Abstract
Improving seed oil yield and quality are central targets in rapeseed (Brassica napus) breeding. The primary goal of our study was to examine and compare the potential and the limits of marker-assisted selection and genome-wide prediction of six important seed quality traits of B. napus. Our study is based on a bi-parental population comprising 202 doubled haploid lines and a diverse validation set including 117 B. napus inbred lines derived from interspecific crosses between B. rapa and B. carinata. We used phenotypic data for seed oil, protein, erucic acid, linolenic acid, stearic acid, and glucosinolate content. All lines were genotyped with a 60k SNP array. We performed five-fold cross-validations in combination with linkage mapping and four genome-wide prediction approaches in the bi-parental population. Quantitative trait loci (QTL) with large effects were detected for erucic acid, stearic acid, and glucosinolate content, blazing the trail for marker-assisted selection. Despite substantial differences in the complexity of the genetic architecture of the six traits, genome-wide prediction models had only minor impacts on the prediction accuracies. We evaluated the effects of training population size, marker density and phenotyping intensity on the prediction accuracy. The prediction accuracy in the independent and genetically very distinct validation set still amounted to 0.14 for protein content and 0.17 for oil content reflecting the utility of the developed calibration models even in very diverse backgrounds.
Introduction
Rapeseed (Brassica napus L.) is one of the most important oilseed crops worldwide [1]. The breeding goal for rapeseed is high oil yield coupled with excellent oil quality [2–4]. The latter is mainly driven by the composition of the fatty acid components of erucic acid (C22:1), stearic acid (C18:0), oleic acid (C18:1), linoleic acid (C18:2), and linolenic acid (C18:3) [2, 3, 5]. Moreover, protein and glucosinolate content determine to a large extent the quality of the rapeseed meal [6–8]. All of these seed traits are influenced by the environment [9–11], and their precise estimation requires phenotyping in replicated multi-environmental field trials. Moreover, measuring quality traits in rapeseed is often labor-intensive. Therefore, quality traits are interesting targets for genomic-assisted crop improvement.
Genomic-assisted crop improvement can either be based on marker-assisted selection [12] or genome-wide predictions [12, 13]. In marker-assisted selection, the performance of individuals is predicted using a few diagnostic markers associated with the traits under consideration [14]. In contrast, genome-wide prediction exploits many markers without performing marker-specific significance tests [15]. The accuracy of marker-assisted selection and genome-wide predictions depends on the genetic architecture underlying the traits under consideration. Marker-assisted selection is most effective if the trait is controlled by a few genes with large effects. If the genetic architecture is complex, quantitative trait loci (QTL) detection is not reliable and genome-wide prediction is more powerful [16].
The presence of QTL underlying quality traits in rapeseed has been investigated in linkage and linkage disequilibrium mapping studies [1, 3, 9–11, 17–28]. Accumulated information of the QTL accounting for seed quality traits such as seed fatty acid has also been identified in other Brassica species, such as B. oleracea and B. juncea [29, 30], which could provide reference for the comparison between species. However, linkage and linkage disequilibrium mapping, are often afflicted by upwards biased estimates in terms of the proportion of genotypic variance explained by QTL. Therefore, cross- or independent validations have been suggested to obtain unbiased estimates of QTL effects but have been applied only in a limited number of studies in rapeseed [31, 32].
The potential and limits of genome-wide predictions have been examined for several major crops, such as barley [33], wheat [15, 34–36], maize [37–42], rice [43], sunflower [44], forage plants [45], sugar beet [46, 47], and soybean [48, 49]. The results underlined the potential of genome-wide prediction as a powerful tool to accelerate selection gain in plant breeding. Recent studies in rapeseed also highlighted the potential of genome-wide prediction of flowering time [31, 50, 51], plant height, protein content, oil content, glucosinolate content, grain yield [31, 51]. Nevertheless, the benefits of genome-wide prediction compared to marker-assisted selection have not been examined in rapeseed. Moreover, the potential to exploit epistasis to predict seed quality traits has not been investigated, although previous studies suggested that epistatic interactions were important for fatty acid metabolism [11].
This study is based on a published dataset from the bi-parental TN DH population comprising 202 DH lines, which has been intensively used to study the genetic architecture of important agronomic traits [9–11, 22, 23] and were genotyped with an Infinium 60K-SNP array [52] being extensively used in Brassica [24, 53, 54]. The two parents of the TN DH mapping population originated from the European and Chinese genepools and have been used widely for rapeseed breeding programs in both target regions. Our objectives were to (i) test for the presence of QTL exhibiting reliable and large effects using five-fold cross-validations, (ii) investigate the effect of the genetic architecture on the superiority of different genome-wide prediction models, (iii) examine the potential to improve the prediction accuracy by modeling digenic epistatic effects, (iv) validate the prediction accuracy in a genetically independent population, and (v) discuss the consequences for implementing genome-wide predictions in applied rapeseed breeding programs.
Materials and Methods
Plant materials and field trials
A bi-parental DH population of B. napus denoted as TN DH has been developed, comprising 202 unique lines [22]. The DH lines were derived from a microspore culture based on the F1 cross between Tapidor and Ningyou7. The parent Tapidor is a European winter cultivar with low erucic acid and glucosinolate content in the seeds. The parent Ningyou7 is a Chinese semi-winter cultivar with high erucic acid and glucosinolate content in seeds. The TN DH mapping population along with its two parents was grown in 11 winter and semi-winter ecotype environments (S1 Table). The phenotypic data was generated and used in a previous linkage mapping study, which was based on a limited set of markers [9–11, 22, 23]. The experimental design was a randomized complete block design with 3 replications. Every plot comprised three rows with a total plot size of 3.0 to 4.0 m2.
Phenotypic data was collected for six important seed quality traits for each DH line and parent: seed oil content (%) and protein content (%), which were separately defined as the percentages of the oil and protein in the total seed dry weight, respectively; three important components of the fatty acid in the seed oil: the erucic acid content (%), the linolenic acid content (%), and the stearic acid content (%); and the content of glucosinolates in the total seed dry weight (µmol/g). The quality traits were determined based on near infrared reflectance spectroscopy measuring three technical and three biological replicates. The details of the phenotyping are outlined in detail in previous studies [10, 11, 22].
A total of 117 genetically independent B. napus inbred lines were used in this study for validating the prediction accuracy based on the TN DH population. The validation population was developed based on hundreds of crosses between B. rapa and B. carinata accessions [55, 56]. The validation population was grown in one semi-winter environment (Wuhan, China) in 2013–2014 in a trial with three replicates. Every plot comprised two rows with a total plot size of 2.0 to 3.0 m2. Seed oil content and protein content was measured using the same method as that used for the TN DH population.
Phenotypic data analyses
The best linear unbiased estimates (BLUEs) of phenotypic values and variance components were estimated by the following linear mixed model using ASREML-R software [57]:
The genotype effects were treated as fixed effects and the other effects were treated as random. To estimate variance components, all effects were treated as random. Broad-sense heritability was calculated as the ratio of genotypic to phenotypic variance:
where NE refers to the number of environments, NR is the average number of replications per location, is the genotypic variance, is the variance of genotype times environment interaction, and refers to the error variance.
Genotypic data analyses
The 202 DH lines of the TN DH population and the two parents were previously fingerprinted using a 60k SNP array based on an Illumina Infinium assay [52]. Quality control was performed and those markers have been removed which are either monomorphic, have missing values of >5%, a minor allele frequency <5%, or degree of heterozygosity >5% in the DH population. After applying the quality check outlined above, 180 DH lines with 13,678 high-quality SNP markers remained. By aligning the marker sequence of the 13,678 SNPs to the reference “Darmor-bzh” genome of B. napus version 4.1[58] via BLAST analysis, 9,628 SNP markers could be assigned a unique physical position in the genome with the parameters of 100% alignment, E value <10−20 and mismatch <2 (S2 Table). After removing redundant SNPs in full linkage disequilibrium (LD), 1,527 markers representing recombination loci (referred to as representative markers) remained (S2 Table). The 1,527 representative markers included 1,052 representative markers from 1,052 genetic bins and 475 single markers. From each of the genetic bins, one marker with the least missing rate and the best available physical alignment position was selected as representative marker. In this way, a total of 1,527 representative markers were obtained and used for the subsequent analysis. Pairwise LD between markers was calculated as the squared Pearson moment correlation coefficient using R package genetics [59]. The 117 lines of the validation population were genotyped using the same SNP array and the 1,527 representative markers selected in the TN DH population were used for prediction.
QTL mapping and genome-wide prediction
For the QTL mapping, the SNP markers were coded according to the F∞ metric [60]. The genome-wide QTL mapping method is based on the inclusion of cofactors [7] obtained by stepwise multiple linear regressions using the Bayesian information criterion [61]. The genome-wide scan was conducted comparing the full model comprising the SNP and all cofactors versus a reduced model including only cofactors. We used a false-discovery rate (FDR) of P<0.1 to test for significance. The proportion of the phenotypic variance explained (PVE) by all QTLs, was estimated using the adjusted R2 values fitting a multiple regression [62].
We performed a five-fold cross-validation of the QTL mapping in which the total population of 180 DH lines was randomly divided into two groups with 100 replications according to the ratio of 4:1 (one group with 144 lines and the other group with 36 lines). One hundred and forty-four lines were used as the training set and the remaining 36 were used as the test set. QTL mapping was performed in each training set and estimated QTL effects were used to predict the genetic values of the lines of the test set. The prediction accuracy was defined as the correlation between the predicted and observed phenotypic values standardized with the square root of the heritability.
For the genome-wide prediction, four different models were used in this study. We implemented three methods exploiting the additive marker effect: genomic best linear unbiased prediction (GBLUP), ridge regression best linear unbiased prediction (RR-BLUP) [63], and BayesCπ [64]. To accelerate computation speed and eliminate the impact of LD on the prediction accuracy of BayesCπ, we removed SNPs with r2>0.95. For BayesCπ, the Gibbs sampling ran 20,000 times, and the first 6,000 cycles were used as burn in. We also implemented an extended GBLUP model denoted as EG-BLUP, which models digenic epistatic effects as well as additive effects [65]. The accuracies of all these genome-wide prediction methods were determined based on the adjusted entry means for the 180 genotypes applying five-fold cross-validation. Details of the implementation of the models have been described elsewhere [41, 42, 65]. We performed 100 cross-validation runs and estimated the accuracy as the Pearson correlation coefficient between predicted and observed values standardized with the square root of the heritability.
To evaluate the dependence of prediction accuracy on training set size, we applied cross-validation with randomly selected subsets of n (n = 48, 80, 112, 144) lines from the full data to form the training set and used the remaining lines as the test set. To evaluate the dependence of prediction accuracy on marker density, we selected subsets of m (m = 100, 1,000, 5,000, 13,678) evenly distributed markers from the full dataset and applied five-fold cross-validations using all 180 lines. The sampling procedure was randomly repeated 100 times for each scheme, and the prediction accuracies were averaged across the 100 cross-validation runs. We focused in the above outlined analyses of sampling of marker subsets and training set sizes on the traits seed oil content and protein content. The traits were selected because oil content was evaluated in a large number of 11 environments and protein content exhibited a high heritability.
We also evaluated the prediction accuracy using an independent validation population. The marker effects were estimated based on RR-BLUP and the TN DH population. Marker effects were used to predict the performance of the 117 individuals of the validation population. The prediction accuracy was again estimated as the Pearson correlation coefficient between predicted and observed values standardized with the square root of the heritability. Heritability was estimated using the variance components estimated for the TN DH population.
Results
Intensive field evaluation of the TN DH population resulted in high-quality phenotypic data
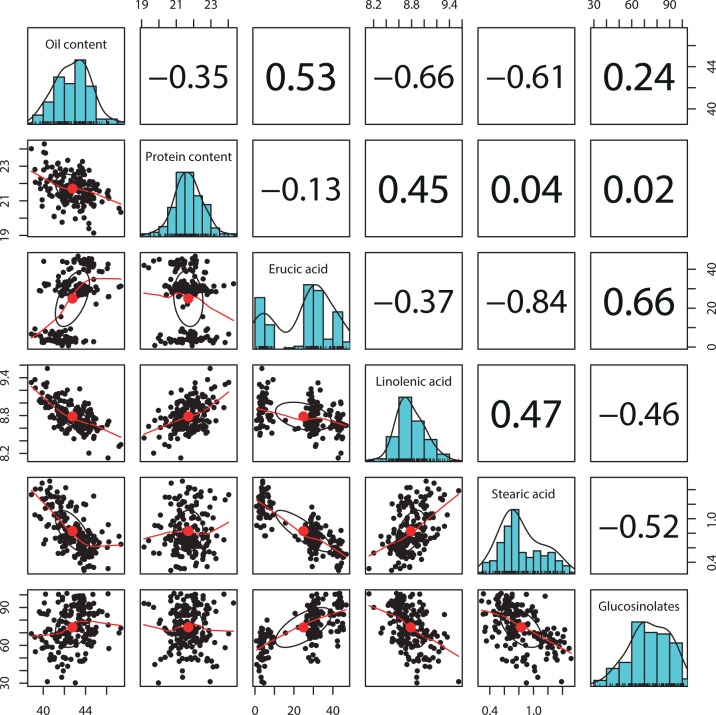
We combined the information on seed protein content with previously published data for other seed quality traits of the TN DH population. We observed a wide variation of BLUEs approximating a normal distribution for most traits, except for erucic acid content (Fig 1, S3 Table). The analyses across environments revealed significant (P<0.001) variances for genotypes, environments, and interactions between genotypes and environments (Table 1). Broad-sense heritability estimates were high for the six traits, ranging from 0.81 for protein content to 0.98 for erucic acid content. Consequently, the intensive phenotyping resulted in high-quality data representing an excellent source for dissecting the genetic basis of the six traits.
Fig 1. Distributions and pairwise correlations for Best Linear Unbiased Estimates of six seed traits evaluated for 202 lines of the TN DH population in multi-environmental field trials.
All correlations passed significance tests with P-values less than 0.001 except for the correlation between protein content and erucic acid, glucosinolates, and stearic acid content.
Table 1. Estimates of variance components (σ2) and broad-sense heritability (h2) for the TN DH population with 202 lines evaluated for six seed traits in multi-environmental field trials.
| Source*/Traits | Oil content | Protein content | Erucic acid content | Linolenic acid content | Stearic acid content | Glucosinolate content |
|---|---|---|---|---|---|---|
| 2.64 | 0.53 | 198.19 | 0.04 | 0.08 | 229.38 | |
| 0.71 | 0.42 | 11.84 | 0.01 | 0 | 120.82 | |
| 1.17 | 0.61 | 7.78 | 0.02 | 0.01 | 54.55 | |
| Heritability | 0.96 | 0.81 | 0.98 | 0.82 | 0.94 | 0.9 |
| Mean | 42.76 | 21.69 | 24.91 | 8.79 | 0.81 | 74.16 |
| Range | 38.87–47.35 | 19.13–24.3 | 0.77–46.76 | 8.13–9.55 | 0.27–1.50 | 30.31–101.17 |
| Nr. of environments | 11 | 5 | 5 | 2 | 2 | 6 |
*All variances pass a significance test with P values less than 0.001.
In total, 80% of the pairwise trait comparisons were significantly (P<0.001) associated with Pearson moment correlation coefficients ranging from -0.84 between erucic acid content and stearic acid content to 0.66 between erucic acid content and glucosinolate content (Fig 1). Interestingly, protein content was only poorly associated with erucic acid, glucosinolate, and stearic acid content. This lack of associations points to independent biochemical pathways and genes controlling the two classes of traits.
Large differences in the complexity of the genetic architecture of the six seed quality traits
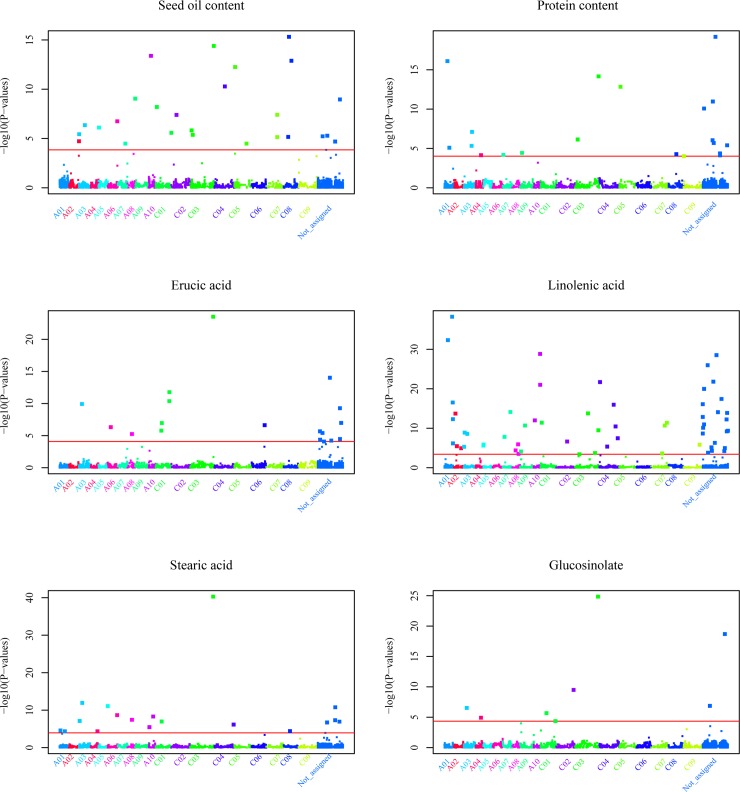
Altogether, 151 SNP markers passed the FDR significance level of P<0.1 in the genome-wide QTL mapping scan (Figs 2 and S1). The QTL numbers for the six traits ranged from 8 to 59 and were distributed across 19 chromosomes of B. napus. Phenotypic variance explained by a single putative QTL exceeded 5% for 27 SNPs and reached 45% for a QTL located on chromosome C03 controlling erucic acid content (Table 2). A second major QTL was detected on chromosome A08 for erucic acid content, explaining 31% of the phenotypic variance. However, the majority of the QTLs, especially those influencing oil and protein content, exhibited only minor effects. Among the detected QTLs, seven were putative pleiotropic QTLs influencing two traits. For instance, the marker “Bn-scaff_15794_1-p347392”, which was physically aligned to C03 and detected as a putative pleiotropic QTL, explained 26% and 45% of the phenotypic variance for stearic acid and erucic acid concentration, respectively.
Fig 2. Manhattan plots based on composite interval QTL mapping for the six seed quality traits.
The x-axis represents the corresponding physical position of each SNP of the 13,678 SNPs across the genome from chromosome A01 to A10 and C01 to C09. Those markers without unique alignment to the reference genome were arranged in the axis noted as “not assigned”. The Y-axis represents the corresponding false-discovery rate (FDR) of each QTL indicating the significance for QTL calling. The PVE, i.e. proportion of the phenotypic variance explained by each QTL, is listed in Table 2.
Table 2. Significant marker-trait associations and the proportion of explained phenotypic variance (PVE) detected in a genome-wide association mapping approach for six quality traits of TN DH population.
| Trait | No. | Marker | P values | PVE | Genetic | Chr. | Physical | Detected in previous studies |
|---|---|---|---|---|---|---|---|---|
| bin code1 | position (bp) 2 | |||||||
| Oil content | 1 | Bn-A10-p5869175 | 4.09E-14 | 4.27 | 612 | A10 | 5499050 | TN-qOC-A10-1 (Jiang et al. 2014)[10] |
| 2 | Bn-A09-p739088 | 9.01E-10 | 2.96 | 551 | A09 | 131541 | ||
| 3 | Bn-A07-p16379135 | 3.29E-05 | 0.01 | 485 | A07 | 18348848 | SG-qOC-A7 (Zhao et al. 2012)[66] | |
| 4 | Bn-A07-p15802174 | 5.96E-06 | 0.7 | single marker | A07 | NA3 | ||
| 5 | Bn-A04-p1684695 | 5.22E-11 | 5.86 | 806 | C04 | 25032235 | ||
| 6 | Bn-A01-p27774666 | 2.64E-06 | 0.5 | single marker | C01 | 38105589 | ||
| 7 | Bn-scaff_15695_1-p294894 | 3.31E-05 | 0.25 | 854 | C05 | 29927748 | ||
| 8 | Bn-scaff_16361_1-p930064 | 0.000142 | 1.23 | 956 | C08 | NA3 | ||
| 9 | Bn-scaff_20942_1-p440106 | 5.21E-06 | 9.49 | 694 | C02 | NA3 | ||
| 10 | Bn-scaff_17637_1-p204439 | 6.89E-06 | 2.74 | 945 | C08 | 12326547 | ||
| 11 | Bn-scaff_16565_1-p1169320 | 4.06E-08 | 1.18 | 698 | C02 | 12445051 | TN-qOC-C2-2 (Jiang et al. 2014)[10] | |
| 12 | Bn-scaff_15838_1-p2253503 | 6.36E-09 | 2.94 | 660 | C01 | 2629345 | TN-qOC-C1-1 (Jiang et al. 2014)[10] | |
| 13 | Bn-A05-p1308471 | 7.76E-07 | 0.06 | 309 | A05 | 1423576 | SG-qOC-A5 (Zhao et al. 2012)[66] | |
| 14 | Bn-Scaffold000217-p20168 | 2.05E-05 | 0.37 | 361 | C05 | NA3 | ||
| 15 | Bn-scaff_20901_1-p1705574 | 5.59E-13 | 4.91 | 839 | C05 | 2309449 | ||
| 16 | Bn-scaff_23761_1-p249628 | 4.09E-15 | 16.44 | single marker | C03 | 57481703 | TN-qOC-C3-3 (Jiang et al. 2014)[10] | |
| 17 | Bn-A02-p27799727 | 1.92E-05 | 0.93 | 139 | A02 | 24756539 | DY-qOC-A2-2 (Delourme et al. 2006)[1]; Z5-qOC-A2-1 (Sun et al. 2012)[67] | |
| 18 | Bn-scaff_16231_1-p2213239 | 1.30E-13 | 5.75 | 949 | C08 | 20090489 | ||
| 19 | Bn-A03-p15397187 | 4.40E-07 | 3.81 | 195 | A03 | 14446606 | TN-qOC-A3-3 (Jiang et al. 2014)[10] | |
| 20 | Bn-scaff_16545_1-p238397 | 4.80E-16 | 6.43 | 508 | C08 | 14155605 | ||
| 21 | Bn-A06-p7949147 | 1.09E-09 | 1.35 | 388 | A06 | NA3 | ||
| 22 | Bn-scaff_16130_1-p1013445 | 3.94E-08 | 1.79 | 911 | C07 | 28755038 | ||
| 23 | Bn-scaff_16130_1-p1039452 | 7.23E-06 | 2.28 | 911 | C07 | 28772215 | ||
| 24 | Bn-A06-p24132842 | 1.79E-07 | 2.49 | 421 | A06 | 23129285 | Z5-qOC-A6-1 (Sun et al. 2012)[67] | |
| 25 | Bn-scaff_22728_1-p357789 | 4.28E-06 | 0.88 | 160 | C03 | 6154024 | TN-qOC-C3-3 (Jiang et al. 2014)[10]; OIL.C3.s.1(Niklas Körber et al.2016)[68] | |
| 26 | Bn-A03-p764274 | 3.67E-06 | 0.06 | 141 | A03 | 632475 | ||
| 27 | Bn-scaff_18936_1-p890286 | 1.50E-06 | 1.48 | 731 | C03 | 3419666 | OIL.C3.s.1(Niklas Körber et al. 2016)[68] | |
| Protein content | 1 | Bn-scaff_15838_3-p256767 | 8.37E-11 | 7.13 | 121 | A02 | NA3 | |
| 2 | Bn-A03-p21225846 | 7.85E-08 | 0.08 | 211 | A03 | 19974471 | ||
| 3 | Bn-A04-p12670129 | 7.18E-05 | 1.96 | 269 | A04 | 13394800 | qThrC-4-2(Xu et al.2015)[69] | |
| 4 | Bn-A03-p20150479 | 4.68E-06 | 2.65 | 209 | A03 | 19014117 | ||
| 5 | Bn-scaff_16361_1-p300435 | 8.14E-06 | 0.5 | 63 | A01 | 11871025 | ||
| 6 | Bn-A09-p5190180 | 3.69E-05 | 0.02 | 556 | A09 | 4862135 | ||
| 7 | Bn-scaff_17526_1-p860459 | 9.60E-05 | 5.21 | 977 | C09 | 1679866 | qMetC-19-9(Xu et al.2015)[69] | |
| 8 | Bn-scaff_16449_1-p251526 | 9.17E-07 | 0.42 | 709 | C02 | NA3 | ||
| 9 | Bn-A09-p33595011 | 1.08E-11 | 2.85 | single marker | NA3 | NA3 | ||
| 10 | Bn-A01-p8058255 | 7.87E-17 | 9.49 | 50 | A01 | 7238500 | qPC-1(Huang et al.2016)[70] | |
| 11 | Bn-A09-p15975138 | 2.03E-06 | 1.91 | 567 | A09 | NA3 | ||
| 12 | Bn-scaff_17119_1-p349622 | 6.35E-20 | 10.98 | 778 | C03 | NA3 | ||
| 13 | Bn-scaff_17119_1-p414142 | 6.82E-15 | 5.37 | 778 | C03 | 57158030 | ||
| 14 | Bn-scaff_27815_1-p367403 | 6.45E-05 | 2.65 | 431 | A07 | 1626003 | ||
| 15 | Bn-A01-p27125649 | 4.28E-05 | 1.35 | 87 | A01 | NA3 | ||
| 16 | Bn-Scaffold000217-p38276 | 7.56E-05 | 1.78 | 361 | C05 | NA3 | ||
| 17 | Bn-scaff_20901_1-p647270 | 1.44E-13 | 10.05 | 840 | C05 | 3389245 | qAlaC-15-4 (Wen et al.2015)[71] | |
| 18 | Bn-scaff_16231_1-p2213239 | 5.32E-05 | 8.59 | 949 | C08 | 20090489 | ||
| 19 | Bn-scaff_23799_1-p6782 | 3.97E-06 | 1.76 | single marker | NA3 | NA3 | ||
| 20 | Bn-scaff_22728_1-p349077 | 7.20E-07 | 2.77 | 160 | C03 | 6162734 | qMetC-13-6 (Xu et al.2015)[69] | |
| Erucic acid | 1 | Bn-scaff_15803_1-p800874 | 1.11E-07 | 0.36 | 58 | C01 | 14815203 | |
| 2 | Bn-scaff_15747_1-p167954 | 2.16E-06 | 0.08 | 675 | C01 | NA3 | ||
| 3 | Bn-scaff_19614_1-p36023 | 1.66E-06 | 0.11 | 675 | C01 | 13532546 | ||
| 4 | Bn-A03-p24897111 | 4.39E-05 | 1.08 | 223 | A03 | NA3 | ||
| 5 | Bn-scaff_18039_1-p206042 | 4.22E-11 | 2.08 | 682 | C01 | 33006005 | ||
| 6 | Bn-scaff_15844_1-p119216 | 1.68E-12 | 0.63 | single marker | C01 | 33345268 | ||
| 7 | Bn-A03-p23609934 | 3.90E-06 | 0.1 | 219 | A03 | NA3 | ||
| 8 | Bn-A01-p3664698 | 8.21E-05 | 1.95 | 33 | A01 | NA3 | ||
| 9 | Bn-scaff_16397_1-p21961 | 2.35E-07 | 0.38 | 885 | C06 | 32884939 | ERA.C6.s.1(Niklas Körber et al.2016)[68] | |
| 10 | Bn-scaff_15794_1-p347392 | 2.97E-24 | 45.35 | 775 | C03 | 55942754 | qC3-3(Wang et al.2015)[11] | |
| 11 | Bn-A09-p19718581 | 9.53E-15 | 0.63 | 568 | A09 | NA3 | ||
| 12 | Bn-scaff_17984_1-p123918 | 5.88E-05 | 3.54 | 569 | A09 | NA3 | ||
| 13 | Bn-A08-p13221380 | 5.64E-06 | 31.31 | 510 | A08 | 10967853 | qA8-5(Wang et al.2015)[11] | |
| 14 | Bn-C14160250-p3687 | 5.39E-10 | 0.98 | 509 | A08 | NA3 | ||
| 15 | Bn-A06-p7636729 | 4.83E-07 | 0.11 | 388 | A06 | 7278355 | ||
| 16 | Bn-A06-p7459428 | 3.40E-05 | 0.69 | single marker | A06 | NA3 | ||
| 17 | Bn-A03-p14811204 | 1.05E-07 | 0.03 | 191 | A03 | NA3 | ||
| 18 | Bn-A03-p8177695 | 1.19E-10 | 1.91 | 173 | A03 | 7472584 | ||
| Linoleic acid | 1 | Bn-A02-p1890913 | 1.40E-13 | 0.23 | 655 | NA3 | NA3 | |
| 2 | Bn-A02-p2451470 | 7.90E-17 | 1.76 | 655 | NA3 | NA3 | ||
| 3 | Bn-A02-p7105435 | 1.89E-14 | 0.22 | single marker | A02 | 4150237 | ||
| 4 | Bn-A10-p510846 | 1.01E-12 | 8.55 | 610 | A10 | 2998077 | ||
| 5 | Bn-A10-p1193336 | 2.13E-09 | 3.29 | 609 | A10 | NA3 | ||
| 6 | Bn-A10-p2092612 | 7.56E-11 | 0.11 | single marker | A10 | NA3 | ||
| 7 | Bn-A09-p3546619 | 1.99E-22 | 4.68 | single marker | C04 | 1322839 | ||
| 8 | Bn-A02-p10850012 | 3.55E-06 | 9.07 | 118 | A02 | 7665679 | ||
| 9 | Bn-A02-p12145607 | 1.07E-11 | 0.12 | 123 | A02 | NA3 | ||
| 10 | Bn-A09-p20863459 | 1.06E-20 | 1.99 | single marker | NA3 | NA3 | ||
| 11 | Bn-A09-p1631944 | 7.56E-05 | 5.13 | 554 | A09 | 2294691 | LIA.A9.w.1(Niklas Körber et al.2016)[68] | |
| 12 | Bn-scaff_22749_1-p250319 | 2.31E-07 | 0.6 | 129 | C02 | 26414418 | ||
| 13 | Bn-A07-p16846624 | 7.65E-15 | 1.68 | 485 | A07 | 18775516 | ||
| 14 | Bn-A01-p27968584 | 1.06E-26 | 0.93 | single marker | NA3 | NA3 | ||
| 15 | Bn-A02-p18438691 | 0.000162 | 3.28 | single marker | A02 | NA3 | ||
| 16 | Bn-A02-p19070958 | 1.17E-05 | 1.08 | 131 | A02 | 18106292 | ||
| 17 | Bn-scaff_18855_1-p795432 | 1.72E-14 | 0.06 | 757 | C03 | 31378910 | ||
| 18 | Bn-scaff_16135_1-p196922 | 0.00032 | 0.03 | 532 | A08 | 15018355 | ||
| 19 | Bn-A10-p15742689 | 1.45E-29 | 2.59 | 648 | A10 | 15807427 | ||
| 20 | Bn-A05-p18147040 | 2.03E-11 | 1.09 | 248 | A09 | 12040388 | ||
| 21 | Bn-scaff_16372_1-p19665 | 0.000194 | 1.27 | 769 | C03 | 48510330 | qC3-2(Wang et al.2015)[11] | |
| 22 | Bn-scaff_20294_1-p438293 | 6.85E-06 | 3.77 | 887 | C06 | NA3 | ||
| 23 | Bn-A10-p15442975 | 9.97E-22 | 2.37 | 652 | A10 | 16049734 | ||
| 24 | Bn-scaff_17799_1-p2773426 | 1.55E-06 | ~0.00 | 990 | C09 | 39884740 | ||
| 25 | Bn-A09-p33542334 | 5.40E-05 | 0.83 | single marker | A09 | NA3 | ||
| 26 | Bn-A01-p22016353 | 5.40E-39 | 5.44 | 76 | A01 | 18645502 | ||
| 27 | Bn-A05-p114598 | 2.39E-06 | 0.01 | single marker | A05 | 128946 | ||
| 28 | Bn-A01-p9810552 | 4.72E-33 | 0.04 | single marker | A01 | 8432723 | ||
| 29 | Bn-A01-p8108178 | 1.57E-22 | 1.96 | 50 | A01 | NA3 | ||
| 30 | Bn-scaff_17821_1-p21053 | 3.33E-10 | 3.01 | 777 | C03 | 56695853 | qC3-3(Wang et al.2015)[11]; qC18:2-13-5(Wen et al.2015)[71] | |
| 31 | Bn-A09-p19688476 | 5.00E-07 | 0.02 | 568 | A09 | NA3 | ||
| 32 | Bn-A05-p472271 | 1.39E-06 | 0.37 | 307 | A05 | 583644 | ||
| 33 | Bn-scaff_15838_1-p2253503 | 3.85E-12 | 7.51 | 660 | C01 | 2629345 | qC18:2-11-3(Wen et al.2015)[71] | |
| 34 | Bn-scaff_15585_1-p1020764 | 3.37E-08 | 0.96 | 279 | C04 | 44431942 | ||
| 35 | Bn-scaff_15676_1-p341508 | 2.92E-29 | 2.23 | 858 | C05 | NA3 | ||
| 36 | Bn-scaff_19170_1-p1107619 | 4.39E-06 | 0.08 | 10 | C04 | 18803084 | ||
| 37 | Bn-scaff_19170_1-p588356 | 8.41E-15 | 0.85 | 10 | C04 | NA3 | ||
| 38 | Bn-A10-p3329131 | 1.54E-08 | 3.3 | 433 | A07 | 4431854 | ||
| 39 | Bn-Scaffold000164-p120459 | 6.78E-07 | 0.22 | 83 | A01 | 20722278 | ||
| 40 | Bn-scaff_21956_1-p160710 | 3.46E-11 | 0.72 | 821 | C04 | 39249761 | ||
| 41 | Bn-scaff_16876_1-p171510 | 3.75E-18 | 0.11 | 817 | NA3 | NA3 | ||
| 42 | Bn-scaff_16876_1-p303006 | 1.11E-16 | 0.46 | 816 | C04 | 34584781 | ||
| 43 | Bn-A01-p24830111 | 5.08E-13 | 0.43 | 82 | A01 | 20561797 | ||
| 44 | Bn-A08-p16562035 | 1.20E-06 | 0.28 | 522 | A08 | 14030898 | qA8-5(Wang et al.2015)[11]; LIA.A8.w.1(Niklas Körber et al.2016)[68] | |
| 45 | Bn-scaff_16069_1-p3780494 | 4.12E-12 | 0.04 | 929 | C07 | 40184750 | ||
| 46 | Bn-scaff_16545_1-p110342 | 4.09E-05 | 0.01 | 508 | A08 | 7961925 | qA8-6(Wang et al.2015)[11] | |
| 47 | Bn-scaff_27204_1-p1544 | 6.30E-05 | 0.42 | 1045 | C07 | NA3 | ||
| 48 | Bn-A08-p11212494 | 1.06E-05 | 0.47 | 509 | A08 | NA3 | ||
| 49 | Bn-scaff_16130_1-p1039452 | 0.000253 | 0.07 | 911 | C07 | 28772215 | ||
| 50 | Bn-scaff_15705_1-p1818177 | 2.15E-11 | 0.29 | 918 | C07 | 35089587 | ||
| 51 | Bn-A02-p2962298 | 5.75E-13 | 0.09 | single marker | A02 | NA3 | ||
| 52 | Bn-A03-p10883930 | 6.17E-10 | 0.08 | 1051 | A03 | NA3 | ||
| 53 | Bn-scaff_19111_1-p177344 | 0.000399 | 0.2 | 744 | C03 | 10482820 | ||
| 54 | Bn-A03-p9098773 | 2.74E-09 | 0.39 | 173 | A03 | 8405389 | ||
| 55 | Bn-scaff_23799_1-p6782 | 1.32E-14 | 0.88 | single marker | NA3 | NA3 | ||
| 56 | Bn-A03-p2491346 | 1.29E-09 | 0.24 | 150 | A03 | 2036063 | ||
| 57 | Bn-A01-p24020451 | 2.95E-17 | 0.15 | single marker | A01 | 20087415 | ||
| 58 | Bn-A03-p764274 | 5.12E-06 | 0.73 | 141 | A03 | 632475 | ||
| 59 | Bn-A02-p2287712 | 4.39E-10 | 1.09 | single marker | NA3 | NA3 | ||
| Stearic acid | 1 | Bn-A10-p9436205 | 5.05E-09 | 1.5 | single marker | A10 | 10869232 | qA10-2(Wang et al.2015)[11] |
| 2 | Bn-A10-p1919293 | 3.42E-06 | 0.04 | 607 | A10 | 1780462 | ||
| 3 | Bn-scaff_15747_1-p396080 | 1.09E-07 | 0.85 | 677 | C01 | 14488446 | ||
| 4 | Bn-A01-p2688662 | 2.70E-05 | 7.74 | 27 | A01 | 2194542 | qA1-5(Wang et al.2015)[11] | |
| 5 | Bn-scaff_17423_1-p100318 | 0.000119 | 0.48 | single marker | A09 | NA3 | ||
| 6 | Bn-A01-p15497190 | 4.41E-05 | 5 | 57 | A01 | 12941895 | qA1-5(Wang et al.2015)[11] | |
| 7 | Bn-Scaffold000178-p33587 | 1.95E-07 | 1.45 | single marker | A09 | NA3 | ||
| 8 | Bn-scaff_15794_1-p347392 | 5.21E-41 | 25.67 | 775 | C03 | 55942754 | qC3-3(Wang et al.2015)[11] | |
| 9 | Bn-A04-p16528010 | 4.43E-05 | 0.47 | 288 | A04 | 16689032 | ||
| 10 | Bn-A04-p17358519 | 7.05E-07 | 2.44 | single marker | C04 | 46402393 | ||
| 11 | Bn-A06-p112339 | 4.94E-08 | 4.83 | 364 | A06 | NA3 | ||
| 12 | Bn-Scaffold000217-p6025 | 8.40E-12 | 6.06 | 361 | A05 | 22747105 | ||
| 13 | Bn-scaff_16614_1-p373513 | 1.71E-11 | 0.59 | single marker | NA3 | NA3 | ||
| 14 | Bn-A03-p2297079 | 7.65E-08 | 1.94 | single marker | A03 | 1863826 | ||
| 15 | Bn-A01-p28047872 | 1.18E-07 | 1.78 | single marker | NA3 | NA3 | ||
| 16 | Bn-A08-p13239816 | 3.77E-08 | 21.16 | 510 | A08 | 10991898 | qA8-5(Wang et al.2015)[11] | |
| 17 | Bn-scaff_15699_1-p577914 | 4.02E-05 | 0.49 | 509 | C08 | 16728901 | ||
| 18 | Bn-A06-p23865356 | 2.19E-09 | 1.55 | 420 | A06 | 22856806 | ||
| 19 | Bn-A03-p8924852 | 1.16E-12 | 3.39 | 173 | A03 | 8233061 | ||
| Glucosinolate | 1 | Bn-scaff_15747_1-p108596 | 2.11E-06 | 13 | 676 | C01 | 14196095 | TN-q.mcG-C1d(Feng et al.2012)[9] |
| 2 | Bn-scaff_19168_1-p31612 | 4.39E-05 | 1.42 | 79 | C01 | 36005339 | ||
| 3 | Bn-A04-p12259499 | 1.21E-05 | 0.24 | 269 | A04 | 13248240 | TN-q.mcG-A4c(Feng et al.2012)[9] | |
| 4 | Bn-A04-p13930713 | 1.36E-07 | 1.16 | single marker | A04 | NA3 | ||
| 5 | Bn-scaff_15918_1-p229987 | 3.20E-10 | 2.86 | 722 | C02 | 42160463 | TN-q.mcG-C2b(Feng et al.2012)[9] | |
| 6 | Bn-scaff_15794_1-p437864 | 1.45E-25 | 18.07 | 774 | C03 | 55837809 | TN-q.mcG-C3c(Feng et al.2012)[9] | |
| 7 | Bn-C14160250-p3687 | 2.05E-19 | 17.14 | 509 | A08 | NA3 | ||
| 8 | Bn-A03-p7838070 | 2.94E-07 | 3.97 | 170 | A03 | 7130138 | TN-cqS-Aro-GST-A3a(Feng et al.2012)[9] | |
| Total/average | 151 | 1.94E-05 | 3.221133333 |
1 More detailed information of each genetic bin is listed in S2 Table.
2 The physical position is presented by the start position of each SNP with unique position to the reference genome of B. napus, Darmor-bzh 4.1, and more information is also available in S2 Table.
3 Not available because of absent of alignment or multiple alignment positions.
The FDR (false-discovery rate) significance level is P<0.1for the detection of associated markers.
We used five-fold cross-validation to reliably estimate the potential of marker-assisted selection (MAS). The average accuracy of MAS ranged from 0.47 for protein content to 0.81 for erucic acid content (Table 3). These values were substantially lower compared to the non-cross-validated results (Table 2), underlining the need to validate findings of linkage mapping.
Table 3. Average prediction accuracy of four genomic selection methods and marker assisted selection (MAS) for six seed quality traits of the TN DH population.
| Marker Type | Method | Oil content | Protein content | Erucic acid | Linolenic acid | Stearic acid | Gluco-sinolates | Average |
|---|---|---|---|---|---|---|---|---|
| 13,678 SNPs | RR-BLUP | 0.74 | 0.65 | 0.81 | 0.73 | 0.78 | 0.69 | 0.73 |
| BayesCπ | 0.75 | 0.62 | 0.89 | 0.49 | 0.61 | 0.79 | 0.69 | |
| EG-BLUP | 0.72 | 0.62 | 0.79 | 0.74 | 0.75 | 0.67 | 0.72 | |
| GBLUP | 0.72 | 0.61 | 0.77 | 0.73 | 0.75 | 0.67 | 0.71 | |
| MAS | 0.52 | 0.47 | 0.81 | 0.51 | 0.74 | 0.64 | 0.62 | |
| 1,527 representative SNPs* | RR-BLUP | 0.76 | 0.66 | 0.83 | 0.75 | 0.81 | 0.72 | 0.76 |
| BayesCπ | 0.76 | 0.64 | 0.88 | 0.45 | 0.61 | 0.79 | 0.69 | |
| EG-BLUP | 0.75 | 0.64 | 0.81 | 0.76 | 0.79 | 0.71 | 0.74 | |
| GBLUP | 0.76 | 0.64 | 0.82 | 0.75 | 0.8 | 0.72 | 0.75 | |
| MAS | 0.59 | 0.45 | 0.74 | 0.54 | 0.68 | 0.62 | 0.6 | |
| 1,527 random SNPs* | RR-BLUP | 0.72 | 0.63 | 0.81 | 0.72 | 0.77 | 0.69 | 0.72 |
*The 1,527 representative SNPs are specifically selected from the 1,527 individual genetic bins of the TN DH population, while the 1,527 random SNPs are randomly selected from the total 13,678 polymorphic SNPs of the TN DH population. Marker assisted selection (MAS) is based on markers significantly associated with the respective traits outlined in detail in the Material and Methods.
Accuracies of genome-wide prediction in the TN DH population
We used four different models to investigate the efficiency of genome-wide prediction for the six seed quality traits. Genomic selection significantly showed higher prediction accuracies than MAS for all traits, with the most pronounced differences observed for linolenic acid, oil, and protein content (Table 3). The average prediction accuracy of RR-BLUP was the highest, while BayesCπ performed best for erucic acid and glucosinolate content. The most complex model comprising main and epistatic effects, EG-BLUP, performed best for linolenic acid content. In general, traits with high heritability could be predicted with higher accuracy compared to traits with low heritability.
As expected for a bi-parental mapping population, a large number of markers were in tight LD and could thus be grouped into genetic bins because of the absence of recombination events. We reduced the co-linearity among markers and removed redundant markers in full linkage disequilibrium, resulting in a subset of 1,527 SNP markers (S2 Table, S2 Fig). Prediction accuracy increased on average by 3% using the reduced 1,527 representative marker set compared to genomic selection based on all SNPs (Table 3).
Effects of marker density, training population size, and number of environments on prediction accuracy
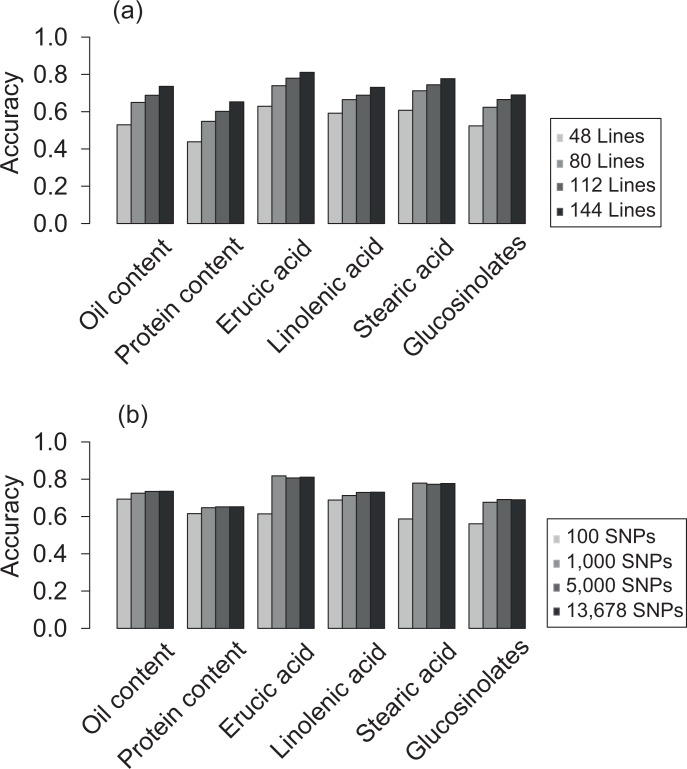
Genome-wide prediction based on RR-BLUP performed best on average and, in addition, was computationally efficient. Therefore, we conducted comprehensive analyses on the factors driving the accuracy in genome-wide prediction exclusively based on RR-BLUP. We varied the training population size and marker density and examined the accuracy of genome-wide predictions in our study. The accuracy remained in the range of 0.44 to 0.67 for all traits using only 48 lines as the training set (Fig 3). Interestingly, prediction accuracy reached a peak with 1,000 randomly selected markers and decreased only marginally for a subset of 100 markers. The prediction accuracy increased by ~4% for all six traits when using a representative set of markers compared to the 1,527 random evenly distributed markers (Table 2). Thus, our results indicated that to improve the accuracy of genome-wide prediction in a bi-parental population, the population size is more important than the density of markers.
Fig 3.
Average prediction accuracy of genomic selection applying RR-BLUP based on (a) varying training population sizes and (b) number of markers.
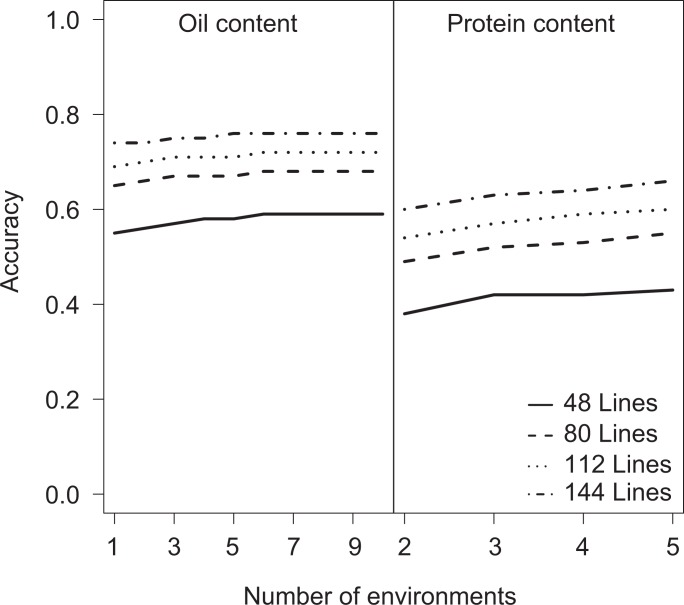
We further studied the effects of the number of environments and training population size on the accuracy of genomic selection by focusing on oil and protein content. The traits were selected because oil content was evaluated in a large number of 11 environments and protein content exhibited a high heritability. We randomly selected training sets comprising n = 48, 80, 112, and 144 lines evaluated for oil content evaluated in subsets of environments (k = 2, 3,…, 11 for oil content; k = 2, 3, 4, 5 for protein content). The accuracy was estimated as the Pearson moment correlation coefficient between predicted genotypic values and the adjusted entry means of all remaining lines evaluated across all environments. This type of cross-validation allows for the study of the prediction accuracy assuming reduced phenotyping intensity. As the test set was not evaluated in any of the environments, their performance could not be estimated by phenotypic correlations between environments. The prediction accuracies based on phenotypic data from only two environments were 0.73 for oil content and 0.60 for protein content (Fig 4). Compared to the accuracy evaluated with the full dataset, the accuracy decreased only in the range of 3% to 6%. The accuracy remained at 0.55 for oil content when only 48 lines and 2 environments were used.
Fig 4. Prediction accuracy of oil content and protein content using marker data for 1,527 representative SNPs according to different numbers of environments and training set size.
Accuracies of genome-wide prediction for seed oil content and protein content validated in a diverse population of 117 B. napus lines
A panel of 117 diverse lines was genotyped and phenotyped in one environment in order to validate the prediction accuracies of seed oil content and protein content. A total of 1148 common genetic bin markers across the AC genome, were screened for the two populations. Since we observed the highest accuracies for RR-BLUP in the TN DH population, we also used this method for prediction. The prediction accuracy amounted to 0.14 for protein content and 0.17 for oil content based on the genetic bin markers.
Discussion
Erucic acid, stearic acid, and glucosinolate content are promising targets for marker-assisted selection
Understanding the genetic basis of seed oil yield and quality is important for efficient rapeseed breeding [10]. Previous studies revealed differences in the complexity of the genetic architecture of the six quality traits examined in our study [1, 3, 9–11, 17, 20, 22, 72], which were further substantiated using five-fold cross-validations (Table 2; S3 Table). Oil, protein, and linolenic content are characterized by the absence of a reliable large-effect QTL, while erucic acid, stearic acid, and glucosinolate content are to a large degree controlled by a few QTL exhibiting large effects. For instance, the major QTL located in A08 and C03 (Table 2) totally explained 76.66% of the phenotypic variance for erucic acid, which has been widely identified previously in TN DH population and other mapping populations of B. napus [21, 22, 24, 73]. The major QTL located on C03 and explaining 16.44% of the phenotypic variance for oil content was identified in both of TN DH population and KN DH population [74]. The QTL with large genetic effects for total seed glucosinolates located in A08, C01 and C03, were also identified previously in this and other mapping populations [9, 19, 75]. These QTLs are interesting targets for marker-assisted selection, which can be applied in rapeseed breeding in combination with the enrichment of target alleles for F2 populations prior to producing DH populations. Besides of the consistent identified QTL, we also detected several new QTLs accounting for these seed quality traits with minor effects in TN DH population compared to the previous QTL identification in this population [10, 11, 22], which possibly because of the improved detection power using the high density SNP markers compared to the previous QTL identification using the relatively low-density markers. For example, the QTL “Bn-Scaffold000217-p20168” in C05, “Bn-scaff_16130_1-p1013445” and “Bn-scaff_16130_1-p1039452” located in C07 was newly identified for seed oil content of this population compared to that detected in Jiang et al., (2014). It is important to note that due to the absence of a physical position of 2,828 SNPs without alignment to the reference genome, we could not compare those QTL without unique physical position with previous studies.
Genetic architecture marginally impacts the choice of the genome-wide prediction model
Previous simulation studies revealed that equal shrinkage of marker effects as applied in RR-BLUP can be inappropriate for traits influenced by QTLs exhibiting large effects [13, 76]. In these cases, Bayesian models such as BayesB or BayesCπ, which allow specific shrinkage of every marker [77], are expected to outperform RR-BLUP. The superiority of BayesB over RR-BLUP has been reported for glucosinolate content in a previous genome-wide prediction study based on a diverse panel of 391 rapeseed lines derived from nine families [31]. Superiority of Bayes models versus RR-BLUP has also been observed for flowering time in the TN DH population [50]. In accordance with this observation, prediction accuracies for erucic acid and glucosinolate content were maximized when applying BayesCπ, with improvements of 8–10% compared to RR-BLUP (Table 2). In contrast, for stearic acid, RR-BLUP outperformed BayesCπ despite the presence of large-effect QTL. This is most likely due to two reasons. First, the ratio between the phenotypic variance explained by the two large-effect QTL versus that explained by the remaining small-effect QTL is approximately 1 to 1 for stearic acid content, while this ratio is 5 to 1 for erucic acid and glucosinolate content. Second, one large-effect QTL controlling stearic acid is reflected in several marker-trait associations with SNPs being in tight linkage disequilibrium (r2 >0.8), while the QTLs are reflected by only a limited number of SNPs for erucic acid and glucosinolate content.
Epistasis, the interaction between genes [78], is an additional potential force influencing the choice of the biometrical model for genome-wide prediction [65]. Previous linkage and linkage disequilibrium mapping studies in rapeseed indicated that epistatic effects are involved in fatty acid metabolism [11, 47]. Consequently, we implemented EG-BLUP for genome-wide prediction, which explicitly considers digenic additive by additive epistatic effects [65]. We observed, however, higher prediction accuracies of EG-BLUP compared to the other genome-wide prediction models only for linolenic acid content (Table 2). Moreover, the gains in prediction accuracy were only marginal. These negligible benefits are in contrast to the non-cross-validated results of previous linkage and linkage disequilibrium studies [11, 47] and point to the strong need to validate the role of epistatic effects. In summary, the accuracy of genomic selection does not crucially depend on the choice of a suitable genome-wide prediction model and is an attractive alternative to marker-assisted selection.
Implementation of genome-wide prediction in rapeseed breeding
The successful implementation of genome-wide prediction in rapeseed breeding requires that a certain threshold of prediction accuracy is realized [40, 79]. Previous model studies in wheat and maize suggested a threshold for the prediction accuracy of 0.5 [80, 81]. We chose two important traits, oil content and protein content, to illustrate the size of the training population, the number of environments, and the marker density required to reach a prediction accuracy of 0.5 for the bi-parental population.
In accordance with previous studies based on bi-parental populations [82–85], approximately one thousand markers were required before the prediction accuracy plateaued (Fig 3). Increasing the number of markers introduced problems due to collinearities. Prediction accuracies were higher for a reduced a set of 1,527 SNPs, which represented recombination loci in the population, in contrast to the full 13,678-marker set (Table 3). Thus, to improve the accuracy of genome-wide prediction in a bi-parental population, the population size indicating recombination events obtained is more important than the density of markers.
The number of lines has a greater impact on the prediction accuracy than the number of environments (Figs 3 and 4). The prediction accuracy is already stagnating at three environments, and thus it is more efficient to invest in training population size. For protein content, approximately 144 lines evaluated in two environments were needed to reach an accuracy of 0.6. For oil content, prediction accuracy amounted to 0.6 when the training population was decreased to 80 lines and the number of environments reduced to two. These results suggest that genome-wide prediction can be successfully implemented in bi-parental populations even with small training population sizes and is an attractive complement to phenotypic selection to improve seed quality traits.
The prediction accuracy within bi-parental populations is of central importance examining the potential to implement genome-wide prediction in breeding programs exploiting the double-haploid technology. Moreover, it is of interest to study the potential to use the prediction model also in unrelated populations. We examined an extreme validation scenario for the prediction of seed oil and protein content using a genetically diverse sample of 117 lines which were based on crosses between B. rapa and B. carinata accessions [55, 56]. The prediction accuracy in this independent and genetically very distinct validation population still amounted to 0.14 for protein content and 0.17 for oil content. While interpreting the prediction accuracies it has to be considered that the validation population exhibits genome segments from B. rapa/B. carinata. However, the used Brassica 60K-SNP array was developed based on the AC genome sequence of B. rapa, B. olearaca and B. napus. Thus, the lack of unique polymorphisms of B. carinata is expected to impair the prediction accuracies. Taking this into consideration, our independent validation reflects the high quality of the developed calibration models even in very diverse backgrounds highlighting the prospects of genome-wide prediction for routine rapeseed breeding programs.
Supporting Information
The green lines are the -log10 P-values of the linear regression method. The red lines are the -log10 P-values of the stepwise multiple linear regression method. The expected uniform distribution of negative -log10 P-values is indicated by the diagonal line in blue.
(PDF)
Within each physical distance class, marker pairs are clustered into five groups with varying r2 values.
(JPEG)
(DOCX)
(XLSX)
(XLSX)
Acknowledgments
The authors gratefully acknowledge the previous colleagues who have contributed to the collected phenotypes and genotypes of the TN DH population (Dr. Dan Qiu, Dr. Congcong Jiang, Dr. Yan Long, Dr. Ruiyuan Li, Dr. Ji Feng, Dr. Jiaqin Shi and others). We also acknowledge Dr. Bin Yi for his technical help on the SNP chip analysis.
Abbreviations
- QTL
quantitative trait loci
- DH
diploid haploid
- BLUEs
best linear unbiased estimates
- LD
linkage disequilibrium
- FDR
false-discovery rate
- PVE
phenotypic variance explained by all QTL
- GBLUP
genomic best linear unbiased prediction
- RR-BLUP
ridge regression best linear unbiased prediction
- EG-BLUP
extended GBLUP model
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the National Basic Research Program of China (Grant No. 2015CB150200), the National Key Research and Development Program of China (No.2016YFD0101300), and the Natural Science Foundation of Hubei Province Key Program 2014CFA008. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Delourme R, Falentin C, Huteau V, Clouet V, Horvais R, Gandon B, et al. Genetic control of oil content in oilseed rape (Brassica napus L.). Theoretical and Applied Genetics. 2006;113(7):1331–45. 10.1007/s00122-006-0386-z . [DOI] [PubMed] [Google Scholar]
- 2.Möllers C. Potential and future prospects for rapeseed oil In: Gunstone FD (ed) Rapeseed and canola oil—production, processing, properties and uses. Oxford, UK: Blackwell Publishing; 2004. [Google Scholar]
- 3.Zhao JY, Dimov Z, Becker HC, Ecke WG, Mollers C. Mapping QTL controlling fatty acid composition in a doubled haploid rapeseed population segregating for oil content. Mol Breeding. 2008;21(1):115–25. 10.1007/s11032-007-9113-y . [DOI] [Google Scholar]
- 4.Abbadi A, Leckband G. Rapeseed breeding for oil content, quality, and sustainability. Eur J Lipid Sci Tech. 2011;113(10):1198–206. 10.1002/ejlt.201100063 . [DOI] [Google Scholar]
- 5.Velasco L, Becker H. Estimating the fatty acid composition of the oil in intact-seed rapeseed (Brassica napus L.) by near-infrared reflectance spectroscopy. Euphytica. 1998;101(2):221–30. 10.1023/A:1018358707847 [DOI] [Google Scholar]
- 6.Bell JM. Nutrients and toxicants in rapeseed meal: a review. Journal of animal science. 1984;58(4):996–1010. . [DOI] [PubMed] [Google Scholar]
- 7.Liu Z, Hirani AH, McVetty PBE, Daayf F, Quiros CF, Li GY. Reducing progoitrin and enriching glucoraphanin in Brassica napus seeds through silencing of the GSL-ALK gene family. Plant Mol Biol. 2012;79(1–2):179–89. 10.1007/s11103-012-9905-2 . [DOI] [PubMed] [Google Scholar]
- 8.Vageeshbabu HS, Chopra VL. Genetic and biotechnological approaches for reducing glucosinolates from rapeseed-mustard meal Plant Biochemistry and Biotechnology. 1997;6(2):53–62. [Google Scholar]
- 9.Feng J, Long Y, Shi L, Shi JQ, Barker G, Meng JL. Characterization of metabolite quantitative trait loci and metabolic networks that control glucosinolate concentration in the seeds and leaves of Brassica napus. New Phytol. 2012;193(1):96–108. 10.1111/j.1469-8137.2011.03890.x . [DOI] [PubMed] [Google Scholar]
- 10.Jiang CC, Shi JQ, Li RY, Long Y, Wang H, Li DR, et al. Quantitative trait loci that control the oil content variation of rapeseed (Brassica napus L.). Theoretical and Applied Genetics. 2014;127(4):957–68. 10.1007/s00122-014-2271-5 . [DOI] [PubMed] [Google Scholar]
- 11.Wang XD, Long Y, Yin YT, Zhang CY, Gan L, Liu LZ, et al. New insights into the genetic networks affecting seed fatty acid concentrations in Brassica napus. Bmc Plant Biol. 2015;15 10.1186/s12870-015-0475-8 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lande R, Thompson R. Efficiency of marker-assisted selection in the improvement of quantitative traits. Genetics. 1990;124(3):743–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meuwissen TH, Hayes BJ, Goddard ME. Prediction of total genetic value using genome-wide dense marker maps. Genetics. 2001;157(4):1819–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bernardo R. Molecular markers and selection for complex traits in plants: Learning from the last 20 years. Crop Sci. 2008;48(5):1649–64. 10.2135/cropsci2008.03.0131 . [DOI] [Google Scholar]
- 15.Zhao Y, Mette MF, Gowda M, Longin CFH, Reif JC. Bridging the gap between marker-assisted and genomic selection of heading time and plant height in hybrid wheat. Heredity. 2014;112(6):638–45. 10.1038/hdy.2014.1 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heslot N, Jannink JL. An alternative covariance estimator to investigate genetic heterogeneity in populations. Genet Sel Evol. 2015;47:93 10.1186/s12711-015-0171-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burns MJ, Barnes SR, Bowman JG, Clarke MHE, Werner CP, Kearsey MJ. QTL analysis of an intervarietal set of substitution lines in Brassica napus: (i) Seed oil content and fatty acid composition. Heredity. 2003;90(1):39–48. 10.1038/sj.hdy.6800176 . [DOI] [PubMed] [Google Scholar]
- 18.Chen YB, Qi L, Zhang XY, Huang JX, Wang JB, Chen HC, et al. Characterization of the quantitative trait locus OilA1 for oil content in Brassica napus. Theoretical and Applied Genetics. 2013;126(10):2499–509. 10.1007/s00122-013-2150-5 . [DOI] [PubMed] [Google Scholar]
- 19.Gajardo HA, Wittkop B, Soto-Cerda B, Higgins EE, Parkin IAP, Snowdon RJ, et al. Association mapping of seed quality traits in Brassica napus L. using GWAS and candidate QTL approaches. Mol Breeding. 2015;35(6). 10.1007/S11032-015-0340-3 . [DOI] [Google Scholar]
- 20.Hu XY, Sullivan-Gilbert M, Gupta M, Thompson SA. Mapping of the loci controlling oleic and linolenic acid contents and development of fad2 and fad3 allele-specific markers in canola (Brassica napus L.). Theoretical and Applied Genetics. 2006;113(3):497–507. 10.1007/s00122-006-0315-1 . [DOI] [PubMed] [Google Scholar]
- 21.Lu GY, Harper AL, Trick M, Morgan C, Fraser F, O'Neill C, et al. Associative Transcriptomics Study Dissects the Genetic Architecture of Seed Glucosinolate Content in Brassica napus. DNA Res. 2014;21(6):613–25. 10.1093/dnares/dsu024 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qiu D, Morgan C, Shi J, Long Y, Liu J, Li R, et al. A comparative linkage map of oilseed rape and its use for QTL analysis of seed oil and erucic acid content. Theoretical and Applied Genetics. 2006;114(1):67–80. 10.1007/s00122-006-0411-2 . [DOI] [PubMed] [Google Scholar]
- 23.Zou J, Jiang CC, Cao ZY, Li RY, Long Y, Chen S, et al. Association mapping of seed oil content in Brassica napus and comparison with quantitative trait loci identified from linkage mapping. Genome / National Research Council Canada = Genome / Conseil national de recherches Canada. 2010;53(11):908–16. . [DOI] [PubMed] [Google Scholar]
- 24.Li F, Chen BY, Xu K, Wu JF, Song WL, Bancroft I, et al. Genome-Wide Association Study Dissects the Genetic Architecture of Seed Weight and Seed Quality in Rapeseed (Brassica napus L.). DNA Res. 2014;21(4):355–67. 10.1093/dnares/dsu002 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Downey RK, Craig BM. Genetic control of fatty acid biosynthesis in rapeseed (Brassica napus L). J Am Oil Chem Soc. 1964: 41:475–478. [Google Scholar]
- 26.Fourmann M, Barret P, Renard M, Pelletier G, Delourme R, Brunel D. The two genes homologous to Arabidopsis FAE1 co-segregate with the two loci governing erucic acid content in Brassica napus. Theor Appl Genet. 1998:96: 852–858. [Google Scholar]
- 27.Harper AL, Trick M, Higgins J, Fraser F, Clissold L, Wells R, et al. Associative transcriptomics of traits in the polyploid crop species Brassica napus. Nat Biotechnol. 2012; 30(8):798–802. 10.1038/nbt.2302 . [DOI] [PubMed] [Google Scholar]
- 28.Wu G, Wu Y, Xiao L, Li X, Lu C. Zero erucic acid trait of rapeseed (Brassica napus L.) results from a deletion of four base pairs in the fatty acid elongase 1 gene. Theor Appl Genet. 2008: 116(4):491–9. 10.1007/s00122-007-0685-z . [DOI] [PubMed] [Google Scholar]
- 29.Barker GC, Larson TR, Graham IA, Lynn JR, King GJ. Novel insights into seed fatty acid synthesis and modification pathways from genetic diversity and quantitative trait Loci analysis of the Brassica C genome. Plant Physiol. 2007:144(4):1827–42. 10.1104/pp.107.096172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gupta V, Mukhopadhyay A, Arumugam N, Sodhi YS, Pental D, Pradhan AK. Molecular tagging of erucic acid trait in oilseed mustard (Brassica juncea) by QTL mapping and single nucleotide polymorphisms in FAE1 gene. Theor Appl Genet. 2004: 108(4):743–9. 10.1007/s00122-003-1481-z . [DOI] [PubMed] [Google Scholar]
- 31.Wurschum T, Abel S, Zhao YS. Potential of genomic selection in rapeseed (Brassica napus L.) breeding. Plant Breeding. 2014;133(1):45–51. 10.1111/pbr.12137 . [DOI] [Google Scholar]
- 32.Raman H, Raman R, Coombes N, Song J, Prangnell R, Bandaranayake C, et al. Genome-wide association analyses reveal complex genetic architecture underlying natural variation for flowering time in canola. Plant Cell Environ. 2016;39(6):1228–39. 10.1111/pce.12644 in process. [DOI] [PubMed] [Google Scholar]
- 33.Zhong S, Dekkers JC, Fernando RL, Jannink JL. Factors affecting accuracy from genomic selection in populations derived from multiple inbred lines: a Barley case study. Genetics. 2009;182(1):355–64. 10.1534/genetics.108.098277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rutkoski JE, Heffner EL, Sorrells ME. Genomic selection for durable stem rust resistance in wheat. Euphytica. 2011;179(1):161–73. 10.1007/s10681-010-0301-1 . [DOI] [Google Scholar]
- 35.Crossa J, de los Campos G, Perez P, Gianola D, Burgueno J, Araus JL, et al. Prediction of Genetic Values of Quantitative Traits in Plant Breeding Using Pedigree and Molecular Markers. Genetics. 2010;186(2):713–U406. 10.1534/genetics.110.118521 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao YS, Li Z, Liu GZ, Jiang Y, Maurer HP, Wurschum T, et al. Genome-based establishment of a high-yielding heterotic pattern for hybrid wheat breeding. P Natl Acad Sci USA. 2015;112(51):15624–9. 10.1073/pnas.1514547112 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Albrecht T, Wimmer V, Auinger HJ, Erbe M, Knaak C, Ouzunova M, et al. Genome-based prediction of testcross values in maize. Theoretical and Applied Genetics. 2011;123(2):339–50. 10.1007/s00122-011-1587-7 . [DOI] [PubMed] [Google Scholar]
- 38.Bernardo R. Genomewide markers as cofactors for precision mapping of quantitative trait loci. Theoretical and Applied Genetics. 2013;126(4):999–1009. 10.1007/s00122-012-2032-2 . [DOI] [PubMed] [Google Scholar]
- 39.Bernardo R. Genomewide Selection of Parental Inbreds: Classes of Loci and Virtual Biparental Populations. Crop Sci. 2014;54(6):2586–95. 10.2135/cropsci2014.01.0088 . [DOI] [Google Scholar]
- 40.Riedelsheimer C, Czedik-Eysenberg A, Grieder C, Lisec J, Technow F, Sulpice R, et al. Genomic and metabolic prediction of complex heterotic traits in hybrid maize. Nature genetics. 2012;44(2):217–20. 10.1038/ng.1033 . [DOI] [PubMed] [Google Scholar]
- 41.Zhao YS, Zeng J, Fernando R, Reif JC. Genomic Prediction of Hybrid Wheat Performance. Crop Sci. 2013;53(3):802–10. 10.2135/cropsci2012.08.0463 . [DOI] [Google Scholar]
- 42.Zhao YS, Gowda M, Wurschum T, Longin CFH, Korzun V, Kollers S, et al. Dissecting the genetic architecture of frost tolerance in Central European winter wheat. J Exp Bot. 2013;64(14):4453–60. 10.1093/jxb/ert259 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Spindel J, Begum H, Akdemir D, Virk P, Collard B, Redona E, et al. Genomic Selection and Association Mapping in Rice (Oryza sativa): Effect of Trait Genetic Architecture, Training Population Composition, Marker Number and Statistical Model on Accuracy of Rice Genomic Selection in Elite, Tropical Rice Breeding Lines. PLOS Genet. 2015;11(2). ARTN e1004982 10.1371/journal.pgen.1004982 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reif JC, Zhao YS, Wurschum T, Gowda M, Hahn V. Genomic prediction of sunflower hybrid performance. Plant Breeding. 2013;132(1):107–14. 10.1111/pbr.12007 . [DOI] [Google Scholar]
- 45.Hayes BJ, Cogan NOI, Pembleton LW, Goddard ME, Wang JP, Spangenberg GC, et al. Prospects for genomic selection in forage plant species. Plant Breeding. 2013;132(2):133–43. 10.1111/pbr.12037 . [DOI] [Google Scholar]
- 46.Hofheinz N, Borchardt D, Weissleder K, Frisch M. Genome-based prediction of test cross performance in two subsequent breeding cycles. Theoretical and Applied Genetics. 2012;125(8):1639–45. 10.1007/s00122-012-1940-5 . [DOI] [PubMed] [Google Scholar]
- 47.Wurschum T, Reif JC, Kraft T, Janssen G, Zhao Y. Genomic selection in sugar beet breeding populations. BMC genetics. 2013;14:85 10.1186/1471-2156-14-85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bao Y, Vuong T, Meinhardt C, Tiffin P, Denny R, Chen SY, et al. Potential of Association Mapping and Genomic Selection to Explore PI 88788 Derived Soybean Cyst Nematode Resistance. Plant Genome-Us. 2014;7(3). 10.3835/plantgenome2013.11.0039 . [DOI] [Google Scholar]
- 49.Shu YJ, Yu DS, Wang D, Bai X, Zhu YM, Guo CH. Genomic selection of seed weight based on low-density SCAR markers in soybean. Genet Mol Res. 2013;12(3):2178–88. 10.4238/2013.July.3.2 . [DOI] [PubMed] [Google Scholar]
- 50.Li L, Long Y, Zhang LB, Dalton-Morgan J, Batley J, Yu LJ, et al. Genome Wide Analysis of Flowering Time Trait in Multiple Environments via High-Throughput Genotyping Technique in Brassica napus L. PLOS One. 2015;10(3). 10.1371/journal.pone.0119425 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jan HU, Abbadi A, Lucke S, Nichols RA, Snowdon RJ. Genomic Prediction of Testcross Performance in Canola (Brassica napus). PLOS One. 2016;11(1). ARTN e0147769 10.1371/journal.pone.0147769 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang Y, Thomas CL, Xiang JX, Long Y, Wang XH, Zou J, et al. Construction of a high-density SNP-based genetic linkage map in Brassica napus and QTL meta-analysis of root traits under contrasting phosphorus supply in two growth systems. 2016:under review. [DOI] [PMC free article] [PubMed]
- 53.Liu LZ, Qu CM, Wittkop B, Yi B, Xiao Y, He YJ, et al. A High-Density SNP Map for Accurate Mapping of Seed Fibre QTL in Brassica napus L. PLOS One. 2013;8(12). 10.1371/journal.pone.0083052 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Qian LW, Qian W, Snowdon RJ. Sub-genomic selection patterns as a signature of breeding in the allopolyploid Brassica napus genome. Bmc Genomics. 2014;15 10.1186/1471-2164-15-1170 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xiao Y, Chen L, Zou J, Tian E, Xia W, Meng J. Development of a population for substantial new type Brassica napus diversified at both A/C genomes. Theor. Appl. Genet. 2010;121, 1141–1150. 10.1007/s00122-010-1378-6 . [DOI] [PubMed] [Google Scholar]
- 56.Zou J, Zhu J, Huang S, Tian E, Xiao Y, Fu D, et al. Broadening the avenue of intersubgenomic heterosis in oilseed Brassica. Theor. Appl. Genet. 2010;120, 283–290. 10.1007/s00122-009-1201-4 . [DOI] [PubMed] [Google Scholar]
- 57.Butler D, Cullis B, Gilmour A, Gogel B. ASREML-R, Reference Manual Version 3 Queensland Department of Primary Industries and Fisheries: Brisbane. 2009.
- 58.Chalhoub B, Denoeud F, Liu SY, Parkin IAP, Tang HB, Wang XY, et al. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science. 2014;345(6199):950–3. 10.1126/science.1253435 . [DOI] [PubMed] [Google Scholar]
- 59.Warnes GR. “The Genetics Package,” R News; 2003.
- 60.Falconer D, Mackay T. Introduction to Quantitative Genetics 4th edn Longman: Harlow: 1996. [Google Scholar]
- 61.Schwarz G. Estimating the Dimension of a Model. Ann Statist. 1978;6:461–4. [Google Scholar]
- 62.Utz HF, Melchinger AE, Schon CC. Bias and Sampling Error of the Estimated Proportion of Genotypic Variance Explained by Quantitative Trait Loci Determined From Experimental Data in Maize Using Cross Validation and Validation With Independent Samples. Genetics. 2000;154(3):1839–49. [PMC free article] [PubMed] [Google Scholar]
- 63.Whittaker JC, Thompson R, Denham MC. Marker-assisted selection using ridge regression. Genet Res. 2000;75(2):249–52. . [DOI] [PubMed] [Google Scholar]
- 64.Habier D, Fernando RL, Kizilkaya K, Garrick DJ. Extension of the bayesian alphabet for genomic selection. BMC bioinformatics. 2011;12:186 10.1186/1471-2105-12-186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jiang Y, Reif JC. Modeling Epistasis in Genomic Selection. Genetics. 2015;201(2):759–+. 10.1534/genetics.115.177907 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhao J, Huang J, Chen F, Xu F, Ni X, Xu H, et al. Molecular mapping of Arabidopsis thaliana lipid-related orthologous genes in Brassica napus. Theor Appl Genet. 2012;124(2):407–21. 10.1007/s00122-011-1716-3 . [DOI] [PubMed] [Google Scholar]
- 67.Sun M, Hua W, Liu J, Huang S, Wang X, Liu G, et al. Design of new genome- and gene-sourced primers and identification of QTL for seed oil content in a specially high-oil Brassica napus cultivar. PLOS One. 2012;7(10):e47037 10.1371. PMCID: PMC3470593. 10.1371/journal.pone.0047037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Körber N, Bus A, Li J, Parkin IA, Wittkop B, Snowdon RJ, et al. Agronomic and Seed Quality Traits Dissected by Genome-Wide Association Mapping in Brassica napus. Front Plant Sci. 2016;7:386 10.3389. PMCID: PMC4814720. 10.3389/fpls.2016.00386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xu JF, Long Y, Wu JG, Xu HM, Wen J, Meng J, et al. QTL mapping and analysis of the embryo and maternal plant for three limiting amino acids in rapeseed meal. Eur Food Res Technol.2015,240:147–158. 10.1007/s00217-014-2316-7 [DOI] [Google Scholar]
- 70.Huang XQ, Huang T, Hou GZ, Li L, Hou Y, Lu YH. Identification of QTLs for seed quality traits in rapeseed (Brassica napus L.) using recombinant inbred lines (RILs). Euphytica (2016) 210:1–16. 10.1007/s10681-016-1675-5 [DOI] [Google Scholar]
- 71.Wen J, Xu JF, Long Y, Wu JG, Xu HM, Meng JL, et al. QTL mapping based on the embryo and maternal genetic systems for non-essential amino acids in rapeseed (Brassica napus L.) meal. J Sci Food Agric. 2016;96(2):465–73. doi: 10.1002 . [DOI] [PubMed] [Google Scholar]
- 72.Zhao JY, Becker HC, Zhang DQ, Zhang YF, Ecke W. Oil content in a European x Chinese rapeseed population: QTL with additive and epistatic effects and their genotype-environment interactions. Crop Sci. 2005;45(1):51–9. [Google Scholar]
- 73.Smooker AM, Wells R, Morgan C, Beaudoin F, Cho K, Fraser F, et al. The identification and mapping of candidate genes and QTL involved in the fatty acid desaturation pathway in Brassica napus. TAG Theoretical and applied genetics Theoretische und angewandte Genetik. 2011;122(6):1075–90. 10.1007/s00122-010-1512-5 . [DOI] [PubMed] [Google Scholar]
- 74.Wang XD, Wang H, Long Y, Li D, Yin Y, Tian J, et al. Identification of QTLs associated with oil content in a high-oil Brassica napus cultivar and construction of a high-density consensus map for QTLs comparison in B. napus. PLOS One. 2013:8(12):e80569 10.1371/journal.pone.0080569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fu Y, Lu K, Qian LW, Mei JQ, Wei DY, Peng XH, et al. Development of genic cleavage markers in association with seed glucosinolate content in canola. Theoretical and Applied Genetics. 2015;128(6):1029–37. 10.1007/s00122-015-2487-z . [DOI] [PubMed] [Google Scholar]
- 76.Xu R. Measuring explained variation in linear mixed effects models. Statistics in medicine. 2003;22(22):3527–41. 10.1002/sim.1572 . [DOI] [PubMed] [Google Scholar]
- 77.Gianola D, de los Campos G, Hill WG, Manfredi E, Fernando R. Additive genetic variability and the Bayesian alphabet. Genetics. 2009;183(1):347–63. 10.1534/genetics.109.103952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Park YC. Theory for the number of genes affecting quantitative characters: II. Biases from drift, dominance, inequality of gene effects, linkage disequilibrium and epistasis. TAG Theoretical and applied genetics Theoretische und angewandte Genetik. 1977;50(4):163–72. 10.1007/BF00277737 . [DOI] [PubMed] [Google Scholar]
- 79.Albrecht T, Auinger HJ, Wimmer V, Ogutu JO, Knaak C, Ouzunova M, et al. Genome-based prediction of maize hybrid performance across genetic groups, testers, locations, and years. Theoretical and Applied Genetics. 2014;127(6):1375–86. 10.1007/s00122-014-2305-z . [DOI] [PubMed] [Google Scholar]
- 80.Heffner EL, Lorenz AJ, Jannink JL, Sorrells ME. Plant Breeding with Genomic Selection: Gain per Unit Time and Cost. Crop Sci. 2010;50(5):1681–90. 10.2135/cropsci2009.11.0662 . [DOI] [Google Scholar]
- 81.Longin CFH, Mi XF, Wurschum T. Genomic selection in wheat: optimum allocation of test resources and comparison of breeding strategies for line and hybrid breeding. Theoretical and Applied Genetics. 2015;128(7):1297–306. 10.1007/s00122-015-2505-1 . [DOI] [PubMed] [Google Scholar]
- 82.Lorenzana RE, Bernardo R. Accuracy of genotypic value predictions for marker-based selection in biparental plant populations. Theoretical and Applied Genetics. 2009;120(1):151–61. 10.1007/s00122-009-1166-3 . [DOI] [PubMed] [Google Scholar]
- 83.Heffner EL, Jannink JL, Iwata H, Souza E, Sorrells ME. Genomic Selection Accuracy for Grain Quality Traits in Biparental Wheat Populations. Crop Sci. 2011;51(6):2597–606. 10.2135/cropsci2011.05.0253 . [DOI] [Google Scholar]
- 84.Hickey JM, Dreisigacker S, Crossa J, Hearne S, Babu R, Prasanna BM, et al. Evaluation of Genomic Selection Training Population Designs and Genotyping Strategies in Plant Breeding Programs Using Simulation. Crop Sci. 2014;54(4):1476–88. 10.2135/cropsci2013.03.0195 . [DOI] [Google Scholar]
- 85.Zhang X, Perez-Rodriguez P, Semagn K, Beyene Y, Babu R, Lopez-Cruz MA, et al. Genomic prediction in biparental tropical maize populations in water-stressed and well-watered environments using low-density and GBS SNPs. Heredity. 2015;114(3):291–9. 10.1038/hdy.2014.99 . [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The green lines are the -log10 P-values of the linear regression method. The red lines are the -log10 P-values of the stepwise multiple linear regression method. The expected uniform distribution of negative -log10 P-values is indicated by the diagonal line in blue.
(PDF)
Within each physical distance class, marker pairs are clustered into five groups with varying r2 values.
(JPEG)
(DOCX)
(XLSX)
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.