Summary
Despite decades of intense study, the functions of sleep are still shrouded in mystery. The difficulty in understanding these functions can be at least partly attributed to the varied manifestations of sleep in different animals. Daily sleep duration can range from 4–20 hrs among mammals, and sleep can manifest throughout the brain, or it can alternate over time between cerebral hemispheres, depending on the species. Ecological factors are likely to have shaped these and other sleep behaviors during evolution by altering the properties of conserved arousal circuits in the brain. Nonetheless, core functions of sleep are likely to have arisen early and to have persisted to the present day in diverse organisms. This review will discuss the evolutionary forces that may be responsible for phylogenetic differences in sleep and the potential core functions that sleep fulfills.
Introduction
Sleep limits defense against predation, foraging for food and mating. Therefore, there is a strong selective pressure not to sleep. Nevertheless, since sleep is highly conserved across evolution, an even greater selection pressure must exist to maintain this behavior. Such a conclusion is supported by numerous studies demonstrating that inadequate sleep detracts from health and functionality. For example, in mammals chronic sleep deprivation leads to lapses in attention, emotional instability, increased sensitivity to pain, metabolic and cardiovascular disorders, immune dysfunction and, in extreme cases, death [1–9]. What needs does sleep fulfill that might prevent these pathophysiological changes? And why do some animals sleep so much more or less than others? To address these and other questions this review will consider the possible core functions of sleep, the ecological and intrinsic factors that shaped sleep differently among diverse animal species, and the neural circuitry on which these factors may have acted over the course of evolution.
Defining and measuring sleep
As its name suggests, the sleep/wake cycle is characterized by behavioral states that differ in their levels of brain arousal. Wakefulness can be simply described as a high arousal state that evolved to optimize interactions of animals with their environment. However, it is harder to describe the sleeping state since its functions are unknown, and it may be driven by internal physiological needs. Across the animal kingdom sleep satisfies most, though not necessarily all, of the following criteria: (1) decreased brain arousal and its behavioral correlate, decreased responsiveness to an animal’s surroundings, which distinguishes sleep from immobile wakefulness (also known as rest); (2) electrical changes in the brain’s activity patterns relative to the waking state; (3) behavioral quiescence, often accompanied by a preferred location and characteristic posture; (4) rapid reversibility, which distinguishes sleep from hibernation, anesthesia and coma; (5) homeostatic regulation, in which lost episodes of behavioral quiescence and low arousal are followed by compensatory (rebound) episodes [10].
In mammals and birds, polygraphic measurements are the gold standard for assessing sleep behavior. One such measurement involves electroencephalography (EEG), in which electrodes are placed on the scalp to measure underlying synchronous electrical activity [11]. The relative contributions of different frequencies to this activity can be determined by calculating the power spectral density (PSD) of the EEG. For example, during waking and rapid eye movement (REM) sleep, functional connectivity within the brain is high, leading to desynchronized brain activity [12]. Waking and REM sleep thus produce a low amplitude EEG signal that translates into a PSD with most of its power in the high frequency range. In contrast, during non-rapid eye movement (NREM) sleep, effective connectivity within the brain is more restricted, especially within the cortex [12]. Furthermore, recurrent feedback with the thalamus helps entrain intrinsic cortical oscillations, leading to a high amplitude signal in the EEG known as slow waves and a corresponding PSD with most of its power in the same low frequency range as thalamocortical activity, ~.5–4 Hz [13]. The transformation of this signal in the PSD is known as slow wave activity (SWA) or delta power, which increases with the duration of prior wakefulness and decreases with time spent asleep. Thus, delta power reflects the time course of changes in sleep need [14, 15].
Along with the EEG, additional polygraphic measurements are often employed to distinguish between waking and the two major sleep states in mammals and birds. Such measurements involve electromyography (EMG), in which electrodes are placed over a skeletal muscle to measure underlying muscle tone, and electrooculography (EOG), in which electrodes are placed near the eyes to measure underlying eye movement [11]. While both waking and REM sleep generate a desynchronized EEG signal accompanied by EOG activity, descending brainstem systems silence motor neurons selectively during REM sleep, leaving this state identifiable by EMG due to muscle atonia [16]. In contrast, NREM sleep produces a highly synchronized EEG signal, minimal EOG activity, and an intermediate EMG signature resulting from reduced but still extant muscle tone.
Despite their utility, these polygraphic measurements are not absolutely required to define sleep in all animals. Invertebrates such as fruit flies, roundworms, sea slugs and crayfish all lack the brain structures responsible for the EEG signatures of REM and NREM sleep. REM and NREM activity have not been detected electrophysiologically in zebrafish either. All these organisms also possess insufficient peripheral and ocular musculature with which to generate an EMG and EOG. Nevertheless, based on various combinations of the five criteria listed above, these organisms sleep [17–26]. For example, they all exhibit rapidly reversible periods of behavioral quiescence and reduced arousal which, if prevented, are homeostatically upregulated at a later time. All of these organisms also exhibit differences in nervous system activity corresponding to differences in arousal state. These observations illustrate that the five criteria listed above are correlated during sleep, and thus prolonged behavioral quiescence is sufficient to estimate sleep in invertebrates and zebrafish.
However, it is more challenging for behavioral criteria alone to discriminate between differences in intensity, or depth, of sleep. Such differences can be measured acutely based on responsiveness to graded sensory stimuli; more intense stimuli are required to arouse animals from more intense sleep [27, 28]. In contrast, sleep intensity is determined chronically by using an EEG to measure SWA, which increases with depth of NREM sleep [27]. Unfortunately numerous sleep studies, including many covered in this review, address only duration of sleep. This qualification is important to keep in mind since sleep duration and intensity both contribute to fulfilling sleep need and may thus represent properties on which evolutionary forces act.
Hypotheses about ecological and intrinsic determinants of sleep behavior
Insights revealed by unihemispheric slow wave sleep and sleep suppression
A combination of measurements is also useful considering that exceptions to any of the five major sleep criteria can be found in nature. For example, following transitions from waking to sleep, terrestrial mammals exhibit bilateral changes in brain activity that are thought to reduce sensory responsiveness and cause behavioral quiescence (thus satisfying criteria 1–3 above) [29]. But in some sleeping birds and marine mammals these changes in brain activity can take the form of NREM sleep that alternates between the two cerebral hemispheres. The resulting aptly named unihemispheric slow wave sleep (USWS) leaves half the brain aroused, which is sufficient to maintain vigilance against predators, to sustain flight over long distances in certain birds, and to navigate oceanic obstacles and remain afloat in the case of marine mammals [30–32]. There are also exceptions to homeostatic regulation of sleep. For example, white-crowned sparrows can reduce their sleep by over 60% for weeks during the migratory season [33]. Similarly, male polygynous pectoral sandpipers can suppress sleep during mating season [34]. Bottlenose dolphins can maintain continuous vigilance to an auditory task for up to 5 days, and in this species and in killer whales, calves and their mothers exhibit little typical sleep behavior for at least one month after birth [35–37](though see [38, 39] for different interpretations of this phenomenon).
The examples above also suggest that multiple ecological determinants shaped the evolution of sleep behavior. Some likely determinants are listed in Table 1. For example, the survival of migrating songbirds depends on traversing unknown and expansive territory as quickly as possible to avoid unfamiliar dangers. Similarly, survival of newborn cetaceans probably depends on continued vigilance by both mother and calf until the latter is able to care for itself [37]. Thus, sleep may be sacrificed to protect against predation. In the case of polygynous pectoral sandpipers, individual birds that remain awake longer than their cohorts are more successful at mating [34]. Thus, in this case it seems that sleep may be sacrificed to promote reproductive fitness. In these contexts it is notable that protracted sleep suppression often occurs for only part of an animal’s life cycle. Such an extreme, adaptive change may thus offer only a short-term benefit to fitness. Chronic sleep suppression may even invoke stress responses that supersede short-term compensatory mechanisms and thus prevent sleep homeostasis. This process, called allostasis, leads to pathophysiological changes in mammals in which it has been studied, including cardiovascular, immune and endocrine dysfunction, which may explain why naturally occurring chronic sleep suppression is so rarely encountered in the animal kingdom [40–43].
Table 1.
Hypothetical determinants of sleep.
| Factor | Promotes (↑) or suppresses (↓) sleep |
|---|---|
| Ecological | |
| Protection against predation | ↓ |
| Enhanced mating success | ↓ |
| Incompatibility with swimming | ↓ |
| Thermoregulation | ↓ |
| Need to forage for food | ↓ |
| Adaptive inactivity | ↑ |
| Intrinsic | |
| Enhancement of memory consolidation | ↑ |
| Complementation/compensation for NREM | ↑ |
| Activity-dependent rewiring of CNS | ↑ |
| Energy conservation | ↑ |
| Metabolic clearance | ↑ |
| Sensorimotor tuning | ↑ |
| Synaptic homeostasis | ↑ |
Evidence for additional ecological variables as drivers of sleep evolution can be found in comparisons of sleep behaviors among cetaceans (whales, dolphins and porpoises), sirenia (manatees) and pinnipeds (phocids, or earless seals; and otariids, including eared seals, walruses and sea lions). The most striking similarity between these marine mammals is that, except for phocids, they all exhibit USWS and little to no REM sleep in the water. As discussed above, it is likely that USWS in these mammals evolved because of the conflicting aquatic needs for NREM sleep and for remaining vigilant against predation. The same argument can be made for REM sleep, but in addition, REM sleep is incompatible with swimming since REM sleep causes muscle atonia. REM sleep is also accompanied by reduced thermoregulatory control [44]. Thus, selection pressure to reduce REM sleep may have arisen from the need to maintain core body temperature and thus optimal performance in the cold waters in which many marine mammals live. Thermoregulatory need might also contribute to total sleep suppression in cetacean neonates, which have reduced insulating blubber relative to adults [29].
Sleep hypotheses evaluated by correlated evolutionary analysis
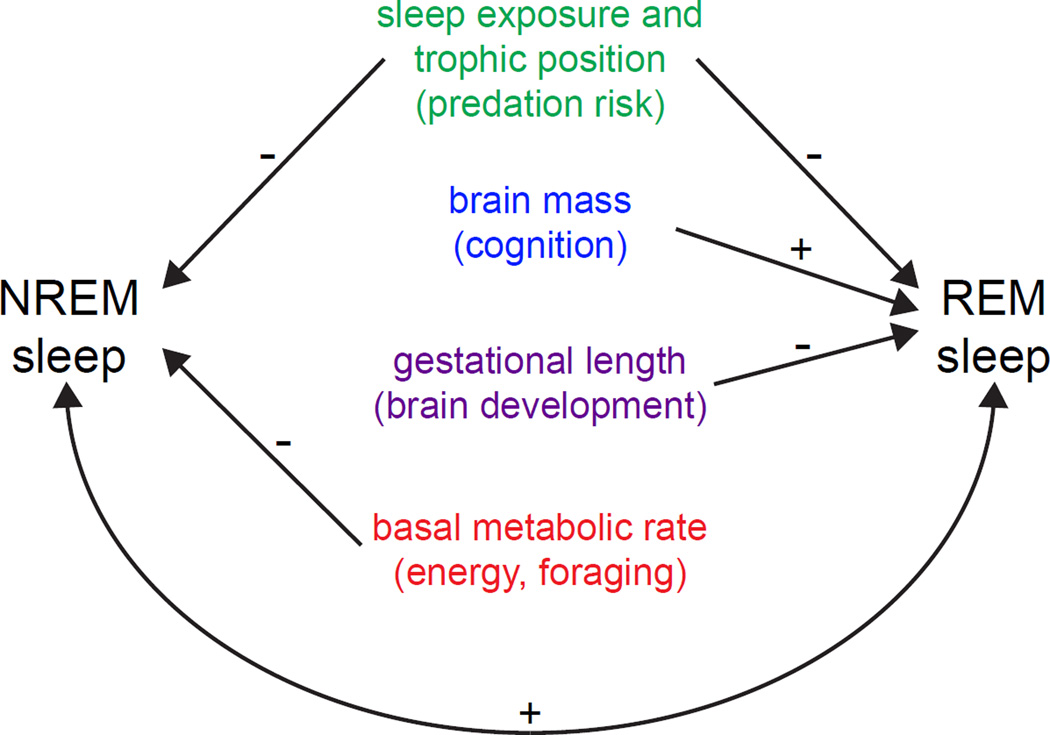
Mammalian sleep duration also varies tremendously among different species, with elephants and armadillos representing opposite extremes at 4 and 20 hrs/day, respectively [45–47]. It is unclear whether this variation reflects evolutionary differences in intrinsic sleep need or, as alluded to above, trade-offs between sleep need and ecological influences. One approach to understanding the evolutionary factors that shape sleep is to correlate sleep measurements with ecological or physiological variables across a wide range of animals. In such studies one must always be aware that unknown factors can be responsible for two otherwise unrelated correlated measurements. However, the statistical power of such studies also has the potential to identify mechanistic relationships that persist across evolution. For example, several recent studies have shown that sleep is negatively correlated with exposure of sleep sites across diverse mammalian taxa (Figure 1) [48–50]. Sleep is also negatively correlated with trophic position, a measure of a mammal’s herbivorous tendencies and position within the food chain [48, 50]. One interpretation of these findings is that they support the previously stated hypothesis that vulnerability to predation suppresses sleep. Because herbivores additionally spend more time than carnivores in search of food, these data also suggest that sleep may be sacrificed to promote foraging [48, 50, 51].
Figure 1. NREM and REM sleep are correlated with quantifiable ecological and physiological variables across evolution.
Positive and negative correlations are marked accordingly. Measured variables are listed centrally in color, with the impacted hypothesis for sleep function listed immediately below in parentheses. Except for brain mass, which was correlated with percent REM sleep, all variables were correlated with NREM or REM sleep durations. The unlabeled bottom loop shows that NREM and REM sleep are positively correlated with each other. Only mammals were used in these studies [48–51]. In cases in which one study was unable to detect a correlation, the positive outcome of another study is still shown based on the notion that correlations are inherently difficult to detect. Not shown: positive correlation between total sleep duration and ratio of cortical density to cortical surface area [75], which has been hypothesized to support a role for sleep in metabolic clearance from the brain [74].
In addition to ecological determinants of sleep evolution that favor waking, intrinsic physiological properties of organisms may hold clues about underlying processes that promote sleep. One such property is brain size, which, when adjusted for body mass (i.e. relative brain size), may reflect differences in cognitive abilities between species [52–54]. Interestingly, correlational analyses across dozens of mammalian species reveal that the percentage of sleep devoted to REM is positively associated with brain mass (Figure 1). Since this effect must come at the expense of NREM sleep, it suggests that REM sleep is selectively important for relatively large-brained animals [48, 49]. This would be consistent with the hypothesis that REM sleep may facilitate the consolidation of memories formed during waking [55–57]. This hypothesis is based in part on the finding that disruption of REM sleep prevents sleep-dependent retention of learning. This interpretation has been criticized for overlooking the effects of nonspecific stress caused by sleep deprivation [58, 59]. However, it has been buttressed by the recent finding that blocking hippocampal oscillatory activity selectively during REM sleep erases subsequent memories of recently learned tasks even in the absence of altered sleep [60]. A more intractable observation undermining the above hypothesis is that most antidepressants suppress REM sleep but not memory [58, 59].
Correlations have also been reported for durations of total REM and NREM sleep, suggesting that the two sleep states may be functionally coupled (Figure 1) [48, 51]. Interestingly, sleep deprivation initially leads to rebound in NREM sleep, and ambient temperatures outside the range of autonomic insensitivity suppress REM sleep more strongly than NREM sleep, thus suggesting that NREM sleep is more important than REM sleep [29]. These observations are consistent with the hypothesis that REM sleep may complement or compensate for NREM sleep, possibly by facilitating periodic upregulation of noradrenergic receptors in preparation for waking [61]. This hypothesis seems difficult to reconcile with other data, however, including the dependence of REM rebound on prior loss of REM sleep rather than extension of NREM sleep [62].
Another explanation for REM sleep comes from the ontogenetic hypothesis, which posits that REM sleep may drive activity-dependent wiring of neural circuits in the developing brain [63]. This hypothesis is based on the observation that juvenile mammals engage in much more REM sleep than their adult counterparts [27]. The ontogenetic hypothesis is supported by studies demonstrating that total REM sleep time is negatively correlated with both gestation period and precocial birth (Figure 1) [48, 49]. One obvious interpretation of this data is that mammals that mature more in the womb are born with a reduced requirement for REM sleep, presumably because the brains of these animals need to undergo less subsequent development. Conversely, higher levels of REM sleep are needed by altricial mammals, which are born with relatively shorter gestational time and thus require relatively more brain development to reach adulthood [48, 49]. These findings are complemented by studies demonstrating that sleep in roundworms and fruit flies is also correlated with development of the nervous system [20, 64, 65]. The debate over the ontogenetic hypothesis continues, however, as some researchers have questioned the reliability of some previous mammalian neonatal sleep studies that did not take into account other defining features of REM sleep such as muscle twitches [66]. The questionable reliability of certain neonatal REM sleep measurements may also explain why other studies have not detected a correlation between relative neonatal brain mass and REM sleep [51].
Correlational analyses have also tackled NREM sleep. Early findings indicated that NREM sleep duration is positively correlated with basal metabolic rate (BMR) in mammals [67], consistent with a reduced metabolic demand in the brain during NREM sleep relative to quiet waking and REM sleep [68–72]. Collectively, both findings support the hypothesis that NREM sleep may facilitate energy conservation [73]. However, when taxa are weighted more appropriately to reflect phylogenetic relatedness, it appears instead that NREM sleep duration is negatively correlated with BMR and uncorrelated with brain mass – findings that do not support the energy hypothesis (Figure 1) [48, 49, 51]. Such a conclusion would be premature though since brain metabolism is clearly reduced during NREM sleep, and this effect might be obscured in whole body BMR measurements. Additional support for a role for NREM sleep in energy conservation is discussed below.
Recent evidence also suggests that the mammalian brain undergoes large increases in interstitial space during sleep in which clearance of extracellular wastes and toxins is enhanced [74]. Interestingly, in mammals the ratio of cortical density to surface area shows a strong positive correlation with total sleep duration, a finding that is consistent with the evolution of high neuronal density to facilitate perfusion by the CSF [75]. These data thus support the hypothesis that sleep may have evolved to facilitate metabolic clearance.
In summary, recent correlational analyses that adjust for evolutionary relatedness among taxa suggest that sleep may be suppressed by various niche-specific ecological factors such as the need to protect against predation and forage for food. The same studies also provide mixed evidence to support roles for REM sleep and NREM sleep in neural plasticity and metabolic functions. It will be especially interesting to see how these findings, which only apply to terrestrial mammals, are affected by the addition of datasets from birds, which also exhibit REM and NREM sleep. Correlations that persist with the expansion of phylogenetic datasets would strengthen arguments for core functions of sleep.
Drivers of sleep: Adaptive inactivity and energy allocation hypotheses
As we have seen, ecological factors are likely to be major determinants of sleep behavior. Generally these seem to favor waking since only in this aroused state is an organism able to exploit its environment to feed, mate and protect itself. Countervailing forces – i.e. those that promote sleep – seem likely to arise from intrinsic physiological needs that have yet to be identified. However, neither type of variable seems sufficient to explain sleep behavior in certain animals. For example, armadillos sleep up to 20 hrs/day, whereas their closest relatives, the sloths, sleep for about half that amount. Yet it is difficult to imagine that armadillos need to sleep 10 hrs more than sloths or 16 hrs more than elephants [45–47, 76].
In some cases these differences may be attributable to species-specific windows of available food and safety. For example, the diet of armadillos largely consists of insects, many of which are most active at dawn and dusk [77]. Since armadillos also occupy cool, deep burrows that offer protection from predators and hot surface temperatures [45], it would thus be potentially dangerous and certainly energetically wasteful to wander around unnecessarily. Related reasoning has also been applied to explain high sleep in bats [78]. Even more extreme situations apply to animals that undergo daily torpor or seasonal hibernation when food sources becomes limiting. In these cases metabolism and arousal are reduced even further than in sleep. Thus, sleep can be thought of as a midpoint in a continuum of active and dormant states. That is, sleep provides the benefit of reduced metabolism but at the cost of reduced arousal [78, 79]. Considering the prevalence of the sleep/wake cycle, it seems likely that optimal performance in most organisms requires a balance between those two variables. The evolution of reduced muscle tone and complete atonia in NREM and REM sleep, respectively, would also be consistent with an adaptive role for inactivity in energy conservation during sleep. The hypothesis that sleep may have evolved as a state of adaptive inactivity does not exclude other proposed functions for sleep [78]. Instead it suggests that those functions evolved, perhaps differently for different species, by co-opting the inactive state that lies at the core of sleep. However, others have argued that this hypothesis is difficult to reconcile with the apparently maladaptive tendency of sleep to eventually force itself on an animal regardless of vulnerability at a given sleeping site [79].
Some researchers have also argued that inactivity isn’t the core function of sleep as much as it is a product of that function. This view draws inspiration from the ontogenetic hypothesis but focuses on the muscle twitching that is a cardinal feature of REM sleep. According to this view REM sleep may have evolved as an internal tuning mechanism for sensorimotor circuits in the absence of conflicting signals from the environment [80]. These circuits are not fully formed in juveniles, so they require more peripheral tuning than they do in adults, thus accounting for decreased REM sleep by adulthood. Muscle twitching during REM sleep in adults has also been interpreted another way. Recent research indicates that twitching does not occur randomly but follows a temporal pattern governed by specific brainstem circuits that drive motor neuron activity. This pattern causes twitching to manifest with increasing frequency as each REM sleep episode progresses. Since REM sleep often immediately precedes wakefulness, some researchers have hypothesized that REM sleep may have evolved to facilitate feedback from motor neurons to promote arousal in preparation for waking [81]. Note that this hypothesis is a more specific version of the previously stated hypothesis that REM sleep may have evolved to complement or compensate for NREM sleep.
The energy allocation (EA) hypothesis takes another unique perspective on sleep’s functions by suggesting that they are energetic in nature but not tied strictly to inactivity the way other hypotheses have proposed. Instead, the EA hypothesis suggests that sleep may allow partitioning of competing biological processes according to behavioral state in order to meet the overall energetic needs of an organism [79]. That is, some processes may be more efficiently performed during waking and others may be performed more efficiently during sleep. Importantly, EA addresses major criticisms of earlier restorative or energy-related hypotheses, namely that it was never clear what sleep actually restores. Certainly proposed brain energy reserves (e.g. glycogen, ATP) and whole-body metabolism don’t change much across the 24 hr cycle [79, 82, 83]. Thus, these findings actually support EA, which argues that metabolic processes are partitioned across the sleep/wake cycle to maximize efficiency of energy consumption. EA also accounts for sleep homeostasis as the result of sleep deprivation causing sleep-related energy-consuming processes to shift to a time when they conflict with wake-related energy-consuming processes, thus increasing sleep drive [79].
EA also explains reduced sleep in migratory songbirds, newborn cetaceans and male polygynous pectoral sandpipers as a temporary adaptive feature that redirects essential biological processes to waking that normally occur during sleep. Animals that cannot accomplish this redirection efficiently suffer negative consequences as a result. Thus, songbirds perform worse on operant tasks when sleep-deprived during the non-migratory compared to the migratory season due to lack of seasonal adaptation to sustained waking [84]. Similarly, EA suggests that more energy-consuming processes have been allocated away from sleep in elephants than in armadillos. Thus, EA posits a core role for sleep in energy conservation, but the particular processes and the timing of their usage may vary across species [79].
Additional hypotheses about sleep’s functions: NREM-mediated memory consolidation and synaptic homeostasis
Current evidence also suggests that NREM sleep may contribute to memory consolidation [57, 85]. This process seems to be initiated by NREM oscillatory activity in the hippocampus called sharp wave ripples. Ripples trigger replay of CA1 place cell firing in a pattern that previously occurred during waking, suggesting that sleep reactivates coherent representations of waking experience [86–88]. NREM sleep may also lower the barrier for replay to transfer temporary hippocampal information to the cortex for long-term storage or to reinforce weak cortical connections established during waking [89–91]. Importantly, NREM sleep facilitates certain forms of memory [92, 93]; its enhancement boosts declarative memory [94]; and specifically blocking NREM sharp wave ripples after learning reduces memory consolidation [95]. Enhancement and deprivation of sleep respectively potentiates and impairs memory in fruit flies as well, suggesting that conserved core functions of sleep may include memory consolidation [28, 96–98].
An influential alternative to the model above is known as the synaptic homeostasis hypothesis (SHY) [99–101]. It posits that desynchronized activity during waking increases functional synaptic connectivity and net potentiation of synapses across the brain. If this process were to continue unabated it would consume energy and saturate synaptic strength. According to SHY, NREM sleep reverses the effects of synaptic potentiation during waking by producing SWA. The low frequency activity pattern of this phenomenon mimics the kind of brain activity that causes synaptic depression. Consequently, the average synaptic strength across the brain remains within a physiologically relevant range for modulation. At the same time this process still preserves the relative increase in strength of synapses that have been reinforced by recent experience [99–101].
Much evidence exists to support this hypothesis including increased effective cortical connectivity and slopes of cortical local field potentials during waking [12, 102]; changes in phosphorylation of glutamate receptors across the sleep/wake cycle that are expected for depression/potentiation of synapses [102]; improvements in performance that are predicted by increased SWA in brain regions involved in a learned task [103]; and the ability of artificially imposed SWA to enhance memories of recently learned events [94]. Support for this hypothesis also comes from invertebrates. Although these organisms do not possess the brain structures to support electrophysiological correlates of NREM sleep, increased synaptic markers of potentiation have been detected in the brains of fruit flies with high sleep pressure, thus suggesting functional conservation of sleep regulatory processes [104–107]. However, SHY may not apply to all sleep-driven forms of synaptic plasticity, especially in light of the requirement for LTP during NREM sleep to facilitate ocular dominance plasticity [108]. SHY also lacks an experimentally verified molecular framework to support general mechanistic predictions of the hypothesis [109].
Molecular indicators of sleep’s functions
Species-specific variations in sleep must be attributable to underlying molecular differences, and many of these were probably shaped by the ecological determinants discussed above. Conversely, core functions of sleep must reflect underlying molecular similarities between species that were retained by evolution because they fulfill shared intrinsic needs. Attempts have been made to identify core functions of sleep by determining the molecular changes that occur with behavioral state. For example, microarray studies have demonstrated that extended waking in animals as distantly related as flies, birds and mammals causes upregulation of brain transcripts involved in energy homeostasis, cellular stress responses and synaptic plasticity in the brain [110]. In contrast, messenger RNAs involved in synaptic depression and macromolecular biosynthesis are downregulated during the same period relative to sleep [110, 111]. A subset of these changes are absent in adrenalectomized compared to control mice, however, suggesting that they are caused by the stress of forced waking rather than waking per se [112].
Collectively, these findings reflect the underlying belief by some researchers that sleep is “of the brain, by the brain, and for the brain” [113]. Indeed, insufficient sleep in mammals most obviously leads to dysfunction in the nervous system, especially in neural circuits that control attention [1], emotional stability [3, 9], sensitivity to pain [6, 7], and learning and memory [57, 114, 115] (which are also affected in flies [28, 96, 97, 116–118]). However, it is clear that transcriptional changes specific to sleep or waking occur in other tissues as well. For example, one study identified sleep-specific changes in three and six percent of all transcripts in the heart and lung, respectively [119]. Another study revealed that three times as many sleep-dependent transcriptional changes occur in the liver relative to the brain, thus suggesting that sleep can selectively affect non-neural tissues [120]. Furthermore, no selective brain pathology has been reported in rats, flies or worms that have been sleep deprived until they die, although those animals showed signs of increased oxidative stress [121, 122]. Lastly, DAF-16/FOXO is required in muscle for normal responses to sleep deprivation in worms [123]. Thus, so far studies have not tied essential functions of sleep definitively to any selective biochemical property of the nervous system, though at the ultrastructural level several studies suggest sleep may modulate spine formation or stability [124, 125].
The genetic tractability of model organisms such as fruit flies, roundworms and zebrafish has led to expanded efforts to mine these organisms for molecules that regulate sleep. These molecules have been covered in other reviews but continue to grow in number [126]. Some probably reflect species-specific effects of the ecological determinants discussed above. However, others may reflect conserved, core functions of sleep. For example, some molecules implicated in sleep regulation may act as gain control mechanisms for brain activity, which could support the hypothesis that sleep’s core function is related to metabolism or energy conservation. These include GABAA receptors and voltage-gated potassium channels, which reduce excitability and promote sleep in flies, zebrafish and mice [117, 127–133]. Neuroligins regulate synaptic transmission, though with opposing effects on sleep in flies and mice [96, 134]. The sleep-promoting SLEEPLESS (SSS) protein suppresses excitability and synaptic transmission in wake-promoting cholinergic neurons of flies, and some of its mammalian homologs can substitute for SSS in vivo [135]. More direct connections between sleep and energy conservation exist with neuromedin U and orexin (also known as hypocretin). In fish and mice these molecules promote waking or locomotion, and in mice they both affect feeding [136–139].
Molecular evidence also supports a function for sleep in reducing cellular stress in the brain. For example, markers of oxidative and ER stress are elevated after sleep loss in mammals and flies [110, 121, 122, 140]. Additionally, cell stress-mitigating chaperones are increased by heat stress, a known inducer of sleep in flies and worms, and these molecules reduce the lethal effects of sleep loss in flies [141–143].
Sleep has also been linked to immune function across evolution. For example, sleep deprivation and infection each increase circulating levels of cytokines such as tumor necrosis factor alpha and interleukin-1 beta, which promote sleep in mammals [144, 145]. Infection also promotes sleep in flies [18, 146].
Other molecules that regulate sleep in diverse organisms are involved in control of the circadian clock. These include cycle and Clock in flies and their homologs BMAL1 and Clock in mice [147–149]. A mutation in Dec2, which encodes a transcriptional regulator of these molecules in mammals, causes reduced sleep in humans and mice, an effect that is mimicked by overexpression of the mutant gene in flies [150]. These findings suggest that sleep may enhance a fundamental function of the circadian clock, such as regulation of metabolism. But the clock regulates the timing of many bodily functions, including behavior. Thus, these findings do not specifically support any one hypothesis about sleep.
Neurotransmitter systems and neural circuitry that regulate the sleep/wake cycle
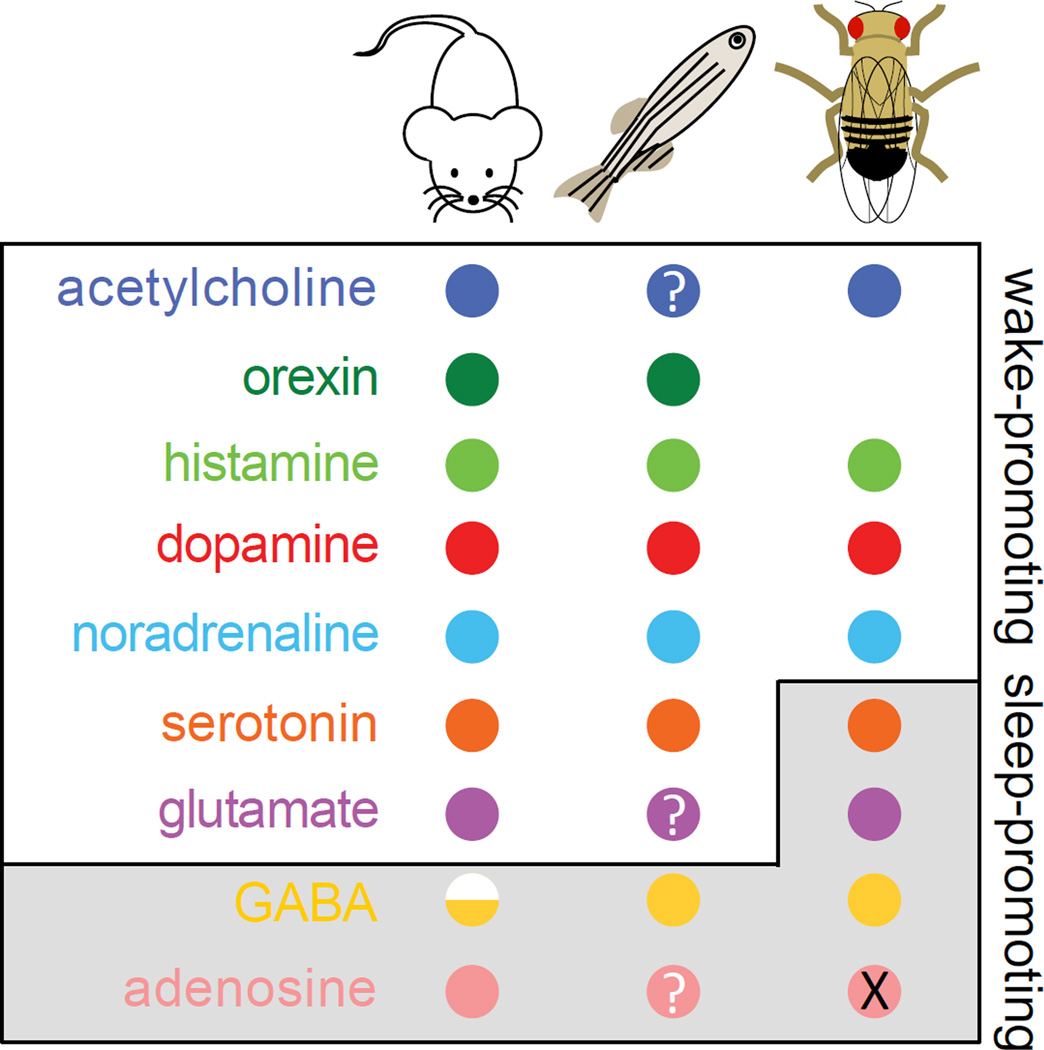
Sleep regulatory mechanisms are probably even more conserved than results from limited molecular and genetic studies have so far revealed. Evidence to support this hypothesis is abundant in studies of sleep pharmacology. For example, antihistamines and GABAergic agonists promote sleep, whereas caffeine promotes waking or locomotion, in flies and zebrafish, just as in mammals [17, 18, 127, 128, 151–153]. Similarly, volatile general anesthetics such as isoflurane and halothane, which are thought to co-opt arousal circuitry in the brain, achieve behavioral endpoints, including unconsciousness, at the same concentrations in fruit flies and mammals [154, 155]. These results suggest that basic arousal circuits with defined molecular identities evolved very early, then elaborated to fulfill various needs of complex organisms without losing core functions, some of which may be mediated by sleep.
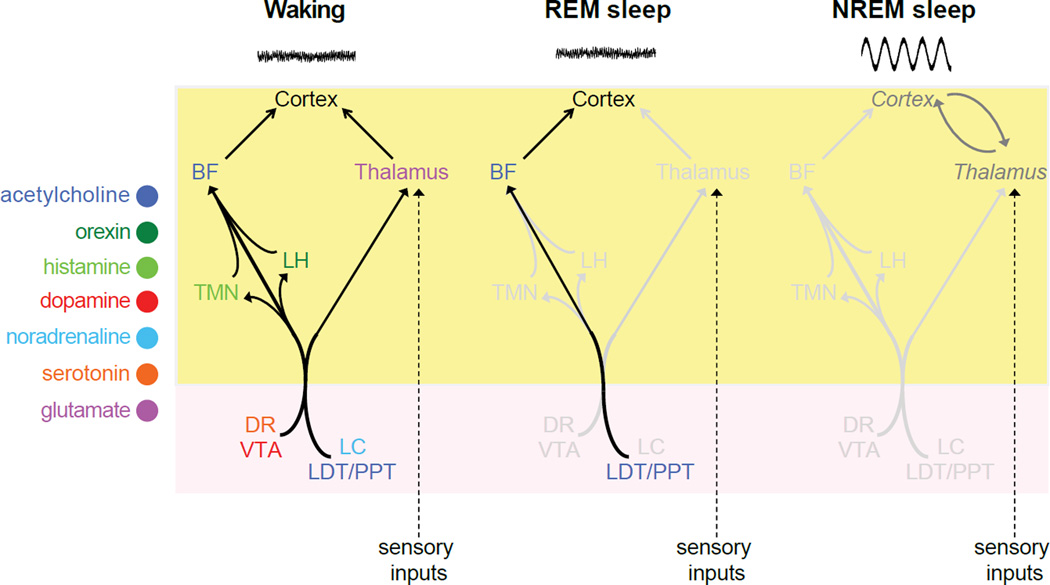
Arousal circuits in model organisms have been identified by various means, including genetic, pharmacological, electrophysiological, lesioning and optogenetic manipulations [156, 157]. Remarkably, the neurotransmitter systems used by these circuits are highly conserved (Figure 2). In mammals these include aminergic and cholinergic projections from the brainstem to dorsal and ventral regions of the forebrain. The dorsal pathway excites the thalamus, which facilitates transmission of sensory information to the cortex. The ventral pathway activates multiple regions of the forebrain that collectively excite the cortex but do not present it with sensory information. Waking ensues when these various systems are active in both pathways [156, 157].
Figure 2. Conserved neurotransmitter systems that control sleep.
Colored circles represent pharmacological, histological or genetic confirmation of the existence of neurotransmitter systems in mice, zebrafish and flies. The invertebrate neurotransmitter equivalent of norepinephrine is octopamine. Question marks label neurotransmitter systems that have not yet been tested for roles in regulating sleep in zebrafish. X indicates that in flies the single known adenosine receptor does not regulate sleep, though it does not exclude the possible involvement of additional adenosine receptors that have yet to be identified.
Although some GABAergic neurons contribute to waking as well, others promote sleep, consistent with the overall effect of anti-insomnia drugs that potentiate GABA-A receptor signaling [132]. For example, sleep-promoting functions have been ascribed to subpopulations of GABAergic neurons in the basal forebrain and the ventrolateral and median preoptic nuclei (VLPO and MnPO) [158–161]. Among these, however, only the VLPO has so far been shown to be required for sleep [162]. The sleep-promoting effect of GABAergic signaling is thought to result from inhibition of neurons in the arousal pathways described above. Thus, during NREM sleep both aminergic and cholinergic activity is reduced. In contrast, during REM sleep cholinergic signaling through the ventral forebrain pathway persists. As a result of this difference, during REM sleep the cortex remains excited, as in waking, but without being able to receive sensory information through the thalamus that allows the brain to make sense of the world (Figure 3) [156, 157].
Figure 3. Neuroanatomical pathways by which neurotransmitter systems regulate arousal in the mammalian brain.
The cell bodies of each neurotransmitter system are located in brain regions whose names are colored and abbreviated as follows: BF (basal forebrain), LH (lateral hypothalamus), TMN (tuberomamillary nucleus), DR (dorsal raphe nucleus), VTA (ventral tegmental area), LC (locus coeruleus), LDT/PPT (laterodorsal tegmental and pedunculopontine nuclei). Not shown: GABAergic inhibition of most of these brain regions by the ventrolateral preoptic nucleus to promote sleep. During waking, the cortex is excited by ventral and dorsal pathways through the basal forebrain and thalamus, respectively. During REM sleep, aminergic signaling is reduced, thus blocking sensory throughput at the level of the thalamus, but the persistence of cholinergic signaling through the ventral pathway continues to excite the cortex. During NREM sleep, aminergic and cholinergic signaling are both reduced, leading to lowered cortical activation, the appearance of SWA, and its entrainment by thalamocortical loops. Yellow and pink areas represent the forebrain and brainstem, respectively.
Histological studies have confirmed that zebrafish aminergic, cholinergic and GABAergic neurons are located in brain regions that resemble those of their mammalian counterparts. Moreover, when these systems have been manipulated pharmacologically or genetically in zebrafish, they have been shown to regulate arousal in much the same way as they do in mammals (Figure 2) [163–167]. Similarly, in flies, activation of aminergic and cholinergic neurons promotes arousal, though some serotonergic neurons may suppress it [28, 168–170]. As in mammals and zebrafish, GABAergic neurons also suppress arousal in flies [117, 127, 128]. Although the hierarchical brainstem and forebrain pathways that control vertebrate arousal are lacking in invertebrates, flies and worms can achieve similar ends with their own unique brain structures. For example, in flies the fan-shaped body of the central complex promotes sleep [98], and the large ventral lateral neurons of the circadian clock promote waking [171]. In contrast, the mushroom bodies, which are important for associative learning and memory, promote both sleep and waking using different populations of neurons [172–174]. Surprisingly, among various sensory systems in flies, only mechanosensitive neurons drive sustained waking [28]. Neurons that respond to mechanical perturbations promote arousal in worms as well. Furthermore, their activity is suppressed during sleep, thus functioning somewhat akin to the thalamic sensory gate of mammals [21, 175]. Central control of sleep in worms is mediated by two classes of peptidergic interneurons. RIA/RIS neurons promote developmental sleep, whereas the single ALA neuron promotes a form of sleep in adults that is induced by cellular stress [65].
The signals that are responsible for switching between sleep and wake states are unknown, but in all animals switching is followed by stabilization of the subsequent behavioral state, thus suggesting that the underlying mechanisms may be conserved. This stability is important since it prevents frequent transitions between the conscious and unconscious states. In mammals this stability has been attributed in part to mutual inhibition by wake-promoting aminergic arousal nuclei and the sleep-promoting VLPO [176]. Neurons in the ventral forebrain pathway enhance the stability of the waking state by exciting aminergic nuclei through release of the neuropeptide orexin. Humans, rodents and dogs with impaired orexin signaling exhibit frequent REM sleep intrusions into the waking state in a condition known as narcolepsy with cataplexy [177].
Orexin signaling has not been described in invertebrates, but these organisms have clearly evolved means for maintaining stability of behavioral states. Flies can remain either in the wake or the sleep state continuously for hours, and this behavior is genetically controlled. For example, disruption of narrow abdomen, which encodes a cation channel, increases sleep and fragments both sleeping and waking periods in flies, similar to symptoms of narcoleptic patients [155]. Furthermore, stabilization of the waking and sleep states seems to be distinct. Loss-of-function mutations in the RNA-editing gene Adar disrupt the waking state without affecting the duration of sleep bouts [107]. Behavioral state stability is also thought to account for hysteretic dissociation of induction and emergence from anesthesia, in which a “barrier” has been proposed to separate behavioral states. This barrier can be raised and lowered by perturbations in known sleep-regulating genes such as Dopamine beta hydroxylase in mice and narrow abdomen and sleepless in flies. The barrier can also be raised by sleep deprivation in flies, suggesting that sleep homeostasis may act in part through feedback mechanisms that stabilize sleeping states [155, 178].
The origins of sleep homeostasis
Since sleep homeostasis compensates for lost sleep by making an animal tired and thus more likely to slumber after extended waking, this process is thought to be intimately tied to sleep need and thus to the functions that sleep fulfills. Our current understanding of sleep homeostasis has its origins in a longstanding model that describes the sleep/wake cycle as the sum of two processes. Process C represents sinusoidal rhythms in arousal arising from the circadian clock, whereas process S represents an unknown source of sleep pressure that rises with waking time and dissipates with sleep [14, 15]. The homeostatic drive to sleep represented by process S can be quantified by depriving animals of sleep and measuring subsequent rebound. As described above, in mammals and birds, sleep homeostasis is often approximated by its electrophysiological surrogate, delta power, which reflects SWA during NREM sleep. Some researchers have argued that SWA is actually an epiphenomenon that reflects an unknown physiological process that drives sleep [179]. Certainly SWA can be induced with muscarinic antagonists in an animal that is awake and responsive [180]. However, under normal conditions it is the best available electrophysiological correlate for the drive to sleep.
Because of its hypothesized linkage to sleep need, which must be related to the functions sleep fulfills, sleep homeostasis has been studied for its underlying mechanisms. Some of these studies have been informed by existing hypotheses about sleep function. For example, one variation of the energy hypothesis predicts that high brain metabolism during waking leads to the ATP breakdown product, adenosine, which is released by neurons as a cotransmitter [181]. Consistent with this hypothesis, extracellular adenosine rises in the basal forebrain during normal waking, continues with sleep deprivation, and declines during sleep [182, 183]. The increase in adenosine in the basal forebrain may inhibit wake-promoting neurons and thus increase drive to sleep [157]. Indeed, experimental elevation of adenosine in the same brain region induces sleep [182], and knockout of the A1 adenosine receptor abrogates the increase in delta power that follows sleep deprivation [184]. Adenosine signaling also explains caffeine’s effects as a stimulant in mammals. Caffeine is a potent antagonist at adenosine A2A receptors, which suggests that endogenous adenosine signaling normally suppresses arousal [185]. An evolutionarily conserved role for adenosine signaling in sleep regulation is unlikely though since knockout of the single known adenosine receptor in flies has no effect on baseline or rebound sleep [186].
Other researchers have looked for markers of sleep homeostasis by examining quantitative trait loci for delta power, which appears to be genetically controlled [187]. Indeed, changes in rebound sleep have been reported following various genetic perturbations in mice and flies. But definitive roles for molecules in sleep homeostasis have been difficult to ascertain since candidates are rare and they sometimes affect baseline sleep. In these cases it is thus conceivable that homeostatic responsiveness is masked by dominant changes in arousal. Recent progress in disentangling baseline sleep from sleep homeostasis has come from flies, but in this case the emphasis has been on the underlying neural circuits. For example, activation of neurons that release acetylcholine, dopamine or octopamine (the fly equivalent of noradrenaline in vertebrates) keeps flies awake when they would normally be asleep, but surprisingly only waking induced by activation of cholinergic neurons causes subsequent rebound sleep [28]. In this study it was suggested that the relevant cholinergic neurons represent rare, peripheral inputs into an unidentified homeostatic sleep circuit. The proximal source of sleep homeostasis has recently been suggested to comprise R2 ring neurons of the ellipsoid body [188]. Activity in these neurons is both necessary and sufficient for generating rebound sleep. Neurons in the sleep-promoting fan-shaped bodies appear to be outputs of this circuit since their activity is increased with sleep deprivation [19], and they are required to translate activity in the ellipsoid body into rebound sleep [188].
It is unclear if such a distinct homeostatic sleep circuit exists in mammals. On the one hand, basal forebrain release of adenosine and the activity of cortical interneurons that express nNOS and neurokinin 1 seem to increase with prior waking time, thus suggesting that sleep need could originate in a limited set of neurons [189]. On the other hand, sleep need seems to manifest at multiple scales in the mammalian brain. For example, synchronous SWA across long distances can coexist with nearby desynchronized activity in the cortex [190]. Electrophysiological correlates of SWA are even apparent following induction of activity of dissociated neurons in culture, suggesting that these effects are intrinsic to cortical neurons [191]. In other words, homeostatic properties may exist within individual neurons and local circuits throughout the cortex, and thalamocortical circuits may bind their collective activities to generate coherent SWA, delta power and sleep drive.
Evolution of NREM and REM sleep
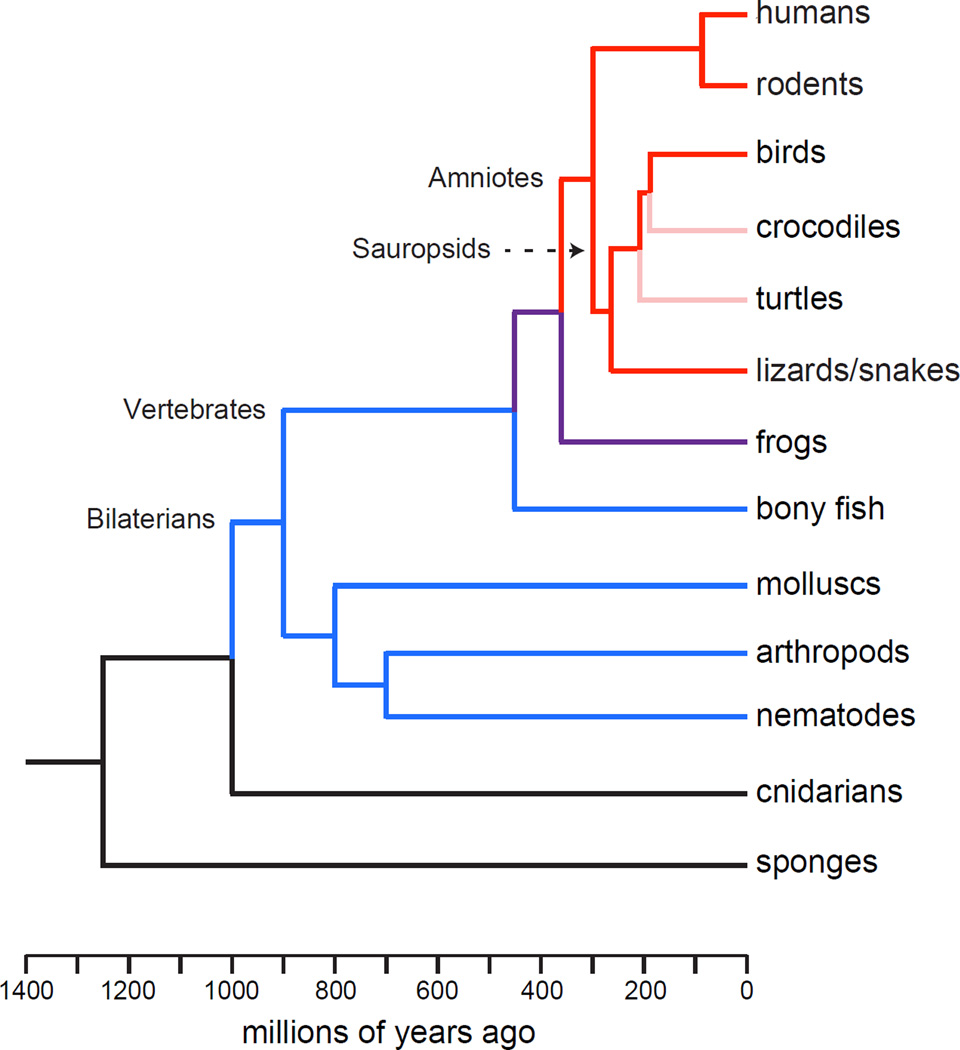
For many years it has been known that birds and mammals possess REM and NREM sleep, whereas only a hypothetical predecessor to NREM sleep was believed to exist in reptiles, a closer avian relative than mammals. These findings led to the hypothesis that distinct sleep states evolved convergently in birds and mammals, perhaps related to ecological factors that also drove their shared endothermy [192]. However, recent EEG recordings show that ectothermic lizards also possess NREM and REM sleep. Furthermore, like their endothermic counterparts, lizards possess an oscillatory circuit that controls the balance between the two sleep states [193]. Thus, even though NREM and REM sleep have been observed in the isolated mammalian forebrain and brainstem, respectively [194], the two sleep states may have existed as coupled mechanisms at least as far back as a common ancestor in the amniote lineage of all three groups of vertebrates (Figure 4) [193]. Such ancient coupling is also supported by the positive correlation between NREM and REM sleep durations across evolution (Figure 1) and the shared developmental origin of populations of NREM-, REM- and wake-promoting neurons in the brainstem [51, 195].
Figure 4. Cladogram of proposed evolutionary relationships across the animal kingdom.
Red lines represent clades in which NREM and REM sleep have been detected by EEG. Pink lines represent taxa in which both sleep states are expected based on relatedness to taxa in red. Purple lines represent taxa with uncertain sleep outcomes. Blue lines represent taxa in which multiple features of sleep have been confirmed but neither NREM nor REM sleep has been detected. Evolutionary relationships were replotted from [193, 209, 210].
If NREM and REM sleep truly evolved together it would suggest that they could serve related needs. However, the phylogenetic origins of these two brains states is unclear. Certainly the earlier developmental onset of REM sleep and its electrophysiological similarity to waking suggest that REM could have evolved first from minor modifications that turned the brain’s attention inward away from sensory stimuli [63]. Superficially, a later origin for NREM sleep also seems plausible considering that SWA involves synchronous activity in the neocortex, a relatively late evolutionary adaptation. The existence of NREM sleep in sauropsids (reptiles and birds) does not truly challenge this view since the neural circuitry and even the molecules that distinguish the neocortex from other mammalian brain structures seem to be intact in analogous regions of sauropsid brains called the dorsal pallium (DP) and dorsal ventricular ridge (DVR) (Figure 5a) [196–198]. SWA is also regulated by the thalamus [199, 200], and sauropsids possess this structure as well. However, in teleost (i.e. bony) fish, which share a common ancestor with amniotes, the constraints on SWA are more severe. For example, zebrafish have a DP and DVR, but they lack the thalamocortical loops that control the frequency and synchrony of SWA [201]. Consistent with this finding, SWA has not been detected in these animals, though neither has REM sleep, thus suggesting that electrophysiological correlates of sleep may not be easily recognizable in non-amniotes [202]. But clues to ancestral sleep-regulating systems may still exist in these animals. For example, in teleost fish the thalamus projects to the pallial equivalent of the mammalian hippocampus [201]. As discussed above, the hippocampus is a source of sharp wave ripples that may facilitate memory consolidation during NREM sleep. Thus, ripples could be rudimentary, spatially restricted forms of NREM oscillatory activity that were superseded by true SWA following the evolution of thalamocortical loops in amniotes (Figure 5a).
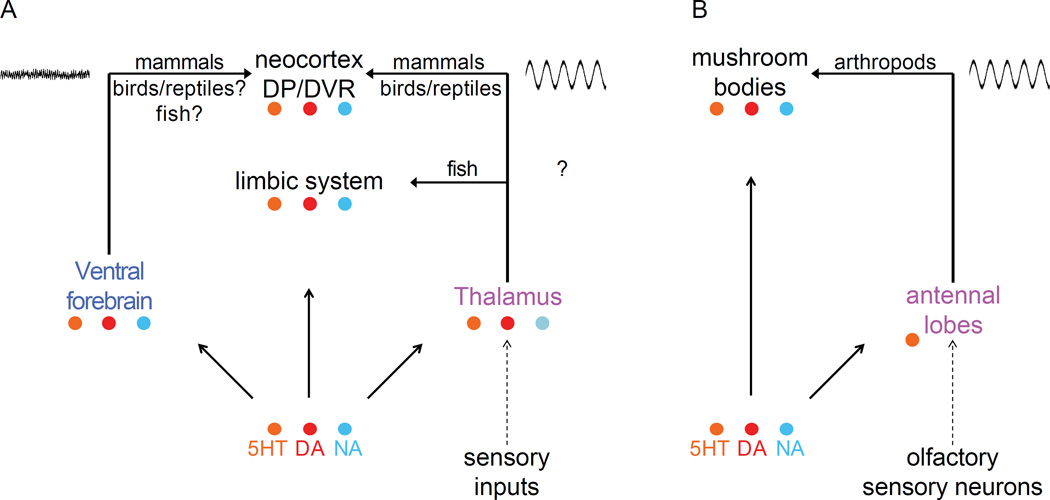
Figure 5. Homology between vertebrates and invertebrates in neuroanatomical control of arousal.
(A) The basic circuit for vertebrate arousal involves aminergic excitation of the ventral forebrain and thalamus, which in turn excite the cortex. This circuit is well-established in mammals and is probably similar in birds and reptiles. Arousal-controlling nuclei are also found in homologous locations in fish, but in these animals the thalamus projects to the limbic system, including the brain region that is thought to function like the mammalian hippocampus. Thus, fish do not possess the thalamocortical loops that allow for entrainment of SWA, and in fact this form of NREM sleep has not been detected in fish. Instead it is possible that hippocampal sharp wave ripples, which have been detected in fish, serve a rudimentary related function. (B) A homologous circuit can be found in insects and other invertebrates that possess mushroom bodies (MBs), which are believed to be derived from an ancestral circuit that gave rise to the vertebrate cerebral cortex. Like the cortex, MBs also undergo oscillations that are thought to be important for memory. Abbreviations: DP (dorsal pallium); DVR (dorsal ventricular ridge); 5HT (serotonin); DA (dopamine); NA (noradrenaline or its invertebrate equivalent, octopamine).
Remarkably, structural and functional homologs of such oscillatory systems may even exist in invertebrates. For example, in insects, olfactory sensory information is processed bilaterally by the antennal lobes, which amplify salient information through oscillations that are transmitted to the mushroom bodies (MBs) (Figure 5b) [203]. Based on conservation of expressed transcripts and shared anatomical developmental patterns, the MBs are thought to have evolved from brain structures that are also ancestral to the mammalian cerebral cortex [204]. Like the cortex, MBs are required for sleep and for certain forms of memory that involve neuronal oscillations [205]. Furthermore, just as in mammals, olfactory-induced reactivation of memories occurs during sleep in insects [206]. Thus, neuronal oscillations in ancestral circuitry may represent ancient forms of signal amplification that serve to enhance information storage, with NREM sleep possibly having evolved to co-opt this mechanism to enhance its function.
How far back in time are basic mechanisms underlying control of sleep likely to have arisen? The conservation of sleep in vertebrates, molluscs, arthropods and nematodes suggests that such mechanisms were already present in the common ancestor of all bilaterally symmetric animals. But there are hints that they may have originated even earlier. For example, one study reported that jellyfish in the wild undergo extended periods of behavioral quiescence at night that can be acutely reversed with certain sensory stimuli [207]. Jellyfish belong to the phylum Cnidaria, which also includes anemones, corals and hydra (Figure 4). Cnidarians are radially symmetric, and they represent the most phylogenetically ancient animals to possess a nervous system [208]. Thus, if sleep can be confirmed in cnidarians then it may have co-evolved with neurons, perhaps to perform functions related to sensory amplification and information storage suggested above. It is doubtful that sleep can be tracked further back in evolution than Cnidaria, though, since the only undisputed more ancient animal phylum, Porifera, consists of organisms such as sponges, which do not have nervous systems and thus cannot exhibit essential features of sleep.
Concluding remarks
It seems likely that ecological factors have shaped sleep differently across phylogeny. Although the underlying molecular changes are unknown, they need not have been numerous providing that the affected molecules altered the activity of arousal-controlling neurons in a way that was adaptive to the species. Core functions of sleep continue to be at least as elusive, with existing hypotheses based largely on correlative data. The variety of these correlations suggests that either sleep serves many functions or that a singular core function is so fundamental that it has profoundly widespread effects on physiology and behavior. Recent progress in identifying components of the sleep homeostat may point to molecular and ultimately physiological needs that sleep fulfills, but these will have to be verified in disparate species to conclude that they are truly universal.
Acknowledgments
I thank Meilin Wu and David Raizen for offering helpful suggestions to improve this manuscript. This work was supported by grants from the NIH (R01 NS072431) and the Department of Defense (W81XWH-13-1-0102).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Basner M, Rao H, Goel N, Dinges DF. Sleep deprivation and neurobehavioral dynamics. Curr Opin Neurobiol. 2013;23:854–863. doi: 10.1016/j.conb.2013.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gottlieb DJ, Punjabi NM, Newman AB, Resnick HE, Redline S, Baldwin CM, Nieto FJ. Association of sleep time with diabetes mellitus and impaired glucose tolerance. Arch Intern Med. 2005;165:863–867. doi: 10.1001/archinte.165.8.863. [DOI] [PubMed] [Google Scholar]
- 3.Gujar N, Yoo SS, Hu P, Walker MP. Sleep deprivation amplifies reactivity of brain reward networks, biasing the appraisal of positive emotional experiences. J Neurosci. 2011;31:4466–4474. doi: 10.1523/JNEUROSCI.3220-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Irwin MR, Opp MR. Sleep-Health: Reciprocal Regulation of Sleep and Innate Immunity. Neuropsychopharmacology. 2016 doi: 10.1038/npp.2016.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rechtschaffen A, Gilliland MA, Bergmann BM, Winter JB. Physiological correlates of prolonged sleep deprivation in rats. Science. 1983;221:182–184. doi: 10.1126/science.6857280. [DOI] [PubMed] [Google Scholar]
- 6.Roehrs T, Hyde M, Blaisdell B, Greenwald M, Roth T. Sleep loss and REM sleep loss are hyperalgesic. Sleep. 2006;29:145–151. doi: 10.1093/sleep/29.2.145. [DOI] [PubMed] [Google Scholar]
- 7.Schuh-Hofer S, Wodarski R, Pfau DB, Caspani O, Magerl W, Kennedy JD, Treede RD. One night of total sleep deprivation promotes a state of generalized hyperalgesia: a surrogate pain model to study the relationship of insomnia and pain. Pain. 2013;154:1613–1621. doi: 10.1016/j.pain.2013.04.046. [DOI] [PubMed] [Google Scholar]
- 8.Tobaldini E, Costantino G, Solbiati M, Cogliati C, Kara T, Nobili L, Montano N. Sleep, sleep deprivation, autonomic nervous system and cardiovascular diseases. Neurosci Biobehav Rev. 2016 doi: 10.1016/j.neubiorev.2016.07.004. [DOI] [PubMed] [Google Scholar]
- 9.Yoo SS, Gujar N, Hu P, Jolesz FA, Walker MP. The human emotional brain without sleep--a prefrontal amygdala disconnect. Curr Biol. 2007;17:R877–R878. doi: 10.1016/j.cub.2007.08.007. [DOI] [PubMed] [Google Scholar]
- 10.Campbell SS, Tobler I. Animal sleep: a review of sleep duration across phylogeny. Neurosci Biobehav Rev. 1984;8:269–300. doi: 10.1016/0149-7634(84)90054-x. [DOI] [PubMed] [Google Scholar]
- 11.Campbell IG. EEG recording and analysis for sleep research. Curr Protoc Neurosci. 2009;Chapter 10(Unit10):12. doi: 10.1002/0471142301.ns1002s49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Massimini M, Ferrarelli F, Huber R, Esser SK, Singh H, Tononi G. Breakdown of cortical effective connectivity during sleep. Science. 2005;309:2228–2232. doi: 10.1126/science.1117256. [DOI] [PubMed] [Google Scholar]
- 13.Amzica F, Steriade M. Electrophysiological correlates of sleep delta waves. Electroencephalogr Clin Neurophysiol. 1998;107:69–83. doi: 10.1016/s0013-4694(98)00051-0. [DOI] [PubMed] [Google Scholar]
- 14.Borbely AA. A two process model of sleep regulation. Hum Neurobiol. 1982;1:195–204. [PubMed] [Google Scholar]
- 15.Daan S, Beersma DG, Borbely AA. Timing of human sleep: recovery process gated by a circadian pacemaker. Am J Physiol. 1984;246:R161–R183. doi: 10.1152/ajpregu.1984.246.2.R161. [DOI] [PubMed] [Google Scholar]
- 16.Vetrivelan R, Fuller PM, Tong Q, Lu J. Medullary circuitry regulating rapid eye movement sleep and motor atonia. J Neurosci. 2009;29:9361–9369. doi: 10.1523/JNEUROSCI.0737-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hendricks JC, Finn SM, Panckeri KA, Chavkin J, Williams JA, Sehgal A, Pack AI. Rest in Drosophila is a sleep-like state. Neuron. 2000;25:129–138. doi: 10.1016/s0896-6273(00)80877-6. [DOI] [PubMed] [Google Scholar]
- 18.Shaw PJ, Cirelli C, Greenspan RJ, Tononi G. Correlates of sleep and waking in Drosophila melanogaster. Science. 2000;287:1834–1837. doi: 10.1126/science.287.5459.1834. [DOI] [PubMed] [Google Scholar]
- 19.Donlea JM, Pimentel D, Miesenbock G. Neuronal machinery of sleep homeostasis in Drosophila. Neuron. 2014;81:860–872. doi: 10.1016/j.neuron.2013.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raizen DM, Zimmerman JE, Maycock MH, Ta UD, You YJ, Sundaram MV, Pack AI. Lethargus is a Caenorhabditis elegans sleep-like state. Nature. 2008;451:569–572. doi: 10.1038/nature06535. [DOI] [PubMed] [Google Scholar]
- 21.Cho JY, Sternberg PW. Multilevel modulation of a sensory motor circuit during C. elegans sleep and arousal. Cell. 2014;156:249–260. doi: 10.1016/j.cell.2013.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Turek M, Besseling J, Spies JP, Konig S, Bringmann H. Sleep-active neuron specification and sleep induction require FLP-11 neuropeptides to systemically induce sleep. Elife. 2016;5 doi: 10.7554/eLife.12499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramon F, Hernandez-Falcon J, Nguyen B, Bullock TH. Slow wave sleep in crayfish. Proc Natl Acad Sci U S A. 2004;101:11857–11861. doi: 10.1073/pnas.0402015101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vorster AP, Krishnan HC, Cirelli C, Lyons LC. Characterization of sleep in Aplysia californica. Sleep. 2014;37:1453–1463. doi: 10.5665/sleep.3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhdanova IV, Wang SY, Leclair OU, Danilova NP. Melatonin promotes sleep-like state in zebrafish. Brain Res. 2001;903:263–268. doi: 10.1016/s0006-8993(01)02444-1. [DOI] [PubMed] [Google Scholar]
- 26.Naumann EA, Kampff AR, Prober DA, Schier AF, Engert F. Monitoring neural activity with bioluminescence during natural behavior. Nat Neurosci. 2010;13:513–520. doi: 10.1038/nn.2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kryger MH, Roth T, Dement WC. Principles and practice of sleep medicine. 5th. St. Louis, Mo.: Elsevier/Saunders; 2011. [Google Scholar]
- 28.Seidner G, Robinson JE, Wu M, Worden K, Masek P, Roberts SW, Keene AC, Joiner WJ. Identification of Neurons with a Privileged Role in Sleep Homeostasis in Drosophila melanogaster. Curr Biol. 2015;25:2928–2938. doi: 10.1016/j.cub.2015.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Siegel JM. Clues to the functions of mammalian sleep. Nature. 2005;437:1264–1271. doi: 10.1038/nature04285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rattenborg NC, Amlaner CJ, Lima SL. Behavioral, neurophysiological and evolutionary perspectives on unihemispheric sleep. Neurosci Biobehav Rev. 2000;24:817–842. doi: 10.1016/s0149-7634(00)00039-7. [DOI] [PubMed] [Google Scholar]
- 31.Lyamin OI, Manger PR, Ridgway SH, Mukhametov LM, Siegel JM. Cetacean sleep: an unusual form of mammalian sleep. Neurosci Biobehav Rev. 2008;32:1451–1484. doi: 10.1016/j.neubiorev.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rattenborg NC, Voirin B, Cruz SM, Tisdale R, Dell'Omo G, Lipp HP, Wikelski M, Vyssotski AL. Evidence that birds sleep in mid-flight. Nat Commun. 2016;7:12468. doi: 10.1038/ncomms12468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rattenborg NC, Mandt BH, Obermeyer WH, Winsauer PJ, Huber R, Wikelski M, Benca RM. Migratory sleeplessness in the white-crowned sparrow (Zonotrichia leucophrys gambelii) PLoS Biol. 2004;2:E212. doi: 10.1371/journal.pbio.0020212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lesku JA, Rattenborg NC, Valcu M, Vyssotski AL, Kuhn S, Kuemmeth F, Heidrich W, Kempenaers B. Adaptive sleep loss in polygynous pectoral sandpipers. Science. 2012;337:1654–1658. doi: 10.1126/science.1220939. [DOI] [PubMed] [Google Scholar]
- 35.Ridgway S, Carder D, Finneran J, Keogh M, Kamolnick T, Todd M, Goldblatt A. Dolphin continuous auditory vigilance for five days. J Exp Biol. 2006;209:3621–3628. doi: 10.1242/jeb.02405. [DOI] [PubMed] [Google Scholar]
- 36.Ridgway S, Keogh M, Carder D, Finneran J, Kamolnick T, Todd M, Goldblatt A. Dolphins maintain cognitive performance during 72 to 120 hours of continuous auditory vigilance. J Exp Biol. 2009;212:1519–1527. doi: 10.1242/jeb.027896. [DOI] [PubMed] [Google Scholar]
- 37.Lyamin O, Pryaslova J, Lance V, Siegel J. Animal behaviour: continuous activity in cetaceans after birth. Nature. 2005;435(1177) doi: 10.1038/4351177a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gnone G, Moriconi T, Gambini G. Sleep behaviour: activity and sleep in dolphins. Nature. 2006;441:E10–E11. doi: 10.1038/nature04899. discussion E11. [DOI] [PubMed] [Google Scholar]
- 39.Sekiguchi Y, Arai K, Kohshima S. Sleep behaviour: sleep in continuously active dolphins. Nature. 2006;441:E9–E10. doi: 10.1038/nature04898. discussion E11. [DOI] [PubMed] [Google Scholar]
- 40.Deurveilher S, Rusak B, Semba K. Time-of-day modulation of homeostatic and allostatic sleep responses to chronic sleep restriction in rats. Am J Physiol Regul Integr Comp Physiol. 2012;302:R1411–R1425. doi: 10.1152/ajpregu.00678.2011. [DOI] [PubMed] [Google Scholar]
- 41.Juster RP, McEwen BS. Sleep and chronic stress: new directions for allostatic load research. Sleep Med. 2015;16:7–8. doi: 10.1016/j.sleep.2014.07.029. [DOI] [PubMed] [Google Scholar]
- 42.Kim Y, Laposky AD, Bergmann BM, Turek FW. Repeated sleep restriction in rats leads to homeostatic and allostatic responses during recovery sleep. Proc Natl Acad Sci U S A. 2007;104:10697–10702. doi: 10.1073/pnas.0610351104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rechtschaffen A, Bergmann BM, Gilliland MA, Bauer K. Effects of method, duration, and sleep stage on rebounds from sleep deprivation in the rat. Sleep. 1999;22:11–31. doi: 10.1093/sleep/22.1.11. [DOI] [PubMed] [Google Scholar]
- 44.Parmeggiani PL. Thermoregulation and sleep. Front Biosci. 2003;8:s557–s567. doi: 10.2741/1054. [DOI] [PubMed] [Google Scholar]
- 45.Prudom AE, Klemm WR. Electrographic correlates of sleep behavior in a primitive mammal, the armadillo Dasypus novemcinctus. Physiol Behav. 1973;10:275–282. doi: 10.1016/0031-9384(73)90310-7. [DOI] [PubMed] [Google Scholar]
- 46.Affanni JM, Cervino CO, Marcos HJ. Absence of penile erections during paradoxical sleep. Peculiar penile events during wakefulness and slow wave sleep in the armadillo. J Sleep Res. 2001;10:219–228. doi: 10.1046/j.1365-2869.2001.00259.x. [DOI] [PubMed] [Google Scholar]
- 47.Tobler I. Behavioral sleep in the Asian elephant in captivity. Sleep. 1992;15:1–12. [PubMed] [Google Scholar]
- 48.Lesku JA, Roth TC, 2nd, Amlaner CJ, Lima SL. A phylogenetic analysis of sleep architecture in mammals: the integration of anatomy, physiology, and ecology. Am Nat. 2006;168:441–453. doi: 10.1086/506973. [DOI] [PubMed] [Google Scholar]
- 49.Lesku JA, Roth TC, Rattenborg NC, Amlaner CJ, Lima SL. Phylogenetics and the correlates of mammalian sleep: a reappraisal. Sleep Med Rev. 2008;12:229–244. doi: 10.1016/j.smrv.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 50.Capellini I, Nunn CL, McNamara P, Preston BT, Barton RA. Energetic constraints, not predation, influence the evolution of sleep patterning in mammals. Funct Ecol. 2008;22:847–853. doi: 10.1111/j.1365-2435.2008.01449.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Capellini I, Barton RA, McNamara P, Preston BT, Nunn CL. Phylogenetic analysis of the ecology and evolution of mammalian sleep. Evolution. 2008;62:1764–1776. doi: 10.1111/j.1558-5646.2008.00392.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Benson-Amram S, Dantzer B, Stricker G, Swanson EM, Holekamp KE. Brain size predicts problem-solving ability in mammalian carnivores. Proc Natl Acad Sci U S A. 2016;113:2532–2537. doi: 10.1073/pnas.1505913113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kotrschal A, Rogell B, Bundsen A, Svensson B, Zajitschek S, Brannstrom I, Immler S, Maklakov AA, Kolm N. Artificial selection on relative brain size in the guppy reveals costs and benefits of evolving a larger brain. Curr Biol. 2013;23:168–171. doi: 10.1016/j.cub.2012.11.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Reader SM, Hager Y, Laland KN. The evolution of primate general and cultural intelligence. Philos Trans R Soc Lond B Biol Sci. 2011;366:1017–1027. doi: 10.1098/rstb.2010.0342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Walker MP, Stickgold R. Sleep-dependent learning and memory consolidation. Neuron. 2004;44:121–133. doi: 10.1016/j.neuron.2004.08.031. [DOI] [PubMed] [Google Scholar]
- 56.Diekelmann S, Wilhelm I, Born J. The whats and whens of sleep-dependent memory consolidation. Sleep Med Rev. 2009;13:309–321. doi: 10.1016/j.smrv.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 57.Rasch B, Born J. About sleep's role in memory. Physiol Rev. 2013;93:681–766. doi: 10.1152/physrev.00032.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Siegel JM. The REM sleep-memory consolidation hypothesis. Science. 2001;294:1058–1063. doi: 10.1126/science.1063049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vertes RP. Memory consolidation in sleep; dream or reality. Neuron. 2004;44:135–148. doi: 10.1016/j.neuron.2004.08.034. [DOI] [PubMed] [Google Scholar]
- 60.Boyce R, Glasgow SD, Williams S, Adamantidis A. Causal evidence for the role of REM sleep theta rhythm in contextual memory consolidation. Science. 2016;352:812–816. doi: 10.1126/science.aad5252. [DOI] [PubMed] [Google Scholar]
- 61.Siegel JM, Rogawski MA. A function for REM sleep: regulation of noradrenergic receptor sensitivity. Brain Res. 1988;472:213–233. doi: 10.1016/0165-0173(88)90007-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rechtschaffen A. Current perspectives on the function of sleep. Perspect Biol Med. 1998;41:359–390. doi: 10.1353/pbm.1998.0051. [DOI] [PubMed] [Google Scholar]
- 63.Roffwarg HP, Muzio JN, Dement WC. Ontogenetic development of the human sleep-dream cycle. Science. 1966;152:604–619. doi: 10.1126/science.152.3722.604. [DOI] [PubMed] [Google Scholar]
- 64.Kayser MS, Yue Z, Sehgal A. A critical period of sleep for development of courtship circuitry and behavior in Drosophila. Science. 2014;344:269–274. doi: 10.1126/science.1250553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Trojanowski NF, Raizen DM. Call it Worm Sleep. Trends Neurosci. 2016;39:54–62. doi: 10.1016/j.tins.2015.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cirelli C, Tononi G. Cortical development, electroencephalogram rhythms, and the sleep/wake cycle. Biol Psychiatry. 2015;77:1071–1078. doi: 10.1016/j.biopsych.2014.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zepelin H, Rechtschaffen A. Mammalian sleep, longevity, and energy metabolism. Brain Behav Evol. 1974;10:425–470. doi: 10.1159/000124330. [DOI] [PubMed] [Google Scholar]
- 68.Buchsbaum MS, Gillin JC, Wu J, Hazlett E, Sicotte N, Dupont RM, Bunney WE., Jr Regional cerebral glucose metabolic rate in human sleep assessed by positron emission tomography. Life Sci. 1989;45:1349–1356. doi: 10.1016/0024-3205(89)90021-0. [DOI] [PubMed] [Google Scholar]
- 69.Kennedy C, Gillin JC, Mendelson W, Suda S, Miyaoka M, Ito M, Nakamura RK, Storch FI, Pettigrew K, Mishkin M, et al. Local cerebral glucose utilization in non-rapid eye movement sleep. Nature. 1982;297:325–327. doi: 10.1038/297325a0. [DOI] [PubMed] [Google Scholar]
- 70.Maquet P, Dive D, Salmon E, Sadzot B, Franco G, Poirrier R, von Frenckell R, Franck G. Cerebral glucose utilization during sleep-wake cycle in man determined by positron emission tomography and [18F]2-fluoro-2-deoxy-D-glucose method. Brain Res. 1990;513:136–143. doi: 10.1016/0006-8993(90)91099-3. [DOI] [PubMed] [Google Scholar]
- 71.Nofzinger EA, Buysse DJ, Miewald JM, Meltzer CC, Price JC, Sembrat RC, Ombao H, Reynolds CF, Monk TH, Hall M, et al. Human regional cerebral glucose metabolism during non-rapid eye movement sleep in relation to waking. Brain. 2002;125:1105–1115. doi: 10.1093/brain/awf103. [DOI] [PubMed] [Google Scholar]
- 72.Wisor JP, Rempe MJ, Schmidt MA, Moore ME, Clegern WC. Sleep slow-wave activity regulates cerebral glycolytic metabolism. Cereb Cortex. 2013;23:1978–1987. doi: 10.1093/cercor/bhs189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Berger RJ, Phillips NH. Energy conservation and sleep. Behav Brain Res. 1995;69:65–73. doi: 10.1016/0166-4328(95)00002-b. [DOI] [PubMed] [Google Scholar]
- 74.Xie L, Kang H, Xu Q, Chen MJ, Liao Y, Thiyagarajan M, O'Donnell J, Christensen DJ, Nicholson C, Iliff JJ, et al. Sleep drives metabolite clearance from the adult brain. Science. 2013;342:373–377. doi: 10.1126/science.1241224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Herculano-Houzel S. Decreasing sleep requirement with increasing numbers of neurons as a driver for bigger brains and bodies in mammalian evolution. Proc Biol Sci. 2015;282:20151853. doi: 10.1098/rspb.2015.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Voirin B, Scriba MF, Martinez-Gonzalez D, Vyssotski AL, Wikelski M, Rattenborg NC. Ecology and neurophysiology of sleep in two wild sloth species. Sleep. 2014;37:753–761. doi: 10.5665/sleep.3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.McDonough CM, Loughry WJ. Influences on activity patterns in a population of nine-banded armadillos. J Mammal. 1997;78:932–941. [Google Scholar]
- 78.Siegel JM. Sleep viewed as a state of adaptive inactivity. Nat Rev Neurosci. 2009;10:747–753. doi: 10.1038/nrn2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schmidt MH. The energy allocation function of sleep: a unifying theory of sleep, torpor, and continuous wakefulness. Neurosci Biobehav Rev. 2014;47:122–153. doi: 10.1016/j.neubiorev.2014.08.001. [DOI] [PubMed] [Google Scholar]
- 80.Blumberg MS, Gall AJ, Todd WD. The development of sleep-wake rhythms and the search for elemental circuits in the infant brain. Behav Neurosci. 2014;128:250–263. doi: 10.1037/a0035891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Brooks PL, Peever J. A Temporally Controlled Inhibitory Drive Coordinates Twitch Movements during REM Sleep. Curr Biol. 2016;26:1177–1182. doi: 10.1016/j.cub.2016.03.013. [DOI] [PubMed] [Google Scholar]
- 82.Scharf MT, Naidoo N, Zimmerman JE, Pack AI. The energy hypothesis of sleep revisited. Prog Neurobiol. 2008;86:264–280. doi: 10.1016/j.pneurobio.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Haddad GG. Does the brain gain back energy during sleep? But what does it mean? Sleep. 2011;34:835–836. doi: 10.5665/SLEEP.1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jones SG, Paletz EM, Obermeyer WH, Hannan CT, Benca RM. Seasonal influences on sleep and executive function in the migratory White-crowned Sparrow (Zonotrichia leucophrys gambelii) BMC Neurosci. 2010;11:87. doi: 10.1186/1471-2202-11-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Abel T, Havekes R, Saletin JM, Walker MP. Sleep, plasticity and memory from molecules to whole-brain networks. Curr Biol. 2013;23:R774–R788. doi: 10.1016/j.cub.2013.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Buzsaki G. Hippocampal sharp wave-ripple: A cognitive biomarker for episodic memory and planning. Hippocampus. 2015;25:1073–1188. doi: 10.1002/hipo.22488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wilson MA, McNaughton BL. Reactivation of hippocampal ensemble memories during sleep. Science. 1994;265:676–679. doi: 10.1126/science.8036517. [DOI] [PubMed] [Google Scholar]
- 88.Davidson TJ, Kloosterman F, Wilson MA. Hippocampal replay of extended experience. Neuron. 2009;63:497–507. doi: 10.1016/j.neuron.2009.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Schwindel CD, McNaughton BL. Hippocampal-cortical interactions and the dynamics of memory trace reactivation. Prog Brain Res. 2011;193:163–177. doi: 10.1016/B978-0-444-53839-0.00011-9. [DOI] [PubMed] [Google Scholar]
- 90.Sadowski JH, Jones MW, Mellor JR. Sharp-Wave Ripples Orchestrate the Induction of Synaptic Plasticity during Reactivation of Place Cell Firing Patterns in the Hippocampus. Cell Rep. 2016;14:1916–1929. doi: 10.1016/j.celrep.2016.01.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Maingret N, Girardeau G, Todorova R, Goutierre M, Zugaro M. Hippocampo-cortical coupling mediates memory consolidation during sleep. Nat Neurosci. 2016;19:959–964. doi: 10.1038/nn.4304. [DOI] [PubMed] [Google Scholar]
- 92.Aeschbach D, Cutler AJ, Ronda JM. A role for non-rapid-eye-movement sleep homeostasis in perceptual learning. J Neurosci. 2008;28:2766–2772. doi: 10.1523/JNEUROSCI.5548-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Landsness EC, Crupi D, Hulse BK, Peterson MJ, Huber R, Ansari H, Coen M, Cirelli C, Benca RM, Ghilardi MF, et al. Sleep-dependent improvement in visuomotor learning: a causal role for slow waves. Sleep. 2009;32:1273–1284. doi: 10.1093/sleep/32.10.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Marshall L, Helgadottir H, Molle M, Born J. Boosting slow oscillations during sleep potentiates memory. Nature. 2006;444:610–613. doi: 10.1038/nature05278. [DOI] [PubMed] [Google Scholar]
- 95.Girardeau G, Benchenane K, Wiener SI, Buzsaki G, Zugaro MB. Selective suppression of hippocampal ripples impairs spatial memory. Nat Neurosci. 2009;12:1222–1223. doi: 10.1038/nn.2384. [DOI] [PubMed] [Google Scholar]
- 96.Li Y, Zhou Z, Zhang X, Tong H, Li P, Zhang ZC, Jia Z, Xie W, Han J. Drosophila neuroligin 4 regulates sleep through modulating GABA transmission. J Neurosci. 2013;33:15545–15554. doi: 10.1523/JNEUROSCI.0819-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Dissel S, Melnattur K, Shaw PJ. Sleep, Performance, and Memory in Flies. Curr Sleep Med Rep. 2015;1:47–54. doi: 10.1007/s40675-014-0006-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Donlea JM, Thimgan MS, Suzuki Y, Gottschalk L, Shaw PJ. Inducing sleep by remote control facilitates memory consolidation in Drosophila. Science. 2011;332:1571–1576. doi: 10.1126/science.1202249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tononi G, Cirelli C. Sleep and synaptic homeostasis: a hypothesis. Brain Res Bull. 2003;62:143–150. doi: 10.1016/j.brainresbull.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 100.Tononi G, Cirelli C. Sleep function and synaptic homeostasis. Sleep Med Rev. 2006;10:49–62. doi: 10.1016/j.smrv.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 101.Tononi G, Cirelli C. Sleep and the price of plasticity: from synaptic and cellular homeostasis to memory consolidation and integration. Neuron. 2014;81:12–34. doi: 10.1016/j.neuron.2013.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Vyazovskiy VV, Cirelli C, Pfister-Genskow M, Faraguna U, Tononi G. Molecular and electrophysiological evidence for net synaptic potentiation in wake and depression in sleep. Nat Neurosci. 2008;11:200–208. doi: 10.1038/nn2035. [DOI] [PubMed] [Google Scholar]
- 103.Huber R, Ghilardi MF, Massimini M, Tononi G. Local sleep and learning. Nature. 2004;430:78–81. doi: 10.1038/nature02663. [DOI] [PubMed] [Google Scholar]
- 104.Donlea JM, Ramanan N, Shaw PJ. Use-dependent plasticity in clock neurons regulates sleep need in Drosophila. Science. 2009;324:105–108. doi: 10.1126/science.1166657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gilestro GF, Tononi G, Cirelli C. Widespread changes in synaptic markers as a function of sleep and wakefulness in Drosophila. Science. 2009;324:109–112. doi: 10.1126/science.1166673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bushey D, Tononi G, Cirelli C. Sleep and synaptic homeostasis: structural evidence in Drosophila. Science. 2011;332:1576–1581. doi: 10.1126/science.1202839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Robinson JE, Paluch J, Dickman DK, Joiner WJ. ADAR-mediated RNA editing suppresses sleep by acting as a brake on glutamatergic synaptic plasticity. Nat Commun. 2016;7:10512. doi: 10.1038/ncomms10512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Aton SJ, Seibt J, Dumoulin M, Jha SK, Steinmetz N, Coleman T, Naidoo N, Frank MG. Mechanisms of sleep-dependent consolidation of cortical plasticity. Neuron. 2009;61:454–466. doi: 10.1016/j.neuron.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Frank MG. Erasing synapses in sleep: is it time to be SHY? Neural Plast. 2012;2012:264–378. doi: 10.1155/2012/264378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Cirelli C. Cellular consequences of sleep deprivation in the brain. Sleep Med Rev. 2006;10:307–321. doi: 10.1016/j.smrv.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 111.Mackiewicz M, Zimmerman JE, Shockley KR, Churchill GA, Pack AI. What are microarrays teaching us about sleep? Trends Mol Med. 2009;15:79–87. doi: 10.1016/j.molmed.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mongrain V, Hernandez SA, Pradervand S, Dorsaz S, Curie T, Hagiwara G, Gip P, Heller HC, Franken P. Separating the contribution of glucocorticoids and wakefulness to the molecular and electrophysiological correlates of sleep homeostasis. Sleep. 2010;33:1147–1157. doi: 10.1093/sleep/33.9.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hobson JA. Sleep is of the brain, by the brain and for the brain. Nature. 2005;437:1254–1256. doi: 10.1038/nature04283. [DOI] [PubMed] [Google Scholar]
- 114.Born J, Wilhelm I. System consolidation of memory during sleep. Psychol Res. 2012;76:192–203. doi: 10.1007/s00426-011-0335-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Diekelmann S, Born J. The memory function of sleep. Nat Rev Neurosci. 2010;11:114–126. doi: 10.1038/nrn2762. [DOI] [PubMed] [Google Scholar]
- 116.Berry JA, Cervantes-Sandoval I, Chakraborty M, Davis RL. Sleep Facilitates Memory by Blocking Dopamine Neuron-Mediated Forgetting. Cell. 2015;161:1656–1667. doi: 10.1016/j.cell.2015.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Haynes PR, Christmann BL, Griffith LC. A single pair of neurons links sleep to memory consolidation in Drosophila melanogaster. Elife. 2015;4 doi: 10.7554/eLife.03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Dissel S, Angadi V, Kirszenblat L, Suzuki Y, Donlea J, Klose M, Koch Z, English D, Winsky-Sommerer R, van Swinderen B, et al. Sleep restores behavioral plasticity to Drosophila mutants. Curr Biol. 2015;25:1270–1281. doi: 10.1016/j.cub.2015.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Anafi RC, Pellegrino R, Shockley KR, Romer M, Tufik S, Pack AI. Sleep is not just for the brain: transcriptional responses to sleep in peripheral tissues. BMC Genomics. 2013;14:362. doi: 10.1186/1471-2164-14-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Maret S, Dorsaz S, Gurcel L, Pradervand S, Petit B, Pfister C, Hagenbuchle O, O'Hara BF, Franken P, Tafti M. Homer1a is a core brain molecular correlate of sleep loss. Proc Natl Acad Sci U S A. 2007;104:20090–20095. doi: 10.1073/pnas.0710131104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Everson CA, Laatsch CD, Hogg N. Antioxidant defense responses to sleep loss and sleep recovery. Am J Physiol Regul Integr Comp Physiol. 2005;288:R374–R383. doi: 10.1152/ajpregu.00565.2004. [DOI] [PubMed] [Google Scholar]
- 122.Everson CA, Thalacker CD, Hogg N. Phagocyte migration and cellular stress induced in liver, lung, and intestine during sleep loss and sleep recovery. Am J Physiol Regul Integr Comp Physiol. 2008;295:R2067–R2074. doi: 10.1152/ajpregu.90623.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Driver RJ, Lamb AL, Wyner AJ, Raizen DM. DAF-16/FOXO regulates homeostasis of essential sleep-like behavior during larval transitions in C. elegans. Curr Biol. 2013;23:501–506. doi: 10.1016/j.cub.2013.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Maret S, Faraguna U, Nelson AB, Cirelli C, Tononi G. Sleep and waking modulate spine turnover in the adolescent mouse cortex. Nat Neurosci. 2011;14:1418–1420. doi: 10.1038/nn.2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Yang G, Lai CS, Cichon J, Ma L, Li W, Gan WB. Sleep promotes branch-specific formation of dendritic spines after learning. Science. 2014;344:1173–1178. doi: 10.1126/science.1249098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Sehgal A, Mignot E. Genetics of sleep and sleep disorders. Cell. 2011;146:194–207. doi: 10.1016/j.cell.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Agosto J, Choi JC, Parisky KM, Stilwell G, Rosbash M, Griffith LC. Modulation of GABAA receptor desensitization uncouples sleep onset and maintenance in Drosophila. Nat Neurosci. 2008;11:354–359. doi: 10.1038/nn2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Parisky KM, Agosto J, Pulver SR, Shang Y, Kuklin E, Hodge JJ, Kang K, Liu X, Garrity PA, Rosbash M, et al. PDF cells are a GABA-responsive wake-promoting component of the Drosophila sleep circuit. Neuron. 2008;60:672–682. doi: 10.1016/j.neuron.2008.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Cirelli C, Bushey D, Hill S, Huber R, Kreber R, Ganetzky B, Tononi G. Reduced sleep in Drosophila Shaker mutants. Nature. 2005;434:1087–1092. doi: 10.1038/nature03486. [DOI] [PubMed] [Google Scholar]
- 130.Yelin-Bekerman L, Elbaz I, Diber A, Dahary D, Gibbs-Bar L, Alon S, Lerer-Goldshtein T, Appelbaum L. Hypocretin neuron-specific transcriptome profiling identifies the sleep modulator Kcnh4a. Elife. 2015;4:e08638. doi: 10.7554/eLife.08638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Douglas CL, Vyazovskiy V, Southard T, Chiu SY, Messing A, Tononi G, Cirelli C. Sleep in Kcna2 knockout mice. BMC Biol. 2007;5:42. doi: 10.1186/1741-7007-5-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Winsky-Sommerer R. Role of GABAA receptors in the physiology and pharmacology of sleep. Eur J Neurosci. 2009;29:1779–1794. doi: 10.1111/j.1460-9568.2009.06716.x. [DOI] [PubMed] [Google Scholar]
- 133.Espinosa F, Marks G, Heintz N, Joho RH. Increased motor drive and sleep loss in mice lacking Kv3-type potassium channels. Genes Brain Behav. 2004;3:90–100. doi: 10.1046/j.1601-183x.2003.00054.x. [DOI] [PubMed] [Google Scholar]
- 134.El Helou J, Belanger-Nelson E, Freyburger M, Dorsaz S, Curie T, La Spada F, Gaudreault PO, Beaumont E, Pouliot P, Lesage F, et al. Neuroligin-1 links neuronal activity to sleep-wake regulation. Proc Natl Acad Sci U S A. 2013;110:9974–9979. doi: 10.1073/pnas.1221381110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wu MN, Joiner WJ, Dean T, Yue Z, Smith CJ, Chen D, Hoshi T, Sehgal A, Koh K. SLEEPLESS, a Ly-6/neurotoxin family member, regulates the levels, localization and activity of Shaker. Nat Neurosci. 2010;13:69–75. doi: 10.1038/nn.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Chiu CN, Rihel J, Lee DA, Singh C, Mosser EA, Chen S, Sapin V, Pham U, Engle J, Niles BJ, et al. A Zebrafish Genetic Screen Identifies Neuromedin U as a Regulator of Sleep/Wake States. Neuron. 2016;89:842–856. doi: 10.1016/j.neuron.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Hanada R, Nakazato M, Murakami N, Sakihara S, Yoshimatsu H, Toshinai K, Hanada T, Suda T, Kangawa K, Matsukura S, et al. A role for neuromedin U in stress response. Biochem Biophys Res Commun. 2001;289:225–228. doi: 10.1006/bbrc.2001.5945. [DOI] [PubMed] [Google Scholar]
- 138.Li J, Hu Z, de Lecea L. The hypocretins/orexins: integrators of multiple physiological functions. Br J Pharmacol. 2014;171:332–350. doi: 10.1111/bph.12415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Semjonous NM, Smith KL, Parkinson JR, Gunner DJ, Liu YL, Murphy KG, Ghatei MA, Bloom SR, Small CJ. Coordinated changes in energy intake and expenditure following hypothalamic administration of neuropeptides involved in energy balance. Int J Obes (Lond) 2009;33:775–785. doi: 10.1038/ijo.2009.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ramanathan L, Gulyani S, Nienhuis R, Siegel JM. Sleep deprivation decreases superoxide dismutase activity in rat hippocampus and brainstem. Neuroreport. 2002;13:1387–1390. doi: 10.1097/00001756-200208070-00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Naidoo N, Giang W, Galante RJ, Pack AI. Sleep deprivation induces the unfolded protein response in mouse cerebral cortex. J Neurochem. 2005;92:1150–1157. doi: 10.1111/j.1471-4159.2004.02952.x. [DOI] [PubMed] [Google Scholar]
- 142.Naidoo N, Casiano V, Cater J, Zimmerman J, Pack AI. A role for the molecular chaperone protein BiP/GRP78 in Drosophila sleep homeostasis. Sleep. 2007;30:557–565. doi: 10.1093/sleep/30.5.557. [DOI] [PubMed] [Google Scholar]
- 143.Shaw PJ, Tononi G, Greenspan RJ, Robinson DF. Stress response genes protect against lethal effects of sleep deprivation in Drosophila. Nature. 2002;417:287–291. doi: 10.1038/417287a. [DOI] [PubMed] [Google Scholar]
- 144.Krueger JM. The role of cytokines in sleep regulation. Curr Pharm Des. 2008;14:3408–3416. doi: 10.2174/138161208786549281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Imeri L, Opp MR. How (and why) the immune system makes us sleep. Nat Rev Neurosci. 2009;10:199–210. doi: 10.1038/nrn2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Kuo TH, Pike DH, Beizaeipour Z, Williams JA. Sleep triggered by an immune response in Drosophila is regulated by the circadian clock and requires the NFkappaB Relish. BMC Neurosci. 2010;11:17. doi: 10.1186/1471-2202-11-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Hendricks JC, Lu S, Kume K, Yin JC, Yang Z, Sehgal A. Gender dimorphism in the role of cycle (BMAL1) in rest, rest regulation, and longevity in Drosophila melanogaster. J Biol Rhythms. 2003;18:12–25. doi: 10.1177/0748730402239673. [DOI] [PubMed] [Google Scholar]
- 148.Laposky A, Easton A, Dugovic C, Walisser J, Bradfield C, Turek F. Deletion of the mammalian circadian clock gene BMAL1/Mop3 alters baseline sleep architecture and the response to sleep deprivation. Sleep. 2005;28:395–409. doi: 10.1093/sleep/28.4.395. [DOI] [PubMed] [Google Scholar]
- 149.Naylor E, Bergmann BM, Krauski K, Zee PC, Takahashi JS, Vitaterna MH, Turek FW. The circadian clock mutation alters sleep homeostasis in the mouse. J Neurosci. 2000;20:8138–8143. doi: 10.1523/JNEUROSCI.20-21-08138.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.He Y, Jones CR, Fujiki N, Xu Y, Guo B, Holder JL, Jr, Rossner MJ, Nishino S, Fu YH. The transcriptional repressor DEC2 regulates sleep length in mammals. Science. 2009;325:866–870. doi: 10.1126/science.1174443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Renier C, Faraco JH, Bourgin P, Motley T, Bonaventure P, Rosa F, Mignot E. Genomic and functional conservation of sedative-hypnotic targets in the zebrafish. Pharmacogenet Genomics. 2007;17:237–253. doi: 10.1097/FPC.0b013e3280119d62. [DOI] [PubMed] [Google Scholar]
- 152.Maximino C, Lima MG, Olivera KR, Picanco-Diniz DL, Herculano AM. Adenosine A1, but not A2, receptor blockade increases anxiety and arousal in Zebrafish. Basic Clin Pharmacol Toxicol. 2011;109:203–207. doi: 10.1111/j.1742-7843.2011.00710.x. [DOI] [PubMed] [Google Scholar]
- 153.Nishimura Y, Okabe S, Sasagawa S, Murakami S, Ashikawa Y, Yuge M, Kawaguchi K, Kawase R, Tanaka T. Pharmacological profiling of zebrafish behavior using chemical and genetic classification of sleep-wake modifiers. Front Pharmacol. 2015;6:257. doi: 10.3389/fphar.2015.00257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Allada R, Nash HA. Drosophila melanogaster as a model for study of general anesthesia: the quantitative response to clinical anesthetics and alkanes. Anesth Analg. 1993;77:19–26. doi: 10.1213/00000539-199307000-00005. [DOI] [PubMed] [Google Scholar]
- 155.Joiner WJ, Friedman EB, Hung HT, Koh K, Sowcik M, Sehgal A, Kelz MB. Genetic and anatomical basis of the barrier separating wakefulness and anesthetic-induced unresponsiveness. PLoS Genet. 2013;9:e1003605. doi: 10.1371/journal.pgen.1003605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Pace-Schott EF, Hobson JA. The neurobiology of sleep: genetics, cellular physiology and subcortical networks. Nat Rev Neurosci. 2002;3:591–605. doi: 10.1038/nrn895. [DOI] [PubMed] [Google Scholar]
- 157.Brown RE, Basheer R, McKenna JT, Strecker RE, McCarley RW. Control of sleep and wakefulness. Physiol Rev. 2012;92:1087–1187. doi: 10.1152/physrev.00032.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Alam MA, Kumar S, McGinty D, Alam MN, Szymusiak R. Neuronal activity in the preoptic hypothalamus during sleep deprivation and recovery sleep. J Neurophysiol. 2014;111:287–299. doi: 10.1152/jn.00504.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Jones BE, Hassani OK. The role of Hcrt/Orx and MCH neurons in sleep-wake state regulation. Sleep. 2013;36:1769–1772. doi: 10.5665/sleep.3188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Luppi PH, Peyron C, Fort P. Not a single but multiple populations of GABAergic neurons control sleep. Sleep Med Rev. 2016 doi: 10.1016/j.smrv.2016.03.002. [DOI] [PubMed] [Google Scholar]
- 161.Xu M, Chung S, Zhang S, Zhong P, Ma C, Chang WC, Weissbourd B, Sakai N, Luo L, Nishino S, et al. Basal forebrain circuit for sleep-wake control. Nat Neurosci. 2015;18:1641–1647. doi: 10.1038/nn.4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Lu J, Greco MA, Shiromani P, Saper CB. Effect of lesions of the ventrolateral preoptic nucleus on NREM and REM sleep. J Neurosci. 2000;20:3830–3842. doi: 10.1523/JNEUROSCI.20-10-03830.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Chiu CN, Prober DA. Regulation of zebrafish sleep and arousal states: current and prospective approaches. Front Neural Circuits. 2013;7:58. doi: 10.3389/fncir.2013.00058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Singh A, Subhashini N, Sharma S, Mallick BN. Involvement of the alpha1-adrenoceptor in sleep-waking and sleep loss-induced anxiety behavior in zebrafish. Neuroscience. 2013;245:136–147. doi: 10.1016/j.neuroscience.2013.04.026. [DOI] [PubMed] [Google Scholar]
- 165.Singh C, Oikonomou G, Prober DA. Norepinephrine is required to promote wakefulness and for hypocretin-induced arousal in zebrafish. Elife. 2015;4:e07000. doi: 10.7554/eLife.07000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Parker MO, Brock AJ, Walton RT, Brennan CH. The role of zebrafish (Danio rerio) in dissecting the genetics and neural circuits of executive function. Front Neural Circuits. 2013;7:63. doi: 10.3389/fncir.2013.00063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Sundvik M, Kudo H, Toivonen P, Rozov S, Chen YC, Panula P. The histaminergic system regulates wakefulness and orexin/hypocretin neuron development via histamine receptor H1 in zebrafish. FASEB J. 2011;25:4338–4347. doi: 10.1096/fj.11-188268. [DOI] [PubMed] [Google Scholar]
- 168.Liu Q, Liu S, Kodama L, Driscoll MR, Wu MN. Two dopaminergic neurons signal to the dorsal fan-shaped body to promote wakefulness in Drosophila. Curr Biol. 2012;22:2114–2123. doi: 10.1016/j.cub.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Yuan Q, Joiner WJ, Sehgal A. A sleep-promoting role for the Drosophila serotonin receptor 1A. Curr Biol. 2006;16:1051–1062. doi: 10.1016/j.cub.2006.04.032. [DOI] [PubMed] [Google Scholar]
- 170.Pooryasin A, Fiala A. Identified Serotonin-Releasing Neurons Induce Behavioral Quiescence and Suppress Mating in Drosophila. J Neurosci. 2015;35:12792–12812. doi: 10.1523/JNEUROSCI.1638-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Sheeba V, Fogle KJ, Kaneko M, Rashid S, Chou YT, Sharma VK, Holmes TC. Large ventral lateral neurons modulate arousal and sleep in Drosophila. Curr Biol. 2008;18:1537–1545. doi: 10.1016/j.cub.2008.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Joiner WJ, Crocker A, White BH, Sehgal A. Sleep in Drosophila is regulated by adult mushroom bodies. Nature. 2006;441:757–760. doi: 10.1038/nature04811. [DOI] [PubMed] [Google Scholar]
- 173.Pitman JL, McGill JJ, Keegan KP, Allada R. A dynamic role for the mushroom bodies in promoting sleep in Drosophila. Nature. 2006;441:753–756. doi: 10.1038/nature04739. [DOI] [PubMed] [Google Scholar]
- 174.Sitaraman D, Aso Y, Jin X, Chen N, Felix M, Rubin GM, Nitabach MN. Propagation of Homeostatic Sleep Signals by Segregated Synaptic Microcircuits of the Drosophila Mushroom Body. Curr Biol. 2015;25:2915–2927. doi: 10.1016/j.cub.2015.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Schwarz J, Lewandrowski I, Bringmann H. Reduced activity of a sensory neuron during a sleep-like state in Caenorhabditis elegans. Curr Biol. 2011;21:R983–R984. doi: 10.1016/j.cub.2011.10.046. [DOI] [PubMed] [Google Scholar]
- 176.Saper CB, Scammell TE, Lu J. Hypothalamic regulation of sleep and circadian rhythms. Nature. 2005;437:1257–1263. doi: 10.1038/nature04284. [DOI] [PubMed] [Google Scholar]
- 177.Dauvilliers Y, Arnulf I, Mignot E. Narcolepsy with cataplexy. Lancet. 2007;369:499–511. doi: 10.1016/S0140-6736(07)60237-2. [DOI] [PubMed] [Google Scholar]
- 178.Friedman EB, Sun Y, Moore JT, Hung HT, Meng QC, Perera P, Joiner WJ, Thomas SA, Eckenhoff RG, Sehgal A, et al. A conserved behavioral state barrier impedes transitions between anesthetic-induced unconsciousness and wakefulness: evidence for neural inertia. PLoS One. 2010;5:e11903. doi: 10.1371/journal.pone.0011903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Davis CJ, Clinton JM, Jewett KA, Zielinski MR, Krueger JM. Delta wave power: an independent sleep phenotype or epiphenomenon? J Clin Sleep Med. 2011;7:S16–S18. doi: 10.5664/JCSM.1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Qiu MH, Chen MC, Lu J. Cortical neuronal activity does not regulate sleep homeostasis. Neuroscience. 2015;297:211–218. doi: 10.1016/j.neuroscience.2015.03.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Benington JH, Heller HC. Restoration of brain energy metabolism as the function of sleep. Prog Neurobiol. 1995;45:347–360. doi: 10.1016/0301-0082(94)00057-o. [DOI] [PubMed] [Google Scholar]
- 182.Porkka-Heiskanen T, Strecker RE, Thakkar M, Bjorkum AA, Greene RW, McCarley RW. Adenosine: a mediator of the sleep-inducing effects of prolonged wakefulness. Science. 1997;276:1265–1268. doi: 10.1126/science.276.5316.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Bjorness TE, Greene RW. Adenosine and sleep. Curr Neuropharmacol. 2009;7:238–245. doi: 10.2174/157015909789152182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Bjorness TE, Kelly CL, Gao T, Poffenberger V, Greene RW. Control and function of the homeostatic sleep response by adenosine A1 receptors. J Neurosci. 2009;29:1267–1276. doi: 10.1523/JNEUROSCI.2942-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Huang ZL, Qu WM, Eguchi N, Chen JF, Schwarzschild MA, Fredholm BB, Urade Y, Hayaishi O. Adenosine A2A, but not A1, receptors mediate the arousal effect of caffeine. Nat Neurosci. 2005;8:858–859. doi: 10.1038/nn1491. [DOI] [PubMed] [Google Scholar]
- 186.Wu MN, Ho K, Crocker A, Yue Z, Koh K, Sehgal A. The effects of caffeine on sleep in Drosophila require PKA activity, but not the adenosine receptor. J Neurosci. 2009;29:11029–11037. doi: 10.1523/JNEUROSCI.1653-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Franken P, Chollet D, Tafti M. The homeostatic regulation of sleep need is under genetic control. J Neurosci. 2001;21:2610–2621. doi: 10.1523/JNEUROSCI.21-08-02610.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Liu S, Liu Q, Tabuchi M, Wu MN. Sleep Drive Is Encoded by Neural Plastic Changes in a Dedicated Circuit. Cell. 2016;165:1347–1360. doi: 10.1016/j.cell.2016.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Morairty SR, Dittrich L, Pasumarthi RK, Valladao D, Heiss JE, Gerashchenko D, Kilduff TS. A role for cortical nNOS/NK1 neurons in coupling homeostatic sleep drive to EEG slow wave activity. Proc Natl Acad Sci U S A. 2013;110:20272–20277. doi: 10.1073/pnas.1314762110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Vyazovskiy VV, Olcese U, Hanlon EC, Nir Y, Cirelli C, Tononi G. Local sleep in awake rats. Nature. 2011;472:443–447. doi: 10.1038/nature10009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Hinard V, Mikhail C, Pradervand S, Curie T, Houtkooper RH, Auwerx J, Franken P, Tafti M. Key electrophysiological, molecular, and metabolic signatures of sleep and wakefulness revealed in primary cortical cultures. J Neurosci. 2012;32:12506–12517. doi: 10.1523/JNEUROSCI.2306-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Rial RV, Akaarir M, Gamundi A, Nicolau C, Garau C, Aparicio S, Tejada S, Gene L, Gonzalez J, De Vera LM, et al. Evolution of wakefulness, sleep and hibernation: from reptiles to mammals. Neurosci Biobehav Rev. 2010;34:1144–1160. doi: 10.1016/j.neubiorev.2010.01.008. [DOI] [PubMed] [Google Scholar]
- 193.Shein-Idelson M, Ondracek JM, Liaw HP, Reiter S, Laurent G. Slow waves, sharp waves, ripples, and REM in sleeping dragons. Science. 2016;352:590–595. doi: 10.1126/science.aaf3621. [DOI] [PubMed] [Google Scholar]
- 194.Villablanca JR. Counterpointing the functional role of the forebrain and of the brainstem in the control of the sleep-waking system. J Sleep Res. 2004;13:179–208. doi: 10.1111/j.1365-2869.2004.00412.x. [DOI] [PubMed] [Google Scholar]
- 195.Hayashi Y, Kashiwagi M, Yasuda K, Ando R, Kanuka M, Sakai K, Itohara S. Cells of a common developmental origin regulate REM/non-REM sleep and wakefulness in mice. Science. 2015;350:957–961. doi: 10.1126/science.aad1023. [DOI] [PubMed] [Google Scholar]
- 196.Dugas-Ford J, Rowell JJ, Ragsdale CW. Cell-type homologies and the origins of the neocortex. Proc Natl Acad Sci U S A. 2012;109:16974–16979. doi: 10.1073/pnas.1204773109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Karten HJ. Neocortical evolution: neuronal circuits arise independently of lamination. Curr Biol. 2013;23:R12–R15. doi: 10.1016/j.cub.2012.11.013. [DOI] [PubMed] [Google Scholar]
- 198.Karten HJ. Vertebrate brains and evolutionary connectomics: on the origins of the mammalian 'neocortex'. Philos Trans R Soc Lond B Biol Sci. 2015;370 doi: 10.1098/rstb.2015.0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.David F, Schmiedt JT, Taylor HL, Orban G, Di Giovanni G, Uebele VN, Renger JJ, Lambert RC, Leresche N, Crunelli V. Essential thalamic contribution to slow waves of natural sleep. J Neurosci. 2013;33:19599–19610. doi: 10.1523/JNEUROSCI.3169-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Lemieux M, Chen JY, Lonjers P, Bazhenov M, Timofeev I. The impact of cortical deafferentation on the neocortical slow oscillation. J Neurosci. 2014;34:5689–5703. doi: 10.1523/JNEUROSCI.1156-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Mueller T. What is the Thalamus in Zebrafish? Front Neurosci. 2012;6:64. doi: 10.3389/fnins.2012.00064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Arnason BB, Thornorsteinsson H, Karlsson KAE. Absence of rapid eye movements during sleep in adult zebrafish. Behav Brain Res. 2015;291:189–194. doi: 10.1016/j.bbr.2015.05.017. [DOI] [PubMed] [Google Scholar]
- 203.Stopfer M. Central processing in the mushroom bodies. Curr Opin Insect Sci. 2014;6:99–103. doi: 10.1016/j.cois.2014.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Tomer R, Denes AS, Tessmar-Raible K, Arendt D. Profiling by image registration reveals common origin of annelid mushroom bodies and vertebrate pallium. Cell. 2010;142:800–809. doi: 10.1016/j.cell.2010.07.043. [DOI] [PubMed] [Google Scholar]
- 205.Stopfer M, Bhagavan S, Smith BH, Laurent G. Impaired odour discrimination on desynchronization of odour-encoding neural assemblies. Nature. 1997;390:70–74. doi: 10.1038/36335. [DOI] [PubMed] [Google Scholar]
- 206.Zwaka H, Bartels R, Gora J, Franck V, Culo A, Gotsch M, Menzel R. Context odor presentation during sleep enhances memory in honeybees. Curr Biol. 2015;25:2869–2874. doi: 10.1016/j.cub.2015.09.069. [DOI] [PubMed] [Google Scholar]
- 207.Seymour JE, Carrette TJ, Sutherland PA. Do box jellyfish sleep at night? Med J Aust. 2004;181:707. doi: 10.5694/j.1326-5377.2004.tb06529.x. [DOI] [PubMed] [Google Scholar]
- 208.Watanabe H, Fujisawa T, Holstein TW. Cnidarians and the evolutionary origin of the nervous system. Dev Growth Differ. 2009;51:167–183. doi: 10.1111/j.1440-169X.2009.01103.x. [DOI] [PubMed] [Google Scholar]
- 209.Hiragaki S, Suzuki T, Mohamed AA, Takeda M. Structures and functions of insect arylalkylamine N-acetyltransferase (iaaNAT); a key enzyme for physiological and behavioral switch in arthropods. Front Physiol. 2015;6:113. doi: 10.3389/fphys.2015.00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Ureta-Vidal A, Ettwiller L, Birney E. Comparative genomics: genome-wide analysis in metazoan eukaryotes. Nat Rev Genet. 2003;4:251–262. doi: 10.1038/nrg1043. [DOI] [PubMed] [Google Scholar]