Abstract
Hedgehog (Hh) signaling plays a central role in development and diseases. Hh activates its signal transducer and GPCR-family protein Smoothened (Smo) by inducing Smo phosphorylation but whether Smo is activated through other post-translational modifications remains unexplored. Here we show that sumoylation acts in parallel with phosphorylation to promote Smo cell surface expression and Hh signaling. We find that Hh stimulates Smo sumoylation by dissociating it from a de-sumoylation enzyme Ulp1. Sumoylation of Smo in turn recruits a deubiquitinase UBPY/USP8 to antagonize Smo ubiquitination and degradation, leading to its cell surface accumulation and elevated Hh pathway activity. We also provide evidence that Shh stimulates sumoylation of mammalian Smo (mSmo) and that sumoylation promotes ciliary localization of mSmo and Shh pathway activity. Our findings reveal a conserved mechanism whereby the SUMO pathway promotes Hh signaling by regulating Smo subcellular localization and shed light on how sumoylation regulates membrane protein trafficking.
Graphical Abstract
Introduction
The Hedgehog (Hh) family of secreted proteins plays critical roles in embryonic development and adult tissue homeostasis (Briscoe and Therond, 2013; Ingham and McMahon, 2001; Jiang and Hui, 2008). Aberrant Hh signaling activity has been implicated in many human disorders including birth defects and cancer (Jiang and Hui, 2008; Taipale and Beachy, 2001; Villavicencio et al., 2000). Hh exerts its biological influence through a conserved signal transduction pathway that culminates in the activation of latent transcription factors Cubitus interruptus (Ci)/Gli proteins (Briscoe and Therond, 2013; Hui and Angers, 2011; Jiang and Hui, 2008; Wilson and Chuang, 2010). The core reception system for Hh signal constitutes two transmembrane proteins Patched (Ptc) and Smoothened (Smo). Hh binding to Ptc alleviates the inhibition of Smo by Ptc, leading to Smo phosphorylation and activation (Chen and Jiang, 2013). Activated Smo then transduce the Hh signal through intracellular signaling complexes to convert Ci/Gli proteins from their repressor forms (CiR/GliR) to activator forms (CiA/GliA).
In Drosophila, Smo is phosphorylated by multiple kinases including PKA, CK1, and Gprk2/GRK2, and phosphorylation promotes Smo cell surface accumulation and active conformation (Apionishev et al., 2005; Chen et al., 2010; Cheng et al., 2010; Jia et al., 2004; Li et al., 2014; Zhang et al., 2004; Zhao et al., 2007). In mammals, Smo is phosphorylated by CK1 and GRK2, and phosphorylation promotes ciliary localization and an active conformation of Smo (Chen et al., 2011). Although phospho-mimetic mutations in both Drosophila and mammalian Smo rendered constitutive pathway activity, these phospho-mimetic Smo variants did not possess full pathway activity and were still stimulated by Hh (Chen et al., 2010; Chen et al., 2011; Jia et al., 2004). It is possible that full activation of Smo by Hh involves additional phosphorylation events (Zhang et al., 2004). Indeed, several other kinases including CK2, aPKC, and Gish/CK1γ have been shown to phosphorylate Smo (Jia et al., 2010; Jiang et al., 2014; Li et al., 2016). However, it remains possible that phosphorylation-independent mechanism(s) may act in parallel with these kinases to promote Smo activation in response to Hh.
Sumoylation is a reversible covalent modification that controls many cellular, physiological, and pathological processes by regulating the fate of modified proteins (Geiss-Friedlander and Melchior, 2007; Johnson, 2004). Attachment of SUMO molecules to a target protein involves activation by a SUMO-activation enzyme (E1) and subsequent transfer by an SUMO-conjugating enzyme E2 (Ubc9). Typically, E2 associates with a SUMO ligase (E3) that recognizes the target protein. Sumoylation is a dynamic process and removal of SUMO molecules (de-sumoylation) from target proteins is accomplished by SUMO-specific isopeptidases, such as Ubiquitin-like protease 1 (Ulp1) in Drosophila and SENP family members in mammals (Johnson, 2004). The majority of SUMO targets are nuclear proteins including transcription factors and cofactors (Seeler and Dejean, 2003). With few exceptions, sumoylation mostly represses transcription (Gill, 2005; Lyst and Stancheva, 2007). Whether sumoylation regulates other cellular components such as transmembrane receptors has remained poorly explored.
In a genetic modifier screen, we identified several components of the SUMO pathway that exhibit genetic interactions with a dominant negative form of Smo. Here we provide evidence that Hh induces sumoylation of Smo and that sumoylation promotes Hh pathway activity by increasing Smo cell surface expression. Mechanistically, we show that Ulp1 forms a complex with Smo to prevent its sumoylation, and that Hh stimulates Smo sumoylation by disrupting Smo/Ulp1 association independent of Smo phosphorylation by PKA/CK1. Furthermore, we find that sumoylation of Smo recruits the deubiquitinase UBPY/USP8 to antagonize its ubiquitination, leading to cell surface accumulation of Smo. Finally, we provide evidence that Shh stimulates sumoylation of mammalian Smo (mSmo) and that sumoylation regulates mSmo ciliary localization and Shh pathway activity.
Results
The SUMO pathway positively regulates Hh signaling activity
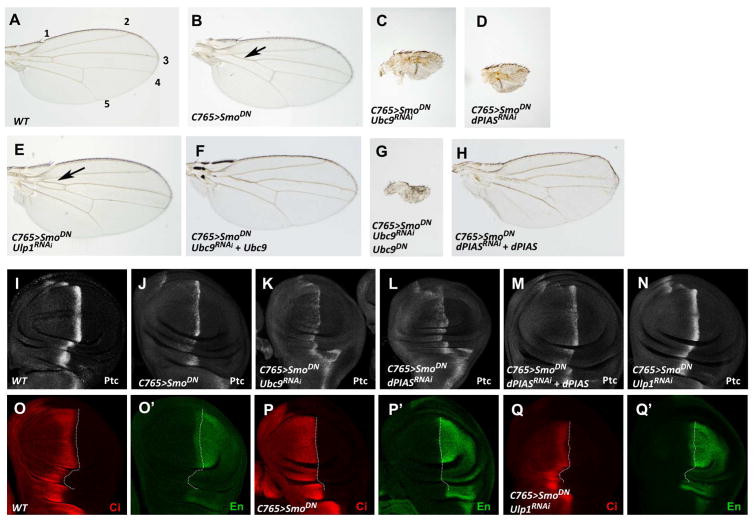
To identify additional Hh pathway regulators, we carried out a genetic modifier screen to identify enhancers or suppressors of a fused wing phenotype caused by expressing a dominant negative form of Smo (SmoDN/Smo-PKA12) with a wing specific Gal4 driver such as C765 (C765>SmoDN) (Fig. 1B) (Jia et al., 2004). Several kinases including Gprk2 and CK2 were identified as enhancers (Chen et al., 2010; Jia et al., 2010). The same screen also identified several components of the SUMO pathway: RNAi of the Drosophila SUMO E2 (Ubc9) encoded by lesswright (lwr)(Huang et al., 2005), or the Drosophila SUMO E3 (dPIAS) encoded by Su(var)2-10 (Hari et al., 2001), enhanced C765>SmoDN induced wing phenotype (Fig. 1C–D compared with 1B). On the other hand, RNAi of the de-sumoylation enzyme Ulp1 suppressed the “fused wing” phenotype induced by C765>SmoDN (Fig. 1E compared with 1B). To exclude the possible off-target effect caused by RNAi, UAS-Ubc9 and UAS-dPIAS transgenes were coexpressed with the corresponding transgenic RNAi. We found that overexpression of Ubc9 or dPIAS reversed the phenotype caused by their RNAi (Fig. 1F, H). By contrast, overexpression of a catalytically dead form of Ubc9 (Ubc9DN) exacerbated the phenotype caused by Ubc9 RNAi (Fig. 1G), consistent with its being a dominant negative form (Huang et al., 2005).
Figure 1. The SUMO pathway positively regulates Hh signaling activity.
(A) A control adult wing with longitudinal veins indicated by the numbers. (B–H) Adult wings of the indicated phenotypes: expression of UAS-SmoDN alone (B), or in combination with UAS-Ubc9-RNAi (C), UAS-dPIAS-RNAi (D), UAS-Ulp1-RNAi (E), UAS-Ubc9-RNAi and UAS-Ubc9 (F), Ubc9-RNAi and UAS-Ubc9DN (G), or UAS-dPIAS-RNAi and UAS-dPIAS (H) using the C765 Gal4 driver. Arrow in (B) indicates the fusion of vein 3 and 4 in the proximal region of the wing blade, which is suppressed by Ulp1 RNAi (arrow in E). (I–Q′) Late third instar larval wing discs from the control (I, O, O′) or indicated genotypes (J–N and P–Q′) were immunostained with Ptc antibody (I–N) or Ci and En (O–Q′) antibodies. The A/P boundary is marked by the dotted lines based on Ci staining (O–Q′). See also Figure S1.
To determine whether the SUMO pathway modified C765>SmoDN induced wing phenotype by regulating Hh pathway activity, we examined Hh target gene expression. In control late third instar wing imaginal discs, Hh induced ptc expression in A-compartment cells near the A/P boundary (Fig. 1I). In C765>SmoDN wing discs, ptc expression at the A/P boundary was greatly reduced (Fig. 1J). Ubc9 or dPIAS RNAi further reduced whereas Ulp1 RNAi restored ptc expression in C765>SmoDN wing discs (Fig. 1K, L, 1N). The effect of dPIAS RNAi on ptc expression was reversed by coexpression of UAS-dPIAS (Fig. 1M). It has been shown that high levels of Hh convert Ci into labile CiA so that A-compartment cells abutting the A/P boundary exhibited low levels of Ci and expressed en, a high threshold Hh target gene (Fig. 1O, O′)(Ohlmeyer and Kalderon, 1998). In C765>SmoDN wing discs, however, A-compartment cells abutting the A/P boundary accumulated high levels of Ci and failed to express en (Fig. 1P, P′), suggesting that Ci to CiA conversion was compromised. Ulp1 RNAi in C765>SmoDN discs restored en expression in A-compartment cells abutting the A/P boundary that also exhibited low levels of Ci staining (Fig. 1Q, Q′), suggesting that inactivation of Ulp1 facilitated Ci activation in C765>SmoDN wing discs.
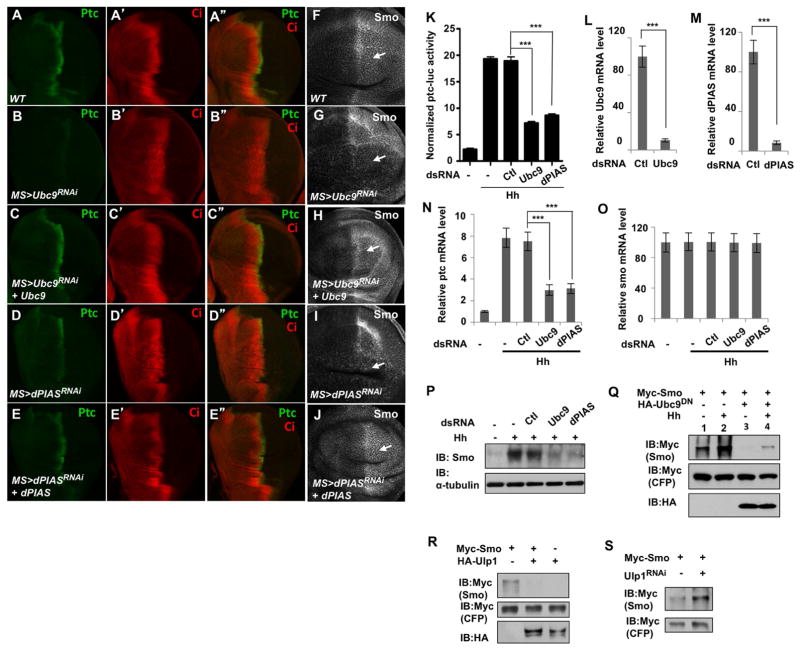
To further explore the role of sumoylation in Hh signal transduction, we examined Hh target gene expression both in vivo and in vitro where SUMO pathway components were inactivated. Expression of either UAS-Ubc9-RNAi or UAS-dPIAS-RNAi using a strong wing specific Gal4 driver, MS1096, which exhibits higher expression in dorsal compartment cells than in ventral compartment cells (Wang et al., 1999), resulted in diminished ptc expression in A-compartment cells near the A/P boundary, most notably in the dorsal region (Fig. 2B, D compared with 2A). Consistent with less Ci was converted into labile CiA in Ubc9 or dPIAS RNAi discs, Ci level was elevated near the A/P boundary in these discs (Fig. 2B′, D′ compared with 2A′). The defects caused by Ubc9 and dPIAS RNAi were suppressed by coexpression of their corresponding transgenes (Fig. 2C-C″, E-E″). Furthermore, we found that knockdown of either Ubc9 or dPIAS inhibited Hh induced ptc-luciferase reporter activity as well as the expression of endogenous ptc in cl8 cells (Fig. 2K–N). Taken together, these results suggest that blocking protein sumoylation inhibits Hh pathway activity.
Figure 2. The SUMO pathway regulates Hh target gene expression and Smo accumulation.
(A–J) Wing discs of wild type control (A-A″, F) or expressing UAS-Ubc9-RNAi (B-B″, G), UAS-Ubc9-RNAi and UAS-Ubc9 (C-C″, H), UAS-dPIAS-RNAi (D–D″, I), or UAS-dPIAS-RNAi and UAS-dPIAS (E-E″, J) with the MS1096 Gal4 driver were immunostained with Ptc and Ci antibodies (A–E″) or Smo (F–J) antibody. (K) ptc-luciferase assay in cl8 cells treated with the corresponding double-strand RNA (dsRNA) in the absence or presence of Hh-conditioned medium. Data are mean ± SEM of normalized ptc-luc activity from three independent experiments. ***, P<0.001 (student’s t-test). (L–O) RT-qPCR to show the relative mRNA level of Ubc9 (L), dPIAS (M), ptc (N), or smo (O) in cl8 cells treated without or with the indicated dsRNAs. GFP dsRNA was used as control (Ctl). Data are mean ± SD from three independent experiments. ***, P<0.001 (student’s t-test). (P) Western blot to shown endogenous Smo protein level in in cl8 cells treated without or with the indicated dsRNAs. (Q–S) Western blot analysis of Myc-Smo expressed in S2 cells with or without HA-Ubc9DN cotransfection and treated with or without Hh-conditioned medium (Q), with or without HA-Ulp1 cotransfection (R), or in the presence of absence of Ulp1 dsRNA (S). Myc-CFP was cotransfected with Myc-Smo to serve as an internal control. See also Figure S2.
Hh induces sumoylation of Ci
The majority of SUMO targets characterized so far are transcription factors. Indeed, two previous studies revealed that mammalian Gli proteins (Gli1, Gli2 and Gli3) were modified by sumoylation at multiple sites (Cox et al., 2010; Han et al., 2012). Inspection of Ci sequence identified four Lys residues with their surrounding amino acid sequence matching the sumoylation consensus site: ψ-K-x-E, where ψ stands for an aliphatic branched amino acid, K is the SUMO receptor, and x stands for any amino acid (Fig. S1A) (Johnson, 2004), suggesting that Ci is likely a SUMO target protein. To test this possibility, S2 cells were transfected with Myc-tagged Ci (Myc-Ci) and HA-tagged SUMO (HA-SUMO) and with or without Aos1 (E1), Ubc9 (E2) and dPIAS (E3). After treatment with Hh-conditioned medium or control medium, Myc-Ci was immunoprecipitated from cell extracts and the presence of sumoylated Ci was determined by western blot analysis with an HA antibody. In the absence of Hh, sumoylation of Ci was not detected even in the presence E1/E2/E3 overexpression (Fig. S1B, lane 1). Sumoylation of Ci was detected after Hh stimulation in conjunction with E1/E2/E3 overexpression (Fig. S1B, lane 4). Adding N-Ethylmaleimide (NEM), a SUMO protease inhibitor (Smith et al., 2011), to cell lysates allowed detection of sumoylated Ci in the absence of E1/E2/E3 overexpression (Fig. S1C, lanes 2–3). Mutating the four putative sumoylation sites (Ci4KR) diminished Hh-stimulated Ci sumoylation (Fig. S1B, lane 5; Fig. S1C, lane 4).
To determine whether sumoylation regulates Ci activity, we compared the activity of Myc-Ci with that of Myc-Ci4KR in S2 cell in the presence of Sufu cotransfection. Under this condition, Myc-Ci exhibited low basal activity measured by the ptc-luc reporter assay but was activated upon Hh stimulation (Fig. S1D)(Han et al., 2015). Coexpression of SUMO and E1/E2/E3 further stimulated whereas Ulp1 suppressed Hh-induced Ci activity (Fig. S1D). Myc-Ci4KR consistently exhibited lower activity than Myc-Ci (Fig. S1D). However, when expressed in wing discs, Myc-Ci4KR and Myc-Ci activated similar levels of ptc-lacZ expression in the P-compartment cells where there was no endogenous Ci, even when the UAS transgenes were driven by a weak Gal4 driver C765 (Fig. S1E–F). It is possible that sumoylation of Ci only modestly altered its activity, which could not be detected by our in vivo assay.
The SUMO pathway regulates Smo accumulation
We noticed that the activity of Myc-Ci4KR in S2 cells was still modulated by coexpression of the sumoylation enzymes or the de-sumoylation enzyme (Fig. S1D), suggesting that the sumoylation pathway may regulate additional Hh pathway component(s). Interestingly, we found that Ubc9 or dPIAS RNAi in wing discs blocked the cell surface accumulation of Smo, which is normally induced by Hh in P compartment cells as well as in A-compartment cells near the A/P boundary (Fig. 2G, 2I compared with 2F) (Denef et al., 2000). Coexpression of Ubc9 or dPIAS transgene with their corresponding RNAi transgenes restored Smo cell surface expression (Fig. 2H, 2J). In addition, dPIAS mutant wing discs exhibited reduced Smo accumulation as well as attenuated Hh target gene expression compared with wild type discs (Fig. S2). These results suggest that the sumoylation pathway may regulate Hh signaling by controlling Smo trafficking and degradation.
To further explore how the SUMO pathway influence Smo protein level, we conducted experiments in cultured cells where protein sumoylation was either blocked or enhanced. Knockdown of Ubc9 or dPIAS in cl8 cells inhibited Hh-induced accumulation of endogenous Smo although smo mRNA abundance was not affected (Fig. 2O–P). When expressed in S2 cells, Myc-Smo was stabilized upon Hh stimulation (Fig. 2Q, lanes 1 and 2). Coexpression of the dominant negative form of Ubc9 (HA-Ubc9DN) greatly suppressed the basal as well as Hh-stimulated Smo accumulation (Fig. 2Q, lanes 3 and 4). Coexpression of Ulp1 decreased whereas Upl1 RNAi increased Smo protein levels (Fig. 2R–S). These results demonstrate that the SUMO pathway promotes Smo protein accumulation.
Hh stimulates sumoylation of Smo
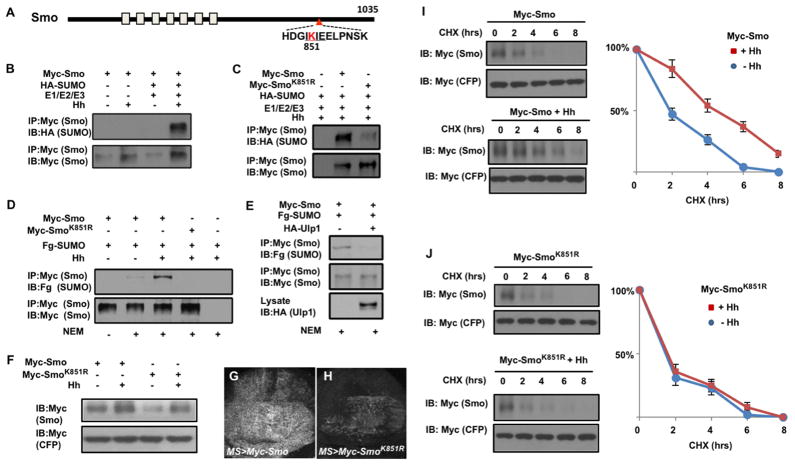
To determine whether Smo is a direct SUMO target, we performed the cell-based sumoylation assay described in Fig. S1B–C. In the absence of Hh stimulation, HA-SUMO conjugation to Myc-Smo was not detected even when the SUMO E1, E2 and E3 enzymes were co-expressed; however, treating cells with Hh-conditioned medium stimulated HA-SUMO conjugation to Myc-Smo in the presence E1/E2/E3 (Fig. 3B). Adding NEM into cell lysates allowed detection of Hh-stimulated HA-SUMO conjugation to Myc-Smo in the absence of E1/E2/E3 overexpression (Fig. 3D, lane 3). Weak SUMO-conjugation to Myc-Smo was observed in the absence of Hh upon NEM treatment (Fig. 3D, lane 2), which was abolished when Ulp1 was overexpressed (Fig. 3E), suggesting that low levels of Smo sumoylation occurred in unstimulated condition. The C-terminal tail of Smo harbors a putative SUMO acceptor site (K851) that conforms the sumoylation consensus site (Fig. 3A). Conversion of K851 to R (Myc-SmoK851R) diminished both the basal and Hh-induced sumoylation on Smo (Fig. 3C, D), indicating that Smo is mainly sumoylated at K851.
Figure 3. Hh stimulates sumoylation of Smo at K851 to regulate its abundance.
(A) Schematic drawing of Smo. (B–C) Western blots to detect SUMO conjugated Smo derived from S2 cells transfected with Myc-Smo (B, C) or Myc-SmoK851R (C) together with HA-SUMO and the SUMO E1/E2/E3 enzymes in the absence or presence of Hh-conditioned medium. (D–E) Western blots to detect SUMO conjugated Smo derived from S2 cells transfected with Myc-Smo or Myc-SmoK851R and Fg-SUMO in the absence or presence of Hh-conditioned medium (D) or HA-Ulp1 cotransfection (E). (F) Western blot analysis of cell extracts from S2 cells transfected with either Myc-Smo or Myc-SmoK851R and treated with Hh-conditioned or control medium. (G–H) Wing discs expressing Myc-Smo (G) or Myc-SmoK851R (H) under the control of MS1096 in conjunction with Gal80ts. Larvae were grown at 18°C until late third instar, then shifted to 30°C for 16 hours before immunostained with anti-Myc antibody. (I–J) Protein stability assay of Myc-Smo and Myc-SmoK851R transfected into S2 cells. Quantification of Myc-Smo levels at different time points was shown to the right. Data are mean ± SD from three independent experiments. Signal intensities at t=0 were defined as 100%. Of note, to ensure relatively equal level of Smo protein at t=0, more Myc-SmoK851R DNA than that of Myc-Smo was transfected and more DNA was transfected in the absence of Hh treatment compared with that treated with Hh-conditioned medium. See also Figure S3.
Sumoylation of Smo regulates its abundance and activity
When expressed in S2 cells, SmoK851R exhibited diminished steady state levels in both unstimulated and Hh-stimulated conditions compared with Myc-Smo (Fig. 3F). In addition, SmoK851R exhibited diminished cell surface expression in response to Hh stimulation (Fig. S3). After a pulse expression in wing discs for 16 hours, Myc-Smo accumulated at much higher levels than Myc-SmoK851R (Fig. 3G–H), suggesting that sumoylation of Smo is essential for its accumulation.
We next carried out protein stability assay to determine how sumoylation altered the half-life of Smo. S2 cells were transfected with Myc-Smo or Myc-SmoK851R in the absence or presence of Hh-conditioned medium and treated with cycloheximide (CHX) for different periods of time, followed by western blot analysis. As shown in Fig. 3I–J, Myc-SmoK851R exhibited reduced half-life compared with Myc-Smo in response to Hh stimulation.
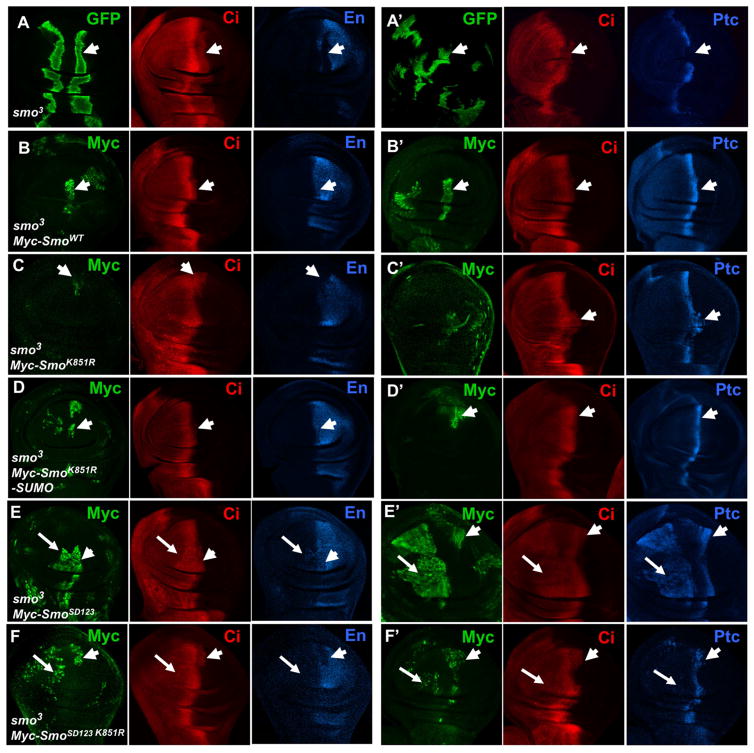
To further explore the biological significance of Smo sumoylation, we compared the activity of wild type and sumoylation deficient forms of Smo in wing discs containing smo mutant clones. We employed the MARCM system to generate mutant clones for a smo null allele, smo3, in which UAS-Myc-Smo or UAS-Myc-SmoK851R was expressed using a weak Gal4 driver C765 (Li et al., 2014). As shown in Fig 4, C765>Myc-Smo completely rescued the expression of ptc and en in smo mutant clones located near the A/P boundary (Fig. 4B, B′ compared with 4A, A′). By contrast, C765>Myc-SmoK851R failed to rescue en and only partially rescued ptc (Fig. 4C-C′). Myc staining indicated that C765>Myc-SmoK851R produced less Smo than C765>Myc-Smo (Fig. 4C compared with 4B), which could contribute to the reduced activity associated with C765>Myc-SmoK851R. Consistent with this, expression of UAS-Myc-SmoK851R using a strong Gal4 driver tub-Gal4 (Li et al., 2014), rescued the expression of both ptc and en (Fig. S4).
Figure 4. Sumoylation of Smo at K851 is required for its optimal activity in vivo.
(A-A′) Wing imaginal discs of late third instar larvae containing smo3 clones induced by the MARCM system were immunostained to show the expression of GFP, Ci, En (blue in A) or Ptc (blue in A′). smo3 clones are marked by the GFP expression (arrowheads). (B–F′) Wing discs containing smo3 clones that express Myc-Smo (B-B′), Myc-SmoK851R (C-C′), Myc-SmoK851R-SUMO (D-D′), Myc-SmoSD123 (E-E′), or Myc-SmoSD123 K851R (F-F′) driven by the C765 Gal4 driver were immunostained with Myc, Ci, En or Ptc antibodies. smo3 clones expressing Smo transgenes are marked by the Myc expression. Arrows and arrowheads indicate clones distant from or close to the A/P compartment boundary, respectively. See also Figure S4.
To confirm the defect associated with SmoK851R was due to the lack of SUMO conjunction, we fused a SUMO moiety to the C-terminus of Myc-SmoK851R to generate Myc-SmoK851R-SUMO. C765>Myc-SmoK851R-SUMO fully rescued the expression of ptc and en in smo3 clones (Fig. 4D, D′). Myc staining indicated that Myc-SmoK851R-SUMO was more abundant than Myc-SmoK851R (Fig. 4D compared with 4C) and reached levels similar to Myc-Smo derived from C765>Myc-Smo (Fig. 4D compared with 4B). Hence blocking sumoylation of Smo reduced its activity most likely by interfering with its cell surface accumulation.
Sumoylation regulates Smo independent of its phosphorylation
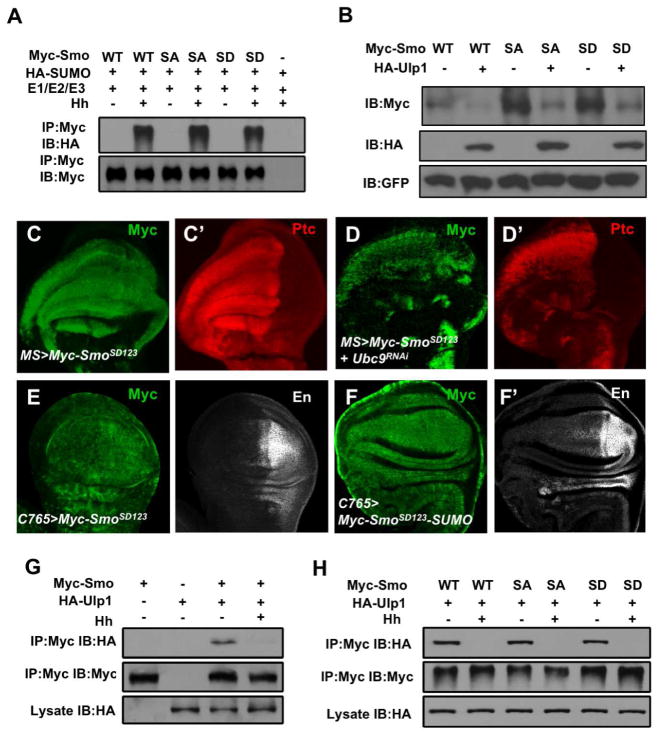
Hh-induced Smo phosphorylation by PKA and CK1 also promotes Smo cell surface accumulation (Jia et al., 2004); however, a phosphorylation-deficient form of Smo (Myc-SmoSA123) with three PKA sites (S667, S687, and S740) mutated to Ala, or a phosphorylation-mimicking form of Smo (Myc-SmoSD123) with the three PKA sites and adjacent CK1 sites mutated to Asp (Jia et al., 2004), was sumoylated equally well in response to Hh stimulation compared with the wild type Smo (Myc-SmoWT) (Fig. 5A), suggesting that Hh induced Smo sumoylation is independent of its phosphorylation by PKA and CK1. Consistent with this notion, overexpression of Ulp1 downregulated the levels of Myc-SmoWT, Myc-SmoSA123, and Myc-SmoSD123 (Fig. 5B). Furthermore, coexpression of UAS-UBC9-RNAi with Myc-SmoSD123 in wing discs using MS1096 reduced Myc-SmoSD123 protein level and diminished the ectopic expression of ptc induced by Myc-SmoSD, suggesting that sumoylation regulates Smo abundance independent of its phosphorylation by PKA and CK1. We also found that the K851R mutation did not affect Smo phosphorylation in P-compartment cells of wing discs (Fig. S5). These results suggest that phosphorylation and sumoylation on Smo occur parallely in response to Hh stimulation.
Figure 5. Hh stimulates sumoylation of Smo independent of its phosphorylation.
(A) Western blot analysis of SUMO-conjugated Myc-Smo of wild type (WT), phosphor-deficient (SA), or phosphor-mimetic (SD) forms transfected into S2 together with HA-SUMO and the SUMO E1/E2/E3 enzymes in the absence or presence of Hh-conditioned medium. (B) Western blot analysis of cell extracts from S2 transfected with the indicated Smo constructs with or without HA-Ulp1cotransfection. Myc-GFP was cotransfected as an internal control. (C-D′) Wing discs expressing UAS-Myc-SmoSD123 either alone (C, C′) or together with UAS-Ubc9-RNAi (D, D′) under the control of MS1096 were immunostained with Myc (green) and Ptc (red) antibodies. (E-F′) Wing discs expressing UAS-Myc-SmoSD123 (E, E′) or UAS-Myc-SmoSD123-SUMO (F, F′) under the control of C765 were immunostained with Myc and En antibodies. (G–H) Hh inhibits Smo/Ulp1 association independent of Smo phosphorylation. See also Figures S5 and S6.
To further explore the relationship between Smo phosphorylation and sumoylation, we either introduced the K851R mutation into Myc-SmoSD123 to generate Myc-SmoSD123 K851R or fused the SUMO moiety to Myc-SmoSD123 to generate Myc-SmoSD123-SUMO. C765>Myc-SmoSD123 not only rescued both ptc and en expression in smo3 clones near the A/P boundary (arrowheads in Fig. 4E, E′) but also induced their ectopic expression (arrows in Fig. 4E, E′). By contrast, C765>Myc-SmoSD123 K851 induced little if any ectopic ptc expression and only partially rescued ptc expression in smo3 clones (arrows and arrowheads in Fig. 4F′). In addition, C765>Myc-SmoSD123 K851 failed rescue en expression in smo3 clones (arrowheads in Fig. 4F). On the other hand, expression of Myc-SmoSD123-SUMO using C765 induced ectopic expression of en in more anteriorly localized cells than Myc-SmoSD123 (Fig. 5E′, 5F′). Myc staining indicated that Myc-SmoSD123 was less stable than Myc-SmoSD123-SUMO (Fig. 5E, 5F) but more stable than Myc-SmoSD123 K851R (Fig. 4E–F′). Taken together, these results suggest that sumoylation acts in parallel with phosphorylation to promote Smo accumulation and activity.
Hh promotes Smo sumoylation by dissociating Ulp1 from Smo
We next determine how Hh stimulates sumoylation of Smo. It has been reported that SUMO substrates can interact with E2 and E3 and in some cases these interactions are regulated (Gareau and Lima, 2010; Geiss-Friedlander and Melchior, 2007). However, our co-immunoprecipitation (Co-IP) experiments failed to detect an interaction of Myc-Smo with either Ubc9 or dPIAS regardless of Hh stimulation (data no shown). Nevertheless, Myc-Smo could pull down HA-Ulp1 when coexpressed in S2 cells in the Co-IP assay (Fig. 5G). Interestingly, Hh stimulation diminished the amounts of Ulp1 coimmunoprecipitated with Myc-Smo (Fig. 5G), suggesting that Hh may stimulate sumoylation of Smo, at least in part, by dissociating Ulp1 from Smo. Interaction between Smo and Ulp1 appears to be independent of Smo phosphorylation by PKA/CK1, as both the phosphorylation-deficient (Myc-SmoSA123) and phosphorylation-mimetic forms of Smo (Myc-SmoSD123) coimmunoprecipitated HA-Ulp1, and their association with Ulp1 was abolished by Hh (Fig. 5H).
HA-Ulp1 interacted strongly with Smo C-tail (Myc-SmoCT) as well as the distal half of the Smo C-tail (Myc-Smo730-1035) but poorly with a truncated form of Smo lacking the C-tail (Myc-SmoΔCT) or the membrane-proximal half of the C-tail (Myc-Smo556-730) (Fig. S6A–B), suggesting that Ulp1 binds to the C-terminal half of the Smo C-tail. However, interaction of HA-Ulp1 with Myc-SmoCT was no longer downregulated upon Hh stimulation (Fig. S6C). The Fused (Fu) kinase acts both downstream and upstream of Smo to regulate Hh signaling (Liu et al., 2007). Interestingly, replacing endogenous Fu with a kinase dead form Fu (FuKR) abrogated the regulation of Smo/Ulp1 interaction by Hh (Fig. S6D), suggesting that Hh inhibits Ulp1 binding to Smo through the Fu kinase activity.
Sumoylation of Smo antagonizes its ubiquitination
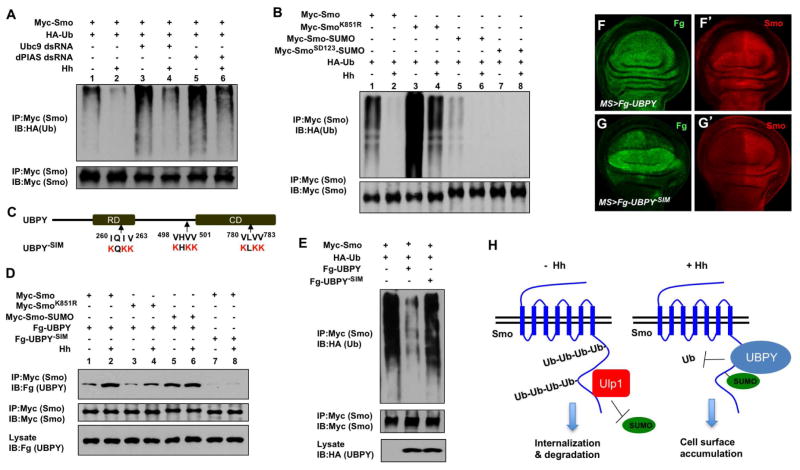
Previous studies suggest that ubiquitination of Smo leads to its internalization and degradation by both proteasome- and lysosome-dependent mechanisms (Li et al., 2012; Xia et al., 2012). Therefore, we determined whether sumoylation promotes Smo cell surface accumulation by antagonizing its ubiquitination. Using a cell-based ubiquitination assay described previously (Li et al., 2012), we found that Ubc9 or dPIAS RNAi increased Smo ubiquitination both in the absence and presence of Hh (Fig. 6A, lanes 3–6 compared with lanes 1–2). The SUMO-deficient Smo (Myc-SmoK851R) also exhibited elevated levels of ubiquitination both in the absence and presence of Hh (Fig. 6B, lanes 3–4 compared with lanes 1–2). On the other hand, fusion of a SUMO moiety to C-terminus of Smo (Myc-Smo-SUMO) that mimics sumoylation greatly reduced the basal ubiquitination of Smo (Fig. 6B, lane 5 compared with lane 1), suggesting that sumoylation of Smo inhibits its ubiquitination. In addition, ubiquitination of Myc-Smo-SUMO was further inhibited by Hh treatment (Fig. 6B, lane 6 compared with lane 5). Consistent with its diminished ubiquitination associated, Myc-Smo-SUMO exhibited high levels of cell surface accumulation (Fig. S3) and increased protein stability (Fig. S7).
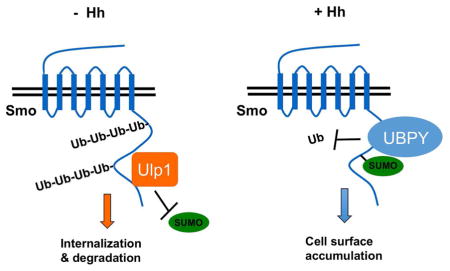
Figure 6. Sumoylation of Smo recruits UBPY to antagonize its ubiquitination.
(A–B) Cell based ubiquitination assays to determine the effect of blocking sumoylation on Smo ubiquitination. (C) Schematic drawing of UBPY with the regulatory domain (RD) and catalytic domain (CD) represented by filled boxes. The arrows indicate the position of the putative SUMO-interacting motifs (SIM). (D) Western blot analysis to show that Hh stimulates UBPY/Smo association through Smo sumoylation. (E) Ubiquitination assay of extracts from S2 cells transfected with the indicated constructs. (F–G′) Wing discs expressing Fg-UBPY (F-F′) or Fg-UBPY-SIM (G-G′) were immunostained with Flag (Fg) and Smo antibodies. (H) Hh inhibits Smo ubiquitination and degradation through stimulating its sumoylation. See also Figure S7.
The observation that ubiquitination of both the SUMO-deficient and SUMO-mimetic forms of Smo was still regulated by Hh suggests that Hh could inhibit ubiquitination of Smo through a mechanism in addition to stimulating its sumoylation. Previous studies suggest that Hh-stimulated Smo phosphorylation inhibited its ubiquitination (Li et al., 2012; Xia et al., 2012). Indeed, introducing the phospho-mimetic mutations into Smo-SUMO (Myc-SmoSD-SUMO) further decreased Smo ubiquitination (Fig. 6B, lane 7 compared with lane 5), suggesting that phosphorylation and sumoylation of Smo act in parallel to inhibit its ubiquitination.
Sumoylation recruits UBPY to inhibit Smo ubiquitination
Hh inhibits Smo ubiquitination depending on the de-ubiquitination enzyme UBPY/USP8, which interacts with Smo C-tail (Li et al., 2012; Xia et al., 2012). We noticed that UBPY contains three putative SUMO-interacting motifs (SIMs) that match the SIM consensus sequence: V/I-X-V/I-V/I (Fig. 6C)(Song et al., 2004), raising an interesting possibility that sumoylation of Smo may recruit UBPY to inhibit its ubiquitination. Indeed, co-immunoprecipitation experiments revealed that Hh stimulated the binding of UBPY to Smo when NEM was added to the cell lysates to preserve SUMO-conjugated Smo (Fig. 6D, lanes 1–2). Mutating the SUMO site in Smo (K851R) diminished its binding to UBPY whereas fusion of SUMO to Smo (Smo-SUMO) resulted in increased binding to UBPY (Fig. 6D, lanes 3–6). Furthermore, mutating the SIM motifs in UBPY (UBPY-SIM; Fig. 6C) diminished its binding to Smo (Fig. 6D, lanes 7 and 8). UBPY-SIM exhibited diminished activity toward inhibiting Smo ubiquitination in S2 cells (Fig. 6E) or stabilizing Smo in wing discs compared with wild type UBPY (Fig. 6F–G′). Taken together, these results suggest that sumoylation recruits UBPY to inhibit Smo ubiquitination.
Sumoylation of mSmo regulates Shh signaling
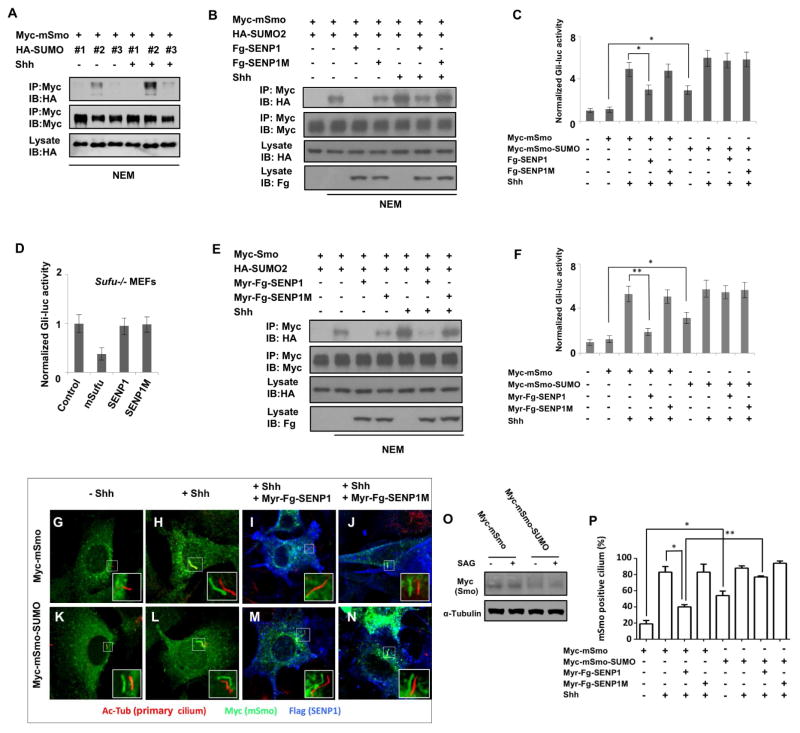
We next determined whether sumoylation regulates mSmo and Shh signaling. There are three SUMO isoforms in mammals: SUMO1, SUMO2, and SUMO3. We found that HA-SUMO2 was most effectively conjugated to Myc-mSmo in NIH3T3 cells and that SUMO conjugation was stimulated by Shh (Fig. 7A). Overexpression of a wild type form of mammalian de-sumoylation enzyme SENP1 (Flag-SENP1) but not its catalytically dead from (Flag-SENP1M) inhibited both the basal and Shh-stimulated sumoylation of mSmo (Fig. 7B).
Figure 7. Sumoylation regulates mSmo ciliary accumulation and Shh signaling.
(A) Western blots to detect SUMO conjugated mSmo derived from NIH3T3 cells transfected with Myc-mSmo and HA-tagged SUMO1, SUMO2 or SUMO3 and treated with or without Shh. (B, E) Western blots to detect SUMO-conjugated mSmo derived from NIH3 cells transfected with the indicated constructs and treated with or without Shh. (C, F) Gli-luc reporter assay in NIH3T3 cells transfected with the indicated constructs and treated with or without Shh. Data are means ± SD from three independent experiments. * P<0.05, ** P<0.01. (D) Gli-luc reporter assay in Sufu−/− MEFs transfected with the indicated constructs. (G–N) NIH 3T3 cells stably expressing Myc-mSmo (G–J) or Myc-mSmo-SUMO (K–N) were transfected with or without Myr-Fg-SENP1 or Myr-Fg-SENP1M and treated with or without Shh, followed by immunostaining with antibodies against acetylated tubulin (primary cilium, red), Myc tag (mSmo, green) and Flag tag (SENP1, blue). The insets show the enlarged view of the selected regions with shifted overlays of primary cilium (red) and mSmo (green) immunofluorescence signals. (O) Western blot analysis of cell extracts form Myc-mSmo or Myc-mSmo-SUMO expressing cells treated with or without SAG. (P) Quantification of mSmo positive primary cilia as indicated by the percentage of cells (50 cells were examined for each group) exhibiting ciliary Myc-mSmo signals. Data are means ± SD from two independent experiments. * P<0.05, ** P<0.01.
A previous study revealed that overexpression of a dominant negative form of Pias1 blocked Shh target gene expression in chick neural tubes (Cox et al., 2010). To determine whether sumoylation regulates Shh signaling at the level of Smo, we carried out Gli-luc reporter assay. We found that SENP1 but not SENP1M inhibited Shh-stimulated Gli-luc reporter activity in NIH3 cells transfected by a wild type form of Smo (Myc-mSmo) (Fig. 7C). However, SENP1 did not affect Gli-luc reporter activity in Sufu mutant MEF cells (Fig. 7D), suggesting that the SUMO pathway regulates Shh signaling upstream of Sufu. If the SUMO pathway regulates Shh signaling by targeting mSmo, then fusion of a SUMO moiety to mSmo (Myc-mSmo-SUMO) may enhance its signaling activity and render it resistant to SENP1-mediated inhibition. Indeed, when transfected into NIH3T3 cells, Myc-mSmo-SUMO exhibited higher basal as well as Shh-induced activity than Myc-mSmo (Fig. 7C). Furthermore, SENP1 failed to inhibit Shh pathway activity elicited by Myc-mSmo-SUMO (Fig. 7C). We also generated membrane-tethered forms of SENP1 and SENP1M by adding a myristoylation signal at their N-termini (Myr-SENP1 and Myr-SENP1M). We found that Myr-SENP1 is more potent than SENP1 in inhibiting mSmo sumoylation and Hh pathway activity, and this inhibition was abolished by mutating the SENP1 catalytic site (Myr-SENP1M) or by conjugating a SUMO moiety to mSmo (Myc-mSmo-SUMO) (Fig. 7E, F). Taken together, these results suggest that the SUMO pathway regulates Shh signaling, at least in part, by targeting mSmo.
Sumoylation of mSmo regulates its ciliary accumulation
In vertebrates, Hh signal transduction occurs at the primary cilium (Huangfu and Anderson, 2006). In response to Hh, Smo is accumulated to the primary cilium where it converts Gli proteins from transcriptional repressors to activators (Chen et al., 2011; Corbit et al., 2005; Rohatgi et al., 2007). To determine whether sumoylation regulates Smo ciliary accumulation, we generated NIH3T3 cell lines that stably express low levels of Myc-mSmo or Myc-mSmo-SUMO using the lentiviral system. Western blot analysis of cell lysates from these cell lines indicated that Myc-mSmo and Myc-mSmo-SUMO were expressed at comparable levels (Fig. 7O). mSmo-expressing cells were transfected without or with Myr-SENP1 or Myr-SENP1M, and treated without or with Shh. Consistent with previous findings, Myc-mSmo was rarely found in the primary cilia in the absence of Shh stimulation, and only ~20% of the cells (12/50) contained ciliary Myc-mSmo signals (Fig. 7G, P). After treatment with Shh, ~80% of the cells exhibited ciliary Myc-mSmo signals (Fig. 7H, P). Expression of Myr-SENP1 but not Myr-SENP1M greatly inhibited Shh-induced mSmo ciliary localization (Fig. 7I, J, P). Addition of SUMO moiety to mSmo increased its basal ciliary localization as Myc-mSmo-SUMO was accumulated to the primary cilium in ~60% of the cells in the absence of Shh (Fig. 7K, P). In response to Shh stimulation, >80% of the cells accumulated Myc-mSmo-SUMO at the primary cilia (Fig. 7L, P). More importantly, ciliary accumulation of Myc-mSmo-SUMO was resistant to Myr-SENP1-mediated inhibition (Fig. 7M, N, P), suggesting that Myr-SENP1 inhibits mSmo ciliary localization by blocking its sumoylation.
Discussion
It has been well documented that Hh activates Smo by inducing its phosphorylation through multiple kinases (Chen and Jiang, 2013); however, whether Smo is also activated through other post-translation modifications has remained largely unexplored. In this study, we demonstrate that Smo is regulated by SUMO conjugation of its C-tail in a manner stimulated by Hh. We find that sumoylation of Smo C-tail is required for its cell surface accumulation and Hh signaling activity. Mechanistically, we demonstrate that sumoylation promotes Smo cell surface expression by recruiting the deubiquitination enzyme UBPY to antagonize its ubiquitination (Fig. 6H), which is known to cause its endocytosis and degradation (Li et al., 2012; Xia et al., 2012). Finally, we provide evidence that sumoylation plays a conserved role in the mammalian Hh pathway and regulates mSmo ciliary localization.
Sumoylation and phosphorylation: two parallel mechanisms for Smo activation
As an obligated signal transducer of the Hh pathway, Smo activity is tightly controlled by multiple mechanisms to ensure appropriated levels of pathway activity in response graded Hh signals. One mechanism is the regulation of Smo conformation: in response to Hh stimulation, Smo switches from a closed and inactivate conformation to an open and active conformation, resulting in dimerization/oligomerization of its C-tail (Zhao et al., 2007). Another level of regulation is Smo subcellular localization: in response to Hh, Smo is stabilized on the cell surface in Drosophila and localized to the primary cilium in vertebrates (Corbit et al., 2005; Denef et al., 2000; Rohatgi et al., 2007). Both processes are regulated by multi-site phosphorylation on Smo C-tail (Chen and Jiang, 2013); however, phospho-mimetic mutations of Smo do not fully activate Smo (Chen et al., 2011; Jia et al., 2004), raising a possibility that Smo is activated through additional mechanisms. Through a combination of genetic and biochemical studies, we provide several lines of evidence that the SUMO pathway promotes Hh signaling by regulating Smo subcellular localization and that Smo is a direct target of SUMO conjugation: 1) inactivation of sumoylation enzymes inhibits Hh pathway activity and block Smo cell surface accumulation; 2) Smo is sumoylated on K851 in a manner stimulated by Hh; 3) blocking Smo sumoylation (K851R) attenuates its cell surface accumulation and Hh pathway activity; and 4) the defects associated with the K851R mutation is rescued by fusion of a SUMO moiety to Smo. Interestingly, we find that Hh stimulates Smo sumoylation independent of its phosphorylation by PKA/CK1 because the sumoylation of both the phosphorylation-deficient (SmoSA123) and phosphorylation-mimetic (SmoSD123) forms was regulated by Hh in the same way as the wild type Smo (Fig. 5). On the other hand, blocking sumoylation of Smo does not appear to inhibit its phosphorylation by PKA/CK1 (Fig. S5). Furthermore, blocking sumoylation in the context of SmoSD123 compromised its activity whereas engineered SUMO conjugation to SmoSD123 enhanced its activity (Fig. 4E–F′; Fig. 5C–F′). Thus, our results suggest that sumoylation and phosphorylation are two parallel and independent events that both contribute to Smo activation.
Inhibition of Smo sumoylation either by inactivating the SUMO enzymes Ubc9 and dPIAS or by mutating the SUMO target site (K851R) on Smo blocks Smo cell surface accumulation (Fig. 2 and Fig. S3), suggesting that sumoylation may regulate Smo activity by promoting its cell surface expression. Consistent with this notion, expression of SmoK851R at high levels can fully rescue smo mutant phenotypes (Fig. S4). However, we cannot rule out the possibility that sumoylation may also regulate Smo conformation and/or its interaction with other components of the Hh pathway. It is also likely that the SUMO pathway regulates Hh signaling by targeting multiple components of the pathway. Indeed, we find that Hh also stimulates sumoylation of Ci and that blocking Ci sumoylation (Ci4KR) attenuated its activation by Hh in cultured cells (Fig. S1). However, blocking Ci sumoylation did not significantly alter its activity in wing discs (Fig. S1). Similarly, when knocked into the endogenous locus, a SUMO-deficient form of Gli2 exhibited Hh pathway activity similar to that of wild type Gli2 in neural tubes (Han et al., 2012).
The Drosophila de-sumoylation enzyme Ulp1 interacts with Smo C-tail in a manner inhibited by Hh (Fig. 5G), which may explain why sumoylation of Smo is elevated in the presence of Hh. The regulation of Smo/Ulp1 interaction by Hh is independent of Smo phosphorylation by PKA/CK1(Fig. 5H) but depends on the Fu kinase activity (Fig. S6D). Previous study revealed that Fu and Costal2 (Cos2) form a complex with Smo even in the absence of Hh (Jia et al., 2003; Lum et al., 2003; Ruel et al., 2003). We speculate that Hh may induce a conformational change of Smo transmembrane helixes, which allows Fu to phosphorylate Ulp1, Smo C-tail, or other proteins, leading to the dissociation of Ulp1 from Smo. Further study will determine the precise mechanism by which Hh inhibits Smo/Ulp1 interaction.
Sumoylation, ubiquitination, and receptor trafficking
Although most of the SUMO substrates are nuclear proteins, several membrane proteins in the nervous system have been shown to be regulated by sumoylation (Geiss-Friedlander and Melchior, 2007). For example, kainate stimulates sumoylation of GluR6 to facilitate receptor endocytosis although the underlying molecular mechanism remains unknown (Martin et al., 2007). Our finding that sumoylation regulates Smo trafficking and cell surface expression suggests that sumoylation may play a more general role in the regulation of receptor trafficking. In addition, our study provides insight into how sumoylation regulates cell surface expression of membrane proteins. We find that Smo sumoylation increases its association with UBPY (Fig. 6), a de-ubiquitination (dub) that deubiquitinates Smo (Li et al., 2012; Xia et al., 2012). Because ubiquitination of Smo results in its internalization and degradation (Li et al., 2012; Xia et al., 2012), antagonizing ubiquitination by sumoylation-mediated recruitment of UBPY should promote Smo cell surface expression (Fig. 6H).
The mechanism by which sumoylation antagonizes ubiquitination we uncover here is distinct from the mechanisms described in other systems which often involve the competition of the same Lys residues for sumoylation and ubiquitination (Desterro et al., 1998; Wilkinson and Henley, 2010). Because UBPY regulates ubiquitination of many other proteins including the Wnt receptor Frizzled (Fz) (Mukai et al., 2010), we speculate that the antagonistic mechanism uncovered here may be employed elsewhere.
Of note, Smo sumoylation may occur at sub-stoichiometric levels as we were unable to detect SUMO-conjugated Smo by direct western blot analysis, raising the question of how a small fraction of sumoylated Smo can lead to significant stabilization of Smo. We offer the following explanation. First, because Smo forms dimer/oligomers under unstimulated condition and form high-order multimers in response to Hh stimulation (Shi et al., 2013; Zhao et al., 2007), we think that even a small fraction of Smo was sumoylated, UBPY recruited by sumoylated protomers could influence the ubiquitination of unsumoylated protomers within the same Smo multimeric complexes and thus will have significant impact on Smo stability at the population level. In other words, Smo multimerization may amplify the effect of sumoylation. Second, our study suggests that sumoylation facilitates the recruit of UBPY to Smo C-tail. Even after de-sumoylation, UBPY already bound to a Smo C-tail could be “trapped” there by SUMO-independent interaction with multiple Smo C-tails within a Smo multimer, thus exerting a long-lasting effect on Smo ubiquitination.
Sumoylation and Smo ciliary localization
Drosophila and Vertebrate Hh pathways diverge significantly with the vertebrate Hh pathway relies on primary cilia whereas Drosophila Hh signal transduction occurs in cells without cilia (Huangfu and Anderson, 2006). However, our recent findings suggest mSmo and Drosophila Smo (dSmo) are regulated by analogous mechanisms including phosphorylation-regulated changes in Smo conformation and subcellular localization (Chen et al., 2011). Here we extend this similarity by showing that Hh stimulates sumoylation of both dSmo and mSmo, and that sumoylation regulates the trafficking of both dSmo and mSmo with sumoylation promotes cell surface expression of dSmo and ciliary accumulation of mSmo. Unlike dSmo that contains a sumoylation consensus site, we did not identify a similar site on mSmo, suggesting that non-consensus sites might be involved in SUMO conjugation of mSmo as occurs in other proteins (Johnson, 2004). Nevertheless, our findings that engineered SUMO conjugation to mSmo (mSmo-SUMO) facilitates its ciliary localization and that SENP1 inhibits ciliary accumulation of wild type mSmo but not mSmo-SUMO strongly argue that sumoylation of mSmo promotes its ciliary localization. A recent study suggests that sumoylation of adenylyl cyclase 3 (AC3) regulates its ciliary localization, suggesting that the SUMO pathway may play a broader role in the regulation of ciliary trafficking of transmembrane proteins. Therefore, it would be interesting to explore the precise mechanism by which sumoylation regulates mSmo ciliary accumulation and whether ubiquitination of mSmo is also involved. Because aberrant activation of Smo has been implicated in many types of human cancer, our finding that sumoylation regulates Smo trafficking and Hh signaling raises an interesting possibility that inhibition of Smo sumoylation may provide an avenue for cancer drug development.
Materials and Methods
Fly stocks and Transgenes
smo3 is a null allele of smo (Chen and Struhl, 1998). dPIAS mutants: Su(var)2-1003697 and Su(var)2-102 are embryonic and larval lethal mutations of dPIAS, respectively (Flybase). Su(var)2-10 transheterozygous third-instar larvae contained melanotic tumors (Hari et al., 2001). Transgenic flies used are: UAS-Ubc9-RNAi (VDRC# 33685, VDRC #104007), UAS-dPIAS-RNAi (VDRC# 30709), UAS-Ulp1-RNAi (VDRC# 31744; VDRC #106625), UAS-Ubc9 (BL# 9324), UAS-Ubc9DN (BL# 9318), UAS-dPIAS (3XHA tag) and UAS-Ulp1 (3XHA tag) were generated by inserting the coding sequence for dPIAS or Ulp1 in a pUAST containing 3 copies of HA tags (Tong and Jiang, 2007). Transgenic flies carrying UAS-dPIAS (3XHA tag) were generated by P element-mediated transformation. The phiC31 integration system was used to generate Smo transgenes into the 75B1 attP locus (Bischof et al., 2007; Jia et al., 2009). For making UAS-Myc-SmoWT, UAS-Myc-SmoK851R, UAS-Myc-SmoK851R-SUMO, UAS-Myc-SmoSD123, UAS-Myc-SmoSDK851R, UAS-Myc-SmoSD123-SUMO, the corresponding coding sequences were subcloned between NotI and XhoI sites in a modified pUAST vector with an attB sequence inserted upstream and six tandem Myc tags inserted downstream of the UAS sites (Jia et al., 2009; Tong and Jiang, 2007). Smo deletion constructs including SmoΔCT, SmoCT, Smo556-730 and Smo730-1035 were previously described (Jia et al., 2003). HA-Fu and HA-FuKR (K33 to R substitution) expression constructs were generated in the pUAST vector and RNAi-insensitive Fu constructs were generated by changing the DNA sequence using chemically synthesized DNA fragment with alternative codon usage for each amino acid in the targeted region (aa 324–434). Fg-SENP1 and Fg-SENP1M were described (Cheng et al., 2007). A myristoylation signal from Src was inserted at the N-terminus to generated Myr-Fg-SNEP1 and Myr-Fg-SNEP1M. To construct UAS-Fg-UBPY and UAS-Fg-UBPY-SIM, the corresponding coding sequences were subcloned between NotI and KpnI sites in a modified pUAST vector with an attB sequence inserted upstream and one copy of Flag tag inserted downstream of the UAS sites. smo mutant clones with or without smo transgene expression were generated by the MARCM (mosaic analysis with a repressible cell marker) system as previously described (Lee and Luo, 2001) using the following genotypes: yw hs-FLP; tubGal80 FRT40/smo3 FRT40; C765 or tub-Gal4/UAS-Myc-Smo, UAS-Myc-SmoK851R, UAS-Myc-SmoK851R-SUMO, UAS-Myc-SmoSD, or UAS-Myc-SmoSD K851R. Immunostaining of wing discs was carried out as previously described (Jiang and Struhl, 1995).
Cell Culture, transfection, immunostaining immunoprecipitation, and western blot analysis
S2 cells were cultured in Schneider’s Drosophila Medium (Life Technologies) with 10% fetal bovine serum (GE Healthcare), penicillin (100 U/ml; Life Technologies), and streptomycin (100 mg/ml; Life Technologies) at 24°C. Cl8 cells were c ultured in Shields and Sang M3 Insect Medium (Sigma) with 2.5% fetal bovine serum, 2.5% fly extract, insulin (0.125 IU/ml; 0.5 mg/ml) (Sigma), penicillin (100 U/ml), and streptomycin (100 mg/ml) at 24°C. Transfection of S2 cells was performed using Calcium Phosphate Transfection Kit (Specialty Media) following the instructions of the manufacturer. Hh-conditioned medium treatment, immunostaining, immunoprecipitation and western blot analysis were carried out as previously described (Jia et al., 2004; Jiang and Struhl, 1995; Zhang et al., 2005). NIH 3T3 cells and Sufu−/− MEFs (kindly provided by Dr. Chi-Chuang Hui) were cultured in DMEM (Sigma-Aldrich) containing 10% BCS (ATCC), and transfected using GenJet Plus in vitro DNA transfection kit (SignaGen). HEK293T cells were transfected with lentiviral plasmid FUXW containing either Myc-mSmo or Myc-mSmo-SUMO together with packaging vectors psPAX2 and pMD2.G. After 48 hours, the medium was collected, filtered and added to pre-seeded NIH 3T3 cells. The resultant lentiviral transduced cell lines were frozen for future analysis. Lentiviral transduced NIH3T3 cells were transiently transfected with Myr-Fg-SENP1 or Myr-Fg-SENP1M. 6 to 8 hour post transfection, a recombinant mouse Shh N-terminal fragment (R&D systems Cat #464-SH) was added at the final concentrate 1μg/ml for overnight. Cells were then fixed and stained with mouse Anti-acetylated tubulin antibody (Sigma Aldrich #T7451), rabbit anti-Myc antibody (Abcam Ab9106) and goat anti-Flag antibody (Abcam ab1257). Other antibodies used in this study are: rabbit and mouse anti-Flag (Sigma), rabbit and mouse anti-Myc (Santa Cruz), mouse anti-HA (Santa Cruz), mouse anti-SmoN (DSHB), rat anti-Ci (2A1; DSHB), mouse anti-Ptc (DSHB), mouse anti-En (DSHB), rabbit anti-GFP (Life Technologies), rabbit anti-pSmo (Fan et al., 2012).
Ubiquitination and sumoylation assays
Ubiquitination assays were carried out based on the protocol described previously (Li et al., 2012; Zhang et al., 2006). For SUMOylation assay, S2 or NIH3T3 cells were transiently transfected with Smo variants plus HA-SUMO or Fg-SUMO for 48 hours and treated with 50 μM MG132 (Calbiochem) for 4 hours before harvesting the cells. Cells were then lysed for 1 hour in the lysis buffer containing 50 mM Tri-HCl pH 7.5, 150mM NaCl, 0.5% sodium deoxycholate, 3mM sodium orthovanadate, 100mM NaF, 0.5 mM EDTA, 1 mM DTT, without or with 20 mM N-ethylmaleimide (NEM) to inhibit desumoylation. SDS was then added to the lysates to a final concentration of 1% and the lysates were incubated for 5 min at 100°C. After 10-fold dilution with cell lysis buffer, the lysates were subjected to immunoprecipitation and western blot analysis.
RNAi and dual-luciferase assay
The dsRNAs were generated using the MEGAscript High Yield Transcription Kit (Ambion). The following primers were used for generating the dsRNA targeting Ubc9: 5′-GAATTAATACGACTCACTATAGGGAGACGCGAATTCGACCCGCCAAGAACCCTGAC-3′ and 5′-GAATTAATACGACTCACTATAGGGAGAGCGTCTAGACGCACGCGCTTCTCGTACTC-3′; dPIAS: 5′-GAATTAATACGACTCACTATAGGGAGACGCGAATTCTGGTGCAGATGCTTCGAG TGGT-3′ and 5′-GAATTAATACGACTCACTATAGGGAGAGCGTCTAGATGGCTGTTGATGCT GTGTGTGTG-3′. The dsRNA targeting Smo 5′ UTR has been previously described (Li et al., 2014). Fu dsRNA targets the coding sequence between amino acid (aa) 324–434. The dsRNA targeting the coding sequence of GFP was used as a negative control. Dual-luciferase activity was measured with Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer’s instructions. Briefly, Cl8 cells were treated with corresponding dsRNA in serum-free culture medium for 8 hour at 24°C, then fetal bovine serum was added to a final concentration of 2.5%. Cells were cultured for 16 hours before transfected with the constructs of ptc-firefly luciferase and Pol III-renilla luciferase. For the cells needed for Hh treatment, two third of the medium was replaced with Hh-conditioned medium, and cells were incubated for 24 hours before harvest. Each sample was measured in triplicate using a FLUOstar OPTIMA plate reader (BMG Labtech).
Supplementary Material
Acknowledgments
We thank Drs. Jianhang Jia, Xunlei Kang, Edward Yeh, Pascal Therond, Steve Cohen, and Chi-Chuang Hui for providing reagents, Bloomington and VDRC stock centers for fly stocks, and DSHB for antibodies. This work was supported by grants from NIH (GM067045 and GM118063) and Welch Foundation (I-1603) to Jin Jiang (J.J.) J.J. is a Eugene McDermott Endowed Scholar in Biomedical Science at UTSW.
Footnotes
Author contributions
G.M., S.Li., Y.H., SX.L., Y.T., and B.W. performed the experiments. G.M., S.Li., Y.H. and J.J. analyzed the data, G.M. and J.J. designed the experiments. J.J. wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Apionishev S, Katanayeva NM, Marks SA, Kalderon D, Tomlinson A. Drosophila Smoothened phosphorylation sites essential for Hedgehog signal transduction. Nat Cell Biol. 2005;7:86–92. doi: 10.1038/ncb1210. [DOI] [PubMed] [Google Scholar]
- Bischof J, Maeda RK, Hediger M, Karch F, Basler K. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc Natl Acad Sci U S A. 2007;104:3312–3317. doi: 10.1073/pnas.0611511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briscoe J, Therond PP. The mechanisms of Hedgehog signalling and its roles in development and disease. Nat Rev Mol Cell Biol. 2013;14:418–431. doi: 10.1038/nrm3598. [DOI] [PubMed] [Google Scholar]
- Chen Y, Jiang J. Decoding the phosphorylation code in Hedgehog signal transduction. Cell Res. 2013;23:186–200. doi: 10.1038/cr.2013.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Li S, Tong C, Zhao Y, Wang B, Liu Y, Jia J, Jiang J. G protein-coupled receptor kinase 2 promotes high-level Hedgehog signaling by regulating the active state of Smo through kinase-dependent and kinase-independent mechanisms in Drosophila. Genes Dev. 2010;24:2054–2067. doi: 10.1101/gad.1948710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Sasai N, Ma G, Yue T, Jia J, Briscoe J, Jiang J. Sonic Hedgehog dependent phosphorylation by CK1alpha and GRK2 is required for ciliary accumulation and activation of smoothened. PLoS Biol. 2011;9:e1001083. doi: 10.1371/journal.pbio.1001083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Struhl G. In vivo evidence that Patched and Smoothened constitute distinct binding and transducing components of a Hedgehog receptor complex. Development. 1998;125:4943–4948. doi: 10.1242/dev.125.24.4943. [DOI] [PubMed] [Google Scholar]
- Cheng J, Kang X, Zhang S, Yeh ET. SUMO-specific protease 1 is essential for stabilization of HIF1alpha during hypoxia. Cell. 2007;131:584–595. doi: 10.1016/j.cell.2007.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S, Maier D, Neubueser D, Hipfner DR. Regulation of smoothened by Drosophila G-protein-coupled receptor kinases. Dev Biol. 2010;337:99–109. doi: 10.1016/j.ydbio.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbit KC, Aanstad P, Singla V, Norman AR, Stainier DY, Reiter JF. Vertebrate Smoothened functions at the primary cilium. Nature. 2005;437:1018–1021. doi: 10.1038/nature04117. [DOI] [PubMed] [Google Scholar]
- Cox B, Briscoe J, Ulloa F. SUMOylation by Pias1 regulates the activity of the Hedgehog dependent Gli transcription factors. PloS one. 2010;5:e11996. doi: 10.1371/journal.pone.0011996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denef N, Neubuser D, Perez L, Cohen SM. Hedgehog induces opposite changes in turnover and subcellular localization of patched and smoothened. Cell. 2000;102:521–531. doi: 10.1016/s0092-8674(00)00056-8. [DOI] [PubMed] [Google Scholar]
- Desterro JM, Rodriguez MS, Hay RT. SUMO-1 modification of IkappaBalpha inhibits NF-kappaB activation. Mol Cell. 1998;2:233–239. doi: 10.1016/s1097-2765(00)80133-1. [DOI] [PubMed] [Google Scholar]
- Fan J, Liu Y, Jia J. Hh-induced Smoothened conformational switch is mediated by differential phosphorylation at its C-terminal tail in a dose- and position-dependent manner. Developmental biology. 2012;366:172–184. doi: 10.1016/j.ydbio.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gareau JR, Lima CD. The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nature reviews Molecular cell biology. 2010;11:861–871. doi: 10.1038/nrm3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiss-Friedlander R, Melchior F. Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol. 2007;8:947–956. doi: 10.1038/nrm2293. [DOI] [PubMed] [Google Scholar]
- Gill G. Something about SUMO inhibits transcription. Curr Opin Genet Dev. 2005;15:536–541. doi: 10.1016/j.gde.2005.07.004. [DOI] [PubMed] [Google Scholar]
- Han L, Pan Y, Wang B. Small ubiquitin-like Modifier (SUMO) modification inhibits GLI2 protein transcriptional activity in vitro and in vivo. J Biol Chem. 2012;287:20483–20489. doi: 10.1074/jbc.M112.359299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Shi Q, Jiang J. Multisite interaction with Sufu regulates Ci/Gli activity through distinct mechanisms in Hh signal transduction. Proc Natl Acad Sci U S A. 2015;112:6383–6388. doi: 10.1073/pnas.1421628112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hari KL, Cook KR, Karpen GH. The Drosophila Su(var)2-10 locus regulates chromosome structure and function and encodes a member of the PIAS protein family. Genes Dev. 2001;15:1334–1348. doi: 10.1101/gad.877901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Ohsako S, Tanda S. The lesswright mutation activates Rel-related proteins, leading to overproduction of larval hemocytes in Drosophila melanogaster. Dev Biol. 2005;280:407–420. doi: 10.1016/j.ydbio.2005.02.006. [DOI] [PubMed] [Google Scholar]
- Huangfu D, Anderson KV. Signaling from Smo to Ci/Gli: conservation and divergence of Hedgehog pathways from Drosophila to vertebrates. Development. 2006;133:3–14. doi: 10.1242/dev.02169. [DOI] [PubMed] [Google Scholar]
- Hui CC, Angers S. Gli proteins in development and disease. Annual review of cell and developmental biology. 2011;27:513–537. doi: 10.1146/annurev-cellbio-092910-154048. [DOI] [PubMed] [Google Scholar]
- Ingham PW, McMahon AP. Hedgehog signaling in animal development: paradigms and principles. Genes Dev. 2001;15:3059–3087. doi: 10.1101/gad.938601. [DOI] [PubMed] [Google Scholar]
- Jia H, Liu Y, Xia R, Tong C, Yue T, Jiang J, Jia J. Casein kinase 2 promotes Hedgehog signaling by regulating both smoothened and Cubitus interruptus. The Journal of biological chemistry. 2010;285:37218–37226. doi: 10.1074/jbc.M110.174565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia H, Liu Y, Yan W, Jia J. PP4 and PP2A regulate Hedgehog signaling by controlling Smo and Ci phosphorylation. Development. 2009;136:307–316. doi: 10.1242/dev.030015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia J, Tong C, Jiang J. Smoothened transduces Hedgehog signal by physically interacting with Costal2/Fused complex through its C-terminal tail. Genes Dev. 2003;17:2709–2720. doi: 10.1101/gad.1136603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia J, Tong C, Wang B, Luo L, Jiang J. Hedgehog signalling activity of Smoothened requires phosphorylation by protein kinase A and casein kinase I. Nature. 2004;432:1045–1050. doi: 10.1038/nature03179. [DOI] [PubMed] [Google Scholar]
- Jiang J, Hui CC. Hedgehog signaling in development and cancer. Dev Cell. 2008;15:801–812. doi: 10.1016/j.devcel.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Struhl G. Protein kinase A and hedgehog signaling in Drosophila limb development. Cell. 1995;80:563–572. doi: 10.1016/0092-8674(95)90510-3. [DOI] [PubMed] [Google Scholar]
- Jiang K, Liu Y, Fan J, Epperly G, Gao T, Jiang J, Jia J. Hedgehog-regulated atypical PKC promotes phosphorylation and activation of Smoothened and Cubitus interruptus in Drosophila. Proc Natl Acad Sci U S A. 2014 doi: 10.1073/pnas.1417147111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson ES. Protein modification by SUMO. Annu Rev Biochem. 2004;73:355–382. doi: 10.1146/annurev.biochem.73.011303.074118. [DOI] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends Neurosci. 2001;24:251–254. doi: 10.1016/s0166-2236(00)01791-4. [DOI] [PubMed] [Google Scholar]
- Li S, Chen Y, Shi Q, Yue T, Wang B, Jiang J. Hedgehog-regulated ubiquitination controls smoothened trafficking and cell surface expression in Drosophila. PLoS biology. 2012;10:e1001239. doi: 10.1371/journal.pbio.1001239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Li S, Han Y, Tong C, Wang B, Chen Y, Jiang J. Regulation of Smoothened Phosphorylation and High-Level Hedgehog Signaling Activity by a Plasma Membrane Associated Kinase. PLoS Biol. 2016;14:e1002481. doi: 10.1371/journal.pbio.1002481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Ma G, Wang B, Jiang J. Hedgehog induces formation of PKA-Smoothened complexes to promote Smoothened phosphorylation and pathway activation. Sci Signal. 2014;7:ra62. doi: 10.1126/scisignal.2005414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Cao X, Jiang J, Jia J. Fused-Costal2 protein complex regulates Hedgehog-induced Smo phosphorylation and cell-surface accumulation. Genes Dev. 2007;21:1949–1963. doi: 10.1101/gad.1557407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lum L, Zhang C, Oh S, Mann RK, von Kessler DP, Taipale J, Weis-Garcia F, Gong R, Wang B, Beachy PA. Hedgehog signal transduction via Smoothened association with a cytoplasmic complex scaffolded by the atypical kinesin, Costal-2. Mol Cell. 2003;12:1261–1274. doi: 10.1016/s1097-2765(03)00426-x. [DOI] [PubMed] [Google Scholar]
- Lyst MJ, Stancheva I. A role for SUMO modification in transcriptional repression and activation. Biochem Soc Trans. 2007;35:1389–1392. doi: 10.1042/BST0351389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin S, Nishimune A, Mellor JR, Henley JM. SUMOylation regulates kainate-receptor-mediated synaptic transmission. Nature. 2007;447:321–325. doi: 10.1038/nature05736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukai A, Yamamoto-Hino M, Awano W, Watanabe W, Komada M, Goto S. Balanced ubiquitylation and deubiquitylation of Frizzled regulate cellular responsiveness to Wg/Wnt. EMBO J. 2010;29:2114–2125. doi: 10.1038/emboj.2010.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlmeyer JT, Kalderon D. Hedgehog stimulates maturation of Cubitus interruptus into a labile transcriptional activator. Nature. 1998;396:749–753. doi: 10.1038/25533. [DOI] [PubMed] [Google Scholar]
- Rohatgi R, Milenkovic L, Scott MP. Patched1 regulates hedgehog signaling at the primary cilium. Science. 2007;317:372–376. doi: 10.1126/science.1139740. [DOI] [PubMed] [Google Scholar]
- Ruel L, Rodriguez R, Gallet A, Lavenant-Staccini L, Therond PP. Stability and association of Smoothened, Costal2 and Fused with Cubitus interruptus are regulated by Hedgehog. Nat Cell Biol. 2003;5:907–913. doi: 10.1038/ncb1052. [DOI] [PubMed] [Google Scholar]
- Seeler JS, Dejean A. Nuclear and unclear functions of SUMO. Nat Rev Mol Cell Biol. 2003;4:690–699. doi: 10.1038/nrm1200. [DOI] [PubMed] [Google Scholar]
- Shi D, Lv X, Zhang Z, Yang X, Zhou Z, Zhang L, Zhao Y. Smoothened oligomerization/higher order clustering in lipid rafts is essential for high Hedgehog activity transduction. J Biol Chem. 2013;288:12605–12614. doi: 10.1074/jbc.M112.399477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M, Mallin DR, Simon JA, Courey AJ. Small ubiquitin-like modifier (SUMO) conjugation impedes transcriptional silencing by the polycomb group repressor Sex Comb on Midleg. J Biol Chem. 2011;286:11391–11400. doi: 10.1074/jbc.M110.214569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Durrin LK, Wilkinson TA, Krontiris TG, Chen Y. Identification of a SUMO-binding motif that recognizes SUMO-modified proteins. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:14373–14378. doi: 10.1073/pnas.0403498101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taipale J, Beachy PA. The Hedgehog and Wnt signalling pathways in cancer. Nature. 2001;411:349–354. doi: 10.1038/35077219. [DOI] [PubMed] [Google Scholar]
- Tong C, Jiang J. Using immunoprecipitation to study protein-protein interactions in the Hedgehog-signaling pathway. Methods Mol Biol. 2007;397:215–229. doi: 10.1007/978-1-59745-516-9_15. [DOI] [PubMed] [Google Scholar]
- Villavicencio EH, Walterhouse DO, Iannaccone PM. The sonic hedgehog-patched-gli pathway in human development and disease. Am J Hum Genet. 2000;67:1047–1054. doi: 10.1016/s0002-9297(07)62934-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Wang B, Jiang J. Protein kinase A antagonizes Hedgehog signaling by regulating both the activator and repressor forms of Cubitus interruptus. Genes Dev. 1999;13:2828–2837. doi: 10.1101/gad.13.21.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson KA, Henley JM. Mechanisms, regulation and consequences of protein SUMOylation. Biochem J. 2010;428:133–145. doi: 10.1042/BJ20100158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson CW, Chuang PT. Mechanism and evolution of cytosolic Hedgehog signal transduction. Development. 2010;137:2079–2094. doi: 10.1242/dev.045021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia R, Jia H, Fan J, Liu Y, Jia J. USP8 promotes smoothened signaling by preventing its ubiquitination and changing its subcellular localization. PLoS biology. 2012;10:e1001238. doi: 10.1371/journal.pbio.1001238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Williams EH, Guo Y, Lum L, Beachy PA. Extensive phosphorylation of Smoothened in Hedgehog pathway activation. Proc Natl Acad Sci U S A. 2004;101:17900–17907. doi: 10.1073/pnas.0408093101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Zhang L, Wang B, Ou CY, Chien CT, Jiang J. A hedgehog-induced BTB protein modulates hedgehog signaling by degrading Ci/Gli transcription factor. Dev Cell. 2006;10:719–729. doi: 10.1016/j.devcel.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Zhang W, Zhao Y, Tong C, Wang G, Wang B, Jia J, Jiang J. Hedgehog-regulated costal2-kinase complexes control phosphorylation and proteolytic processing of cubitus interruptus. Dev Cell. 2005;8:267–278. doi: 10.1016/j.devcel.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Tong C, Jiang J. Hedgehog regulates smoothened activity by inducing a conformational switch. Nature. 2007;450:252–258. doi: 10.1038/nature06225. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.