Abstract
Current treatment options for castration-resistant prostate cancer (CRPC) are limited. In this study, a high-throughput screen of 4910 drugs and drug-like molecules was performed to identify antiproliferative compounds in androgen ablated prostate cancer cells. The effect of compounds on cell viability was compared in androgen ablated LNCaP prostate cancer cells and in LNCaP cells grown in presence of androgens as well as in two non-malignant prostate epithelial cells (RWPE-1 and EP156T). Validation experiments of cancer specific anti-proliferative compounds indicated pinosylvin methyl ether (PSME) and tanshinone IIA as potent inhibitors of androgen ablated LNCaP cell proliferation. PSME is a stilbene compound with no previously described anti-neoplastic activity whereas tanshinone IIA is currently used in cardiovascular disorders and proposed as a cancer drug. To gain insights into growth inhibitory mechanisms in CRPC, genome-wide gene expression analysis was performed in PSME- and tanshinone IIA-exposed cells. Both compounds altered the expression of genes involved in cell cycle and steroid and cholesterol biosynthesis in androgen ablated LNCaP cells. Decrease in androgen signalling was confirmed by reduced expression of androgen receptor and prostate specific antigen in PSME- or tanshinone IIA-exposed cells. Taken together, this systematic screen identified a novel anti-proliferative agent, PSME, for CRPC. Moreover, our screen confirmed tanshinone IIA as well as several other compounds as potential prostate cancer growth inhibitors also in androgen ablated prostate cancer cells. These results provide valuable starting points for preclinical and clinical studies for CRPC treatment.
Introduction
Androgen deprivation, surgery and radiation therapy are the main treatment options for prostate cancer patients. However, hormonal therapy is not curative and leads to the development of castration-resistant prostate cancer (CRPC). The median survival time for CRPC is around 2 years (Tannock et al. 2004). Several mechanisms underlying CRPC development have been described: androgen receptor (AR) overexpression sensitizing cancer cells to low levels of androgens, AR mutations enabling non-androgenic ligands to activate the receptor and intra-tumoral steroidogenesis in which androgens are produced in cancer cells (Locke et al. 2008, Taichman et al. 2007). Thus, AR and its co-regulators are potent drug targets for CRPC treatment. Recently, novel promising anti-androgens and chemotherapeutic agents such as Enzalutamide, have been developed to target CRPC (Aragon-Ching 2012, Berruti et al. 2012, Dhingra et al. 2013, Schrader et al. 2013). Although these anti-androgens and chemotherapeutics have shown significant increase in patient survival by lengthening survival time by several months, there is a need for better treatment options to further improve the outcome of patients suffering from CRPC.
We have previously applied high-throughput screening to systematically explore most currently marketed drugs and drug-like molecules for their efficacy against a panel of prostate cells (Iljin et al. 2009). Here, we performed a cell-based viability screen with a library of 4910 drug-like small molecule compounds in LNCaP prostate cancer cells grown either in presence or in absence of androgens (Culig et al. 1999). The androgen-independent derivative of the androgen-dependent LNCaP prostate cancer cell line used in this study had been previously generated by long-term androgen deprivation (Culig et al. 1999). The screening results from LNCaP cells were compared to the previous cell viability results in non-malignant prostate epithelial cells RWPE-1 and EP156T to identify cancer-selective compounds (Iljin et al. 2009). Out of the growth inhibitory compounds identified in LNCaP cells, pinosylvin methyl ether (PSME) and tanshinone IIA were chosen for further analysis due to their cancer selective anti-proliferative effect in androgen ablated prostate cancer cells.
Materials and Methods
Cells
Parental (LNCaP-par) and androgen ablated LNCaP (LNCaP-abl) prostate carcinoma cell lines were obtained from Zoran Culig (Culig et al. 1999) and grown in RPMI-1640 medium containing 10% FBS (LNCaP-par) or 10% charcoal:dextran stripped FBS (LNCaP-abl) supplemented with 1% glutamate and 1% penicillin-streptomycin.
Compounds
Tanshinone IIA was purchased from Apin Chemicals Ltd. (Abingdon, UK) and pinosylvin methyl ether was purchased from Gentaur (London, UK). Both compounds were dissolved in DMSO.
High-throughput screening (HTS)
A high-throughput compound screening was performed twice in LNCaP-par and LNCaP-abl cells using cell viability as the endpoint. The results were compared to the ones from our previous screen performed in non-malignant RWPE-1 and EP156T cells with the same compound libraries and concentrations used in this study (Iljin et al. 2009). Briefly, 4910 compounds, including experimental compounds, most of the existing Food and Drug Administration–approved drugs, kinase and phosphatase inhibitors as well as natural products, were screened with at least two different concentrations. The cell viability was determined after a 3-day incubation (1500 cells per well in 384-well format) with the compounds using a CellTiter-Glo (CTG) fluorescent cell viability assay (Promega, Inc.). The compounds that qualified as hits inhibited cell viability (loess-score) by at least three standard deviations from the median of the controls.
Cell viability and apoptosis assays
Cell viability and apoptosis assays were performed on 384-well plates (Falcon) by plating 2,000 cells per well in 35 μl of their respective growth media and left to attach overnight. Next, compound dilutions were added to the cells, plates were incubated for 48 hours and cell viability was determined using CellTiter-Blue (CTB) or CTG cell viability assay (Promega, Inc.) according to the manufacturer’s instructions. Induction of caspase-3 and 7 activities was detected with homogenous Apo-ONE assay (Promega, Madison, WI). The fluorometric signal from CTB (excitation FITC 485 nm, emission FITC 535 nm) or luminescence signals (700 nm) from CTG and apoptosis assays were quantified using Envision Multilabel Plate Reader (Perkin-Elmer, Massachusetts, MA).
Gene expression analysis using bead-arrays
Ablated and parental LNCaP cells were grown into approximately 70% confluence and treated with PSME or tanshinone IIA for 24 h before harvesting. Total RNA was extracted using RNeasy (Qiagen) according to the manufacturer’s protocol and the integrity of the RNA was monitored prior to hybridization using a Bioanalyzer 2100 (Agilent, Santa Clara, CA) according to manufacturer’s instructions. 500 ng of purified RNA was amplified with the TotalPrep Kit (Ambion, Austin, TX) and the biotin labelled cRNA was hybridized to Sentrix HumanRef-8 vs.3 Expression BeadChips (Illumina, San Diego, CA) while the arrays were scanned with the BeadArray Reader (Illumina).
Statistical analysis of gene expression data
The raw gene expression data were quantile-normalized and analyzed with the R / Bioconductor software as previously described (Gentleman et al. 2004, He et al. 2014). Differentially expressed genes from microarray hybridizations were identified using the significance analysis of microarrays program (SAM), with a false discovery rate set to zero and a minimum fold change of ≥ 1.4. Hierarchical clustering and multidimensional scaling with principal coordinates analysis was performed to visualize the distribution of prostate cell lines based on their compound responses. The functional gene ontology and pathway annotations were analyzed for differentially expressed genes (R>0.5 and p<0.001) using Ingenuity Pathway Analysis (IPA) Software (Ingenuity Systems Inc., Redwood City, CA, USA).
Quantitative reverse transcriptase PCR
Reverse transcription using 500 ng of total RNA was performed with Applied Biosystem´s cDNA synthesis kit. TaqMan gene expression probes and primers from the Universal Probe Library (Roche Diagnostics, Espoo, Finland) were used to study androgen receptor (AR), prostate specific antigen (PSA) and β-actin mRNA expression. Primer sequences are listed in Supplementary Table 1. Real-time quantitative PCR was performed using ABI Prism 7900 (Applied Biosystems, Foster City, CA). Quantitation was carried out using the ΔΔCT method with the RQ manager 1.2 software (Applied Biosystems). β-actin was used as an endogenous control. Average expression of the control samples was considered for the calculation of the fold changes. Two to four replicate samples were studied for quantitation of mRNA expression.
Western blot analysis
LNCaP-par and LNCaP-abl cells were plated at 70 % confluency and left to attach over night before treatments with indicated compounds. 10 µg of total protein was denatured at 95 °C for 5 min in Laemmli buffer, separated on 10 % precast SDS-polyacrylamide gel (Lonza, Basel Switzerland) and transferred to Protran nitrocellulose transfer membrane (Schleicher & Schuell, Niedersachsen, Germany). Western blot analysis was performed using specific antibodies against AR (1:1000 dilution, mouse monoclonal, Labvision, Fremont, CA), prostate specific antigen (PSA, 1:1000, rabbit polyclonal, DakoCytomation, Denmark), and β-actin (1:4000 dilution, mouse monoclonal, Becton Dickinson, Franklin Lakes, NJ). Signal was detected with 1:4000 dilutions of appropriate HRP-conjugated secondary antibodies (all from Invitrogen Molecular Probes, Carlsbad, CA) followed by visualization with the enhanced chemiluminescence reagent (Amersham Biosciences, Little Chalfont, UK).
Results
Identification of selective antineoplastic compounds for castrate-resistant prostate cancer
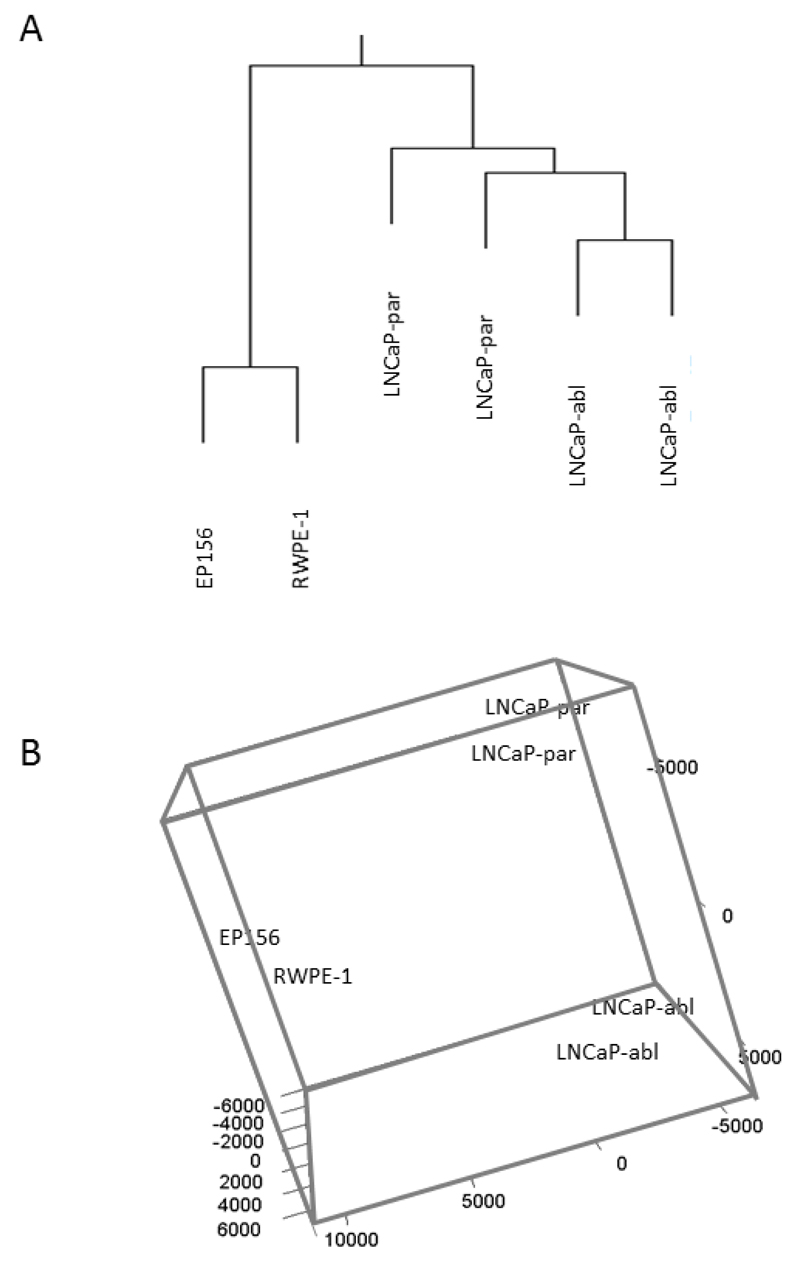
To identify novel selective antineoplastic compounds and to explore targetable molecular pathways for castrate-resistant prostate cancer, we carried out a cell-based high-throughput screen (HTS) in LNCaP cells grown in absence (LNCaP-abl) and presence (LNCaP-par) of androgens. Altogether, the effect of 4910 marketed drugs and small molecule compounds on cell viability was measured after 3-day incubation with compounds. Cell viability results were compared to the results from previous screens performed in RWPE-1 and EP156T non-malignant prostate epithelial cells (Iljin et al., 2009). The anti-proliferative hit compounds inhibited cell viability (loess-score) by at least three standard deviations from the median of the controls. To visualize the distribution of prostate cell lines based on their compound responses, hierarchical clustering and multidimensional scaling with principal component analysis was performed. The results indicated that normal epithelium-derived cell lines formed one group and the prostate cancer cell lines formed another group where separate clusters could be found for LNCaP –abl and LNCaP-par cells (Figure 1A and B).
Figure 1.
The distribution of prostate cells based on their compound responses demonstrated by using (A) hierarchical clustering and (B) multidimensional scaling with principal component analysis. Normal epithelium-derived cell lines formed one group and LNCaP prostate cancer cells grown in presence (LNCaP-par) and absence of androgens (LNCaP-abl) formed separate clusters.
In total, 44 cancer cell selective anti-proliferative compounds qualified as hits in both independent LNCaP-abl screens (Supplementary Table 2). Most of these compounds reduced cell viability also in LNCaP-par cells at least in one of the two screens. These compounds included drugs such as methotrexate and methotrexate hydrate, currently used in prostate cancer chemotherapy (Straus et al. 1982) as well as Jun N-terminal kinase inhibitor (SP600125) and PI3-kinase inhibitors (LY294002 and Wortmannin) previously shown to induce apoptosis in prostate cancer cells (Bennett et al. 2001, Lin et al. 1999, Tyagi et al. 2003). In addition, hypericin, an Hsp90 and HIF-1α degradator, known to reduce prostate cancer cell growth in vitro and in vivo (Barliya et al. 2011, Colasanti et al. 2000, Xie et al. 2001), was identified among LNCaP-abl anti-proliferative hits. Moreover, aldehyde dehydrogenase (ALDH) inhibitor disulfiram, antibiotic monensin, fungicide thiram and HDAC inhibitor tricostatin A (TSA), that we previously identified as cancer-selective compounds using a panel of cultured prostate cells, also inhibited the viability of LNCaP-abl prostate cancer cells (Iljin et al. 2009). Furthermore, HDAC inhibitor SAHA, previously shown to inhibit ERG-positive prostate cancer cell growth (Bjorkman et al. 2008), reduced LNCaP-abl cell proliferation as well. Interestingly, one of the anti-proliferative agents identified in both LNCaP-abl and LNCaP-par screens was microtubule polymerization interfering agent Nocodazole. It has been shown to block nuclear translocation of AR and ablate dihydrotestosterone induced PSA expression in LNCaP cells (Zhu et al. 2010). Altogether, identification of these compounds among our hits indicate that drugs with known antiproliferative effects in prostate cancer cells, including AR targeting agents, are also able to reduce the viability of both LNCaP-par and LNCaP-abl cells in our screen. This suggests that targeting AR is a potent way to also reduce LNCaP-abl cell viability.
Different molecular pathways are targeted by LNCaP-abl-selective compounds
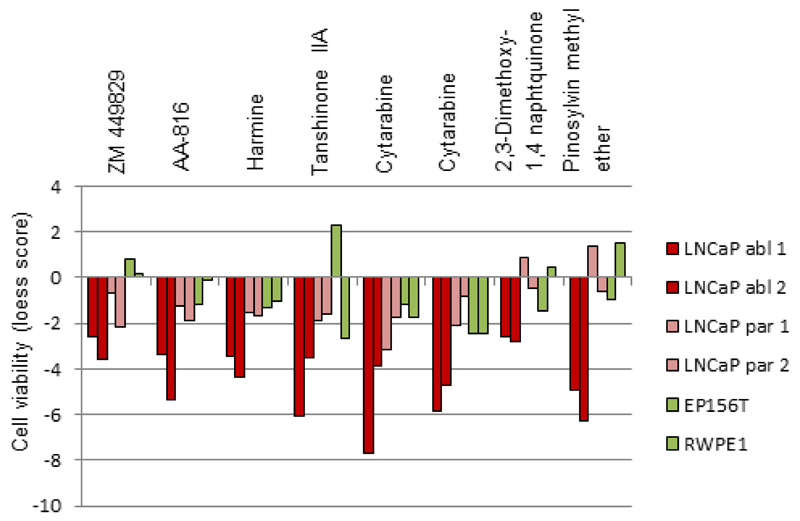
We chose the seven most potent LNCaP-abl selective anti-proliferative compounds for further validation based on the HTS results. The cell viability results for these compounds, AA-816, harmine, tanshinone IIA, pinosylvin methyl ether, 2,3-dimethoxy-1,4-naphthoquinone (Alexan), cytosine-1-beta-D-arabinofuranoside hydrochlorid (Cytarabine) and ZM449829 in LNCaP-abl, LNCaP-par and non-malignant prostate epithelial cells (RWPE1 and EP156T) are presented in Figure 2. Interestingly, analysis of known mechanisms of action for these compounds indicated that they target different molecular pathways. For example, AA-816 is a 5-lipoxygenase inhibitor that blocks arachidonic acid induced prostate cancer cell proliferation (Steele et al. 1999) whereas harmine, a natural compound in medicinal plants (Peganum harmala and Eurycoma longifolia), regulates VEGF, tissue inhibitor metalloprotease (TIMP), matrix metalloproteases MMP-2 and MMP-9 and interleukin 2 (IL-2) expression in breast cancer cells, induces apoptosis in hepatocellular carcinoma cells as well as sensitizes breast cancer cells to mitoxantrone and camptothecin (Cao et al. 2011, Chen et al. 2005, Dai et al. 2012, Hamsa & Kuttan 2010, Li et al. 2011). Harmine has also been patented as an androgen receptor inhibitor (US Patent 8119660). Other LNCaP-abl-selective hit compounds identified in our screen included the DNA damaging agent Alexan that has been used as a tool to study the effect of reactive oxygen species (Morgan 1995), a chemotherapy agent cytarabine that has been studied in phase II trials for treatment of castration-resistant prostate cancer (Dhani et al. 2012) as well as ZM 449829, a Janus kinase (JAK) and epidermal growth factor receptor (EGFR) inhibitor (Luo & Laaja 2004).
Figure 2.
The normalized cell viability (loess score) values presented for seven cancer cell selective anti-proliferative hit compounds identified in both high-throughput screens performed in androgen ablated (LNCaP-abl) cells. For comparison, the corresponding cell viability results are presented also for LNCaP cells grown in the presence of androgens (LNCaP-par) as well as for two non-malignant prostate epithelial cells (EP156T and RWPE-1).
PSME, tanshinone IIA and cytarabine are the most effective anti-proliferative hits in LNCaP-abl cells
To investigate the antiproliferative effect of the seven selected compounds in more detail, we first determined the EC50 values for each of the compounds in both LNCaP-abl and LNCaP-par cells. The results confirmed that all seven compounds inhibited LNCaP-abl cell viability (Table 1). However, only PSME, tanshinone IIA and cytarabine inhibited LNCaP-abl cell growth at low nanomolar concentrations. Since cytarabine is already in clinical trials for androgen independent prostate cancer, we selected PSME and Tanshinone IIA for further analyses.
Table 1.
EC50 values of seven compounds in LNCaP-abl and LNCaP-par cells. The high-throughput screen identified these compounds among cancer cell selective anti-proliferative hits in LNCaP-abl cells.
| Compound | LNCaP-par EC50 (nM) | LNCaP-abl EC50 (nM) |
|---|---|---|
| AA-816 | >10 000 | 5300 |
| Harmine | >10 000 | 4950 |
| Tanshinone IIA | 360 | 440 |
| 2,3-Dimethoxy-1,4-naphthoquinone (Alexan) | 4500 | 2460 |
| Cytosine-1-beta-D-arabinofuranoside Hydrochlorid (Cytarabine) | 5200 | 320 |
| ZM 449829 | >10 000 | 2100 |
| Pinosylvin methyl ether (PSME) | >1000 | 250 |
PSME and tanshinone IIA induce apoptosis in LNCaP-abl cells
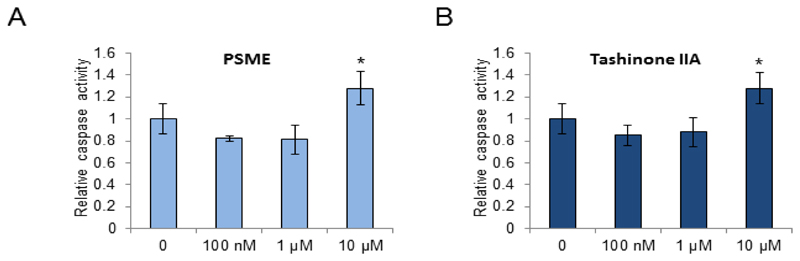
Next, we determined whether the observed decrease in cell viability in response to PSME and tanshinone IIA exposure in LNCaP-abl cells is due to induction of apoptosis. Caspase 3 and 7 activities were determined in response to 24-hour PSME or tanshinone IIA exposure using an ApoONE assay (Promega). The results indicated that PSME and tanshinone IIA induce caspase 3 and 7 activities at 10 µM concentrations in LNCaP-abl cells (Figure 3A and B).
Figure 3.
Apoptosis assay to measure induction of caspase 3 and 7 activity in PSME- and tanshinone IIA-exposed LNCaP-abl cells.
PSME and Tanshinone IIA effects on AR and PSA
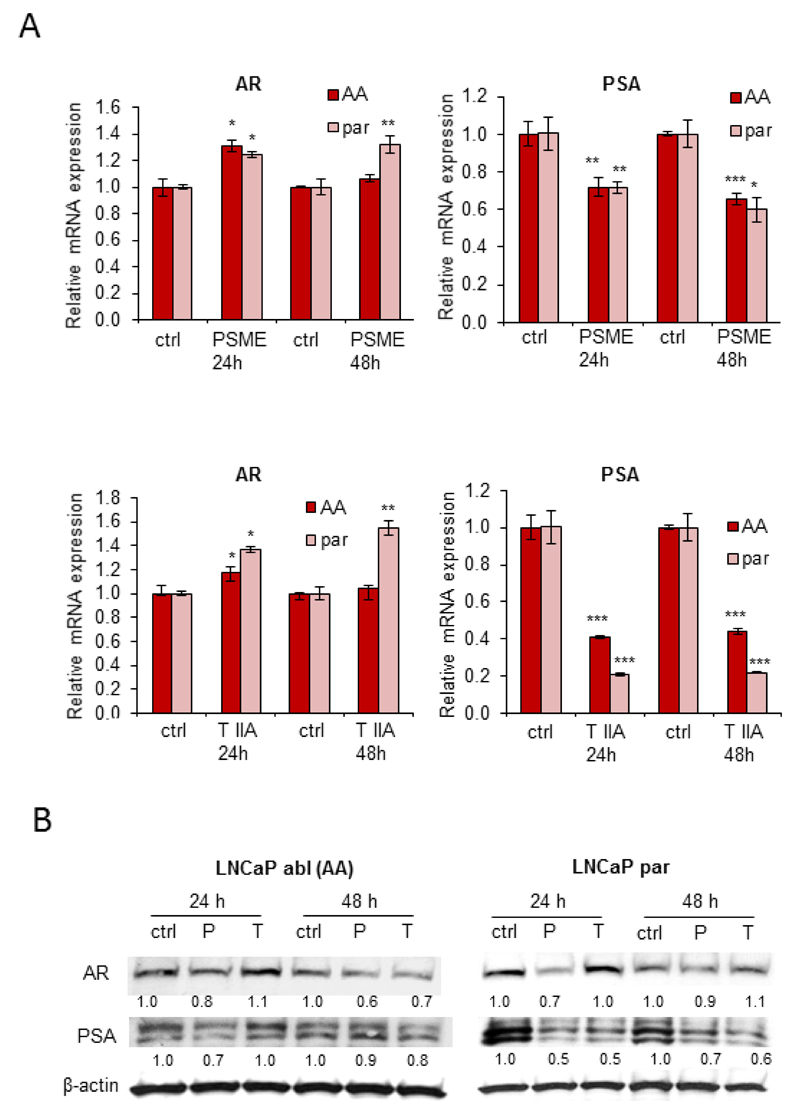
To find out whether PSME or tanshinone IIA affect AR and prostate specific antigen (PSA) expression, LNCaP-abl and LNCaP-par cells were exposed to 1 µM concentration of compounds for 24 or 48 hours. Results from quantitative RT-PCR indicated that instead of a decrease, a small increase in AR mRNA expression was seen in response to PSME and tanshinone IIA at the 24-hour time point in both LNCaP-abl and LNCaP-par cells (30% and 18% increase in LNCaP-abl cells, p < 0.05 and 22% and 38% increase in LNCaP-par cells, p < 0.05, respectively). At the 48-hour time point, PSME induced a statistically significant increase in AR mRNA expression in LNCaP-par cells (30% increase, p<0.01) whereas in LNCaP-abl cells, AR mRNA expression was not altered compared to vehicle control. A similar change was observed in AR mRNA expression in response to 48-hour tanshinone IIA exposure in LNCaP-par cells (55% increase, p<0.01). Surprisingly, however, a significant decrease in PSA mRNA expression in response to PSME and tanshinone IIA was seen in both LNCaP-abl and LNCaP-par cells at both 24-hour and 48-hour time points; PSME reduced PSA mRNA expression by 35% and 40% and tanshinone IIA by 60 % and 80 % at both time points in LNCaP-abl and LNCaP-par cells, respectively (Figure 4A).
Figure 4.
Analysis of PSME- and tanshinone IIA-induced effects on androgen receptor (AR) and prostate specific antigen (PSA) expression in LNCaP cells grown in androgen ablation (AA) or in presence of androgens (par). (A) Analysis of AR and PSA mRNA expression by quantitative PCR. (B) Analysis of AR and PSA protein expression using Western blot analysis. β-actin was used as an endogenous control.
To find out whether PSME and tanshinone IIA induce similar alterations in AR and PSA expression at protein level, protein lysates were prepared and ran on SDS-PAGE gels followed by Western blot analysis. The results indicated that PSME reduced both AR and PSA protein expression in both LNCaP-abl and LNCaP-par cells at the 48-hour time point (Figure 4B) whereas tanshinone IIA-reduced AR protein expression was only seen after a 48-hour exposure in LNCaP-abl cells. Interestingly, the PSME-induced decrease in AR expression was stronger in LNCaP–abl cells as compared to the LNCaP-par cells, indicating that differences in AR protein expression may at least partly explain the PSME-induced increase in sensitivity seen in LNCaP-abl cells, compared to the sensitivity in LNCaP-par cells. Results from tanshinone IIA-exposed LNCaP cells further support this hypothesis as a decrease in AR protein expression was only seen in tanshinone IIA-sensitive LNCaP-abl cells at the 48 hour timepoint.
PSME and tanshinone IIA alter the expression of genes involved in multiple cellular processes
To gain further insights into PSME and tanshinone IIA induced alterations in prostate cancer cells and to better understand the differential sensitivity of LNCaP-abl and LNCaP- par cells to the compounds, genome-wide gene expression analysis of PMSE and tanshinone IIA-exposed LNCaP-abl and LNCaP-par cells was performed. The gene expression in PSME and tanshinone IIA vs. DMSO- (vehicle control) exposed cells (24 hours) were compared and the most significantly altered biological processes were determined using the Ingenuity Pathway Analysis software. The analysis of PSME and tanshinone IIA modulated pathways indicated that the most differentially expressed genes in response to PSME or tanshinone IIA were involved in steroid and cholesterol metabolism (p-values of 1,10E-06 to 1,64E-03) as well as cell cycle regulation and stress response pathways (p-values of 1.81E-09 to 2.79E-04) (Supplementary Table 3). Moreover, cellular processes such as cellular movement and development were altered both in response to PSME and tanshinone IIA, indicating that these compounds may contribute to multiple biological processes in addition to cell viability and apoptosis.
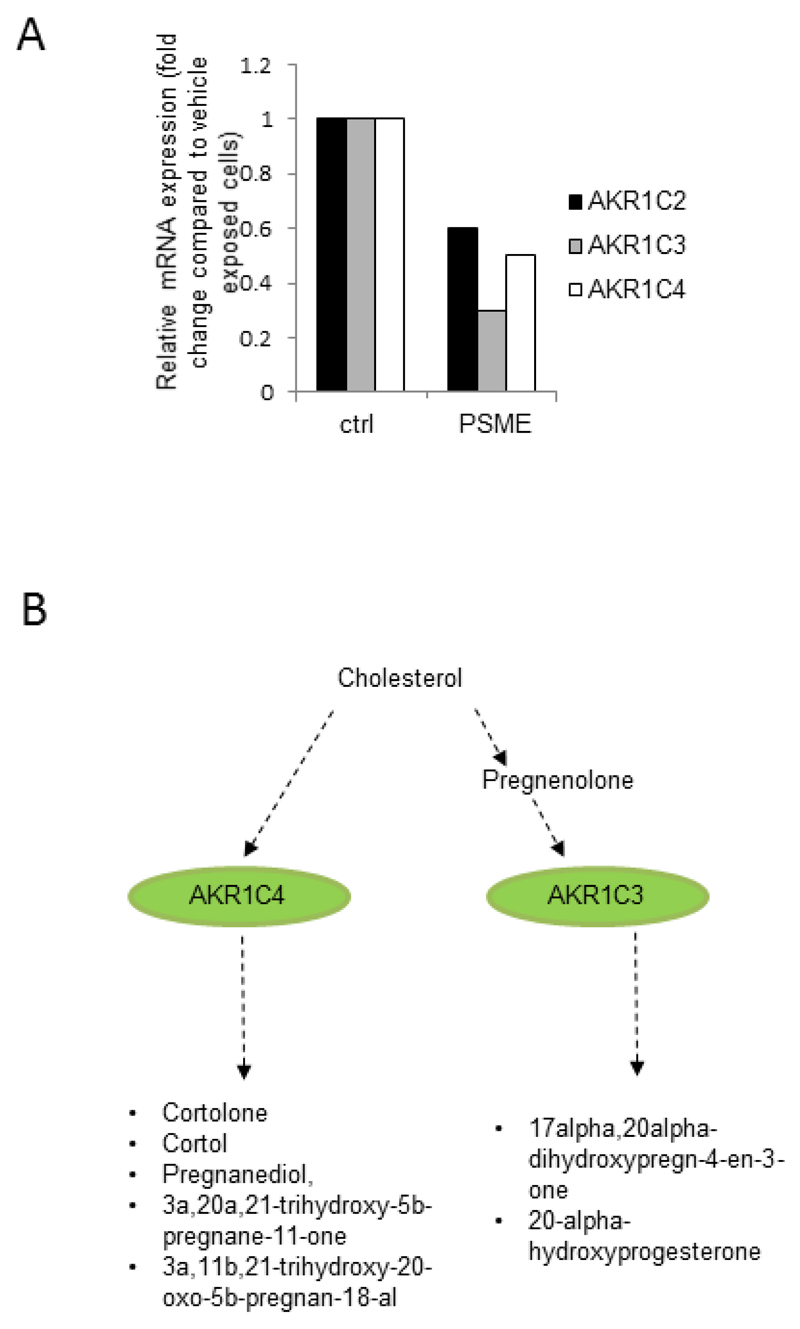
Our results from AR mRNA expression analysis indicated that AR mRNA levels did not decrease in response to PSME or tanshinone IIA exposure in LNCaP cells, indicating that the decrease in AR protein expression is likely to be controlled by other means than by direct regulation of AR transcription. Also, there was no clear relationship between AR and PSA protein levels in response to PSME and tanshinone IIA exposure, indicating that AR signalling measured by PSA does not directly correlate with AR protein expression. To get clues on putative modulators of AR signalling, we focussed our analysis on lipid metabolism-related changes in PSME- and tanshinone IIA-exposed LNCaP-abl cells. Indeed, lipid metabolism related changes were identified in both tanshinone IIA- and PSME-exposed cells, as evidenced by altered expression of aldo-keto reductases AKR1C2, AKR1C3 and AKR1C4 in response to PSME, and Methylsterol Monooxygenase 1 (MSMO1/ SC4MOL), Squalene Monooxygenase (SQLE), ATP-Binding Cassette Sub-Family G Member 1 (ABCG1) and proprotein convertase subtilisin kexin 9 (PCSK9) in response to tanshinone IIA in LNCaP-abl cells. Interestingly, similar results were not seen in LNCaP-par cells where most enriched pathways included cell cycle regulation and stress pathways (Supplementary Table 3). Previous studies have indicated that MSMO1 and SQLE are involved in cholesterol metabolism and also ABCG1 expression is regulated by cholesterol and plays an important role in macrophage lipid homeostasis (Kennedy et al. 2005, Out et al. 2008, Souchek et al. 2014, Vaughan & Oram 2005). PCSK9 is a recently discovered protein, whose high activity results in hypercholesterolemia and has been suggested as a novel drug target for invasive cancers (Seidah 2013). PSME-induced changes in lipid metabolism were linked to reduced aldo-keto reductase expression (AKR1C2, AKR1C3 and AKR1C4) in LNCaP-abl cells (Figure 5A). Aldo-keto reductases are enzymes involved in steroidogenesis and catalyze the conversion of aldehydes and ketones to their corresponding alcohols by utilizing NADH and/or NADPH as cofactors (Figure 5B). Aldo-keto reductases also regulate testosterone levels in prostate cancer (Penning & Byrns 2009) and aldo-keto reductase AKR1C3 is a known cofactor of the androgen receptor (AR) and inducer of AR signaling in prostate cancer (Yepuru et al. 2013). Taken together, analysis of tanshinone IIA- and PSME-induced changes in LNCaP-abl cells using genome-wide gene expression profiling revealed alterations in multiple biological processes and provided insights into tanshinone IIA- and PSME-induced changes in steroido-genesis.
Figure 5.
Analysis of aldo-keto reductase (AKCR) mRNA expression and steroidogenesis pathway alterations in PSME-exposed LNCaP abl cells. (A) Gene expression alterations in aldo-ketoreductase AKR1C2-4 mRNA levels in LNCaP-abl cells. Relative AKR1C2, AKR1C3 and AKR1C4 mRNA expression is presented as fold change to vehicle exposed cells. (B) AKRC enzymes in steroid hormone metabolism pathway. Green: downregulation of AKR1C3 and AKR1C4 gene expression.
Discussion
Prostate cancer remains the most common cancer in the western male population. Although several novel drugs improving prostate cancer patient survival have been identified, there is no cure for prostate cancer patients suffering from a castration-resistant disease. Here, we utilized a cell-based high-throughput chemical biology screen to identify anti-proliferative compounds for castration-resistant prostate cancer (CRPC) and to explore the key targetable molecular pathways in CRPC. As a model, we used parental (par) and androgen ablated (abl) LNCaP cells and compared drug effects into those in non-malignant RWPE-1 and EP156T prostate cells screened previously (Iljin et al. 2009). A total of 44 cancer-selective anti-proliferative hits were identified in LNCaP-abl cells and out of these, 12 compounds were found to be already, either in clinical use, in clinical trials or preclinical studies in prostate cancer, validating the functionality of our screen. Seven most potent anti-proliferative compounds were selected for further validation and two of these, pinosylvin methyl ether (PSME) and tanshinone IIA, were identified as potent inhibitors of LNCaP-abl cell viability.
Tanshinone IIA is a compound originally extracted from Danshen (Radix Salviae Miltiorrhizae). It has been used in traditional Chinese medicine for cardiovascular diseases. In addition to our results in prostate cancer cells, Tanshinone IIA has been shown to have anti-neoplastic potential in leukemic, breast, colon and hepatocellular carcinoma cells (Cheng & Su 2010, Chiu & Su 2010, Jiao & Wen 2011, Su & Lin 2008, Wang et al. 2005). Recently, tanshinone IIA was also shown to reduce prostate cancer cell growth in PC-3 and LNCaP cells both in vitro and in vivo (Won et al. 2010, Won et al. 2012, Zhang et al. 2012). Our results indicate that tanshinone IIA inhibits also androgen ablated LNCaP cell proliferation at nanomolar concentrations, suggesting that this compound may also have therapeutic potential in CRPC.
Pinosylvin methyl ether (PSME) is a stilbene compound that is isolated from green alder (Bryant et al. 1983). It has a similar structure to resveratrol, a widely studied antineoplastic compound studied in different cancers (ElAttar & Virji 1999, Hsieh & Wu 1999, Jang et al. 1997, Jang & Pezzuto 1999, Lu & Serrero 1999). However, although resveratrol was among the screened compounds, it did not come up as an anti-proliferative hit in our screen. To our knowledge PSME has not been previously studied nor proposed as an anti-proliferative compound in any cancer.
Importantly, both tanshinone IIA and PSME decreased AR protein expression as well as PSA mRNA and protein expression in LNCaP-abl cells. Genome-wide gene expression analyses indicated that in addition to cell cycle regulation, both tanshinone IIA and PSME may modulate multiple biological processes such as the steroid / cholesterol metabolism. Interestingly, the expression of aldo-keto reductases AKR1C2, AKR1C3 and AKR1C4 was decreased in castration-resistant LNCaP cells in response to PSME exposure. Aldo-keto reductases induce steroidogenesis and AKR1C3 is a biomarker and therapeutic target for CRPC (Adeniji et al. 2013, Hamid et al. 2012). Recently, other pinosylvin derivatives have been proposed to be potential AKR1C3 inhibitors, in a virtual screening of a fragment library (Brozic et al. 2012). This further supports our hypothesis of PSME as a novel AKR1C3 inhibitor. Thus, PSME is of interest while developing novel treatment options for CRPC targeting AKR1C3-induced AR signalling.
Taken together, this systematic screen identified a novel anti-proliferative agent, PSME, for CRPC. PSME reduces androgen signaling and intracellular steroidogenesis evidenced by downregulation of AR and PSA protein levels and aldo-keto reductase expression in CRPC cells in vitro. Moreover, our screen confirmed tanshinone IIA as well as several other compounds as potential prostate cancer growth inhibitors in androgen ablated prostate cancer cells. These results provide interesting starting points for the design of preclinical and clinical approaches for CRPC treatment.
Supplementary Material
Acknowledgements
We thank Finnish DNA Microarray Centre for running the Illumina experiments. This research was supported by Marie Curie Canceromics (MEXT-CT-2003-2728), EU-PRIMA project (contract # LSHC-CT-204-504587), EU-GENICA project (FP7-HEALTH-2007-A), Academy of Finland and the Cancer Organizations of Finland and Sigrid Juselius stiftelse.
Footnotes
Competing interests
None
Authors’ contributions
KK carried out the high-throughput screening, designed and prepared experiments, analysed gene expression and pathway data and prepared the manuscript. MV prepared qPCR and western blot analyses. PK normalized and participated in analysing the high-throughput screening data. VF normalized and participated in analysing the gene expression data. ZC, KI and OK participated in the study design, coordination and revising the manuscript. All authors read and approved the final manuscript.
References
- Adeniji AO, Chen M, Penning TM. AKR1C3 as a target in castrate resistant prostate cancer. J Steroid Biochem Mol Biol. 2013;137:136–149. doi: 10.1016/j.jsbmb.2013.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aragon-Ching JB. Enzalutamide (formerly MDV3100) as a new therapeutic option for men with metastatic castration-resistant prostate cancer. Asian J Androl. 2012;14:805–806. doi: 10.1038/aja.2012.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barliya T, Mandel M, Livnat T, Weinberger D, Lavie G. Degradation of HIF-1alpha under hypoxia combined with induction of Hsp90 polyubiquitination in cancer cells by hypericin: a unique cancer therapy. PloS one. 2011;6:e22849. doi: 10.1371/journal.pone.0022849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett BL, Sasaki DT, Murray BW, O'Leary EC, Sakata ST, Xu W, Leisten JC, Motiwala A, Pierce S, Satoh Y, Bhagwat SS, et al. SP600125, an anthrapyrazolone inhibitor of Jun N-terminal kinase. Proc Natl Acad Sci U S A. 2001;98:13681–13686. doi: 10.1073/pnas.251194298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berruti A, Generali D, Tampellini M. Enzalu-tamide in prostate cancer after chemotherapy. N Engl J Med. 2012;367:2448. doi: 10.1056/NEJMc1212940. [DOI] [PubMed] [Google Scholar]
- Bjorkman M, Iljin K, Halonen P, Sara H, Kaivanto E, Nees M, Kallioniemi OP. Defining the molecular action of HDAC inhibitors and synergism with androgen deprivation in ERG-positive prostate cancer. Int J Cancer. 2008;123:2774–2781. doi: 10.1002/ijc.23885. [DOI] [PubMed] [Google Scholar]
- Brožič P, Turk S, Adeniji AO, Konc J, Janežič D, Penning TM, Lanišnik Rižner T, Gobec S. Selective inhibitors of aldo-keto reductases AKR1C1 and AKR1C3 discovered by virtual screening of a fragment library. J Med Chem. 2012;55:7417–7424. doi: 10.1021/jm300841n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant JP, Wieland GD, Reichardt PB, Lewis VE, McCarthy MC. Pinosylvin methyl ether deters snowshoe hare feeding on green alder. Science. 1983;222:1023–1025. doi: 10.1126/science.222.4627.1023. [DOI] [PubMed] [Google Scholar]
- Cao MR, Li Q, Liu ZL, Liu HH, Wang W, Liao XL, Pan YL, Jiang JW. Harmine induces apoptosis in HepG2 cells via mitochondrial signaling pathway. Hepatobiliary Pancreat Dis Int. 2011;10:599–604. doi: 10.1016/s1499-3872(11)60102-1. [DOI] [PubMed] [Google Scholar]
- Chen Q, Chao R, Chen H, Hou X, Yan H, Zhou S, Peng W, Xu A. Antitumor and neurotoxic effects of novel harmine derivatives and structure activity relationship analysis. Int J Cancer. 2005;114:675–682. doi: 10.1002/ijc.20703. [DOI] [PubMed] [Google Scholar]
- Cheng CY, Su CC. Tanshinone IIA may inhibit the growth of small cell lung cancer H146 cells by up-regulating the Bax/Bcl-2 ratio and decreasing mitochondrial membrane potential. Mol Med Rep. 2010;3:645–650. doi: 10.3892/mmr_00000310. [DOI] [PubMed] [Google Scholar]
- Chiu TL, Su CC. Tanshinone IIA induces apoptosis in human lung cancer A549 cells through the induction of reactive oxygen species and decreasing the mitochondrial membrane potential. Int J Mol Med. 2010;25:231–236. [PubMed] [Google Scholar]
- Colasanti A, Kisslinger A, Liuzzi R, Quarto M, Riccio P, Roberti G, Tramontano D, Villani F. Hypericin photosensitization of tumor and metastatic cell lines of human prostate. J Photochem Photobiol B. 2000;54:103–107. doi: 10.1016/s1011-1344(99)00149-9. [DOI] [PubMed] [Google Scholar]
- Culig Z, Hoffmann J, Erdel M, Eder IE, Hobisch A, Hittmair A, Bartsch G, Utermann G, Schneider MR, Parczyk K, Klocker H. Switch from antagonist to agonist of the androgen receptor bicalutamide is associated with prostate tumour progression in a new model system. Br J Cancer. 1999;81:242–251. doi: 10.1038/sj.bjc.6690684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai F, Chen Y, Song Y, Huang L, Zhai D, Dong Y, Lai L, Zhang T, Li D, Pang X, Liu M, et al. A natural small molecule harmine inhibits angiogenesis and suppresses tumour growth through activation of p53 in endothelial cells. PLoS One. 2012;7:e52162. doi: 10.1371/journal.pone.0052162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhani NC, Emmenegger U, Adams L, Jongstra J, Tannock IF, Sridhar SS, Knox JJ, Day JR, Groskopf J, Joshua AM. Phase II study of cytarabine in men with docetaxel-refractory, castration-resistant prostate cancer with evaluation of TMPRSS2-ERG and SPINK1 as serum biomarkers. BJU Int. 2012;110:840–845. doi: 10.1111/j.1464-410X.2011.10922.x. [DOI] [PubMed] [Google Scholar]
- Dhingra R, Sharma T, Singh S, Sharma S, Tomar P, Malhotra M, Bhardwaj TR. Enzalutamide: A novel anti-androgen with prolonged survival rate in CRPC patients. Mini Rev Med Chem. 2013;13:1475–1486. doi: 10.2174/13895575113139990003. [DOI] [PubMed] [Google Scholar]
- ElAttar TM, Virji AS. Modulating effect of resveratrol and quercetin on oral cancer cell growth and proliferation. Anticancer Drugs. 1999;10:187–193. doi: 10.1097/00001813-199902000-00007. [DOI] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamid AR, Pfeiffer MJ, Verhaegh GW, Schaafsma E, Brandt A, Sweep FC, Sedelaar JP, Schalken JA. Aldo-keto reductase family 1 member C3 (AKR1C3) is a biomarker and therapeutic target for castration-resistant prostate cancer. Mol Med. 2013;18:1449–1455. doi: 10.2119/molmed.2012.00296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamsa TP, Kuttan G. Harmine inhibits tumour specific neo-vessel formation by regulating VEGF, MMP, TIMP and pro-inflammatory mediators both in vivo and in vitro. Eur J Pharmacol. 2010;649:64–73. doi: 10.1016/j.ejphar.2010.09.010. [DOI] [PubMed] [Google Scholar]
- He T, Haapa-Paananen S, Kaminskyy VO, Kohonen P, Fey V, Zhivotovsky B, Kallioniemi O, Perälä M. Inhibition of the mitochondrial pyrimidine bio-synthesis enzyme dihydroorotate dehydrogenase by doxorubicin and brequinar sensitizes cancer cells to TRAIL-induced apoptosis. Oncogene. 2014;33:3538–3549. doi: 10.1038/onc.2013.313. [DOI] [PubMed] [Google Scholar]
- Hsieh TC, Wu JM. Differential effects on growth, cell cycle arrest, and induction of apoptosis by resveratrol in human prostate cancer cell lines. Exp Cell Res. 1999;249:109–115. doi: 10.1006/excr.1999.4471. [DOI] [PubMed] [Google Scholar]
- Iljin K, Ketola K, Vainio P, Halonen P, Kohonen P, Fey V, Grafström RC, Perälä M, Kallioniemi O. High-throughput cell-based screening of 4910 known drugs and drug-like small molecules identifies disulfiram as an inhibitor of prostate cancer cell growth. Clin Cancer Res. 2009;15:6070–6078. doi: 10.1158/1078-0432.CCR-09-1035. [DOI] [PubMed] [Google Scholar]
- Jang M, Cai L, Udeani GO, Slowing KV, Thomas CF, Beecher CW, Fong HH, Farnsworth NR, Kinghorn AD, Mehta RG, Moon RC, et al. Cancer chemopreventive activity of resveratrol, a natural product derived from grapes. Science. 1997;275:218–220. doi: 10.1126/science.275.5297.218. [DOI] [PubMed] [Google Scholar]
- Jang M, Pezzuto JM. Cancer chemopreventive activity of resveratrol. Drugs Exp Clin Res. 1999;25:65–77. [PubMed] [Google Scholar]
- Jiao JW, Wen F. Tanshinone IIA acts via p38 MAPK to induce apoptosis and the down-regulation of ERCC1 and lung-resistance protein in cisplatinresistant ovarian cancer cells. Oncol Rep. 2011;25:781–788. doi: 10.3892/or.2010.1107. [DOI] [PubMed] [Google Scholar]
- Kennedy MA, Barrera GC, Nakamura K, Baldán A, Tarr P, Fishbein MC, Frank J, Francone OL, Edwards PA. ABCG1 has a critical role in mediating cholesterol efflux to HDL and preventing cellular lipid accumulation. Cell Metab. 2005;1:121–131. doi: 10.1016/j.cmet.2005.01.002. [DOI] [PubMed] [Google Scholar]
- Li Y, Sattler R, Yang EJ, Nunes A, Ayukawa Y, Akhtar S, Ji G, Zhang PW, Rothstein JD. Harmine, a natural beta-carboline alkaloid, upregulates astroglial glutamate transporter expression. Neuropharmacology. 2011;60:1168–1175. doi: 10.1016/j.neuropharm.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Adam RM, Santiestevan E, Freeman MR. The phosphatidylinositol 3'-kinase pathway is a dominant growth factor-activated cell survival pathway in LNCaP human prostate carcinoma cells. Cancer Res. 1999;59:2891–2897. [PubMed] [Google Scholar]
- Locke JA, Guns ES, Lubik AA, Adomat HH, Hendy SC, Wood CA, Ettinger SL, Gleave ME, Nelson CC. Androgen levels increase by intratumoral de novo steroidogenesis during progression of castrationresistant prostate cancer. Cancer Res. 2008;68:6407–6415. doi: 10.1158/0008-5472.CAN-07-5997. [DOI] [PubMed] [Google Scholar]
- Lu R, Serrero G. Resveratrol, a natural product derived from grape, exhibits antiestrogenic activity and inhibits the growth of human breast cancer cells. J Cell Physiol. 1999;179:297–304. doi: 10.1002/(SICI)1097-4652(199906)179:3<297::AID-JCP7>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Luo C, Laaja P. Inhibitors of JAKs/STATs and the kinases: a possible new cluster of drugs. Drug Discov Today. 2004;9:268–275. doi: 10.1016/S1359-6446(03)03014-9. [DOI] [PubMed] [Google Scholar]
- Morgan WA. Naphthoquinone-induced DNA damage in the absence of oxidative stress. Biochem Soc Trans. 1995;23:225S. doi: 10.1042/bst023225s. [DOI] [PubMed] [Google Scholar]
- Out R, Hoekstra M, Habets K, Meurs I, de Waard V, Hildebrand RB, Wang Y, Chimini G, Kuiper J, Van Berkel TJ, Van Eck M. Combined deletion of macrophage ABCA1 and ABCG1 leads to massive lipid accumulation in tissue macrophages and distinct atherosclerosis at relatively low plasma cholesterol levels. Arterioscler Thromb Vasc Biol. 2008;28:258–264. doi: 10.1161/ATVBAHA.107.156935. [DOI] [PubMed] [Google Scholar]
- Penning TM, Byrns MC. Steroid hormone transforming aldo-keto reductases and cancer. Ann N Y Acad Sci. 2009;1155:33–42. doi: 10.1111/j.1749-6632.2009.03700.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader AJ, Boegemann M, Ohlmann CH, Schnoeller TJ, Krabbe LM, Hajili T, Jentzmik F, Stoeckle M, Schrader M, Herrmann E, Cronauer MV. Enzalutamide in castration-resistant prostate cancer patients progressing after docetaxel and abiraterone. Eur Urol. 2014;65:30–36. doi: 10.1016/j.eururo.2013.06.042. [DOI] [PubMed] [Google Scholar]
- Seidah NG. Proprotein convertase subtilisin kexin 9 (PCSK9) inhibitors in the treatment of hypercholesterolemia and other pathologies. Curr Pharm Des. 2013;19:3161–3172. doi: 10.2174/13816128113199990313. [DOI] [PubMed] [Google Scholar]
- Souchek JJ, Baine MJ, Lin C, Rachagani S, Gupta S, Kaur S, Lester K, Zheng D, Chen S, Smith L, Lazenby A, et al. Unbiased analysis of pancreatic cancer radiation resistance reveals cholesterol biosynthesis as a novel target for radiosensitisation. Br J Cancer. 2014;111:1139–1149. doi: 10.1038/bjc.2014.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele VE, Holmes CA, Hawk ET, Kopelovich L, Lubet RA, Crowell JA, Sigman CC, Kelloff GJ. Lipoxygenase inhibitors as potential cancer chemopreventives. Cancer Epidemiol Biomarkers Prev. 1999;8:467–483. [PubMed] [Google Scholar]
- Straus MJ, Fleit JP, Engelking C. Treatment of advanced prostate cancer with cyclophosphamide, doxorubicin, and methotrexate. Cancer Treat Rep. 1982;66:1797–1802. [PubMed] [Google Scholar]
- Su CC, Lin YH. Tanshinone IIA down-regulates the protein expression of ErbB-2 and up-regulates TNF-alpha in colon cancer cells in vitro and in vivo. Int J Mol Med. 2008;22:847–851. [PubMed] [Google Scholar]
- Taichman RS, Loberg RD, Mehra R, Pienta KJ. The evolving biology and treatment of prostate cancer. J Clin Invest. 2007;117:2351–2361. doi: 10.1172/JCI31791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tannock IF, de Wit R, Berry WR, Horti J, Pluzanska A, Chi KN, Oudard S, Théodore C, James ND, Turesson I, Rosenthal MA, et al. Docetaxel plus prednisone or mitoxantrone plus prednisone for advanced prostate cancer. N Engl J Med. 2004;351:1502–1512. doi: 10.1056/NEJMoa040720. [DOI] [PubMed] [Google Scholar]
- Tyagi A, Agarwal R, Agarwal C. Grape seed extract inhibits EGF-induced and constitutively active mitogenic signaling but activates JNK in human prostate carcinoma DU145 cells: possible role in antiproliferation and apoptosis. Oncogene. 2003;22:1302–1316. doi: 10.1038/sj.onc.1206265. [DOI] [PubMed] [Google Scholar]
- Vaughan AM, Oram JF. ABCG1 redistributes cell cholesterol to domains removable by high density lipoprotein but not by lipid-depleted apolipoproteins. J Biol Chem. 2005;280:30150–30157. doi: 10.1074/jbc.M505368200. [DOI] [PubMed] [Google Scholar]
- Wang X, Wei Y, Yuan S, Liu G, Lu Y, Zhang J, Wang W. Potential anticancer activity of tanshinone IIA against human breast cancer. Int J Cancer. 2005;116:799–807. doi: 10.1002/ijc.20880. [DOI] [PubMed] [Google Scholar]
- Won SH, Lee HJ, Jeong SJ, Lee HJ, Lee EO, Jung DB, Shin JM, Kwon TR, Yun SM, Lee MH, Choi SH, et al. Tanshinone IIA induces mitochondria dependent apoptosis in prostate cancer cells in association with an inhibition of phosphoinositide 3-kinase/AKT pathway. Biol Pharm Bull. 2010;33:1828–1834. doi: 10.1248/bpb.33.1828. [DOI] [PubMed] [Google Scholar]
- Won SH, Lee HJ, Jeong SJ, Lu J, Kim SH. Activation of p53 signaling and inhibition of androgen receptor mediate tanshinone IIA induced G1 arrest in LNCaP prostate cancer cells. Phytother Res. 2012;26:669–674. doi: 10.1002/ptr.3616. [DOI] [PubMed] [Google Scholar]
- Xie X, Hudson JB, Guns ES. Tumor-specific and photodependent cytotoxicity of hypericin in the human LNCaP prostate tumor model. Photochem Photobiol. 2001;74:221–225. doi: 10.1562/0031-8655(2001)074<0221:tsapco>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Yepuru M, Wu Z, Kulkarni A, Yin F, Barrett CM, Kim J, Steiner MS, Miller DD, Dalton JT, Narayanan R. Steroidogenic enzyme AKR1C3 is a novel androgen receptor-selective coactivator that promotes prostate cancer growth. Clin Cancer Res. 2013;19:5613–5625. doi: 10.1158/1078-0432.CCR-13-1151. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Won SH, Jiang C, Lee HJ, Jeong SJ, Lee EO, Zhang J, Ye M, Kim SH, Lü J. Tanshinones from Chinese medicinal herb Danshen (Salvia miltiorrhiza Bunge) suppress prostate cancer growth and androgen receptor signaling. Pharm Res. 2012;29:1595–1608. doi: 10.1007/s11095-012-0670-3. [DOI] [PubMed] [Google Scholar]
- Zhu ML, Horbinski CM, Garzotto M, Qian DZ, Beer TM, Kyprianou N. Tubulin-targeting chemotherapy impairs androgen receptor activity in prostate cancer. Cancer Res. 2010;70:7992–8002. doi: 10.1158/0008-5472.CAN-10-0585. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.