanol Affects Biopolyester Molecular Weight in Recombinant Escherichia coli. ACS Chem Biol. 2013;8:2568–2576. doi: 10.1021/cb400465p. [DOI] [PubMed] [Google Scholar]
- Iram SH, Cronan JE. The β-oxidation systems of Escherichia coli and Salmonella enterica are not functionally equivalent. J Bacteriol. 2006;188:599–608. doi: 10.1128/JB.188.2.599-608.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iuchi S, Ec L. arcA (dye), a global regulatory gene in Escherichia coli mediating repression of enzymes in aerobic pathways. Proc Natl Acad Sci. 1988;85:1888–1892. doi: 10.1073/pnas.85.6.1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iuchi S, Lin EC. Purification and phosphorylation of the Arc regulatory components of Escherichia coli. J Bacteriol. 1992;174:5617–5623. doi: 10.1128/jb.174.17.5617-5623.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins LS, Nunn WD. Genetic and molecular characterization of the genes involved in short-chain fatty acid degradation in Escherichia coli: the ato system. J Bacteriol. 1987;169:42–52. doi: 10.1128/jb.169.1.42-52.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jishage M, Iwata A, Ueda S, Ishihama A. Regulation of RNA polymerase sigma subunit synthesis in Escherichia coli: intracellular levels of four species of sigma subunit under various growth conditions. J Bacteriol. 1996;178:5447–5451. doi: 10.1128/jb.178.18.5447-5451.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kameda K, Nunn WD. Purification and characterization of acyl coenzyme A synthetase from Escherichia coli. J Biol Chem. 1981;256:5702–5707. [PubMed] [Google Scholar]
- Kaur G. Strategies for Large-scale Production of Polyhydroxyalkanoates. Chem Biochem Eng Q. 2015;29:157–172. doi: 10.15255/CABEQ.2014.2255. [DOI] [Google Scholar]
- Kyriakidis DA, Tiligada E. Signal transduction and adaptive regulation through bacterial two-component systems: the Escherichia coli AtoSC paradigm. Amino Acids. 2009;37:443–458. doi: 10.1007/s00726-009-0241-z. [DOI] [PubMed] [Google Scholar]
- Langenbach S, Rehm BHA, Steinbüchel A. Functional expression of the PHA synthase gene phaC1 from Pseudomonas aeruginosa in Escherichia coli results in poly(3-hydroxyalkanoate) synthesis. FEMS Microbiol Lett. 1997;150:303–309. doi: 10.1016/S0378-1097(97)00142-0. [DOI] [PubMed] [Google Scholar]
- Laycock B, Halley P, Pratt S, Werker A, Lant P. The chemomechanical properties of microbial polyhydroxyalkanoates. Prog Polym Sci. 2013;38:536–583. doi: 10.1016/j.progpolymsci.2012.06.003. [DOI] [Google Scholar]
- Lepore BW, Indic M, Pham H, Hearn EM, Patel DR, van den Berg B. Ligand-gated diffusion across the bacterial outer membrane. Proc Natl Acad Sci USA. 2011;108:10121–10126. doi: 10.1073/pnas.1018532108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine AC, Sparano A, Twigg FF, Numata K, Nomura CT. Influence of cross-linking on the physical properties and cytotoxicity of polyhydroxyalkanoate (PHA) scaffolds for tissue engineering. ACS Biomater Sci Eng. 2015;1:567–576. doi: 10.1021/acsbiomaterials.5b00052. [DOI] [PubMed] [Google Scholar]
- Levine AC, Heberlig GW, Nomura CT. Use of thiol-ene click chemistry to modify mechanical and thermal properties of polyhydroxyalkanoates (PHAs) Int J Biol Macromol. 2016;83:358–365. doi: 10.1016/j.ijbiomac.2015.11.048. [DOI] [PubMed] [Google Scholar]
- Liu X, Wulf PD. Probing the arca-P modulon of Escherichia coli by whole genome transcriptional analysis and sequence recognition profiling. J Biol Chem. 2004;279:12588–12597. doi: 10.1074/jbc.M313454200. [DOI] [PubMed] [Google Scholar]
- Liu Q, Luo G, Zhou XR, Chen G-Q. Biosynthesis of poly(3-hydroxydecanoate) and 3-hydroxydodecanoate dominating polyhydroxyalkanoates by β-oxidation pathway inhibited Pseudomonas putida. Metab Eng. 2011;13:11–17. doi: 10.1016/j.ymben.2010.10.004. [DOI] [PubMed] [Google Scholar]
- Losen M, Frölich B, Pohl M, Büchs J. Effect of oxygen limitation and medium composition on Escherichia coli fermentation in shake-flask cultures. Biotechnol Prog. 2004;20:1062–1068. doi: 10.1021/bp034282t. [DOI] [PubMed] [Google Scholar]
- Lu J, Tappel RC, Nomura CT. Mini-Review: biosynthesis of Poly(hydroxyalkanoates) Polym Rev. 2009;49(226):248. [Google Scholar]
- Lundgren BR, Thornton W, Dornan MH, Villegas-Peñaranda LR, Boddy CN, Nomura CT. Gene PA2449 is essential for glycine metabolism and pyocyanin biosynthesis in Pseudomonas aeruginosa PAO1. J Bacteriol. 2013;195:2087–2100. doi: 10.1128/JB.02205-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch AS, Lin EC. Transcriptional control mediated by the ArcA two-component response regulator protein of Escherichia coli: characterization of DNA binding at target promoters. J Bacteriol. 1996;178:6238–6249. doi: 10.1128/jb.178.21.6238-6249.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubara M, Mizuno T. EnvZ-independent phosphotransfer signaling pathway of the OmpR-mediated osmoregulatory expression of OmpC and OmpF in Escherichia coli. Biosci Biotechnol Biochem. 1999;63:408–414. doi: 10.1271/bbb.63.408. [DOI] [PubMed] [Google Scholar]
- Matsumoto K, Takase K, Aoki E, Doi Y, Taguchi S. Synergistic effects of Glu130Asp substitution in the type II polyhydroxyalkanoate (PHA) synthase: enhancement of PHA production and alteration of polymer molecular weight. Biomacromolecules. 2005;6:99–104. doi: 10.1021/bm049650b. [DOI] [PubMed] [Google Scholar]
- Membrillo-Hernandez J, Echave P, Cabiscol E, Tamarit J, Ros J, Lin EC. Evolution of the adhE gene product of Escherichia coli from a functional reductase to a dehydrogenase. Genetic and biochemical studies of the mutant proteins. J Biol Chem. 2000;275:33869–33875. doi: 10.1074/jbc.M005464200. [DOI] [PubMed] [Google Scholar]
- Mizuno T, Mizushima S. Signal transduction and gene regulation through the phosphorylation of two regulatory components: the molecular basis for the osmotic regulation of the porin genes. Mol Microbiol. 1990;4:1077–1082. doi: 10.1111/j.1365-2958.1990.tb00681.x. [DOI] [PubMed] [Google Scholar]
- Oshima T, Aiba H, Masuda Y, Kanaya S, Sugiura M, Wanner BL, Mori H, Mizuno T. Transcriptome analysis of all two-component regulatory system mutants of Escherichia coli K-12. Mol Microbiol. 2002;46:281–291. doi: 10.1046/j.1365-2958.2002.03170.x. [DOI] [PubMed] [Google Scholar]
- Partridge JD, Scott C, Tang Y, Poole RK, Green J. Escherichia coli Transcriptome Dynamics during the Transition from Anaerobic to Aerobic Conditions. J Biol Chem. 2006;281:27806–27815. doi: 10.1074/jbc.M603450200. [DOI] [PubMed] [Google Scholar]
- Peña-Sandoval GR, Georgellis D. The ArcB sensor kinase of Escherichia coli autophosphorylates by an intramolecular reaction. J Bacteriol. 2010;192:1735–1739. doi: 10.1128/JB.01401-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto A, Ciesla JH, Palucci A, Sutliff BP, Nomura CT. Chemically intractable no more. In vivo incorporation of “Click”-ready fatty acids into poly-[(R)-3-hydroxyalkanoates] in Escherichia coli. ACS Macro Lett. 2016;5:215–219. doi: 10.1021/acsmacrolett.5b00823. [DOI] [PubMed] [Google Scholar]
- Qi Q, Rehm BHA, Steinbüchel A. Synthesis of poly(3-hydroxyalkanoates) in Escherichia coli expressing the PHA synthase gene phaC2 from Pseudomonas aeruginosa: comparison of PhaC1 and PhaC2. FEMS Microbiol Lett. 1997;157:155–162. doi: 10.1111/j.1574-6968.1997.tb12767.x. [DOI] [PubMed] [Google Scholar]
- Qi Q, Steinbüchel A, Rehm BHA. Metabolic routing towards polyhydroxyalkanoic acid synthesis in recombinant Escherichia coli (fadR): inhibition of fatty acid β-oxidation by acrylic acid. FEMS Microbiol Lett. 1998;167:89–94. doi: 10.1111/j.1574-6968.1998.tb13212.x. [DOI] [PubMed] [Google Scholar]
- Ruijter JM, Ramakers C, Hoogaars WMH, Karlen Y, Bakker O, van den Hoff MJB, Moorman AFM. Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 2009;37:e45–e45. doi: 10.1093/nar/gkp045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook JJ, Russell DW. Molecular cloning: A laboratory manual. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Sarwar Z, Lundgren BR, Grassa MT, Wang MX, Gribble M, Moffat JF, Nomura CT (2016) GcsR, a TyrR-like enhancer-binding protein, regulates expression of the glycine cleavage system in Pseudomonas aeruginosa PAO1. mSphere 1:e00020-16. doi: 10.1128/mSphere.00020-16 [DOI] [PMC free article] [PubMed]
- Sato S, Kanazawa H, Tsuge T. Expression and characterization of (R)-specific enoyl coenzyme A hydratases making a channeling route to polyhydroxyalkanoate biosynthesis in Pseudomonas putida. Appl Microbiol Biotechnol. 2011;90:951–959. doi: 10.1007/s00253-011-3150-5. [DOI] [PubMed] [Google Scholar]
- Shalel-Levanon S, San K-Y, Bennett GN. Effect of ArcA and FNR on the expression of genes related to the oxygen regulation and the glycolysis pathway in Escherichia coli under microaerobic growth conditions. Biotechnol Bioeng. 2005;92:147–159. doi: 10.1002/bit.20583. [DOI] [PubMed] [Google Scholar]
- Silhavy TJ, Pratt LA (1995) Porin Regulon of Escherichia coli. In: Silhavy TJ, Hoch JA (eds) Two-Component Signal Transduction. American Society of Microbiology, pp 105–127
- Spratt SK, Ginsburgh CL, Nunn WD. Isolation and genetic characterization of Escherichia coli mutants defective in propionate metabolism. J Bacteriol. 1981;146:1166–1169. doi: 10.1128/jb.146.3.1166-1169.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takase K, Taguchi S, Doi Y. Enhanced synthesis of poly(3-hydroxybutyrate) in recombinant Escherichia coli by means of error-prone PCR mutagenesis, saturation mutagenesis, and in vitro recombination of the type II polyhydroxyalkanoate synthase gene. J Biochem (Tokyo) 2003;133:139–145. doi: 10.1093/jb/mvg015. [DOI] [PubMed] [Google Scholar]
- Takase K, Matsumoto K, Taguchi S, Doi Y. Alteration of substrate chain-length specificity of type II synthase for polyhydroxyalkanoate biosynthesis by in vitro evolution: in vivo and in vitro enzyme assays. Biomacromolecules. 2004;5:480–485. doi: 10.1021/bm034323+. [DOI] [PubMed] [Google Scholar]
- Tappel RC, Kucharski JM, Mastroianni JM, Stipanovic AJ, Nomura CT. Biosynthesis of poly[(R)-3-hydroxyalkanoate] copolymers with controlled repeating unit compositions and physical properties. Biomacromolecules. 2012;13:2964. doi: 10.1021/bm301043t. [DOI] [PubMed] [Google Scholar]
- Tappel RC, Wang Q, Nomura CT. Precise control of repeating unit composition in biodegradable poly(3-hydroxyalkanoate) polymers synthesized by Escherichia coli. J Biosci Bioeng. 2012;113:480–486. doi: 10.1016/j.jbiosc.2011.12.004. [DOI] [PubMed] [Google Scholar]
- Theodorou MC, Panagiotidis CA, Panagiotidis CH, Pantazaki AA, Kyriakidis DA. Involvement of the AtoS-AtoC signal transduction system in poly-(R)-3-hydroxybutyrate biosynthesis in Escherichia coli. Biochim Biophys Acta. 2006;1760:896–906. doi: 10.1016/j.bbagen.2006.01.020. [DOI] [PubMed] [Google Scholar]
- Tsuge T, Taguchi K, Doi Y, et al. Molecular characterization and properties of (R)-specific enoyl-CoA hydratases from Pseudomonas aeruginosa: metabolic tools for synthesis of polyhydroxyalkanoates via fatty acid β-oxidation. Int J Biol Macromol. 2003;31:195–205. doi: 10.1016/S0141-8130(02)00082-X. [DOI] [PubMed] [Google Scholar]


