SUMMARY
Binding of the transcriptional co-activator YAP with the transcription factor TEAD stimulates growth of the heart and other organs. YAP overexpression potently stimulates fetal cardiomyocyte (CM) proliferation, but YAP's mitogenic potency declines post-natally. While investigating factors that limit YAP's postnatal mitogenic activity, we found that the CM-enriched TEAD1 binding protein VGLL4 inhibits CM proliferation by inhibiting TEAD1-YAP interaction and by targeting TEAD1 for degradation. Importantly, VGLL4 acetylation at lysine 225 negatively regulated its binding to TEAD1. This developmentally regulated acetylation event critically governs postnatal heart growth, since overexpression of an acetylation-refractory VGLL4 mutant enhanced TEAD1 degradation, limited neonatal CM proliferation, and caused CM necrosis. Our study defines an acetylation-mediated, VGLL4-dependent switch that regulates TEAD stability and YAP-TEAD activity. These insights may improve targeted modulation of TEAD-YAP activity in applications from cardiac regeneration to cancer.
INTRODUCTION
Control of organ growth is fundamental to animal development and organ homeostasis. Unrestrained activity of growth-promoting pathways leads to cancer, whereas targeted activation of these pathways may be a strategy for organ regeneration (Pan, 2010; Lin and Pu, 2015). One area with great need for advances in regenerative medicine is heart disease. Heart failure is the world's leading cause of death, and its prevalence is expected to further increase as the population ages (Heidenreich et al., 2013). Cardiomyocyte (CM) loss is a central pathogenic mechanism in heart failure, but limited endogenous regenerative capacity in the adult heart has precluded development of therapeutic approaches to efficiently replace these lost CMs (Lin and Pu, 2014). Unlike adult CMs, fetal CMs robustly proliferate to match the rapid growth of the embryo. This transition from CM proliferation to cell-cycle exit occurs in the first week of life in the mouse (Walsh et al., 2010). The mechanisms that regulate this neonatal cell-cycle exit are poorly understood, and greater insights would inform efforts to enhance cardiac regeneration.
The transcriptional co-activator YAP (Yes-associated protein) is a key driver of organ growth (Pan, 2010). YAP binds to TEA domain (TEAD)-containing transcription factors (TEAD1–TEAD4) to activate transcription of cell-cycle and cell-survival genes and thereby promotes organ growth. The potent growth-promoting activity of the YAP-TEAD complex is closely regulated through incompletely understood signaling pathways. Among the most studied regulatory mechanisms is the Hippo kinase cascade, which phosphorylates YAP, leading to its nuclear exclusion (Huang et al., 2005). Both YAP and its regulation by the Hippo kinase cascade have been shown to be essential for normal heart development (Heallen et al., 2011; von Gise et al., 2012; Xin et al., 2011). YAP was necessary for fetal CM proliferation, and its activation through overexpression or Hippo inhibition was sufficient to drive massive fetal cardiac overgrowth. YAP activation likewise stimulated neonatal as well as adult CM proliferation, but the level of CM cell-cycle activity achieved diminished with postnatal age (Lin et al., 2014; Xin et al., 2013; Heallen et al., 2013). These data show that regulation of YAP activity is crucial for normal cardiac growth control. Moreover, they suggest that unknown mechanisms suppress YAP mitogenic activity in the postnatal heart.
In addition to the Hippo kinase pathway, Hippo-independent YAP regulatory mechanisms also exist. For example, α-catenin, a cellular adhesion molecule, binds YAP under high cell density conditions, promoting its cytoplasmic sequestration by limiting its dephosphorylation (Schlegelmilch et al., 2011; Li et al., 2014). Recently, a new level of YAP-TEAD regulation was described. In Drosophila, the orthologs of YAP and TEAD are named Yorkie (Yki) and Scalloped (Sd), respectively. The gene Tgi was discovered in Drosophila screens for Yki-Sd antagonists (Koontz et al., 2013). TGI protein contains two TEAD-binding regions, named Tondu (TDU) domains, and competes with YKI for SD binding. By reducing YKI-SD activity and the transcription of YKI-SD target genes, TGI inhibited growth. In mammals, there are four TDU domain-containing proteins, vestigial-like 1 to 4 (VGLL1–VGLL4), with VGLL4 being the most closely related to TGI. Massive liver overgrowth driven by YAP was suppressed by VGLL4 (Koontz et al., 2013), indicating that VGLL4 is a potent inhibitor of YAP in mammalian cells.
Profiling of VGLL4 across mouse tissues showed that it was most highly expressed in heart (Chen et al., 2004), suggesting that VGLL4 potentially suppresses postnatal cardiac YAP activity. Here we studied VGLL4 function in regulating cardiac YAP-TEAD activity and neonatal cardiac growth and function. We found that VGLL4 regulates both TEAD stability and its interaction with YAP. Moreover, VGLL4 acetylation at a key residue within the TDU domain regulates its binding to TEAD, revealing a novel YAP-TEAD regulatory mechanism. Acetylation of VGLL4 in neonatal heart was essential to limit its activity and thereby permit normal heart growth and function.
RESULTS
The Major Cardiac Interaction Partner of TEAD1 Changes with Postnatal Age
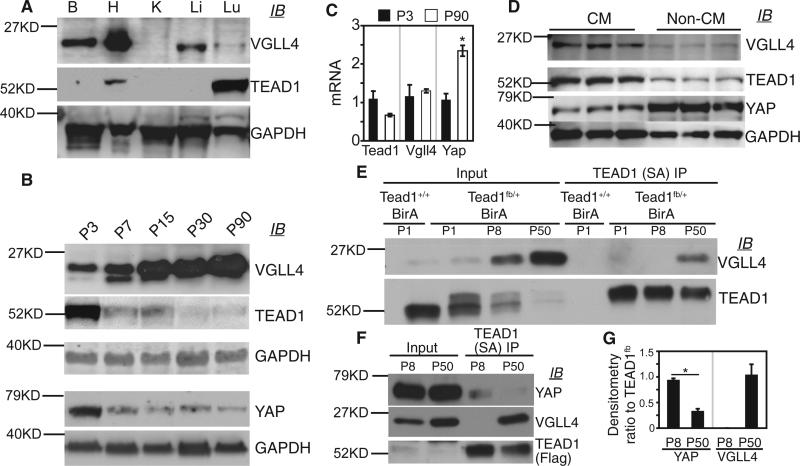
During heart development, the Hippo-YAP pathway regulates CM proliferation (Heallen et al., 2011; von Gise et al., 2012; Xin et al., 2011). YAP and TEAD1 are terminal effectors of the Hippo-YAP pathway. VGLL4, a TEAD1-binding protein that antagonizes overexpressed YAP in the liver (Koontz et al., 2013), was previously reported to have cardiac-restricted RNA expression (Chen et al., 2004). To better understand YAP, TEAD1, and VGLL4 function in regulating organ growth, we measured their expression in several adult mouse tissues. YAP was widely expressed, as we demonstrated previously (von Gise et al., 2012). We detected robust VGLL4 expression in the heart, with lower levels also present in the brain, liver, and lung (Figure 1A). TEAD1 protein was abundant in the lung, less expressed in the heart, and undetectable in the other organs examined (Figure 1A). Focusing on the postnatal heart, we measured expression of these proteins at several different ages. Interestingly, VGLL4 expression increased from low levels in the newborn heart to high levels in the adult heart (Figure 1B). TEAD1 and YAP levels were anti-correlated with VGLL4 and decreased with age (Figure 1B). These changes in cardiac protein level did not correlate with changes in their corresponding transcripts (Figure 1C), indicating that the expression of these proteins is regulated post-transcriptionally.
Figure 1. Developmental Changes in VGLL4-TEAD1 and YAP-TEAD1 Interaction in the Mouse Heart.
(A) Immunoblot (IB) of protein extracts from adult mouse brain (B), heart (H), kidney (K), liver (Li), and lung (Lu).
(B) Immunoblot (IB) of heart protein extracts from mice with the indicated postnatal (P) age in days. GAPDH internal control belonging to respective immunoblots is shown.
(C) qRT-PCR measurement of Vgll4, Tead1, and Yap mRNA level in 3-day-old (P3) and 90-day-old (P90) hearts. *p < 0.05, n = 3.
(D) VGLL4, TEAD1, and YAP expression in CMs and non-CMs. Adult hearts were dissociated by collagenase perfusion and then separated into CM and non-CM fractions. Protein extracts were immunoblotted with the indicated antibodies.
(E) Age-dependent association of VGLL4 and TEAD1 in mouse heart. Tead1fb/+;R26BirA/+ heart extract was incubated with immobilized streptavidin (SA). Co-precipitated VGLL4 and TEAD1 were measured by immunoblotting. Tead1+/+;R26BirA/+ heart extract was used as a negative control.
(F) Age-dependent association of YAP and TEAD1 in mouse heart. TEAD1 was precipitated from protein from P1, P8, or P50 mouse heart as in (D). Co-precipitated proteins were detected by immunoblotting.
(G) Relative YAP or VGLL4 coIP with TEAD1, determined by quantification of (E). Precipitated proteins were normalized to TEAD1fb. *p < 0.05, n = 3.
All error bars represent the SEM.
To determine whether these proteins were expressed in CMs or non-CMs in adult heart, we dissociated hearts and isolated purified cell populations. VGLL4 and TEAD1 were mainly expressed in CMs rather than non-CMs, whereas YAP was predominantly expressed in non-CMs (Figure 1D).
Based on these protein expression changes, we hypothesized that TEAD1's primary interaction partner changes from YAP to VGLL4 between newborn and adult heart. To test this hypothesis, we generated a Tead1 knockin allele, Tead1fb, which contains the FLAG and AviTag (Bio) (He et al., 2012) epitope tags fused to the Tead1 C terminus (Tead1fb; Figures S1A and S1B). The Escherichia coli enzyme BirA specifically recognizes and biotinylates the Bio tag (de Boer et al., 2003), permitting high-affinity pull-down on immobilized streptavidin (SA). Tead1fb/fb; Rosa26BirA/BirA mice survived normally to weaning, had no overt phenotype (Figures S1C and S1D), and expressed biotinylated TEAD1fb (Figures S1E and S1F). We then precipitated TEAD1fb and its interacting proteins on SA from postnatal day 1 (P1), P8, and adult (P50) hearts (Figures 1E–1G). This revealed that TEAD1 and VGLL4 strongly interacted in the adult but not the neonatal (P1 or P8) heart (Figures 1E–1G). TEAD1 and YAP interaction showed the opposite pattern, with strong interaction detected in the neonatal heart and weaker interaction in the adult heart (Figures 1F and 1G).
Precocious VGLL4 Overexpression Did Not Suppress Neonatal Cardiac Growth
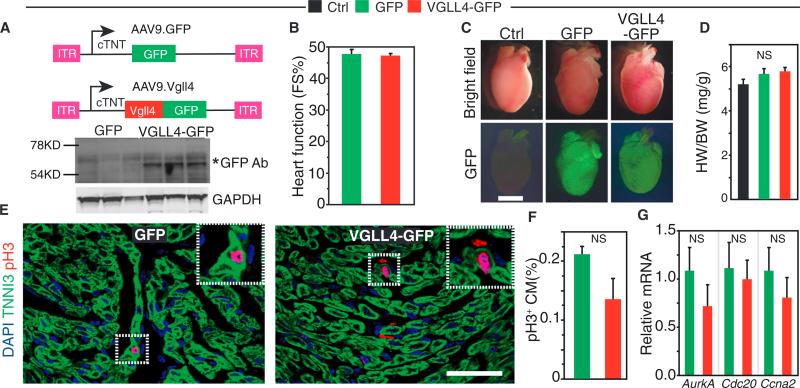
To further test the hypothesis that VGLL4 limits CM proliferation by reducing TEAD1-YAP interaction, we overexpressed VGLL4 in the newborn heart using adeno-associated virus serotype 9 (AAV9), an efficient cardiac gene delivery vector (Lin et al., 2014). We generated AAV9.VGLL4-GFP (AAV9.VGLL4) and AAV9.GFP, which express VGLL4-GFP fusion protein or GFP, respectively, from the cardiomyocyte-specific chicken cardiac troponin T (cTNT) promoter (Figure 2A), and injected them into P1 wild-type pups. Hearts were analyzed 7 days later. Immunoblots confirmed cardiac VGLL4-GFP expression (Figure 2A). Unexpectedly, AAV9.VGLL4 did not significantly change heart function or size compared with untreated (Ctrl) or AAV9.GFP-treated hearts (Figures 2B–2D). Staining for phospho-histone H3 (pH3), an M-phase cell-cycle marker, suggested that CM cell-cycle activity was not significantly changed by AAV9.VGLL4 compared with AAV-GFP (Figures 2E and 2F). Consistent with this observation, the expression of cell-cycle genes Aurka, Cdc20, and Ccna2 did not differ significantly between groups (Figure 2G).
Figure 2. VGLL4 Overexpression Did Not Suppress Neonatal Cardiac Growth.
P1 pups were injected subcutaneously with AAV9.GFP or AAV9.VGLL4-GFP. Control (Ctrl) mice were untreated. Hearts were analyzed at P8.
(A) AAV9 expression constructs. AAV9.GFP and AAV9.VGLL4-GFP incorporate the cardiac troponin T (cTNT) promoter to drive selective CM expression. Heart protein immunoblots probed with GFP antibody demonstrated VGLL4-GFP fusion protein expression (asterisk).
(B) Echocardiographic assessment of neonatal heart function. FS%, fractional shortening. n = 4.
(C) Whole-mount images of hearts showing lack of substantial differences between groups. Scale bar, 2 mm.
(D) Heart to body weight ratio was not significantly different (NS) between groups. n = 3.
(E) Representative pH3 staining results. Scale bar, 50 μm.
(F) Quantitation of pH3+ CMs. n = 3. NS, not significant.
(G) Cell-cycle gene expression from P8 ventricular myocardium after treatment with AAV9.GFP or AAV9.VGLL4-GFP. Gene expression was measured by qRT-PCR and normalized to the AAV9.GFP group. n = 4. NS, not significant.
All error bars represent the SEM.
We investigated the effect of AAV9.VGLL4 on YAP-TEAD1 interaction. In adult heart, where TEAD1 robustly interacted with endogenous VGLL4, we confirmed that VGLL4-GFP bound TEAD1 (Figure S2A). This result indicated that the GFP tag did not disrupt VGLL4-TEAD1 interaction. However, we did not detect VGLL4-GFP binding to TEAD1 in the neonatal heart (Figure S2B), suggesting that the lack of robust interaction between VGLL4 and TEAD1 in neonatal heart was not solely due to lower neonatal VGLL4 expression. Rather, the finding that forced expression of VGLL4-GFP in neonatal heart was not sufficient to drive its interaction with TEAD1 led us to hypothesize that additional factors regulate this interaction. These additional factors would limit interaction of overexpressed VGLL4-GFP and TEAD1 and thereby could account for the lack of phenotype in AAV9.VGLL4-treated heart.
VGLL4 Activity Is Regulated by Its TDU Domain Acetylation
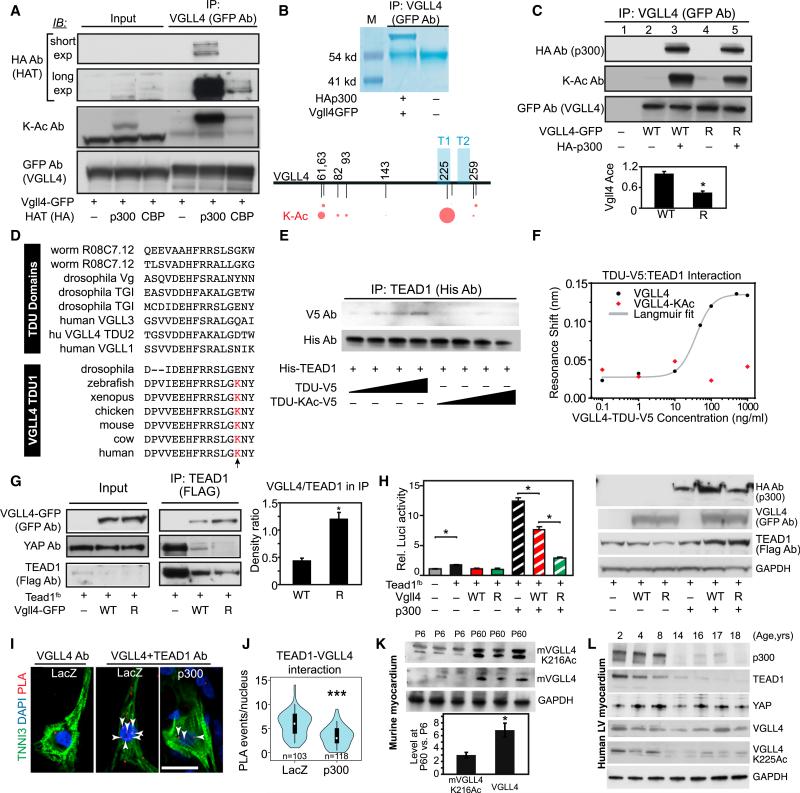
Post-translational modification is one potential regulatory mechanism that might govern VGLL4-TEAD1 interaction in the neonatal heart. Adopting a candidate strategy, we first investigated VGLL4 acetylation. Histone acetyltransferases such as p300 or CBP acetylate lysine resides of non-histone proteins, including transcription factors, in addition to histones (Chan and La Thangue, 2001). In co-immunoprecipitation (coIP) as-says, VGLL4 robustly interacted with p300, whereas its interaction with CBP was considerably weaker (Figure 3A). Moreover, p300 but not CBP heavily acetylated VGLL4 (Figure 3A). To identify VGLL4 acetylation sites, we co-expressed VGLL4-GFP fusion protein and p300 in HEK293T cells. Immunoprecipitated VGLL4-GFP was then analyzed by mass spectrometry. Several acetylation sites were identified, with the highest fraction of acetylated residues occurring at lysine 225 (K255; Figure 3B). To confirm this observation, we mutated VGLL4 K225 to arginine (VGLL4[R]), which is structurally similar to lysine but cannot be acetylated. In p300 co-transfected cells, compared with wild-type VGLL4, VGLL4[R] acetylation was significantly reduced (Figure 3C, ratio of KAc to total VGLL4 in lane 5 versus 3), consistent with VGLL4 being acetylated predominantly but not solely at K225. Vestigial-like family members interact with TEAD through their Tondu (TDU) domains (Koontz et al., 2013), and VGLL4 contains two TDU domains. K225 is located in the first TDU domain of human VGLL4 and is conserved among vertebrate VGLL4 proteins but not in TDU domains from other proteins (Figure 3D).
Figure 3. VGLL4 TDU Domain Acetylation Decreased VGLL4-TEAD1 Interaction.
(A) p300 bound and acetylated VGLL4. HEK293T cells were transfected with the indicated GFP and histone acetyltransferase (HAT; HA-tagged) expression plasmids. Proteins that co-precipitated with GFP were detected by immunoblotting (IB). K-Ac Ab, acetylated lysine-specific antibody.
(B) VGLL4-K225 is the major VGLL4 acetylation site. VGLL4-GFP was overexpressed in HEK293T cells in the presence of p300, immunoprecipitated with GFP antibody, and analyzed by mass spectrometry. The area of the red circles is proportional to the fraction of peptides detected that contain the acetyl lysine residue indicated by the corresponding number. T1 and T2 represent the two TDU domains of VGLL4.
(C) VGLL4 K225R mutation decreased VGLL4 acetylation. Wild-type (WT) or K255R mutated (R) VGLL4-GFP were co-expressed in HEK293T with p300, as indicated. VGLL4-GFP acetylation was detected by immunoprecipitation and western blot. Acetylation of VGLL4 K225R was normalized to wild-type VGLL4. *p < 0.05, n = 4.
(D) Alignment of TDU domains from different proteins (top group) or from the first TDU domain of VGLL4 from different species (bottom group). The residue aligned with K225 of human VGLL4 is highlighted in red and indicated by an arrow. This residue is conserved in vertebrate VGLL4 but is not conserved across TDU domains.
(E and F) VGLL4 K225 acetylation decreased VGLL4-TEAD1 interaction in vitro. Interaction between recombinant His-TEAD1[211–427] and synthetic, un-acetylated, or K225-acetylated VGLL4 TDU domain peptides was detected by immunoprecipitation and western blot (E) or by a nanoscale photonic interaction assay (F).
(G) VGLL4[R] increased VGLL4-TEAD1 and decreased YAP-TEAD1 interaction in cultured cells. TEAD1fb and VGLL4-GFP expression plasmids were co-transfected into 293T cells. TEAD1 CoIP was carried out using FLAG antibody. The ratio between VGLL4 and TEAD1 in the immunoprecipitate was quantified by densitometry. *p < 0.05, n = 3.
(H) p300 effect on YAP-TEAD1 transcriptional activity. 293T cells were co-transfected with the indicated plasmids plus pRL-TK. Left: 24 hr after transfection, cells were collected for luciferase activity measurement. Firefly luciferase activity was normalized to Renilla luciferase. *p < 0.05, n = 4. Right: western blot showing the expression of p300, Vgll4, and TEAD1fb in transduced cells.
(I and J) Effect of VGLL4 acetylation on VGLL4-TEAD1 interaction in NRVM. The PLA was used to detect endogenous VGLL4-TEAD1 interaction in cultured NRVMs. Representative image (I) and quantification of TEAD1-VGLL4 interaction events in the nucleus (J) are shown. Each red dot was counted as an interaction event. Scale bar, 20 μm. ***p < 0.0001, Wilcoxon.
(K) Endogenous levels of mVGLL4 (murine VGLL4) and mVGLL4-K216Ac (which corresponds to human K225Ac) in P6 and P60 heart. Hearts were lysed with denaturing buffer containing 2% SDS, and 100 μg of total protein was immunoblotted for total VGLL4 or VGLL4-K216Ac. Fold change of protein levels between P60 and P6 was determined by densitometry. *p < 0.05.
(L) Endogenous levels of VGLL4 and VGLL4-K225Ac in human left ventricular myocardium, obtained from unused transplant donor hearts of the indicated ages.
All error bars represent the SEM.
Based on its location within the TEAD1-binding domain of VGLL4, we hypothesized that acetylation of K225 modulates VGLL4-TEAD interaction. To test this hypothesis, we synthesized peptides corresponding to the VGLL4 TDU domains, with or without K225 acetylation (Figure S3A). These V5 epitope-tagged VGLL4 peptides were co-incubated with recombinant, His-tagged TEAD1 (residues 211–427), which contains the YAP and VGLL4 interaction domains (Jiao et al., 2014) (Figures S3B and S3C). The interaction between VGLL4 peptide and His-TEAD1 [211–427] was measured by immunoprecipitation followed by western blotting (Figure 3E). The synthetic non-acetylated VGLL4-TDU domain peptide bound TEAD1 in a dose-dependent manner. In contrast, the acetylated VGLL4-TDU peptide did not detectably interact with TEAD1. To quantify this result, we used a recently developed nanoscale photonic interaction assay (Yang et al., 2014). His-TEAD1[211–427] was immobilized on a nanobeam sensor and then incubated with increasing concentrations of the VGLL4-TDU peptide. The non-acetylated VGLL4-TDU peptide induced a concentration-dependent resonance shift of the sensor (Figure 3F), indicative of binding to TEAD1. Fitting the curve to the Langmuir equation yielded a VGLL4-TEAD1 interaction affinity of 3.1 ± 1.3 nM. In contrast, the acetylated VGLL4-TDU peptide did not induce a resonance shift up to a peptide concentration of 1 μg/mL (Figure 3F). Together these results indicate that VGLL4 acetylation at K225 strongly impedes its binding to TEAD1.
To study the effect of VGLL4 acetylation on VGLL4-TEAD1 interaction in a cellular context, we co-expressed VGLL4-GFP or VGLL4[R]-GFP with Tead1fb in 293T cells and evaluated their interaction by coIP. Even though TEAD1 level was lower in VGLL4[R]- than in VGLL4-expressing cells (see following section), TEAD1 co-precipitated significantly more VGLL4[R] (Figure 3G), suggesting that acetylation at K225 reduces VGLL4-TEAD1 interaction in vivo. Consistent with this result, VGLL4[R] inhibited TEAD1-YAP interaction with greater potency than wild-type VGLL4 (Figure 3G). We measured the functional effect of VGLL4 and its acetylation on TEAD1-YAP transcriptional activity using 8xGTIIC-Luci (Dupont et al., 2011), a luciferase reporter driven by a multimerized TEAD1 binding site. TEAD1 alone weakly stimulated reporter activity, and this was inhibited by both VGLL4 and VGLL4[R] (Figure 3H). Consistent with its broad role as a transcriptional co-activator, p300 strongly stimulated TEAD1 transcriptional activity. VGLL4 partially blocked this stimulation, as expected based on its antagonism of TEAD1-YAP interaction (Figure 3G). Compared with VGLL4, VGLL4[R] more potently blocked p300 stimulation (Figure 3H), in agreement with the more potent disruption of TEAD1-YAP interaction by VGLL4[R] (Figure 3G).
To determine whether VGLL4 acetylation affects VGLL4 and TEAD1 interaction in CMs, we used the proximity ligation assay (PLA) (Söderberg et al., 2006) to study the in situ interaction between VGLL4 and TEAD1 in cultured neonatal rat ventricular cardiomyocytes (NRVMs), with or without the overexpression of p300. NRVMs stained with TEAD1 or VGLL4 antibodies individually showed that TEAD1 was localized in the nucleus, while VGLL4 was located in both cytoplasm and nucleus (Figure S3D). The PLA showed in situ TEAD1-VGLL4 interaction primarily in the nucleus (Figure 3I). p300 overexpression significantly reduced TEAD1-VGLL4 interaction (p < 0.001; Figures 3I and 3J). These data indicate that acetylation of VGLL4 decreases its interaction with TEAD1 in CMs.
To determine whether VGLL4 is acetylated endogenously in CMs in vivo, we developed an antibody that recognized VGLL4 acetylated at K225 but not VGLL4 lacking this modification (Figures S3E and S3F). We used the antibody to assess acetylation of the corresponding residue of VGLL4 in murine heart (murine K216 corresponds to K225 of human VGLL4). Immunoblotting of mouse hearts showed that mVGLL4-K216Ac and total VGLL4 level increased 3- and 6-fold, respectively, between P6 and P60 (Figure 3K), indicating that the ratio of mVGLL4-K216Ac to total VGLL4 decreases with age. Consistent with the decrease in the fraction of acetylated VGLL4, p300 expression in the mouse heart also decreased with age (Figure S3G).
To assess the potential relevance of developmental changes in VGLL4, VGLL4 K225Ac, TEAD1, YAP, and p300 to the human heart, we examined their expression in normal human myocardium at different postnatal ages (Figure 3L). TEAD1 protein levels declined with age, as we observed in mouse. Unlike mouse, YAP expression and VGLL4 expression were relatively constant in 2- to 18-year-old hearts. However, VGLL4-K225Ac strongly decreased with postnatal age, as did expression of p300, paralleling our observations in mouse. These results suggest that developmentally regulated protein expression and VGLL4-K225 acetylation contribute to regulation of YAP-TEAD activity in the human heart. Whereas VGLL4 expression may contribute to stage-specific regulation of VGLL4-TEAD interaction, VGLL4 acetylation appears to predominate in humans.
We conclude that K225 acetylation of VGLL4 within its TDU domain inhibits its binding to TEAD1 in the heart. Mutation of VGLL4 K225 to arginine abrogates this inhibitory acetylation and promotes TEAD1-VGLL4 interaction.
VGLL4 Suppresses YAP Activity Partially by Promoting TEAD1 Degradation
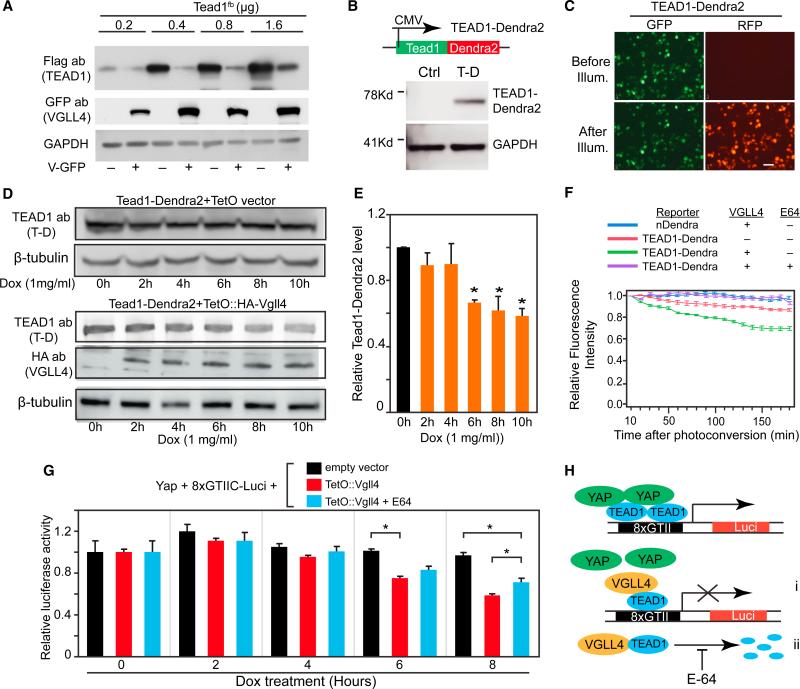
While investigating VGLL4-TEAD1 interaction, we noticed that VGLL4 overexpression reduced TEAD1 protein level (e.g., Figure 3G). To expand on this observation, we studied the effect of VGLL4 on different levels of transfected TEAD1. Consistently, VGLL4 reduced the steady-state level of TEAD1 (Figure 4A), leading to the hypothesis that VGLL4 regulates TEAD1's degradation rate. To test this hypothesis, we expressed TEAD1 fused to Dendra2, a fluorescent protein that converts from green to red fluorescence upon transient illumination with 405-nm light (Zhang et al., 2007) (Figures 4B and 4C). This allows the degradation of photoconverted protein to be monitored in real time, independent of ongoing protein synthesis. To allow rapid expression of VGLL4, we used an inducible expression system in which addition of doxycycline to the culture medium rapidly induced VGLL4 expression (Figures S4A and S4B). In control cells that did not express VGLL4, doxycycline treatment did not significantly affect steady-state TEAD1 levels over a 10-hr period (Figure 4D). In contrast, in cells that expressed VGLL4 upon doxycycline addition, steady-state TEAD1 levels declined by approximately 50% over the same period (p < 0.05; Figures 4D and 4E). VGLL4 did not reduce the mRNA level of Tead1-Dendra2 (Figure S4C), indicating that the effect of VGLL4 was post-transcriptional.
Figure 4. VGLL4 Overexpression Decreased TEAD1 Stability.
(A) VGLL4 overexpression decreased TEAD1 protein level. Different doses of TEAD1 plasmids (indicated in μg) were co-transfected with 1.6 μg of VGLL4-GFP plasmid. Cells were collected for western blot 24 hr after transfection.
(B and C) Generation and validation of TEAD1-Dendra2 construct. Tead1-Dendra2 plasmid was transfected into 293T cells. Western blot confirmed expression of TEAD1-Dendra2 fusion protein (B). TEAD1-Dendra2 merge fusion protein was green before illumination with 405 nm light. After 30 s of illumination, a fraction of TEAD1-Dendra2 exhibited red fluorescence (C).
(D and E) Doxycycline (Dox)-inducible expression of VGLL4 caused TEAD1-Dendra2 degradation. pTEAD1-Dendra2 and pEF1a-rtTA were co-transfected into 293T cells along with pTetO empty vector (upper panel) or pTetO:HA-VGLL4 (lower panel). Twenty-four hours after transfection, Dox was added. Cells were analyzed at the indicated time points (D). Quantification of TEAD1-Dendra2 protein level is shown in (E). *p < 0.05, n = 3.
(F) Time-lapse imaging of TEAD1-Dendra2 or Dendra2 proteins. Indicated plasmids were transfected into 293T cells. Twenty-four hours later, cells were treated with Dox. Four hours later, Dendra2 was photoconverted with 405-nm light. Relative red fluorescence intensity (RFI) was monitored for 3 hr by taking one image per minute. RFI was normalized to the value immediately after photoconversion. Plot shows average RFI signal over 10 min. n = 10. Experiment is representative of three independent repeats.
(G) Dual luciferase assay of YAP-TEAD1 transcriptional activity. 293T cells were co-transfected with YAP[S127A], EF1a:rtTA, 8xGIITC-luciferase, pRL-TK internal control, and either TetO empty vector or TetO-VGLL4 as indicated. E64 was added as indicated. Twenty-four hours after transfection, cells were treated with Dox for the indicated number of hours, when cell extracts were analyzed for Firefly and Renilla luciferase activity. Relative luciferase activity was the ratio of Firefly to Renilla luciferase, normalized to empty vector at time 0. *p < 0.05, n = 4.
(H) Model of VGLL4 regulation of YAP-TEAD1 activity. In the absence of VGLL4, YAP binds to TEAD1 to activate target gene expression. VGLL4 overexpression suppressed YAP-TEAD1 activity by both inhibiting TEAD1 transcriptional activity (i) and promoting TEAD1 degradation (ii).
All error bars represent the SEM.
We then combined doxycycline-induced VGLL4 expression with live cell imaging of TEAD1-Dendra2 to specifically probe the effect of VGLL4 on TEAD1 stability. Four hours after doxycycline treatment, TEAD1-Dendra2 was pulse-labeled by photoconversion. In the absence of VGLL4, TEAD1-Dendra2 protein fluorescence intensity dropped by 10% during the 3-hr imaging process, reflecting the basal degradation rate of TEAD1. In contrast, in the presence of VGLL4 the fluorescence intensity of photoconverted TEAD1-Dendra2 declined 30% over the same period (Figure 4F), indicating that VGLL4 accelerates TEAD1-Dendra2 degradation. VGLL4 did not affect the stability of Dendra2 itself (Figure 4F), indicating that the loss of TEAD1-Dendra2 was specific to the TEAD1 component. These data demonstrate that VGLL4 strongly influences TEAD1 stability.
Major pathways for protein degradation are the proteasome and cysteine, serine, and threonine peptidases. To determine whether these candidate pathways are involved in VGLL4-mediated TEAD1 degradation, we studied the effect of MG132 (a universal proteasome inhibitor [Lee et al., 1998]), leupeptin (an inhibitor of cysteine, serine, and threonine peptidases [Umezawa, 1976]), and E64 (an inhibitor specific to cysteine proteases [Barrett et al., 1982]) on VGLL4-mediated TEAD1 degradation. In 293T cells co-transfected with TEAD1 and VGLL4, leupeptin, and E64, but not MG132, reduced VGLL4-mediated reduction of TEAD1 steady-state levels (Figure S4D), suggesting that VGLL4 triggers TEAD1 degradation through cysteine proteases and not the proteosome. We used pulse-labeled TEAD1-Dendra2 and live cell imaging to confirm that E64 reduced the destabilizing effect of VGLL4 on TEAD1-Dendra2 (Figure 4F). Together, our data indicate that VGLL4 is sufficient to stimulate TEAD1 degradation through a cysteine protease-dependent pathway.
Previously, VGLL4 was shown to antagonize YAP-TEAD1 transcriptional activity by competitively binding to TEAD, displacing YAP (Koontz et al., 2013). Our data suggest an additional mechanism, in which VGLL4 binding to TEAD1 promotes its degradation and thereby reduces the amount available to interact with YAP. To test this hypothesis, we used the TEAD1 luciferase reporter 8xGTIIC-Luci (Dupont et al., 2011) to monitor TEAD1-YAP transcriptional activity. 293T cells were co-transfected with 8xGTIIC-Luci, doxycycline-inducible VGLL4, and YAP expression constructs. One day after transfection, luciferase reporter activity was measured 0–8 hr after doxycycline treatment. In the control group lacking doxycycline-inducible VGLL4, reporter activity was relatively stable after the addition of doxycycline (Figure 4G). In contrast, in cells with doxycycline-induced VGLL4, luciferase activity declined significantly over this time period (p < 0.05, Figure 4G). To probe the role of protein degradation in the effect of VGLL4, we treated cells with E64. This significantly increased reporter activity 8 hr after doxycycline treatment (p < 0.05; Figure 4G), suggesting that VGLL4 stimulation of TEAD1 degradation contributes to the decrease in transcriptional activity observed after VGLL4 induction. However, E64 did not completely abrogate the reduction of TEAD1 transcriptional activity caused by VGLL4 induction. In part this reflects incomplete E64 protection of TEAD1 from VGLL4-mediated degradation (Figure S4D), as well as the additional inhibitory effect of VGLL4 on YAP recruitment to TEAD1 (Figure 4H). We conclude that VGLL4 is sufficient to stimulate TEAD1 degradation through cysteine-dependent proteases, and that VGLL4 antagonizes TEAD1-YAP transcriptional activity by stimulating TEAD1 degradation in addition to disrupting TEAD1-YAP interaction.
Precocious Overexpression of VGLL4[R] in Neonatal Heart Leads to Heart Failure
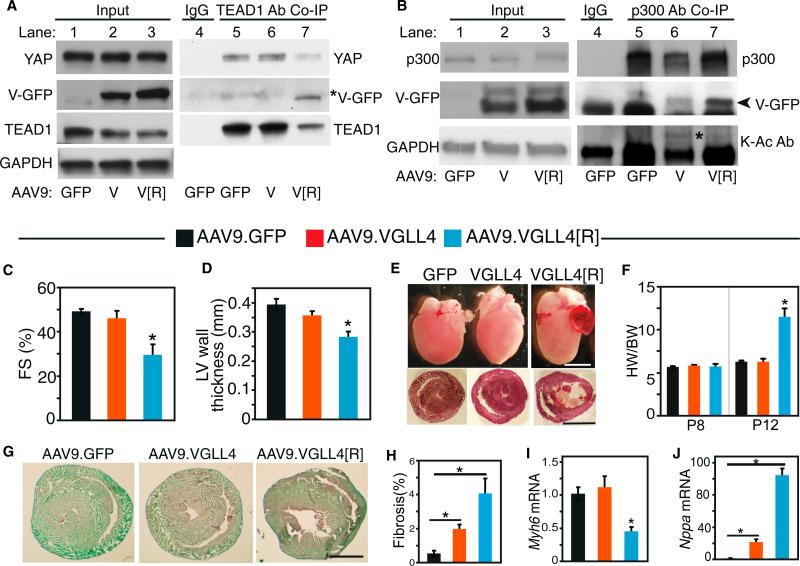
VGLL4 overexpressed in the neonatal heart did not interact with TEAD1 and did not significantly affect neonatal heart growth or function (Figure 2). However, VGLL4-K225 acetylation in the neonatal heart reduced VGLL4 effect on TEAD1-YAP and thus may have masked VGLL4's biological activity. Because the VGLL4[R] mutant is refractory to inhibition by K225 acetylation, we hypothesized that this single amino acid substitution would reveal the cardiac activity of overexpressed VGLL4. To test this hypothesis, we introduced the mutant protein into the neonatal heart by developing and administering AAV9.VGLL4[R]. AAV9.GFP and AAV9.VGLL4 were used as negative controls. TEAD1-interacting proteins were detected by coIP. Consistent with our prior results, we did not detect significant interaction between TEAD1 and VGLL4 (Figure 5A, lane 6 versus lane 5). Accordingly, AAV9.VGLL4 did not affect TEAD1 level or TEAD1-YAP interaction. In contrast, VGLL4[R] did interact with TEAD1 (Figure 5A, lane 7 versus lanes 5 and 6). Consistent with this interaction, TEAD1 level was reduced by VGLL4[R] (Figure 5A, lane 7 versus lanes 5 and 6), and the balance between TEAD1's interaction partners was altered, with greater binding to VGLL4[R] and reduced binding to YAP. Together, these results support our model that VGLL4 acetylation at K225 governs its interaction with TEAD1 in the neonatal heart. By blocking K225 acetylation, the K225R mutation reveals the potent effect of VGLL4 on neonatal heart function.
Figure 5. Abrogation of VGLL4-K225 Acetylation Unmasked Disruptive Effects of VGLL4 on YAP-TEAD Interaction and Neonatal Heart Maturation.
P1 pups were treated with AAV9.VGLL4, AAV9.VGLL4[R] (containing the K225R mutation), or AAV.GFP. Hearts were examined at P8 or P12, as indicated.
(A) Assay of cardiac TEAD1-interacting proteins. TEAD1 and its associated proteins were immunoprecipitated, and indicated proteins were detected by western blotting. Asterisk indicates the VGLL4-GFP band.
(B) Endogenous p300 interacts with and acetylates VGLL4 in the neonatal heart. AAV9.GFP, AAV9.VGLL4-GFP, or AAV9.VGLL4[R]-GFP were administered to P1 mouse pups. p300 was immunoprecipitated from P8 heart extracts and probed with indicated antibodies. K-Ac Ab, acetyl lysine-specific antibody. Asterisk indicates acetylated VGLL4-GFP. Arrowhead indicates the VGLL4-GFP band, which runs just above the immunoglobulin heavy chain.
(C and D) Echocardiographic measurement of left ventricular (LV) systolic function (fractional shortening, FS) (C) and diastolic LV wall thickness (D) at P8. *p < 0.05 compared with GFP control, n = 4.
(E) Whole-mount (upper panels) and H&E-stained short-axis sections of AAV-transduced hearts at P12. Scale bars, 2 mm.
(F) Heart to body weight ratio of AAV-transduced hearts at P8 or P12. *p < 0.05, n = 4.
(G and H) Cardiac fibrosis was visualized by pirosirius red/fast green staining. Representative images (G) and quantification (H). Scale bar, 1 mm. *p < 0.05, n = 3.
(I and J) qRT-PCR measurement of heart failure marker gene transcripts Myh6 and Nppa. Levels were normalized to GAPDH and expressed relative to the AAV9.GFP control group. *p < 0.05, n = 4.
All error bars represent the SEM.
Next, we addressed the role of p300 in VGLL4 K225R acetylation in the neonatal heart. p300 level was not affected by overexpression of VGLL4 or VGLL4[R] (Figure 5B, lanes 2 and 3 versus lane 1). VGLL4 and VGLL4[R] both co-immunoprecipitated with p300 (Figure 5B, lanes 6 and 7 versus lane 5) in neonatal heart, while this interaction was not detected in adult heart (Figure S5A). Co-precipitated VGLL4 was acetylated, whereas VGLL4[R] was not detectably acetylated (Figure 5B, lane 6 versus lane 7). This result validated that the K225R mutation reduced VGLL4 acetylation, and suggested that p300 mediates VGLL4 acetylation in vivo.
Given that VGLL4 K225R mutation increases TEAD1-VGLL4 interaction in neonatal heart at the expense of TEAD1-YAP, we next examined the biological effect of this single amino acid substitution. We delivered AAV9.VGLL4[R] to P1 mice. Litter-mates treated with AAV9.VGLL4 (wild-type) and AAV9.GFP-treated were used as negative controls. At P8, all three groups had similar heart and body weights (Figure S5B), and, as we observed previously, heart function was no different between AAV9.VGLL4 and AAV-GFP groups (Figure 5C). However, AAV9.VGLL4[R] induced severe myocardial dysfunction and myocardial wall thinning (Figures 5C and 5D). The AAV9.VGLL4[R]-treated mice failed to grow normally, and 30% died prior to a planned necropsy date at P12 (Figures S5B and S5C). Those that survived to P12 had striking ventricular and atrial enlargement that was not observed in either negative control group (Figure 5E). These mice had lower body weight and higher heart weight than the other two groups (Figures 5F, S5B, and S5C). Staining of heart sections with picrosirius red showed that AAV9.VGLL4[R] hearts had extensive fibrosis (Figures 5G and 5H). Interestingly, AAV9.VGLL4 hearts also had mildly but significantly increased fibrosis compared with AAV9.GFP. AAV9. Gene expression measurements by qRT-PCR showed that VGLL4[R] induced cardiac upregulation of Nppa and downregulation of Myh6, changes frequently observed in heart failure (Figures 5I and 5J).
Together, these results show that K225R acetylation regulates VGLL4 activity in the neonatal heart. Blocking K225R uncovered potent VGLL4 activity to disrupt TEAD1-YAP interaction and neonatal heart development and function.
Activation of VGLL4 in the Neonatal Heart Suppresses Cardiomyocyte Proliferation by Disrupting the YAP-TEAD1 Complex
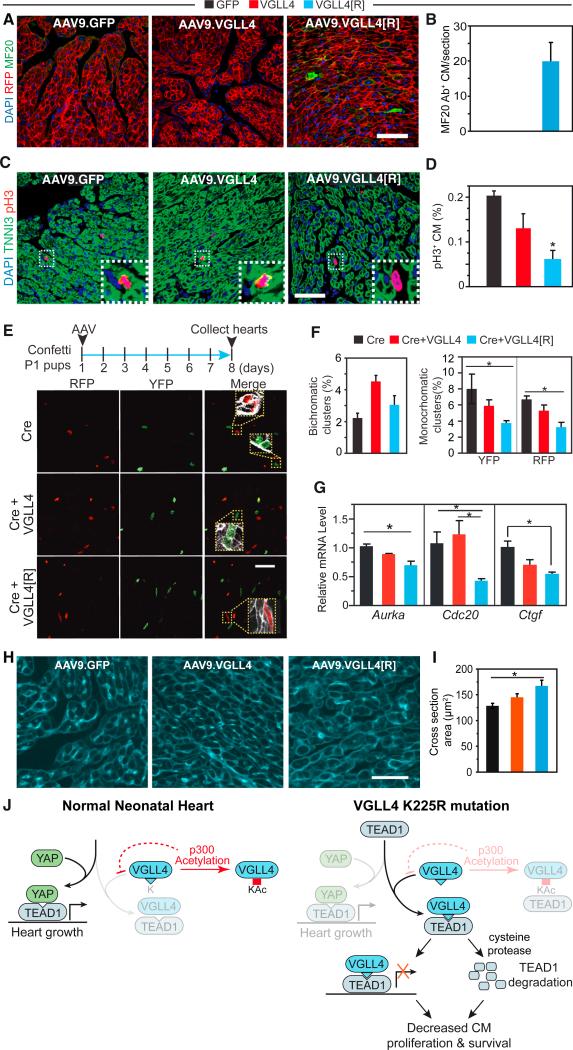
The YAP-TEAD complex promotes CM proliferation (von Gise et al., 2012; Xin et al., 2011; Heallen et al., 2011; Lin et al., 2014), and loss of this activity caused heart failure at least in part due to reduced CM number (Del Re et al., 2013). In addition, YAP-TEAD has been implicated in regulating CM survival (Del Re et al., 2013). To understand the cellular mechanisms underlying VGLL4[R]-induced heart failure, we investigated processes that might alter CM survival (apoptosis or necrosis) or production (proliferation). VGLL4[R] did not significantly induce CM apoptosis, as measured by TUNEL staining (Figure S6A). CMs undergoing necrosis have plasma membranes that are abnormally permeable to macromolecules such as antibodies. Therefore, uptake of injected anti-myosin antibody by CMs has been used as an assay of CM necrosis (Nakayama et al., 2007). To determine whether AAV9.VGLL4[R] caused CM necrosis, we injected mouse anti-myosin antibody (MF20) into mouse pups at P7, and hearts were analyzed at P8. In both negative control groups, MF20+ CMs were rarely observed, whereas MF20+ CMs were readily observed in the AAV9.VGLL4[R] group (Figures 6A and 6B).
Figure 6. Acetylation-Deficient VGLL4[R] Decreased Cardiomyocyte Proliferation and Survival.
AAV9.GFP, AAV9.VGLL4, or AAV9.VGLL4[R] were delivered to P1 pups, and hearts were examined at P8.
(A and B) Measurement of CM necrosis. Rosa26mTmG (membrane localized RFP) P1 pups were treated with AAV9. Anti-myosin antibody MF20 was injected into mice at P7. At P8, mice were collected and intracellular MF20 antibody was detected by immunofluorescent staining. Representative images (A) and quantification (B). Scale bar, 50 μm.
(C and D) Measurement of CM proliferation using pH3 immunofluorescence staining. (C) Representative images with boxed regions magnified in insets. Scale bar, 50 μm. (D) Quantification of pH3+ CMs. *p < 0.05, n = 3.
(E and F) Clonal assay for CM proliferation. Confetti P1 pups were treated with mosaic dose of AAV9.Cre to label individual CMs, in addition to treatment with VGLL4 or control AAVs at the usual dose, which transduces nearly all CMs. Hearts were examined at P8. (E) Representative images with boxed regions magnified in insets. Scale bar, 50 μm. (F) quantification of clusters of adjacent, labeled CMs containing one color (monochromatic, potentially arising from proliferation) or two colors (bichromatic, arising from adjacent labeling events). Scale bar, 50 μm. *p < 0.05, n = 4.
(G) qRT-PCR measurement of relative levels of YAP-TEAD canonical targets gene transcripts Aurka, Cdc20, and Ctgf in P12 heart. *p < 0.05, n = 3.
(H and I) Measurement of cardiomyocyte cross-sectional area. CMs were outlined by wheat germ agglutinin staining. Representative images (H) and quantification of cross-sectional area (I). *p < 0.05, n = 3.
(J) Model of VGLL4 regulation of heart growth. In normal newborn heart, predominant YAP-TEAD1 stimulates CM proliferation. The interaction between VGLL4 and TEAD1 is blunted by p300-mediated VGLL4 acetylation. Inhibition of VGLL4 acetylation, as in the K225R mutant, suppresses cardiac growth by both inhibiting YAP-TEAD1 interaction and decreasing TEAD1 stability.
All error bars represent the SEM.
To determine whether VGLL4[R] reduced CM proliferation, we performed quantitative pH3 staining. VGLL4[R] strongly decreased the fraction of pH3+ CMs compared with VGLL4 or GFP (Figure 6C). Because CM multinucleation or polyploidization can dissociate CM M-phase activity from CM number, we undertook a clonal analysis to more directly probe the effect of VGLL4 on CM proliferation. As we described previously (Lin et al., 2014), by pulse-labeling a low fraction of CMs and later counting the number of CMs in individual labeled clusters, one can assess the extent of productive CM cell-cycle activity. Use of the multi-color Confetti reporter mouse (Snippert et al., 2010), in which Cre stochastically activates expression of one of four fluorescent proteins, further enhances this strategy by allowing one to distinguish chance labeling of two neighboring cells (potentially yielding multichromatic clusters) from expansion of a single CM (monochromatic clusters). AAV9.cTNT::Cre (AAV9.Cre) was injected into the P1 Brainbow pups at a low dose to irreversibly label a small fraction of CMs with one of the four different fluorescent proteins: CFP, RFP, nuclear GFP (nGFP), and YFP. For technical reasons, we only considered RFP and YFP readouts. In pilot experiments, we determined the optimal dose of AAV9.Cre to achieve the desired CM labeling rate (Figures S6B–S6D). We delivered this dose of AAV9.Cre to P1 Confetti mouse pups. At the same time, we delivered AAV9.VGLL4fb or AAV9.VGLL4 [R]fb, in which the GFP tag has been replaced by the FLAG-Bio tag, at doses which should transduce greater than 90% of cardiomyocytes. In P8 hearts, we determined the frequency of bichromatic and monochromatic cell clusters, where a cluster was defined as two or more labeled, adjacent cells (Figure 6E). As expected, bichromatic clusters, representing independent labeling of neighboring cells, occurred at similar frequency in AAV9.Cre, AAV9.Cre+AAV9.VGLL4fb, and AAV9.Cre+AAV9.VGLL4[R]fb groups (Figure 6F). In contrast, monochromatic clusters, representing productive CM proliferation, were less common in the VGLL4 [R] group (Figure 6F), supporting our hypothesis that VGLL4[R] overexpression reduces neonatal CM proliferation. Consistent with these measurements of CM proliferation, expression of cell-cycle genes Aurka and Cdc20, as well as the canonical TEAD-YAP target gene Ctgf, was reduced in P12 hearts treated with AAV9.VGLL4[R], compared with AAV9.GFP (Figure 6G).
Both reduction of CM cell-cycle activity and CM survival would reduce CM number, leading to increased workload for remaining CMs. Stressed CMs undergo compensatory hypertrophy, and indeed we observed that AAV9.VGLL4[R]-treated CMs are larger than CMs in the GFP-negative control group (Figures 6H and 6I). In combination with increased cardiac fibrosis, these changes may account for the increased weight of AAV9.VGLL4[R]-treated hearts (Figures 5F and S5C).
We conclude that blocking VGLL4 K225 acetylation reveals the potent effect of VGLL4 on CM proliferation and survival in the neonatal heart, at least in part through disruption of TEAD1-YAP interaction and destabilization of TEAD1.
DISCUSSION
Our work identifies VGLL4 as an important negative regulator of cardiac growth driven by YAP-TEAD1, defines VGLL4 acetylation as a novel mechanism to regulate Hippo-YAP signaling and organ growth, and reveals VGLL4 regulation of TEAD1 stability as an additional mechanism whereby VGLL4 modulates YAP-TEAD activity. Although our studies focused on the effect of VGLL4 on YAP-TEAD activity, it is possible that VGLL4 has other important biological functions in CMs. For instance, the Hippo-YAP pathway is closely intertwined with Wnt/β-catenin signaling (Heallen et al., 2011; Varelas et al., 2010), and in future studies it will be invaluable to assess the impact of VGLL4 on this pathway.
Developmental Regulation of TEAD-Interacting Proteins
The Hippo-YAP pathway controls the growth of mitotic organs, such as liver (Dong et al., 2007), intestine (Camargo et al., 2007), skin (Schlegelmilch et al., 2011), and fetal heart (Heallen et al., 2011; von Gise et al., 2012; Xin et al., 2011). In the adult heart, YAP activation induced limited CM proliferation and was insufficient to cause cardiomegaly (Lin et al., 2014). We investigated mechanisms that restrain YAP activity in the adult heart, and our data identify VGLL4 as a potential negative regulatory factor. Among the organs that we studied, VGLL4 was most highly expressed in the heart. As proliferative neonatal CMs transitioned to become postmitotic adult CMs, VGLL4 protein levels increased markedly, whereas YAP and TEAD1 protein levels declined. Interestingly, these protein level changes were post-transcriptionally mediated through uncharacterized mechanisms. Consistent with these changes in protein expression, the main interaction partner of TEAD1 switched from YAP in the neonatal heart to VGLL4 in the adult heart. This developmentally regulated switch in interaction partners is crucial for normal heart maturation, since precocious formation of TEAD1-VGLL4 complex in the neonatal heart caused cardiac hypoplasia, CM necrosis, and lethal heart failure. We are now developing essential reagents to test whether blocking VGLL4 activity in the adult heart will boost YAP-TEAD1 mitogenic activity.
Regulation of YAP-TEAD1 Activity through VGLL4 Acetylation
Many studies have examined the regulatory pathways that converge on YAP to regulate its cellular localization and transcriptional activity, often through YAP post-translational modifications that include phosphorylation, acetylation, and methylation (Pan, 2010; Hata et al., 2012; Oudhoff et al., 2013). As a transcriptional co-activator, YAP transcriptional activity depends upon its binding to a partner DNA-binding transcription factor. On a genome-wide scale, TEAD is the major transcription factor partner of YAP (Zanconato et al., 2015; Galli et al., 2015), and we previously showed that TEAD1-YAP interaction is essential for fetal heart growth (von Gise et al., 2012). TEAD1 likely also has additional roles in the regulation of muscle gene expression (Yoshida, 2008). Nevertheless, little attention has been directed at mechanisms that control TEAD1 activity and stability.
We found that VGLL4-TEAD1 interaction is developmentally regulated. Although this is partially explained by developmentally regulated changes in protein expression, the lack of interaction between overexpressed VGLL4 and TEAD1 in the neonatal heart pointed to additional regulatory mechanisms. We discovered a second mechanism, VGLL4 acetylation at K225, which normally impedes VGLL4-TEAD interaction in the neonatal heart (Figure 6J). Abrogation of acetylation at this residue in the VGLL4 [R] mutant precociously impaired VGLL4-TEAD1 interaction, thereby reducing YAP-TEAD1 mitogenic and pro-survival activity (Figure 6J). Thus, our data show that this post-translational modification is a critical regulatory switch that is essential for neonatal CM proliferation and survival.
One of the major acetyltransferases that acetylates VGLL4 is p300. We readily detected interaction between endogenous p300 and VGLL4 in the neonatal but not adult heart, indicating that this interaction is also developmentally regulated. Decline of p300 levels, alteration of its primary interaction partners, or destabilization of the p300-VGLL4 complex (e.g., due to changes in the composition of each protein's interacting partner complexes) may all contribute to this developmentally reduced interaction. Decreased p300-VGLL4 interaction between neonatal and adult CMs correlated with a decrease in the fraction of VGLL4 that is acetylated. However, VGLL4 acetylation was still present in the adult heart, and indeed the absolute level of acetylated VGLL4 was higher. This suggests that a different acetyltransferase may acetylate VGLL4 in adult CMs. Alternatively p300 may still acetylate VGLL4, albeit with reduced efficiency that reflects the decline of p300-VGLL4 interaction detectable by coIP. Understanding the mechanisms that regulate VGLL4 acetylation may lead to therapeutic strategies to enhance VGLL4 acetylation and thereby mitigate its inhibitory effects on YAP-TEAD activity in mature CMs.
Interestingly, VGLL4 appeared to be regulated differently in human and mouse hearts. Whereas mouse VGLL4 increased markedly with postnatal age, its expression in human heart was relatively constant. On the other hand, VGLL4-K225Ac declined with age in human heart. In mouse it increases, but total VGLL4 increases even more, so that the proportion of VGLL4-K225Ac decreases. This suggests that developmental regulation of VGLL4 activity may be achieved in different species by varying combinations of regulated expression or acetylation.
Precise YAP-TEAD regulation is often achieved through signaling pathways that either add or remove post-translational modifications. For instance, the Hippo kinase cascade phosphorylates YAP to trigger its cytoplasmic sequestration (Huang et al., 2005). This is counterbalanced by YAP dephosphorylation by the protein phosphatase PP2A, a process regulated by YAP interaction with α-catenin (Schlegelmilch et al., 2011). Lysine acetylation is also a reversible post-translational modification that can be removed by deacetylases. Thus in future work it will be interesting to consider whether deacetylases counterbalance VGLL4 acetylation.
VGLL4 Regulates TEAD1 Stability
Previous work suggested that VGLL4 suppresses YAP activity by competing for TEAD binding (Zhang et al., 2014; Koontz et al., 2013). Our data support this mechanism, and uncovered an additional previously unrecognized mechanism by which VGLL4 regulates YAP-TEAD1 activity. We found that VGLL4 interaction with TEAD1 accelerated TEAD1 protein degradation through cysteine peptidases (Figure 6J). Cysteine peptidase inhibition by E64 only partially rescued YAP-TEAD1 transcriptional activity, suggesting that VGLL4 regulates YAP-TEAD1 activity by both causing TEAD1 degradation and inhibiting YAP-TEAD1 interaction. Future work is required to define the mechanism by which VGLL4 enhances TEAD1 degradation, and the specific cysteine peptidases that degrade TEAD1.
VGLL4 Affects Postnatal Cardiac Growth and Maturation by Suppressing CM Proliferation and Inducing Necrosis
We found that VGLL4 acetylation at K225 blocks its activity in the neonatal heart. Overriding VGLL4 acetylation by mutating this residue to arginine unmasked the potent effect of VGLL4 gain of function in the neonatal heart, resulting in destabilization of the TEAD1-YAP complex and reduced CM proliferation and YAP target gene expression. Moreover, VGLL4[R] induced CM necrosis but not apoptosis. As a result of reduced CM proliferation and increased necrosis, AAV9.VGLL4[R]-transduced pups developed heart failure.
These data identify an additional, previously unreported role of YAP-TEAD1 in CMs to suppress necrosis. This function of YAP may cross cell types, as YAP deficiency predisposed hepatocytes to undergo necrosis after bile duct ligation (Bai et al., 2012). CM necrosis is an important mechanism of CM loss following experimental myocardial infarction and genetically induced CM calcium overload (Kajstura et al., 1998; Nakayama et al., 2007). It will be interesting to dissect the mechanisms by which the balance between VGLL4-TEAD and YAP-TEAD governs CM necrosis.
Conclusions
This study identified novel mechanisms that regulate YAP-TEAD activity. Normal deployment of these mechanisms is crucial for neonatal cardiac growth and maturation. Our study suggests that manipulation of the balance between VGLL4, TEAD, and YAP activity, and/or suppressing VGLL4-mediated TEAD1 degradation, may be of use for therapeutic cardiac regeneration or repair. Alternatively, augmenting the inhibitory action of VGLL4 or reducing its acetylation may be useful strategies for the control of oncogenic growth driven by excessive YAP-TEAD activity.
EXPERIMENTAL PROCEDURES
For detailed methods, please refer to Supplemental Experimental Procedures.
Animal Experiments
Animal procedures were approved by the Boston Children's Hospital Animal Care and Use Committee. Tead1fb knockin mice (MMRRC:037514-JAX) were generated by targeting the C terminus of Tead1 in murine embryonic stem cells to introduce FLAG and Bio epitope tags.
Human Myocardium
Human left ventricular myocardium from unused donor hearts was snap-frozen within 2 hr of organ harvest, under protocols approved by the Institutional Review Boards of the University of Sydney and St. Vincent's Hospital.
AAV9 Vector Generation and Delivery
Vgll4 or Vgll4[R] were cloned into ITR-containing AAV plasmid (Penn Vector Core P1967) harboring the chicken cardiac TNT promoter (Prasad et al., 2011). AAV9 was packaged as described by Lin et al. (2014) and delivered by subcutaneous injection.
Proteins and Peptides
Murine TEAD1 residues 211–427, containing the YAP and VGLL4 binding domain, was fused to a polyhistidine tag, expressed in bacteria, and purified on Ni-nitrilotriacetic acid agarose (Qiagen). Peptides VGLL4-TDU-V5 and VGLL4-TDU-KAc-V5 (Figure S3) were chemically synthesized. VGLL4-TEAD1 binding affinity was determined using photonic crystal nanobeam sensors (Yang et al., 2014).
Cell or tissue-soluble protein extracts for coIP were immunoprecipitated using protein A or streptavidin Dynabeads (Life Technologies). Acetylation was analyzed using pan-acetylated lysine antibody (Cell Signaling Technology, 9441S) or mass spectrometry (Harvard Medical School Taplin Biological Mass Spectrometry Facility). Murine VGLL4-K216Ac specific antibody was raised in rabbits and isolated by affinity purification.
Co-immunoprecipitation and In Vitro VGLL4 Acetylation Analysis
Cell or tissue-soluble protein extracts for coIP were prepared in lysis buffer (20 mM Tris-HCl [pH 8], 137 mM NaCl, 10% glycerol, 1% Triton X-100, and 2 mM EDTA). Protease inhibitor cocktail (Roche) was added to the lysis buffer immediately before use. The protein solution was diluted with 1 volume of immunoprecipitation buffer (lysis buffer without glycerol) and pre-cleared with protein A Dynabeads (Life Technologies, 10008D). Antibody or immunoglobulin G was added to the pre-cleared extract, and antibody bound protein complexes were pulled down with pre-equilibrated protein A Dynabeads. After three washes, the immunoprecipitated proteins were eluted with 1× SDS loading buffer.
For analysis of VGLL4 acetylation, 293T cells were co-transfected with VGLL4-GFP and other indicated plasmids and cultured for 48 hr. Two hours before harvest, cells were treated with 5 μM trichostatin A (TSA; Cayman Chemical, CAS 58880-19-6). GFP antibody was used to pull down VGLL4-GFP in the presence of 5 μM TSA. Acetylation was analyzed using pan-acetylated lysine antibody (Cell Signaling Technology, 9441S) or mass spectrometry (Harvard Medical School Taplin Biological Mass Spectrometry Facility).
Cardiomyocyte Isolation and Culture
Neonatal rat ventricle cardiomyocytes (NRVMs) were isolated from 2-day-old Wistar rats (Charles River) using the Neomyts cardiomyocyte dissociation kit (Cellutron). Isolated cardiomyocytes were initially cultured for 24 hr in the presence of 10% fetal bovine serum. Cardiomyocytes were then cultured on fibronectin-coated glasses in 1% horse serum medium and cultured for 24 hr before 4% paraformaldehyde (PFA) fixation.
Duolink In Situ PLA
PFA (4%)-fixed NRVMs were permeabilized with PBS containing 0.1% Triton X-100. Duolink PLA was carried out using the Sigma Duolink In Situ Red Starter Kit Mouse/Rabbit (Duo92101-1KT). To visualize cardiomyocytes, we co-stained cells with goat-originated cardiac troponin I antibody (Abcam). In brief, NRVMs were incubated at 4°C overnight with primary antibodies against VGLL4, TEAD1, and cardiac troponin I. On the second day, PLA was carried out following the kit protocol. Alexa 488-conjugated donkey anti-goat secondary antibody was added at the signal amplification step.
Statistics
Values are expressed as mean ± SEM. Student's t test or ANOVA with Tukey's honest significant difference post hoc test was used to test for statistical significance involving two or more than two groups, respectively.
Source Data
Original immunoblot data have been deposited at Mendeley Data with the following digital object identifiers, corresponding to Figures 1, 2, 3, 4, and 5 and Figures S1–S6, respectively: 10.17632/36rcz2ndks.1, 10.17632/8mg7zn6k9n.1, 10.17632/s9x6x6shp3.1, 10.17632/xn4xhbfms9.1, 10.17632/yxmwb5r5j7.1, and 10.17632/jfdxk7nnhs.1.
Supplementary Material
Highlights.
TEAD-YAP stimulates organ growth, but its regulation is incompletely understood
p300-mediated acetylation of VGLL4 K225 regulates VGLL4 antagonism of TEAD-YAP
VGLL4-TEAD1 interaction promotes TEAD1 degradation
VGLL4-K225Ac regulates TEAD1 activity to allow normal heart development
ACKNOWLEDGMENTS
We thank Ross Tomaino from Taplin Biological Mass Spectrometry Facility for mass spectroscopy analysis of VGLL4 post-translational modifications. W.T.P. was supported by NIH HL116461, U01 HL100401, and UM1 HL098166, by an AHA Established Investigator Award (12EIA8440003), and by charitable contributions from Gail Federici Smith and Dr. and Mrs. Edwin A. Boger. Z.L. was supported by an American Heart Association Scientist Development Grant.
Footnotes
AUTHOR CONTRIBUTIONS
Z.L. designed and performed the study and wrote the manuscript. W.T.P. analyzed the data and co-wrote the manuscript. H.G., S.Z., Y.C., N.V.D., Y.G., Q.M., P.Z., J.Z., A.H., P.R.v.G., and S.S. generated reagents and acquired data. F.L. and Q.Q. performed nanoscale photonic interaction assays. A.L. and C.d.R. provided human heart samples. V.J.B. analyzed the Dendra2 time-lapse imaging data.
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Experimental Procedures and six figures and can be found with this article online at http://dx.doi.org/10.1016/j.devcel.2016.09.005.
REFERENCES
- Bai H, Zhang N, Xu Y, Chen Q, Khan M, Potter JJ, Nayar SK, Cornish T, Alpini G, et al. Yes-associated protein regulates the hepatic response after bile duct ligation. Hepatology. 2012;56:1097–1107. doi: 10.1002/hep.25769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett AJ, Kembhavi AA, Brown MA, Kirschke H, Knight CG, Tamai M, Hanada K. L-trans-Epoxysuccinyl-leucylamido(4-guanidino) butane (E-64) and its analogues as inhibitors of cysteine proteinases including cathepsins B, H and L. Biochem. J. 1982;201:189–198. doi: 10.1042/bj2010189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camargo FD, Gokhale S, Johnnidis JB, Fu D, Bell GW, Jaenisch R, Brummelkamp TR. YAP1 increases organ size and expands un-differentiated progenitor cells. Curr. Biol. 2007;17:2054–2060. doi: 10.1016/j.cub.2007.10.039. [DOI] [PubMed] [Google Scholar]
- Chan HM, La Thangue NB. p300/CBP proteins: HATs for transcriptional bridges and scaffolds. J. Cell Sci. 2001;114:2363–2373. doi: 10.1242/jcs.114.13.2363. [DOI] [PubMed] [Google Scholar]
- Chen HH, Mullett SJ, Stewart AF. Vgl-4, a novel member of the vestigial-like family of transcription cofactors, regulates alpha1-adrenergic activation of gene expression in cardiac myocytes. J. Biol. Chem. 2004;279:30800–30806. doi: 10.1074/jbc.M400154200. [DOI] [PubMed] [Google Scholar]
- de Boer E, Rodriguez P, Bonte E, Krijgsveld J, Katsantoni E, Heck A, Grosveld F, Strouboulis J. Efficient biotinylation and single-step purification of tagged transcription factors in mammalian cells and transgenic mice. Proc. Natl. Acad. Sci. USA. 2003;100:7480–7485. doi: 10.1073/pnas.1332608100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Re DP, Yang Y, Nakano N, Cho J, Zhai P, Yamamoto T, Zhang N, Yabuta N, Nojima H, et al. Yes-associated protein isoform 1 (Yap1) promotes cardiomyocyte survival and growth to protect against myocardial ischemic injury. J. Biol. Chem. 2013;288:3977–3988. doi: 10.1074/jbc.M112.436311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Feldmann G, Huang J, Wu S, Zhang N, Comerford SA, Gayyed MF, Anders RA, Maitra A, Pan D. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120–1133. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, Zanconato F, Le Digabel J, Forcato M, et al. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474:179–183. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- Galli GG, Carrara M, Yuan W-C, Valdes-Quezada C, Gurung B, Pepe-Mooney B, Zhang T, Geeven G, Gray NS, et al. YAP drives growth by controlling transcriptional pause release from dynamic enhancers. Mol. Cell. 2015;60:328–337. doi: 10.1016/j.molcel.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata S, Hirayama J, Kajiho H, Nakagawa K, Hata Y, Katada T, Furutani-Seiki M, Nishina H. A novel acetylation cycle of transcription co-activator Yes-associated protein that is downstream of Hippo pathway is triggered in response to SN2 alkylating agents. J. Biol. Chem. 2012;287:22089–22098. doi: 10.1074/jbc.M111.334714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He A, Shen X, Ma Q, Cao J, von Gise A, Zhou P, Wang G, Marquez VE, Orkin SH, Pu WT. PRC2 directly methylates GATA4 and represses its transcriptional activity. Genes Dev. 2012;26:37–42. doi: 10.1101/gad.173930.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heallen T, Zhang M, Wang J, Bonilla-Claudio M, Klysik E, Johnson RL, Martin JF. Hippo pathway inhibits Wnt signaling to restrain cardiomyocyte proliferation and heart size. Science. 2011;332:458–461. doi: 10.1126/science.1199010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heallen T, Morikawa Y, Leach J, Tao G, Willerson JT, Johnson RL, Martin JF. Hippo signaling impedes adult heart regeneration. Development. 2013;140:4683–4690. doi: 10.1242/dev.102798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidenreich PA, Albert NM, Allen LA, Bluemke DA, Butler J, Fonarow GC, Ikonomidis JS, Khavjou O, Konstam MA, et al. Forecasting the impact of heart failure in the United States: a policy statement from the American Heart Association. Circ. Heart Fail. 2013;6:606–619. doi: 10.1161/HHF.0b013e318291329a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Wu S, Barrera J, Matthews K, Pan D. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila homolog of YAP. Cell. 2005;122:421–434. doi: 10.1016/j.cell.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Jiao S, Wang H, Shi Z, Dong A, Zhang W, Song X, He F, Wang Y, Zhang Z, et al. A peptide mimicking VGLL4 function acts as a YAP antagonist therapy against gastric cancer. Cancer Cell. 2014;25:166–180. doi: 10.1016/j.ccr.2014.01.010. [DOI] [PubMed] [Google Scholar]
- Kajstura J, Leri A, Finato N, Di Loreto C, Beltrami CA, Anversa P. Myocyte proliferation in end-stage cardiac failure in humans. Proc. Natl. Acad. Sci. USA. 1998;95:8801–8805. doi: 10.1073/pnas.95.15.8801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koontz LM, Liu-Chittenden Y, Yin F, Zheng Y, Yu J, Huang B, Chen Q, Wu S, Pan D. The Hippo effector Yorkie controls normal tissue growth by antagonizing scalloped-mediated default repression. Dev. Cell. 2013;25:388–401. doi: 10.1016/j.devcel.2013.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Shioi T, Kasahara H, Jobe SM, Wiese RJ, Markham BE, Izumo S. The cardiac tissue-restricted homeobox protein Csx/ Nkx2.5 physically associates with the zinc finger protein GATA4 and cooperatively activates atrial natriuretic factor gene expression. Mol. Cell Biol. 1998;18:3120–3129. doi: 10.1128/mcb.18.6.3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Gao E, Vite A, Yi R, Gomez L, Goossens S, van Roy F, Radice G. Alpha-catenins control cardiomyocyte proliferation by regulating yap activity. Circ. Res. 2014;116:70–79. doi: 10.1161/CIRCRESAHA.116.304472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z, Pu WT. Strategies for cardiac regeneration and repair. Sci. Transl. Med. 2014;6:239rv1. doi: 10.1126/scitranslmed.3006681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z, Pu WT. Releasing YAP from an a-catenin trap increases cardiomyocyte proliferation. Circ. Res. 2015;116:9–11. doi: 10.1161/CIRCRESAHA.114.305496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z, von Gise A, Zhou P, Gu F, Ma Q, Jiang J, Yau AL, Buck JN, Gouin KA, et al. Cardiac-specific YAP activation improves cardiac function and survival in an experimental murine MI model. Circ. Res. 2014;115:354–363. doi: 10.1161/CIRCRESAHA.115.303632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama H, Chen X, Baines CP, Klevitsky R, Zhang X, Zhang H, Jaleel N, Chua BHL, Hewett TE, et al. Ca2+- and mitochondrial-dependent cardiomyocyte necrosis as a primary mediator of heart failure. J. Clin. Invest. 2007;117:2431–2444. doi: 10.1172/JCI31060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oudhoff MJ, Freeman SA, Couzens AL, Antignano F, Kuznetsova E, Min PH, Northrop JP, Lehnertz B, Barsyte-Lovejoy D, et al. Control of the hippo pathway by Set7-dependent methylation of Yap. Dev. Cell. 2013;26:188–194. doi: 10.1016/j.devcel.2013.05.025. [DOI] [PubMed] [Google Scholar]
- Pan D. The hippo signaling pathway in development and cancer. Dev. Cell. 2010;19:491–505. doi: 10.1016/j.devcel.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad K-MR, Xu Y, Yang Z, Acton T, French A. Robust cardiomyocyte-specific gene expression following systemic injection of AAV: in vivo gene delivery follows a Poisson distribution. Gene Ther. 2011;18:43–52. doi: 10.1038/gt.2010.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlegelmilch K, Mohseni M, Kirak O, Pruszak J, Rodriguez JR, Zhou D, Kreger BT, Vasioukhin V, Avruch J, et al. Yap1 acts downstream of alpha-catenin to control epidermal proliferation. Cell. 2011;144:782–795. doi: 10.1016/j.cell.2011.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snippert HJ, van der Flier LG, Sato T, van Es JH, van den Born M, Kroon-Veenboer C, Barker N, Klein AM, van Rheenen J, et al. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell. 2010;143:134–144. doi: 10.1016/j.cell.2010.09.016. [DOI] [PubMed] [Google Scholar]
- Söderberg O, Gullberg M, Jarvius M, Ridderstråle K, Leuchowius K-J, Jarvius J, Wester K, Hydbring P, Bahram F, et al. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat. Methods. 2006;3:995. doi: 10.1038/nmeth947. [DOI] [PubMed] [Google Scholar]
- Umezawa H. Structures and activities of protease inhibitors of microbial origin. Methods Enzymol. 1976;45:678–695. doi: 10.1016/s0076-6879(76)45058-9. [DOI] [PubMed] [Google Scholar]
- Varelas X, Miller BW, Sopko R, Song S, Gregorieff A, Fellouse FA, Sakuma R, Pawson T, Hunziker W, et al. The Hippo pathway regulates Wnt/beta-catenin signaling. Dev. Cell. 2010;18:579–591. doi: 10.1016/j.devcel.2010.03.007. [DOI] [PubMed] [Google Scholar]
- von Gise A, Lin Z, Schlegelmilch K, Honor LB, Pan GM, Buck JN, Ma Q, Ishiwata T, Zhou B, et al. YAP1, the nuclear target of Hippo signaling, stimulates heart growth through cardiomyocyte proliferation but not hypertrophy. Proc. Natl. Acad. Sci. USA. 2012;109:2394–2399. doi: 10.1073/pnas.1116136109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh S, Ponten A, Fleischmann BK, Jovinge S. Cardiomyocyte cell cycle control and growth estimation in vivo—an analysis based on cardiomyocyte nuclei. Cardiovasc. Res. 2010;86:365–373. doi: 10.1093/cvr/cvq005. [DOI] [PubMed] [Google Scholar]
- Xin M, Kim Y, Sutherland LB, Qi X, McAnally J, Schwartz RJ, Richardson JA, Bassel-Duby R, Olson EN. Regulation of insulin-like growth factor signaling by Yap governs cardiomyocyte proliferation and embryonic heart size. Sci. Signal. 2011;4:ra70. doi: 10.1126/scisignal.2002278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin M, Kim Y, Sutherland LB, Murakami M, Qi X, McAnally J, Porrello ER, Mahmoud AI, Tan W, et al. Hippo pathway effector Yap promotes cardiac regeneration. Proc. Natl. Acad. Sci. USA. 2013;110:13839–13844. doi: 10.1073/pnas.1313192110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D, Kita S, Liang F, Wang C, Tian H, Ji Y, Lončar M, Quan Q. High sensitivity and high Q-factor nanoslotted parallel quadrabeam photonic crystal cavity for real-time and label-free sensing. Appl. Phys. Lett. 2014;105:063118. [Google Scholar]
- Yoshida T. MCAT elements and the TEF-1 family of transcription factors in muscle development and disease. Arterioscler. Thromb. Vasc. Biol. 2008;28:8–17. doi: 10.1161/ATVBAHA.107.155788. [DOI] [PubMed] [Google Scholar]
- Zanconato F, Forcato M, Battilana G, Azzolin L, Quaranta E, Bodega B, Rosato A, Bicciato S, Cordenonsi M, Piccolo S. Genome-wide association between YAP/TAZ/TEAD and AP-1 at enhancers drives oncogenic growth. Nat. Cell Biol. 2015;17:1218–1227. doi: 10.1038/ncb3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZD, Paccanaro A, Fu Y, Weissman S, Weng Z, Chang J, Snyder M, Gerstein MB. Statistical analysis of the genomic distribution and correlation of regulatory elements in the ENCODE regions. Genome Res. 2007;17:787–797. doi: 10.1101/gr.5573107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, von Gise A, Liu Q, Hu T, Tian X, He L, Pu W, Huang X, He L, Cai CL, et al. Yap1 is required for endothelial to mesenchymal transition of the atrioventricular cushion. J. Biol. Chem. 2014;289:18681–18692. doi: 10.1074/jbc.M114.554584. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.