Abstract
Background
Weight loss may decrease risk of colorectal cancer in obese individuals, yet its effect in the colorectum is not well understood. We used integrative network modeling, Passing Attributes between Networks for Data Assimilation, to estimate transcriptional regulatory network models from mRNA expression levels from rectal mucosa biopsies measured pre- and post-weight loss in 10 obese, pre-menopausal women.
Results
We identified significantly greater regulatory targeting of glucose transport pathways in the post-weight loss regulatory network, including “regulation of glucose transport” (FDR = 0.02), “hexose transport” (FDR = 0.06), “glucose transport” (FDR = 0.06) and “monosaccharide transport” (FDR = 0.08). These findings were not evident by gene expression analysis alone. Network analysis also suggested a regulatory switch from NFKB1 to MAX control of MYC post-weight loss.
Conclusions
These network-based results expand upon standard gene expression analysis by providing evidence for a potential mechanistic alteration caused by weight loss.
Introduction
Obesity occurs in over one third of the American population and is associated with increased risk of colorectal cancer [1,2]. However, it is not clear if one must be of normal weight throughout life to be protected against colorectal cancer or if weight loss decreases risk in obese individuals. It has been hypothesized that since weight loss decreases systemic inflammation [3], weight loss may mediate anti-cancer effects in the colorectum. Weight loss interventions have been successful among individuals with high colorectal cancer risk [4]. However, there is no defined mechanism of action for this commonly prescribed cancer prevention and lifestyle intervention.
Most human studies on the effect of body mass in colorectal cancer have focused on the association between disease and weight gain, and even those findings are inconsistent. For example, different studies have found an increased risk for colorectal cancer in response to weight gain (1) among men but not women [5], (2) only among men that were overweight at baseline [6], or (3) among both men and women [7]. Most recently, Song, et al. [8] observed a significant 64% increased risk for colorectal cancer among men who gained ≥20kg in adulthood versus weight stable men, and a trend for a negative association between adult weight loss and colorectal cancer risk among men. Similar, but weaker associations were also observed among women. Recent meta-analyses on weight gain and colorectal cancer studies demonstrates an increased risk of colon cancer among men, but not women, who gain weight in adulthood [9–11].
Similarly, the few studies examining the effects of weight loss are also inconsistent and often null. Although there are several reports of an association of intentional weight loss with reduced colorectal cancer [12] [13][14], many studies have been unable to find this association [15] [16] [17] [18]. These inconsistent results suggest the effects of weight loss may be BMI-, time-, dose- and even person-dependent and demonstrates a need to better understand the impact of weight loss in the colorectum of obese and overweight individuals. Further, these studies do not allow for the disentanglement of effects due to weight loss versus effects due to extreme changes in diet.
Obesity is a chronic inflammatory state, resulting in an increase in circulating insulin, adipokines, and other hormones and leading to changes in glucose transport and activation of the PI3K/mTOR pathway. Subsequently, colorectal epithelial cells damaged by inflammation with access to plenty of circulating glucose at their disposal are signaled to grow and potentially undergo malignant transformation [19][20]. A prevailing assumption is that the opposite mechanism is at work under lower body fat, post-weight loss conditions. Indeed, those who lose weight display more normal glucose regulation [21,22]. Circulating insulin and hemoglobin A1C levels are also positively associated with colorectal cancer risk [23]. While literature reviews describe limited but suggestive evidence that weight loss decreases circulating biomarkers of inflammation [24], there is a lack of rigorous mechanistic and epidemiological evidence linking intentional weight loss itself to decreased colorectal cancer risk.
Modeling gene regulation as a complex network is an important way to characterize and explore the regulatory mechanisms mediating cellular processes [25]. Although there are many approaches for modeling networks, it has become increasingly obvious that integrative approaches combining multiple sources of evidence produce the most informative and accurate networks [26]. PANDA (Passing Attributes between Networks for Data Assimilation; [27–30]) uses an integrative message-passing approach to reconstruct gene regulatory networks. What distinguishes PANDA from other approaches is its focus on information flow when estimating regulatory relationships. Specifically, PANDA does not derive edges in the network from direct correlation in expression patterns between a transcription factor and a downstream target gene, but rather shared patterns of co-expression between common targets of a transcription factor.
In this study we used PANDA to integrate publicly available gene expression data from rectal mucosal biopsy samples pre- and post-diet induced weight loss in 10 women. With a prior regulatory map derived from existing transcription factor binding motif information, we built models representing the gene regulatory network of the colorectum both pre- and post-weight loss (Pendyala, Neff, Suarez-Farinas, & Holt, 2011; GSE20931). We then compared these networks to characterize the effects of weight loss on cellular pathways. Specifically, we identified MYC- and glucose transport-related regulatory shift post-weight loss. Our results complement standard gene expression analyses by providing additional information and allowing us to hypothesize on alterations in transcription factor regulation-mediated expression changes. These changes can be used to develop hypotheses on the biological processes of the colorectum that are most affected in response to weight loss.
Results
Gene regulatory network models at baseline and end-of-study
We used PANDA to build network models of the gene regulatory structure by combining transcription factor motif information with gene expression data from paired mucosal biopsies collected from 10 pre-menopausal women both before (baseline) and after weight loss (end-of-study; Figure 1). Finding obese but otherwise healthy participants who are able to adhere to a very low calories diet is challenging and obtaining paired rectal biopsies from subjects is rare. While the number of samples in this gene expression dataset is fairly small, we have previously used PANDA to model gene regulatory networks in other systems with limited samples [22] and found that the algorithm is able to estimate networks whose structure provides insight into the underlying biology. It is this structure, and how it changes as a result of weight-loss, that we investigate here.
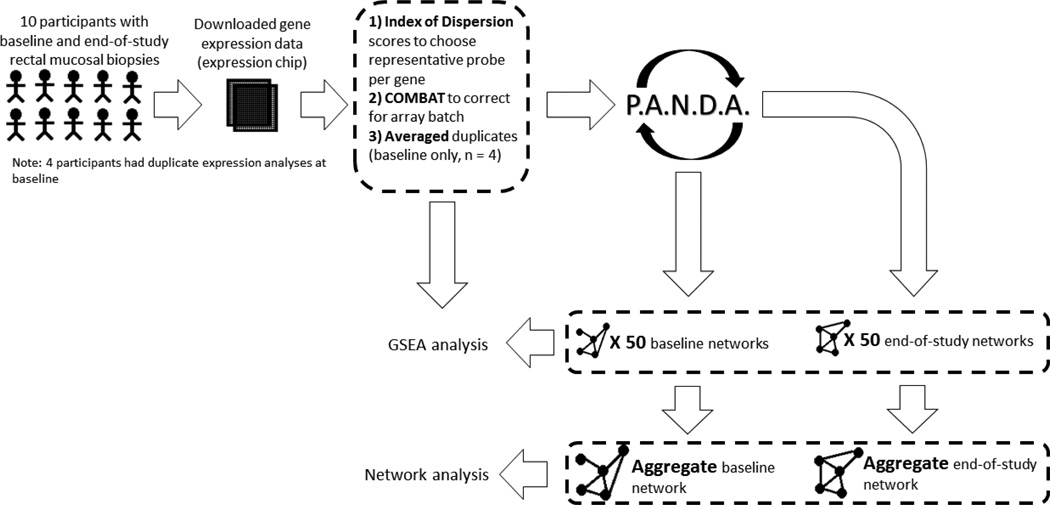
Figure 1. Schematic overview of the analyses performed.
In a previous study (Pendyala, Neff, Suarez-Farinas, & Holt, 2011) 10 obese, pre-menopausal women provided baseline rectal mucosal biopsies, followed a very low calorie diet (<800 kcals/day) to achieve >8% body weight loss and then provided an end-of-study rectal mucosal biopsy sample. Gene expression data from these biopsy samples was downloaded from the Gene Expression Omnibus database (GEO; GSE20931). Processing of this data included (1) collapsing probes representing the same gene by selecting the probe with the highest Index of Dispersion across its expression values to represent that given gene’s expression levels, (2) removing batch effects between the two expression chips by running COMBAT (Johnson and Rabinovic, 2007), and (3) averaging four duplicate baseline gene expression measurements to obtain two expression samples for each participant, one at baseline and one at end of study. Due to the small sample size, we randomly chose five participants from the ten total participants (without replacement). We did this multiple times to create fifty subsamples of individuals. We then applied Passing Attributes between Networks for Data Assimilation (PANDA; Glass, et al. 2013) to the gene expression data from individuals within each subsample in order to estimate fifty paired baseline and end-of-study gene regulatory networks. Both the processed gene expression data and these 100 networks were input into GSEA to identify gene sets with either increased expression or targeting at end-of-study versus baseline. Finally, the baseline and end-of-study networks’ edge values were averaged to create one averaged, aggregate baseline network and one averaged, aggregate end-of-study network, respectfully. “High-confidence” edges were identified within these aggregate networks and used for network-based analysis including change in targeting (change out-degree) for transcription factors.
To begin, we downloaded gene expression data from the Gene Expression Omnibus (GEO; GSE20931), corrected for batch effect, merged replicate samples and selected the probe with the highest index of dispersion to represent each gene. PANDA was then used to integrate the pairwise co-expression levels of genes (estimated using the Pearson correlation) in either the baseline and end-of-study samples with a prior regulatory network constructed by scanning promoter regions (defined as [+750, −250] around the TSS) for transcription factor binding sites [32]. Because of the relatively small number of samples, we used a jack-knifing approach to build fifty networks based on the baseline samples and fifty network based on the end-of-study samples [25]. We did this by creating 50 random subsamples of 5 participants each. Each subsample contained an individual no more than one time but individuals were represented in multiple subsamples. For each subsample, we identified the pre- and post-weight loss expression data corresponding to the individuals in the subsample and applied PANDA to estimate a paired baseline and end-of-study regulatory gene network. We did this for all fifty subsamples, resulting in fifty baseline and fifty end-of-study networks. Because we selected samples from the same individuals when creating each baseline and end-of-study network, these can be thought of as paired sets of networks, which can help us account for between-subject heterogeneity in our analysis. For more information on the expression and motif data processing and network modeling, please see Methods.
We compared the fifty baseline and fifty end-of-study networks to identify biological processes and pathways that are robustly differentially-targeted between the pre- and post-weight loss states. To do this, we ran Gene Set Enrichment Analysis (GSEA; Mootha et al., 2003; Subramanian et al., 2005) using the in-degree of genes (number of transcription factors targeting that gene) in our 50 baseline and 50 end-of-study networks as an input [30]. Although no gene sets were significantly enriched for increased targeting at baseline compared to end-of-study, we observe a slight trend for carbohydrate biosynthetic processes (Table 1; S1 Table; S2 Table). On the other hand, four glucose transport pathways were among the top significantly enriched pathways at end-of-study (Table 1; S2 Table): “regulation of glucose transport” (FDR = 0.02), “hexose transport” (FDR = 0.06), “glucose transport” (FDR = 0.06) and “monosaccharide transport” (FDR = 0.08). This, juxtaposed with a suggestion of biosynthesis at baseline (monosaccharide and hexose biosynthesis, FDR q-value = 0.25 and 0.28, respectively), is evidence that there is a shift away from carbohydrate synthesis and an increase in glucose transport regulation in response to weight loss. To evaluate whether these shifts in local network structure around glucose transport genes would be evident from a standard gene expression-based analysis, gene expression values were input into GSEA from our baseline and end-of-study samples. Consistent with our network-based results, we did not find any gene sets enriched at baseline (data not shown). However, we found many processes related to mitochondrial function and cellular respiration significantly enriched at end-of study, as well as chemokine (FDR = 0.002) and cytokine activity (FDR = 0.038) (S3 Table). These latter two processes were also identified by GSEA analysis of the gene expression data in the previous analysis of this data, which used an older Gene Ontology dataset for GSEA analyses [31]. It is interesting to note, however, that neither the original analysis of the expression data, nor our re-analysis, identified significant differential-expression of the glucose transport genes.
Table 1.
Top 10 most significant gene sets (pathways) with <75 members in baseline and end-of-study based on a differential-targeting analysis using GSEA. Abbreviations: Enrichment score (ES), Normalize ES (NES), False Discovery Rate (FDR), biological process (BP), cellular component (CC), and molecular function (MF).
| Enriched at | Gene sets (pathways) | ES | NES | FDR Q-val |
|---|---|---|---|---|
| Baseline | Oxidoreductase activity, acting on NADH or NADPH, quinone or similar compound as acceptor (MF) |
0.484 | 1.716 | 0.209 |
| Establishment of protein localization to peroxisome (BP) | 0.618 | 1.717 | 0.211 | |
| Peroxisome organization (BP) | 0.563 | 1.767 | 0.212 | |
| Protein targeting to peroxisome (BP) | 0.618 | 1.722 | 0.215 | |
| Nucleobase, nucleoside, nucleotide and nucleic acid transmembrane transporter activity (MF) |
0.613 | 1.72 | 0.215 | |
| Regulation of lipid transport (BP) | 0.495 | 1.749 | 0.215 | |
| Microvillus membrane (CC) | 0.61 | 1.711 | 0.216 | |
| Response to fatty acid (BP) | 0.575 | 1.745 | 0.217 | |
| L-amino acid transport (BP) | 0.613 | 1.731 | 0.218 | |
| Tetrapyrrole metabolic process (BP) | 0.515 | 1.757 | 0.218 | |
| End-of-study | Receptor tyrosine kinase binding (MF) | −0.635 | −2.176 | 0.016 |
| Translational initiation (BP) | −0.565 | −2.131 | 0.018 | |
| Regulation of glucose transport (BP) | −0.505 | −2.179 | 0.022 | |
| Hexose transport (BP) | −0.445 | −1.910 | 0.058 | |
| Glucose transport (BP) | −0.445 | −1.886 | 0.062 | |
| Monosaccharide transport (BP) | −0.437 | −1.854 | 0.079 | |
| Nuclear pore (CC) | −0.430 | −1.831 | 0.086 | |
| Transcription termination, DNA-dependent (BP) | −0.418 | −1.826 | 0.086 | |
| Somatic stem cell division (BP) | −0.620 | −1.838 | 0.088 | |
| Nuclear-transcribed mrna catabolic process, nonsense- mediated decay (BP) |
−0.510 | −1.795 | 0.101 |
Key transcription factors alter targeting from baseline to end-of-study
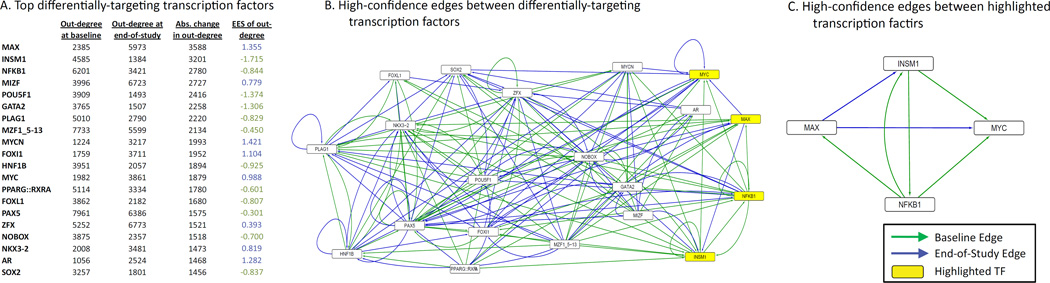
We next investigated which transcription factors might be driving the changes in the networks between baseline and end-of-study. To begin, we created a single aggregate baseline and a single aggregate end-of-study network by averaging the 50 baseline and 50 end-of-study networks, respectively. Next, we limited these aggregate networks to only include “high-confidence” edges, which we identified based on a combined probability score that represents both the likelihood that a given edge exists and that is stronger in baseline compared to end-of-study, or vice versus (see Methods; Supplementary data). To identify large-scale patterns that might define biologically meaningful differences between these two “high-confidence” subnetworks, we compared the change in out-degree (number of gene targets) of each transcription factor [28]. Figure 2A lists the twenty transcription factors with the greatest absolute change in out-degree (targeting) between the baseline and end-of-study high-confidence subnetworks and their corresponding “edge-enrichment score” [28]. For more information see Methods.
Figure 2. Changes in transcription factor targeting between high-confidence baseline and end-of-study subnetworks.
(A) The 20 transcription factors with the largest change in absolute (abs.) out-degree between aggregate subnetworks of high confidence edges from baseline to end-of-study. (B) Visualization of the high-confidence edges that extend between any pair of these transcription factors. Note that not every transcription factor motif had a corresponding gene name (e.g. PPARG::RXRA) and thus some nodes in this network only have out-going edges (i.e. not all transcription factors are also gene targets). Three transcription factors (NFKB1, MAX and INSM1) with the largest change in targeting (e.g. out-degree) and Myc (a key oncogenic factor and regulatory associate of these transcription factors) are highlighted in yellow. (C) The high confidence edges between these four highlighted transcription factors describe a shift in regulatory control from NFKB1 to MAX after weight loss. Of interest, these genes are mediators in glucose transport regulatory pathways.
The three transcription factors with the largest change in gene targeting were MAX, which had 3,588 more gene targets in the end-of-study aggregate subnetwork compared to baseline, INSM1 which had 3,201 fewer, and NFKB1 which had 2,780 fewer (Figure 2A). MYC (an oncogenic factor and regulatory associate of these transcription factors; Fernandez et al., 2003a; Gu, Cechova, Tassi, & Dalla-Favera, 1993; F. La Rosa, Pierce, & Sonenshein, 1994; Lahoz, Xu, Schreiber-Agus, & DePinho, 1994) also had a greatly increased level of gene targeting post weight loss. However, the MYC::MAX heterodimer had overall decreased targeting (data not shown), suggesting that although both MYC and MAX are both targeting more genes post weight loss, this likely is not the result of these two proteins working together in a protein complex.
To gain a better understanding of the changes in the local regulatory network around the top twenty transcription factors with altered regulatory partners, we visualized high-confidence edges from the baseline and end-of-study aggregate subnetworks that extend between any pair of these top 20 transcription factors (Figure 2B). It is important to note that these subnetworks likely contain some false-positive edges; however, it is also interesting that we observe a high-level of regulatory activity around these transcription factors, with 75 high-confidence edges at baseline to 80 at end-of-study.
We next limited our view to the transcription factors with the largest changes in gene targeting in the aggregate subnetwork of high confidence edges (MAX, INSM1 and NFKB1) and a shared transcription factor target of key importance in carcinogenesis and cellular growth (MYC; Figure 2C) [39]. We observed a shift from reliance on NFKB1 and INSM1 cross-talk to modulate MYC at baseline to MAX modulating both INSM1 and MYC at end-of-study. This shift was observed despite the fact that the average log-fold change values in expression for MAX, INSM1 and NFKB1 were only 0.010, −0.026, and −0.001, respectively. We also saw changes in co-expression levels among the targets of these transcription factors, providing additional evidence of their importance in mediating changes induced by weight loss, and helping to explain this gene regulatory shift (S1 Figure).
A MYC-related mechanism for the shift towards glucose transport post-weight loss
GSEA analysis of our network models indicated that genes involved in glucose transport are differentially-targeted between baseline and end-of-study (Table 1). Thus, we next examined the relationship between this finding and the network-rewiring we observed occurring around MYC, MAX NFKB1 and INSM1 in our aggregate models. To do this we identified the subset of genes annotated to the regulation of glucose transport, hexose transport, monosaccharide transport and/or glucose transport gene sets used in the GSEA analysis. We then selected high-confidence edges that target at least one of these genes, resulting in a subnetwork of 7,949 high-confidence glucose transport-specific edges at baseline and a subnetwork of 8,264 high-confidence glucose transport-specific edges at end-of-study.
As in the aggregate network of high-confidence edges, we determined the number of genes targeted by each transcription factor in these glucose transport-specific subnetworks. We observe that three of our previously-identified transcription factors (NFKB1, MAX and INSM1) have some of the largest changes in targeting between baseline and end-of-study (Table 2). This confirms the notion that the shift from INSM1 and NFKB1 to MAX control (Figure 2C) is a potential mediator of the shift in glucose transport regulation post-weight loss.
Table 2.
Top 10 transcription factors with the largest change in out-degree of high confidence edges from baseline to end-of-study in the aggregate network restricted to only genes involved in glucose transport pathways. Glucose transport pathway genes includes genes annotated to at least one of four Gene Ontology categories: “glucose transport”, “hexose transport”, “monosaccharide transport” and “regulation of glucose transport”. Abbreviations: Edge Enrichment Score (EES)
| Transcription factor |
Baseline out-degree |
End-of-study out-degree |
Change in out-degree |
Absolute change in out-degree |
Log2(EES) of out-degree |
|---|---|---|---|---|---|
| NFKB1 | 108 | 42 | −66 | 66 | −1.419 |
| MAX | 33 | 90 | 57 | 57 | 1.391 |
| MZF1_5−13 | 130 | 80 | −50 | 50 | −0.757 |
| INSM1 | 67 | 19 | −48 | 48 | −1.874 |
| MIZF | 67 | 107 | 40 | 40 | 0.619 |
| NFIL3 | 29 | 69 | 40 | 40 | 1.194 |
| E2F1 | 40 | 79 | 39 | 39 | 0.926 |
| FOXD3 | 23 | 61 | 38 | 38 | 1.351 |
| SOX17 | 36 | 74 | 38 | 38 | 0.983 |
| PAX5 | 127 | 90 | −37 | −37 | −0.553 |
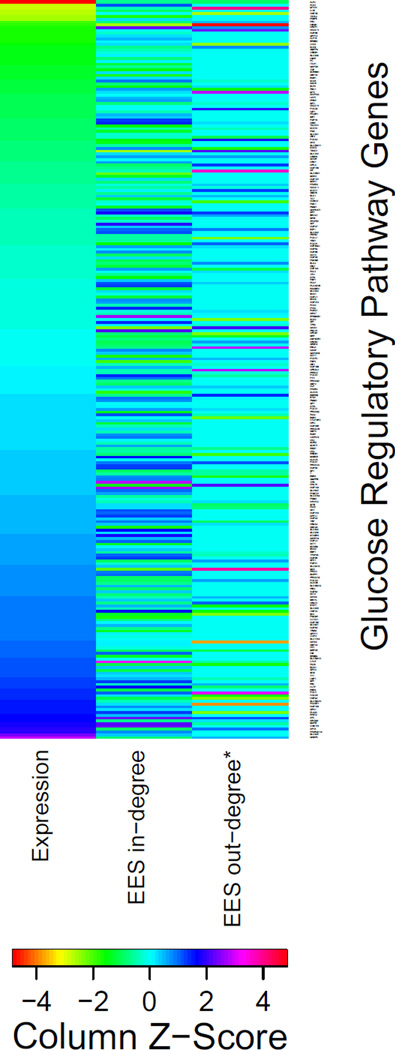
Our network analysis was able to discern changes in targeting around glucose transport genes that were not identified in a differential-expression analysis. We therefore were also curious about the relationship between the re-wiring we observed around these glucose pathway genes and their differential-expression between baseline and end-of-study. To investigate this further, we directly compared the log-fold change in expression from baseline to end-of-study with the “edge enrichment score” (EES) for in- and out-degree for each gene and transcription factor in the glucose transport-specific subnetwork (Figure 3; note that genes that are not also transcription factors will have a nominal out-degree EES of zero). Expression (log-fold change), EES in-degree and EES out-degree all provide different information about genes in the glucose regulatory pathway, emphasizing the importance of investigating the information highlighted in each of the three data analyses.
Figure 3. Comparison of changes in expression and targeting for genes in glucose transport regulatory pathways.
The three columns of the heatmap show the (1) log2-fold change gene expression levels between baseline and end-of-study (“Expression”), (2) the Edge Enrichment Scores (EES) calculated based on change in gene in-degree between the aggregate subnetworks when restricted to genes involved in glucose transport (see Table 2), and (3) the EES calculated based on change in gene out-degree between the aggregate subnetworks when restricted to genes involved in glucose transport (see Table 2). Clear differences between the expression data and the EES are likely a result of PANDA’s focus on integrating gene co-expression information with a regulatory motif prior (see S1 Figure). A value of zero indicates no change in expression, EES in-degree or EES out-degree for a given gene from baseline to end-of-study. *There are many fewer changes observed for EES out-degree as compared to in-degree because many genes are targets and thus have in-degrees, but many fewer genes also serve as transcription factors. Thus there are many genes with zero out-degree at both baseline and end-of-study.
Discussion
Network-based gene expression analysis of rectal mucosa biopsy samples from 10 obese, pre-menopausal women before and after supervised, diet-induced weight loss suggests that weight loss leads to changes in glucose/carbohydrate transport via a shift from INSM1 and NFKB1, to MAX gene regulatory control. These results complement earlier observations of a decrease in NFKB1-related inflammation, and a decrease in fasting glucose (meanbaseline = 95 mg/dL, meanend-of-study = 85 mg/dL), and triglycerides (meanbaseline = 122 mg/dL, meanend-of-study = 93 mg/dL) in these women [31]. Additionally, our findings also describe a MAX-based mechanism for the observed increase in glucose mobilization and/or use along with less biosynthesis.
Obesity-associated colorectal cancer has been hypothesized to be mediated by exposure of the colorectum to chronic inflammatory insults in the presence of abundant glucose [21–24,40] but the mechanisms of weight loss on the colorectum that mediate colorectal cancer risk are not well understood. A standard GSEA analysis of gene expression levels in these participants, along with measuring biomarkers of inflammation, previously provided evidence that weight loss induces a decrease in inflammatory related genes (JUN and FOS), inflammatory pathways (cytokine activity, chemokine receptor binding, chemokine activity, etc.) and led to the hypothesis that these changes were modulated by TNF-α, IL-6, IL-1, and IL-8 [31].
The PANDA-based network approach we describe herein not only identified many of these same changes in inflammatory pathways, but also highlighted an important shift in the targeting of glucose regulatory pathways. Additionally, this network-based analysis allowed us to identify a possible mechanism by which weight loss decreases inflammation and alters glucose transport in rectal mucosa. Although little change in mRNA expression levels of NFKB1, INSM1 and MAX was observed, our integrative network models depict a striking decrease in the number of genes targeted by NFKB1 and INSM1 post-weight loss, while MAX increased the number of genes it targeted. To our knowledge this is the first description of a shift in gene regulatory control post-weight loss to MAX.
INSM1, NFKB1 and MAX are all involved in glucose metabolism and/or inflammation. Perhaps the best studied of the three is NFKB1, which is activated by pro-inflammatory pathways observed in obese persons, confers a selective growth advantage [41] and promotes epithelial to mesynchymal transition of colorectal cells [42]. It is also thought to contribute to the risk of colorectal cancer [43]. While targets of INSM1 in the rectum are unknown, INSM1 has been shown to target the AKT/PI3K pathway in the pancreas [44] which itself regulates glucose transport via mTOR [45]. MAX’s involvement in glucose regulation is likely mediated through its MYC-related mechanisms described below [37,38].
The oncogenic, growth promoting transcription factor MYC is mediated by all three genes of interest. Specifically, NFKB1 promotes MYC expression [36], while INSM1 is a target of MYC [35], and MAX heterodimerizes with MYC to form MYC::MAX [37,38]. However, as we observed a large increase in MAX targeting genes from baseline to end-of-study, we simultaneously observed a large decrease in MAX::MYC targeting. Thus instead of dimerizing with MYC after weight loss, MAX likely forms a MAX::MAX homodimer which has been shown in vitro to repress MYC-induced cell growth and malignant transformation [37,38].
In addition to being a prolific regulator of cell growth and oncogenesis, MYC also regulates a majority of the genes that regulate glycolysis [46]. However, in part due to the key role MYC plays in normal cell functions, designing drugs to target MYC has been incredibly challenging [39].We suggest that it may be reasonable to combine already available NFKB inhibiting drugs [47], and/or to develop agents that inhibit INSM1 or promote MAX in order to induce the anti-inflammatory and glucose regulatory changes that weight loss induces for cancer prevention or, potentially, weight loss induction itself.
Integrative network modeling has previously been used to describe the tissue-specific effects of weight loss in adipose tissue [48]. Our application of PANDA similarly demonstrates how network approaches help to build upon previous gene expression-based findings [31]. However, we recognize that our results are limited by the inability to assign cause to weight loss versus extreme dietary change since they occurred concurrently in these participants, a lack of protein measurements, biases in the available transcription factor motif datasets, and the potential for false positives due to the small number of gene expression samples used to construct the networks. One reason we chose to model networks using this particular dataset was that is contained paired samples from the same individual, minimizing the effects of the underlying heterogeneity across individuals in our analysis and allowing us to focus on changes that are most likely a result of weight loss. By subsampling, we also minimized the effect of changes in gene expression that are specific to only one individual (outliers). In addition, to mitigate the influence of false-positive edges in our networks, we chose to focus on large-scale changes, such as alterations in transcription factor degree, or in the targeting patterns around a set of pathway genes. Despite taking these precautions, we recognize that future studies will be needed to confirm our proposed NFKB1/INSM1 to MAX regulatory shift mechanism in model systems by measuring protein levels.
In summary, using network modeling, we identified a significant change in targeting of glucose transport genes in the rectal mucosa of overweight women who underwent intentional weight. These changes were explained by a putative mechanism whereby NFKB1 and INSM1 decrease their gene targeting activity and MAX takes over to alter MYC and other glucose and inflammatory gene expression levels. Although these findings are highly promising, we recognize that this mechanism needs to be confirmed in model systems before moving towards targeting of these transcription factors as potential inducers of weight loss and/or cancer preventative therapies.
Materials and Methods
Participants and data source
In a previous study [31] ten obese, pre-menopausal women (mean age = 43; Figure 1) were enrolled in a weight loss trial. Briefly, exclusion criteria for participation included not being weight stable (≥ 6 months), history of cancer, current weight loss treatment, history of intestinal surgery, history “suggestive of malabsorption”, other major medical concerns, use of anti-inflammatory medications or medications with contraindications for severe weight loss/low calorie diet. Participants were put on a closely supervised, low calorie diet (<800 kcals/day) until they lost >8% of body weight. Mean body weight loss was 10.1% and mean time on study was 46.5 days. Gene expression was measured on mucosal biopsies taken at baseline and end-of-study. These gene expression data were then deposited in the Gene Expression Omnibus (GEO; GSE20931) as anonymous data after all identifiers were removed. Additional participant characteristics and weight-related biomarker changes related to this dataset can be found in (Pendyala et. al., 2011). This weight loss study was originally approved by the Institutional Review Board of The Rockefeller University (New York, NY), where written informed consent was obtained prior to study participation.
Expression data processing
We downloaded *.soft files containing normalized mRNA expression levels from the mucosal biopsies (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE20931). This data included a total of 24 anonymous gene expression samples, with 10 baseline and 10 end-of-study samples (one baseline and one end-of-study for each participant), plus replicates for four of the baseline samples. A *.geo and *.annot file containing the keys to convert Illumina probe IDs to gene symbols for each gene were also downloaded from GEO and used to annotate the gene expression files.
We corrected for batch effects using COMBAT (R package, Johnson, Li, & Rabinovic, 2007) in R (RStudio version 0.98.994, RStudio Inc., Boston, MA; Figure 1). For genes with multiple probes, the probe with the highest index of dispersion, defined as the variance divided by the mean, was chosen to represent the expression levels for that gene. Finally, replicate gene expression values (for the four participants with duplicate gene expression samples at baseline) were averaged. This resulted in 10 baseline expression samples and 10 end-of-study expression samples that were used for subsequent analyses.
Analysis using gene expression data
We downloaded human Gene Ontology annotation information from www.geneontology.org and built a *.gmt file containing sets of genes annotated to 15033 different Gene Ontology (GO) categories. To evaluate the differential-expression of these gene sets between baseline and end-of-study, we ran Gene Set Enrichment Analysis (GSEA; Broad Institute, Cambridge, MA; http://www.broadinstitute.org/gsea/index.jsp; Mootha et al., 2003; Subramanian et al., 2005), comparing the expression levels in the 10 baseline and 10 end-of-study expression samples.
Network model development
Networks were derived using Passing Attributes between Networks for Data Assimilation (PANDA; Glass, Huttenhower, Quackenbush, & Yuan, 2013; Figure 1). PANDA combines co-expression of genes based on Pearson correlations and a prior regulatory network. To minimize the effect of outliers in our networks built on a smaller sample size (n = 10), five participants were chosen at random (without replacement) to form subsamples. 50 subsamples were formed such that no participants were in the same subsample twice but participants could be in multiple different subsamples. Gene expression from these subsamples of participants was used to reconstruct 50 baseline gene regulatory networks and 50 end-of-study gene regulatory networks. Namely, gene expression data for each group of five participants was integrated with a prior network structure using PANDA with the alpha parameter set at 0.25 (indicating a high degree of message passing).
The prior regulatory network was estimated by scanning the human genome for 130 position weight matrixes (PWM) from the JASPAR core vertebrate transcription factor database [32]. To determine locations for each motif, each sequence S was given a score equal to log [P(S|M)/P(S|B)], where P(S|M) is the probability of observing sequence S given motif M and P(S|B) is the probability of observing sequence S given the genome background B. The background distribution of motif scores was determined by randomly sampling the genome 106 times. Motif sites that fell within the promoter region ([−750,+250] base-pairs around the transcriptional start site) of one of the genes measured in the expression data with a significance less than 10−5 were used to define edges between a transcription factor and gene in the regulatory network prior.
In total 50 baseline and 50 end-of-study (100 total) directed, fully connected networks were derived. The nodes in these networks are genes (either transcription factors, gene targets or both) and the edges each have an associated Z-score weight indicating the probability that the edge exists. 17511 genes were included in the expression data and were targeted by at least one TF motif in our regulatory prior, resulting in weight values for 2,276,429 edges in each network. Note that not all transcription factors from the regulatory motif prior were measured on the expression chip.
Network analysis
Gene set targeting analysis
We evaluated differential-targeting of gene sets (pathways) as in [30]. Namely, for each of the 100 reconstructed networks, we calculated the weighted in-degree of each target gene by summing the Z-score weights for all edges to that gene. Then, to identify gene sets that are differentially-targeted between baseline and end-of-study, we ran GSEA comparing the weighted in-degree values across the 50 baseline and 50 end-of-study networks.
Defining Subnetworks and Evaluating TF/Gene Edge Enrichment
We also generated a single, aggregate baseline and single, aggregate end-of-study network by averaging edge weights across the 50 baseline networks and 50 end-of-study networks, respectively (Supplementary data). We evaluated these aggregate networks as in [28] and defined an edge confidence score (EC) for each network:
where is the z-score weight of the edge between node i and j in the baseline network, is the z-score edge-weight in the end-of-study network and CDF−1 is the inverse cumulative distribution function of a normal distribution. We then identified “high-confidence edges” as those with EC>.25 (~24% of all edges met this criterion). High-confidence edges at baseline (ECb>0.25) can be interpreted as edges that are both likely to exist in the baseline network and that have increased evidence in the baseline as opposed to the end-of-study network; the inverse is true of high-confidence edges at end-of-study. These edges define distinct subnetworks for baseline and end-of-study. When the aggregate networks were restricted only to edges of high-confidence, the baseline network had 548,736 edges, while the end-of-study network had 540,701 edges.
We quantified differences in gene targeting between these high-confidence-edge subnetworks by calculating the change in degree (either in-degree or out-degree) for each gene (i) and using an Edge Enrichment Score (EES [23]):
Where and are the degree of high-confidence edges for gene i in the end-of-study and baseline subnetworks, respectively, and Ne and Nb are the total number of edges that make up the end-of-study and baseline subnetworks. Note that the EES will be positive for edge-enrichment around a particular gene in the end-of-study subnetwork, and negative for edge-enrichment around a gene in the baseline subnetwork.
Supplementary Material
Highlights.
Despite previous evidence of their importance, genes in glucose transport pathways were not significantly differentially-expressed upon diet-induced weight-loss. However network analysis showed increased regulatory activity around these pathways post-weight loss.
Network analysis also identified a potential putative mechanism of action for weight loss in the colorectum. Specifically, our models indicated a regulatory shift around glucose pathways genes that includes a switch from NFKB1 to MAX control of MYC.
Overall, our results demonstrate the importance of using network-based approaches to complement the findings of standard gene expression analysis.
Acknowledgments
AV was supported by the Cancer Prevention Fellowship Program, National Cancer Institute. JQ and KG were supported by a grant from the National Heart, Lung, and Blood Institute of the US National Institutes of Health (1R01 HL111759)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of interest: No conflicts of interests to report for any author
References
- 1.Moghaddam AA, Woodward M, Huxley R. Obesity and risk of colorectal cancer: a meta-analysis of 31 studies with 70,000 events. Cancer Epidemiol Biomarkers Prev. 2007;16:2533–2547. doi: 10.1158/1055-9965.EPI-07-0708. [DOI] [PubMed] [Google Scholar]
- 2.WCRF/AICR. Food, Nutrition, Physical Activity and the Prevention of Cancer: a Global Perspective [Internet] 2007 [Google Scholar]
- 3.Nicklas BJ, Ambrosius W, Messier SP, Miller GD, Penninx BWJH, Loeser RF, et al. Diet-induced weight loss, exercise, and chronic inflammation in older, obese adults: a randomized controlled clinical trial 1 – 3. 2004:544–551. doi: 10.1093/ajcn/79.4.544. [DOI] [PubMed] [Google Scholar]
- 4.Anderson AS, Craigie AM, Caswell S, Treweek S, Stead M, Macleod M, et al. The impact of a bodyweight and physical activity intervention (BeWEL) initiated through a national colorectal cancer screening programme: randomised controlled trial. BMJ. 2014;348:g1823. doi: 10.1136/bmj.g1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bassett JK, Severi G, English DR, Baglietto L, Krishnan K, Hopper JL, et al. Body size, weight change, and risk of colon cancer. Cancer Epidemiol Biomarkers Prev. 2010;19:2978–2986. doi: 10.1158/1055-9965.EPI-10-0543. [DOI] [PubMed] [Google Scholar]
- 6.Laake I, Thune I, Selmer R, Tretli S, Slattery ML, Veierød MB. A prospective study of body mass index, weight change, and risk of cancer in the proximal and distal colon. Cancer Epidemiol Biomarkers Prev. 2010;19:1511–1522. doi: 10.1158/1055-9965.EPI-09-0813. [DOI] [PubMed] [Google Scholar]
- 7.Sedjo RL, Byers T, Levin TR, Haffner SM, Saad MF, Tooze Ja, et al. Change in body size and the risk of colorectal adenomas. Cancer Epidemiol Biomarkers Prev. 2007;16:526–531. doi: 10.1158/1055-9965.EPI-06-0229. [DOI] [PubMed] [Google Scholar]
- 8.Song M, Hu FB, Spiegelman D, Chan AT, Wu K, Ogino S, Fuchs CS, Willett WC, Giovannucci EL. Adulthood weight change and risk of colorectal cancer in the Nurse's Health Study and Health Professionals Follow-up Study. Canc Prev Res. 2015;8:620–627. doi: 10.1158/1940-6207.CAPR-15-0061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keum N, Greenwood DC, Lee D Hoon, Kim R, Aune D, Ju W, et al. Adult weight gain and adiposity-related cancers: a dose-response meta-analysis of prospective observational studies. J Natl Cancer Inst. 2015;107:1–14. doi: 10.1093/jnci/djv088. [DOI] [PubMed] [Google Scholar]
- 10.Chen Q, Wang J, Yang J, Jin Z, Shi W, Qin Y, et al. Association between adult weight gain and colorectal cancer: A dose-response meta-analysis of observational studies. Int J Cancer. 2015;136:2880–2889. doi: 10.1002/ijc.29331. [DOI] [PubMed] [Google Scholar]
- 11.Schlesinger S, Lieb W, Koch M, Fedirko V, Dahm CC, Pischon T, et al. Body weight gain and risk of colorectal cancer: A systematic review and meta-analysis of observational studies. Obes Rev. 2015;16:607–619. doi: 10.1111/obr.12286. [DOI] [PubMed] [Google Scholar]
- 12.Parker ED, Folsom a R. Intentional weight loss and incidence of obesity-related cancers: the Iowa Women’s Health Study. Int J Obes Relat Metab Disord. 2003;27:1447–1452. doi: 10.1038/sj.ijo.0802437. [DOI] [PubMed] [Google Scholar]
- 13.Rapp K, Klenk J, Ulmer H, Concin H, Diem G, Oberaigner W, et al. Weight change and cancer risk in a cohort of more than 65,000 adults in Austria. Ann Oncol. 2008;19:641–648. doi: 10.1093/annonc/mdm549. [DOI] [PubMed] [Google Scholar]
- 14.Yamaji Y, Okamoto M, Yoshida H, Kawabe T, Wada R, Mitsushima T, et al. The effect of body weight reduction on the incidence of colorectal adenoma. Am J Gastroenterol. 2008;103:2061–2067. doi: 10.1111/j.1572-0241.2008.01936.x. [DOI] [PubMed] [Google Scholar]
- 15.Laiyemo aO, Doubeni C, Badurdeen DS, Murphy G, Marcus PM, Schoen RE, et al. Obesity, weight change, and risk of adenoma recurrence: a prospective trial. Endoscopy. 2012;44:813–818. doi: 10.1055/s-0032-1309837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Steins Bisschop CN, van Gils CH, Emaus MJ, Bueno-de-Mesquita HB, Monninkhof EM, Boeing H, et al. Weight change later in life and colon and rectal cancer risk in participants in the EPICPANACEA study. Am J Clin Nutr. 2014;99:139–147. doi: 10.3945/ajcn.113.066530. [DOI] [PubMed] [Google Scholar]
- 17.Thygesen LC, Grønbaek M, Johansen C, Fuchs CS, Willett WC, Giovannucci E. Prospective weight change and colon cancer risk in male US health professionals. Int J Cancer. 2008;123:1160–1165. doi: 10.1002/ijc.23612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Renehan AG, Flood A, Adams KF, Olden M, Hollenbeck AR, Cross AJ, et al. Body mass index at different adult ages, weight change, and colorectal cancer risk in the National Institutes of Health-AARP Cohort. Am J Epidemiol. 2012;176:1130–1140. doi: 10.1093/aje/kws192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen J. Multiple signal pathways in obesity-associated cancer. Obes Rev. 2011;12:1063–1070. doi: 10.1111/j.1467-789X.2011.00917.x. [DOI] [PubMed] [Google Scholar]
- 20.Riondino S, Roselli M, Palmirotta R, Della-Morte D, Ferroni P, Guadagni F. Obesity and colorectal cancer: role of adipokines in tumor initiation and progression. World J Gastroenterol. 2014;20:5177–5190. doi: 10.3748/wjg.v20.i18.5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ross R, Dagnone D, Jones PJH, Smith H, Paddags A, Hudson R, et al. Reduction in Obesity and Related Comorbid Conditions after Diet-Induced Weight Loss or Exercise-Induced Weight Loss in Men A Randomized, Controlled Trial. 2014 doi: 10.7326/0003-4819-133-2-200007180-00008. [DOI] [PubMed] [Google Scholar]
- 22.Chae JS, Paik JK, Kang R, Kim M, Choi Y, Lee S-H, et al. Mild weight loss reduces inflammatory cytokines, leukocyte count, and oxidative stress in overweight and moderately obese participants treated for 3 years with dietary modification. Nutr Res. Elsevier Inc. 2013;33:195–203. doi: 10.1016/j.nutres.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 23.Parekh N, Lin Y, Vadiveloo M, Hayes RB, Lu-Yao GL. Metabolic dysregulation of the insulin-glucose axis and risk of obesity-related cancers in the Framingham heart study-offspring cohort (1971–2008) Cancer Epidemiol Biomarkers Prev. 2013;22:1825–1836. doi: 10.1158/1055-9965.EPI-13-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Byers T, Sedjo RL. review article. 2011:1063–1072. [Google Scholar]
- 25.De Smet R, Marchal K. Advantages and limitations of current network inference methods. Nat Rev Microbiol. Nature Publishing Group. 2010;8:717–729. doi: 10.1038/nrmicro2419. [DOI] [PubMed] [Google Scholar]
- 26.Hecker M, Lambeck S, Toepfer S, van Someren E, Guthke R. Gene regulatory network inference: Data integration in dynamic models-A review. BioSystems. 2009;96:86–103. doi: 10.1016/j.biosystems.2008.12.004. [DOI] [PubMed] [Google Scholar]
- 27.Lao T, Glass K, Qiu W, Polverino F, Gupta K, Morrow J, et al. Haploinsufficiency of Hedgehog interacting protein causes increased emphysema induced by cigarette smoke through network rewiring. Genome Med. 2015;7:1–13. doi: 10.1186/s13073-015-0137-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Glass K, Spentzos D, Quackenbush J, Haibe-Kains B, Yuan G. A Network Model for Angiogenesis in Ovarian Cancer. BMC Bioinformatics. 2015;16:1–17. doi: 10.1186/s12859-015-0551-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Glass K, Huttenhower C, Quackenbush J, Yuan G-C. Passing messages between biological networks to refine predicted interactions. PLoS One. 2013;8:e64832. doi: 10.1371/journal.pone.0064832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Glass K, Quackenbush J, Silverman EK, Celli B, Rennard SI, Yuan G, et al. Sexually-dimorphic targeting of functionally- related genes in COPD. 2014:1–17. doi: 10.1186/s12918-014-0118-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pendyala S, Neff LM, Suarez-Farinas M, Holt PR. Diet-induced weight loss reduces colorectal inflammation: implications. 2011:234–242. doi: 10.3945/ajcn.110.002683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mathelier A, Zhao X, Zhang AW, Parcy F, Worsley-Hunt R, Arenillas DJ, et al. JASPAR 2014: An extensively expanded and updated open-access database of transcription factor binding profiles. Nucleic Acids Res. 2014;42:1–6. doi: 10.1093/nar/gkt997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mootha VK, Lindgren CM, Eriksson K-F, Subramanian A, Sihag S, Lehar J, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 34.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette Ma, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fernandez PC, Frank SR, Wang L, Schroeder M, Liu S, Greene J, et al. Genomic targets of the human c-Myc protein. Genes Dev. 2003;17:1115–1129. doi: 10.1101/gad.1067003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.La Rosa F, Pierce JW, Sonenshein GE. Differential regulation of the c-myc oncogene promoter by the NF-kappa B rel family of transcription factors. Mol Cell Biol. 1994;14:1039–1044. doi: 10.1128/mcb.14.2.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gu W, Cechova K, Tassi V, Dalla-Favera R. Opposite regulation of gene transcription and cell proliferation by c-Myc and Max. Proc Natl Acad Sci U S A. 1993;90:2935–2939. doi: 10.1073/pnas.90.7.2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lahoz EG, Xu L, Schreiber-Agus N, DePinho Ra. Suppression of Myc, but not E1a, transformation activity by Max-associated proteins, Mad and Mxi1. Proc Natl Acad Sci U S A. 1994;91:5503–5507. doi: 10.1073/pnas.91.12.5503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li B, Simon MC. Molecular Pathways: Targeting MYC-induced Metabolic Reprogramming and Oncogenic Stress in Cancer Molecular Pathways: Targeting MYC-induced Metabolic Reprogramming and Oncogenic Stress in Cancer. 2013:5835–5841. doi: 10.1158/1078-0432.CCR-12-3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hedges TR, Gerner EW. Ross’ syndrome (tonic pupil plus) Br J Ophthalmol. 1975;59:387–391. doi: 10.1136/bjo.59.7.387. 1975/07/01 ed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Porquet N, Poirier A, Houle F, Pin A-L, Gout S, Tremblay P-L, et al. Survival advantages conferred to colon cancer cells by E-selectin-induced activation of the PI3K-NFκB survival axis downstream of Death receptor-3. BMC Cancer. BioMed Central Ltd. 2011;11:285. doi: 10.1186/1471-2407-11-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gopalakrishnan N, Devaraj N, Devaraj H. Synergistic association of Notch and NFκB signaling and role of Notch signaling in modulating epithelial to mesenchymal transition in colorectal adenocarcinoma. Biochimie. Elsevier B.V. 2014:1–9. doi: 10.1016/j.biochi.2014.09.020. [DOI] [PubMed] [Google Scholar]
- 43.Dolcet X, Llobet D, Pallares J, Matias-Guiu X. NF-kB in development and progression of human cancer. Virchows Arch. 2005;446:475–482. doi: 10.1007/s00428-005-1264-9. [DOI] [PubMed] [Google Scholar]
- 44.Zhang T, Chen C, Breslin MB, Song K, Lan MS. Extra-nuclear activity of INSM1 transcription factor enhances insulin receptor signaling pathway and Nkx6.1 expression through RACK1 interaction. Cell Signal. Elsevier Inc. 2014;26:740–747. doi: 10.1016/j.cellsig.2013.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Buller CL, Loberg RD, Fan M-H, Zhu Q, Park JL, Vesely E, et al. A GSK-3/TSC2/mTOR pathway regulates glucose uptake and GLUT1 glucose transporter expression. Am J Physiol Cell Physiol. 2008;295:C836–C843. doi: 10.1152/ajpcell.00554.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim J, Zeller KI, Wang Y, Anil G, Aronow BJ, Donnell KAO, et al. Evaluation of Myc E-Box Phylogenetic Footprints in Glycolytic Genes by Chromatin Immunoprecipitation Assays Evaluation of Myc E-Box Phylogenetic Footprints in Glycolytic Genes by Chromatin Immunoprecipitation Assays †. 2004 [Google Scholar]
- 47.Miller SC, Huang R, Sakamuru S, Shukla SJ, Attene- MS, Shinn P, et al. Identification of Known Drugs that Act as Inhibitors of NF-κB Signaling and their Mechanism of Action. 2011;79:1272–1280. doi: 10.1016/j.bcp.2009.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Montastier E, Villa-Vialaneix N, Caspar-Bauguil S, Hlavaty P, Tvrzicka E, Gonzalez I, et al. System Model Network for Adipose Tissue Signatures Related to Weight Changes in Response to Calorie Restriction and Subsequent Weight Maintenance. PLOS Comput Biol. 2015;11:e1004047. doi: 10.1371/journal.pcbi.1004047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.