Abstract
Purpose
Primary open angle glaucoma (POAG) is a major cause of blindness and visual disability. Several genetic risk factors for POAG and optic nerve features have been identified. Here we measure the relative risk for glaucoma these factors contribute to participants in the Ocular Hypertension Treatment Study (OHTS).
Design
Comparative Series
Participants
1057 (65%) of the 1636 participants of the OHTS were enrolled in this genetics ancillary study.
Methods
DNA samples were available from 1057 OHTS participants. Of these, 209 developed POAG (cases) and 848 did not develop glaucoma (controls) between 1994 and 2009. The frequencies of 13 risk alleles previously associated with POAG or with optic disc features in other cohorts were compared between POAG cases and controls in the OHTS cohort using ANOVA. The two largest subgroups non-Hispanic whites (n = 752; 70.7%) and blacks (n = 249, 23.7%) were also analyzed separately. The probability of developing glaucoma over the course of the OHTS was compared between participants stratified for TMCO1 risk alleles using Kaplan-Meier and Cox proportional hazards analyses.
Main Outcome Measures
Association of POAG with known genetic factors.
Results
No association was detected between the known POAG risk alleles when the OHTS cohort was examined as a whole. However, in the subgroup of non-Hispanic whites, allele frequencies at the TMCO1 locus were statistically different between cases and controls (p = 0.00028). By 13 years, non-Hispanic white participants with TMCO1 risk alleles had a 12% higher cumulative frequency of developing glaucoma than participants with no TMCO1 risk alleles. Moreover, the Cox proportional hazard analysis demonstrated that TMCO1 alleles increase relative risk comparable to that of some previously analyzed clinical measures (i.e. intraocular pressure).
Conclusions
The size of the OHTS cohort and its composition of two large racial subgroups may limit its power to detect some glaucoma risk factors. However, TMCO1 genotype was found to increase risk for developing glaucoma among non-Hispanic whites, the largest racial subgroup in the OHTS cohort, at a magnitude similar to clinical predictors of disease that have long been associated with glaucoma.
INTRODUCTION
Primary open angle glaucoma (POAG) is a common cause of blindness and visual disability. 1–3 POAG is characterized by retinal ganglion cell death that produces a distinctive pattern of optic nerve head damage (cupping) and visual field loss. Four classic risk factors for glaucoma include increasing age, family history of disease, race, and elevated intraocular pressure (IOP).4
Heredity is important in pathogenesis of POAG, however, the genetic basis of glaucoma is complex.5 Some cases of POAG are caused primarily by mutations in a single gene, such as myocilin (MYOC),6 optineurin (OPTN),7 or TANK binding kinase 1 (TBK1).8 Mutations in these glaucoma-causing genes are inherited in an autosomal dominant pattern and have high penetrance. The vast majority of individuals with mutations in these genes develop glaucoma. Consequently, such mutations are only rarely observed in healthy individuals (i.e. those not known to have glaucoma). Mutations in MYOC cause POAG that is characterized by high IOP,9 while mutations in OPTN or TBK1 are associated with POAG that typically occurs at low IOP.7, 8 Together these genes are responsible for approximately 5% of POAG cases.5
Other cases of POAG are caused by the combined actions of many genes and environmental factors. Genome-wide association studies (GWAS) of glaucoma patients have begun to identify these genetic risk factors. The first glaucoma risk factor, CAV1/CAV2, was detected by a large GWAS of POAG patients from Iceland.10 Additional POAG risk factors (CDKN2B-AS1 and TMCO1) were detected by a GWAS that included especially severe cases of POAG.11 Other GWAS confirmed that CAV1/CAV2,12 CDKN2B–AS1,13–16 and TMCO117 are POAG risk alleles and identified more factors including ATOH7,13 SIX1/SIX6,13, 16 and GAS7.17 More recently additional factors (ABCA1,18, 19 AFAP1,18 GMDS,18 PMM2,19 FNDC3B,20, 21 TGFBR3,22 TXNRD2,23 ATXN2,23 and FOXC123) have been detected by GWAS with larger cohort sizes
GWAS of normal tension glaucoma patients have identified additional risk factors for glaucoma that occurs at IOP levels at or below population norms including SRBD1.24 Another potential risk factor, ELOVL5, nearly met genome-wide threshold for significance24 and was later studied in targeted association studies suggesting it may also be a glaucoma risk factor.25 TLR4 has also been identified as risk factor for normal tension glaucoma with some,26, 27 but not all, 28, 29 association studies focused on this gene. Further GWAS of normal tension glaucoma have detected a novel chromosome 8q22 locus15 and have also shown that a locus previously identified with studies of POAG with high IOP, CDKN2B–AS1, is also associated with normal tension glaucoma.15
The genetic basis of quantitative features or endophenotypes of glaucoma has also been investigated using GWAS. Several genes that influence the magnitude of vertical cup-to-disc ratio (CDKN2B-AS1, SIX1/SIX6, SCYL1, CHEK2, ATOH7, and DCLK1) or optic disc area (ATOH7, CDC7/TGFBR3, and SALL1) were identified in early endophenotype studies of glaucoma.30, 31 More recent GWAS have detected additional genetic factors that determine optic disc features.32 Similar studies have also identified many genetic factors associated with other endophenotypes of glaucoma including the magnitude of IOP17, 20 and central corneal thickness.21, 33–35 There is some overlap between the genes identified by GWAS of endophenotypes and GWAS of glaucoma overall.
Elevated IOP in the absence of diagnostic features of glaucoma (optic nerve damage and/or visual field loss) is classified as ocular hypertension (OHT).36 The Ocular Hypertension Treatment Study (OHTS) was a multicenter treatment trial that investigated the efficacy of medical treatment of OHT to prevent development of POAG.36 Of the 1,636 individuals with ocular hypertension that were enrolled in the OHTS, 19% developed POAG at 13 years of follow up between 1994 and 2009.37 Here we report investigating the role of 10 previously reported glaucoma risk alleles and 3 alleles associated with optic nerve features in the participants of the OHTS. We tested OHTS participants at SNPs located in a total of 13 loci. Six of these loci were discovered with genome-wide association studies of POAG with high IOP (ATOH7, CAV1/CAV2, CDKN2B-AS1, GAS7, SIX1/SIX6, and TMCO1). We also tested SNPs in five loci that have been associated with glaucoma that occurs with low IOP (CDKN2B-AS1, Chromosome 8q22, ELOVL5, SRBD1, and TLR4), one of which is also associated with glaucoma with high IOP. Finally, we evaluated SNPs at 6 loci that influence optic disc features (ATOH7, CDC7/TGFBR3, CDKN2B-AS1, CHEK1, SALL1, SIX1/SIX6), three of which are also associated with POAG. Since the onset of our study of these 13 factors, additional POAG risk factors have been detected but were not included in our analysis.
PATIENTS AND METHODS
Patient cohort
Informed consent was obtained from all participants and approval for the study was granted by the Institutional Review Boards of all participating institutions. A total of 1,636 individuals with ocular hypertension were enrolled in the Ocular Hypertension Treatment Study (OHTS) with inclusion and exclusion criteria that have been previously described.36 Briefly, participants were required to have ocular hypertension defined as IOP ≥ 24 mm Hg, but ≤ 32 mm Hg in one eye and IOP ≥ 21 mm Hg, but ≤ 32 mm Hg in the other eye at the time of enrollment after washout of any topical glaucoma medications. OHTS participants were also between 40 and 80 years of age inclusively and had normal and reliable baseline visual fields as determined by Humphrey 30–2 visual field testing and review by the OHTS Visual Field Reading center. Finally, OHTS participants were judged to have normal optic nerve heads by both clinicians and review of stereoscopic optic disc photos by the OHTS Optic Disc Reading Center. At the time of enrollment, all participants were judged to have ocular hypertension and none had been diagnosed with glaucoma. The demographics, including self-reported ethnicity, of the entire OHTS cohort have been previously described.36
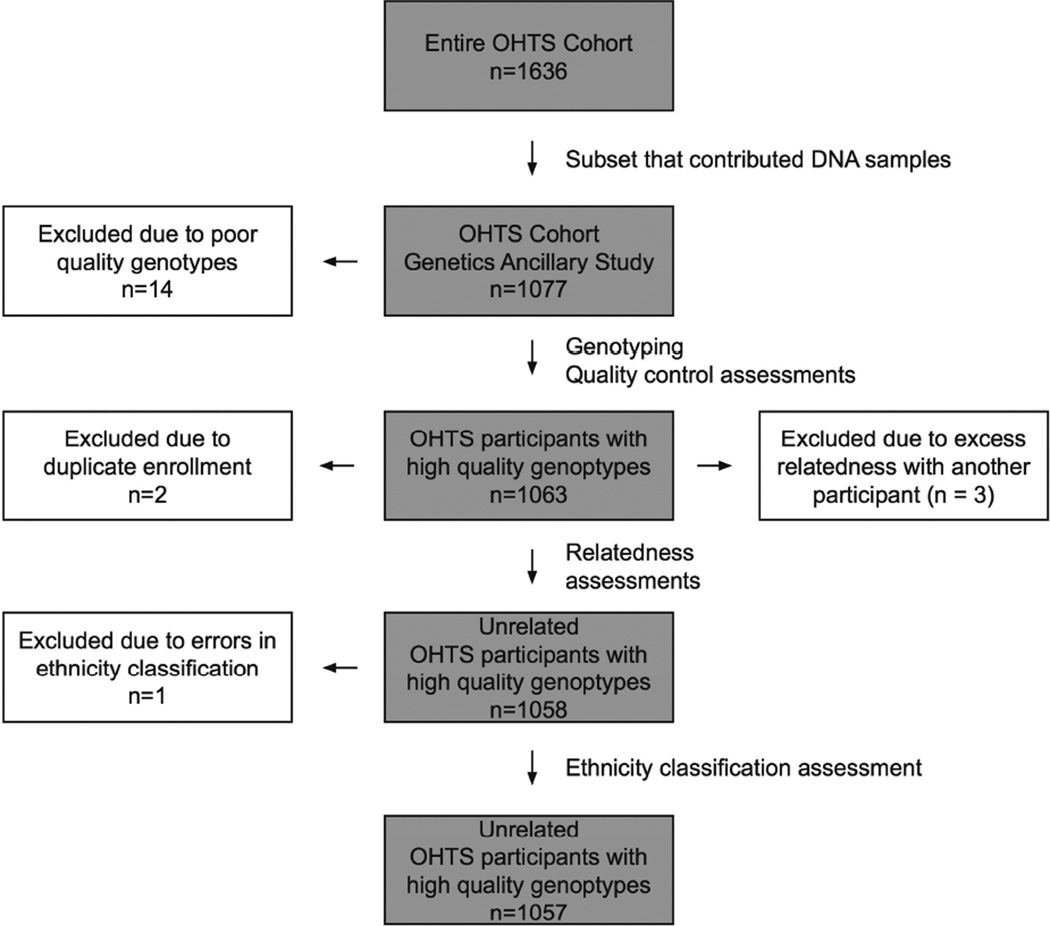
A subset (n=1,077) of the 1,636 subjects of the Ocular Hypertension Treatment Study (OHTS) were enrolled in an ancillary genetics study. Informed consent for genetic studies was obtained from all study participants. DNA was obtained from peripheral blood samples using standard methods38 and high quality DNA samples was obtained from 1,063 (98.7%) of the 1,077 participants. The analysis sample investigated in the current study is described in Figure 1.
Figure 1. Enrollment of OHTS participants in a genetics ancillary studies.
Of the cohort of 1636 OHTS participants, 1077 contributed DNA samples and were part of a genome-wide association study. The genotypes of 1057 of these participants passed quality control metrics and were analyzed in this study.
OHTS participants were examined bi-annually for the development of POAG using a well-described set of criteria for optic nerve damage and/or visual field defects.36 By 2009, 209 (19.7%) of the 1063 participants that contributed DNA samples for genetic study had met study criteria for a diagnosis of glaucoma.
Genotyping
The cohort of 1063 OHTS participants that contributed DNA samples was studied as part of a genome-wide association study (GWAS) to search for genes that determine quantitative features of glaucoma. Participants were genotyped at 1.1 million single nucleotide polymorphisms using Illumina Omni-1M-Quad microarrays (Illumina, San Diego, CA) in collaboration with the Center for Inherited Disease Research (CIDR cidr.jhmi.edu) and the National Human Genome Research Institute (NHGRI genome.gov). An additional 24 masked duplicate samples and 50 control samples from the International HapMap Project were also genotyped to aid with quality control. All DNA samples were genotyped at the same time, on the same genotyping platform, and were plated in random order. Quality control and data cleaning of the genotypic data was conducted in collaboration with CIDR. A total of six participants were excluded from this analysis due to unexpected duplicate enrollment (n=2), ethnicity classification errors (n=1), and unexpected relatedness (n=3) The genotypes generated from the remaining 1057 samples all passed quality control thresholds with overall call rates > 87.9%. Individual SNPs that produced genotypes with call rates < 99% or that produced Hardy-Weinberg disequilibrium < 10−6 were eliminated from the study. Concordance of genotype calls between duplicate samples was 99.99% for masked OHTS duplicates and 99.7% for HapMap controls. Genotypes from 905,636 of the SNPs on the Omni-1M-Quad microarrays satisfied all quality control criteria for the 1057 study participants and were released by CIDR and have been posted at dbGaP on the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov).
Power Calculations
We calculated that our sample of 209 participants who were diagnosed with POAG (cases) and 848 participants who were not diagnosed with POAG has greater than 80% power to detect a 7.2% difference in risk factor allele frequency between cases and controls when the minor allele frequency is 10% among controls. The Bonferroni method was used to correct p-values for multiple measures at an overall significance of 0.05.
Analysis of genotypes
Genotypes of 1057 OHTS participants were further analyzed using GenomeStudio (Illumina, San Diego, CA) and PLINK v1.07. Multiple dimensional scaling was used as a covariate in linear regression to control for possible population stratification and was compared with self-reported ethnicity. Age and gender were also included as covariates.
Of 20 SNPs at 13 loci previously associated with POAG, 10 SNPS were present on the Omni1-Quad microarray and genotypes were readily available for analysis. For the remaining 10 SNPs, an alternate SNP was chosen using the HapMap linkage disequilibrium data (release 27). There were two cases where a single SNP replaced a pair of SNPs in the TLR4 gene. Using these alternates resulted in examining 20 unique SNPs at 13 previously associated loci (Table 2).
Table 2.
Candidate gene association study.
| OHTS cohort | Black subgroup | Non-Hispanic white subgroup | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| POAG | No POAG | POAG | No POAG | POAG | No POAG | ||||||
| n = 209 | n = 848 | n = 70 | n = 179 | n = 124 | n = 628 | ||||||
| Locus / Gene |
Original SNP |
SNP Analyzed |
MAF |
p-value |
MAF |
p-value |
MAF |
p-value |
|||
| ATOH7 | rs1900004 | rs7916697 | 0.426 | 0.357 | 0.66 | 0.307 | 0.279 | 0.95 | 0.290 | 0.255 | 0.27 |
| CAV1/CAV2 | rs4236601 | rs2024211 | 0.318 | 0.325 | 0.94 | 0.336 | 0.344 | 0.76 | 0.315 | 0.326 | 0.77 |
| CDC7/TGFBR3 | rs1192415 | rs1192419 | 0.225 | 0.211 | 0.93 | 0.293 | 0.285 | 0.66 | 0.186 | 0.191 | 0.89 |
| rs1063192 | rs1063192 | 0.263 | 0.369 | 0.02 | 0.086 | 0.084 | 0.93 | 0.371 | 0.459 | 0.0060 | |
| CDKN2B–AS1 | rs2157719 | SNP9–22023366 | 0.263 | 0.364 | 0.03 | 0.086 | 0.078 | 0.77 | 0.371 | 0.452 | 0.012 |
| rs4977756 | rs4977756 | 0.357 | 0.400 | 0.15 | 0.407 | 0.335 | 0.18 | 0.351 | 0.427 | 0.012 | |
| CHEK2 | rs1547014 | rs1547014 | 0.361 | 0.346 | 0.34 | 0.479 | 0.489 | 0.42 | 0.298 | 0.315 | 0.42 |
| Chrom 8q22 | rs284489 | rs284489 | 0.431 | 0.396 | 0.39 | 0.393 | 0.444 | 0.24 | 0.339 | 0.348 | 0.66 |
| ELOVL5 | rs735860 | rs735860 | 0.278 | 0.355 | 0.06 | 0.093 | 0.098 | 0.69 | 0.387 | 0.431 | 0.093 |
| GAS7 | rs11656696 | rs11656696 | 0.323 | 0.348 | 0.79 | 0.236 | 0.249 | 0.69 | 0.367 | 0.377 | 0.85 |
| SALL1 | rs1362756 | rs1345467 | 0.270 | 0.278 | 0.95 | 0.314 | 0.355 | 0.76 | 0.246 | 0.256 | 0.95 |
| SIX1/SIX6 | rs10483727 | rs2057135 | 0.153 | 0.148 | 0.52 | 0.214 | 0.321 | 0.35 | 0.109 | 0.101 | 0.67 |
| SRBD1 | rs3213787 | rs17033801 | 0.022 | 0.027 | 0.90 | 0.000 | 0.008 | NA | 0.020 | 0.030 | 0.46 |
| rs12377632 | rs10759930 | 0.309 | 0.333 | 0.71 | 0.093 | 0.103 | 0.46 | 0.436 | 0.393 | 0.28 | |
| rs2149356 | rs1360094 | 0.452 | 0.423 | 0.41 | 0.257 | 0.254 | 0.68 | 0.290 | 0.336 | 0.20 | |
| TLR4 | rs10759930 | rs10759930 | 0.309 | 0.333 | 0.71 | 0.093 | 0.103 | 0.46 | 0.436 | 0.393 | 0.28 |
| rs1927914 | rs1360094 | 0.452 | 0.423 | 0.41 | 0.257 | 0.254 | 0.68 | 0.290 | 0.336 | 0.20 | |
| rs1927911 | rs1927911 | 0.373 | 0.338 | 0.76 | 0.357 | 0.390 | 0.31 | 0.226 | 0.267 | 0.25 | |
| rs7037117 | rs1927906 | 0.220 | 0.160 | 0.41 | 0.414 | 0.430 | 0.45 | 0.093 | 0.084 | 0.11 | |
| TMCO1 | rs4656461 | rs4656461 | 0.234 | 0.178 | 0.03 | 0.221 | 0.254 | 0.17 | 0.246 | 0.155 | 0.00028 |
The threshold for a significant association corrected for multiple measures is p = 0.0038. The alleles of the SNP rs4656461 at the TMCO1 locus are significantly associated with POAG in the non-Hispanic white subgroup.
Statistical Analysis
The allele frequencies of 20 SNPs at 13 loci were compared between OHTS participants who developed POAG (n = 209) and those who did not develop POAG (n = 848) at 13 years follow-up using a Mantel-Haenszel test to control for race. The allele frequencies of these SNPs in the two largest sub-populations (self-reported blacks n = 249 and non-Hispanic whites n = 752) were also compared separately using a chi square test. A gene-based (n = 13) Bonferroni correction for multiple measures was used to set the threshold for significance at p = 0.05 / 13 = 0.0038.
The risk for glaucoma was assessed using the Kaplan Meier survival analysis and Cox proportional hazards analysis. The proportion of OHTS participants that remained free of glaucoma at 6 month intervals was assessed for those initially randomized to treatment or observation. We calculated the Cox proportional hazard for POAG using the software package R (The R Project for Statistical Computing, http://www.r-project.org) with the same covariates as previously used to assess risk in the OHTS cohort. These covariates include: 1) age at enrollment; 2) IOP at enrollment (average between right and left eyes); 3) central corneal thickness (average between right and left eyes); 4) vertical cup-to-disc ratio at enrollment (average between right and left eyes); 5) pattern standard deviation of Humphrey visual field testing at enrollment (average between right and left eyes). We added randomization to treatment or observation at enrollment; gender; and presence of TMCO1 risk alleles to the Cox proportional hazard model.
RESULTS
Study participants
A total of 1,636 participants were enrolled in the OHTS, and DNA samples were available from a subset of this cohort (n=1077). These 1077 OHTS participants were genotyped at 1,051,295 SNPs as part of a quantitative traits study. After quality control criteria were applied, 905,636 SNPs typed in 1063 participants were released for analysis. An additional 6 participants were removed from the study due to inadvertent duplicate enrollment in the OHTS (n=2); due to excess relatedness (n=3); and due to an ethnicity misclassification (n=1). The remaining 1057 OHTS participants and their genotypes are the focus of this study (Figure 1).
The demographic information from 1057 OHTS subjects in the genetics study is shown in Table 1. The racial composition of this subset of the OHTS is 71.1% non-Hispanic white and 23.6% black and is similar to the entire OHTS cohort as previously described.58 Over the course of 13 years of follow-up examinations, 209 (19.8%) of these 1057 OHTS participants met study criteria for a diagnosis of POAG, while 848 (80.2%) were not diagnosed with glaucoma.
Table 1.
Demographics of the OHTS cohort and the subset in the genetics study.
| Overall OHTS Cohort n = 1636 |
OHTS participants in this study n = 1057 |
p-value |
|
|---|---|---|---|
| Mean age at enrollment (years) | 55.4 | 55.9 | 0.52 |
| Gender | 0.60 | ||
| Male | 705 (43.1%) | 461 (43.6%) | |
| Female | 931 (56.9%) | 596 (56.4) | |
| Self-reported race | 0.51 | ||
| White | 1138 (69.6%) | 752 (71.1%) | |
| Black | 409 (25.0%) | 249 (23.6%) | |
| Hispanic | 59 (3.61%) | 36 (3.41%) | |
| Asian/Pacific Islander | 14 (0.856%) | 9 (0.851%) | |
| Other | 14 (0.856%) | 9 (0.851%) | |
| American Indian / Alaskan | 4 (2.44%) | 2 (0.189%) | |
| Cummulative proportion that developed POAG in the OHTS |
0.91 | ||
| Overall | 279 (17.1%) | 209 (19.8%) | |
| White | 170 (14.9%) | 124 (16.5%) | |
| Black | 91 (22.2%) | 70 (28.1%) | |
| Hispanic | 12 (20.3%) | 10 (27.8%) | |
| Asian/Pacific Islander | 2 (14.3%) | 2 (22.2%) | |
| Other | 3 (21.4%) | 2 (22.2%) | |
| American Indian / Alaskan | 1 (25.0%) | 1 (50.0%) |
The distributions of gender, race, and ethnicity are closely matched between the entire OHTS cohort (first column) and the subset of the cohort available for genetic study (second column).
Analysis of previously reported genetic factors for POAG
Prior studies have reported associations between SNPs at nineteen loci and POAG (including normal tension glaucoma). Many additional loci associated with endophenotypes of glaucoma such as vertical cup-to-disc ratio have also been identified. We examined the role of SNPs at 13 loci that had been discovered at the time of our study’s onset. SNPs at 10 loci associated with POAG were investigated in our study (ATOH7,13 CAV1/CAV2,12 CDKN2B–AS1,13–16 CHR 8q22,15 ELOVL5,24 GAS7,17 SIX1/SIX6,13, 16 SRBD1,24 TLR4,26, 27 and TMCO117) and POAG. Of these 10 loci, CDKN2B–AS1, CHR 8q22, ELOVL5, SRBD1, and TLR4 have been associated with normal tension glaucoma. We also investigated six loci that have been associated with optic disc features (i.e. disc area or vertical cup-to-disk ratio) ATOH7, CDC7/TGFBR3,13, 39 CDKN2B–AS1, CHEK2,40 SALL1,13 and SIX1/SIX6 (three of which are also associated with POAG). Genotype frequencies at each of these 13 loci were compared between 209 OHTS participants that were diagnosed with POAG and 848 OHTS participants who were not diagnosed with glaucoma using linear regression controlling for population stratification, gender, and age as covariates. When the entire OHTS cohort (n=1057) was analyzed (Table 2), no SNPs at these 13 loci were associated with POAG with a threshold for significance corrected for multiple measures (p < 0.0038).
The OHTS cohort includes participants from several racial and ethnic groups (Table 1), however, the two largest subgroups are self-reported Non-Hispanic whites (n=752) and blacks (n=249). When these groups were analyzed separately (Table 2), one locus (TMCO1) contained a SNP (rs4656461) with allele frequencies that were significantly associated with POAG among the Non-Hispanic white subset of participants. The minor allele frequency of rs4656461 was 24.6% in non-Hispanic whites that developed POAG and 15.5% in those that did not develop POAG (p = 0.00028). Numerous clinical studies have established that age, IOP, and CCT are important risk factors for POAG. However, TMCO1 genotypes were not associated with age, IOP, or CCT in the OHTS cohort (p > 0.05), suggesting that TMCO1 is an independent risk factor for POAG. Moreover, the association between TMCO1 and POAG in non-Hispanic white participants remained significant (p = 0.000072) when the data were reanalyzed using age, gender, IOP, and CCT as covariates, further indicating that TMCO1 is independently associated with POAG in the OHTS cohort.
Assessment of risk for POAG conferred by TMCO1 alleles at the rs4656461 locus
At the onset of the OHTS, all participants were determined to be free of glaucoma damage. Over the course of the first 13 years of follow-up, 209 of 1057 participants who contributed DNA samples developed incident cases of POAG. At 13 years, 59 (29.6%) of the 199 OHTS participants with TMCO1 risk alleles still in the study had glaucoma, while 69 (19.1%) of the remaining 362 participants without TMCO1 risk alleles had glaucoma. Overall, the proportion of participants with glaucoma at 13 years was 10.5% greater in OHTS participants with TMCO1 risk alleles than in those with no risk alleles.
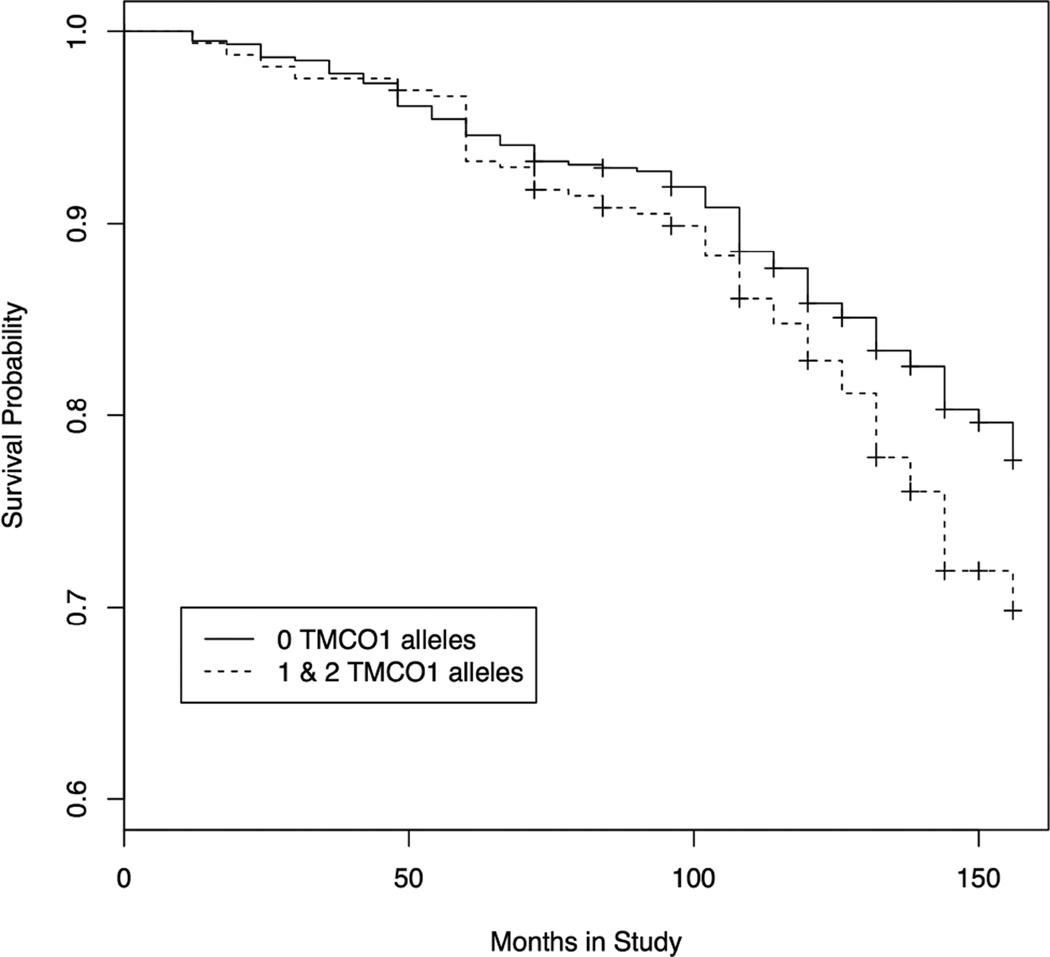
The rate at which POAG was diagnosed was compared between OHTS participants with TMCO1 risk alleles (either one or both alleles) and participants with no risk alleles using a modified Kaplan-Meier analysis (Figure 2A). Carriers of one or two TMCO1 risk alleles were grouped together, because of the relative rarity of participants with two risk alleles. Overall, participants with TMCO1 risk alleles developed glaucoma at a higher rate than participants with no risk alleles (p = 0.0128). By 13 years, OHTS participants with TMCO1 risk alleles have a 30.2% probability of developing glaucoma compared with a probability of 22.3% for those with no TMCO1 risk alleles (Figure 2A). Thus, TMCO1 risk alleles are associated with a 7.9% higher probability for developing glaucoma at 13 years.
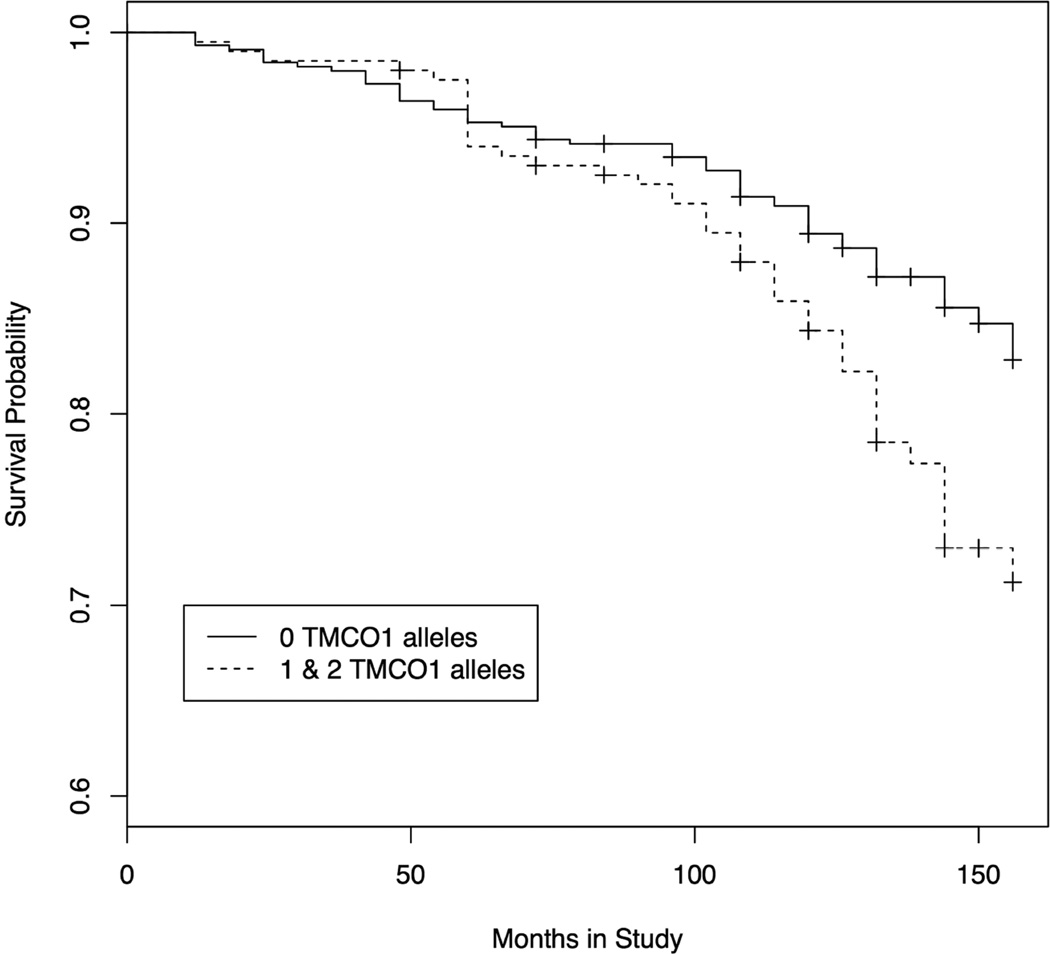
Figure 2. Kaplan-Meier analysis of OHTS participants with TMCO1 glaucoma risk factors.
At the onset of the study, none of the OHTS participants had a diagnosis of glaucoma. At six-month intervals participants were evaluated for onset of glaucoma. Survival probability, which refers to the probability of remaining free of glaucoma, is plotted on the Y-axis. Thus, the probability of developing glaucoma is equal to 1 – the probability of survival. Survival probability for participants with no TMCO1 risk alleles is plotted with a solid line, while survival probability for participants with either 1 or 2 TMCO1 risk alleles is plotted with a dashed line. A. Data plotted from all participants in the genetics ancillary study of the OHTS (n = 1057) B. Data plotted from the non-Hispanic white subset of the ancillary study (n = 752).
Reduction of IOP is known to influence the risk for developing glaucoma. Consequently, we compared TMCO1 risk allele frequencies between the subset of the OHTS participants that was randomized to initially receive IOP lowering medications and the subset that was randomized to initially receive placebo (Table 3). TMCO1 risk allele frequencies are the same in treated and untreated arms of the OHTS (p = 0.35). Moreover, the same trend of increased rates of glaucoma among carriers of TMCO1 risk alleles was observed when either initially treated or untreated subsets of the OHTS participants were investigated separately with Kaplan-Meier analysis (Supplementary Figure 1A and 1B).
Table 3.
Distribution of TMCO1 risk alleles between subsets of the OHTS cohort randomized to initial observation or to initial treatment.
| Number ofTMCO1 risk alleles (rs4656461) |
Whole cohort | Non-Hispanic whites | Blacks | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Total | Initially treated |
Initially observed |
Total |
Initially treated |
Initially observed |
Total |
Initially treated |
Initially observed |
|
| G alleles |
n=1056 |
n=546 |
n=510 |
n=751 |
n=395 |
n=346 |
n=249 |
n=126 |
n=123 |
| 0 | 690 | 367 | 323 | 521 | 273 | 248 | 134 | 78 | 56 |
| 1 | 332 | 164 | 168 | 205 | 108 | 97 | 108 | 47 | 61 |
| 2 | 34 | 15 | 19 | 25 | 14 | 11 | 7 | 1 | 6 |
| p = 0.35 | p = 0.94 | p = 0.011 | |||||||
TMCO1 risk alleles are present at the same frequency among OHTS participants randomized to initial treatment or to initial observation when the whole cohort is analyzed (left columns). The TMCO1 risk alleles are also present at the same frequency in white participants randomized to initial treatment or initial observation (middle columns), while their was a difference between groups among the black participants (right columns) which is likely due to the smaller sample size.
The two largest racial subsets of the OHTS participants (non-Hispanic whites and blacks) were also studied separately with Kaplan-Meier analysis. Non-Hispanic whites that carried one or two TMCO1 risk alleles developed glaucoma at a statistically higher rate than those with no risk alleles (Figure 2B, p = 0.0014). These data show that after 13 years, non-Hispanic whites with high-risk TMCO1 alleles have a 28.8% probability of having glaucoma compared with a probability of 17.1% for those with no TMCO1 risk alleles (Figure 2B). These data suggest that TMCO1 risk alleles are responsible for an 11.7% higher probability for developing glaucoma at 13 years. Further, Kaplan-Meier analysis shows that TMCO1 risk alleles are associated with a higher risk for developing glaucoma in both the initially treated (p = 0.00909) or untreated (p = 0.0304) subgroups of Non-Hispanic whites (Supplemental Figures 1A and 1B). No statistically significant difference in the rate of glaucoma was detected between the smaller cohorts of black OHTS participants with TMCO1 risk alleles and black participants with no risk alleles.
Calculation of Risk
We analyzed the longitudinal OHTS data to quantify the POAG risk from TMCO1 alleles using the Cox proportional hazards model. The original risk calculator for POAG in the OHTS cohort included 1) age, 2) IOP, 3) central corneal thickness, 4) vertical cup-to-disc ratio, and 5) pattern standard deviation of Humphrey visual field tests. We added initial randomization to treatment or observation, gender and TMCO1 genotype to our POAG risk calculator. We focused our analysis on the non-Hispanic white subset of the OHTS participants (n = 752) for which TMCO1 alleles are significantly associated with POAG. In this model, TMCO1 genotype is highly associated with risk for POAG (Table 4). TMCO1 risk alleles have a hazards ratio of 1.73 per allele (p-value = 0.00036) and have an influence on risk for glaucoma that is on par with other clinical and demographic features of glaucoma (Table 4). A similar analysis with the black subset of the OHTS cohort did not produce hazards ratios that were statistically significant (p=0.085).
Table 4.
Cox Proportional Hazards Analysis.
| Hazards Ratio |
95% confidence interval |
p-value |
|
|---|---|---|---|
| Age (decade) | 1.40 | 1.14 – 1.71 | 0.0011 |
| Gender (male) | 1.53 | 1.05 – 2.23 | 0.025 |
| IOP (per 2.7 mm Hg [SE]) | 1.28 | 1.03 – 1.46 | 0.021 |
| CCT (per 37 microns [SE]) | 1.57 | 1.30 – 1.89 | 0.0000028 |
| Vertical cup-to-disc ratio (per 0.19 [SE]) | 1.62 | 1.33 – 1.96 | 0.00000096 |
| Visual Field defect (PSD, per 0.25 [SE]) | 1.20 | 0.99 – 1.45 | 0.070 |
| TMCO1 (per risk allele) | 1.73 | 1.28 – 2.34 | 0.00036 |
The OHTS risk calculator was adapted to include number of TMCO1 risk alleles. Hazards ratios were normalized by standard error (SE).
DISCUSSION
Observational and epidemiological studies have provided strong evidence for genetic contributions to both the overall risk for a clinical diagnosis of glaucoma as well as many of the component quantitative features of disease, such as corneal thickness, IOP, and optic disc morphology.41–45 The first successes in genetic studies of glaucoma were achieved with investigations of Mendelian (single-gene) forms of POAG. Linkage studies of rare pedigrees identified mutations in genes (e.g. MYOC, OPTN, and TBK1) that are each capable of causing glaucoma with little influence from other genes and the environment.5 Risk factors have also been discovered for forms of glaucoma that have a complex basis involving many genes that contribute to pathogenesis but are not causative on their own. For example, the LOXL1 locus has been identified as a powerful risk factor for exfoliation syndrome (odds ratio >20) and exfoliation glaucoma.46 Several POAG risk factors have also been discovered including CAV1/CAV2,10 CDKN2B–AS1,11, 13, 15, 47 TMCO1,11 ATOH7,13 SIX1/SIX6,13 GAS7,17 TLR4,26 SRBD1,25 E LOVL5,25 chromosome 8q22,15 ABCA1,18 AFAP1,18 GMDS,18 P MM2,19 FNDC3B,20, 21 TGFBR3,22 TXNRD2,23 ATXN2,23 and FOXC1.23 Each of these POAG risk factors was discovered with case control association studies that provide estimates of their influence on the development of glaucoma (i.e. odds ratios). These factors each confer relatively modest risk for POAG, which suggests that numerous risk factors (genetic and environmental) must be present for disease to occur and that many more factors remain to be discovered.
The known glaucoma risk factors have been studied and further validated in patient cohorts from prior epidemiological studies and treatment trials. Members of the Nurses Health Study (NHS) and the Health Professionals Follow-up Study (HPFS) were screened for glaucoma and included in multicenter POAG genetic studies (GLAUGEN and NEIGHBOR). Associations between CDKN2B-AS1 and SIX1/SIX6 were validated with these cohorts and a new chromosome 8q22 risk factor locus was discovered.15 The Blue Mountain Eye Study (BMES) examined a cohort of 3,654 predominantly white residents of a suburb of Sydney to determine the prevalence of glaucoma in this population (2.4% definite glaucoma).48 Later DNA samples from BMES participants were used in the initial GWAS study that discovered an association between risk factors TMCO1 and CDKN2B–AS1 and POAG.11 Furthermore a subset of the BMES participants were followed over 10 years for the development of incident cases of POAG. In a recent report, a focused association study of BMES participants confirmed the association between several previously discovered risk factors and incident cases of glaucoma in the BMES cohort. SNPs in CDKN2B–AS1 (rs141829), and SIX1/SIX6 (rs10483727) were each associated with increased risk for developing glaucoma with calculated odds ratios (univariate) of 1.67 and 1.66 respectively.49 A marginal association between TMCO1 and incident glaucoma was also detected with an odds ratio of 1.74 (univariate) but did not survive multiple measures corrections.49
Our analysis of the OHTS cohort identified a significant association between TMCO1 risk alleles and incident glaucoma cases among white participants. It is likely that the association was detected in whites and not in black participants primarily due to larger sample size (Table 1). There is 9.1% difference in TMCO1 minor allele frequency of SNP rs4656461 between non-Hispanic whites with POAG and non-Hispanic whites with no POAG in the OHTS. If the same difference in minor allele frequency were observed in the smaller cohort of black OHTS participants, there would only be 36% power to detect a statistically significant difference at this locus. This low power suggests that small sample size might be one reason that an association was not detected between TMCO1 and glaucoma in the black subset of the OHTS participants. It is also notable that the minor allele at the TMCO1 locus (rs4656461) that is associated with glaucoma in non-Hispanic whites is more common among blacks without glaucoma (controls) than among blacks with glaucoma (Table 2). These data suggest that if there were an association between TMC01 and glaucoma in blacks that could be detectable with a larger cohort, the association would likely be in the opposite direction (i.e. with the major allele at rs4656461). Moreover, some of the other SNPs in our study, such as rs1063192 and SNP9–22023366 at the CDKN2B-AS1 locus, have much lower minor allele frequencies in black participants than in non-Hispanic white participants which may have also challenged our ability to detect statistically significant differences. It is interesting to note, that these same CDKN2B-AS1 SNP alleles that are less common in blacks are “protective” and reduce risk for glaucoma and could be a source of the increased risk for glaucoma among blacks. Finally, other studies of African blacks have suggested that known glaucoma risk alleles may not be as strong in these populations and that a different set of risk factors may be more important.50 Our analysis of black OHTS participants is consistent with this conclusion.
Our failure to detect significant associations with additional known glaucoma risk factors is most likely due to the relatively small size of the OHTS cohort. For example, nominal associations were detected at the CDKN2B-AS1 locus (p = 0.0060) that didn’t survive multiple measures corrections for 13 loci. It is possible that an association between CDKN2B–AS1 or other factors might have been detectable if the OHTS cohort were larger or if less stringent multiple measures corrections were employed.
Although TMCO1 was first associated with POAG,11 it was later identified as a gene that regulates the magnitude of IOP. 17, 51 Consequently, we considered that the association between TMCO1 and glaucoma might be influenced by IOP. However, TMCO1 remains associated with POAG in the OHTS cohort even when baseline IOP is controlled using linear regression. These results suggest that TMCO1 may contribute risk for glaucoma that is independent of its influence on IOP. Since both the POAG cases and controls in the OHTS cohort were required to have high IOP through enrollment criteria, our analyses may be poorly suited to detecting IOP-dependent risk for POAG.
Since this analysis of 13 loci was begun, several additional SNPs associated with POAG have been identified with studies of larger patient cohorts (ABAC1, AFAP1, GMDS, PMM2, FNDC3B, TGFBR3, TXNRD2, ATXN2, and FOXC1). Future studies of these additional genes with the OHTS cohort may be warranted.
The primary goal of the OHTS randomized trial was to determine the safety and efficacy of medical reduction of IOP reduction on the risk for developing glaucoma. Rigorous longitudinal assessments for incident cases of glaucoma were conducted at 6-month intervals for over 13 years and demonstrated that IOP reduction lowers risk for progression of ocular hypertension to POAG. We were able to use the same glaucoma incidence dataset to determine the influence of TMCO1 risk alleles on glaucoma incidence. Analysis of non-Hispanic white OHTS participants demonstrated that those with one or two TMCO1 risk alleles developed glaucoma at a significantly higher rate than those with no risk alleles (p = 0.0014, Figure 2B). After 13 years, 12% more of the non-Hispanic whites with TMCO1 risk alleles developed POAG than non-Hispanic whites without risk alleles. Half of the OHTS participants were randomized to initial observation and half to IOP reduction therapy. TMCO1 alleles influence risk for glaucoma when initially-observed or initially-treated groups are analyzed separately (Supplementary Figure 1A and 1B).
The OHTS cohort TMCO1 genotype data was also examined with Cox proportional hazards models similar to those used in prior studies.52 In addition to the clinical predictors used in the original model, our calculator included three more factors: 1) number of TMCO1 risk alleles, 2) gender, and 3) initial randomization (observation or treatment). The number of TMCO1 risk alleles is highly associated with development of glaucoma (p = 0.00036). In our model each TMCO1 risk allele has a hazard ratio of 1.73 and confers risk for glaucoma greater than the risk associated with a 0.1 larger baseline cup-to-disc ratio or with a 40 micron thinner baseline corneal thickness (Table 4).
Genetic analysis of the OHTS cohort provides evidence that TMCO1 genotype is a strong predictor for the development of glaucoma. This analysis of TMCO1 is based on individuals with ocular hypertension and may not be generalizable to all types of glaucoma suspects, however, its conclusions are supported by Burdon and coworkers recent study of incident cases of POAG in the BMES.49 Both Kaplan-Meier analyses and Cox proportion hazards models suggest that determining TMCO1 genotype might have clinical utility in assessing risk for glaucoma. The potential value of including TMCO1 genotypes in a glaucoma risk calculator is promising, but requires additional validation.
Supplementary Material
Genetic analysis of the Ocular Hypertension Treatment Study (OHTS) detects increased probability of developing primary open angle glaucoma (POAG) among participants with transmembrane and coiled-coil domains 1 (TMCO1) risk alleles.
Acknowledgments
This research was supported in part by NIH grants R01 EY018825 (Fingert), X01 HG005248 (Fingert), R01 EY023512 (Fingert), U10 EY009307 (Kass), UG1 EY025180 (Kass), U10 EY009341 (Gordon), UG1 EY025182 (Gordon).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Sommer A, Tielsch JM, Katz J, et al. Racial differences in the cause-specific prevalence of blindness in east Baltimore. N Engl J Med. 1991;325:1412–1417. doi: 10.1056/NEJM199111143252004. [DOI] [PubMed] [Google Scholar]
- 2.Hyman L, Wu SY, Connell AM, et al. Prevalence and causes of visual impairment in The Barbados Eye Study. Ophthalmology. 2001;108:1751–1756. doi: 10.1016/s0161-6420(01)00590-5. [DOI] [PubMed] [Google Scholar]
- 3.Quigley HA, Broman AT. The number of people with glaucoma worldwide in 2010 and 2020. Br J Ophthalmol. 2006;90:262–267. doi: 10.1136/bjo.2005.081224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kwon YH, Fingert JH, Kuehn MH, Alward WLM. Primary Open-Angle Glaucoma. N Engl J Med. 2009;360:1113–1124. doi: 10.1056/NEJMra0804630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fingert JH. Primary open-angle glaucoma genes. Eye. 2011;25:587–595. doi: 10.1038/eye.2011.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stone EM, Fingert JH, Alward WLM, et al. Identification of a Gene That Causes Primary Open Angle Glaucoma. Science. 1997;275:668–670. doi: 10.1126/science.275.5300.668. [DOI] [PubMed] [Google Scholar]
- 7.Rezaie T, Child A, Hitchings R, et al. Adult-onset primary open-angle glaucoma caused by mutations in optineurin. Science. 2002;295:1077–1079. doi: 10.1126/science.1066901. [DOI] [PubMed] [Google Scholar]
- 8.Fingert JH, Robin AL, Ben R, Roos, et al. Copy number variations on chromosome 12q14 in patients with normal tension glaucoma. Hum Mol Genet. 2011;20:2482–2494. doi: 10.1093/hmg/ddr123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alward WL, Fingert JH, Coote MA, et al. Clinical features associated with mutations in the chromosome 1 open-angle glaucoma gene (GLC1A) N Engl J Med. 1998;338:1022–1027. doi: 10.1056/NEJM199804093381503. [DOI] [PubMed] [Google Scholar]
- 10.Thorleifsson G, Walters GB, Hewitt AW, et al. Common variants near CAV1 and CAV2 are associated with primary open-angle glaucoma. Nat Genet. 2010;42:906–909. doi: 10.1038/ng.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burdon KP, MacGregor S, Hewitt AW, et al. Genome-wide association study identifies susceptibility loci for open angle glaucoma at TMCO1 and CDKN2B–AS1. Nat Genet. 2011;43:574–578. doi: 10.1038/ng.824. [DOI] [PubMed] [Google Scholar]
- 12.Wiggs JL, Kang JH, Yaspan BL, et al. Common variants near CAV1 and CAV2 are associated with primary open-angle glaucoma in Caucasians from the USA. Hum Mol Genet. 2011;20:4707–4713. doi: 10.1093/hmg/ddr382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ramdas WD, van Koolwijk LME, Lemij HG, et al. Common genetic variants associated with open-angle glaucoma. Hum Mol Genet. 2011;20:2464–2471. doi: 10.1093/hmg/ddr120. [DOI] [PubMed] [Google Scholar]
- 14.Nakano M, Ikeda Y, Tokuda Y, et al. Common variants in CDKN2B–AS1 associated with optic-nerve vulnerability of glaucoma identified by genome-wide association studies in Japanese. PLoS ONE. 2012;7:e33389. doi: 10.1371/journal.pone.0033389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wiggs JL, Yaspan BL, Hauser MA, et al. Common variants at 9p21 and 8q22 are associated with increased susceptibility to optic nerve degeneration in glaucoma. PLoS Genet. 2012;8:e1002654. doi: 10.1371/journal.pgen.1002654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Osman W, Low S-K, Takahashi A, et al. A genome-wide association study in the Japanese population confirms 9p21 and 14q23 as susceptibility loci for primary open angle glaucoma. Hum Mol Genet. 2012;21:2836–2842. doi: 10.1093/hmg/dds103. [DOI] [PubMed] [Google Scholar]
- 17.van Koolwijk LME, Ramdas WD, Ikram MK, et al. Common genetic determinants of intraocular pressure and primary open-angle glaucoma. PLoS Genet. 2012;8:e1002611. doi: 10.1371/journal.pgen.1002611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gharahkhani P, Burdon KP, Fogarty R, et al. Common variants near ABCA1, AFAP1 and GMDS confer risk of primary open-angle glaucoma. Nat Genet. 2014;46:1120–1125. doi: 10.1038/ng.3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen Y, Lin Y, Vithana EN, et al. Common variants near ABCA1 and in PMM2 are associated with primary open-angle glaucoma. Nat Genet. 2014;46:1115–1119. doi: 10.1038/ng.3078. [DOI] [PubMed] [Google Scholar]
- 20.Hysi PG, Cheng C-Y, Springelkamp H, et al. Genome-wide analysis of multi-ancestry cohorts identifies new loci influencing intraocular pressure and susceptibility to glaucoma. Nat Genet. 2014;46:1126–1130. doi: 10.1038/ng.3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu Y, Vitart V, Burdon KP, et al. Genome-wide association analyses identify multiple loci associated with central corneal thickness and keratoconus. Nat Genet. 2013;45:155–163. doi: 10.1038/ng.2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Z, Allingham RR, Nakano M, et al. A common variant near TGFBR3 is associated with primary open angle glaucoma. Hum Mol Genet. 2015;24:3880–3892. doi: 10.1093/hmg/ddv128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bailey JNC, Loomis SJ, Kang JH, et al. Genome-wide association analysis identifies TXNRD2, ATXN2 and FOXC1 as susceptibility loci for primary open-angle glaucoma. Nat Genet. 2016;48:189–194. doi: 10.1038/ng.3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meguro A, Inoko H, et al. Writing Committee for the Normal Tension Glaucoma Genetic Study Group of Japan Glaucoma Society. Genome-wide association study of normal tension glaucoma: common variants in SRBD1 and ELOVL5 contribute to disease susceptibility. Ophthalmology. 2010;117:1331–8.e5. doi: 10.1016/j.ophtha.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 25.Mabuchi F, Sakurada Y, Kashiwagi K, et al. Association between SRBD1 and ELOVL5 gene polymorphisms and primary open-angle glaucoma. Invest Ophthalmol Vis Sci. 2011;52:4626–4629. doi: 10.1167/iovs.11-7382. [DOI] [PubMed] [Google Scholar]
- 26.Shibuya E, Meguro A, Ota M, et al. Association of Toll-like receptor 4 gene polymorphisms with normal tension glaucoma. Invest Ophthalmol Vis Sci. 2008;49:4453–4457. doi: 10.1167/iovs.07-1575. [DOI] [PubMed] [Google Scholar]
- 27.Takano Y, Shi D, Shimizu A, et al. Association of Toll-like receptor 4 gene polymorphisms in Japanese subjects with primary open-angle, normal-tension, and exfoliation glaucoma. Am J Ophthalmol. 2012;154:825–832.e1. doi: 10.1016/j.ajo.2012.03.050. [DOI] [PubMed] [Google Scholar]
- 28.Suh W, Kim S, Ki C-S, Kee C. Toll-like receptor 4 gene polymorphisms do not associate with normal tension glaucoma in a Korean population. Mol Vis. 2011;17:2343–2348. [PMC free article] [PubMed] [Google Scholar]
- 29.Analysis of toll-like receptor rs4986790 polymorphism in Saudi patients with primary open angle glaucoma. Ophthalmic Genetics. 2016:1–5. doi: 10.3109/13816810.2016.1151900. [DOI] [PubMed] [Google Scholar]
- 30.Ramdas WD, van Koolwijk LME, Ikram MK, et al. A genome-wide association study of optic disc parameters. PLoS Genet. 2010;6:e1000978. doi: 10.1371/journal.pgen.1000978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.MacGregor S, Hewitt AW, Hysi PG, et al. Genome-wide association identifies ATOH7 as a major gene determining human optic disc size. Hum Mol Genet. 2010;19:2716–2724. doi: 10.1093/hmg/ddq144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Springelkamp H, Mishra A, Hysi PG, et al. Meta-analysis of Genome-Wide Association Studies Identifies Novel Loci Associated With Optic Disc Morphology. Genet Epidemiol. 2015;39:207–216. doi: 10.1002/gepi.21886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu Y, Dimasi DP, Hysi PG, et al. Common genetic variants near the Brittle Cornea Syndrome locus ZNF469 influence the blinding disease risk factor central corneal thickness. PLoS Genet. 2010;6:e1000947. doi: 10.1371/journal.pgen.1000947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vitart V, Bencić G, Hayward C, et al. New loci associated with central cornea thickness include COL5A1, AKAP13 and AVGR8. Hum Mol Genet. 2010;19:4304–4311. doi: 10.1093/hmg/ddq349. [DOI] [PubMed] [Google Scholar]
- 35.Vithana EN, Aung T, Khor CC, et al. Collagen-related genes influence the glaucoma risk factor, central corneal thickness. Hum Mol Genet. 2011;20:649–658. doi: 10.1093/hmg/ddq511. [DOI] [PubMed] [Google Scholar]
- 36.Gordon MO, Kass MA. The Ocular Hypertension Treatment Study: design and baseline description of the participants. Arch Ophthalmol. 1999;117:573–583. doi: 10.1001/archopht.117.5.573. [DOI] [PubMed] [Google Scholar]
- 37.Kass MA, Gordon MO, Gao F, et al. Delaying treatment of ocular hypertension: the ocular hypertension treatment study. Arch Ophthalmol. 2010;128:276–287. doi: 10.1001/archophthalmol.2010.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buffone GJ, Darlington GJ. Isolation of DNA from biological specimens without extraction with phenol. Clin Chem. 1985;31:164–165. [PubMed] [Google Scholar]
- 39.Axenovich T, Zorkoltseva I, Belonogova N, et al. Linkage and association analyses of glaucoma related traits in a large pedigree from a Dutch genetically isolated population. J Med Genet. 2011;48:802–809. doi: 10.1136/jmedgenet-2011-100436. [DOI] [PubMed] [Google Scholar]
- 40.Mabuchi F, Sakurada Y, Kashiwagi K, et al. Association between Genetic Variants Associated with Vertical Cup-to-Disc Ratio and Phenotypic Features of Primary Open-Angle Glaucoma. Ophthalmology. 2012;119:1819–1825. doi: 10.1016/j.ophtha.2012.02.044. [DOI] [PubMed] [Google Scholar]
- 41.Tielsch JM, Katz J, Sommer A, et al. Family history risk of primary open angle glaucoma. The Baltimore Eye Survey. Arch Ophthalmol. 1994;112:69–73. doi: 10.1001/archopht.1994.01090130079022. [DOI] [PubMed] [Google Scholar]
- 42.Becker B, KOLKER AE, ROTH FD. Glaucoma family study. Am J Ophthalmol. 1960;50:557–567. doi: 10.1016/0002-9394(60)90233-6. [DOI] [PubMed] [Google Scholar]
- 43.Alsbirk PH. Corneal thickness II. Environmental and genetic factors. Acta ophthalmologica. 1978;56:105–113. doi: 10.1111/j.1755-3768.1978.tb00472.x. [DOI] [PubMed] [Google Scholar]
- 44.Armaly MF. Genetic determination of cup/disc ratio of the optic nerve. Arch Ophthalmol. 1967;78:35–43. doi: 10.1001/archopht.1967.00980030037007. [DOI] [PubMed] [Google Scholar]
- 45.Armaly MF. The genetic determination of ocular pressure in the normal eye. Arch Ophthalmol. 1967;78:187–192. doi: 10.1001/archopht.1967.00980030189011. [DOI] [PubMed] [Google Scholar]
- 46.Thorleifsson G, Magnusson KP, Sulem P, et al. Common sequence variants in the LOXL1 gene confer susceptibility to exfoliation glaucoma. Science. 2007;317:1397–1400. doi: 10.1126/science.1146554. [DOI] [PubMed] [Google Scholar]
- 47.Fan BJ, Wang DY, Pasquale LR, et al. Genetic variants associated with optic nerve vertical cup-to-disc ratio are risk factors for primary open angle glaucoma in a US Caucasian population. Invest Ophthalmol Vis Sci. 2011;52:1788–1792. doi: 10.1167/iovs.10-6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mitchell P, Smith W, Attebo K, Healey PR. Prevalence of open-angle glaucoma in Australia. The Blue Mountains Eye Study. Ophthalmology. 1996;103:1661–1669. doi: 10.1016/s0161-6420(96)30449-1. [DOI] [PubMed] [Google Scholar]
- 49.Burdon KP, Mitchell P, Lee A, et al. Association of open-angle glaucoma loci with incident glaucoma in the Blue Mountains Eye Study. Am J Ophthalmol. 2015;159:31–6.e1. doi: 10.1016/j.ajo.2014.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu Y, Hauser MA, Akafo SK, et al. Investigation of known genetic risk factors for primary open angle glaucoma in two populations of African ancestry. Invest Ophthalmol Vis Sci. 2013;54:6248–6254. doi: 10.1167/iovs.13-12779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ozel AB, Moroi SE, Reed DM, et al. Genome-wide association study and meta-analysis of intraocular pressure. Hum Genet. 2014;133:41–57. doi: 10.1007/s00439-013-1349-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gordon MO, Beiser JA, Brandt JD, et al. The Ocular Hypertension Treatment Study: baseline factors that predict the onset of primary open-angle glaucoma. Arch Ophthalmol. 2002;120:714–720. doi: 10.1001/archopht.120.6.714. discussion 829–30. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.