Abstract
Vitiligo is an autoimmune disease in which depigmented skin results from destruction of melanocytes1, with epidemiologic association with other autoimmune diseases2. In previous linkage and genome-wide association studies (GWAS1, GWAS2), we identified 27 vitiligo susceptibility loci in patients of European (EUR) ancestry. We carried out a third GWAS (GWAS3) in EUR subjects, with augmented GWAS1 and GWAS2 controls, genome-wide imputation, and meta-analysis of all three GWAS, followed by an independent replication. The combined analyses, with 4,680 cases and 39,586 controls, identified 23 new loci and 7 suggestive loci, most encoding immune and apoptotic regulators, some also associated with other autoimmune diseases, as well as several melanocyte regulators. Bioinformatic analyses indicate a predominance of causal regulatory variation, some corresponding to eQTL at these loci. Together, the identified genes provide a framework for vitiligo genetic architecture and pathobiology, highlight relationships to other autoimmune diseases and melanoma, and offer potential targets for treatment.
In previous genome-wide linkage and association studies, we identified 27 vitiligo susceptibility loci3–6 in EUR subjects, principally encoding immunoregulatory proteins, many of which are associated with other autoimmune diseases7. Several other vitiligo-associated genes encode melanocyte components that regulate normal pigmentary variation8 and in some cases are major vitiligo autoimmune antigens, with an inverse association of variation at these loci with vitiligo versus malignant melanoma4,6. To detect additional vitiligo-associations with lower odds ratios (ORs), as well as uncommon risk alleles with higher ORs, we conducted a third GWAS (GWAS3) of EUR subjects. We augmented the number of population controls in our previous GWAS1 and GWAS2 and performed genome-wide imputation of all three EUR vitiligo GWAS. After quality control procedures, the augmented studies included 1,381 cases and 14,518 controls (GWAS1), 413 cases and 5,209 controls (GWAS2), and 1,059 cases and 17,678 controls (GWAS3), with genomic inflation factors 1.068, 1.059, and 1.013, respectively. We performed a fixed-effects meta-analysis of the three GWAS datasets for 8,966,411 markers (GWAS123; Online Methods). Replication used an additional 1,827 EUR vitiligo cases and 2,181 controls.
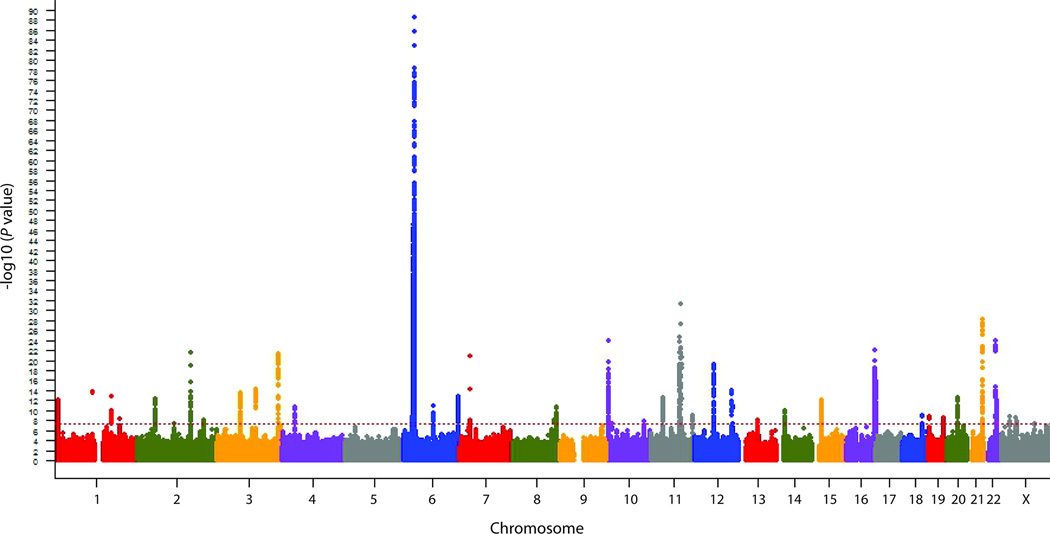
Results for the three individual GWAS, the meta-analysis, and the replication study are presented in Table 1, Supplementary Table 1, and Fig. 1. Twenty-three new loci achieved genome-wide significance (P < 5 × 10−8) for association with vitiligo and demonstrated subsequent replication; of these, 21 are completely novel (FASLG, PTPRC, PPP4R3B, BCL2L11, FARP2-STK25, UBE2E2, FBXO45-NRROS, PPP3CA, IRF4, SERPINB9, CPVL, NEK6, ARID5B, a multigenic segment that includes BAD, TNFSF11, KAT2A-HSPB9-RAB5C, TNFRSF11A, SCAF1-IRF3-BCL2L12, a multigenic segment that includes ASIP, PTPN1, and IL1RAPL1), while two, CTLA4 and TICAM1, were suggestive in our previous studies. One previously significant locus, CLNK, was no longer significant (Supplementary Table 1). Another potential new locus, PVT1, exceeded genome-wide significance in the discovery meta-analysis (P = 7.74 × 10−9), but could not be successfully genotyped in the replication study and so remains uncertain. Two other loci, FLI1 and LOC101060498, exceeded genome-wide significance in the discovery meta-analysis (P = 3.76 × 10−8 and P = 3.60 × 10−11, respectively), but did not demonstrate replication. Seven additional novel loci achieved suggestive significance (P < 10−5) in the discovery meta-analysis (STAT4, PPARGC1B, c7orf72, PARP12, FADS2, CBFA2T3, and a chr17 locus in the vicinity of AFMID) and gave evidence of replication, but failed to achieve genome-wide significance (Supplementary Table 1).
Table 1.
Allelic associations at vitiligo susceptibility loci following GWAS meta-analysis and replication study
| GWAS123 meta-analysis | GWAS3 replication study | GWAS123 & GWAS3 replication study meta-analysis |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Chr. | Variant | Position (Build 37) |
Locus | EA/OA | P value | Odds ratio |
P value | Odds ratio |
P value | Odds ratio (95% CI) |
Heritability explained* (%) |
| 1 | rs301807 | 8484823 | RERE | A/G | 1.84 × 10−12 | 1.22 | 4.09 × 10−04 | 1.17 | 4.14 × 10−15 | 1.21 (1.15–1.27) | 0.003 |
| 1 | rs2476601 | 114377568 | PTPN22 | A/G | 2.21 × 10−14 | 1.39 | 1.08 × 10−05 | 1.36 | 1.21 × 10−18 | 1.38 (1.29–1.49) | 0.003 |
| 1 | rs78037977 | 172715702 | FASLG | G/A | 1.39 × 10−13 | 1.33 | 8.95 × 10−05 | 1.29 | 6.74 × 10−17 | 1.32 (1.24–1.41) | 0.003 |
| 1 | rs16843742 | 198672299 | PTPRC | C/T | 8.84 × 10−09 | 0.82 | 1.87 × 10−02 | 0.88 | 1.02 × 10−09 | 0.83 (0.79–0.88) | 0.002 |
| 2 | rs10200159 | 55845109 | PPP4R3B | C/T | 3.35 × 10−13 | 1.48 | 3.70 × 10−07 | 1.55 | 3.73 × 10−19 | 1.51 (1.38–1.66) | 0.003 |
| 2 | rs4308124 | 112010486 | BCL2L11-MIR4435-2HG | C/T | 4.99 × 10−08 | 1.17 | 1.67 × 10−02 | 1.12 | 3.96 × 10−09 | 1.15 (1.10–1.21) | 0.002 |
| 2 | rs2111485 | 163110536 | IFIH1 | A/G | 2.69 × 10−22 | 0.75 | 8.58 × 10−05 | 0.83 | 6.40 × 10−25 | 0.77 (0.73–0.81) | 0.008 |
| 2 | rs231725 | 204740675 | CTLA4 | A/G | 2.25 × 10−08 | 1.18 | 1.57 × 10−03 | 1.16 | 1.49 × 10−10 | 1.18 (1.12–1.24) | 0.002 |
| 2 | rs41342147 | 242407588 | FARP2-STK25 | A/G | 8.03 × 10−07 | 0.80 | 1.25 × 10−03 | 0.80 | 3.70 × 10−09 | 0.80 (0.74–0.86) | 0.003 |
| 3 | rs35161626 | 23512312 | UBE2E2 | I/D | 7.34 × 10−07 | 0.87 | 1.09 × 10−02 | 0.89 | 3.13 × 10−08 | 0.87 (0.83–0.92) | 0.001 |
| 3 | rs34346645 | 71557945 | FOXP1 | A/C | 6.11 × 10−14 | 0.80 | 4.23 × 10−06 | 0.81 | 7.99 × 10−19 | 0.80 (0.76–0.84) | 0.004 |
| 3 | rs148136154 | 119283468 | CD80-ADPRH | C/T | 5.02 × 10−15 | 1.37 | 1.74 × 10−02 | 1.17 | 4.58 × 10−15 | 1.31 (1.22–1.40) | 0.003 |
| 3 | rs13076312 | 188089254 | LPP | T/C | 3.58 × 10−22 | 1.32 | 3.48 × 10−10 | 1.33 | 1.61 × 10−30 | 1.32 (1.26–1.38) | 0.009 |
| 3 | rs6583331 | 196347253 | FBXO45-NRROS | A/T | 1.39 × 10−07 | 0.86 | 3.62 × 10−02 | 0.91 | 2.53 × 10−08 | 0.87 (0.83–0.92) | 0.002 |
| 4 | rs1031034 | 102223386 | PPP3CA | A/C | 4.78 × 10−06 | 0.86 | 2.14 × 10−03 | 0.86 | 3.43 × 10−08 | 0.86 (0.81–0.91) | 0.001 |
| 6 | rs12203592 | 396321 | IRF4 | T/C | 1.03 × 10−09 | 0.77 | 3.17 × 10−08 | 0.68 | 8.86 × 10−16 | 0.75 (0.70–0.80) | 0.001 |
| 6 | rs78521699 | 2908591 | SERPINB9 | G/A | 3.33 × 10−06 | 0.79 | 2.27 × 10−03 | 0.80 | 2.54 × 10−08 | 0.79 (0.73–0.86) | 0.001 |
| 6 | rs60131261 | 29937335 | HLA-A | D/I | 2.63 × 10−48 | 1.53 | 8.01 × 10−20 | 1.54 | 1.56 × 10−66 | 1.54 (1.46–1.61) | 0.016 |
| 6 | rs9271597 | 32591291 | HLA-DRB1/DQA1 | A/T | 3.15 × 10−89 | 1.77 | nd | nd | nd | nd | 0.042 |
| 6 | rs72928038 | 90976768 | BACH2 | A/G | 1.12 × 10−11 | 1.28 | 2.04 × 10−04 | 1.25 | 1.00 × 10−14 | 1.27 (1.19–1.35) | 0.003 |
| 6 | rs2247314 | 167370230 |
RNASET2-FGFR1OP- CCR6 |
C/T | 1.97 × 10−13 | 0.79 | 1.56 × 10−06 | 0.79 | 1.72 × 10−18 | 0.79 (0.75–0.84) | 0.003 |
| 7 | rs117744081 | 29132279 | CPVL | G/A | 3.74 × 10−22 | 1.95 | 1.88 × 10−06 | 1.66 | 8.72 × 10−26 | 1.84 (1.64–2.06) | 0.004 |
| 8 | rs2687812 | 133931055 | TG-SLA-WISP1 | A/T | 1.98 × 10−11 | 1.21 | 1.69 × 10−03 | 1.15 | 2.19 × 10−13 | 1.19 (1.14–1.25) | 0.007 |
| 9 | rs10986311 | 127071493 | NEK6 | C/T | 5.45 × 10−07 | 1.16 | 5.10 × 10−03 | 1.14 | 1.01 × 10−08 | 1.15 (1.10–1.21) | 0.001 |
| 10 | rs706779 | 6098824 | IL2RA | C/T | 1.30 × 10−24 | 0.74 | 9.25 × 10−05 | 0.84 | 7.20 × 10−27 | 0.77 (0.73–0.81) | 0.012 |
| 10 | rs71508903 | 63779871 | ARID5B | T/C | 1.09 × 10−06 | 1.18 | 1.52 × 10−03 | 1.19 | 6.93 × 10−09 | 1.18 (1.12–1.25) | 0.001 |
| 10 | rs12771452 | 115488331 | CASP7 | A/G | 9.16 × 10−08 | 0.83 | 8.42 × 10−06 | 0.79 | 4.43 × 10−12 | 0.82 (0.78–0.87) | 0.002 |
| 11 | rs1043101 | 35274829 | CD44-SLC1A2 | G/A | 2.08 × 10−13 | 1.24 | 4.20 × 10−06 | 1.24 | 5.26 × 10−18 | 1.23 (1.18–1.29) | 0.003 |
| 11 | rs12421615 | 64021605 |
PPP1R14B-PLCB3-BAD- GPR137-KCNK4-TEX40- ESRRA-TRMT112- PRDX5 |
A/G | 3.38 × 10−06 | 0.87 | 3.78 × 10−03 | 0.87 | 4.81 × 10−08 | 0.87 (0.83–0.91) | 0.001 |
| 11 | rs1126809 | 89017961 | TYR | A/G | 7.13 × 10−32 | 0.67 | 2.54 × 10−13 | 0.68 | 1.16 × 10−43 | 0.67 (0.63–0.71) | 0.012 |
| 11 | rs11021232 | 95320808 | Gene desert | C/T | 1.01 × 10−21 | 1.38 | 3.81 × 10−04 | 1.22 | 2.10 × 10−23 | 1.34 (1.26–1.41) | 0.005 |
| 12 | rs2017445 | 56407072 | IKZF4 | A/G | 3.81 × 10−20 | 1.31 | 1.22 × 10−12 | 1.40 | 6.62 × 10−31 | 1.33 (1.27–1.40) | 0.005 |
| 12 | rs10774624 | 111833788 | SH2B3-ATXN2 | A/G | 1.88 × 10−14 | 0.80 | 1.52 × 10−10 | 0.75 | 6.22 × 10−23 | 0.79 (0.75–0.83) | 0.004 |
| 13 | rs35860234 | 43070206 | TNFSF11 | G/T | 2.82 × 10−06 | 1.16 | 3.45 × 10−04 | 1.20 | 4.76 × 10−09 | 1.17 (1.11–1.23) | 0.001 |
| 14 | rs8192917 | 25102160 | GZMB | C/T | 1.37 × 10−10 | 1.23 | 1.23 × 10−06 | 1.29 | 8.91 × 10−16 | 1.25 (1.18–1.32) | 0.002 |
| 15 | rs1635168 | 28535266 | OCA2-HERC2 | A/C | 6.97 × 10−13 | 1.43 | 7.45 × 10−03 | 1.25 | 8.78 × 10−14 | 1.37 (1.26–1.49) | 0.003 |
| 16 | rs4268748 | 90026512 | MC1R? | C/T | 1.63 × 10−20 | 0.73 | 8.23 × 10−15 | 0.66 | 2.88 × 10−33 | 0.71 (0.67–0.75) | 0.013 |
| 17 | rs11079035 | 40289012 | KAT2A-HSPB9-RAB5C | A/G | 3.20 × 10−06 | 1.18 | 3.19 × 10−05 | 1.28 | 6.77 × 10−10 | 1.21 (1.14–1.29) | 0.001 |
| 18 | rs8083511 | 60028655 | TNFRSF11A | C/A | 9.42 × 10−10 | 1.24 | 3.23 × 10−02 | 1.13 | 2.81 × 10−10 | 1.21 (1.14–1.28) | 0.002 |
| 19 | rs4807000 | 4831878 | TICAM1 | A/G | 1.58 × 10−09 | 1.19 | 2.11 × 10−06 | 1.24 | 1.94 × 10−14 | 1.21 (1.15–1.26) | 0.002 |
| 19 | rs2304206 | 50168871 | SCAF1-IRF3-BCL2L12 | A/G | 6.45 × 10−09 | 0.82 | 4.52 × 10−02 | 0.90 | 2.36 × 10−09 | 0.84 (0.80–0.89) | 0.002 |
| 20 | rs6059655 | 32665748 |
RALY-EIF252-ASIP- AHCY-ITCH |
A/G | 3.58 × 10−13 | 0.63 | 3.08 × 10−08 | 0.57 | 1.04 × 10−19 | 0.61 (0.55–0.68) | 0.004 |
| 20 | rs6012953 | 49123043 | PTPN1 | G/A | 1.18 × 10−07 | 1.16 | 1.74 × 10−02 | 1.11 | 9.47 × 10−09 | 1.15 (1.10–1.20) | 0.002 |
| 21 | rs12482904 | 43851828 | UBASH3A | A/T | 5.74 × 10−29 | 1.43 | 1.16 × 10−03 | 1.18 | 5.84 × 10−29 | 1.35 (1.28–1.43) | 0.010 |
| 22 | rs229527 | 37581485 | C1QTNF6 | A/C | 1.40 × 10−24 | 1.34 | 1.15 × 10−07 | 1.27 | 1.14 × 10−30 | 1.32 (1.26–1.38) | 0.006 |
| 22 | rs9611565 | 41767486 | ZC3H7B-TEF | C/T | 1.99 × 10−12 | 0.78 | 3.34 × 10−04 | 0.82 | 3.13 × 10−15 | 0.79 (0.75–0.84) | 0.003 |
| x | rs73456411 | 29737404 | IL1RAPL1 | T/G | 1.57 × 10−07 | 1.72 | 5.90 × 10−03 | 1.62 | 7.34 × 10−10 | 1.77 (1.47–2.13) | 0.001 |
| x | rs5952553 | 49392721 | CCDC22-FOXP3-GAGE | C/T | 1.81 × 10−08 | 0.85 | 3.48 × 10−02 | 0.92 | 1.05 × 10−09 | 0.86 (0.82–0.90) | 0.001 |
Heritability explained by all independent signals of the locus. Chr., chromosome; CI, confidence interval; nd, not determined; EA, effect allele; OA, other allele. Bold, novel significant vitiligo susceptibility loci. The chromosome 16 association peak spans a large number of genes, including MC1R.
Figure 1.
Genome-wide meta-analysis results. The genome-wide distribution of −log10 (P values) from the Cochran-Mantel-Haenszel meta-analysis for 8,966,411 genotyped and imputed markers from GWAS1, GWAS2, and GWAS3 is shown across the chromosomes. The dotted line indicates the threshold for genome-wide significance (P < 5 × 10−8).
Together, the most significantly associated variants at the 48 loci (Table 1) identified by meta-analyses of the three GWAS account for 17.4% of vitiligo heritability (h2 ~ 0.75). To assess whether additional independent variants at these loci might account for additional vitiligo heritability, we performed logistic regression conditional on the most significant SNP at each locus. Eight loci (FARP2-STK25, IFIH1, IL2RA, LPP, MC1R, SLA/TG, TYR, UBASH3A) and the MHC showed evidence of additional independent associations, accounting for an additional 5.1% of vitiligo heritability, for a total of 22.5%. In general, the ORs for the 23 new confirmed loci were lower than those for loci detected previously6, 1.15 to 1.27, excepting CPVL (OR = 1.84), RALY-EIF252-ASIP-AHCY-ITCH (OR = 1.64), and IL1RAPL1 (OR = 1.77); for these three signals the associated alleles are uncommon (minor allele frequencies 0.03, 0.07, and 0.01, respectively) and thus were not detected in the previous GWAS due to power limitations.
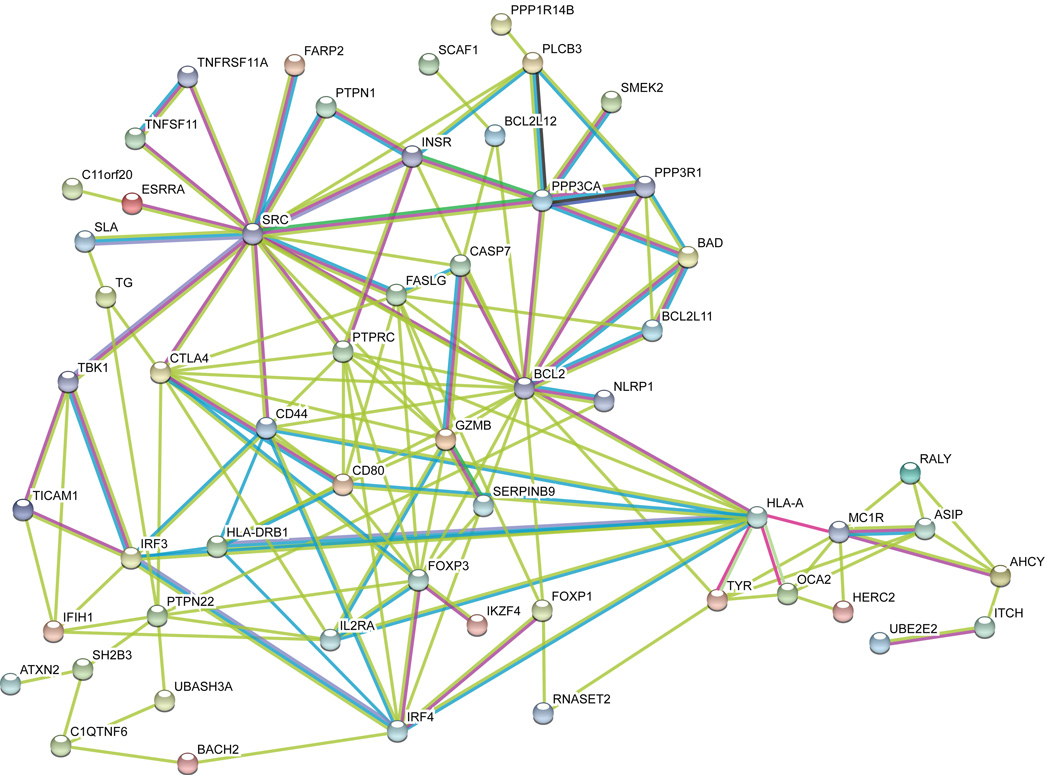
To screen for functional relationships among proteins encoded at the 48 confirmed vitiligo-associated loci, we included all genes under the association peaks at these loci in unsupervised pathway analyses using g:PROFILER9, PANTHER10, and STRING11. PANTHER and gPROFILER identified an enriched network of BioGRID interactions, most significant for the GO categories immune response, immune system process, positive regulation of response to stimulus, positive regulation of biological process, and regulation of response to stimulus. STRING identified a large potential interaction network (Fig. 2), with a predominance of proteins involved in immunoregulation, T-cell receptor repertoire, apoptosis, antigen processing and presentation, and melanocyte function.
Figure 2.
Bioinformatic functional interaction network analysis of proteins encoded by all positional candidate genes at all confirmed and suggestive vitiligo candidate loci. As a first step, unsupervised functional interaction network analysis was carried out using STRING v10.011, considering each protein as a node and permitting ≤ 5 second-order interactions to maximize connectivity. Nodes that shared no edges with other nodes were then excluded from the network. Edge colors are per STRING: teal, interactions from curated databases; purple, experimentally determined interactions; green, gene neighborhood; blue, databases; red, gene fusions; dark blue, gene co-occurrence; pale green, text-mining; black, co-expression; lavender, protein homology. Note that SMEK2 is an alternative name for PPP4R3B.
Considering proteins encoded at the 23 newly confirmed vitiligo candidate loci, at least twelve (CTLA4, TICAM1, PTPRC, FARP2, UBE2E2, NRROS, CPVL, ARID5B, PTPN1, TNFSF11, TNFRSF11A, IRF3, and perhaps also IL1RAPL1) play roles in immune regulation, and PPP3CA may regulate FOXP3 via NFATC2 and is associated with canine lupus12. Six (FASLG, BCL2L11, BCL2L12, SERPINB9, NEK6, BAD) are regulators of apoptosis, particularly involving immune cells. ASIP is a regulator of melanocyte gene expression, and IRF4 is a key transcription factor for both immune cells and melanocytes.
Strikingly, several vitiligo-associated genes encode proteins that interact physically and functionally. BCL2L11 and BAD are binding partners that promote apoptosis13. CD80 binds to CTLA4 to inhibit T cell activation14. BCL2L12 binds to and neutralizes caspase 7 (CASP7)15. SERPINB9 binds to and specifically inhibits granzyme B (GZMB)16. Eos (IKZF4) binds and is an obligatory co-repressor of FOXP3 in regulatory T cells17. RANK (TNFRSF11A) binds to RANKL (TNFSF11) to regulate many aspects of immune cell function, including interactions of T cells and dendritic cells and thymic tolerization18. Agouti signaling protein (ASIP) binds to the melanocortin-1 receptor (MC1R) to down-regulate production of brown-black eumelanin19. IRF4 cooperates with MITF to activate transcription of TYR20. And the vitiligo-associated HLA-A*02:01:01:01 subtype presents peptide antigens derived from several different melanocyte proteins, including tyrosinase (TYR), OCA2, and MC1R4,6,21. Together, these relationships appear to highlight key pathways of vitiligo pathogenesis that are beginning to coalesce.
An unexpected finding from vitiligo GWAS has been an inverse relationship between vitiligo and malignant melanoma risk for genes that encode melanocyte structural and regulatory proteins. TYR, OCA2, and MC1R, encode functional components of the melanocyte and are key vitiligo autoantigens. IRF4 encodes a transcription factor for melanocytes as well as lymphoid, myeloid, and dendritic cells22, controlled by alternative tissue-specific enhancers23. ASIP and PPARGC1B encode paracrine regulators of melanocyte gene expression. All six loci play important roles in normal pigmentary variation8,24, and for all six the specific SNPs associated with vitiligo risk are also associated with melanoma protection, and vice-versa25–27. The inverse genetic relationship of susceptibility to vitiligo versus melanoma suggests that vitiligo may represent enhanced immune surveillance against melanoma27,28, consistent with the threefold reduction in melanoma incidence among vitiligo patients29,30 and prolonged survival of melanoma patients who develop vitiligo during immunotherapy31.
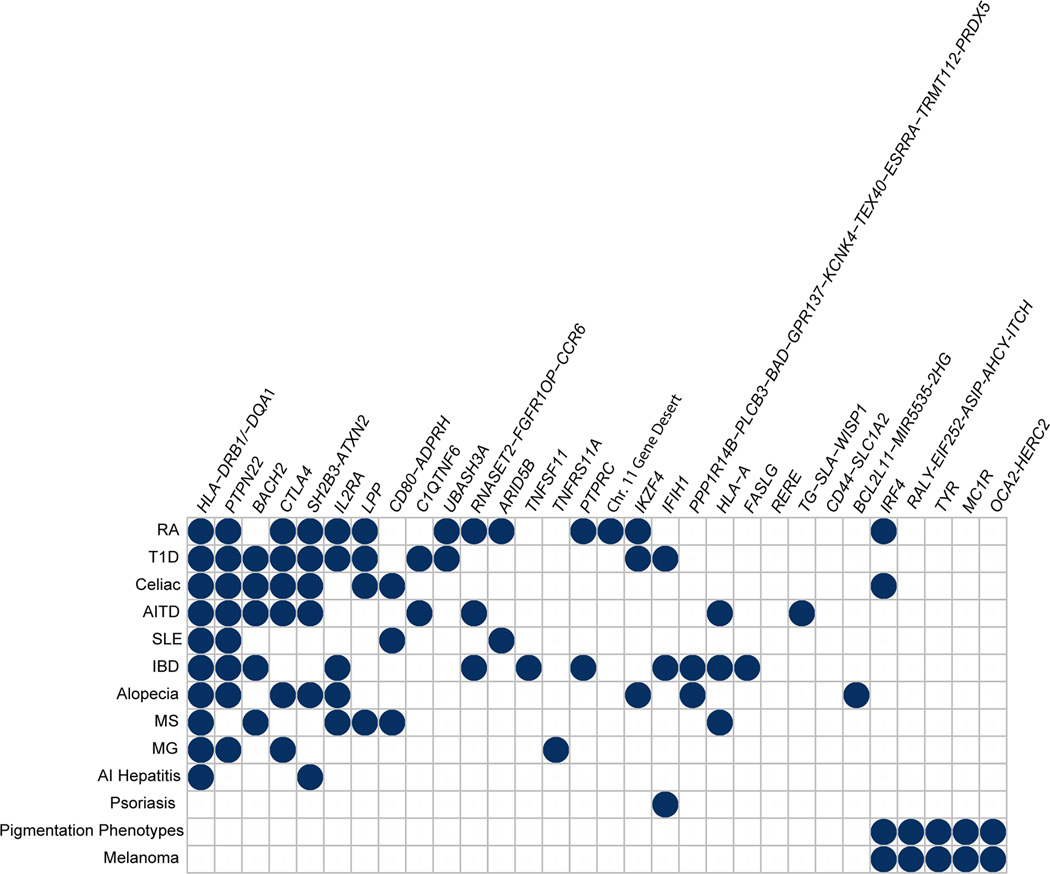
Vitiligo is epidemiologically associated with several other autoimmune diseases, including autoimmune thyroid disease, pernicious anemia, rheumatoid arthritis, adult-onset type 1 diabetes, Addison’s disease, and lupus2,32. We searched the NHGRI-EBI GWAS Catalog and PubMed for the 48 genome-wide significant and 7 suggestive vitiligo susceptibility loci for associations with other autoimmune, inflammatory, and immune-related disorders. As shown in Fig. 3, of the 23 novel genome-wide significant vitiligo loci, FASLG has been associated with celiac disease33 and Crohn’s disease34; PTPRC with ulcerative colitis35; BCL2L11 with primary sclerosing cholangitis36; CTLA4 with alopecia areata37, rheumatoid arthritis38, autoimmune thyroid disease39,40, myasthenia gravis41, and type 1 diabetes autoantibody production42; TNFRSF11A with myasthenia gravis41; and ARID5B with systemic lupus erythematosus43. Of the seven suggestive loci, STAT4 has been associated with Behḉet’s disease44, Sjögren’s syndrome45, and lupus46; and c7orf72 with lupus47. These concordant associations for vitiligo and other autoimmune and inflammatory diseases add to those involving previously identified vitiligo susceptibility loci, which include RERE, PTPN22, IFIH1, CD80, LPP, BACH2, RNASET2-FGFR1OP-CCR6, TG/SLA, IL2RA, CD44, a chr11q21 gene desert, IKZF4, SH2B3-ATXN2, UBASH3A, and C1QTNF64,6. Nevertheless, in most cases it remains uncertain whether apparent shared locus associations for different autoimmune diseases reflect shared or different underlying causal variants.
Figure 3.
Concordant associations for vitiligo and other autoimmune and inflammatory diseases. We searched the NHGRI-EBI GWAS Catalog and PubMed for associations of the 48 genome-wide significant and 7 suggestive vitiligo susceptibility loci with other autoimmune, inflammatory, and immune-related disorders, and for association with normal human pigmentation variation. Only reported associations that achieved genome-wide significance (P < 5 × 10−8) are included. RA, rheumatoid arthritis; T1D, type 1 diabetes mellitus; AITD, autoimmune thyroid disease; SLE, systemic lupus erythematosus; IBD, inflammatory bowel disease; MS, multiple sclerosis; MG, myasthenia gravis; AI hepatitis, autoimmune hepatitis.
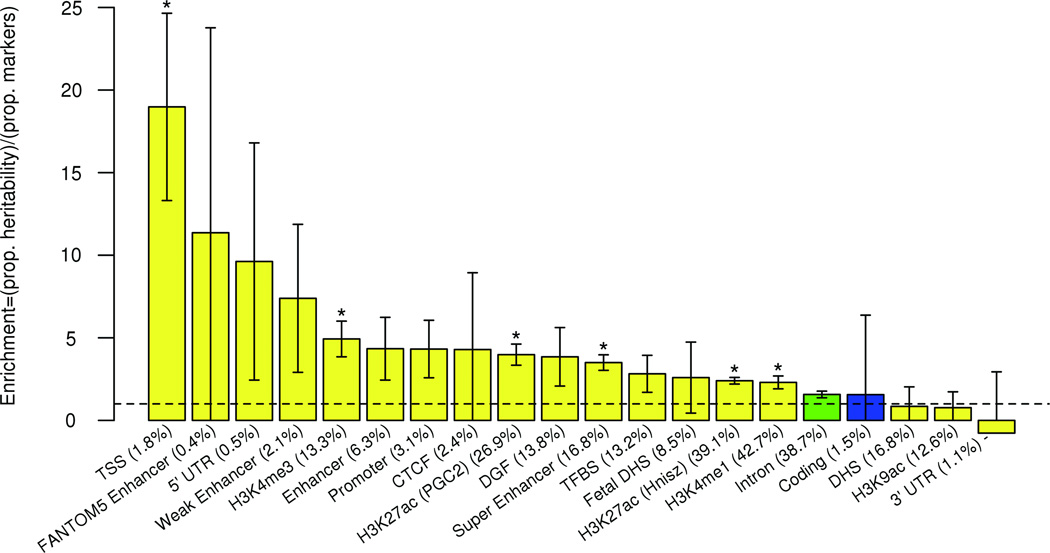
A majority of loci associated with complex traits involve causal variants that are regulatory in nature48–52, often corresponding to apparent expression quantitative trait loci (eQTLs)52. For TYR21, GZMB53, and MC1R7, principal vitiligo risk derives from missense substitutions, whereas for OCA26 and the MHC class I54 and class II55 loci principal vitiligo risk is associated with causal variation in nearby transcriptional regulatory elements. To assess the fraction of vitiligo-associated loci for which causal variation is likely regulatory, we carried out conditional logistic regression analysis of all loci to define independent association signals, and for each signal we compiled all variants that could not be statistically distinguished. All variants were then annotated against all available ENCODE datasets for immune-related and melanocyte-related cells (Supplementary Table 2). Overall, at approximately 58% of loci, the most significant variants (or statistically indistinguishable variants) are within a transcriptional regulatory element predicted by ENCODE data56,57. Only about 15% are in coding regions, several resulting in missense substitutions. To further assess the general functional categories of apparent causal variants for vitiligo, we applied stratified LD score regression51 to the GWAS meta-analysis summary statistics. As shown in Fig. 4, greatest enrichment of heritability was observed for markers in regulatory functional categories, with considerably less enrichment of markers in protein coding regions.
Figure 4.
Enrichment estimates for functional annotations. The combined CMH GWAS123 summary statistics were analyzed using the stratified LD score regression method utilizing the full baseline model51. Regulatory, yellow; protein coding, blue; intron, green. Bar height represents enrichment which is defined to be the proportion of SNP heritability in the category divided by the proportion of SNPs in that category. Error bars represent jackknife standard error around the enrichment. For each category, percentage of the total markers in the category is in parentheses. Dashed line represents a ratio of 1 (no enrichment). Asterisks indicate enrichment significant at P < 0.05 after Bonferroni correction for the 20 categories tested (the categories conserved, repressed, transcribed, and promoter flanking were removed and considered insufficiently specific). CTCF, CCCTC-binding factor; DGF, digital genomic footprint; DHS, DNase hypersensitivity site; TFBS, transcription factor binding site; TSS, transcriptional start site; 5’ and 3’ UTR, 5’ and 3’ untranslated regions. H3K4me1, H3K4me3, H3K9ac, and H3K27ac are regulatory chromatin marks56,57.
We utilized two approaches to assess correspondence of vitiligo association signals with expression of genes in the vicinity. We used PrediXcan58 to predict expression of 11,553 genes in whole blood for each study subject and then tested association of predicted expression of each gene with vitiligo affection status. We used a Bayesian method to assess co-localization of cis eQTL signals in purified blood monocytes with the confirmed vitiligo association signals. The PrediXcan analysis found 83 genes with significant differential predicted expression in vitiligo cases versus controls after Bonferroni correction (Supplementary Table 3); of these, 75 were located within 1 Mb of one of the 48 confirmed vitiligo susceptibility loci, demonstrating highly significant enrichment compared with locations of genes non-significant for PrediXcan (P value < 0.00001). The eQTL analysis found that 8 of the confirmed vitiligo association signals showed significant co-localization with eQTL association signals identified in purified monocytes (Supplementary Fig. 1 and Supplementary Table 4). Of the confirmed vitiligo-associated genes that could be tested using both methods, 6 were significant in both analyses (CASP7, HERC2-OCA2, ZC3H7B-TEF, TICAM1, RERE, RNASET2-FGFR1OP-CCR6). For all of these except CASP7, one or more of the most associated SNPs not distinguishable by logistic regression was located within or very close to an ENCODE element likely to regulate gene expression in immune cell types, melanocytes, or both (Supplementary Table 2).
Like a jigsaw puzzle, the pieces of the vitiligo pathogenome are thus beginning to fit together, revealing a complex network of immunoregulatory proteins, apoptotic regulators, and melanocyte components that mediate both autoimmune targeting of melanocytes in vitiligo and susceptibility to melanoma. For vitiligo as for other complex diseases, there is enrichment of causal variation in regions that regulate gene expression. This may bode well for identifying potential therapeutic targets, as pharmacologic modulation of dysregulated biological pathways may prove more tractable than attempting to target proteins impacted by amino acid substitutions.
ONLINE METHODS
Subjects
The genome-wide portion of this study included unrelated cases from our three generalized vitiligo GWAS: GWAS14 (n = 1514), GWAS26 (n = 450), and the current GWAS3 (n = 1090). All cases were of non-Hispanic-Latino European-derived white ancestry (EUR) from North America and Europe, and met strict clinical criteria for generalized vitiligo59. All controls were EUR individuals not specifically known to have any autoimmune disease or malignant melanoma, for whom genome-wide genotypes were obtained from the NCBI database of Genotypes and Phenotypes (dbGaP; phs000092.v1.p1, phs000125.v1.p1, phs000138.v2.p1, phs000142.v1.p1, phs000168.v1.p1, phs000169.v1.p1, phs000206.v3.p2, phs000237.v1.p1, phs000346.v1.p1, and phs000439.v1.p1 for GWAS1; phs000203.v1.p1, and phs000289.v2.p1 for GWAS2; phs000196.v2.p1, phs000303.v1.p1, phs000304.v1.p1, phs000368.v1.p1, phs000381.v1.p1, phs000387.v1.p1, phs000389.v1.p1, phs000395.v1.p1, phs000408.v1.p1, phs000421.v1.p1, phs000494.v1.p1, and phs000524.v1.p1 for GWAS3). Control datasets were matched to each of the three GWAS case datasets based on platforms used for genotyping. The independent replication study included 1827 unrelated EUR vitiligo cases and 2181 unrelated EUR controls not included in any of the GWAS. All subjects provided written informed consent. This study was carried out under the jurisdiction of each local IRB with overall oversight of the Colorado Multiple Institutional Review Board (COMIRB).
Genome-wide genotyping
Saliva specimens were obtained using a DNA self-collection kit (Oragene, DNA Genotek), and DNA was prepared using either the Maxwell apparatus/16 LEV Blood DNA kit (Promega) or the DNA Genotek Oragene Purifier protocol. DNA concentrations were measured using either the Qubit dsDNA BR Assay kit and Qubit 2.0 Fluorometer (Invitrogen) or the Promega QuantiFluor ONE dsDNA kit and GloMax®-Multi+ Detection System (Promega).
Genome-wide genotyping for the GWAS3 cases was performed for 716,503 variants using Illumina Human OmniExpress BeadChips by the Center for Inherited Disease Research (CIDR). Genotype data for GWAS3 were deposited in dbGaP (phs000224.v3.p1). GWAS14 and GWAS26 have been described previously.
Genome-wide quality control procedures
Quality control filtering of genome-wide genotype data was carried out using PLINK60, version 1.9. For each case/control dataset, DNA strand calls were reversed as needed. Cases were excluded on the basis of SNP call rates <98.5%, discordance between reported and observed sex, or inadvertent subject duplication, and controls were excluded on the basis of SNP call rates < 98%. SNPs were excluded on the basis of genotype missing rate > 2% for SNPs with observed minor allele frequency (MAF) ≥ 0.01, and for SNPs with MAF < 0.01 exclusion criteria were genotype missing rate >1% and < 5 minor alleles observed, or significant (P < 10−4) deviation from Hardy-Weinberg equilibrium. For X chromosome SNPs, Hardy-Weinberg equilibrium tests were performed in females, and SNPs with P < 10−4 were excluded from the final analysis. For each GWAS, only SNPs that existed in all case and control datasets were retained for imputation.
Within each GWAS, subjects were excluded based on cryptic relatedness identified by pairwise identity-by-descent estimations (pi-hat > 0.0625), in which case the individual with lower SNP call rate was excluded. For each of the three GWAS, the cleaned case dataset was combined with one cleaned control dataset at a time and the genotype data of 270 subjects of Phase I and II of the International HapMap Project from 4 populations, and principal components analysis (PCA) was performed with EIGENSOFT59 based on tag-SNPs (within which no pair were correlated with r2 >0.2) selected from genotyped SNPs. The first two eigenvectors were used to produce a PCA plot. A PCA plot was first made for cases and HapMap samples, and cases that were clearly separated from the main cluster of cases and HapMap EUR samples were excluded as outliers. A PCA plot of controls and HapMap samples was then made, and the same x and y coordinates that separated the case outliers from the main cluster of cases and HapMap EUR samples were used to identify control outliers.
After all QC procedures, the final number of genotyped SNPs remaining in GWAS1, GWAS2, and GWAS3 were 464,902, 494,043, and 483,609, respectively. For autosomal analyses, the final numbers of cases and controls in GWAS1, GWAS2, and GWAS3 were 1,381 and 14,518, 413 and 5,209, and 1,059 and 17,678, respectively, whereas for X chromosome analyses, the final numbers of cases and controls in GWAS1, GWAS2, and GWAS3 were 1,380 and 9,439, 413 and 5,209, and 1,059 and 14,220, respectively. This sample size provided at least 85% power to detect associations with OR ≥ 1.22 at genome-wide significance (P = 5 × 10−8) for MAF ≥ 0.25.
Genome-wide Genotype Imputation
For each GWAS, we used SHAPEIT version2 to pre-phase genotypes to produce best-guess haplotypes, and then performed imputation with these estimated haplotypes using IMPUTE2 and the 1000 Genomes Project phase I integrated variant set version 3 (March, 2012) as the reference panel. All cryptic related individuals and outliers from each GWAS were included in the process to improve imputation accuracy, but were removed for the final analyses. Only genotypes with imputation INFO ≥ 0.5 were retained, which were combined with prior SNP genotype data. Imputed genotypes for variants with MAF ≥ 0.01 calculated from all 3 GWAS combined and without significant (P > 10−5) deviation from Hardy-Weinberg equilibrium were used in the final analysis, which included 8,721,242 autosome variants and 245,169 chromosome X variants.
Replication study genotyping and quality control procedures
For the replication study, genotyping was attempted for 379 variants using a custom Illumina Golden Gate array by CIDR. 71 SNPs were excluded on the basis of genotype missing rate > 2% (which includes apparent technical failures), or significant (P < 10−4) deviation from Hardy-Weinberg equilibrium. For X chromosome SNPs, Hardy-Weinberg equilibrium tests were performed in females. Subjects were excluded on the basis of SNP call rates <95%, or discordance between reported and observed sex. Unintended duplicate samples were identified by pairwise identity-by-descent estimations (pi-hat > 0.99), in which case the individual with lower SNP call rate was excluded. The final numbers of remaining cases and controls were 1,827 and 2,181, respectively, providing at least 80% power to replicate associations at P = 0.05 with Bonferroni correction for up to 48 independent tests for OR ≥ 1.23 for MAF ≥ 0.25.
Statistical analyses
To control for the effects of population stratification, we assigned cases and controls of each GWAS to homogenous clusters using GemTools60, and performed Cochran-Mantel- Haenszel (CMH) analysis to test for association for each GWAS and the combined GWAS data, with the cluster variable defined by the case-control clusters from each GWAS. After removing variants within the extended MHC, the genomic inflation factor for GWAS1, GWAS2, and GWAS3 was 1.068, 1.059, and 1.013, respectively. For the combined GWAS1–GWAS2–GWAS3 genotype data for shared SNPs, the genomic inflation factor was 1.019.
For the replication study, after quality control procedures we compared allele frequencies for the remaining 308 SNPs in the remaining 1,827 cases and 2,181 controls using the Cochran-Armitage trend test. ORs and 95% confidence limits were calculated by logistic regression analysis. We used CMH analysis to obtain ORs and P values for the combined GWAS plus the replication study data, with the cluster variable defined by the case-control clusters from each GWAS and the replication study data as one cluster. To analyze X chromosome SNPs, we assumed complete X-inactivation and similar effect size between males and females, with the effect of having an A allele in a male equal to the effect of having two A alleles in a female63. We thus coded males as homozygous for the allele carried for each variant and tested for association by CMH analysis to obtain ORs and P values for each GWAS, the combined GWAS, and the combined GWAS plus the replication study data, and by the Cochran- Armitage trend test for the replication study data.
To test heterogeneity of associations across the three GWAS and the replication study data, we performed the Cochran Q test. The analysis was done with PLINK, version 1.07, using the ORs and standard errors estimated from the CMH analysis of each GWAS, and from logistic regression analysis of the replication study data. The I2 statistic from the Q test quantifies heterogeneity and ranges from 0% to 100%64, with a value of 75% or greater typically taken to indicate a high degree of heterogeneity65.
To test for multiple independent signals at each locus, we performed logistic regression analysis of each locus conditional on the most significantly associated variant, including as covariates in the model the significant principal components for each GWAS derived from GemTools62 to control for population stratification, and used a stepwise procedure to select additional variants, one by one, until no additional variants showed conditional P values ≤ 1.0 × 10−5. If a tested variant and the conditional variant could not improve each other significantly (P ≥ 0.05 when comparing the two SNP model to a single SNP model), then both variants were considered to represent the same signal. We calculated the variance explained by a specific variant or a set of variants from the combined GWAS as the Pseudo R2 of a logistic regression model which included the specific variants tested.
Bioinformatic pathway and functional enrichment analyses
To screen for functional relationships among the vitiligo candidate genes, we carried out pathway analysis of the protein products of all positional candidate genes at all 48 confirmed loci and the seven suggestive loci using g:PROFILER10, PANTHER11, and STRING12. To assess enrichment of association signals in different functional genomic categories contributing to vitiligo heritability, we applied stratified LD score regression51 to the combined CMH GWAS123 summary statistics. The regression model contained 24 overlapping functional categories, including coding, UTR, promoter and intronic regions, annotations for different histone marks, DNase I hypersensitivity site (DHS) regions, combined ChromHMM and Segway predictions, conserved regions in mammals, super-enhancers and FANTOM5 enhancers. For each of the 24 categories, a 500-bp window was used. Linkage disequilibrium data were provided by the LD score software, estimated from the EUR samples in the 1000 Genomes Project Phase 1. Enrichment per category was calculated by the ratio of the estimated proportion of heritability explained by the category over the proportion of the markers in the category.
PrediXcan and Monocyte eQTL Co-Localization analyses
We carried out a gene-based test of association of vitiligo with “imputed” expression profiles for 11,553 autosomal genes in whole blood using PrediXcan58. The analysis included 2,853 cases and 37,412 controls from the combined GWAS. Association testing between expression estimates for each gene and affection status for vitiligo was performed by generalized logistic regression. P values were adjusted for the number of genes tested (n = 11,553). NRROS, ZC3H7B, TNFRSF11A, BCL2L12, RALY, ASIP, OCA2, and TYR were excluded from the PrediXcan analysis due to poor prediction of gene expression in blood cells.
We derived expression quantitative trait loci (eQTLs) in peripheral blood monocytes from 414 EUR subjects with paired genotyping and gene expression data66. SHAPEIT version2 was used to pre-phase genotypes to produce best-guess haplotypes with imputation performed using IMPUTE2 and the 1000 Genomes Project phase I integrated variant set version 3 (March, 2012) as reference panel. We tested for co-localization of eQTL and vitiligo GWAS autosomal association patterns as described67,68. Vitiligo susceptibility loci were defined by windows of robust association plus an added 100 kb buffer on both sides. eQTL probes were selected by choosing probes that resided within these windows. Probe quality annotation was performed using ReMOAT69 and all probes with an annotation of “bad” were removed. After removing non-autosomal loci and duplicate probe IDs, a total of 904 probes remained. All vitiligo susceptibility loci contained at least one probe with the exception of the gene desert 3’ of TYR, for which the only probe that intersected the locus was excluded due to ReMOAT annotation of “bad”. Within each locus window, all SNPs were tested for association with all probes using linear regression. P values, MAF for each SNP and respective sample sizes were used as input to test for co- localization, simultaneously testing five mutually exclusive hypotheses by generating 5 corresponding posterior probabilities (PP):
H0 (PP0): There is no association with either the GWAS or the eQTL.
H1 (PP1): There is association for the GWAS only.
H2 (PP2): There is association for the eQTL only.
H3 (PP3): There is association for both the GWAS and the eQTL, but the associated variants are different for the GWAS and the eQTL.
H4 (PP4): The associated variants are the same for both the GWAS and the eQTL (co-localization).
Posterior probabilities were calculated using the R package “coloc” using default settings for prior probabilities of association. Co-localization was assessed as per Guo et al.68; significant co-localization was PP3+PP4 > 0.99 and PP4:PP3 > 5, and suggestive co-localization was PP3+PP4 > 0.95 and PP4:PP3 > 3.
Supplementary Material
Acknowledgments
We thank the thousands of vitiligo patients and normal control individuals around the world who participated in this study. We thank the Center for Inherited Disease Research (CIDR) for genotyping. This work utilized the Janus supercomputer, which is supported by the National Science Foundation (award number CNS-0821794), the University of Colorado Boulder, the University of Colorado Denver, and the National Center for Atmospheric Research. The Janus supercomputer is operated by the University of Colorado Boulder. This work was supported by grants R01AR045584, R01AR056292, X01HG007484, and P30AR057212 from the U.S. National Institutes of Health and by institutional research funding IUT20-46 from the Estonian Ministry of Education and Research.
Footnotes
URLs. 1000 Genomes Project, http://www.1000genomes.org/; 1000 Genomes Project data, http://www.sph.umich.edu/csg/abecasis/MACH/download/1000G-2010-08.html; NHGRI-EBI GWAS Catalog, http://www.ebi.ac.uk/gwas/; NIH Database of Genotypes and Phenotypes (dbGaP), http://www.ncbi.nlm.nih.gov/gapr; Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/omim; PLINK, http://pngu.mgh.harvard.edu/purcell/plink/; STATA, http://www.stata.com; STRING database, http://string-db.org.
Accession Codes. Genotype and phenotype data for GWAS1, GWAS2, and GWAS 3 have been deposited with the NIH Database of Genotypes and Phenotypes (dbGaP) as phs000224.v1.p1, phs000224.v2.p1, and phs000224.v3.p1, respectively.
AUTHOR CONTRIBUTIONS
Y.J., G.A. and D.Y. performed statistical analyses. J.S. managed computer databases, software, and genotype data. T.M.F., S.B., G.A., and K.M.B. managed DNA samples and contributed to experimental procedures. P.J.H. managed subject coordination. S.A.B., A.H., A.L., R.M.L., A.W., J.P.W.vdV., N.vG., J.L., D.C.B., A.T., K.E., E.H.K., D.J.G., A.P.W., S.K., E.P., K.K., M.K., M.R.W., W.T.M., A.O., S.M., R.C., M.P., N.B.S., M.S., Y.V., I.K., M.B., H.L., I.H., L.Z., and Q.-S.M. provided subject samples and phenotype information. S.A.S., P.R.F. and R.A.S. conceived, oversaw, and managed all aspects of the study. R.A.S. wrote the first draft of the manuscript. All authors contributed to the final paper.
URLs. 1000 Genomes Project, http://www.1000genomes.org/; coloc, https://cran.r-project.org/web/packages/coloc/index.html; GemTools, http://wpicr.wpic.pitt.edu/WPICCompgen/GemTools/GemTools.htm; IMPUTE2, https://mathgen.stats.ox.ac.uk/impute/impute_v2.html; International HapMap Project, http://hapmap.ncbi.nlm.nih.gov/; PrediXcan, https://github.com/hriordan/PrediXcan; SHAPEIT, http://www.shapeit.fr/; REMOAT, http://remoat.sysbiol.cam.ac.uk/transcript.php.
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Picardo M, Taïeb A, editors. Vitiligo. Springer, Heidelberg & New York: 2010. [Google Scholar]
- 2.Alkhateeb A, et al. Epidemiology of vitiligo and associated autoimmune diseases in Caucasian probands and their families. Pigment Cell Res. 2003;16:208–214. doi: 10.1034/j.1600-0749.2003.00032.x. [DOI] [PubMed] [Google Scholar]
- 3.Jin Y, et al. NALP1 in vitiligo-associated multiple autoimmune disease. N. Engl. J. Med. 2007;356:1216–1225. doi: 10.1056/NEJMoa061592. [DOI] [PubMed] [Google Scholar]
- 4.Jin Y, et al. Variant of TYR and autoimmunity susceptibility loci in generalized vitiligo. N. Engl. J. Med. 2010;362:1686–1697. doi: 10.1056/NEJMoa0908547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jin Y, et al. Common variants in FOXP1 are associated with generalized vitiligo. Nat. Genet. 2010;42:576–578. doi: 10.1038/ng.602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jin Y, et al. Genome-wide association study and meta-analysis identifies 13 new melanocyte-specific and immunoregulatory susceptibility loci for generalized vitiligo. Nat. Genet. 2012;44:676–680. doi: 10.1038/ng.2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ricano-Ponce I, Wijmenga C. Mapping of immune-mediated disease genes. Annu. Rev. Genomics. Hum. Genet. 2013;14:325–353. doi: 10.1146/annurev-genom-091212-153450. [DOI] [PubMed] [Google Scholar]
- 8.Liu F, et al. Genetics of skin color variation in Europeans: genome-wide association studies with functional follow-up. Hum Genet. 2015;134:823–835. doi: 10.1007/s00439-015-1559-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reimand J, Arak T, Vilo J. g:Profiler—a web server for functional interpretation of gene lists (2011 update) Nucleic Acids Res. 2011;39:W307–W315. doi: 10.1093/nar/gkr378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mi H, Poudel S, Muruganujan A, Casagrande JT, Thomas PD. PANTHER version 10: expanded protein families and functions, and analysis tools. Nucleic. Acids. Res. 2016;44:D336–D342. doi: 10.1093/nar/gkv1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Szklarczyk D, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilbe M, et al. Multiple changes of gene expression and function reveal genomic and phenotypic complexity in SLE-like disease. PLoS Genet. 2015;11:e1005248. doi: 10.1371/journal.pgen.1005248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang X, Xing D, Liu L, Chen WR. BimL directly neutralizes Bcl-xL to promote Bax activation during UV-induced apoptosis. FEBS Lett. 2009;583:1873–1879. doi: 10.1016/j.febslet.2009.04.045. [DOI] [PubMed] [Google Scholar]
- 14.Linsley PS, et al. Human B7-1 (CD80) and B7-2 (CD86) bind with similar avidities but distinct kinetics to CD28 and CTLA-4 receptors. Immunity. 1994;1:793–801. doi: 10.1016/s1074-7613(94)80021-9. [DOI] [PubMed] [Google Scholar]
- 15.Stegh AH, et al. Bcl2L12 inhibits post-mitochondrial apoptosis signaling in glioblastoma. Genes Dev. 2007;21:98–111. doi: 10.1101/gad.1480007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun J, et al. A cytosolic granzyme B inhibitor related to the viral apoptotic regulator cytokine response modifier A is present in cytotoxic lymphocytes. J. Biol. Chem. 1996;271:27802–27809. doi: 10.1074/jbc.271.44.27802. [DOI] [PubMed] [Google Scholar]
- 17.Pan F, et al. Eos mediates Foxp3-dependent gene silencing in CD4+ regulatory T cells. Science. 2009;325:1142–1146. doi: 10.1126/science.1176077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Akiyama T, Shinzawa M, Akiyama N. RANKL-RANK interaction in immune regulatory systems. World J. Orthop. 2012;3:142–150. doi: 10.5312/wjo.v3.i9.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ollmann MM, Lamoreux ML, Wilson BD, Barsh GS. Interaction of Agouti protein with the melanocortin 1 receptor in vitro and in vivo. Genes Dev. 1998;12:316–330. doi: 10.1101/gad.12.3.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Praetorius C, et al. A polymorphism in IRF4 affects human pigmentation through a tyrosinase-dependent MITF/TFAP2A pathway. Cell. 2013;155:1022–1033. doi: 10.1016/j.cell.2013.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jin Y, et al. Next-generation DNA re-sequencing identifies common variants of TYR and HLA-A that modulate the risk of generalized vitiligo via antigen presentation. J. Invest. Dermatol. 2012;132:1730–1733. doi: 10.1038/jid.2012.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gualco G, Weiss LM, Bacchi CE. MUM1/IRF4. A review. Appl. Imunohistochem. Mol. Morphol. 2010;18:301–310. doi: 10.1097/PAI.0b013e3181cf1126. [DOI] [PubMed] [Google Scholar]
- 23.Visser M, Palstra RJ, Kayser M. Allele-specific transcriptional regulation of IRF4 in melanocytes is mediated by chromatin looping of the intronic rs12203592 enhancer to the IRF4 promoter. Hum. Mol. Genet. 2015;25:2629–2661. doi: 10.1093/hmg/ddv029. [DOI] [PubMed] [Google Scholar]
- 24.Nan H, et al. Genome-wide association study of tanning phenotype in a population of European ancestry. J. Invest. Dermatol. 2009;129:2250–2257. doi: 10.1038/jid.2009.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shoag J, et al. PGC-1 coactivators regulate MITF and the tanning response. Mol. Cell. 2013;49:145–157. doi: 10.1016/j.molcel.2012.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Read J, Wadt KA, Hayward NK. Melanoma genetics. J. Med. Genet. 2016;53:1–14. doi: 10.1136/jmedgenet-2015-103150. [DOI] [PubMed] [Google Scholar]
- 27.Spritz RA. The genetics of generalized vitiligo: autoimmune pathways and an inverse relationship with malignant melanoma. Genome Med. 2010;19:2, 78. doi: 10.1186/gm199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Das PK, van den Wijngaard RMJGJ, Wankowicz-Kalinska A, Le Poole IC. A symbiotic concept of autoimmunity and tumour immunity: lessons from vitiligo. Trends Immunol. 2001;22:130–136. doi: 10.1016/s1471-4906(00)01844-5. [DOI] [PubMed] [Google Scholar]
- 29.Teulings HE, et al. Decreased risk of melanoma and nonmelanoma skin cancer in patients with vitiligo: a survey among 1307 patients and their partners. Br. J. Dermatol. 2013;168:162–171. doi: 10.1111/bjd.12111. [DOI] [PubMed] [Google Scholar]
- 30.Paradisi A, et al. Markedly reduced incidence of melanoma and nonmelanoma skin cancer in a nonconcurrent cohort of 10040 patients with vitiligo. J. Am. Acad. Dermatol. 2014;71:1110–1116. doi: 10.1016/j.jaad.2014.07.050. [DOI] [PubMed] [Google Scholar]
- 31.Teulings HE, et al. Vitiligo-like depigmentation in patients with stage III–IV melanoma receiving immunotherapy and its association with survival: a systematic review and meta-analysis. J. Clin. Onc. 2015;33:773–781. doi: 10.1200/JCO.2014.57.4756. [DOI] [PubMed] [Google Scholar]
- 32.Laberge G, et al. Early disease onset and increased risk of other autoimmune diseases in familial generalized vitiligo. Pigment Cell Res. 2005;18:300–305. doi: 10.1111/j.1600-0749.2005.00242.x. [DOI] [PubMed] [Google Scholar]
- 33.Dubois PC, et al. Multiple common variants for celiac disease influencing immune gene expression. Nat. Genet. 2010;42:295–302. doi: 10.1038/ng.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Franke A, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn's disease susceptibility loci. Nat. Genet. 2010;42:1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Juyal G, et al. Genome-wide association scan in north Indians reveals three novel HLA-independent risk loci for ulcerative colitis. Gut. 2015;64:571–579. doi: 10.1136/gutjnl-2013-306625. [DOI] [PubMed] [Google Scholar]
- 36.Melum E, et al. Genome-wide association analysis in primary sclerosing cholangitis identifies two non-HLA susceptibility loci. Nat. Genet. 2011;43:17–19. doi: 10.1038/ng.728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Petukhova L, et al. Genome-wide association study in alopecia areata implicates both innate and adaptive immunity. Nature. 2010;466:113–117. doi: 10.1038/nature09114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gregersen PK, et al. REL, encoding a member of the NF-kappaB family of transcription factors, is a newly defined risk locus for rheumatoid arthritis. Nat. Genet. 2009;41:820–823. doi: 10.1038/ng.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chu X, et al. A genome-wide association study identifies two new risk loci for Graves' disease. Nat. Genet. 2011;43:897–901. doi: 10.1038/ng.898. [DOI] [PubMed] [Google Scholar]
- 40.Eriksson N, et al. Novel associations for hypothyroidism include known autoimmune risk loci. PloS One. 2012;7:e34442. doi: 10.1371/journal.pone.0034442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Renton AE, et al. A genome-wide association study of myasthenia gravis. JAMA Neurol. 2015;72:396–404. doi: 10.1001/jamaneurol.2014.4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Plagnol V, et al. Genome-wide association analysis of autoantibody positivity in type 1 diabetes cases. PLoS Genet. 2011;7:e1002216. doi: 10.1371/journal.pgen.1002216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang W, et al. Meta-analysis followed by replication identifies loci in or near CDKN1B, TET3, CD80, DRAM1, and ARID5B as associated with systemic lupus erythematosus in Asians. Am. J. Hum. Genet. 2013;92:41–51. doi: 10.1016/j.ajhg.2012.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hou S, et al. Identification of a susceptibility locus in STAT4 for Behcet's disease in Han Chinese in a genome-wide association study. Arth. Rheumat. 2012;64:4104–4113. doi: 10.1002/art.37708. [DOI] [PubMed] [Google Scholar]
- 45.Li Y, et al. A genome-wide association study in Han Chinese identifies a susceptibility locus for primary Sjogren's syndrome at 7q11.23. Nat. Genet. 2013;45:1361–1365. doi: 10.1038/ng.2779. [DOI] [PubMed] [Google Scholar]
- 46.Lee YH, Bae SC, Choi SJ, Ji JD, Song GG. Genome-wide pathway analysis of genome-wide association studies on systemic lupus erythematosus and rheumatoid arthritis. Molec. Biol. Rep. 2012;39:10627–10635. doi: 10.1007/s11033-012-1952-x. [DOI] [PubMed] [Google Scholar]
- 47.Han JW, et al. Genome-wide association study in a Chinese Han population identifies nine new susceptibility loci for systemic lupus erythematosus. Nat. Genet. 2009;41:1234–1237. doi: 10.1038/ng.472. [DOI] [PubMed] [Google Scholar]
- 48.Corradin O, Schcheri PC. Enhancer variants: evaluating functions in common disease. Genome Med. 2014;6:85. doi: 10.1186/s13073-014-0085-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gusev A, et al. Partitioning heritability of regulatory and cell-type-specific variants across 11 common diseases. Am. J. Hum. Genet. 2014;95:535–552. doi: 10.1016/j.ajhg.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Paul DS, et al. Functional interpretation of non-coding sequence variation: concepts and challenges. Bioessays. 2014;36:191–199. doi: 10.1002/bies.201300126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Finucane HK, et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat. Genet. 2015;47:1228–1235. doi: 10.1038/ng.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nicolae D, et al. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet. 2010;6:e1000888. doi: 10.1371/journal.pgen.1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ferrara TM, Jin Y, Gowan K, Fain PR, Spritz RA. Risk of generalized vitiligo is associated with the common 55R-94A-247H variant haplotype of GZMB (encoding Granzyme B) J. Investig. Dermatol. 2013;133:1677–1679. doi: 10.1038/jid.2013.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hayashi M, et al. Autoimmune vitiligo is associated with gain of function by a transcriptional regulator that elevates expression of HLA-A*02:01 in vivo. Proc. Natl. Acad. Sci. U.S.A. 2016;113:1357–1362. doi: 10.1073/pnas.1525001113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cavalli G, et al. MHC class II super-enhancer increases surface expression of HLA-DR and HLA-DQ and affects cytokine production in autoimmune vitiligo. Proc. Natl. Acad. Sci. U.S.A. 2016;113:1363–1368. doi: 10.1073/pnas.1523482113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shlyueva D, Stampfel G, Stark A. Transcriptional enhancers from properties to genome-wide predictions. Nat. Rev. Genet. 2014;15:272–286. doi: 10.1038/nrg3682. [DOI] [PubMed] [Google Scholar]
- 57.Kellis M, et al. Defining functional DNA elements in the human genome. Proc. Natl. Acad. Sci. U.S.A. 2014;111:6131–6138. doi: 10.1073/pnas.1318948111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gamazon ER, et al. A gene-based association method for mapping traits using reference transcriptome data. Nat. Genet. 2015;47:1091–1098. doi: 10.1038/ng.3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only References
- 59.Taïeb A, Picardo M. The definition and assessment of vitiligo: a consensus report of the VitiligoEuropean Task Force. Pigment Cell Res. 2007;20:27–35. doi: 10.1111/j.1600-0749.2006.00355.x. [DOI] [PubMed] [Google Scholar]
- 60.Purcell S, et al. PLINK: a toolset for whole-genome association and population-based linkage analysis. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 62.Lee AB, Luca D, Klei L, Devlin B, Roeder K. Discovering genetic ancestry using spectral graph theory. Genet. Epidemiol. 2010;34:51–59. doi: 10.1002/gepi.20434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chang D, et al. Accounting for eXentricities: analysis of the X chromosome in GWAS reveals X- linked genes implicated in autoimmune diseases. PLoS One. 2014;9:e113684. doi: 10.1371/journal.pone.0113684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Higgins JP, Thompson SP. Quantifying heterogeneity in a meta-analysis. Stat. Med. 2002;21:1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 65.Higgins JP, et al. Measuring inconsistency in meta-analyses. Br. Med. J. 2003;327:557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fairfax BP, et al. Innate immune activity conditions the effect of regulatory variants upon monocyte gene expression. Science. 2014;343:1246949. doi: 10.1126/science.1246949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Giambartolomei C, et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 2014;10:e1004383. doi: 10.1371/journal.pgen.1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Guo H, et al. Integration of disease association and eQTL data using a Bayesian colocalisation approach highlights six candidate causal genes in immune-mediated diseases. Hum. Mol. Genet. 2015;24:3305–3313. doi: 10.1093/hmg/ddv077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Arloth J, Bader DM, Röh S, Altmann A. Re-Annotator: Annotation pipeline for microarray probe sequences. PLoS One. 2015;10:1–13. doi: 10.1371/journal.pone.0139516. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.