The title 8-hydroxyquinoline derivative has an E conformation about the C=C bond, and the quinoline ring system and the benzene ring are inclined to one another by 29.22 (7)°.
Keywords: crystal structure, 8-hydroxyquinoline derivative, vinyl, inversion dimer, hydrogen bonding, inversion dimers, C—H⋯π interactions
Abstract
The title compound, C19H15NO3, was synthesized by a Perkin reaction of 2-methyl-8-hydroxyquinoline and 4-formyl-2-methylbenzoate in acetic anhydride under a nitrogen atmosphere. The molecule has an E conformation about the C=C bond, and the quinoline ring system and the benzene ring are inclined to one another by 29.22 (7)°. There is an intramolecular O—H⋯N hydrogen bond in the 8-hydroxyquinoline moiety. In the crystal, molecules are linked by pairs of O—H⋯O hydrogen bonds, forming inversion dimers with an R 2 2(28) ring motif. The dimers are linked by C—H⋯O hydrogen bonds and C—H⋯π interactions, forming sheets parallel to plane (10-1).
Chemical context
In recent years, 8-hydroxyquinoline and its derivatives have played an important role in coordination chemistry (Albrecht et al., 2008 ▸; Cacciatore et al., 2013 ▸), shown to exhibit biological activity (du Moulinet d’Hardemare et al., 2012 ▸) and have found various applications in the fields of synthetic chemistry (Song et al., 2006 ▸) and organic light-emitting diodes, which have been extensively exploited in the synthesis of luminescent metal complexes (Tang et al., 1987 ▸). It is therefore highly desirable to develop new efficient 8-hydroxyquinoline derivatives for use in luminescent metal complexes. In the present work, we report on the synthesis and crystal structure of a new 8-hydroxyquinoline derivative, synthesized by the Perkin reaction of 2-methyl-8-hydroxyquinoline and 4-formyl-2-methylbenzoate.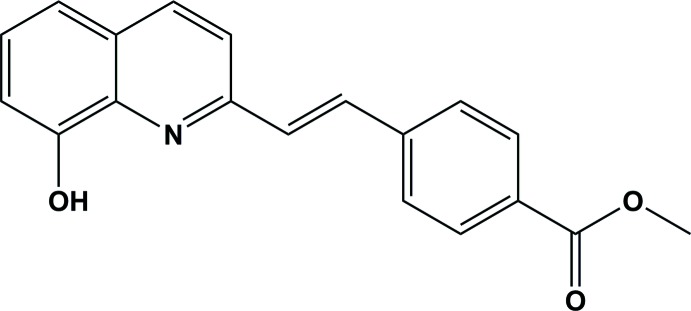
Structural commentary
The molecular structure of the title compound is shown in Fig. 1 ▸. It contains an 8-hydroxyquinoline moiety, with an intramolecular O—H⋯N hydrogen bond (Fig. 1 ▸ and Table 1 ▸), and a methylbenzoate unit. They are linked by the C9=C10 bond [1.321 (2) Å] with an E conformation. The C11—C10 and C6—C9 bond lengths are 1.463 (2) and 1.466 (2) Å, respectively. These distances are shorter than the standard length of a C—C single bond (ca 1.5 Å) because of the conjugate system formed by the C9=C10 bond and the aromatic systems. The quinoline ring system and the benzene ring are inclined to one another by 29.22 (7)°.
Figure 1.
View of the molecular structure of the title compound, showing the atom labelling and 40% probability displacement ellipsoids. The intramolecular O—H⋯N hydrogen bond is shown as a dashed line (see Table 1 ▸).
Table 1. Hydrogen-bond geometry (Å, °).
Cg1, Cg2 and Cg3 are the centroids of rings N1/C11–C14/C19, C3–C8 and C14–C19, respectively.
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O3—H3O⋯N1 | 0.86 (2) | 2.19 (3) | 2.715 (2) | 120 (2) |
| O3—H3O⋯O1i | 0.86 (2) | 2.23 (2) | 2.901 (2) | 136 (2) |
| C5—H5A⋯O3ii | 0.93 | 2.57 | 3.437 (2) | 155 |
| C7—H7A⋯Cg3iii | 0.93 | 2.99 | 3.605 (2) | 125 |
| C8—H8A⋯Cg1iii | 0.93 | 2.93 | 3.559 (2) | 126 |
| C15—H15A⋯Cg2ii | 0.93 | 2.83 | 3.639 (2) | 146 |
Symmetry codes: (i)  ; (ii)
; (ii)  ; (iii)
; (iii)  .
.
Supramolecular features
In the crystal, molecules are linked by pairs of O—H⋯O hydrogen bonds, forming inversion dimers with an  (28) ring motif (Table 1 ▸ and Fig. 2 ▸). The dimers are linked by C—H⋯O hydrogen bonds and C—H⋯π interactions, forming sheets parallel to (10
(28) ring motif (Table 1 ▸ and Fig. 2 ▸). The dimers are linked by C—H⋯O hydrogen bonds and C—H⋯π interactions, forming sheets parallel to (10 ); see Table 1 ▸ and Fig. 3 ▸.
); see Table 1 ▸ and Fig. 3 ▸.
Figure 2.
A view along the a axis of the  (28) ring motifs in the crystal of the title compound. Hydrogen bonds are shown as dashed lines (see Table 1 ▸), and for clarity only H atoms H3O and H5A are included.
(28) ring motifs in the crystal of the title compound. Hydrogen bonds are shown as dashed lines (see Table 1 ▸), and for clarity only H atoms H3O and H5A are included.
Figure 3.
A view along the b axis of the crystal packing of the title compound. Hydrogen bonds are shown as dashed lines (see Table 1 ▸) and, for clarity, only H atoms H3O and H5A are included.
Database survey
A search of the Cambridge Structural Database (CSD, Version 5.37, last update May 2016; Groom et al., 2016 ▸) for the substructure 2-styrylquinolin-8-ol gave 17 hits; however, certain of these involve bis(8-hydroxyquinolines) or a (9-anthryl) moiety. Three compounds are similar to the title compound in the sense that they also have an E conformation about the C=C bond, and in the crystal they also form inversion dimers. They include 2-{2-[4-(trifluoromethyl)phenyl]vinyl}quinolin-8-ol (HUKTOY; Huo et al., 2015 ▸), 2-[2-(4-methoxyphenyl)vinyl]quinolin-8-ol (MIMPOP; Yuan et al., 2013 ▸), and 2-[2-(2,4-dinitrophenyl)vinyl]quinolin-8-ol (WELKEF; Yuan et al., 2013 ▸). In these three compounds, the quinoline and benzene rings are inclined to one another by 36.72 (10) and 16.66 (10)° in HUKTOY (there are two independent molecules in the asymmetric unit), 42.59 (7)° in MIMPOP and 5.63 (6)° in WELKEF, compared to 29.22 (7)° in the title compound.
Synthesis and crystallization
The title compound was prepared following reported procedures (Jing et al., 2006 ▸; Yuan et al., 2012 ▸). A mixture of 2-methy-8-hydroxyquinoline (1.59 g, 10 mmol), 4-formyl-2-methylbenzoate (1.64 g, 10 mmol) and acetic anhydride (20 ml) was stirred for 12 h at 423 K under a nitrogen atmosphere. After cooling it was poured into ice–water (150 ml) and stirred for 1–2 h. Then, the puce solid obtained was filtered and together with triethylamine (1 g, 10 mmol) was dissolved in DMF (30 ml) and the mixture stirred for 3 h at 408 K. After cooling, the reaction mixture was concentrated and purified by chromatography on silica gel (petroleum ether/EtOAc = 3/1). The product obtained was dissolved in ethanol, and on slow evaporation of the solvent yellow crystals were obtained within 2 weeks.
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 2 ▸. The hydroxy-H atom was located in a difference Fourier map and freely refined. The C-bound H atoms were positioned geometrically and allowed to ride on their parent atoms: C—H = 0.93–0.96 Å with U iso(H) = 1.5U eq(C-methyl) and 1.2U eq(C) for other H atoms.
Table 2. Experimental details.
| Crystal data | |
| Chemical formula | C19H15NO3 |
| M r | 305.32 |
| Crystal system, space group | Monoclinic, P21/n |
| Temperature (K) | 296 |
| a, b, c (Å) | 12.0236 (4), 9.7045 (4), 13.2607 (4) |
| β (°) | 96.260 (2) |
| V (Å3) | 1538.07 (9) |
| Z | 4 |
| Radiation type | Mo Kα |
| μ (mm−1) | 0.09 |
| Crystal size (mm) | 0.08 × 0.06 × 0.05 |
| Data collection | |
| Diffractometer | Bruker SMART CCD area-detector |
| Absorption correction | Multi-scan (SADABS; Bruker, 2005 ▸) |
| T min, T max | 0.993, 0.996 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 13068, 3511, 2049 |
| R int | 0.041 |
| (sin θ/λ)max (Å−1) | 0.651 |
| Refinement | |
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.046, 0.137, 1.02 |
| No. of reflections | 3511 |
| No. of parameters | 217 |
| H-atom treatment | H atoms treated by a mixture of independent and constrained refinement |
| Δρmax, Δρmin (e Å−3) | 0.17, −0.18 |
Supplementary Material
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S205698901601210X/su5312sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S205698901601210X/su5312Isup2.hkl
Supporting information file. DOI: 10.1107/S205698901601210X/su5312Isup3.mol
Supporting information file. DOI: 10.1107/S205698901601210X/su5312Isup4.cml
CCDC reference: 859030
Additional supporting information: crystallographic information; 3D view; checkCIF report
Acknowledgments
We gratefully acknowledge the support of the Natural Science Foundation of Zhejiang Province (LY12B01003).
supplementary crystallographic information
Crystal data
| C19H15NO3 | F(000) = 640 |
| Mr = 305.32 | Dx = 1.319 Mg m−3 |
| Monoclinic, P21/n | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2yn | Cell parameters from 1747 reflections |
| a = 12.0236 (4) Å | θ = 2.2–27.6° |
| b = 9.7045 (4) Å | µ = 0.09 mm−1 |
| c = 13.2607 (4) Å | T = 296 K |
| β = 96.260 (2)° | Block, yellow |
| V = 1538.07 (9) Å3 | 0.08 × 0.06 × 0.05 mm |
| Z = 4 |
Data collection
| Bruker SMART CCD area-detector diffractometer | 3511 independent reflections |
| Radiation source: fine-focus sealed tube | 2049 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.041 |
| phi and ω scans | θmax = 27.6°, θmin = 2.2° |
| Absorption correction: multi-scan (SADABS; Bruker, 2005) | h = −15→14 |
| Tmin = 0.993, Tmax = 0.996 | k = −12→12 |
| 13068 measured reflections | l = −17→17 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.046 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.137 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.02 | w = 1/[σ2(Fo2) + (0.0631P)2 + 0.1091P] where P = (Fo2 + 2Fc2)/3 |
| 3511 reflections | (Δ/σ)max < 0.001 |
| 217 parameters | Δρmax = 0.17 e Å−3 |
| 0 restraints | Δρmin = −0.18 e Å−3 |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > 2sigma(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| N1 | 0.87623 (11) | 0.22440 (15) | 0.17162 (11) | 0.0507 (4) | |
| O3 | 1.04463 (12) | 0.41136 (15) | 0.20030 (12) | 0.0678 (4) | |
| C19 | 0.97735 (12) | 0.17971 (18) | 0.21670 (12) | 0.0465 (4) | |
| C3 | 0.26628 (13) | 0.27214 (18) | −0.01100 (13) | 0.0496 (4) | |
| C14 | 0.99928 (13) | 0.04272 (18) | 0.24806 (12) | 0.0481 (4) | |
| C11 | 0.79264 (13) | 0.13449 (18) | 0.15909 (12) | 0.0491 (4) | |
| O2 | 0.07181 (10) | 0.24711 (14) | −0.02756 (10) | 0.0691 (4) | |
| C6 | 0.47912 (13) | 0.17659 (19) | 0.06283 (12) | 0.0508 (4) | |
| C13 | 0.90819 (14) | −0.04932 (19) | 0.23276 (13) | 0.0544 (5) | |
| H13A | 0.9178 | −0.1412 | 0.2517 | 0.065* | |
| C18 | 1.06399 (13) | 0.27912 (19) | 0.23164 (13) | 0.0521 (4) | |
| C12 | 0.80665 (14) | −0.0046 (2) | 0.19061 (13) | 0.0549 (5) | |
| H12A | 0.7464 | −0.0651 | 0.1824 | 0.066* | |
| C2 | 0.15483 (14) | 0.3263 (2) | −0.05173 (14) | 0.0574 (5) | |
| O1 | 0.14069 (11) | 0.42886 (17) | −0.10263 (13) | 0.0939 (5) | |
| C10 | 0.68606 (13) | 0.18748 (19) | 0.11050 (13) | 0.0550 (5) | |
| H10A | 0.6873 | 0.2733 | 0.0795 | 0.066* | |
| C9 | 0.58827 (13) | 0.1246 (2) | 0.10668 (13) | 0.0550 (5) | |
| H9A | 0.5887 | 0.0370 | 0.1352 | 0.066* | |
| C8 | 0.36132 (14) | 0.3354 (2) | −0.03918 (13) | 0.0587 (5) | |
| H8A | 0.3541 | 0.4107 | −0.0828 | 0.070* | |
| C15 | 1.10686 (14) | 0.0073 (2) | 0.29359 (13) | 0.0581 (5) | |
| H15A | 1.1226 | −0.0831 | 0.3136 | 0.070* | |
| C17 | 1.16678 (14) | 0.2423 (2) | 0.27826 (15) | 0.0611 (5) | |
| H17A | 1.2229 | 0.3081 | 0.2901 | 0.073* | |
| C7 | 0.46638 (14) | 0.2885 (2) | −0.00345 (14) | 0.0587 (5) | |
| H7A | 0.5293 | 0.3318 | −0.0237 | 0.070* | |
| C4 | 0.27801 (14) | 0.15980 (19) | 0.05325 (14) | 0.0564 (5) | |
| H4A | 0.2148 | 0.1156 | 0.0721 | 0.068* | |
| C16 | 1.18752 (15) | 0.1057 (2) | 0.30817 (14) | 0.0641 (5) | |
| H16A | 1.2581 | 0.0816 | 0.3387 | 0.077* | |
| C5 | 0.38325 (14) | 0.11293 (19) | 0.08960 (14) | 0.0589 (5) | |
| H5A | 0.3900 | 0.0372 | 0.1329 | 0.071* | |
| C1 | −0.04060 (15) | 0.2875 (3) | −0.06672 (17) | 0.0812 (7) | |
| H1B | −0.0932 | 0.2228 | −0.0445 | 0.122* | |
| H1C | −0.0470 | 0.2886 | −0.1395 | 0.122* | |
| H1D | −0.0561 | 0.3777 | −0.0421 | 0.122* | |
| H3O | 0.9748 (17) | 0.414 (3) | 0.178 (2) | 0.101 (8) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| N1 | 0.0405 (7) | 0.0524 (9) | 0.0578 (9) | 0.0016 (7) | −0.0003 (6) | −0.0028 (7) |
| O3 | 0.0509 (8) | 0.0541 (8) | 0.0935 (10) | −0.0052 (7) | −0.0140 (8) | 0.0047 (7) |
| C19 | 0.0397 (9) | 0.0523 (10) | 0.0470 (9) | 0.0023 (8) | 0.0028 (7) | −0.0005 (8) |
| C3 | 0.0447 (9) | 0.0557 (11) | 0.0471 (9) | 0.0034 (8) | −0.0015 (7) | −0.0011 (8) |
| C14 | 0.0422 (9) | 0.0528 (11) | 0.0495 (9) | 0.0062 (8) | 0.0057 (7) | 0.0010 (8) |
| C11 | 0.0412 (9) | 0.0503 (10) | 0.0549 (10) | 0.0016 (8) | 0.0005 (7) | −0.0059 (8) |
| O2 | 0.0414 (7) | 0.0864 (10) | 0.0774 (9) | 0.0056 (7) | −0.0034 (6) | 0.0080 (7) |
| C6 | 0.0445 (9) | 0.0530 (10) | 0.0529 (10) | 0.0011 (8) | −0.0034 (7) | −0.0051 (8) |
| C13 | 0.0527 (10) | 0.0471 (10) | 0.0631 (11) | 0.0035 (8) | 0.0056 (8) | 0.0009 (8) |
| C18 | 0.0430 (9) | 0.0531 (11) | 0.0590 (10) | 0.0007 (8) | 0.0007 (8) | 0.0001 (9) |
| C12 | 0.0457 (9) | 0.0515 (11) | 0.0663 (11) | −0.0036 (8) | 0.0013 (8) | −0.0061 (9) |
| C2 | 0.0447 (10) | 0.0687 (13) | 0.0575 (11) | 0.0050 (10) | 0.0003 (8) | −0.0010 (10) |
| O1 | 0.0597 (9) | 0.0984 (12) | 0.1208 (13) | 0.0137 (8) | −0.0033 (8) | 0.0450 (11) |
| C10 | 0.0468 (10) | 0.0530 (11) | 0.0637 (11) | 0.0046 (8) | −0.0011 (8) | −0.0015 (9) |
| C9 | 0.0446 (9) | 0.0549 (11) | 0.0635 (11) | 0.0026 (8) | −0.0032 (8) | −0.0027 (9) |
| C8 | 0.0521 (10) | 0.0638 (12) | 0.0586 (11) | 0.0015 (9) | −0.0004 (8) | 0.0133 (9) |
| C15 | 0.0456 (10) | 0.0620 (12) | 0.0661 (11) | 0.0115 (9) | 0.0034 (8) | 0.0087 (9) |
| C17 | 0.0424 (10) | 0.0680 (13) | 0.0713 (12) | −0.0049 (9) | −0.0011 (8) | −0.0012 (10) |
| C7 | 0.0427 (9) | 0.0703 (13) | 0.0624 (11) | −0.0039 (9) | 0.0027 (8) | 0.0070 (10) |
| C4 | 0.0436 (9) | 0.0584 (11) | 0.0657 (11) | −0.0046 (8) | −0.0005 (8) | 0.0035 (9) |
| C16 | 0.0404 (9) | 0.0767 (15) | 0.0731 (12) | 0.0064 (9) | −0.0044 (8) | 0.0075 (11) |
| C5 | 0.0495 (10) | 0.0554 (11) | 0.0691 (12) | −0.0004 (8) | −0.0054 (8) | 0.0110 (9) |
| C1 | 0.0402 (10) | 0.1128 (19) | 0.0881 (15) | 0.0112 (11) | −0.0039 (9) | 0.0050 (13) |
| H3O | 0.039 (13) | 0.12 (2) | 0.14 (2) | −0.025 (14) | −0.010 (13) | 0.009 (16) |
Geometric parameters (Å, º)
| N1—C11 | 1.328 (2) | C18—C17 | 1.368 (2) |
| N1—C19 | 1.3657 (19) | C12—H12A | 0.9300 |
| O3—C18 | 1.361 (2) | C2—O1 | 1.204 (2) |
| O3—H3O | 0.86 (2) | C10—C9 | 1.321 (2) |
| C19—C14 | 1.409 (2) | C10—H10A | 0.9300 |
| C19—C18 | 1.418 (2) | C9—H9A | 0.9300 |
| C3—C4 | 1.381 (2) | C8—C7 | 1.377 (2) |
| C3—C8 | 1.384 (2) | C8—H8A | 0.9300 |
| C3—C2 | 1.485 (2) | C15—C16 | 1.360 (3) |
| C14—C15 | 1.409 (2) | C15—H15A | 0.9300 |
| C14—C13 | 1.411 (2) | C17—C16 | 1.398 (3) |
| C11—C12 | 1.417 (3) | C17—H17A | 0.9300 |
| C11—C10 | 1.463 (2) | C7—H7A | 0.9300 |
| O2—C2 | 1.326 (2) | C4—C5 | 1.381 (2) |
| O2—C1 | 1.448 (2) | C4—H4A | 0.9300 |
| C6—C5 | 1.388 (2) | C16—H16A | 0.9300 |
| C6—C7 | 1.395 (3) | C5—H5A | 0.9300 |
| C6—C9 | 1.466 (2) | C1—H1B | 0.9600 |
| C13—C12 | 1.357 (2) | C1—H1C | 0.9600 |
| C13—H13A | 0.9300 | C1—H1D | 0.9600 |
| C11—N1—C19 | 118.15 (15) | C11—C10—H10A | 117.0 |
| C18—O3—H3O | 105.2 (17) | C10—C9—C6 | 127.66 (18) |
| N1—C19—C14 | 123.90 (15) | C10—C9—H9A | 116.2 |
| N1—C19—C18 | 116.78 (16) | C6—C9—H9A | 116.2 |
| C14—C19—C18 | 119.32 (14) | C7—C8—C3 | 120.90 (17) |
| C4—C3—C8 | 119.02 (15) | C7—C8—H8A | 119.5 |
| C4—C3—C2 | 122.07 (17) | C3—C8—H8A | 119.5 |
| C8—C3—C2 | 118.91 (17) | C16—C15—C14 | 119.79 (18) |
| C15—C14—C19 | 119.34 (16) | C16—C15—H15A | 120.1 |
| C15—C14—C13 | 124.70 (17) | C14—C15—H15A | 120.1 |
| C19—C14—C13 | 115.94 (14) | C18—C17—C16 | 120.02 (17) |
| N1—C11—C12 | 121.65 (14) | C18—C17—H17A | 120.0 |
| N1—C11—C10 | 116.03 (16) | C16—C17—H17A | 120.0 |
| C12—C11—C10 | 122.32 (15) | C8—C7—C6 | 120.51 (17) |
| C2—O2—C1 | 117.01 (16) | C8—C7—H7A | 119.7 |
| C5—C6—C7 | 118.07 (15) | C6—C7—H7A | 119.7 |
| C5—C6—C9 | 118.56 (17) | C5—C4—C3 | 120.21 (17) |
| C7—C6—C9 | 123.36 (17) | C5—C4—H4A | 119.9 |
| C12—C13—C14 | 120.40 (17) | C3—C4—H4A | 119.9 |
| C12—C13—H13A | 119.8 | C15—C16—C17 | 121.54 (16) |
| C14—C13—H13A | 119.8 | C15—C16—H16A | 119.2 |
| O3—C18—C17 | 120.02 (17) | C17—C16—H16A | 119.2 |
| O3—C18—C19 | 120.01 (14) | C4—C5—C6 | 121.28 (17) |
| C17—C18—C19 | 119.96 (17) | C4—C5—H5A | 119.4 |
| C13—C12—C11 | 119.92 (16) | C6—C5—H5A | 119.4 |
| C13—C12—H12A | 120.0 | O2—C1—H1B | 109.5 |
| C11—C12—H12A | 120.0 | O2—C1—H1C | 109.5 |
| O1—C2—O2 | 123.39 (16) | H1B—C1—H1C | 109.5 |
| O1—C2—C3 | 124.26 (18) | O2—C1—H1D | 109.5 |
| O2—C2—C3 | 112.33 (17) | H1B—C1—H1D | 109.5 |
| C9—C10—C11 | 126.00 (18) | H1C—C1—H1D | 109.5 |
| C9—C10—H10A | 117.0 | ||
| C11—N1—C19—C14 | 2.0 (2) | C8—C3—C2—O2 | 173.08 (16) |
| C11—N1—C19—C18 | −178.23 (15) | N1—C11—C10—C9 | −167.28 (17) |
| N1—C19—C14—C15 | 179.78 (15) | C12—C11—C10—C9 | 13.1 (3) |
| C18—C19—C14—C15 | 0.0 (2) | C11—C10—C9—C6 | 177.07 (16) |
| N1—C19—C14—C13 | −1.6 (2) | C5—C6—C9—C10 | −162.92 (18) |
| C18—C19—C14—C13 | 178.58 (15) | C7—C6—C9—C10 | 16.0 (3) |
| C19—N1—C11—C12 | −0.4 (2) | C4—C3—C8—C7 | −0.5 (3) |
| C19—N1—C11—C10 | 179.96 (14) | C2—C3—C8—C7 | −179.86 (16) |
| C15—C14—C13—C12 | 178.19 (17) | C19—C14—C15—C16 | 1.0 (3) |
| C19—C14—C13—C12 | −0.3 (2) | C13—C14—C15—C16 | −177.45 (18) |
| N1—C19—C18—O3 | −0.9 (2) | O3—C18—C17—C16 | −178.39 (18) |
| C14—C19—C18—O3 | 178.95 (16) | C19—C18—C17—C16 | 2.1 (3) |
| N1—C19—C18—C17 | 178.64 (16) | C3—C8—C7—C6 | −0.6 (3) |
| C14—C19—C18—C17 | −1.6 (2) | C5—C6—C7—C8 | 1.3 (3) |
| C14—C13—C12—C11 | 1.8 (3) | C9—C6—C7—C8 | −177.65 (17) |
| N1—C11—C12—C13 | −1.5 (3) | C8—C3—C4—C5 | 0.8 (3) |
| C10—C11—C12—C13 | 178.16 (16) | C2—C3—C4—C5 | −179.85 (16) |
| C1—O2—C2—O1 | 0.9 (3) | C14—C15—C16—C17 | −0.5 (3) |
| C1—O2—C2—C3 | −177.71 (15) | C18—C17—C16—C15 | −1.1 (3) |
| C4—C3—C2—O1 | 175.11 (19) | C3—C4—C5—C6 | −0.1 (3) |
| C8—C3—C2—O1 | −5.6 (3) | C7—C6—C5—C4 | −1.0 (3) |
| C4—C3—C2—O2 | −6.3 (3) | C9—C6—C5—C4 | 178.01 (16) |
Hydrogen-bond geometry (Å, º)
Cg1, Cg2 and Cg3 are the centroids of rings N1/C11–C14/C19, C3–C8 and C14–C19, respectively.
| D—H···A | D—H | H···A | D···A | D—H···A |
| O3—H3O···N1 | 0.86 (2) | 2.19 (3) | 2.715 (2) | 120 (2) |
| O3—H3O···O1i | 0.86 (2) | 2.23 (2) | 2.901 (2) | 136 (2) |
| C5—H5A···O3ii | 0.93 | 2.57 | 3.437 (2) | 155 |
| C7—H7A···Cg3iii | 0.93 | 2.99 | 3.605 (2) | 125 |
| C8—H8A···Cg1iii | 0.93 | 2.93 | 3.559 (2) | 126 |
| C15—H15A···Cg2ii | 0.93 | 2.83 | 3.639 (2) | 146 |
Symmetry codes: (i) −x+1, −y+1, −z; (ii) −x+3/2, y−1/2, −z+1/2; (iii) x−1/2, −y+1/2, z−1/2.
References
- Albrecht, M., Fiege, M. & Osetska, O. (2008). Coord. Chem. Rev. 252, 812–824.
- Bruker (2005). SMART, SAINT and SADABS. Bruker AXS Inc., Madison, Wisconsin, USA.
- Cacciatore, I., Fornasari, E., Baldassare, L., Cornacchia, C., Fulle, S., DiFilippo, E. S., Pietrangelo, T. & Pinnen, F. (2013). Pharmaceuticals, 6, 54–69. [DOI] [PMC free article] [PubMed]
- Groom, C. R., Bruno, I. J., Lightfoot, M. P. & Ward, S. C. (2016). Acta Cryst. B72, 171–179. [DOI] [PMC free article] [PubMed]
- Huo, Y., Wang, C., Lu, J., Hu, S., Li, X. & Zhang, L. (2015). J. Mol. Struct. 1098, 311–317.
- Jing, H.-L., Zeng, H.-P., Zhou, Y.-D., Wang, T.-T., Yuan, G.-Z. & Ouyang, X.-H. (2006). Chin. J. Chem. 24, 966–972.
- Macrae, C. F., Bruno, I. J., Chisholm, J. A., Edgington, P. R., McCabe, P., Pidcock, E., Rodriguez-Monge, L., Taylor, R., van de Streek, J. & Wood, P. A. (2008). J. Appl. Cryst. 41, 466–470.
- Moulinet d’Hardemare, A. du, Gellon, G., Philouze, C. & Serratrice, G. (2012). Inorg. Chem. 51, 12142–12151. [DOI] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Song, K.-C., Kim, J.-S., Park, S.-M., Chung, K.-C., Ahn, S. & Chang, S.-K. (2006). Org. Lett. 8, 3413–3416. [DOI] [PubMed]
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
- Tang, C.-W. & VanSlyke, S. A. (1987). Appl. Phys. Lett. 51, 913–915.
- Yuan, G.-Z., Huo, Y.-P., Rong, L.-L., Nie, X.-L. & Fang, X.-M. (2012). Inorg. Chem. Commun. 23, 90–94.
- Yuan, G.-Z., Rong, L.-L., Qiao, X.-L., Xia, Y.-P., Guo, T. & Wei, X.-W. (2013). Wuji Huaxue Xuebao, 29, 1769.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S205698901601210X/su5312sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S205698901601210X/su5312Isup2.hkl
Supporting information file. DOI: 10.1107/S205698901601210X/su5312Isup3.mol
Supporting information file. DOI: 10.1107/S205698901601210X/su5312Isup4.cml
CCDC reference: 859030
Additional supporting information: crystallographic information; 3D view; checkCIF report





