Abstract
The glycolytic protein, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), has a vast array of extraglycolytic cellular functions, including interactions with nucleic acids. GAPDH has been implicated in the translocation of transfer RNA (tRNA), the regulation of cellular messenger RNA (mRNA) stability and translation, as well as the regulation of replication and gene expression of many single-stranded RNA viruses. A growing body of evidence supports GAPDH–RNA interactions serving as part of a larger coordination between intermediary metabolism and RNA biogenesis. Despite the established role of GAPDH in nucleic acid regulation, it is still unclear how and where GAPDH binds to its RNA targets, highlighted by the absence of any conserved RNA-binding sequences. This review will summarize our current understanding of GAPDH-mediated regulation of RNA function.
INTRODUCTION
Glyceraldehyde-3-phosphate dehydrogenase (GAPDH, E.C. 1.2.1.12) was initially characterized as a homotetrameric oxidoreductase playing a critical role in intermediary metabolism, wherein it catalyzes the sixth step of glycolysis by converting glyceraldehyde-3-phosphate (G3P) into 1,3-bispho-sphoglycerate. GAPDH is a multifunctional protein implicated in a variety of other cellular processes beyond metabolism, including membrane fusion,1 cytoskeletal dynamics,2,3 receptor-mediated cell signaling,4 heme metabolism,5,6 apoptosis,7,8 DNA replication and repair,9-11 and maintenance of genomic integrity.12-14 In addition, GAPDH has a multiplicity of roles in dictating RNA localization, catalysis, translation, and replication (reviewed in Refs 15–22).
Mirroring this multitude of cellular functions, GAPDH has been shown to localize to various cell compartments. GAPDH is predominantly cytoplasmic, often associated with polysomes and cytoskeletal components, but is also found in the nucleus and in the mitochondria.20 Its cytoplasmic distribution is due in part to an exportin 1-binding site (residues 259–271), which directs its nuclear export in the absence of stress.23 Conversely, GAPDH responds to various stresses by associating to effector proteins and translocating to the nucleus, leading to apoptosis.7,8,23-25 Other studies have shown that nuclear localization of GAPDH is regulated by a cell-cycle dependent mechanism, and that nuclear GAPDH may differ from cyto-plasmic GAPDH.12 In addition, dimeric/non-native forms of nuclear GAPDH12,26 have been observed, suggesting that GAPDH functions may be correlated with discrete structural determinants. The mechanisms by which GAPDH switches among these various functions and locations in the cell are unknown, but may depend on cell status, GAPDH posttranslational modifications, and its interaction partners.
An important extraglycolytic function of GAPDH is its binding to nucleic acids. GAPDH binding to single-stranded DNA was first reported in 1977 by Perucho et al.27 Shortly after, GAPDH was found to bind to a wide range of DNAs and RNAs.28–30 Since then, GAPDH has unambiguously emerged as a noncanonical RNA-binding protein (RBP) that binds a multitude of RNA scaffolds, including transfer RNA (tRNA), cellular RNAs, ribo-zymes, and viral RNAs. The diversity of GAPDH-RNA interactions is paralleled by the variety of their effects on RNA function.
In this review, we will survey the RNA-binding function of GAPDH, its possible involvement in the regulation of protein expression, the potential link between cellular energetics and RNA biogenesis, and the possible RNA-binding sites of GAPDH.
MESSENGER RNA-MEDIATED CONTROL OF GENE EXPRESSION
Dynamic control of gene expression levels is essential for cellular responses to internal and external stimuli.31,32 Posttranscriptional control of eukaryotic gene expression comprises several levels of regulation including messenger RNA (mRNA) processing, export, turnover, and translation such that steady-state mRNA levels are not solely determined by the rate of mRNA synthesis. The fate of mRNA transcripts is determined in part by adenine–uridine rich cis-acting regulatory elements (AREs) located within the 3′ untranslated regions (3′ UTRs)33 of cytokines, transcription factors, and proto-oncogenes.34 AREs serve as binding sites for ARE-binding proteins (AUBPs), which are trans-acting factors in the regulation of ARE-dependent mRNA turnover and translation.35-37 AUBP binding can decrease, as in the case of Tristetraprolin (TTP),38 or enhance, as in the case of HuR39,40 and Hsp70,41 mRNA stability and may induce the formation of a complex of proteins at the ARE.42,43 Emerging evidence suggests that some AUBPs, such as AUF-1,44 can either stabilize or destabilize transcripts, depending on the specific mRNA bound.45 In addition, as several AUBPs can target the same AREs, the enhancement or inhibition of mRNA stability and translation may depend on the balance between the different proteins bound.46–49
RBPs are implicated in a number of human diseases, such as cancers, metabolic disorders, neuropathies, and muscular atrophies.50–52 These proteins are characterized by a multitude of functional domains and sequence motifs, including the RNA-binding domain (RBD) or RNA recognition motif (RRM), K-Homology domain, cold shock domain, RGG (Arg-Gly-Gly) box, Sm domain, zinc-finger domain, double-stranded RRM, PUF repeat, and arginine rich motif (reviewed in Refs 53-55). However, many RBPs, especially those with multiple cellular functions, do not possess these motifs and have been identified as noncanonical RBPs,56-59 as is the case for GAPDH. Other metabolic enzymes, including aconitase,60 aldolase,3 lactate dehydrogenase (LDH),43 phosphoglycerate kinase (PGK),61 glucose-6-phosphate dehydrogenase (G6PDH),62 glutamate dehydrogenase (GDH),63 and isocitrate dehydrogen-ase (IDH)64 have been shown to play similar roles (reviewed in Ref 65). For these noncanonical RBPs, little is known about their RNA-binding sites and the mechanisms allowing these enzymes to switch between their metabolic and RNA-binding functions.
GAPDH–RNA INTERACTIONS
Transfer RNA
Nuclear export of tRNAs into the cytoplasm is a carrier-mediated process,66 and GAPDH has been shown to be involved in tRNA translocation, in agreement with its nuclear and cytoplasmic localizations.67 Several mutations and deletions in tRNA impaired GAPDH binding, thus demonstrating the key roles of the T-stem, anticodon, and variable loops in this specific interaction. Binding between GAPDH and in vitro transcribed Escherichia coli tRNAtyr and yeast tRNAser occurred with high affinity (apparent KD of 18 nM), and was disrupted by high concentrations of NAD+, suggesting that the Rossman fold may be part of the tRNA-binding site.67 Raman spectroscopy and circular dichroism characterization of GAPDH-tRNAphe complex also alluded to a potential role of the Rossman fold in the interaction (vide infra).68
Cellular RNAs
There are numerous reports of interactions between GAPDH and cellular RNAs, both in vitro and in cells (Table 1). Many of these studies rely on in vitro qualitative experiments including UV cross-linking, native PAGE, and electrophoretic mobility shift assays (EMSAs), but few report quantitative measurements of binding affinity, for example via fluorescence anisotropy. In cells, characterizations include co-immunoprecipitation, ribonucleoprotein immunoprecipitation, RNA-affinity pull-down, mRNA decay kinetics, and luciferase assays. Only a handful of these studies characterized the effect of GAPDH binding on the RNA structure. The direct biological consequences of many of these interactions have yet to be elucidated. Regardless, the diversity of the RNAs interacting with GAPDH allows for a greater appreciation of the intricacies of this complex enzyme in its cellular environment.
TABLE 1.
Cellular RNAs Shown to Interact with GAPDH
| RNA Stability |
Protein Expression |
||||||
|---|---|---|---|---|---|---|---|
| Translational Efficiency |
Abundance |
||||||
| RNA | Increase | Decrease | Increase | Decrease | Upregulated | Downregulated | Reference |
| tRNA | 67 – 69 | ||||||
| IFN-γ | X | X | 69,70 | ||||
| c-myc | 69 | ||||||
| GM-CSF | 69 | ||||||
| IL-2 | X | X | 69,70 | ||||
| CSF-1 | X | X | 71,72 | ||||
| Cox-2 | X | X | 73 | ||||
| CTGF/CCN-2 | X | X | 74 | ||||
| ET-1 | ? | ? | X | 75 | |||
| AT1R | X | X | 76 | ||||
| TNF-α | 77 | ||||||
| SCN1A | X | X | 78 | ||||
| TNF-α ribozyme | X | 79 | |||||
| TERC | 14 | ||||||
| MyHC | 3 | ||||||
| GLUT-1 | 62 | ||||||
GAPDH, glyceraldehyde-3-phosphate dehydrogenase; tRNA, transfer RNA; IFN-γ, interferon-γ; GM-CSF, granulocyte macrophage colony-stimulating factor; IL-2, interleukin-2; CSF-1, colony-stimulating factor-1; Cox-2, cyclooxygenase-2; CTGF/CCN-2, connective tissue growth factor; ET-1, endothelin-1; AT1R, angiotensin II type I receptor; TNF-α, tumor necrosis factor-α; TERC, telomerase RNA component; MyHC, myosin heavy chain; GLUT-1, glucose transporter.
GAPDH is an AUBP
Nagy and Rigby were the first to demonstrate the proclivity of GAPDH binding to AU-rich RNA.69 GAPDH was shown to bind AU-rich RNA probes derived from the 3′ UTR of interferon-γ (IFN-γ), c-myc, granulocyte macrophage colony-stimulating factor (GM-CSF), and interleukin-2 (IL-2) in vitro.69 Other AU-rich mRNA targets of GAPDH include colony-stimulating factor-1 (CSF-1),71 cyclooxygen-ase-2 (Cox-2),73 connective tissue growth factor (CTGF/CCN-2),74 endothelin-1 (ET-1),75 and angiotensin II type I receptor (AT1R).76 In our laboratory, we have recently shown that GAPDH binds to the core ARE from tumor necrosis factor-α (TNF-α) 3′ UTR mRNA in vitro.77 We showed qualitatively (EMSA) and quantitatively (fluorescence anisotropy) that GAPDH binding to the ARE-probe occurred via a sequential two-step binding mechanism yielding a high-affinity complex (KD1 = 97 nM) and a low-affinity complex (KD2 = 1.4 μM). To our knowledge, this is the first identification of multiple ribonucleoprotein complexes between GAPDH and a cellular mRNA. While the ARE-RNA targets of GAPDH differ in sequence and length, they all contain three to eight AUUUA pentanucleotide motifs, required for strong binding.69 The biological relevance of some of these interactions was confirmed in cells, as discussed below.
(De)stabilization of mRNA Transcripts by GAPDH
GAPDH was found to modulate the stability of mRNA transcripts and regulate protein expression for genes important in cancer (CSF-1, Cox-2, CTGF/CCN-2), in cardiovascular homeostasis (ET-1), and in brain function (SCN1A). GAPDH binding to these mRNAs may either promote disease progression (CSF-1, CCN2), or alleviate disease conditions (Cox-2, ET-1, SCN1A), suggesting that targeting these interactions may have important therapeutic applications.
GAPDH was shown to be overexpressed in ovarian cancer cells and interact with the ARE within the 3′ UTR of CSF-1.71 Small interfering RNA (siRNA)-mediated downregulation of GAPDH resulted in decreased stability of the mRNA transcript, concomitant with decreased CSF-1 mRNA and protein levels. These results suggested that GAPDH binding promoted CSF-1 mRNA stabilization and increased production of CSF-1 protein,72 both of which are associated with progression or recurrence of the disease and poor prognosis. Similarly, GAPDH was shown to increase the stability of the CCN2 mRNA via binding to the cis-acting element of structure-anchored repression (CAESAR).74 Hypoxia-induced gene expression of CCN2 is post-transcriptionally modulated by CAESAR, which is an ARE-destabilizing motif in the 3′ UTR of CCN2 mRNA.80,81 In hypoxic conditions, GAPDH binding to CAESAR leads to CCN2 overexpression and may contribute to its key role in tumor angiogenesis and bone metastasis.
In contrast, GAPDH was shown to destabilize the mRNA of ET-1, Cox-2, and SCN1A. GAPDH associates in vitro and in cells with the 3′ UTR of ET-1 mRNA.75 Partial knockdown of GAPDH via siRNA treatment resulted in increased ET-1 mRNA and protein levels in cells and a modest increase in reporter expression, suggesting that GAPDH mediates the 3′ UTR-dependent downregulation of ET-1 mRNA.82 Notably, ET-1 mRNA half-lives were indirectly determined from luciferase activity, which also depends on translational control. Thus, the exact mechanism by which GAPDH regulates ET-1 expression may need further examination. Regardless, as increased ET-1 levels are associated with many diseases,83,84 controlling ET-1 production via modulation of its mRNA stability offers a promising novel therapeutic strategy.85 Similarly, siRNA-mediated knockdown of GAPDH lead to a moderate increase in Cox-2 mRNA and protein levels after lipopolysac-charide induction, suggesting that GAPDH may destabilize the Cox-2 mRNA transcript.73 Native PAGE gel-shift assays suggested that GAPDH binding to the core ARE of the Cox-2 mRNA 3′ UTR may be regulated by cellular GSH/GSSG ratios and GAPDH localization, however, the biological consequences of this interaction remain to be determined. Finally, in neurons, the voltage-gated sodium channel type 1 a subunit (NaV 1.1) is encoded by the SCN1A gene. Mutations within this gene have been linked to migraine, epilepsy, autism, and Dravet syndrome.86 The C1794U mutation in the 3′ UTR of SCN1A mRNA was shown to affect protein expression and mRNA stability, possibly via changes in predicted mRNA secondary structures. GAPDH was shown to selectively bind to the U-rich mutant 3′ UTR and to negatively regulate protein expression and mRNA stability.78
Translational Regulation by GAPDH
In two cases, GAPDH was found to regulate protein expression at the translational level and not by affecting mRNA stability. GAPDH was found to be a key mediator in the enhancement of effector function of activated T cells via aerobic glycolysis by inhibiting the translation of IFN-γ and IL-2.70 In activated T cells deprived of aerobic glycolysis, GAPDH immu-noprecipitation showed its association with the ARE of the IFN-γ mRNA 3′ UTR. GAPDH binding inhibited IFN-γ protein production and subsequent effector function in T cells.70 These results further suggested that GAPDH may switch between its various functions depending on the energetic status of the cell (vide infra). Similarly, GAPDH was shown to regulate the translation of AT1R, which plays a crucial role in cardiovascular homeostasis. GAPDH binds the AT1R 3′ UTR in vitro and in cells,76 and decreases AT1R expression by inhibition of AT1R translation. In addition, GAPDH mediates the effects of oxidative stress on AT1R expression via its decreased expression and binding affinity to the AT1R 3′ UTR.
Importantly, for both IFN-γ and AT1R, regulating the levels of GAPDH may enhance the efficacy of immunotherapy treatments, help control aberrant inflammation, and restore healthy levels of AT1R after oxidative stress.
Additional Interactions
GAPDH was found to bind the hammerhead ribozyme directed against tumor necrosis factor-α (TNF-α-Rz) in peripheral blood mononuclear extracts. Binding of GAPDH to TNF-α-Rz was much stronger than binding to tRNA or ARE-probes and was inhibited by the NAD+ cofactor.79 This interaction protected TNF-α-Rz from cellular degradation, and enhanced its cleavage activity possibly due to GAPDH unfolding activity.
Under stress, GAPDH has been shown to travel to the nucleus to act as either a protective12,13 or apoptotic agent.7,8,23-25 In the nucleus of noncancerous cells, GAPDH is involved in the maintenance of telomeres, functioning in antiaging and cell protective capacities.12,13 In contrast, in breast cancer cells overexpressing GAPDH, cell senescence occurred via GAPDH inhibition of telomerase.14 GAPDH binding to the telomerase RNA component (TERC) was thought to occur via the Rossman fold, but did not directly cause inhibition. Instead, TERC binding promoted inhibition of the telomerase reverse transcriptase (TERT) by the GAPDH C-terminal domain.14 These results demonstrate that multiple regions of the protein can be utilized in tandem to perform specific tasks.
GAPDH was also shown to interact with the 3′ UTR of myosin heavy chain (MyHC) mRNA, mostly in skeletal muscle cells.3 The role of this interaction has not been established, although it was proposed that it may serve a localization function, as GAPDH is also known to interact with both microtubules2 and actin filaments.87
It is intriguing that GAPDH binding to various cellular mRNAs can result in either stabilization or destabilization of the specific target. A structure–function model has been proposed to explain this apparent discrepancy, whereby the catalytic domain may play an active role in RNA binding.19 In addition, several pathways have been described for the posttranscriptional control of gene expression.49 However, the exact mechanisms by which GAPDH affects mRNA stability and/or protein expression remain unknown. The dual role of GAPDH in regulating mRNA decay is reminiscent of that played by the well-established AUBP, AUF-1.88 It has been proposed that the fate of mRNA depends on the balance between stabilizing and destabilizing AUBPs. This mechanism is very attractive for GAPDH, as other AUBPs bind to the same AREs.46,72
Viral RNAs
GAPDH has been shown to either protect cells against viral infection or to promote virus progression via interaction with the viral genome. While cells possess intrinsic antiviral properties via host protein-viral nucleic acid interactions, many viruses are also known to co-opt host cell factors to assist in gene expression and replication (Table 2).
TABLE 2.
GAPDH Interactions with Viral RNAs
| Virus | Virus Family | Genetic Material | GAPDH Role | Reference |
|---|---|---|---|---|
| BaMV | Alphaflexiviridae | (+) ssRNA | Protective | 89 |
| satBaMV | BaMV satellite | (+) ssRNA | Protective | 89 |
| HAV | Picornaviridae | (+) ssRNA | Protective | 90 – 93 |
| TGEV | Coronoviridae | (+) ssRNA | Protective | 94 |
| HCV | Flaviviridae | (+) ssRNA | Co-opted | 95 |
| JEV | Flaviviridae | (+) ssRNA | Unknown | 96 |
| TBSV | Tombusviridae | (+) ssRNA | Co-opted | 97,98 |
| HDV | HBV satellite | (−) ssRNA | Co-opted | 99 |
| HPIV3 | Paramoxyviridae | (−) ssRNA | Co-opted | 100 |
GAPDH, glyceraldehyde-3-phosphate dehydrogenase; BaMV, bamboo mosaic virus; HAV, hepatitis A virus; TGEV, transmissible gastroenteritis virus; HCV, hepatitis C virus; JEV, Japanese encephalitis virus; TBSV, tomato bushy stunt virus; HDV, hepatitis D virus; HPIV, human parainfluenza virus; ssRNA, single-stranded RNA.
Antiviral Functions of GAPDH
The means by which GAPDH can function in a cell protective capacity varies depending on the specific virus. The majority of RNA viruses with which GAPDH interacts have single-stranded positive sense genomes, which replicate via asymmetric synthesis of positive sense progeny RNA.101 GAPDH shows preferential binding to the 3′ UTR of positive sense progeny RNA of both the bamboo mosaic virus (BaMV) and its satellite virus satBaMV, reducing the positive to negative ratio and diminishing viral propaga-tion.89 Studies with GAPDH knockdown in Nicotiana benthamiana showed that GAPDH binding to positive sense progeny RNA prevents the transcription of negative sense RNA and halts the replication process.89
Additional GAPDH antiviral functions arise from competing with other cellular proteins for RNA binding as seen with the hepatitis A virus (HAV). GAPDH has been found to bind to its 3′ UTR90 and its 5′ UTR and compete with the polypyrimidine tract-binding protein (PTB).91-93 While PTB binding increased gene expression, GAPDH binding decreased gene expression, notably through its ability to destabilize the RNA secondary structure.92 In the case of transmissible gastroenteritis virus (TGEV), siRNA-mediated gene silencing showed a threefold increase in replicon activity following GAPDH knockdown.94 It is not clear whether GAPDH’s protective role arises from competitive binding with other host proteins, including PTB, or preferential binding of the positive sense progeny RNA as seen with BaMV.
Viral Subversion of GAPDH
Many RNA viruses co-opt GAPDH for use in viral progression such as the positive sense single-stranded RNA (ssRNA) hepatitis C virus (HCV),95 Japanese encephalitis virus (JEV),96 and tomato bushy stunt virus (TBSV),97,98 and the negative sense ssRNA hepatitis D virus (HDV),99 and human parainfluenza virus (HPIV).100
In the case of positive sense ssRNA viruses, GAPDH can be co-opted to either stabilize the viral RNA or alter the positive:negative progeny ratio. During HCV infection, GAPDH stabilizes the viral RNA via binding to its 3′ UTR.95 For JEV infection, GAPDH was shown to preferentially bind negative sense RNA, but the biological consequences of the interaction remain unclear.96 For TBSV, GAPDH was shown to sequester negative sense RNA to the replicase complex and promote the release of positive sense progeny to support viral replication.97,98
HDV is a satellite of Hepatitis B virus and one of two negative sense ssRNA viruses known to bind GAPDH via the UC-rich regions of the genomic and antigenomic RNA.99 As seen with TNF-α ribozyme, GAPDH binding was observed to increase the intrinsic ribozyme activity of the HDV RNA.99 Finally, GAPDH was shown to bind the 3′ UTR of the genomic sequence of HPIV3 in vitro and in cells.100 For many of the GAPDH-viral RNA interactions, including BaMV, HAV, JEV, and HDV, several ribonucleoprotein complexes were observed by EMSA. As in the case of GAPDH-TNF-α ARE interactions,77 the biological significance of these multicomplexes has not yet been elucidated.
GAPDH: A MISSING LINK BETWEEN RNA BIOGENESIS AND ENERGY METABOLISM?
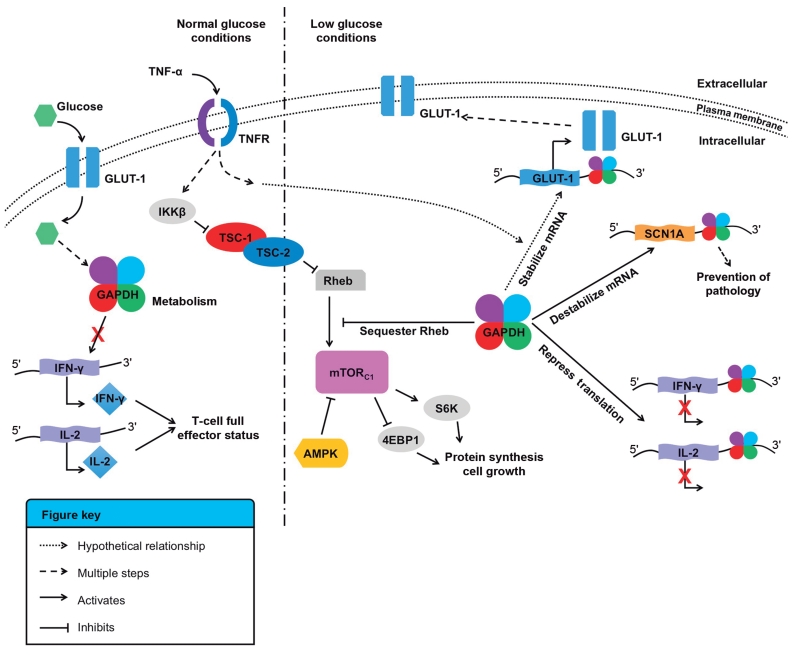
Evidence has emerged to further delineate the complexities of the involvement of GAPDH and other metabolic enzymes in the regulation of RNA translation. The REM (RNA, Enzyme, Metabolite) hypothesis notes that many intermediary metabolic enzymes possess RNA-binding functions. As such, there may be posttranscriptional regulatory networks that exist between metabolism and gene expression based on RNA, enzyme, and metabolite interactions.102 The first example of a link between RNA biogenesis and intermediary metabolism came from the finding that SRN1, a suppressor of RNA processing in yeast, relieves glucose repression.103 Such links have also been made with GAPDH binding to the glucose transporter, GLUT-1, mRNA. The expression of GLUT-1 is regulated via alterations in mRNA turnover following treatment with TNF-α.104,105 GAPDH was shown to bind to the AU-rich region of the GLUT-1 mRNA 3′ UTR in vitro.62 While the biological consequences of this interaction have yet to be determined, it is tempting to speculate that GAPDH could regulate the expression of GLUT-1 in response to nutrient levels (Figure 1). This hypothesis is reinforced by the fact that GAPDH was previously shown to regulate the function of mTOR as a function of glycolytic flux. Under low glucose conditions, GAPDH inhibited mTORC1 by binding to and sequestering the GTPase Rheb, preventing cell growth and proliferation.106
FIGURE 1.
Glyceraldehyde-3-phosphate dehydrogenase (GAPDH), metabolism, and RNA binding. Schematic for the proposed links between GAPDH, metabolism, glucose sensing, and RNA binding. Under normal glucose conditions, GAPDH is involved in glycolysis. IKKβ mediates tumor necrosis factor-α (TNFα)-dependent inhibition of the tuberous sclerosis complex (TSC1-TSC2) via phosphorylation, decreases its activity toward the G-protein Rheb (Ras homolog enriched in the brain), and activates the mTOR (mammalian target of rapamycin) complex 1 (mTORc1), which leads to protein synthesis and cell growth.106 Under low glucose conditions, GAPDH is involved in several pathways. First, GAPDH may stabilize the glucose transporter (GLUT-1) mRNA via binding to its AU-rich 3′ UTR.62 Second, GAPDH binds to the 3′ UTR of interferon-γ (IFN-γ) and interleukin-2 (IL-2) and decreases translation of both cytokines, preventing activated T cells to reach full effector status.70 Third, GAPDH sequesters Rheb and prevents mTORc1 activation.106 Finally, in ketogenic diets, GAPDH may be free from glycolysis and able to destabilize the SCN1A mRNA to reduce translation of the sodium transporter NaV 1.1, and alleviate conditions linked to Dravet syndrome.78
Several other lines of evidence support the hypothesized link between metabolism and mRNA binding by GAPDH, including the fact that ketogenic diets (high in fat, low in carbohydrates, shifting metabolism from glycolysis to ketosis) reduce the effects of Dravet syndrome and epilepsy. GAPDH was shown to bind mutant SCN1A mRNA and decrease the translation of the sodium transporter NaV 1.1 (vide supra).78 It may be that in a ketogenic environment, GAPDH would be free from the demands of glycolysis and could more readily bind to SCN1A transcripts and alter their translation (Figure 1). This hypothesis remains to be tested.
Many normal cells as well as tumor cells preferentially utilize glycolysis over oxidative phosphoryla-tion even under normoxic conditions. This could be due to the increased energy demand of highly proliferating cells, but it could also have additional utilities. Indeed, activated T cells rely on aerobic glycolysis to obtain full effector status via mRNA translation of IFN-γ and IL-2 cytokines (vide supra). In cells not relying on glycolysis, GAPDH binding to IFN-γ and IL-2 mRNA increased and correlated with decreased cytokine production, suggesting that GAPDH binding to the AU-rich 3′ UTR of either mRNA could prevent their translation.70 It is thought that in cells relying on glycolysis or in cells deprived of nutrients by neighboring cancer cells, GAPDH cannot bind to and inhibit cytokine translation. Addition of glucose reverses this effect. In this way, GAPDH would serve as a metabolic sensor to coordinate efficient glycolysis to T cell function70 (Figure 1). This hypothesis requires further examination, as it may provide additional means to avoid aberrant inflammation or to restore T cell effector function.
In summary, GAPDH (and other metabolic enzymes) could provide the missing link between nutrient sensing and regulation of gene expression, crucial to several cellular pathways. However, more studies are required to confirm this hypothesis.
STRUCTURAL DETERMINANTS OF GAPDH
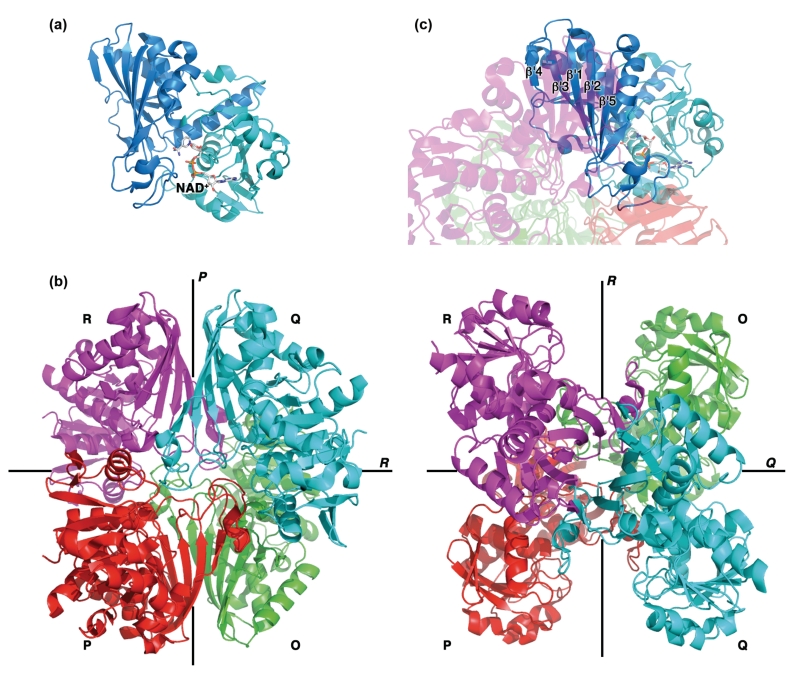
Human GAPDH is expressed as a 36 kDa polypeptide of 335 amino acids that can be divided into two domains: a Rossman fold/cofactor-binding domain (residues 1–150 and 317–335) and a catalytic domain (residues 151–316; Figure 2(a)). The protein is highly conserved across all kingdoms of life and its enzymatic function requires a 144 kDa homotetramer that is best described as a dimer of dimers.107 The four subunits are labeled O, P, Q, and R with the pairs O/P and Q/R each forming a dimer (Figure 2 (b)). The identical subunits are structurally very similar to each other [average root mean squared deviation (RMSD) <0.3 Å] and are related to each other by three twofold symmetry axes P, Q, and R. The dimer interfaces are located along the P axis, while the tetra-mer interface (between dimers O/P and Q/R) is located along the R axis. The catalytic domains form the dimer interface via a five-stranded antiparallel β-sheet (Figure 2(c)), while the NAD+-binding domains participate in the tetrameric interface.
FIGURE 2.
Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) structural overview. (a) Each GAPDH subunit is comprised of two domains, the Rossman fold/cofactor-binding domain (cyan) and the catalytic domain (blue). (b) Tetrameric assembly of GAPDH (PDB code 4WNC). The four subunits (O-R) are related to one another by three twofold symmetry axes (P, Q, and R). (c) The dimer interface is formed by antiparallel five-stranded β-sheets from the R and Q (or P and O) subunits.
The Catalytic Domain and Putative RRM
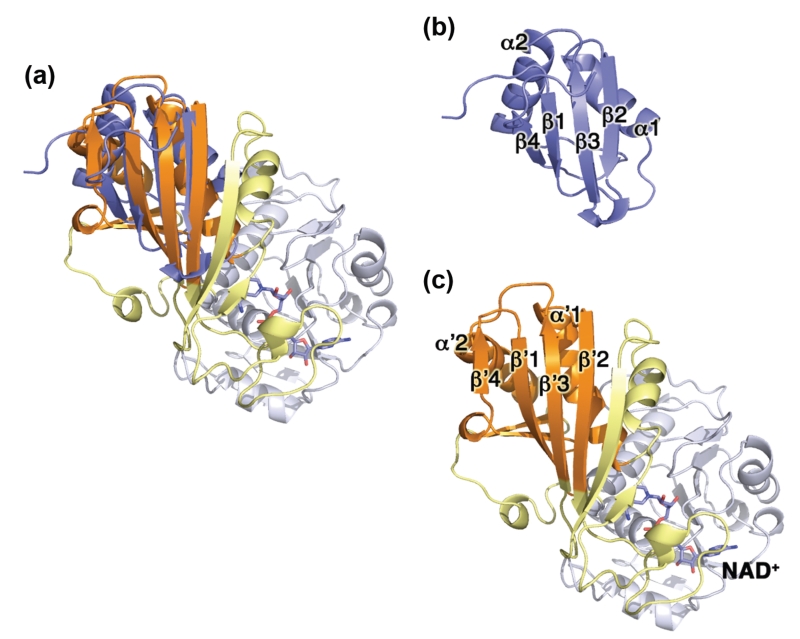
We performed a structural alignment108 between GAPDH (PDB code 4WNC) and the RRM1 from HnRNP L (PDB code 3R27), yielding an overall RMSD of 3.7 Å for 78 amino acids (our unpublished data and Figure 3). The RRM-like structural motif in GAPDH encompasses residues 152–178, 242–267, 274–277, and 291–315, and adopts a similar three-dimensional structure to the canonical RRM (βαββαβ fold), but shares no sequence similarity. Interestingly, GAPDH residues 303–308 were previously proposed as a putative tRNA-binding site.68 The RRM-like structural motif in GAPDH is located at the dimer interface, but is not positively charged or rich in aromatic residues, as seen for other RBPs. Because RRMs can also mediate protein–protein interactions and have been found in proteins that do not bind RNA,109 it remains to be seen whether this motif is involved in GAPDH binding to RNA (vide infra).
FIGURE 3.
Structural comparison of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and heterogeneous nuclear ribonucleoprotein L. (a) Structural overlay of monomeric GAPDH and HnRNP L (blue). The x-ray structures of HnRNP L and GAPDH are shown separately in (b) and (c) for clarity. (b) X-ray structure of the first RNA recognition motif (RRM) domain of HnRNP L (PDB code 3R27) showing the canonical βαββαβ motif. (c) Structure of the GAPDH monomer (PDB code 4WNC) showing the conserved structural motif (β′α′β′β′α′β′) in the catalytic domain. The NAD+-binding domain of GAPDH is shown in white, the catalytic domain is shown in yellow and orange (RRM-like subdomain).
NAD+-Binding Site and Oligomeric Interface
The structures of GAPDH from archaea, bacteria, and eukaryotes have been determined by x-ray crystallography, and show that the NAD+ cofactor is bound, despite not being added during protein purification. In mammalian GAPDH structures, two,110 three,77,107 or even four NAD+ molecules have been observed111-113 per homotetramer. In addition, we recently showed that GAPDH was present as a mixture of NAD+-bound and NAD+-free species in solution by nano-ESI/MS/MS studies.77 These results confirm that GAPDH binds NAD+ with high affinity, and that studies aiming at determining the effect of the cofactor should be done with NAD+-free GAPDH.
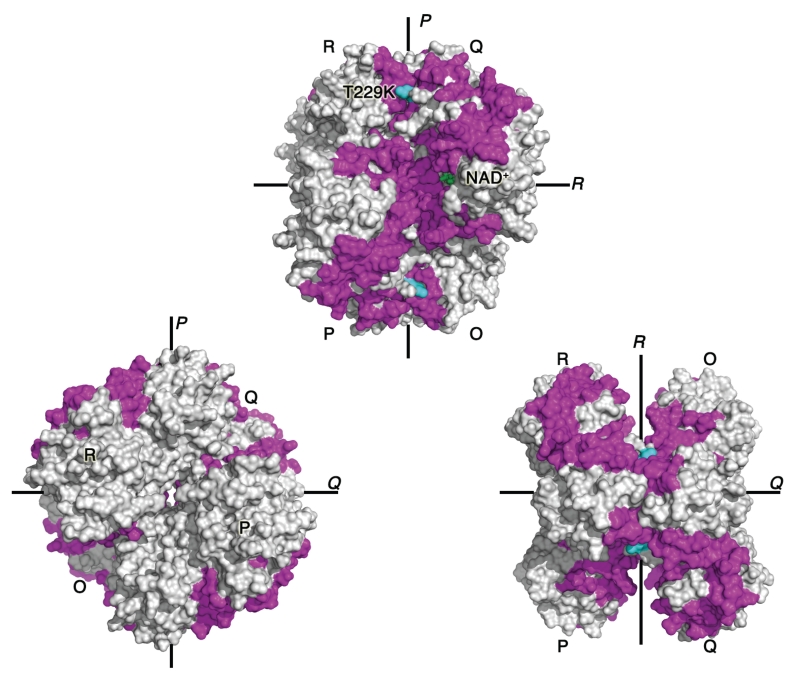
Our recent hydrogen–deuterium exchange mass spectrometry studies showed an unexpected connection between the NAD+-binding site and the dimer interface.77 A single mutation at the dimer interface promoted short-range and long-range structural changes and increased dynamics of protein regions at the dimer and tetramer interfaces, and in the distant NAD+-binding site (Figure 4). In addition, the mutation significantly decreased the amount of NAD+-bound GAPDH species present in solution. These results strongly suggest an unexpected link between GAPDH oligomeric interfaces and the cofactor-binding site.
FIGURE 4.
Interconnection between oligomeric interfaces and NAD+-binding site. Dimer-interface mutation T229K (cyan) induces a series of subtle conformational shifts that propagate throughout the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) tetramer. We showed that regions displaying increased solvent exchange upon mutation (magenta) are clustered along the P axis: dimer and tetramer interfaces, and NAD+-binding site (green sticks).77
THE ELUSIVE SEARCH FOR THE GAPDH RNA-BINDING SITE
GAPDH is undoubtedly an RBP, but it does not contain any canonical RNA-binding sequences. Despite over 30 years of research, the RNA-binding site remains elusive. Several GAPDH regions have been proposed as putative RNA-binding sites, including the Rossman fold, the positively charged substrate-binding groove, and the dimer interface.
The Rossman Fold
The Rossman fold, which binds NAD+, has often been heralded as the putative RNA-binding site of GAPDH (Figure 5(a)). Several lines of evidence support this hypothesis. A multitude of other metabolic enzymes also bind RNA, several of which are NAD+-dependent dehydrogenases with similar mononucleotide-binding domain 1 in the Rossman fold.115 The majority of studies discussed herein employ competition experiments demonstrating the ability of cofactor and substrate to decrease RNA binding in a concentration-dependent manner, implying that a shared binding site exists. This phenomenon has been observed with all RNAs shown to bind to GAPDH,67,69,73-75,79,89,90,92,115 except in one case where increasing NAD+ concentrations increased binding of GAPDH to ccn2 mRNA.74 However, many of these studies rely on UV cross-linking of the ribonucleoprotein complexes prior to EMSA.14,69,73,74,115 Because NAD+, NADH, and ATP strongly absorb at 260 nm, increasing concentrations of these molecules would dramatically reduce the cross-linking efficiency. As a result, the observed decreases in GAPDH-RNA complexes could be artifactual, at least in some of these cases. Other reports showed a similar trend, even in the absence of UV cross-linking,67,75,79 supporting a potential involvement of the Rossman fold in binding the RNA. Nevertheless, all studies used a large excess of NAD+ compared to protein and RNA to observe a significant effect on RNA binding, thus suggesting that the cofactor-binding site may only be part of the RNA-binding site. In addition, in light of our results suggesting a connection between the NAD+-binding site and the GAPDH-oligomeric interfaces,77 these studies would be unable to distinguish between a direct competition or an allosteric effect by NAD+.
FIGURE 5.
Proposed RNA-binding sites in glyceraldehyde-3-phosphate dehydrogenase (GAPDH). (a) Structure of the GAPDH monomer with the Rossman fold/NAD+-binding domain (cyan) and the catalytic domain (blue). RNA was proposed to bind in or near the NAD+-binding site. (b) Electrostatic potential of GAPDH mapped onto the solvent-accessible surface and colored by electrostatic potential with electropositive regions colored blue and electronegative regions colored red. The positively charged substrate grooves span the entire length of the GAPDH tetramer along the P-axis. (c) The dimer interface contains basic and aromatic residues that may play a role in RNA binding. Peptides that were proposed to be involved in RNA binding are highlighted in gray (peptide 252–260)114 and dark blue (residues 306–311).68
To narrow down the RNA-binding sites, several groups have used truncated or mutated GAPDH constructs to perform RNA-EMSAs, with different outcomes. Full length and GST-GAPDH1-151 were shown to interact with TERC, while the C-terminal construct GST-GAPDH148-335 did not.14 Similar results were obtained with the AU-rich IFN-γ 3′ UTR, where GST-GAPDH1-43 was shown to be sufficient for binding.69,115 In contrast, only full-length GAPDH was shown to bind to the AU-rich ET-1 3′ UTR.75 Studies with mutant GAPDH (D35A, Y45A, S51G) confirmed a potential role of the Rossman fold in TERC binding.14 Some of these mutations are key to the oligomeric interfaces, but any effects of the mutations on the structure of the protein were not discussed. In our laboratory, we have observed that several mutations, not directly involved in cofactor binding but present at the oligomeric interfaces, negatively impact GAPDH stability (unpublished data). Thus, RNA-binding activity of GAPDH truncations and mutations that may also affect protein stability, folding, oligomeric assembly, and activity, should be evaluated carefully.
Other NAD+-dependent metabolic enzymes can bind RNA as well,102,116 and some, including LDH, malate dehydrogenase, alcohol dehydrogenase, and G6PDH show similar sequence specificity for AREs in the mRNA 3′ UTR.43,62,104 This suggests that the Rossman fold could serve as a noncanonical RNA-binding site. Recent studies have shown that this fold was underrepresented in the identification of the mRNA interactome in proliferating HeLa cells, possibly because their metabolic status may prevent Rossman-containing proteins to lectively, these results suggest that while the Rossman fold clearly plays a role in RNA binding, its importance may be dependent upon the specific RNA, cell type, and metabolic state.
The Positive Substrate-Binding Groove
GAPDH contains two regions spanning about 70 Å forming the substrate-binding grooves, which are located along the homotetramer P axis and are rich in positively charged amino acids (Figure 5(b)). These positive grooves have been suggested as the sites for RNA (and DNA) binding,13,68 as they are wide enough (~25 Å) to accommodate an 18 Å-wide nucleic acid helix or loop.68 In general, protein–RNA-binding interactions include hydrophobic stacking (affinity), electrostatic interactions (specificity), and hydrogen bonds, as seen in most RNA-binding motifs.53 Aromatic residues, positively charged residues (Arg, Lys), polar residues (Asn, Gln), and negatively charged residues (Asp, Glu) are usually involved in protein interactions with ssRNA,55 while proteins that bind double-stranded DNA (dsDNA) or double-stranded RNA (dsRNA) usually contains extensive positively charged regions on their surface.55 It is not known yet whether GAPDH binds to ssRNA and/or dsRNA, but earlier studies suggested a preference for single-over double-stranded RNA.29 Notably, single-stranded AU-rich RNA generally adopts specific secondary structures that GAPDH could preferentially bind to,34,75,90 while dsRNA may contain specific nucleotide sequences important for GAPDH recognition. Finally, several studies have shown that the GAPDH substrate (G3P) competes with RNA binding,14,70,75 further supporting a role of the positive grooves in RNA binding.
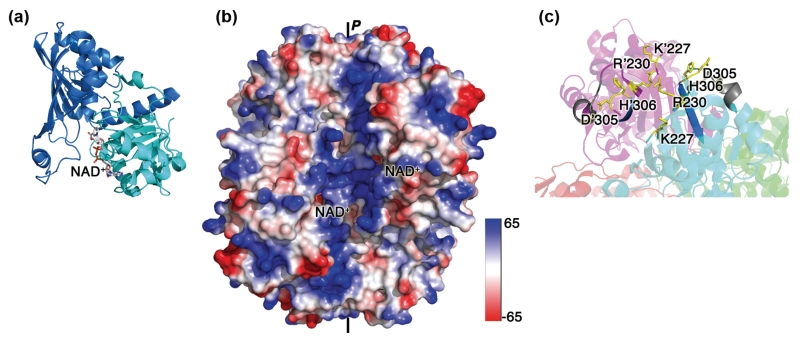
The Dimer Interface
The dimer interface of GAPDH has also been implicated in RNA binding (Figure 5(c)). Several lines of evidence support this hypothesis. First, it contains basic and aromatic residues (K227, R230, H305, F306). Second, the dimer-interface peptide 250–258 from yeast GAPDH (252–260 in human GAPDH) was cross-linked to an AU dinucleotide in cells as shown by mass spectrometry.114 This peptide is adjacent to residues 306–311, proposed to share sequence similarity to the RRM2 RBD,68 and is also located along the positive groove. Third, we have shown that a dimer-interface mutant impaired formation of complex 2 between GAPDH and the TNF-α ARE probe, and affected the structure of bound RNA.77 While more studies are needed to confirm whether the effect of the mutation is direct or indirect, our results suggested an involvement of the dimer interface in RNA binding.
Collectively, these studies suggest that the RNA-binding site in GAPDH would span a large area via a combination of the Rossman fold, the positive groove, and the dimer interface. This hypothesis is strongly supported by the interconnection of these regions in GAPDH.77 In addition, human GAPDH requires at least three AUUUA pentanucleotides for strong binding,69 supporting the hypothesis for an extended RNA-binding site in GAPDH. This is in contrast with the canonical RBDs that bind to shorter RNA sequences (2–8 nucleotides). However, many RBPs combine several RNA-binding motifs to bind longer RNA sequences with high affinity.55,109 Similarly, GAPDH could combine several protein regions to achieve high affinity for RNA binding. Structural studies will be required to unambiguously elucidate the RNA-binding site in GAPDH and the exact stoichiometry of the ribonucleoprotein complex.
POSTTRANSLATIONAL CONTROL OF GAPDH BINDING TO RNA
Posttranslational modifications would offer an elegant mechanism to allow GAPDH to switch between its metabolic and RNA-binding functions. Many studies have linked GAPDH redox sensitivity via the active site cysteine (C152) to regulate its RNA-related functions. For instance, binding of tRNA,69 IFN-γ,69 and CTGF/CCN274 mRNAs was increased in the presence of 2-mercaptoethanol and decreased in the presence of diamide. In addition, glutathionylation was shown to inhibit GAPDH binding to the 3′ UTRs of Cox-273 and ET-1,75 and cysteine oxidation reduced binding to TERC14 and AT1R mRNA,76 but increased binding to tRNA and DNA.26 These results suggest that the GAPDH redox state affects nucleic acid binding. However, studies evaluating possible structural consequences of such modifications that could alter RNA binding are needed, as GAPDH oxidation was shown to lead to disulfide-linked aggregation.118-120
The roles of other posttranslational modifications of GAPDH in its RNA-binding function have not yet been elucidated. Many of these modifications occur at or near the dimer interface, including acetylation,121,122 phosphorylation,123,124 and glyco-sylation.125,126 Thus, studies to determine whether these modifications affect GAPDH binding to RNA in vitro and in cells will be crucial. These modifications would offer GAPDH an elegant mechanism for switching between its various cellular functions as a function of the cell cycle and metabolic state.
In addition to posttranslational modifications, GAPDH switching between its glycolytic and RNA-binding functions could be promoted by its cellular localization, oligomerization state, interactions with other proteins, nutrient concentration (e.g., glucose), and redox and metabolic cell status. More research is necessary to elucidate the regulatory mechanisms allowing this abundant enzyme to perform the multitude of cellular functions described so far (see Introduction).
GAPDH–RNA INTERACTIONS ARE CONSERVED IN OTHER SPECIES
The GAPDH amino-acid sequence is highly conserved across all species. The key role of GAPDH in the regulation of gene expression via RNA binding was also observed in plants and yeast. In plants, cytoplasmic GAPDH was shown to regulate replication of phytopathogenic viruses by binding to their RNA (vide supra). In yeast, several GAPDH isoforms (GAPDH1-3) are present (65% sequence identity with human GAPDH) and all three were shown to bind AU-rich RNA. Early studies showed that the Saccharomyces cerevisiae basic GAPDH isoform (Sc GAPDH1) bound to poly(U) RNA and had a strong nucleic acid-helix destabilizing effect.29 The Sc GAPDH3 protein was shown to weakly bind AU-rich or Poly(A) RNA in vitro (KD of ~110–150 μM), but not Poly(U).127 In contrast to mammalian GAPDH, Sc GAPDH3 showed higher affinity for shorter RNA with only one or two AUUUA motifs. In yeast cells, the GAPDH2 isoform was identified in a cross-link to an AU dinucleotide by mass spectrometry.114 These results confirm that the RNA-binding function of GAPDH is conserved in other organisms.
CONCLUSION
GAPDH binds to a wide array of RNA molecules in vitro and in cells. Many of these interactions appear to be dependent upon cell type and metabolic and redox states. GAPDH-RNA interactions appear to have three distinct outcomes with respect to RNA fate: decreased transcript stability, increased transcript stability, or inhibited translation. The factors that determine these outcomes are not well understood, but may involve preventing or promoting interactions with endonucleases, competition with other RBPs and translation machinery, or binding to and acting on the RNA target itself. These interactions could then elicit a coordinated regulatory response that is intricately tied into the metabolic or redox state of the cell. The interacting RNAs identified thus far are most likely only a small subset of the number of interacting partners, as most studies to date have focused on the binding of one or a few specific RNAs or RNA fragments in vitro. A repository of GAPDH–RNA interactions (GAPDH-RNA interactome) in cells would provide a better understanding of the complexities of GAPDH function in health and disease as well as shed light on the link between RNA biogenesis and intermediary metabolism.
Additional information crucial to the complete understanding of GAPDH-RNA interactions includes the delineation of regulatory mechanisms and the effects of various GAPDH posttranslational modifications on RNA binding. Many of the RNA targets of GAPDH are also targets for canonical RBPs and microRNAs. Understanding the complex interplay between these competing interactions is key to elucidate the regulation of GAPDH-RNA interactions and how they dictate variable levels of posttranscriptional gene regulation.
Many key questions remain unanswered regarding specific GAPDH-RNA interactions, the effect of GAPDH binding on RNA fate and structure, and the stoichiometry of the GAPDH-RNA complex(es). To answer these questions, structural determination of a GAPDH-RNA complex is crucial. With this information, it may be possible to design small molecules, peptides, or microRNAs that mimic or disrupt the interaction between GAPDH and target mRNAs. As GAPDH regulates the expression of several proteins key to pathological conditions, modulation of mRNA levels via GAPDH may offer alternative strategies in the treatment of various diseases.
ACKNOWLEDGMENTS
We thank Richard Karpel for insightful discussions and input on this article. Work on GAPDH in our lab is supported by UMBC SRAIS. MRW is supported by National Institutes of Health Grant T32 GM066706.
Footnotes
Conflict of interest: The authors have declared no conflict of interest for this article.
FURTHER READING
- Muto Y, Yokoyama S. Structural insight into RNA recognition motifs: versatile molecular Lego building blocks for biological systems. WIREs RNA. 2012;3:229–46. doi: 10.1002/wrna.1107. doi:10.1002/wrna.1107. Epub 2012 Jan 25. Review. PubMed PMID: 22278943. [DOI] [PubMed] [Google Scholar]
- Beisang D, Bohjanen PR. Perspectives on the ARE as it turns 25 years old. WIREs RNA. 2012;3:719–731. doi: 10.1002/wrna.1125. doi:10.1002/wrna.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clingman CC, Ryder SP. Metabolite sensing in eukaryotic mRNA biology. WIREs RNA. 2013;4:387–396. doi: 10.1002/wrna.1167. doi:10.1002/wrna.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
REFERENCES
- 1.Glaser PE, Gross RW. Rapid plasmenylethanolamine-selective fusion of membrane bilayers catalyzed by an isoform of glyceraldehyde-3-phosphate dehydrogenase: discrimination between glycolytic and fusogenic roles of individual isoforms. Biochemistry. 1995;34:12193–12203. doi: 10.1021/bi00038a013. [DOI] [PubMed] [Google Scholar]
- 2.Muronetz VI, Wang ZX, Keith TJ, Knull HR, Srivastava DK. Binding constants and stoichiometries of glyceraldehyde 3-phosphate dehydrogenase-tubulin complexes. Arch Biochem Biophys. 1994;313:253–260. doi: 10.1006/abbi.1994.1385. [DOI] [PubMed] [Google Scholar]
- 3.Kiri A, Goldspink G. RNA-protein interactions of the 3′-untranslated regions of myosin heavy chain transcripts. J Muscle Res Cell Motil. 2002;23:119–129. doi: 10.1023/a:1020211729728. [DOI] [PubMed] [Google Scholar]
- 4.Raje CI, Kumar S, Harle A, Nanda JS, Raje M. The macrophage cell surface glyceraldehyde-3-phosphate dehydrogenase is a novel transferrin receptor. J Biol Chem. 2007;282:3252–3261. doi: 10.1074/jbc.M608328200. [DOI] [PubMed] [Google Scholar]
- 5.Hannibal L, Collins D, Brassard J, Chakravarti R, Vempati R, Dorlet P, Santolini J, Dawson JH, Stuehr DJ. Heme binding properties of glyceraldehyde-3-phosphate dehydrogenase. Biochemistry. 2012;51:8514–8529. doi: 10.1021/bi300863a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chakravarti R, Aulak KS, Fox PL, Stuehr DJ. GAPDH regulates cellular heme insertion into inducible nitric oxide synthase. Proc Natl Acad Sci. 2010;107:18004–18009. doi: 10.1073/pnas.1008133107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hara MR, Agrawal N, Kim SF, Cascio MB, Fujimuro M, Ozeki Y, Takahashi M, Cheah JH, Tankou SK, Hester LD, et al. S-nitrosylated GAPDH initiates apop-totic cell death by nuclear translocation following Siah1 binding. Nat Cell Biol. 2005;7:665–674. doi: 10.1038/ncb1268. [DOI] [PubMed] [Google Scholar]
- 8.Sen N, Hara MR, Kornberg MD, Cascio MB, Bae BI, Shahani N, Thomas B, Dawson TM, Dawson VL, Snyder SH, et al. Nitric oxide-induced nuclear GAPDH activates p300/CBP and mediates apoptosis. Nat Cell Biol. 2008;10:866–873. doi: 10.1038/ncb1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baxi MD, Vishwanatha JK. Uracil DNA-glycosylase/glyceraldehyde-3-phosphate dehydrogenase is an Ap4A binding protein. Biochemistry. 1995;34:9700–9707. doi: 10.1021/bi00030a007. [DOI] [PubMed] [Google Scholar]
- 10.Azam S, Jouvet N, Jilani A, Vongsamphanh R, Yang X, Yang S, Ramotar D. Human glyceraldehyde-3-phosphate dehydrogenase plays a direct role in reactivating oxidized forms of the DNA repair enzyme APE1. J Biol Chem. 2008;283:30632–30641. doi: 10.1074/jbc.M801401200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meyer-Siegler K, Mauro DJ, Seal G, Wurzer J, deRiel JK, Sirover MA. A human nuclear uracil DNA glycosylase is the 37-kDa subunit of glyceraldehyde-3-phosphate dehydrogenase. Proc Natl Acad Sci USA. 1991;88:8460–8464. doi: 10.1073/pnas.88.19.8460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sundararaj KP, Wood RE, Ponnusamy S, Salas AM, Szulc Z, Bielawska A, Obeid LM, Hannun YA, Ogretmen B. Rapid shortening of telomere length in response to ceramide involves the inhibition of telo-mere binding activity of nuclear glyceraldehyde-3-phosphate dehydrogenase. J Biol Chem. 2004;279:6152–6162. doi: 10.1074/jbc.M310549200. [DOI] [PubMed] [Google Scholar]
- 13.Demarse NA, Ponnusamy S, Spicer EK, Apohan E, Baatz JE, Ogretmen B, Davies C. Direct binding of glyceraldehyde 3-phosphate dehydrogenase to telomeric DNA protects telomeres against chemotherapy-induced rapid degradation. J Mol Biol. 2009;394:789–803. doi: 10.1016/j.jmb.2009.09.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nicholls C, Pinto A, Li H, Li L, Wang L, Simpson R, Liu JP. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) induces cancer cell senescence by interacting with telomerase RNA component. Proc Natl Acad Sci. 2012;109:13308–13313. doi: 10.1073/pnas.1206672109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sirover MA. New insights into an old protein: the functional diversity of mammalian glyceraldehyde-3-phosphate dehydrogenase. Biochim Biophys Acta. 1999;1432:159–184. doi: 10.1016/s0167-4838(99)00119-3. [DOI] [PubMed] [Google Scholar]
- 16.Sirover MA. New nuclear functions of the glycolytic protein, glyceraldehyde-3-phosphate dehydrogenase, in mammalian cells. J Cell Biochem. 2005;95:45–52. doi: 10.1002/jcb.20399. [DOI] [PubMed] [Google Scholar]
- 17.Sirover MA. On the functional diversity of glyceraldehyde-3-phosphate dehydrogenase: biochemical mechanisms and regulatory control. Biochim Biophys Acta. 2011;1810:741–751. doi: 10.1016/j.bbagen.2011.05.010. [DOI] [PubMed] [Google Scholar]
- 18.Sirover MA. Subcellular dynamics of multifunctional protein regulation: mechanisms of GAPDH intracellular translocation. J Cell Biochem. 2012;113:2193–2200. doi: 10.1002/jcb.24113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sirover MA. Structural analysis of glyceraldehyde-3-phosphate dehydrogenase functional diversity. Int J Biochem Cell Biol. 2014;57:20–26. doi: 10.1016/j.biocel.2014.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tristan C, Shahani N, Sedlak TW, Sawa A. The diverse functions of GAPDH: views from different sub-cellular compartments. Cell Signal. 2011;23:317–323. doi: 10.1016/j.cellsig.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Siedler N. GAPDH: Biological Properties and Diversity Advances in Experimental Medicine and Biology. Springer; New York: 2013. [Google Scholar]
- 22.Nicholls C, Li H, Liu JP. GAPDH: a common enzyme with uncommon functions. Clin Exp Pharmacol Physiol. 2012;39:674–679. doi: 10.1111/j.1440-1681.2011.05599.x. [DOI] [PubMed] [Google Scholar]
- 23.Brown VM, Krynetski EY, Krynetskaia NF, Grieger D, Mukatira ST, Murti KG, Slaughter CA, Park HW, Evans WE. A novel CRM1-mediated nuclear export signal governs nuclear accumulation of glyceralde-hyde-3-phosphate dehydrogenase following genotoxic stress. J Biol Chem. 2004;279:5984–5992. doi: 10.1074/jbc.M307071200. [DOI] [PubMed] [Google Scholar]
- 24.Kodama R, Kondo T, Yokote H, Jing X, Sawada T, Hironishi M, Sakaguchi K. Nuclear localization of glyceraldehyde-3-phosphate dehydrogenase is not involved in the initiation of apoptosis induced by 1-Methyl-4-phenyl-pyridium iodide (MPP+): nuclear GAPDH without apoptosis. Genes Cells. 2005;10:1211–1219. doi: 10.1111/j.1365-2443.2005.00911.x. [DOI] [PubMed] [Google Scholar]
- 25.Chuang DM, Hough C, Senatorov VV. Glyceralde-hyde-3-phosphate dehydrogenase, apoptosis, and neurodegenerative diseases. Annu Rev Pharmacol Toxicol. 2005;45:269–290. doi: 10.1146/annurev.pharmtox.45.120403.095902. [DOI] [PubMed] [Google Scholar]
- 26.Arutyunova EI, Danshina PV, Domnina LV, Pleten AP, Muronetz VI. Oxidation of glyceraldehyde-3-phosphate dehydrogenase enhances its binding to nucleic acids. Biochem Biophys Res Commun. 2003;307:547–552. doi: 10.1016/s0006-291x(03)01222-1. [DOI] [PubMed] [Google Scholar]
- 27.Perucho M, Salas J, Salas M. Identification of mammalian DNA-binding protein P8 as glyceraldehyde-3-phosphate dehydrogenase. Eur J Biochem. 1977;81:557–562. doi: 10.1111/j.1432-1033.1977.tb11982.x. [DOI] [PubMed] [Google Scholar]
- 28.Ryazanov AG. Glyceraldehyde-3-phosphate dehydrogenase is one of the three major RNA-binding proteins of rabbit reticulocytes. FEBS Lett. 1985;192:131–134. doi: 10.1016/0014-5793(85)80058-2. [DOI] [PubMed] [Google Scholar]
- 29.Karpel RL, Burchard AC. A basic isozyme of yeast glyceraldehyde-3-phosphate dehydrogenase with nucleic acid helix-destabilizing activity. Biochim Biophys Acta. 1981;654:256–267. doi: 10.1016/0005-2787(81)90180-5. [DOI] [PubMed] [Google Scholar]
- 30.Milhaud P, Blanchard J-M, Jeanteur P. Hela cytosolic protein isolated by poly (A)-sepharose chromatography is glyceraldehyde-3-P-dehydrogenase. Biochimie. 1978;60:1343–1346. doi: 10.1016/s0300-9084(79)80454-x. [DOI] [PubMed] [Google Scholar]
- 31.Parker R, Song H. The enzymes and control of eukaryotic mRNA turnover. Nat Struct Mol Biol. 2004;11:121–127. doi: 10.1038/nsmb724. [DOI] [PubMed] [Google Scholar]
- 32.Wilusz CJ, Wormington M, Peltz SW. The cap-to-tail guide to mRNA turnover. Nat Rev Mol Cell Biol. 2001;2:237–246. doi: 10.1038/35067025. [DOI] [PubMed] [Google Scholar]
- 33.Grillo G, Turi A, Licciulli F, Mignone F, Liuni S, Banfi S, Gennarino VA, Horner DS, Pavesi G, Picardi E, et al. UTRdb and UTRsite (RELEASE 2010): a collection of sequences and regulatory motifs of the untranslated regions of eukaryotic mRNAs. Nucleic Acids Res. 2010;38:D75–D80. doi: 10.1093/nar/gkp902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barreau C. AU-rich elements and associated factors: are there unifying principles? Nucleic Acids Res. 2005;33:7138–7150. doi: 10.1093/nar/gki1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilson GM, Brewer G. The search for trans-acting factors controlling messenger RNA decay. Prog Nucleic Acid Res Mol Biol. 1999;62:257–291. doi: 10.1016/s0079-6603(08)60510-3. [DOI] [PubMed] [Google Scholar]
- 36.Shim J, Karin M. The control of mRNA stability in response to extracellular stimuli. Mol Cells. 2002;14:323–331. [PubMed] [Google Scholar]
- 37.Gao M, Wilusz CJ, Peltz SW, Wilusz J. A novel mRNA-decapping activity in HeLa cytoplasmic extracts is regulated by AU-rich elements. EMBO J. 2001;20:1134–1143. doi: 10.1093/emboj/20.5.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lai WS, Carballo E, Strum JR, Kennington EA, Phillips RS, Blackshear PJ. Evidence that tristetraprolin binds to AU-rich elements and promotes the deadeny-lation and destabilization of tumor necrosis factor / mRNA. Mol Cell Biol. 1999;19:4311–4323. doi: 10.1128/mcb.19.6.4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Myer VE, Fan XC, Steitz JA. Identification of HuR as a protein implicated in AUUUA-mediated mRNA decay. EMBO J. 1997;16:2130–2139. doi: 10.1093/emboj/16.8.2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peng SS, Chen CY, Xu N, Shyu AB. RNA stabilization by the AU-rich element binding protein, HuR, an ELAV protein. EMBO J. 1998;17:3461–3470. doi: 10.1093/emboj/17.12.3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kishor A, Tandukar B, Ly YV, Toth EA, Suarez Y, Brewer G, Wilson GM. Hsp70 is a novel posttranscriptional regulator of gene expression that binds and stabilizes selected mRNAs containing AU-rich elements. Mol Cell Biol. 2013;33:71–84. doi: 10.1128/MCB.01275-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen CY, Shyu AB. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem Sci. 1995;20:465–470. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- 43.Pioli PA, Jonell Hamilton B, Connolly JE, Brewer G, Rigby WF. Lactate dehydrogenase is an AU-rich element-binding protein that directly interacts with AUF1. J Biol Chem. 2002;277:35738–35745. doi: 10.1074/jbc.M204002200. [DOI] [PubMed] [Google Scholar]
- 44.White E, Brewer G, Wilson G. Post-transcriptional control of gene expression by AUF1: mechanisms, physiological targets, and regulation. Biochim Biophys Acta. 2013;1829:680–688. doi: 10.1016/j.bbagrm.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yoon J-H, De S, Srikantan S, Abdelmohsen K, Grammatikakis I, Kim J, Kim KM, Noh JH, White EJF, Martindale JL, et al. PAR-CLIP analysis uncovers AUF1 impact on target RNA fate and genome integrity. Nat Commun. 2014;5:5248. doi: 10.1038/ncomms6248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Woo H-H, Baker T, Laszlo C, Chambers SK. Nucleolin mediates microRNA-directed CSF-1 mRNA deadenylation but increases translation of CSF-1 mRNA. Mol Cell Proteomics. 2013;12:1661–1677. doi: 10.1074/mcp.M112.025288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blackshear PJ. Tristetraprolin and other CCCH tandem zinc-finger proteins in the regulation of mRNA turnover. Biochem Soc Trans. 2002;30:945–952. doi: 10.1042/bst0300945. [DOI] [PubMed] [Google Scholar]
- 48.Raineri I. Roles of AUF1 isoforms, HuR and BRF1 in ARE-dependent mRNA turnover studied by RNA interference. Nucleic Acids Res. 2004;32:1279–1288. doi: 10.1093/nar/gkh282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wilusz CJ, Wilusz J. Bringing the role of mRNA decay in the control of gene expression into focus. Trends Genet. 2004;20:491–497. doi: 10.1016/j.tig.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 50.Bassell GJ, Kelic S. Binding proteins for mRNA localization and local translation, and their dysfunction in genetic neurological disease. Curr Opin Neurobiol. 2004;14:574–581. doi: 10.1016/j.conb.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 51.Musunuru K. Cell-specific RNA-binding proteins in human disease. Trends Cardiovasc Med. 2003;13:188–195. doi: 10.1016/s1050-1738(03)00075-6. [DOI] [PubMed] [Google Scholar]
- 52.Blaxall BC, Dwyer-Nield LD, Bauer AK, Bohlmeyer TJ, Malkinson AM, Port JD. Differential expression and localization of the mRNA binding proteins, AU-rich element mRNA binding protein (AUF-1) and Hu antigen R (HuR), in neoplastic lung tissue. Mol Carcinog. 2000;28:76–83. [PubMed] [Google Scholar]
- 53.Burd C, Dreyfuss G. Conserved functions and diversity structures of RNA-binding proteins. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- 54.Messias AC, Sattler M. Structural basis of single-stranded RNA recognition. Acc Chem Res. 2004;37:279–287. doi: 10.1021/ar030034m. [DOI] [PubMed] [Google Scholar]
- 55.Auweter SD, Oberstrass FC, Allain FH. Sequence-specific binding of single-stranded RNA: is there a code for recognition? Nucleic Acids Res. 2006;34:4943–4959. doi: 10.1093/nar/gkl620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee I, Hong W. RAP - a putative RNA binding domain. Trends Biochem Sci. 2004;29:567–570. doi: 10.1016/j.tibs.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 57.Niessing D, Huttelmaier S, Zenklusen D, Singer RH, Burley SK. She2p is a novel RNA binding protein with a basic helical hairpin motif. Cell. 2004;119:491–502. doi: 10.1016/j.cell.2004.10.018. [DOI] [PubMed] [Google Scholar]
- 58.Rammelt C, Bilen B, Zavolan M, Keller W. PAPD5, a noncanonical poly(A) polymerase with an unusual RNA-binding motif. RNA. 2011;17:1737–1746. doi: 10.1261/rna.2787011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zalfa F, Adinolfi S, Napoli I, Kühn-Hölsken E, Urlaub H, Achsel T, Pastore A, Bagni C. Fragile X mental retardation protein (FMRP) binds specifically to the brain cytoplasmic RNAs BC1/BC200 via a novel RNA-binding motif. J Biol Chem. 2005;280:33403–33410. doi: 10.1074/jbc.M504286200. [DOI] [PubMed] [Google Scholar]
- 60.Klausner RD, Rouault TA. A double life: cytosolic aconitase as a regulatory RNA binding protein. Mol Biol Cell. 1993;4:1–5. doi: 10.1091/mbc.4.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shetty S, Muniyappa H, Halady PK, Idell S. Regulation of urokinase receptor expression by phosphogly-cerate kinase. Am J Respir Cell Mol Biol. 2004;31:100–106. doi: 10.1165/rcmb.2003-0104OC. [DOI] [PubMed] [Google Scholar]
- 62.McGowan K, Pekala PH. Dehydrogenase binding to the 3′-untranslated region of GLUT1 mRNA. Biochem Biophys Res Commun. 1996;221:42–45. doi: 10.1006/bbrc.1996.0541. [DOI] [PubMed] [Google Scholar]
- 63.Preiss T, Hall AG, Lightowlers RN. Identification of bovine glutamate dehydrogenase as an RNA-binding protein. J Biol Chem. 1993;268:24523–24526. [PubMed] [Google Scholar]
- 64.Elzinga SD, Bednarz AL, van Oosterum K, Dekker PJ, Grivell L. Yeast mitochondrial NAD+-dependent isocitrate dehydrogenase is an RNA-binding protein. Nucleic Acids Res. 1993;21:5328–5331. doi: 10.1093/nar/21.23.5328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ciesla J. Metabolic enzymes that bind RNA: yet another level of cellular regulatory network? Acta Biochim Pol. 2006;53:11–32. [PubMed] [Google Scholar]
- 66.Zasloff M. tRNA transport from the nucleus in a eukaryotic cell: carrier-mediated translocation process. Proc Natl Acad Sci USA. 1983;80:6436–6440. doi: 10.1073/pnas.80.21.6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Singh R, Green MR. Sequence-specific binding of transfer RNA by glyceraldehyde-3-phosphate dehydrogenase. Science. 1993;259:365–368. doi: 10.1126/science.8420004. [DOI] [PubMed] [Google Scholar]
- 68.Carmona P, Rodriguez-Casado A, Molina M. Conformational structure and binding mode of glyceraldehyde-3-phosphate dehydrogenase to tRNA studied by Raman and CD spectroscopy. Biochim Bio-phys Acta. 1999;1432:222–233. doi: 10.1016/s0167-4838(99)00113-2. [DOI] [PubMed] [Google Scholar]
- 69.Nagy E, Rigby WF. Glyceraldehyde-3-phosphate dehydrogenase selectively binds AU-rich RNA in the NAD (+)-binding region (Rossmann Fold) J Biol Chem. 1995;270:2755–2763. doi: 10.1074/jbc.270.6.2755. [DOI] [PubMed] [Google Scholar]
- 70.Chang CH, Curtis JD, Maggi LB, Jr, Faubert B, Villarino AV, O’Sullivan D, Huang SC, van der Windt GJ, Blagih J, Qiu J, et al. Posttranscriptional control of T cell effector function by aerobic glycolysis. Cell. 2013;153:1239–1251. doi: 10.1016/j.cell.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bonafé N, Gilmore-Hebert M, Folk NL, Azodi M, Zhou Y, Chambers SK. Glyceraldehyde-3-phosphate dehydrogenase binds to the AU-Rich 3′ untranslated region of colony-stimulating factor-1 (CSF-1) messenger RNA in human ovarian cancer cells: possible role in CSF-1 posttranscriptional regulation and tumor phenotype. Cancer Res. 2005;65:3762–3771. doi: 10.1158/0008-5472.CAN-04-3954. [DOI] [PubMed] [Google Scholar]
- 72.Zhou Y, Yi X, Stoffer JB, Bonafe N, Gilmore-Hebert M, McAlpine J, Chambers SK. The multifunctional protein glyceraldehyde-3-phosphate dehydrogenase is both regulated and controls colony-stimulating factor-1 messenger RNA stability in ovarian cancer. Mol Cancer Res. 2008;6:1375–1384. doi: 10.1158/1541-7786.MCR-07-2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ikeda Y, Yamaji R, Irie K, Kioka N, Murakami A. Glyceraldehyde-3-phosphate dehydrogenase regulates cyclooxygenase-2 expression by targeting mRNA stability. Arch Biochem Biophys. 2012;528:141–147. doi: 10.1016/j.abb.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 74.Kondo S, Kubota S, Mukudai Y, Nishida T, Yoshihama Y, Shirota T, Shintani S, Tagigawa M. Binding of glyceraldehyde-3-phosphate dehydrogenase to the cis-acting element of structure-anchored repression in ccn2 mRNA. Biochem Biophys Res Commun. 2011;405:382–387. doi: 10.1016/j.bbrc.2011.01.034. [DOI] [PubMed] [Google Scholar]
- 75.Rodriguez-Pascual F, Redondo-Horcajo M, Magan-Marchal N, Lagares D, Martinez-Ruiz A, Kleinert H, Lamas S. Glyceraldehyde-3-phosphate dehydrogenase regulates endothelin-1 expression by a novel, redox-sensitive mechanism involving mRNA stability. Mol Cell Biol. 2008;28:7139–7155. doi: 10.1128/MCB.01145-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Backlund M, Paukku K, Daviet L, De Boer RA, Valo E, Hautaniemi S, Kalkkinen N, Ehsan A, Kontula KK, Lehtonen JY. Posttranscriptional regulation of angiotensin II type 1 receptor expression by glyceraldehyde 3-phosphate dehydrogenase. Nucleic Acids Res. 2009;37:2346–2358. doi: 10.1093/nar/gkp098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.White MR, Khan MM, Deredge D, Ross CR, Quintyn R, Zucconi BE, Wysocki VH, Wintrode PL, Wilson GM, Garcin ED. A dimer interface mutation in glyceraldehyde-3-phosphate dehydrogenase regulates its binding to AU-rich RNA. J Biol Chem. 2015;290:1770–1785. doi: 10.1074/jbc.M114.618165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zeng T, Dong ZF, Liu SJ, Wan RP, Tang LJ, Liu T, Zhao QH, Shi YW, Yi YH, Liao WP, Long YS. A novel variant in the 3′ UTR of human SCN1A gene from a patient with Dravet syndrome decreases mRNA stability mediated by GAPDH’s binding. Hum Genet. 2014;133:801–811. doi: 10.1007/s00439-014-1422-8. [DOI] [PubMed] [Google Scholar]
- 79.Sioud M, Jespersen L. Enhancement of hammerhead ribozyme catalysis by glyceraldehyde-3-phosphate dehydrogenase. J Mol Biol. 1996;257:775–789. doi: 10.1006/jmbi.1996.0201. [DOI] [PubMed] [Google Scholar]
- 80.Kubota S, Hattori T, Nakanishi T, Takigawa M. Involvement of cis-acting repressive element(s) in the 3′-untranslated region of human connective tissue growth factor gene. FEBS Lett. 1999;450:84–88. doi: 10.1016/s0014-5793(99)00480-9. [DOI] [PubMed] [Google Scholar]
- 81.Kubota S, Kondo S, Eguchi T, Hattori T, Nakanishi T, Pomerantz RJ, Takigawa M. Identification of an RNA element that confers post-transcriptional repression of connective tissue growth factor/hypertrophic chondrocyte specific 24 (ctgf/hcs24) gene: similarities to retroviral RNA-protein interactions. Oncogene. 2000;19:4773–4786. doi: 10.1038/sj.onc.1203835. [DOI] [PubMed] [Google Scholar]
- 82.Reimunde FM, Castanares C, Redondo-Horcajo M, Lamas S, Rodriguez-Pascual F. Endothelin-1 expression is strongly repressed by AU-rich elements in the 3′-untranslated region of the gene. Biochem J. 2005;387:763–772. doi: 10.1042/BJ20041687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ross B, D’Orleans-Juste P, Giaid A. Potential role of endothelin-1 in pulmonary fibrosis: from the bench to the clinic. Am J Respir Cell Mol Biol. 2010;42:16–20. doi: 10.1165/rcmb.2009-0175TR. [DOI] [PubMed] [Google Scholar]
- 84.Uguccioni M, Pulsatelli L, Grigolo B, Facchini A, Fasano L, Cinti C, Fabbri M, Gasbarrini G, Meliconi R. Endothelin-1 in idiopathic pulmonary fibrosis. J Clin Pathol. 1995;48:330–334. doi: 10.1136/jcp.48.4.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Eberhardt W, Doller A, Akool el-S, Pfeilschifter J. Modulation of mRNA stability as a novel therapeutic approach. Pharmacol Ther. 2007;114:56–73. doi: 10.1016/j.pharmthera.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 86.Frosk P, Mhanni AA, Rafay MF. SCN1A mutation associated with intractable myoclonic epilepsy and migraine headache. J Child Neurol. 2012;28:389–391. doi: 10.1177/0883073812443309. [DOI] [PubMed] [Google Scholar]
- 87.Schmitz HD, Bereiter-Hahn J. Glyceraldehyde-3-phosphate dehydrogenase associates with actin filaments in serum deprived NIH 3 T3 cells only. Cell Biol Int. 2002;26:155–164. doi: 10.1006/cbir.2001.0819. [DOI] [PubMed] [Google Scholar]
- 88.Gratacós FM, Brewer G. The role of AUF1 in regulated mRNA decay. WIREs RNA. 2010;1:457–473. doi: 10.1002/wrna.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Prasanth KR, Huang YW, Liou MR, Wang RY, Hu CC, Tsai CH, Meng M, Lin NS, Hsu YH. Glyceraldehyde 3-phosphate dehydrogenase negatively regulates the replication of bamboo mosaic virus and its associated satellite RNA. J Virol. 2011;85:8829–8840. doi: 10.1128/JVI.00556-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Dollenmaier G, Weitz M. Interaction of glyceraldehyde-3-phosphate dehydrogenase with secondary and tertiary RNA structural elements of the hepatitis A virus 3′ translated and non-translated regions. J Gen Virol. 2003;84:403–414. doi: 10.1099/vir.0.18501-0. [DOI] [PubMed] [Google Scholar]
- 91.Chang KH, Brown EA, Lemon SM. Cell type-specific proteins which interact with the 5′ nontranslated region of hepatitis A virus RNA. J Virol. 1993;67:6716–6725. doi: 10.1128/jvi.67.11.6716-6725.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Schultz DE, Hardin CC, Lemon SM. Specific interaction of glyceraldehyde 3-phosphate dehydrogenase with the 5′-nontranslated RNA of hepatitis A virus. Biochemistry. 1996;271:14134–14142. doi: 10.1074/jbc.271.24.14134. [DOI] [PubMed] [Google Scholar]
- 93.Yi M, Schultz DE, Lemon SM. Functional significance of the interaction of hepatitis A virus RNA with glyceraldehyde 3-phosphate dehydrogenase (GAPDH): opposing effects of GAPDH and polypyrimidine tract binding protein on internal ribosome entry site function. J Virol. 2000;74:6459–6468. doi: 10.1128/jvi.74.14.6459-6468.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Galan C, Sola I, Nogales A, Thomas B, Akoulitchev A, Enjuanes L, Almazán F. Host cell proteins interacting with the 3′ end of TGEV coronavirus genome influence virus replication. Virology. 2009;391:304–314. doi: 10.1016/j.virol.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Petrik J, Parker H, Alexander GJ. Human hepatic glyceraldehyde-3-phosphate dehydrogenase binds to the poly(U) tract of the 3′ non-coding region of hepatitis C virus genomic RNA. J Gen Virol. 1999;80:3109–3113. doi: 10.1099/0022-1317-80-12-3109. [DOI] [PubMed] [Google Scholar]
- 96.Yang SH, Liu ML, Tien CF, Chou SJ, Chang RY. Gly-ceraldehyde-3-phosphate dehydrogenase (GAPDH) interaction with 3′ ends of Japanese encephalitis virus RNA and colocalization with the viral NS5 protein. J Biomed Sci. 2009;16:1–10. doi: 10.1186/1423-0127-16-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang RY, Nagy PD. Tomato bushy stunt virus coopts the RNA-binding function of a host metabolic enzyme for viral genomic RNA synthesis. Cell Host Microbe. 2008;3:178–187. doi: 10.1016/j.chom.2008.02.005. [DOI] [PubMed] [Google Scholar]
- 98.Huang TS, Nagy PD. Direct inhibition of tombusvirus plus-strand RNA synthesis by a dominant negative mutant of a host metabolic enzyme, glyceraldehyde-3-phosphate dehydrogenase, in yeast and plants. J Virol. 2011;85:9090–9102. doi: 10.1128/JVI.00666-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lin SS, Chang SC, Wang YH, Sun CY, Chang MF. Specific interaction between the hepatitis delta virus RNA and glyceraldehyde 3-phosphate dehydrogenase: an enhancement on ribozyme catalysis. Virology. 2000;271:46–57. doi: 10.1006/viro.2000.0302. [DOI] [PubMed] [Google Scholar]
- 100.De BP, Gupta S, Zhao H, Drazba JA, Banerjee AK. Specific interaction in vitro and in vivo of glyceraldehyde-3-phosphate dehydrogenase and LA protein with cis-acting RNAs of human parainfluenza virus type 3. J Biol Chem. 1996;271:24728–24735. doi: 10.1074/jbc.271.40.24728. [DOI] [PubMed] [Google Scholar]
- 101.Buck KW. Comparison of the replication of positive-stranded RNA viruses of plants and animals. Adv Virus Res. 1996;47:159–251. doi: 10.1016/S0065-3527(08)60736-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hentze MW, Preiss T. The REM phase of gene regulation. Trends Biochem Sci. 2010;35:423–426. doi: 10.1016/j.tibs.2010.05.009. [DOI] [PubMed] [Google Scholar]
- 103.Tung KS, Norbeck LL, Nolan SL, Atkinson NS, Hopper AK. SRN1, a yeast gene involved in RNA processing, is identical to HEX2/REG1, a negative regulator in glucose repression. Mol Cell Biol. 1992;12:2673–2680. doi: 10.1128/mcb.12.6.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Stephens JM, Carter BZ, Pekala PH, Malter JS. Tumor necrosis factor a-induced glucose transporter (GLUT-1) mRNA stabilization in 3T3-L1 preadipocytes. J Biol Chem. 1992;267:8336–8341. [PubMed] [Google Scholar]
- 105.Cornelius P, Marlowe M, Lee MD, Pekala PH. The growth factor-like effects of tumor necrosis factor-alpha. Stimulation of glucose transport activity and induction of glucose transporter and immediate early gene expression in 3T3-L1 preadipocytes. J Biol Chem. 1990;265:20506–20516. [PubMed] [Google Scholar]
- 106.Lee MN, Ha SH, Kim J, Koh A, Lee CS, Kim JH, Jeon H, Kim DH, Su PG, Ryu SH. Glycolytic flux signals to mTOR through glyceraldehyde-3-phosphate dehydrogenase-mediated regulation of Rheb. Mol Cell Biol. 2009;29:3991–4001. doi: 10.1128/MCB.00165-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jenkins JL, Tanner JJ. High-resolution structure of human D-glyceraldehyde-3-phosphate dehydrogenase. Acta Crystallogr D Biol Crystallogr. 2006;62:290–301. doi: 10.1107/S0907444905042289. [DOI] [PubMed] [Google Scholar]
- 108.Shindyalov IN, Bourne PE. Protein structure alignment by incremental combinatorial extension (CE) of the optimal path. Protein Eng. 1998;11:739–747. doi: 10.1093/protein/11.9.739. [DOI] [PubMed] [Google Scholar]
- 109.Maris C, Dominguez C, Allain FH. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression: the RRM domain, a plastic RNA-binding platform. FEBS J. 2005;272:2118–2131. doi: 10.1111/j.1742-4658.2005.04653.x. [DOI] [PubMed] [Google Scholar]
- 110.Cowan-Jacob SW, Kaufmann M, Anselmo AN, Stark W, Grütter MG. Structure of rabbit-muscle glyceraldehyde-3-phosphate dehydrogenase. Acta Crystallogr D Biol Crystallogr. 2003;59:2218–2227. doi: 10.1107/s0907444903020493. [DOI] [PubMed] [Google Scholar]
- 111.Ismail SA, Park HW. Structural analysis of human liver glyceraldehyde-3-phosphate dehydrogenase. Acta Crystallogr D Biol Crystallogr. 2005;61:1508–1513. doi: 10.1107/S0907444905026740. [DOI] [PubMed] [Google Scholar]
- 112.Chaikuad A, Shafqat N, Al-Mokhtar R, Cameron G, Clarke AR, Brady RL, Oppermann U, Frayne J, Yue WW. Structure and kinetic characterization of human sperm-specific glyceraldehyde-3-phosphate dehydrogenase, GAPDS. Biochem J. 2011;435:401–409. doi: 10.1042/BJ20101442. [DOI] [PubMed] [Google Scholar]
- 113.Baker BY, Shi W, Wang B, Palczewski K. High-resolution crystal structures of the photoreceptor gly-ceraldehyde 3-phosphate dehydrogenase (GAPDH) with three and four-bound NAD molecules: structures of the GAPDH Photoreceptor. Protein Sci. 2014;23:1629–1639. doi: 10.1002/pro.2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kramer K, Sachsenberg T, Beckmann BM, Qamar S, Boon KL, Hentze MW, Kohlbacher O, Urlaub H. Photo-cross-linking and high-resolution mass spectrometry for assignment of RNA-binding sites in RNA-binding proteins. Nat Methods. 2014;11:1064–1074. doi: 10.1038/nmeth.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Nagy E, Henics T, Eckert M, Miseta A, Lightowlers RN, Kellermayer M. Identification of the NAD (+)-binding fold of glyceraldehyde-3-phosphate dehydrogenase as a novel RNA-binding domain. Biochem Biophys Res Commun. 2000;275:253–260. doi: 10.1006/bbrc.2000.3246. [DOI] [PubMed] [Google Scholar]
- 116.Hentze MW. Enzymes as RNA-binding proteins: a role for (di)nucleotide-binding domains? Trends Biochem Sci. 1994;19:101–103. doi: 10.1016/0968-0004(94)90198-8. [DOI] [PubMed] [Google Scholar]
- 117.Castello A, Fischer B, Eichelbaum K, Horos R, Beck-mann BM, Strein C, Davey NE, Humphreys DT, Preiss T, Steinmetz LM, Hentze MW. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell. 2012;149:1393–1406. doi: 10.1016/j.cell.2012.04.031. [DOI] [PubMed] [Google Scholar]
- 118.Samson AL, Knaupp AS, Kass I, Kleifeld O, Marijanovic EM, Hughes VA, Lupton CJ, Buckle AM, Bottomley SP, Medcalf RL. Oxidation of an exposed methionine instigates the aggregation of glyceraldehyde-3-phosphate dehydrogenase. J Biol Chem. 2014;289:26922–26936. doi: 10.1074/jbc.M114.570275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Nakajima H, Amano W, Fujita A, Fukuhara A, Azuma YT, Hata F, Inui T, Takeuchi T. The active site cysteine of the proapoptotic protein glyceraldehyde-3-phosphate dehydrogenase is essential in oxidative stress-induced aggregation and cell death. J Biol Chem. 2007;282:26562–26574. doi: 10.1074/jbc.M704199200. [DOI] [PubMed] [Google Scholar]
- 120.Nakajima H, Amano W, Kubo T, Fukuhara A, Ihara H, Azuma YT, Tajima H, Inui T, Sawa A, Takeuchi T. Glyceraldehyde-3-phosphate dehydrogenase aggregate formation participates in oxidative stress-induced cell death. J Biol Chem. 2009;284:34331–34341. doi: 10.1074/jbc.M109.027698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ventura M, Mateo F, Serratosa J, Salaet I, Carujo S, Bachs O, Pujol MJ. Nuclear translocation of glyceraldehyde-3-phosphate dehydrogenase is regulated by acetylation. Int J Biochem Cell Biol. 2010;42:1672–1680. doi: 10.1016/j.biocel.2010.06.014. [DOI] [PubMed] [Google Scholar]
- 122.Seo J, Jeong J, Kim YM, Hwang N, Paek E, Lee KJ. Strategy for comprehensive identification of post-translational modifications in cellular proteins, including low abundant modifications: application to glyceraldehyde-3-phosphate dehydrogenase. J Proteome Res. 2008;7:587–602. doi: 10.1021/pr700657y. [DOI] [PubMed] [Google Scholar]
- 123.Yogalingam G, Hwang S, Ferreira JC, Mochly-Rosen D. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) phosphorylation by protein kinase C delta (8PKC) inhibits mitochondrial elimination by lysosomal-like structures following ischemia and reoxygenation-induced injury. J Biol Chem. 2013;288:18947–18960. doi: 10.1074/jbc.M113.466870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Huang Q, Lan F, Zheng Z, Xie F, Han J, Dong L, Xie Y, Zheng F. Akt2 kinase suppresses glyceral-dehyde-3-phosphate dehydrogenase (GAPDH)-mediated apoptosis in ovarian cancer cells via phosphorylating GAPDH at threonine 237 and decreasing its nuclear translocation. J Biol Chem. 2011;286:42211–42220. doi: 10.1074/jbc.M111.296905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Park J, Han D, Kim K, Kang Y, Kim Y. O-GlcNAcylation disrupts glyceraldehyde-3-phosphate dehydrogenase homo-tetramer formation and mediates its nuclear translocation. Biochim Biophys Acta. 2009;1794:254–262. doi: 10.1016/j.bbapap.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 126.Gao X, Wang X, Pham TH, Feuerbacher LA, Lubos ML, Huang M, Olsen R, Mushegian A, Slawson C, Hardwidge PR. NleB, a bacterial effector with glyco-syltransferase activity, targets GAPDH function to inhibit NF-κB activation. Cell Host Microbe. 2013;13:87–99. doi: 10.1016/j.chom.2012.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Shen H, Wang H, Liu Q, Liu H, Teng M, Li X. Structural insights into RNA recognition properties of glyceraldehyde-3-phosphate dehydrogenase 3 from Saccharomyces cerevisiae. IUBMB Life. 2014;66:631–638. doi: 10.1002/iub.1313. [DOI] [PubMed] [Google Scholar]