Abstract
Desminopathy is a subtype of myofibrillar myopathy caused by desmin mutations and characterized by protein aggregates accumulating in muscle fibers. The aim of this study was to assess the protein composition of these aggregates. Aggregates and intact myofiber sections were obtained from skeletal muscle biopsies of five desminopathy patients by laser microdissection and analyzed by a label-free spectral count-based proteomic approach. We identified 397 proteins with 22 showing significantly higher spectral indices in aggregates (ratio >1.8, p < 0.05). Fifteen of these proteins not previously reported as specific aggregate components provide new insights regarding pathomechanisms of desminopathy. Results of proteomic analysis were supported by immunolocalization studies and parallel reaction monitoring. Three mutant desmin variants were detected directly on the protein level as components of the aggregates, suggesting their direct involvement in aggregate-formation and demonstrating for the first time that proteomic analysis can be used for direct identification of a disease-causing mutation in myofibrillar myopathy. Comparison of the proteomic results in desminopathy with our previous analysis of aggregate composition in filaminopathy, another myofibrillar myopathy subtype, allows to determine subtype-specific proteomic profile that facilitates identification of the specific disorder.
Keywords: Myofibrillar myopathy, Desminopathy, Filaminopathy, Laser microdissection, Mass spectrometry, Parallel reaction monitoring
1. Introduction
Myofibrillar myopathies (MFM) are genetically and clinically heterogeneous muscle disorders characterized by focal myofibrillar destruction predominantly at the Z-discs and accumulation of massive aggregates of degradation products within muscle fibers [1,2]. Desminopathy is an MFM subtype caused by mutations in DES, the gene encoding desmin [3]. Desmin is a 53-kDa intermediate filament (IF) type III protein most abundant in skeletal, cardiac and smooth muscle cells [4]. It forms a cytoskeletal network that maintains the structural and functional integrity of the muscle [1,5]. Desmin cytoskeleton connects myofibrils to each other, the nuclear lamina and the sarcolemma [5].
More than 60 different disease-causing mutations in DES have been reported since the first description of desminopathy by Goldfarb et al. in 1998 [3,4]. The pattern of inheritance is autosomal dominant in most cases but autosomal recessive pattern of inheritance and sporadic forms have also been reported (see [6,4] for review). The age of onset is variable but in the majority of patients first symptoms occur between the 2nd and 4th decades of life [6–8]. Progressive muscle weakness involves distal and proximal limb muscles, truncal, neck, facial, bulbar and in some cases respiratory muscles [6,7]. Cardiac disease manifestations observed in about three-quarters of patients comprise cardiomyopathy, cardiac conduction defects and arrhythmias and are the major causes of premature death [4,6,7,9].
In vitro assembly analyses and transfection studies performed in muscle and non-muscle cell lines revealed that mutant desmin is unable to form stable IF networks. There is also evidence that it induces mitochondrial pathology and affects protein quality control (see [4] for review). Immunohistochemical studies of skeletal muscle biopsies from desminopathy patients demonstrated that the abnormal intracellular aggregates contain Z-disc and Z-disc associated proteins and those involved in protein degradation [10–13]. However, hypothesis-free detailed analysis of the composition of the protein aggregates has not been attempted. It is expected that more complete knowledge of the aggregate components would provide insights into pathomechanisms of this disease and help to identify specific biomarker candidates and therapeutic targets.
Over the past years, proteomic studies of myopathies were mainly aimed at the identification of protein biomarker candidates for diseases such as Duchenne muscular dystrophy (for review see [14]), hypokalemic myopathy [15], and sporadic inclusion body myositis [16]. But these studies used non-targeted total crude muscle protein extracts or soluble cytosolic protein fractions swamped with components that are irrelevant to the disease-related mechanisms and therefore blurred the results of proteomic analysis.
We set up a combined laser microdissection and label-free proteomic approach that enables identification and relative quantitation of proteins in abnormal aggregates selectively collected from skeletal muscle sections of MFM patients. We tested this approach in a study of filaminopathy, another subtype of MFM caused by FLNC mutations [17–20]. In the filaminopathy study, we were able to detect about 400 proteins, of which thirty-one were statistically significantly over-represented in protein aggregate samples from abnormal fibers with a ratio >1.8 to samples from clean unaffected regions. Among these proteins, filamin C (FLNC) showed the highest spectral index; many other aggregating protein components were newly identified [20]. This provided new information about disease-relevant proteins whose role in the pathogenesis of filaminopathy is being further examined by biochemical and functional studies.
We present here a differential proteomic study performed in five desminopathy patients. Our combined laser microdissection and mass spectrometry approach was applied to unravel the composition of protein aggregates that occur within affected muscle fibers of these patients and compared the results with our previous findings in filaminopathy. In addition, we searched the mass spectrometric data for mutant desmin peptides to see if this approach would allow to identify disease-causing mutations directly on the protein level.
2. Material and methods
2.1. Patients
Skeletal muscle samples from five desminopathy patients carrying different DES-mutations were identified as suited for laser microdissection and to be included in this study. In each patient the diagnosis was confirmed by genetic analysis and histological and immunofluorescence studies on skeletal muscle sections as described [21,22]. One patient carried a p.N116S mutation [9] in the α-helical segment 1A of desmin and one patient a p.E245D mutation in the α-helical segment 1B [23]. Two patients harbored desmin mutations in the central α-helical-core 2B-region (p.L392P [21] and p.D399Y [24]). The remaining patient carried the p.P419S mutation [21] in the tail-domain of desmin. Detailed information on the patients is provided in Table 1. Informed consent was obtained from each patient (in accordance with The Code of Ethics of the World Medical Association (Declaration of Helsinki) for experiments involving humans with the approval of the ethics committee of the Ruhr-University Bochum ([#4368-12]).
Table 1.
Overview of desminopathy patients included in this study.
| ID | Gender | Age at biopsy [years] |
Muscle |
DES mutation |
|---|---|---|---|---|
| 1 | Female | 19 | Vastus lateralis | p.N116S |
| 2 | Female | 73 | Vastus lateralis | p.E245D |
| 3 | Female | 34 | Triceps brachii | p.L392P |
| 4 | Female | 38 | Medial gastrocnemius | p.D399Y |
| 5 | Female | 59 | Anterior tibialis | p.P419S |
2.2. Laser microdissection
Tissue excised at muscle biopsy was cut into pieces of about 0.5 cm3, embedded into Tissue-Freezing Medium® (Leica Microsystems, Wetzlar, Germany) and frozen in liquid nitrogen. An established combined laser microdissection and label-free proteomic approach was applied for sample analysis as described [20]. Immunofluorescence staining was performed to detect areas of protein aggregation in muscle fibers: 10 µm frozen sections of muscle samples were incubated with a primary antibody directed against myotilin (mouse monoclonal, RS034, Novocastra/Leica Microsystems, Wetzlar, Germany) in 1:20 dilution in PBS (0.1 M NaCl + 3 mM KCl + 1 mM KH2PO4 + 5 mM Na2HPO4, pH 7.4) for 1 h at room temperature (RT) followed by incubation with a secondary antibody conjugated with DyLight 488 (goat anti-mouse IgG antibody, Dianova, Hamburg, Germany, 1:1000 in PBS) for 45 min at RT. A total area of 250,000 µm2 of aggregate-containing sections in abnormal fibers, thereafter referred to as “aggregate samples”, and an area of 250,000 µm2 of aggregate-free sections in normally looking muscle fibers, referred to as “controls”, were collected into tubes with 40 µl formic acid (FA; 98–100%) by laser microdissection (LMD 6500, Leica Microsystems, Wetzlar, Germany). After incubation for 1 h and sonication for 5 min (RK31, BANDELIN electronic, Berlin, Germany) the samples were centrifuged (5 min, 10,000 rpm, RT) and frozen at −80 °C.
2.3. In-solution digestion
For the tryptic in-solution digestion FA was removed from samples by vacuum vaporization (Rotational-Vacuum-Concentrator RVC2-25CDplus, Martin Christ GmbH, Osterode am Harz, Germany) and the collected tissue material was diluted in 50 mM ammonium bicarbonate (pH 7.8) to a final volume of 74.25 µl. The samples were reduced for 20 min at 56 °C in 0.25 µl 2 M DTT followed by alkylation with 0.55 M iodoacetamide for 15 min in the dark at RT. The pH was adjusted to 7.4 before adding 1 µl of 1% Trypsin Enhancer ProteaseMAX™ Surfactant (Promega, Mannheim, Germany) in 50 mM NH4HCO3 (pH 7.8). The digestion was initiated by adding 1.8 µl of a trypsin (Serva Electrophoresis GmbH, Heidelberg, Germany) solution (1 µg/µl 50 mM acetic acid), performed overnight at 37 °C and stopped by the addition of 5.25 µl 10% TFA. After purification with OMIX C18 tips (Varian, Agilent Technologies, Boeblingen, Germany) and concentration for 5 min in vacuum the final volume was adjusted to 63 µl with 1% TFA. 15 µl were used for each mass spectrometric analysis.
2.4. Nano-HPLC and mass spectrometry
Nano-HPLC was performed on an UltiMate 3000 RSLCnano LC system (Dionex, Idstein, Germany). Samples were loaded on a trap column (Dionex, 75 µm × 2 cm, particle size 3 µm, pore size 100 Å) with a flow rate of 10 µl/min with 0.1% TFA. After washing, the trap column was serially connected with an analytical C18 column (Dionex, 75 µm × 25 cm, particle size 2 µm, pore size 100 Å). The peptides were separated with a flow rate of 400 nl/min using the following solvent system: (A) 95% ACN, 0.1% FA; (B) 80% ACN, 0.1% FA. After a first gradient from 4% A to 40% B about 95 min a second gradient from 40% B to 95% B (2 min) and finally a gradient from 95% to 5% B were applied.
The HPLC system was on-line connected to the nano-electrospray ionization source of a LTQ Orbitrap Velos mass spectrometer (Thermo Fisher Scientific, Dreieich, Germany). In the ESI-MS/MS-analysis MS spectra were scanned between 300 and 2000 m/z with a resolution of 30,000 and a maximal acquisition time of 500 ms. Lock mass polydimethylcyclosiloxane (m/z 445.120) was used for internal recalibration. The m/z values initiating MS/MS were set on a dynamic exclusion list for 35 s and the 20 most intensive ions (charge > 1) were selected for MS/MS-fragmentation in the ion trap. Fragments were generated by low-energy collision-induced dissociation (CID) on isolated ions with collision energy of 35% and maximal acquisition time of 50 ms.
2.5. PRM analysis on a quadrupole-orbitrap mass spectrometer
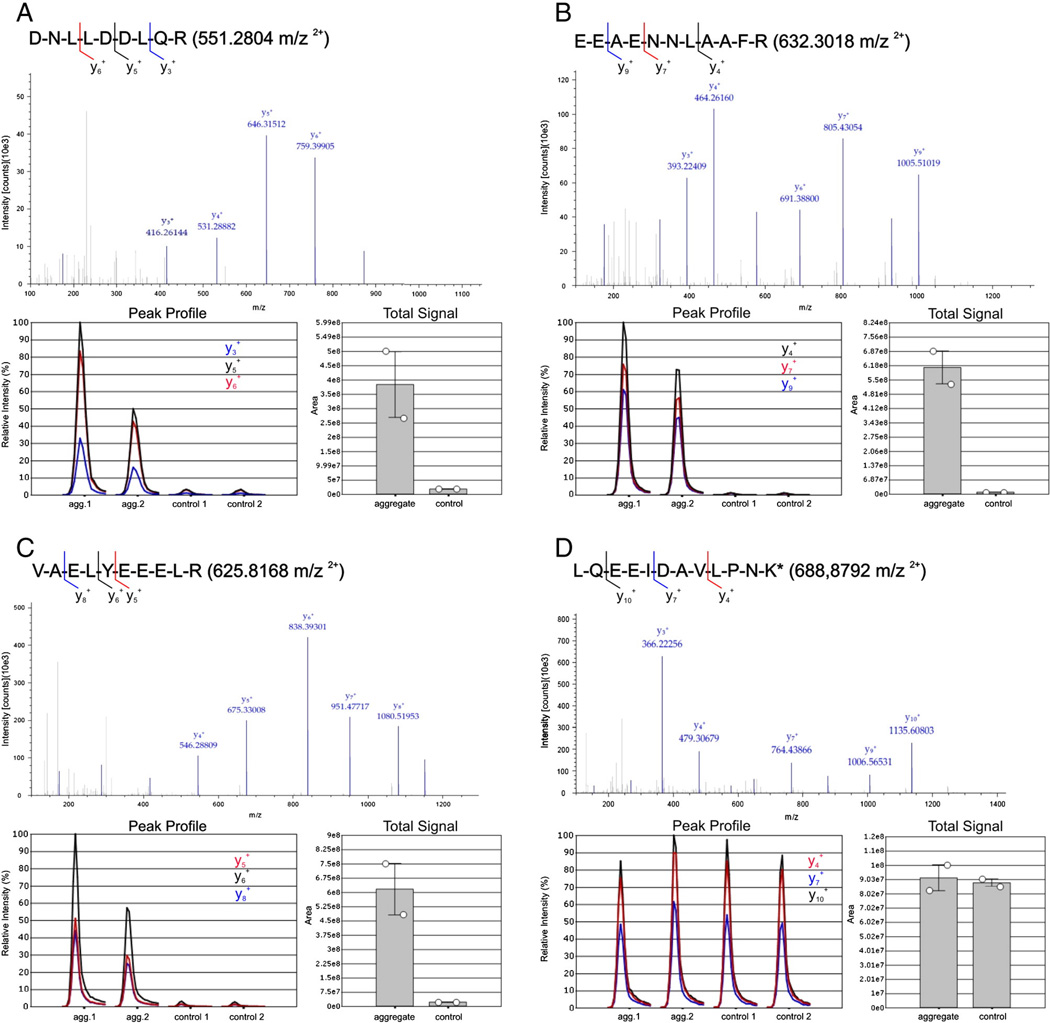
Parallel reaction monitoring (PRM) analyses were performed on two replicates of aggregate and control samples from one desminopathy patient (ID 4, more details are provided in Table 1). Two different proteins, desmin and FLNC, respectively three unique peptides for each protein were chosen for PRM: for desmin the peptides DNLLDDLQR (551.2804 m/z, 2+), EEFAENNLAAFR (632.3081 m/z, 2+) and VAELYEEELR (625.8168 m/z, 2+) and for FLNC the peptides GAGTGGLGLAIEGPSEAK (792.9150 m/z, 2+), SLTATGGNHVTAR (642.8364 m/z, 2+) and VHVQPAVDTSGVK (668.8646 m/z, 2+). The analyses were performed using a Q-Exactive mass spectrometer (Thermo Fisher Scientific) as described previously with some modification [25,26]. In brief using a nano-electrospray source ionization was performed with a spray voltage of 1500 V and capillary temperature of 250 °C. In all experiments, a full mass spectrum at 70,000 resolution at m/z 200 (AGC target 1 × 106, 250 ms maximum infection time, m/z 350–1400) was followed by a PRM scan at 35,000 resolution at m/z 200 (AGC target 2 × 105, 120 ms maximum injection time, isolation window of ±2) whereas all selected precursors from the inclusion list were scanned individually in one PRM scan. Ion dissociation was performed at normalized collision energy of 25% and a fixed first mass at 100 m/z.
For the analysis of the 6 peptides, LC separation was performed as described above with a trap column (Dionex, 300 µm × 5 mm, particle size 5 µm, pore size 100 Å) and an analytical C18 column (Dionex, 75 µm × 50 cm, particle size 2 µm, pore size 100 Å). Prior to nano-LC separation, the digested samples were spiked with a known concentration (100 fmol) of a heavy labeled peptide (LQEEIDAVLPN(KLYS(+8 Da)), Heavy Peptide™, AQUA Ultimate, Thermo Fisher Scientific) which provides an internal standard as quality control.
2.6. Data processing, database search and detection of desmin mutants on the protein level
For database search, the raw files were transformed into *.mgf-files (ProteomDiscoverer 1.3, Thermo Fisher Scientific), imported in ProteinScape™ (version 2.1, Bruker Daltonics, Bremen, Germany), analyzed using Mascot (Matrixscience, London UK), and searched against DecoyDatabaseBuilder [27] containing the entire Uniprot/Swissprot (release 2011/6, 529056 entries) with one additional shuffled decoy for each protein. The following search parameters were used: peptide mass tolerance of 10 ppm, fragment mass tolerance of 0.5 Da, one allowed missed cleavage and carbamidomethylation (C), oxidation (M) as well as phosphorylation (S, T, Y) as variable modifications.
For the detection of desmin mutants on protein level the used database was extended by protein sequences of all known desmin mutations. Searches were done against the extended database with the same search parameters as above. The quality of MS/MS-spectra of mutant peptides was monitored manually (Xcalibur™, thermo Fisher Scientific) by theoretical fragmentation using the MS-Product software tool (ProteinProspector™ 5.10.4, University of California, San Francisco, USA). The charge states of all abundant fragment ions were determined from their isotopic pattern and compared to theoretical ions.
2.7. Protein quantitation by spectral index calculation
After peptide identification, an algorithm similar to the ProteinExtractor in ProteinScape which is using a given minimal peptide score (minPepScore) and minimal peptide count per protein (minNrPeps) was applied as described [20]. A minNrPeps of 2 was used for the given data of aggregate and control samples to exclude “one hit wonders”, which yielded a minPepScore of 20 for the comparison between aggregate and control desminopathy samples. A minPepScore of 21 was calculated for the comparison of aggregate samples from desminopathy and filaminopathy patients. Among the proteins in these lists, every peptide spectrum match (PSM) was extracted.
These PSMs were further processed using the PivotTable function of Microsoft Excel resulting in a table representing spectral counts for every peptide belonging to a certain protein. A spectral index (SI) based on spectral and peptide counts was calculated as described previously [28,29] and subsequently used as basis for label-free quantitation. In brief, spectral index calculation was performed for each sample by summing up all spectral counts belonging to the respective protein and the multiplication of this value by the normalization factor for the individual sample. This factor was calculated by dividing the mean of total spectral counts (considering all aggregate and control samples) by the total spectral counts in a given sample. Because of protein sequence homologies some identified peptides may have been assigned to more than one protein. To avoid this, peptides were considered only if unique within the complete dataset. To identify proteins that are over- or under-represented in desminopathy aggregates, the ratio between the averaged spectral index in aggregate and control samples was calculated for each detected protein. Furthermore, results of proteomic analysis of aggregate samples were compared with data from our previous study in filaminopathy [20] to identify differences in proteomic profiles. In both analyses an unpaired two-tailed t-test (equal variances assumed) was conducted for each protein. A protein was considered as over-represented when the aggregate–control ratio reached >1.8 and p-value was <0.05.
2.8. Relative quantitation based on PRM analysis
Data analysis was performed using Proteome Discoverer (version 1.3, Thermo Fisher Scientific) and Pinpoint (version 1.2, Thermo Fisher Scientific). For all analyzed peptides the areas under the curve (AUC) of all PRM transitions were computed by the software (Suppl. Fig. 3). Relative quantitation of the selected peptides for desmin and FLNC were based on the peak area of the three most intensive fragment ions. The total signal was calculated by normalizing all peak areas of selected fragments in a sample group.
2.9. Validation of proteomic findings by immunofluorescence studies
Double immunofluorescence studies were performed for validation of the proteomic data. 6-µm-thick frozen skeletal muscle sections from patients with desminopathy were incubated with primary antibodies directed against the following selected proteins identified as significantly over-represented or decreased in aggregate samples: desmin, Xin actin-binding repeat-containing protein 2 (Xirp2), αB-crystallin, Nebulin-related-anchoring protein (N-RAP), Xin actin-binding repeat-containing protein 1 (Xin), vimentin, four and a half LIM domains protein 1 (FHL1), sarcosynapsin, versican core protein, tubulin beta chain, myopalladin, supervillin, cold shock domain-containing protein A (CSDA), flotillin-1, FRAP-related protein 1, myomesin-1 and myomesin-2. Antibodies against FLNC and myotilin were used as sensitive tools to detect areas of pathological protein aggregation in desminopathies [4]. A complete list of primary antibodies used in the current study, including information about clones, sources and dilutions, is provided in Supplemental Table 1.
Sections were incubated overnight at 4 °C with primary antibodies, followed by three washing steps with PBS for 5 min and an incubation with isotype specific secondary antibodies conjugated with DyLight 488 (Dianova, Hamburg, Germany, dilution 1:1000) or Texas Red (Dianova, Hamburg, Germany, dilution 1:400) for 45 min at RT. The staining procedure was concluded by three washing steps with PBS for 5 min.
3. Results
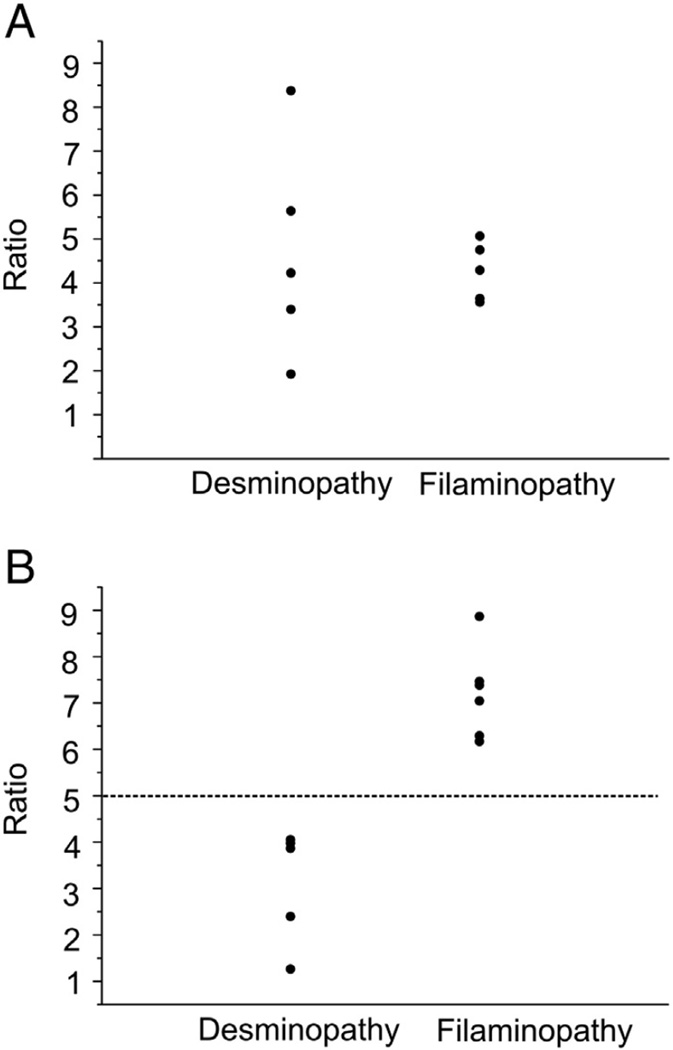
A typical histopathological hallmark of desminopathy is a massive protein aggregation in amuscle fiber. The composition of these protein aggregates is largely unknown. To understand which proteins are involved in aggregate formation we applied an established combined laser microdissection and label-free proteomic approach [20]. This highly sensitive approach was used to analyze both aggregate and aggregate-free sections of skeletal muscle biopsies from patients which enable quantitation of proteins by spectral index calculation. Abnormal fibers harboring protein aggregates were detected by immunofluorescence staining using an antibody directed against myotilin, a widely used marker for protein aggregates in desminopathies. In each muscle biopsy the number of abnormal fibers was sufficient to collect a combined aggregate area of 250,000 µm2. In this material, a total of 397 different proteins have been identified and quantified. Using significance level parameters of p-value <0.05 (unpaired two-tailed t-test, equal variances assumed) and regulation factor >1.8 (if applicable), we identified 22 proteins with a significantly higher spectral index in aggregate samples compared to controls (Table 2) and 12 proteins significantly under-represented in aggregate samples (Table 3). Proteins listed in Tables 2 and 3 are ordered by the mean sum of unique peptides detected in protein aggregates and control samples normalized by the number of total counts. To estimate similarities and differences in the composition of protein aggregates in desminopathy and filaminopathy and to identify potential differential biomarkers specific of desminopathy, we compared the results of label-free quantitation for desminopathy with the results of filaminopathy from our previous proteomic study [20] (Table 4). This comparison revealed that the averaged spectral index of desmin was higher in desminopathy samples (ratio 1.7) but the result was not statistically significant (p-value 0.15). Estimates of individual variability among desminopathy patients (aggregate samples compared to intraindividual controls) were in a wide range (ratios from 1.9 to 8.3) and showed an overlap with findings in filaminopathy patients (ratios from 3.6 to 5.0) (Fig. 1A). Averaged spectral indices of FLNC and its binding partner Xin were significantly higher in filaminopathy than in desminopathy (Table 4). Individual ratios of FLNC (spectral index in aggregate samples compared to intraindividual controls) ranged from 1.3 to 4.0 in desminopathy patients and from 6.2 to 8.9 in filaminopathy patients (Fig. 1B).
Table 2.
Results of label-free quantitation of proteins significantly over-represented in desminopathy aggregate samples.
| Acc. | Protein | SC | Pep. a | Aggregate (SI) |
Control (SI) |
Ratio | p-Value |
|---|---|---|---|---|---|---|---|
| P17661 | Desmin | 73.2 | 27 | 238.5 | 47.7 | 5.0 | 0.03 |
| Q14315 | Filamin C | 43.2 | 89 | 93.9 | 30.6 | 3.1 | 0.01 |
| A4UGR9 | Xin actin-binding repeat-containing protein 2 (Xirp2) | 28.0 | 4 | 82.7 | 0.0 | n.a. | 0.004 |
| P02511 | αB-Crystallin | 57.7 | 9 | 65.9 | 6.3 | 10.4 | 0.03 |
| Q86VF7 | Nebulin-related-anchoring protein (N-RAP) | 34.9 | 65 | 44.6 | 0.8 | 56.5 | 0.001 |
| Q702N8 | Xin actin-binding repeat-containing protein 1 (Xin) | 15.5 | 29 | 16.3 | 0.2 | 72.8 | 0.003 |
| P04792 | Heat shock protein beta-1 (Hsp27/HspB1) | 50.2 | 10 | 16.1 | 0.9 | 18.6 | 0.045 |
| P08670 | Vimentin | 21.9 | 6 | 8.3 | 1.7 | 4.7 | 0.02 |
| P33176 | Kinesin-1 heavy chain | 0.2 | 2 | 4.0 | 0.6 | 6.8 | 0.046 |
| Q13642 | Four and a half LIM domains protein 1 (FHL1) | 11.8 | 4 | 3.1 | 1.6 | 1.9 | 0.045 |
| P08123 | Collagen α-2(I) chain | 3.1 | 3 | 3.1 | 1.2 | 2.6 | 0.04 |
| P28070 | Proteasome subunit beta type-4 | 2.8 | 7 | 3.0 | 0.0 | n.a. | 0.03 |
| Q15773 | Myeloid leukemia factor 2 | 8.1 | 2 | 2.7 | 0.6 | 4.6 | 0.01 |
| Q2TBA0 | Sarcosynapsin | 6.0 | 5 | 1.7 | 0.2 | 9.8 | 0.04 |
| P13611 | Versican core protein precursor | 0.1 | 2 | 1.4 | 0.0 | n.a. | 0.007 |
| Q7Z4W1 | l-Xylulose reductase | 16.0 | 4 | 1.4 | 0.0 | n.a. | 0.04 |
| P07437 | Tubulin beta chain | 17.1 | 2 | 1.2 | 0.0 | n.a. | 2.3 × 10−5 |
| Q86TC9 | Myopalladin | 3.6 | 5 | 1.2 | 0.0 | n.a. | 0.03 |
| O95425 | Supervillin | 2.6 | 4 | 1.2 | 0.2 | 6.1 | 0.04 |
| P16989 | Cold shock domain-containing protein A (CSDA) | 13.7 | 2 | 1.0 | 0.0 | n.a. | 0.01 |
| O75955 | Flotillin-1 | 7.0 | 2 | 0.8 | 0.0 | n.a. | 0.048 |
| Q13535 | FRAP-related protein 1 | 0.8 | 3 | 0.6 | 0.0 | n.a. | 0.047 |
Acc. = UniProtKB accession number, SC = sequence coverage (%), SI = spectral index, n.a. = not applicable.
Number of different peptides.
Table 3.
Results of label-free quantitation of proteins significantly under-represented in desminopathy aggregate samples.
| Acc. | Protein | SC | Pep. a | Aggregate (SI) |
Control (SI) |
Ratio | p-Value |
|---|---|---|---|---|---|---|---|
| P54296 | Myomesin-2 | 27.6 | 45 | 14.4 | 32.9 | 0.4 | 0.002 |
| P52179 | Myomesin-1 | 27.3 | 51 | 12.1 | 29.4 | 0.4 | 0.007 |
| Q08043 | Α-actinin-3 | 42.8 | 20 | 1.1 | 12.7 | 0.1 | 0.04 |
| Q96HC4 | PDZ and LIM domain protein 5 | 16.8 | 7 | 2.5 | 6.7 | 0.4 | 0.003 |
| P00403 | Cytochrome c oxidase subunit 2 | 11.5 | 2 | 0.9 | 3.6 | 0.3 | 0.009 |
| Q9NRC6 | Spectrin beta chain, non-erythrocytic 5 | 0.8 | 2 | 0.2 | 2.0 | 0.1 | 0.002 |
| Q9Y3A5 | Ribosome maturation protein SBDS | 6.0 | 2 | 0.4 | 1.8 | 0.2 | 0.002 |
| P56385 | ATP synthase subunit e | 26.1 | 2 | 0.0 | 1.6 | 0.0 | 0.0001 |
| Q14004 | Cyclin-dependent kinase 13 | 1.4 | 1 | 0.2 | 1.6 | 0.1 | 0.01 |
| Q8IVF4 | Dynein heavy chain 10, axonemal | 0.6 | 2 | 0.0 | 0.6 | 0.0 | 0.04 |
| Q92526 | T-complex protein 1 subunit zeta-2 | 0.3 | 1 | 0.0 | 0.6 | 0.0 | 0.04 |
| Q92576 | PHD finger protein 3 | 1.0 | 2 | 0.0 | 0.6 | 0.0 | 0.04 |
Acc. = UniProtKB accession number, SC = sequence coverage (%), SI = spectral index.
Number of different peptides.
Table 4.
Results of the label-free quantitation of desminopathy and filaminopathy aggregate samples (for proteins listed in Table 2)
| Acc. | Protein | Aggregate Des a (SI) | Aggregate FLNCb (SI) | Ratio | p-Value |
|---|---|---|---|---|---|
| P17661 | Desmin | 236.5 | 136.6 | 1.73 | 0.15 |
| Q14315 | Filamin C | 93.9 | 240.9 | 0.39 | 0.0003 |
| A4UGR9 | Xirp2 | 82.4 | 75.3 | 1.09 | 0.73 |
| P02511 | αB-Crystallin | 66.0 | 37.8 | 1.75 | 0.20 |
| Q86VF7 | N-RAP | 43.9 | 58.2 | 0.75 | 0.13 |
| Q702N8 | Xin | 16.2 | 34.3 | 0.47 | 0.002 |
| P04792 | Hsp27 | 15.9 | 15.7 | 1.01 | 0.97 |
| P08670 | Vimentin | 7.8 | 5.8 | 1.34 | 0.34 |
| Q13642 | FHL1 | 3.0 | 3.1 | 0.96 | 0.80 |
| P08123 | Collagen alpha-2(I) chain | 3.1 | 2.7 | 1.15 | 0.68 |
| P28070 | Proteasome subunit beta type-4 | 3.0 | 1.4 | 2.11 | 0.20 |
| Q15773 | Myeloid leukemia factor 2 | 2.5 | 1.9 | 1.34 | 0.36 |
| Q2TBA0 | Sarcosynapsin | 1.6 | 2.7 | 0.57 | 0.07 |
| Q86TC9 | Myopalladin | 1.4 | 4.0 | 0.35 | 0.01 |
| P13611 | Versican core protein | 1.2 | 0.9 | 1.40 | 0.41 |
| P07437 | Tubulin beta chain | 1.2 | 1.5 | 0.78 | 0.32 |
| O95425 | Supervillin | 1.1 | 1.9 | 0.56 | 0.41 |
| P16989 | CSDA | 0.9 | 0.6 | 1.66 | 0.36 |
| O75955 | Flotillin-1 | 0.7 | 1.0 | 0.69 | 0.46 |
| Q13535 | FRAP-related protein 1 | 0.6 | 0.0 | n.a. | 0.029 |
Acc. = UniProtKB accession number, SI = averaged spectral index.
Aggregate samples of five desminopathy patients.
Aggregate samples of six filaminopathy patients.
Fig. 1.
Individual ratios between spectral indices of aggregate and control samples for desmin and filamin C in desminopathy and filaminopathy patients. A: Ratios for desmin between aggregate and control samples in five desminopathy and six filaminopathy patients. The ratios were calculated for each individual using spectral index calculation by summing up all spectral counts belonging to the respective protein. B: Ratios for filamin C between aggregate and control samples in five desminopathy and six filaminopathy patients.
3.1. Validation of proteomic data
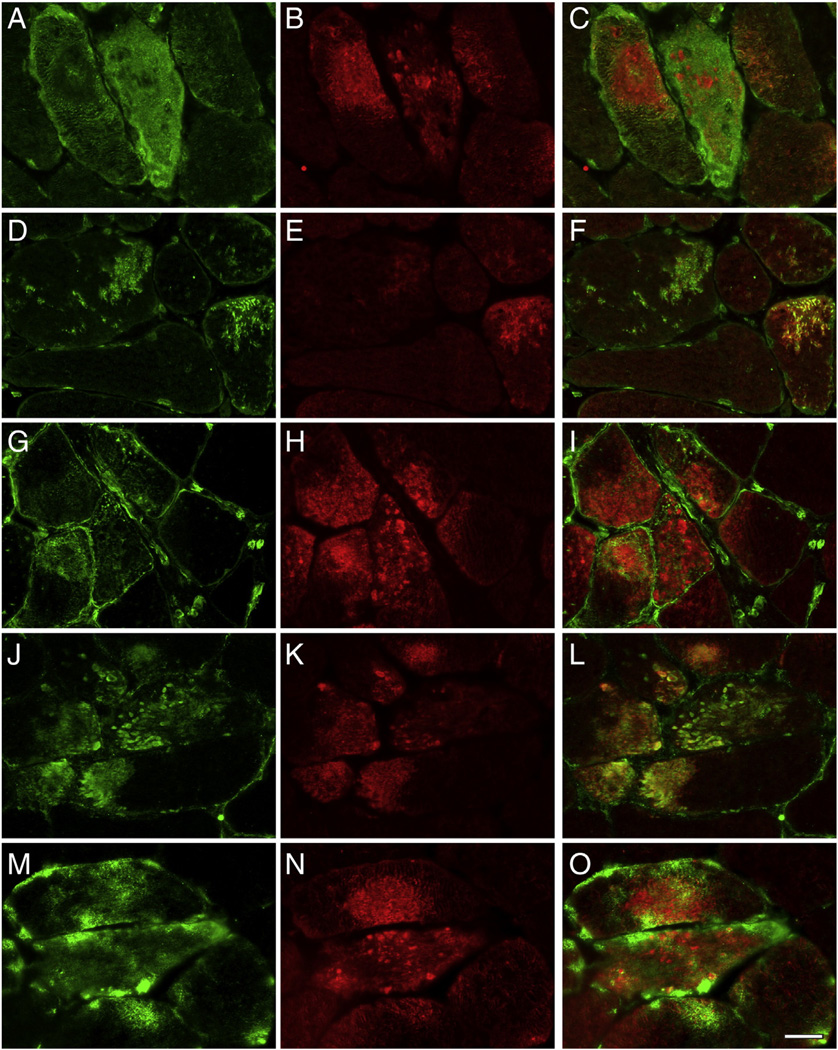
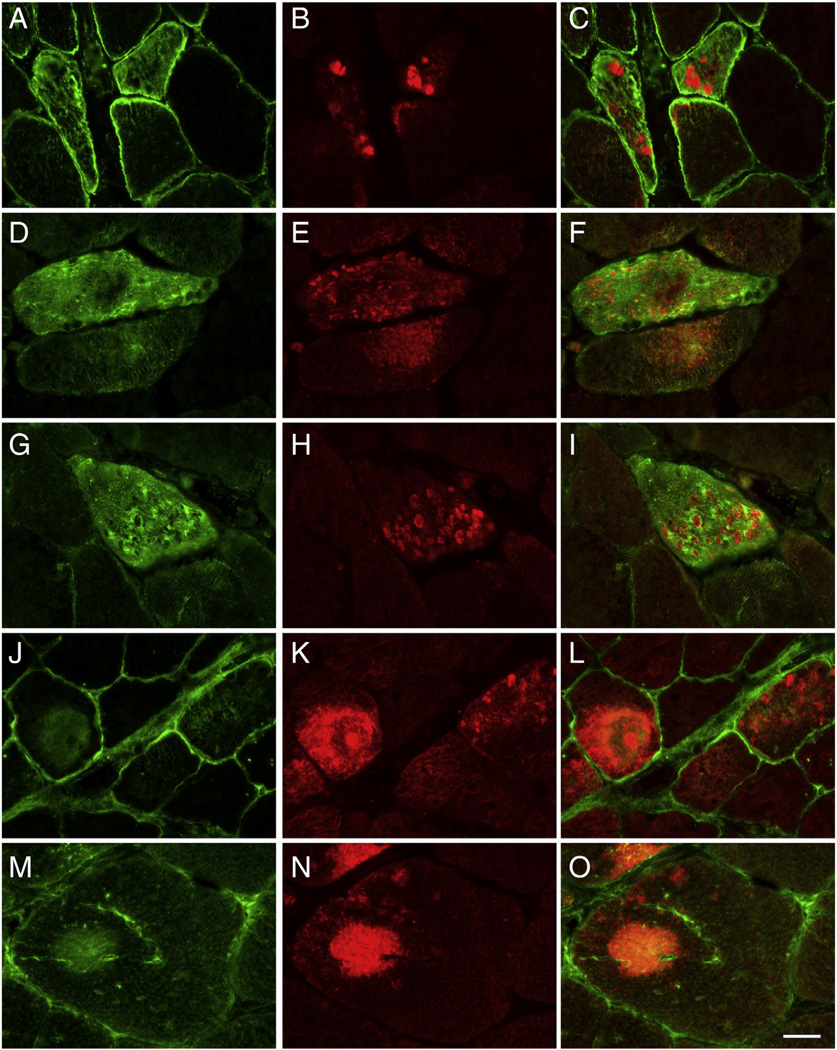
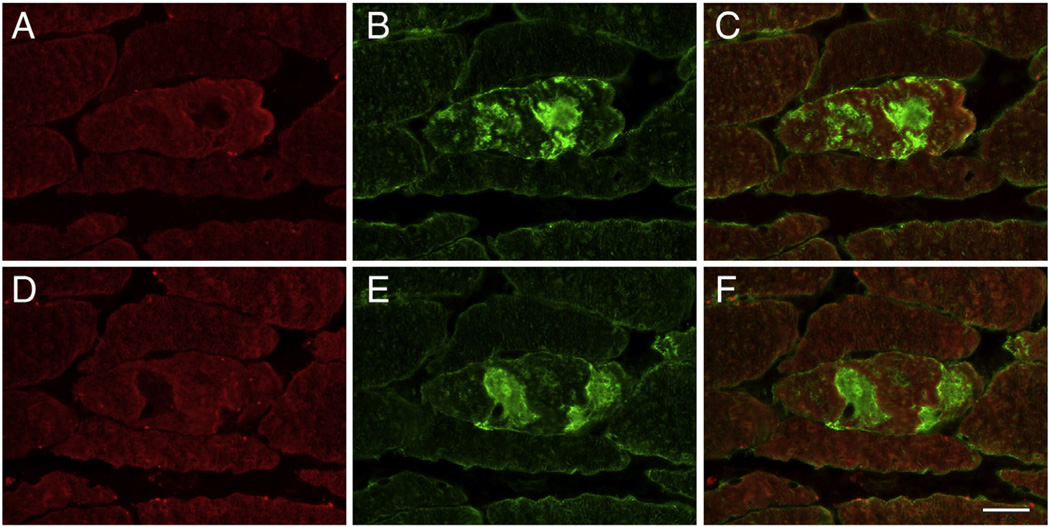
The results for 15 proteins with a significantly higher (Table 2) and two with significantly lower spectral indices in aggregates (Table 3) were validated by double immunofluorescence staining. To visualize abnormal fibers harboring aggregates, cryosections of skeletal muscle samples were immunostained with antibodies directed against myotilin or FLNC. As expected, aggregates displayed an increased immunoreactivity for the disease causing protein desmin (Suppl. Fig. 1), which was identified as a significantly enriched protein in the desminopathy aggregates. Abnormal fibers also showed increased immunoreactivity for the proteins myopalladin, N-RAP, tubulin beta chain, FRAP-related protein 1, CSDA, flotillin-1, sarcosynapsin, supervillin, versican core protein, FHL1, FLNC, αB-crystallin, Xin, Xirp2, and vimentin (Figs. 2, 3, Suppl. Fig. 1), consistent with the proteomic data. In addition, aggregates showed a decreased immunoreactivity for the M-band proteins myomesin-1 and myomesin-2 (Fig. 4) corresponding to a decrease seen on proteomic assessment.
Fig. 2.
Validation of proteomic data for the proteins N-RAP, tubulin beta, FRAP-related protein-1, and CSDA by immunolocalization studies performed on skeletal muscle sections of desminopathy patients. Abnormal muscle fibers were detected by immunostaining with primary antibodies against myotilin (B, E, H, K, N). Consistent with the results of proteomic analysis, abnormal fibers show increased immunoreactivity for myopalladin (A), N-RAP (D), tubulin beta (G), FRAP-related protein-1 (J), and CSDA (M). C, F, I, L, O: merged images. Scale bar = 25 µm.
Fig. 3.
Validation of proteomic data for the proteins flotillin-1, sarcosynapsin, supervillin, versican core protein and FHL1 by immunolocalization studies performed on skeletal muscle section of desminopathy patients. Abnormal muscle fibers were detected by immunostaining with primary antibodies against myotilin (B, E, H, K, N). Corresponding to the results of proteomic analysis, abnormal fibers show an increased immunoreactivity for flotillin-1 (A), sarcosynapsin (D), supervillin (G), versican core protein (J) and FHL1 (M). C, F, I, L, O: merged images. Scale bar = 25 µm.
Fig. 4.
Validation of proteomic data for the proteins myomesin-1 and myomesin-2 by immunolocalization studies performed on serial skeletal muscle sections of a desminopathy patient. Areas of protein aggregation in abnormal fibers detected by immunostaining with antibodies recognizing filamin C (B, E) show a decreased immunoreactivity for myomesin-1 (A) and myomesin-2 (B). C, F: merged images. Scale bar = 25 µm.
In the PRM analysis, three desmin specific and three FLNC specific peptides were analyzed. A spiked-in heavy labeled peptide with a known concentration provided an internal standard as quality control. The selected fragment ions of the targeted peptides for desmin and FLNC showed markedly higher intensities in aggregate samples compared to controls (Fig. 6, Suppl. Fig. 2). These findings are consistent with the results of relative quantitation by spectral index calculation. The fragment ions of the internal standard showed similar intensities in all samples. This underlines the reproducibility of our PRM analyses.
Fig. 6.
Quantitation of three desmin specific peptides (A–C) and one heavy labeled peptide (D) in two replicates of aggregate and control samples from a desminopathy patient with p.L392P mutation. The four targeted peptides were analyzed by PRM individually in one scan. Chosen fragment ions for quantitation are indicated in the peptide sequence respectively. Exemplified for all peptides MS/MS spectra of one aggregate sample are shown. Relative quantitation of the selected peptides was performed on the peak area of the three most intensive fragment ions shown in the peak profile. The ratio was calculated between the total signal area of those fragment ions for each peptide in the group of aggregate samples and in the group of control samples. The total signal areas were computed by normalizing all peak areas of the most intensive fragment ions within one sample group. (A–C) Consistent with the results of the label-free analysis by spectral index calculation the fragment ions of all desmin specific peptides showed a significantly higher signal area in the group of aggregate samples than in the control group. (D) The labeled standard peptide was spiked in with a concentration of 100 fmol as a quality control for the analysis. The peak profile of the extracted three fragment ions as well as the total signal area showed no significant differences between the samples, proving a high reproducibility of the measurements.
3.2. Detection of mutant desmin in protein aggregates
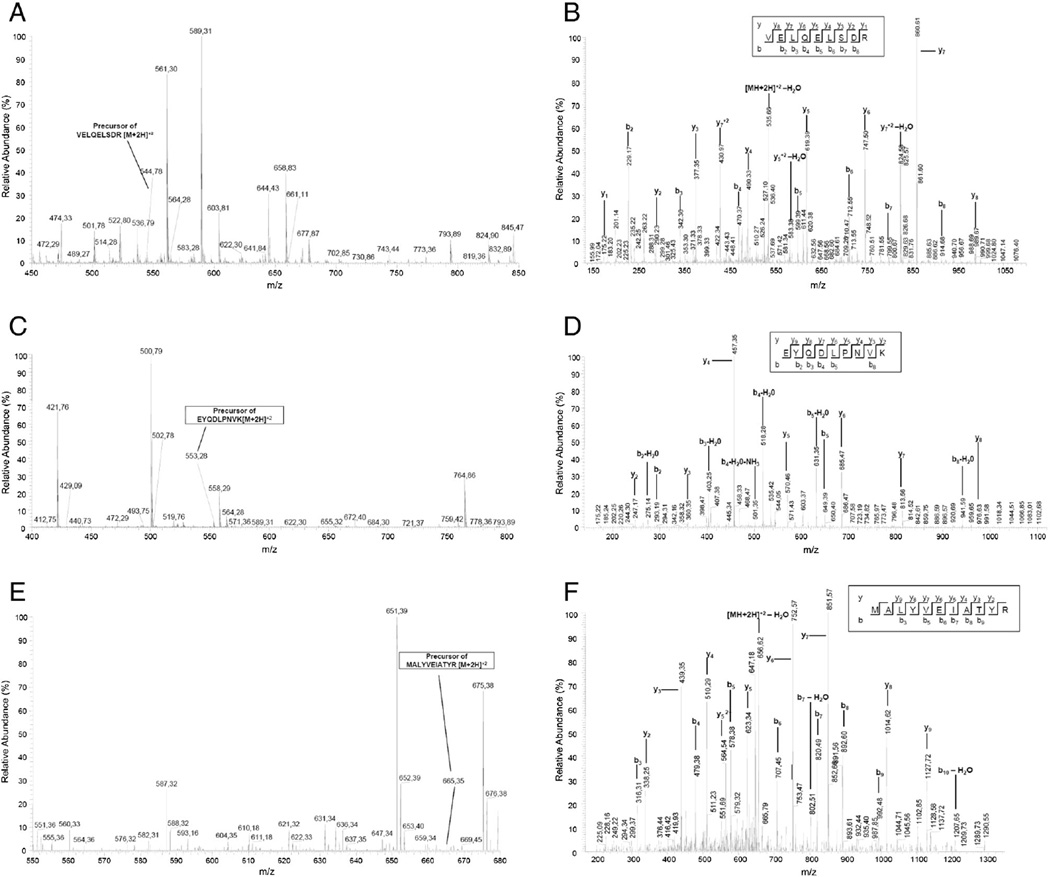
In order to demonstrate expression of mutant desmin and its presence in protein aggregates we searched the mass spectrometric data against an extended protein database. The identification of mutant desmin variants as components of aggregates supports their involvement in mechanisms leading to aggregate formation and gives us new important insights in the overall disease pathomechanisms. We identified mutant peptides corresponding to the causative gene mutation in 3 patients with desminopathy: for p.N116S the mutation-specific peptide VELQELSDR, for p.L392P the mutation-specific peptide EYQDLPNVK, and for p.D399Y the peptide MALYVEIATYR (Table 5). The quality of MS/MS-spectra (Fig. 5) of mutation-specific peptides was validated manually by theoretical fragmentation. The charge states and m/z of all abundant fragment ions were determined from their isotopic pattern and compared to theoretical ions. For the mutation-specific peptide VELQELSDR of the DES-mutation p.N116S the masses of the b-type fragment ions b2 to b8 and of the y-type fragment ions y1 to y8 could be assigned (Fig. 5 A, B). Peptide EYQDLPNVK including DES-mutation p.L392P was unequivocally identified by assignment of masses of the b-type fragment ions b2 and b3 to b8 respectively the y-type fragment ions y2 to y9 (Fig. 5 C, D). The mutation-specific peptide MALYVEIATYR of the DES-mutation p.D399Y has been confirmed by assignment of the b-type-fragment ions b3 and b5 to b9 as well as the y-type fragment ions y2 to y9 (Fig. 5 E, F).
Table 5.
Detection of mutant desmin on the protein level.
| DES mutation | Protein domain | Peptide sequencea | Sequence range | m/z | References |
|---|---|---|---|---|---|
| p.N116S | 1A helix | VELQELSDR | 110–118 | 544.78 | Klauke et al. (2010) |
| p.L392P | 2B helix | EYQDLPNVK | 387–395 | 553.28 | Vrabrie et al. (2005) |
| p.D399Y | 2B helix | MALYVEIATYR | 396–406 | 665.35 | Olivé et al. (2007) |
Sequence of mutant desmin peptides.
Fig. 5.
Detection of desmin mutations p.N116S, p.L392P and p.D399Y on protein level by nanoLC–ESI-MS/MS-analysis of mutation-specific peptides. ESI-MS/MS-analysis was performed on a LTQ Orbitrap Velos mass spectrometer. MS spectra were scanned between 300 and 2000 m/z with a resolution of 30,000 and a maximal acquisition time of 500 ms. Fragments were generated in the ion trap of the mass spectrometer by low-energy collision-induced dissociation (CID) on isolated ions with collision energy of 35% and maximal acquisition time of 50 ms. The quality of the MS/MS-spectrum was monitored manually by theoretical fragmentation. The charge states of all abundant fragment ions of the b- and y-series were determined from their isotopic pattern and compared to theoretical ions. (A) MS-spectrum of the precursor ion (m/z 544.78; z + 2) and (B) MS/MS-spectrum of peptide VELQELSDR, specific for desmin mutation p.N116S. (C) MS-spectrum of the precursor ion (m/z 553.28; z + 2) and (D) MS/MS-spectrum of peptide EYQDLPNVK, specific for desmin mutation p.L392P. (E) MS-spectrum of the precursor ion (m/z 665.35; z + 2) and (F) MS/MS-spectrum of the peptide MALYVEIATYR, specific for desmin mutation p.D399Y.
4. Discussion
Focal protein aggregation in the affected skeletal muscle fibers is a histological hallmark of myofibrillar myopathies. This is also the case in desminopathy, a subtype of MFM caused by mutations in DES, the gene encoding desmin. Previous immunolocalization studies revealed an accumulation of many proteins in the abnormal aggregates seen in muscle fibers of desminopathy patients but these analyses were restricted to pre-selected proteins [10]. We now applied a newly established proteomic approach [20] to get a better view of aggregate composition. The main goal of our study was to detect disease-relevant proteins that may provide new insights into pathomechanisms and identify a specific proteomic profile that may help to distinguish desminopathy from other MFM subtypes. An additional aim of our study was to detect mutant desmin directly on the protein level to confirm its expression in muscle fibers and localization in aggregates. The localization of mutant desmin peptides within the aggregates is not only important for unraveling the mechanisms leading to aggregate formation and the clarification of pathomechanisms involved in desminopathy but also for providing tools for the identification of MFM-relevant mutations in genetically unresolved MFM cases.
We analyzed skeletal muscle samples from five patients with different DES mutations. Considering the rareness of the disease that clearly limits the availability of muscle samples, this is a respectable number. Age at biopsy, disease duration and the site of biopsy varied between the patients but our previous study in filaminopathy, another subtype of MFM, suggested that these variables do not significantly influence the composition of protein aggregates [20]. Laser microdissection was used to collect aggregate samples from abnormal fibers and aggregate-free control samples from normally looking muscle fibers. A great advantage of this method compared to the use of homogenates is that it allows to separate affected tissues from normal instead of dealing with a complex mixture of different cell types. Mass spectrometric analysis and subsequent spectral index calculation enabled a relative quantitation of proteins. To estimate the abundance of detected proteins and reduce the risk of false positive results, we defined a cutoff value of a minimum ratio of 1.8 for proteins to be considered as over-represented in aggregates.
In total, 397 proteins have been identified in the patients' samples, 22 of them showing a statistically significant higher spectral index in aggregate samples compared to control samples. Most of these proteins were three or more times more abundant or exclusively present in aggregates. Only seven of these 22 proteins, namely desmin and its binding partners αB-crystallin [30] and vimentin [31], and the Z-disc protein FLNC and its interaction partners Xin [32], Xirp2 [20] and Hsp27 [20], were previously identified as aggregate components by immunohistochemistry [10,33]. This study adds 15 further proteins accumulating in aggregates, most of them validated by double immunofluorescence and PRM studies, and thus provide new information about proteins involved in aggregate formation and therefore relevant to the pathogenesis of desminopathy. As an example, two of the newly identified aggregate proteins, N-RAP and myopalladin, are associated with the Z-disc or its developmental precursors which corresponds well with findings from ultrastructural studies in MFM showing that myofibrillar pathology starts from the Z-disc region [1,2,18]. At the same time, immunolocalization studies have shown that not all known Z-disc proteins are involved in this process [10,20]. Indeed, our proteomic data show that the two Z-disc proteins α-actinin-3 and the α-actinin binding partner PDZ and LIM domain protein 5 [34] are significantly decreased in aggregate samples of desminopathy patients. These and many other results derived from proteomic analysis provide new information about Z-disc protein involvement in the pathogenesis of desminopathy.
Our proteomic and immunolocalization data confirmed that, consistent with previous findings, desmin binding partners αB-crystallin and vimentin accumulate in aggregates [10]. Desmin is not part of the Z-disc but colocalizes and interacts with Z-disc proteins. It also interacts with myomesin-1 [35], a protein that crosslinks myosin filaments at the sarcomeric M-band. However, as previously observed in filaminopathy patients [20], the M-band components myomesin-1 and myomesin-2 were significantly under-represented in aggregates, as was validated by our immunolocalization analysis. These findings underline that only a subset of sarcomeric proteins and not all binding partners of mutated MFM proteins are involved in aggregate formation.
Dysfunctions in protein quality control and protein degradation via the ubiquitin-proteasome system (UPS) and the autophagic-lysosomal pathway seem to play an essential pathogenic role in desminopathy (see [4] for review) and other MFM subtypes [12,36]. Immunolocalization studies in desminopathy, myotilinopathy and filaminopathy demonstrated that abnormal muscle fibers show a markedly increased immunoreactivity for a number of UPS and autophagy proteins [36–38]. Expanding these findings, our proteomic analysis revealed that tubulin beta that is involved in regulation of autophagy [39], and proteasome subunit beta type-4 also accumulates in desminopathy aggregates.
Dysfunction of the UPS can induce apoptosis [40] and there is evidence for crosstalk between autophagy and apoptosis [41,42]. This suggests that apoptosis may be a relevant mechanism of muscle fiber death in MFM. Indeed, nuclear apoptosis was found in MFM subtypes αB-crystallinopathy and Bag3-myopathy but not in a series of six MFM patients with unknown causative mutation [43–45]. The role of apoptosis in desminopathy is unclear but it is known that desmin interacts with apoptosis-related proteins (see [4] for review). In this context, it is noteworthy that the accumulation of the apoptosis-associated FRAP-related protein 1 and CSDA in abnormal fibers in desminopathy muscle samples is in agreement with previous findings [46–48]. CSDA was previously shown to be upregulated during hypoxia upon damage of human skeletal muscle cells and induction of apoptosis [49].
Comparing our findings in desminopathy with the results of our previous proteomic study in filaminopathy [20], we find an overlap of proteins accumulating in aggregates. Desmin, FLNC, Xirp2, αB-crystallin, N-RAP, Xin, Hsp27, vimentin and myopalladin showed a significant accumulation in both MFM subtypes. This indicates that these diseases share similar pathomechanisms and expands information obtained in previous immunolocalization studies [10,20,22,36]. But we also found significant differences in protein composition suggesting that proteomic profiling may allow the differentiation of MFM subtypes. This would be helpful in differential diagnosis, particularly in view of the fact that clinical and histopathological features of various MFM subtypes overlap and therefore are usually not specific [2].
Ranking the proteins that are significantly enriched in aggregates by their spectral indices reveals that desmin is the most prominent protein accumulating in desminopathy as FLNC is in filaminopathy [20]. However, this is only a semi-quantitative evaluation with known limitations [20]. Although the mean spectral index of desmin was higher in desminopathy than in filaminopathy aggregates, the difference was not statistically significant. Furthermore, individual desmin ratios varied considerably between desminopathy patients. This variability might be related to the domain location of the causative desmin mutations: they were previously demonstrated to have extremely diverse biological effects in in vitro assembly studies [48–50]. In contrast, the difference in spectral indices of FLNC between aggregate samples of desminopathy and filaminopathy patients was significant and the individual FLNC ratios in filaminopathy patients showed no overlap compared to the data of desminopathy patients which indicates that the FLNC ratio may be a suitable diagnostic biomarker to distinguish between these two MFM subtypes. A recently reported proteomic study in a patient with MFM-causing FHL1 mutation compared to an MFM patient carrying a non-FHL1 mutation found the FLNC ratio to be below 2 in both patients [51]. This suggests that the FLNC ratio may be used to differentiate filaminopathy also from other MFM subtypes. However, a direct comparison of the data from the study of Feldkirchner et al. [51] with our results presented here is difficult due to the fact that different proteomic approaches were applied.
Three further proteins showed significantly different spectral indices in desminopathy and filaminopathy aggregates. Xin and myopalladin were more abundant in filaminopathy and FRAP-related protein 1 in desminopathy. These findings suggest differences in pathomechanisms of distinct MFM subtypes and indicate that specific proteomic profiles may be helpful in differential diagnosis of MFM.
PRM is a recently established method for high resolution and high mass accuracy (HR/AM) quantitative, targeted proteomics. We used this new technique for quantitation of selected proteins. To our knowledge, it is the first time that PRM was used to analyze human muscle samples. Our findings indicate that PRM qualifies as a new and powerful validation method for relative protein quantitation based on spectral index calculation: PRM also revealed that desmin and FLNC are highly over-represented in desminopathy aggregates and therefore confirmed our results from label-free analysis by spectral index calculation and immunolocalization studies. The PRM technique has clear advantages over other quantitation methods like single reaction monitoring (SRM) as PRM spectra are highly specific because all potential transitions of a peptide are available to confirm the identity of a peptide [26]. Moreover, no further knowledge is needed with regard to preselection of targeted transitions before analysis [26].
We also report for the first time that mutant desmin is expressed and present in the abnormal aggregates, which supports the assumption that mutant misfolded desmin molecules are directly involved in aggregate formation. The detection of mutant desmin peptides not only illustrates the specificity and sensitivity of our approach, but also suggests for the first time that proteomic analysis enables a direct identification of the causative gene mutations in still unresolved subsets of MFM patients without extensive genetic searches.
Conclusions
The use of a highly sensitive proteomic approach helped to discover a number of new disease-relevant proteins accumulating in abnormal muscle fibers of desminopathy patients. This provides new insights into pathomechanisms of aggregate formation, one of the basic phenomena in the pathogenesis of desminopathy. The established significant differences between subtype-specific proteomic profiles may be helpful in differential diagnostics of patients with protein aggregation myopathies. The detection of mutant desmin peptides in the aggregates demonstrates for the first time that proteomic analysis enables a direct identification of the pathogenic mutation in a pathologically defined subset of MFM patients.
Supplementary Material
Biological significance.
Our proteomic analysis provides essential new insights in the composition of pathological protein aggregates in skeletal muscle fibers of desminopathy patients. The results contribute to a better understanding of pathomechanisms in myofibrillar myopathies and provide the basis for hypothesis-driven studies. The detection of specific proteomic profiles in different myofibrillar myopathy subtypes indicates that proteomic analysis may become a useful tool in differential diagnosis of protein aggregate myopathies.
Acknowledgments
This research was supported by grants from the Ruhr-University Bochum (FoRUM F680-2009, F599R-2008 and F755-2012), the German Research Foundation (KL 2487/1-1, FOR1228 and FOR1352), and the Heimer Foundation. This work was supported in part by the Intramural Program of the National Institute of Neurological Disorders and Stroke, NIH, Bethesda, MD, USA. We thank the patients for participation in this study.
Abbreviations
- AUC
area under the curve
- BSA
bovine serum albumin
- CSDA
cold shock domain protein A
- DES
desmin
- FA
formic acid
- FDR
false discovery rate
- FLNC
filamin C
- FHL1
four and a half LIM domains protein 1
- LMD
laser microdissection
- Hsp
heat shock protein
- MFM
myofibrillar myopathy
- N-RAP
Nebulin-related-anchoring protein
- RBM
reducing body myopathy
- SI
spectral index
- RT
room temperature
- PET
polyethylene terephthalate
- PSM
peptide spectrum match
- PRM
parallel reaction monitoring
- SRM
single reaction monitoring
- Xin
Xin actin-binding repeat-containing protein 1
- Xirp2
Xin actin-binding repeat-containing protein 2
Footnotes
This article is part of a Special Issue entitled: From Genome to Proteome: Open Innovations.
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.jprot.2013.04.026.
Conflict of interest
All the authors declare that they do not have any conflict of interest to disclose regarding this manuscript.
REFERENCES
- 1.Olivé M, Odgerel Z, Martínez A, Poza JJ, Bragado FG, Zabalza RJ, et al. Clinical and myopathological evaluation of early- and late-onset subtypes of myofibrillar myopathy. Neuromuscul Disord. 2011;21:533–542. doi: 10.1016/j.nmd.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Selcen D. Myofibrillar myopathies. Neuromuscul Disord. 2011;21:161–171. doi: 10.1016/j.nmd.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goldfarb LG, Park KY, Cervenáková L, Gorokhova S, Lee HS, Vasconcelos O, et al. Missense mutations in desmin associated with familial cardiac and skeletal myopathy. Nat Genet. 1998;19:402–403. doi: 10.1038/1300. [DOI] [PubMed] [Google Scholar]
- 4.Clemen CS, Herrmann H, Strelkov SV, Schröder R. Desminopathies: pathology and mechanisms. Acta Neuropathol. 2013;125(1):47–75. doi: 10.1007/s00401-012-1057-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Herrmann H, Aebi U. Intermediate filaments: molecular structure, assembly mechanism, and integration into functionally distinct intracellular Scaffolds. Annu Rev Biochem. 2004;73:749–789. doi: 10.1146/annurev.biochem.73.011303.073823. [DOI] [PubMed] [Google Scholar]
- 6.Goldfarb LG, Vicart P, Goebel HH, Dalakas MC. Desmin myopathy. Brain. 2004;127:723–734. doi: 10.1093/brain/awh033. [DOI] [PubMed] [Google Scholar]
- 7.Maloyan A, Robbins J. Autophagy in desmin-related cardiomyopathy: thoughts at the halfway point. Autophagy. 2010;6 doi: 10.1161/CIRCRESAHA.109.212639. [DOI] [PubMed] [Google Scholar]
- 8.Schröder R, Schoser B. Myofibrillar myopathies: a clinical and myopathological guide. Brain Pathol. 2009;19:483–492. doi: 10.1111/j.1750-3639.2009.00289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klauke B, Kossmann S, Gaertner A, Brand K, Stork I, Brodehl A, et al. De novo desmin-mutation N116S is associated with arrhythmogenic right ventricular cardiomyopathy. Hum Mol Genet. 2010;19:4595–4607. doi: 10.1093/hmg/ddq387. [DOI] [PubMed] [Google Scholar]
- 10.Claeys KG, van der Ven PFM, Behin A, Stojkovic T, Eymard B, Dubourg O, et al. Differential involvement of sarcomeric proteins in myofibrillar myopathies: a morphological and immunohistochemical study. Acta Neuropathol. 2009;117:293–307. doi: 10.1007/s00401-008-0479-7. [DOI] [PubMed] [Google Scholar]
- 11.Ferrer I, Olivé M. Molecular pathology of myofibrillar myopathies. Expert Rev Mol Med. 2008;10:e25. doi: 10.1017/S1462399408000793. [DOI] [PubMed] [Google Scholar]
- 12.Olivé M. Extralysosomal protein degradation in myofibrillar myopathies. Brain Pathol. 2009;19:507–515. doi: 10.1111/j.1750-3639.2009.00288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Selcen D, Ohno K, Engel AG. Myofibrillar myopathy: clinical, morphological and genetic studies in 63 patients. Brain. 2004;127:439–451. doi: 10.1093/brain/awh052. [DOI] [PubMed] [Google Scholar]
- 14.Lewis C, Carberry S, Ohlendieck K. Proteomic profiling of x-linked muscular dystrophy. J Muscle Res Cell Motil. 2009;30:267–269. doi: 10.1007/s10974-009-9197-6. [DOI] [PubMed] [Google Scholar]
- 15.Thongboonkerd V, Kanlaya R, Sinchaikul S, Parichatikanond P, Chen ST, Malasit P. Proteomic identification of altered proteins in skeletal muscle during chronic potassium depletion: implications for hypokalemic myopathy. J Proteome Res. 2006;5:3326–3335. doi: 10.1021/pr060136h. [DOI] [PubMed] [Google Scholar]
- 16.Li J, Yin C, Okamoto H, Jaffe H, Oldfield EH, Zhuang Z, et al. Proteomic analysis of inclusion body myositis. J Neuropathol Exp Neurol. 2006;65:826–833. doi: 10.1097/01.jnen.0000228204.19915.69. [DOI] [PubMed] [Google Scholar]
- 17.Vorgerd M, van der Ven PFM, Bruchertseifer V, Löwe T, Kley RA, Schröder R, et al. A mutation in the dimerization domain of filamin C causes a novel type of autosomal dominant myofibrillar myopathy. Am J Hum Genet. 2005;77:297–304. doi: 10.1086/431959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fürst DO, Goldfarb LG, Kley RA, Vorgerd M, Olivé M, van der Ven PFM. Filamin C-related myopathies: pathology and mechanisms. Acta Neuropathol. 2013;125(1):33–34. doi: 10.1007/s00401-012-1054-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shatunov A, Olivé M, Odgerel Z, Stadelmann-Nessler C, Irlbacher K, van LF, et al. In-frame deletion in the seventh immunoglobulin-like repeat of filamin C in a family with myofibrillar myopathy. Eur J Hum Genet. 2009;17:656–663. doi: 10.1038/ejhg.2008.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kley RA, Maerkens A, Leber Y, Theiss V, Schreiner A, van der Ven PFM, et al. A combined laser microdissection and mass spectrometry approach reveals new disease relevant proteins accumulating in aggregates of filaminopathy patients. Mol Cell Proteomics. 2012 doi: 10.1074/mcp.M112.023176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Olivé M, Armstrong J, Miralles F, Pou A, Fardeau M, Gonzalez L, et al. Phenotypic patterns of desminopathy associated with three novel mutations in the desmin gene. Neuromuscul Disord. 2007;17:443–450. doi: 10.1016/j.nmd.2007.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kley RA, Hellenbroich Y, van der Ven PFM, Fürst DO, Huebner A, Bruchertseifer V, et al. Clinical andmorphological phenotype of the filamin myopathy: a study of 31 German patients. Brain. 2007;130:3250–3264. doi: 10.1093/brain/awm271. [DOI] [PubMed] [Google Scholar]
- 23.Vrabie A, Goldfarb LG, Shatunov A, Nägele A, Fritz P, Kaczmarek I, et al. The enlarging spectrum of desminopathies: new morphological findings, eastward geographic spread, novel exon 3 desmin mutation. Acta Neuropathol. 2005;109:411–417. doi: 10.1007/s00401-005-0980-1. [DOI] [PubMed] [Google Scholar]
- 24.Goudeau B, Rodrigues-Lima F, Fischer D, Casteras-Simon M, Sambuughin N, de Visser M, et al. Variable pathogenic potentials of mutations located in the desmin alpha-helical domain. Hum Mutat. 2006;27:906–913. doi: 10.1002/humu.20351. [DOI] [PubMed] [Google Scholar]
- 25.Gallien S, Duriez E, Crone C, Kellmann M, Moehring T, Domon B. Targeted proteomic quantification on quadrupole-orbitrap mass spectrometer. Mol Cell Proteomics. 2012;11(12):1709–1723. doi: 10.1074/mcp.O112.019802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peterson AC, Russell JD, Bailey DJ, Westphall MS, Coon JJ. Parallel reaction monitoring for high resolution and high mass accuracy quantitative, targeted proteomics. Mol Cell Proteomics. 2012;11(11):1475–1488. doi: 10.1074/mcp.O112.020131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reidegeld KA, Eisenacher M, Kohl M, Chamrad D, Korting G, Bluggel M, et al. An easy-to-use Decoy Database Builder software tool, implementing different decoy strategies for false discovery rate calculation in automated MS/MS protein identifications. Proteomics. 2008;8:1129–1137. doi: 10.1002/pmic.200701073. [DOI] [PubMed] [Google Scholar]
- 28.Spitzer P, Klafki HW, Blennow K, Buee L, Esselmann H, Herruka SK, et al. cNEUPRO: novel biomarkers for neurodegenerative diseases. Int J Alzheimers Dis. 2010;2010 doi: 10.4061/2010/548145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Müller T, Loosse C, Schrötter A, Schnabel A, Helling S, Egensperger R, et al. The AICD interacting protein DAB1 is up-regulated in Alzheimer frontal cortex brain samples and causes deregulation of proteins involved in gene expression changes. Curr Alzheimer Res. 2011;8:573–582. doi: 10.2174/156720511796391827. [DOI] [PubMed] [Google Scholar]
- 30.Bennardini F, Wrzosek A, Chiesi M. Alpha B-crystallin in cardiac tissue. Association with actin and desmin filaments. Circ Res. 1992;71:288–294. doi: 10.1161/01.res.71.2.288. [DOI] [PubMed] [Google Scholar]
- 31.Steinert PM, Chou YH, Prahlad V, Parry DA, Marekov LN, Wu KC, et al. A high molecular weight intermediate filament-associated protein in BHK-21 cells is nestin, a type VI intermediate filament protein. Limited co-assembly in vitro to form heteropolymers with type III vimentin and type IV alpha-internexin. J Biol Chem. 1999;274:9881–9890. doi: 10.1074/jbc.274.14.9881. [DOI] [PubMed] [Google Scholar]
- 32.van der Ven PFM, Ehler E, Vakeel P, Eulitz S, Schenk JA, Milting H, et al. Unusual splicing events result in distinct Xin isoforms that associate differentially with filamin c and Mena/VASP. Exp Cell Res. 2006;312:2154–2167. doi: 10.1016/j.yexcr.2006.03.015. [DOI] [PubMed] [Google Scholar]
- 33.Schröder R, Vrabie A, Goebel HH. Primary desminopathies. J Cell Mol Med. 2007;11:416–426. doi: 10.1111/j.1582-4934.2007.00057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Niederländer N, Fayein NA, Auffray C, Pomiès P. Characterization of a new human isoform of the enigma homolog family specifically expressed in skeletalmuscle. Biochem Biophys Res Commun. 2004;325:1304–1311. doi: 10.1016/j.bbrc.2004.10.178. [DOI] [PubMed] [Google Scholar]
- 35.Price MG. Skelemins: cytoskeletal proteins located at the periphery of M-discs in mammalian striated muscle. J Cell Biol. 1987;104:1325–1336. doi: 10.1083/jcb.104.5.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kley RA, Serdaroglu-Oflazer P, Leber Y, Odgerel Z, van der Ven PFM, Olivé M, et al. Pathophysiology of protein aggregation and extended phenotyping in filaminopathy. Brain. 2012;135:2642–2660. doi: 10.1093/brain/aws200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ferrer I, Martín B, Castaño JG, Lucas JJ, Moreno D, Olivé M. Proteasomal expression, induction of immunoproteasome subunits, and local MHC class I presentation in myofibrillar myopathy and inclusion body myositis. J Neuropathol Exp Neurol. 2004;63:484–498. doi: 10.1093/jnen/63.5.484. [DOI] [PubMed] [Google Scholar]
- 38.Olivé M, van Leeuwen FW, Janué A, Moreno D, Torrejón-Escribano B, Ferrer I. Expression ofmutant ubiquitin (UBB + 1) and p62 in myotilinopathies and desminopathies. Neuropathol Appl Neurobiol. 2008;34:76–87. doi: 10.1111/j.1365-2990.2007.00864.x. [DOI] [PubMed] [Google Scholar]
- 39.Geeraert C, Ratier A, Pfisterer SG, Perdiz D, Cantaloube I, Rouault A, et al. Starvation-induced hyperacetylation of tubulin is required for the stimulation of autophagy by nutrient deprivation. J Biol Chem. 2010;285:24184–24194. doi: 10.1074/jbc.M109.091553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hack L. On how to retain experienced RNs. Can Nurse. 1998;94:9–10. [PubMed] [Google Scholar]
- 41.Nishida K, Yamaguchi O, Otsu K. Crosstalk between autophagy and apoptosis in heart disease. Circ Res. 2008;103:343–351. doi: 10.1161/CIRCRESAHA.108.175448. [DOI] [PubMed] [Google Scholar]
- 42.Platini F, Pérez-Tomás R, Ambrosio S, Tessitore L. Understanding autophagy in cell death control. Curr Pharm Des. 2010;16:101–113. doi: 10.2174/138161210789941810. [DOI] [PubMed] [Google Scholar]
- 43.Selcen D, Engel AG. Myofibrillar myopathy caused by novel dominant negative alpha B-crystallin mutations. Ann Neurol. 2003;54:804–810. doi: 10.1002/ana.10767. [DOI] [PubMed] [Google Scholar]
- 44.Selcen D, Muntoni F, Burton BK, Pegoraro E, Sewry C, Bite AV, et al. Mutation in BAG3 causes severe dominant childhood muscular dystrophy. Ann Neurol. 2009;65:83–89. doi: 10.1002/ana.21553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Amato AA, Jackson CE, Lampkin S, Kagan-Hallet K. Myofibrillar myopathy: no evidence of apoptosis by TUNEL. Neurology. 1999;52:861–863. doi: 10.1212/wnl.52.4.861. [DOI] [PubMed] [Google Scholar]
- 46.Tibbetts RS, Cortez D, Brumbaugh KM, Scully R, Livingston D, Elledge SJ, et al. Functional interactions between BRCA1 and the checkpoint kinase ATR during genotoxic stress. Genes Dev. 2000;14:2989–3002. doi: 10.1101/gad.851000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Unkrüer B, Pekcec A, Fuest C, Wehmeyer A, Balda MS, Horn A, et al. Cellular localization of Y-box binding protein 1 in brain tissue of rats, macaques, and humans. BMC Neurosci. 2009;10:28. doi: 10.1186/1471-2202-10-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kohno K, Izumi H, Uchiumi T, Ashizuka M, Kuwano M. The pleiotropic functions of the Y-box-binding protein, YB-1. Bioessays. 2003;25:691–698. doi: 10.1002/bies.10300. [DOI] [PubMed] [Google Scholar]
- 49.Saito Y, Nakagami H, Azuma N, Hirata S, Sanada F, Taniyama Y, et al. Critical roles of cold shock domain protein A as an endogenous angiogenesis inhibitor in skeletal muscle. Antioxid Redox Signal. 2011;15:2109–2120. doi: 10.1089/ars.2010.3714. [DOI] [PubMed] [Google Scholar]
- 50.Bär H, Mücke N, Kostareva A, Sjöberg G, Aebi U, Herrmann H. Severe muscle disease-causing desmin mutations interfere with in vitro filament assembly at distinct stages. Proc Natl Acad Sci U S A. 2005;102:15099–15104. doi: 10.1073/pnas.0504568102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Feldkirchner S, Schessl J, Müller S, Schoser B, Hanisch F-G. Patient-specific protein aggregates in myofibrillar myopathies: laser microdissection and differential proteomics for identification of plaque components. Proteomics. 2012;12(23–24):3598–3609. doi: 10.1002/pmic.201100559. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.