SUMMARY
The blastema is a mass of progenitor cells that enables regeneration of amputated salamander limbs or fish fins. Methodology to label and track blastemal cell progeny has been deficient, restricting our understanding of appendage regeneration. Here, we created a system for clonal analysis and quantitative imaging of hundreds of blastemal cells and their respective progeny in living adult zebrafish undergoing fin regeneration. Amputation stimulates resident cells within a limited recruitment zone to reset proximodistal (PD) positional information and assemble the blastema. Within the newly formed blastema, the spatial coordinates of connective tissue progenitors are predictive of their ultimate contributions to regenerated skeletal structures, indicating early development of an approximate PD pre-pattern. Calcineurin regulates size recovery by controlling the average number of progeny divisions without disrupting this pre-pattern. Our longitudinal clonal analyses of regenerating zebrafish fins provide evidence that connective tissue progenitors are rapidly organized into a scalable blueprint of lost structures.
Graphical abstract
INTRODUCTION
The defining event in regeneration of an amputated salamander limb or teleost fin is the creation of a blastema, a proliferative mass of lineage-restricted progenitor cells [1, 2]. Recent reports using genetic fate-mapping strategies have indicated that teleost fin, salamander limb, and mouse digit tip blastemas are composed of subsets of progenitor cells that do not cross lineage boundaries [3-7]. These studies have provided tissue-level resolution of the blastema but have not addressed how the cumulative potential to restore an entire adult tissue lineage is encoded within a pool of individual cells. Ectopic transplantation has traditionally been performed to interrogate the developmental properties of blastemal tissue [8-12], yet this technique provides a limited sampling and is not designed to interpret contributions of individual cells in their endogenous contexts.
Clonal analysis is a powerful prerequisite to capture the endogenous developmental potentials of progenitor cells at single-cell resolution. While this methodology has been applied to many contexts of morphogenesis and regeneration to define the nature and variability of cell contributions [13, 14], formation and organization of the appendage blastema have not been assessed. This omission is mainly due to challenges of accessing individual appendage progenitors with permanent cell labeling technology. Among model systems for regeneration, zebrafish, and their fins, have attributes likely to surmount these challenges [15]. Fins consist of several segmented bony rays that each form a blastema within a few days of amputation, before vigorously regenerating lost structures. Models for fin regeneration indicate the maintenance of a zone of proliferation and patterning events at the distal tip of each regenerating fin ray, a region that progressively diminishes as regenerative events culminate. Critically, genetic fate mapping techniques are available for studies of regeneration in adult zebrafish, plus the transparency of fins facilitates live imaging, making it feasible to track the contributions of blastemal cells in real time.
Here, we perform a longitudinal clonal analysis of regenerating zebrafish fins. By tracking contributions of hundreds of individual fin cells in living zebrafish, we visualize and quantify at unprecedented resolution how the blastema is formed and the basis for its ability to regenerate an entire connective tissue compartment. We find that fibroblast progenitors of the fin blastema have unexpected, profound heterogeneity in the extent and PD patterns of their contributions. Some cells give rise to exclusively proximal regions, some to exclusively medial structures, and some to only distal regions, whereas the progeny of other cells might span across multiple regions. By probability calculations and direct visualization, this heterogeneity is explained in part by the early establishment of a pre-pattern in the blastema, compartmentalized based on preferential contributions to regenerating PD structures. We also use clonal analysis to define a function for Calcineurin in scaling regeneration, through control of blastemal cell progeny division without affecting organization of the pre-pattern. These experiments provide a high-resolution view of the blastema that can inform strategies for enhancing complex tissue regeneration.
RESULTS AND DISCUSSION
tph1b Regulatory Sequences Label Connective Tissue Progenitors within the Zebrafish Fin Blastema
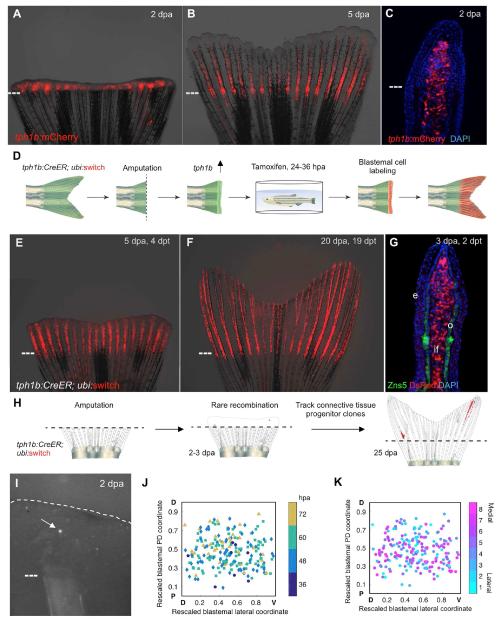
To create a strategy for genetic clonal analysis, we first examined transcriptome datasets for genes with sharp increases in mRNA levels during fin regeneration [16]. tryptophan hydroxylase 1b (tph1b), which encodes the rate-limiting enzyme in serotonin synthesis, was induced 30-fold at 4 days post-amputation (dpa), whereas its paralog tph1a and the related gene tph2 showed little or no change (Figures S1A and S1B). To visualize tph1b expression, we used a transgenic reporter line harboring 5 kb of sequences upstream of the tph1b translation start site fused to an mCherry cassette [17]. Uninjured tph1b:mCherry fins showed limited expression prior to injury (data not shown). By contrast, tph1b regulatory sequences induced mCherry fluorescence in blastemal tissue upon amputation, evident by 1 dpa and stronger by 2 dpa (Figure 1A). At 5 dpa, transgenic reporter expression was diminished distally but remained high in proximal regenerated structures (Figure 1B). Longitudinal sections through 2 dpa regenerating rays revealed tph1b-driven expression in a large medial compartment of the blastema, excluding lateral positions occupied by early osteoblasts that have not begun to deposit bone matrix (Figure 1C).
Figure 1. tph1b Regulatory Sequences Permit Clonal Analysis of Blastemal Cells.
(A and B) tph1b:mCherry reporter expression in regenerating fins at 2 (A) and 5 (B) days post amputation (dpa).
(C) Section of tph1b:mCherry fin at 2 dpa.
(D) Design of lineage tracing experiments in (E and F).
(E and F) tph1b:CreER; ubi:switch fins were treated with 4 μM tamoxifen from 24-36 hours post amputation (hpa) and imaged at 5 (E) and 20 (F) dpa.
(G) Section through 3 dpa lineage-labeled tph1b:CreER; ubi:switch fin, stained for osteoblasts (green) using the Zns5 antibody as well as nuclei (DAPI, blue). e, epidermis; o, osteoblasts, if, intraray fibroblasts.
(H) Design of clonal analysis experiments in (I to K).
(I) Representative image of a single labeled blastemal cell (arrow) in a 2 dpa blastema.
(J) Two-D projection of all initial labeling events onto a rescaled virtual blastema, with rescaled lateral coordinates (D, dorsal; V, ventral) and initial PD coordinates (amputation plane, P, proximal; distal tip, D, distal), color-coded by time of detection of the initial labeling event.
(K) Two-D projection of all initial labeling events onto a rescaled virtual blastema, color-coded by ray of origin (1 indicates lateral-most ray, 8 indicates the medial ray). Dashed lines, amputation planes.
See also Figure S1.
To permanently label tph1b+ blastemal cells and determine the identity of their progeny during regeneration, we fused tph1b regulatory sequences upstream of a tamoxifen-inducible Cre recombinase and generated a transgenic line with this construct. Upon crossing tph1b:CreER zebrafish with animals transgenic for a ubi:switch reporter construct [18], we detected little or no Cre-released expression in embryos or adults (Figures S1C and S1D; data not shown). However, after fin amputation and transient incubation with tamoxifen, strong ubi-driven mCherry fluorescence was induced in regenerating structures. This lineage trace remained stable throughout regeneration, where it exclusively marked intra- and interray connective tissue fibroblasts, representing up to 81.5% of all non-osteoblast cells of the blastema (Figures 1D–1G). Fibroblasts are the predominant cell type in fins and thus comprise the major share of regenerated tissue. Pectoral fin fibroblasts express a code of patterning transcription factor genes that is distinct for each fin ray, indicating that connective tissue possesses positional information that is instructive during regeneration [19]. Thus, the tph1b:CreER transgene enables permanent marking of blastemal connective tissue progenitors, corroborating previous conclusions that fibroblasts and other cell types like epidermis or osteoblasts have distinct, lineage-restricted precursors in regenerating fins [20].
To establish conditions for clonal analysis, we amputated tph1b:CreER; ubi:switch fins and examined Cre-mediated recombination events in the presence of decreasing levels of tamoxifen or no tamoxifen. We found that amputated fins from animals treated with no tamoxifen displayed rare, spontaneous blastemal cell labeling events, evident as singlets, doublets, or occasional tight quadruplets between 1.5 and 3 days dpa (Figures 1H and 1I). The calculated rate of blastemal labeling was 1.87 events per fin, or 10.99% of blastemas labeled, and we estimated the probability of an additional labeling event occurring in the same blastema to be very low (1.21%). Labeling of actively regenerating tissue beyond 4 dpa was extremely rare (13 distinct events in 2567 rays, or 0.51% of rays, and never in the most distal regions). We determined the position of the 216 labeling events that retained expression during regeneration (of 282 total events, or 76.6% of detected initial labeling events) and projected relative coordinates of labeled cells onto a rescaled virtual blastema. Labeling events occurred in blastemas of all fin rays, and all PD areas of the blastema were labeled (Figures 1J, 1K, S1E, and Table S1). Labeling events were distributed throughout rays with a slight skew towards the proximal regions of blastemas (Figure S1F). The distribution across the dorsoventral axis of blastemas was symmetrical, inclusive of all regions except for the lateral-most portions (Figures S1F and S1G). Neither the time of first detected labeling or ray of origin correlated with the spatial coordinates of labeling (Figures 1J, 1K, and S1E).
Blastemal Cells Display Marked Heterogeneity in Contributions to Regeneration
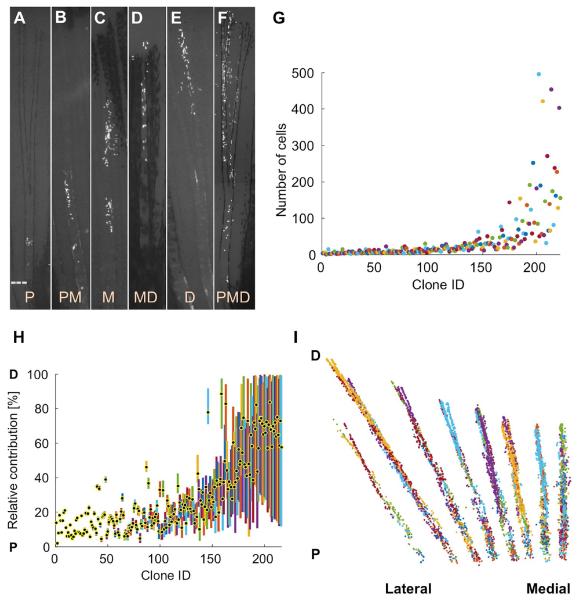
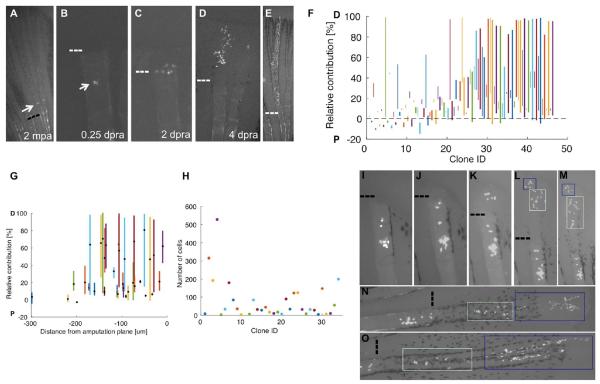
To monitor the output of blastemal labeling events, we imaged the tagged progeny of over 200 clones, i.e. the whole progeny of a common labeling event, twice daily until 7 dpa, and then every few days over the ensuing 3-4 weeks until completion of regeneration (Figures 2A–2F). Using epifluorescence and brightfield microscopy we identified cell positions, amputation sites, and the distal and lateral boundaries of each clone at all timepoints. Generally, cell progeny did not remain adherent in tight clusters, enabling reliable quantification of the numbers of cells in each clone using a fluorescence dissecting microscope. We confirmed this method of quantification by comparison with confocal imaging using optical slices through the entire regenerate. Our results indicate that the counts from fluorescence dissecting microscope images amount to on average 95.8% of the counts obtained from confocal imaging (Figures S2E and S2F). The range of final clone sizes in this dataset was striking, from one cell to 496 cells (Figures 2G; Figures S2A and S2B). Equally remarkable was the range of contributions of connective tissue clones to different PD regions of regenerated rays, with clones often occupying a limited region of the fin ray.
Figure 2. Extreme Heterogeneity in Blastemal Cell Clone Size and PD Contribution.
(A–F) Connective tissue clones initially labeled at 1.5-3 dpa and imaged at 28 dpf, showing examples of proximal (P; A), proximomedial (PM; B), medial (M; C), mediodistal (MD; D), and distal (D; E) clones, as well as clones spanning the PD axis (PMD; F). Dashed line, amputation plane.
(G) Final number of cells in each clone.
(H) Plot of the relative position along the PD axis in which the clone progeny are distributed, normalized for regenerated ray length (amputation plane to distal tip) as a percentage. The centroid position of each clone is indicated with a black and yellow dot.
(I) Cumulative 2-D projection of 216 clones onto a virtual fin lobe at the final timepoint. P, proximal; D, distal.
We identified 6 distinct classes referring to their occupied portion(s) of regenerated fin rays: proximal (P), proximomedial (PM), medial (M), mediodistal (MD), distal (D), and clones with progeny in P, M, and D regions (PMD) (Figures 2A–2F, 2H, and S2C and S2D). To illustrate the contributions of all 216 clones at each timepoint, we projected the rescaled cell coordinates onto a virtual fin lobe also viewable as frames in a virtual movie of fin regeneration (Figure 2I and Movie S1). These projections indicated a combined near-complete coverage of all fin rays from our panel of clones, except for the distal tips of the most lateral rays.
Arguably the most unexpected contributions of the clones we observed were those limited to medial and/or distal structures, which displayed notable behaviors. In some cases, blastemal cell progeny remained stationed in the distal portion of the regenerating ray throughout all imaging timepoints, where they contributed uniformly to the regenerated structures (Figures S2G and S2H). In other cases, progeny showed evidence of specific morphogenetic decisions. For example, in multiple clones a subset of progeny separated from clonal partners to contribute to the connective tissue compartment of only one of two branched ray components. (Figures S2I and S2J). Additionally, we noted that distally restricted clones were characterized by a few labeled cells retained in the distal tip of the regenerate, with progeny expansion beginning relatively late (~10–14 dpa) in the process of regeneration (Figures 3E, S2K and S2L). Similar to this, in other examples we observed that cells in the vicinity of the distal tip could retain their position and make contributions to distal regions late in regeneration that were spatially well separated from those of their clonal partners (Figures S2M–S2O). These behaviors were consistent with histological snapshot assays of cell proliferation during early and middle stages of regeneration that indicate low proliferation indices in the distal-most region of the mesenchymal compartment [21, 22]. Our direct observations of cells in this region suggest that this distal region, which expresses the Wnt inhibitor Dkk1b [23], might function to maintain a cell reservoir for the progenitors of distal skeletal elements that finalize regeneration.
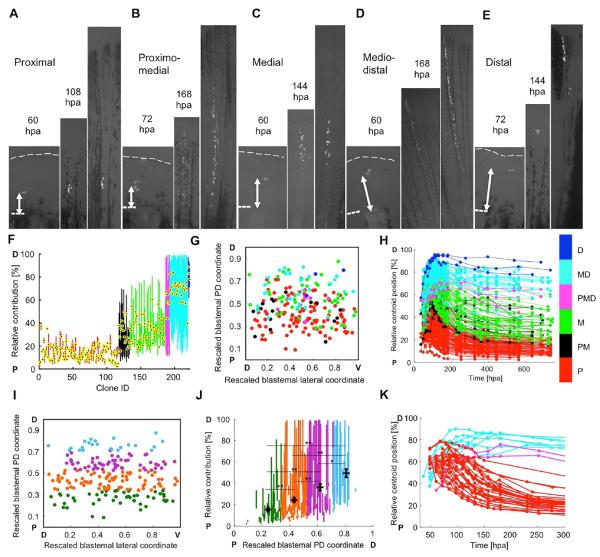
Figure 3. Evidence that Blastema Connective Tissue Progenitors Assemble a PD Pre-Pattern.
(A–E) Examples of clone expansion over time in hours post amputation (hpa), for proximal (A), proximomedial (B), medial (C), mediodistal (D), and distal (E) clones. The final image in each panel is the final timepoint, collected at 25-29 dpa. Dashed lines, amputation planes.
(F) The relative positions of clones in Fig. 2G grouped and color-coded by final PD contribution, used for retrospective assessment of initial labeling position: proximal (red), proximomedial (black), medial (green), mediodistal (cyan), distal (blue), and spanning the PD axis (magenta). The centroid position of each clone is indicated with a black and yellow dot.
(G) Two-dimensional projection of all initial labeling events onto a rescaled virtual blastema, with rescaled lateral (D, dorsal; V, ventral) and PD (P, proximal; D, distal) coordinates, color-coded by final PD occupancy as in (F).
(H) The relative position of each clone in the regenerating ray over time, color-coded by PD contribution. Relative position is defined as the mean distance to the distal plane, divided by the mean distance from amputation plane to distal tip, multiplied by 100.
(I) Two-dimensional projection of all initial labeling events onto a rescaled virtual blastema, grouped and color-coded by initial cell location in the blastema: 15-33.75% (green, bin 1), 33.75-52.5% (orange, bin 2), 52.5-71.25% (purple, bin 3), and 71.25-90% (light blue, bin 4).
(J) Final relative position of each clone, including the mean calculated centroid. Clones are grouped and color-coded by their initial cell location in the blastema, as in (I). *P < 0.01, **P < 0.005; Mann-Whitney two-tailed test, n = 216 clones total; n = 47 (bin 1), n = 90 (bin 2), n = 56 (bin 3), and n = 23 (bin 4).
(K) Cells initially labeled in the proximal half of the blastema that contribute to mediodistal regions, and cells initially labeled in the distal half of the blastema that contribute to proximal regenerate tissue, were identified and their relative centroid positions were plotted over time, color-coded by final PD contribution: proximal (red), mediodistal (cyan), and spanning the PD axis (magenta). Presence of these clones indicate a capacity to mix or non-conform to the pre-pattern. Dashed lines, amputation planes.
See also Figure S3.
To assess whether a second strategy yielded similar results, we crossed tph1:CreER with a different loxP sequence-flanked reporter line, priZm, which employs a β-actin2 promoter-driven reporter transgene [24]. While priZm is a multicolor, Brainbow-based reporter, we assessed only the green channel, as connective tissue cells showed variable levels of expression (Figure S2P). We treated tph1b:CreER; priZm animals with 2 μM tamoxifen at 24-36 hpa and observed infrequent labeling that was suitable for clonal analysis. Clones could not be tracked past 7 dpa, possibly due to some form of transgene silencing, and thus we examined a limited number of clones (35) at 7 dpa. We identified a wide range of 2 to 60 cells in these clones (Figure S2Q), and a wide range of contributions to different PD regions of 7 dpa rays (Figures S2R – S2T). Thus, our data indicate a high degree of heterogeneity in the contributions by blastemal progenitors of the connective tissue lineage during zebrafish fin regeneration. They are inconsistent with a mechanism in which tph1b+ blastemal cells are uniform in their developmental potentials and progressively change as a population during the course of regeneration.
Blastemal Connective Tissue Progenitors Assemble an Approximate Pre-Pattern
Upon visual inspection of examples of P, PM, M, MD, or D clones in tph1b:CreER; ubi:switch regenerates, we noticed many cases of blastemal cells with proximal coordinates generating P clones, as well as blastemal cells with distal coordinates generating D clones (Figures 3A–3E). To quantitatively test this association, we retrospectively classified the initial labeling events of all clones in our dataset directly by the final positions of clones (Figures 3F and 3G). Our data revealed that the majority of P and PM clones were generated by cells labeled in the proximal half of the blastema (74% proximal half; 26% distal half) whereas more distal MD and D clones had a much higher probability of a source in the distal hemi-blastema (24% proximal half; 76% distal half) (Figures 3G and S3A). Longitudinal monitoring and calculation of the centroid of each clone indicated that most clones maintain their relative positions throughout the regenerative process, from source to destination (Figures 3H and S3B).
To complement this approach, we performed a prospective analysis of blastemal cell contributions by binning the PD positions of initial cell labeling events (Figure 3I). Based on calculation of centroids of the clones resulting from these progenitors, we found an increasing likelihood of proximal labeling events to contribute more proximal clones, with distal blastemal cells contributing more distal clones (Figure 3J). Thus, both retrospective and prospective analyses of our data indicate a mechanism in which connective tissue progenitors are organized by 2-3 dpa into blastemal compartments that have distinct likelihoods of contributing to different PD regions. We refer to this organization as a “pre-pattern”, in that the position of cells in the blastema impacts their likelihood to behave in a certain way.
Notably, several blastemal cells we tracked in these experiments did not obey predictions of a strict pre-pattern, as their progeny changed position relative to surrounding cells during regeneration (Figure 3K). Roughly one-quarter of clones originating in the distal half of the blastema contributed to proximal tissue, and a similar percentage of cells originating in the proximal half of the blastema contributed to mediodistal tissue (Figures 3K and S3B). These clones showed morphological characteristics more aligned with those of other clones in a neighboring final position rather than retaining features of most clones originating in the same area of the blastema (Figures S3C–S3K). We interpret this to mean that, while our evidence reveals what we define as a blastemal pre-pattern, this is not a rigid or absolute structure. Blastemal cell fates remain subject to noise or plasticity that can enable differential migration or propulsion, mixing, and cell division during fin regeneration.
Cell Recruitment and Proliferation Dynamics of Resident Cells
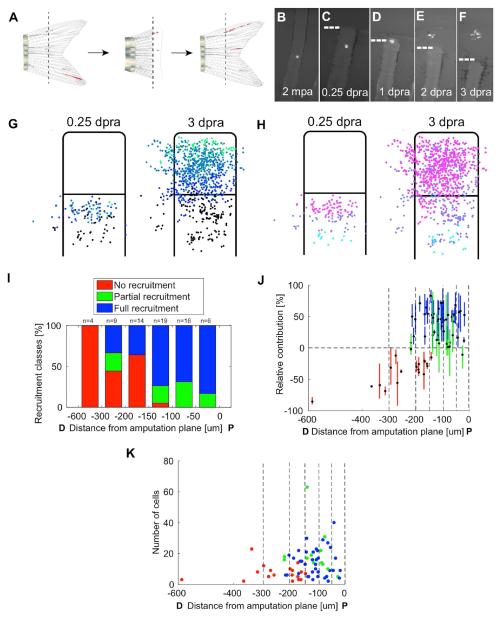
Zebrafish fins regenerate following repeated amputations [25]. To extend our clonal analysis to the contributions of resident differentiated fin cells, we re-amputated slightly distal to established P clones in fins that had completed at least 30 days of regeneration (Figure 4A). Following amputation, resident cells in these clones often, but not always, contributed cells to the blastema (Figures 4B–4F). To visualize how the blastema is built and organized from recruited cells, we first tracked 68 labeled clones in re-amputated rays in which cell progeny could be reliably tracked as they assembled the blastema. This sampling of cells contributed to most areas of the 3 days post re-amputation (dpra) blastema (Figures 4G and 4H; Movies S2 and S3). We performed retrospective analysis of these clones, as well as a prospective analysis that binned cells into 3 equal regions proximal to the amputation plane (Figures 4G and 4H). Probability calculations indicated that proximity to the amputation plane is a general predictor of whether resident cells will contribute to the blastema (Figures 4G–4J; Movies S2 and S3). Intra- and interray cells could be recruited from a limited range to form the ray blastema, up to approximately 300 μm proximal to the amputation plane (usually within one segment, and rarely a second segment; Figure 4I).
Figure 4. Recruitment Dynamics of Resident Cells during Blastema Formation.
(A) Diagram of re-amputation experiment visualizing recruitment of labeled resident cells to form a new blastema and regenerated structures.
(B–F) Re-amputation experiment visualizing recruitment of labeled cells to form a new blastema, shown at 2 months post initial amputation (mpa) (B), and 0.25 (C), 1 (D), 2 (E), and 3 days post re-amputation (dpra) (F).
(G and H) Retrospective (G) and prospective (H) analyses of labeled cell recruitment following amputation injury, shown as cumulative 2-D projections of cells from 68 clones onto a rescaled virtual stump and blastema at 0.25 or 3 dpa, as indicated. For retrospective analyses, the clone centers of masses were binned by their relative positions at 3 dpra, color-coded from proximal (below the amputation plane) to distal (tip of blastema) as black, blue, light blue, and green (G). For prospective analyses, clones’ center of masses were binned by their relative positions at 0.25 dpra, color-coded from furthest to closest to amputation plane as cyan, purple, or magenta (H). Resident cells are recruited towards the injury site and proliferate to occupy most areas of the blastema.
(I) Probabilities for recruitment to the blastema for clones binned by their initial distance (in μm) from the amputation plane. Cell contributions were categorized as fully recruited to the blastema with no progeny below the amputation plane (full recruitment, blue), progeny contributing both to the blastema and below the amputation plane (partial recruitment, green), or contribution to the blastema (no recruitment, red). Generally, cells near the amputation plane have a higher probability of contributing to the blastema, although the distance from amputation plane is not indicative of extent of contribution.
(J) The relative contributions of progeny of below the amputation plane (below 0%) or to the blastema (above 0%) at 3 dpra, relative to the cells initial distance (in μm) from the amputation plane. Cell contributions are color-coded by full recruitment (blue), partial recruitment (green), or no recruitment (red). Clones were binned by their initial distance (in μm) from the amputation plane (vertical dashed lines). Cells generally have a higher probability of recruitment to the blastema if they are closer to the site of injury, although the distance from the amputation plane does not determine the extent or final PD position of their contributions.
(K) The final number of cells in each clone generated by resident cells at 3 dpra, relative to the initial distance (in μm) of the resident cell from the amputation plane. Cell contributions are color-coded by full recruitment (blue), partial recruitment (green), or no recruitment (red). Clones were binned by their initial distance (in μm) from the amputation plane (vertical dashed lines). Cells that contribute to the blastema proliferate to different extents, with no evidence of a direct association with distance of the resident cell from the amputation plane.
Interestingly, the extent and patterns of contribution to 3 dpra blastemas were not faithfully predicted by proximity to the amputation plane (Figures 4G–4K and S4Q–S4S). Even for cells at the same position, participation in blastema formation was variable in number and PD region of the blastema (Figures 4J and 4K; Figures S4A–S4P). For example, cells from within the most proximal 150 μm of the amputation plane could contribute to any region in the entire PD and DV spans of the 3 dpra blastema (Figures 4I–K). If not contributing to the blastema, as for many cells located 200-300 μm from the amputation plane, other observed behaviors proximal to the amputation plane were both lateral and PD mobility, cell proliferation events, and cell morphology changes (Figures 4J and 4K; Figures S4A–S4P). These experiments indicate that amputation stimulates a range of behaviors of established cells spared by the injury, including recruitment of cells within a limited distance to make non-uniform contributions to the regeneration blastema.
To assess how fin cells created during an initial regeneration event respond after a second amputation injury, we amputated fins with established clones occupying proximal regions of the fin and quantified progeny contributions after 3-4 weeks of regeneration. Notably, established proximal cells could contribute to structures extending throughout the PD axis of the regenerate (Figures 5A–5E). This observation indicates that resident cells reset positional information as they are recruited to form the blastema. Even if the progeny of a single blastemal cell are limited to proximal contributions during one regeneration event, a second injury enables contributions by subsequent cell generations to even the most distal portions of a regenerating fin. Established cells recruited to form the blastema displayed marked heterogeneity in their contributions to the second regenerate, analogous to our initial labeling experiments (Figures 5F–5H).
Figure 5. Amputation Resets the Positional Information of Resident Cells.
(A–E) Re-amputation experiment visualizing recruitment of labeled resident cells to form a new blastema and regenerated structures, shown at 2 months post initial amputation (mpa), and 0 (A), 0.25 (B), 2 (C), 4 (D), and 28 days post re-amputation (dpra) (E).
(F) Comparison of respective first and second clones’ relative PD contributions to regenerated ray lengths. Second clones’ relative contributions plotted as bold lines. Cells that had been part of established proximal clones could contribute to structures extending throughout the PD axis of the second regenerate.
(G) Distance of the labeled cell proximal to the second amputation plane (in μm) plotted against the PD contribution of the resultant clone after the second amputation (in μm). Distance of recruited cells to the amputation plane does not directly correlate with final PD contribution to regenerate (linear regression, R = −0.2586, P = 0.0826).
(H) Relative PD contribution and position of clone centroid (black dot) attributable to single resident cells after re-amputation, plotted by the distance of the cell (in μm) from the second amputation plane. Recruited cells varied in their relative contributions to the regenerate, and these contributions did not correlate with their distance from the amputation plane (linear regression, relative contribution R = 0.18, P = 0.298; centroid R = 0.25, P = 0.147).
(I–O) Re-amputation experiment visualizing recruitment of labeled connective tissue cells from a P clone to form a new blastema, shown at 0.5 (I), 1 (J), 2 (K), 3 (L), 4 (M), 6 (N), and 9 (O) days post re-amputation (dpra). Blue and green boxes track progeny of cells from the 3 dpra blastema. Dashed lines, amputation planes.
See also Figure S5.
Re-amputation experiments yielded many fins that had multiple cells labeled within the subsequent blastema. This sample set permitted visualization of how cells with different PD coordinates within a single blastema behaved with respect to each other. In multiple examples, two clones generated within a blastema maintained relative PD positions as their progeny divided throughout regeneration (Figures 5I–5O; Figures S5A-X). Thus, lineage tracing not only of newly labeled blastemal cells, but also of resident cells recruited into the blastema, indicate that the fin blastema is a heterogeneous cell mass that sorts early into a PD pre-pattern.
Calcineurin Regulates Clone Size without Disrupting Blastemal Organization
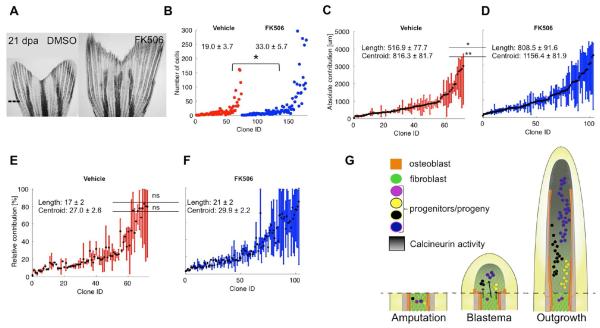
To understand how a pro-regenerative stimulus affects the proliferation dynamics of blastemal cells, we manipulated the activity of Calcineurin, a known regulator of fin regeneration, and performed clonal analysis. Low Calcineurin activity levels correspond to high rates of fin regeneration, and Calcineurin inhibition during regeneration produces greatly lengthened fin rays while retaining overall fin shape (Figure 6A) [26]. We amputated tph1b:CreER; ubi:switch caudal fins to permit rare labeling before incubating animals in either FK506 or vehicle beginning at 3 dpa, after the blastema forms and labeling occurs (Figure 6A). At 21 dpa, we imaged clones, which contributed to a range of regenerated PD structures in both groups (Figures S5A–S5D). On average, connective tissue clones of FK506-treated zebrafish had 74% more cells than control clones (Figure 6B). Notably, clones of FK506-treated animals contributed significantly longer (56%) regions of fins and had significantly more distalized (+41.7%) centroids than control clones (Figures 6C, 6D, and S5E). Length differences were not due to a preponderance of large PMD clones, which were comparable between groups (Figure S5D). In fact, the relative contributions of blastemal cells to different structures along the PD axis, and the mean relative centroid positions of clones, did not differ significantly overall between groups (Figures 6E, 6F, and S5E).
Figure 6. Calcineurin Enhances Regeneration by Scaling Clone Size.
(A) Representative images of 21 dpa tph1b:CreER; ubi:switch fins treated with vehicle (DMSO) or FK506 from 3 to 21 dpa. Dashed line, amputation plane.
(B) Final numbers of cells in clones from vehicle- (red) or FK506-treated (blue) zebrafish at 21 dpa. *P < 0.05, Mann-Whitney one-tailed test, n = 73 (vehicle), n = 104 (FK506).
(C and D) Absolute positions along the PD axis (in μm) indicating clone contributions, for clones from vehicle- (C) or FK506-treated (D) fish. The centroid position of each clone is indicated with a black dot. Clone length is significantly increased in clones from FK506-treated fish, *P < 0.05, Mann-Whitney one-tailed test, n = 73 (vehicle), n = 104 (FK506). Centroid position is significantly distalized in clones from FK506-treated fish, *P < 0.005, Mann-Whitney one-tailed test, n = 73 (vehicle), n = 104 (FK506).
(E and F) Relative position of each clone, including the mean calculated centroid of clones from vehicle- (E) or FK506-treated (F) fish. Clone length and centroid positions are not significantly changed between the two groups, ns, not significant.
(G) Model for blastemal proliferation dynamics. (Left) Cells are recruited to form the blastema from regions proximal to the amputation plane. (Middle) By 3 dpa, blastemal connective tissue progenitors establish preferences to contribute to distinct PD regions based on their coordinates. (Right) The connective tissue compartment regenerates from progenitors in the blastemal approximate pre-pattern. Calcineurin activity increases as regeneration proceeds (dark gradient) and progressively lowers proliferation of fibroblast progeny. Blocking Calcineurin activity with FK506 releases the brake on cell division and clone cell number, scaling up the size of regenerated skeletal elements.
See also Figure S6.
Taken together, these results indicate that Calcineurin does not regulate the heterogeneity of connective tissue clones, the proportions of clones contributing to different regions, or the assembly of an approximate pre-pattern. Rather, Calcineurin less discriminately controls the number of rounds of cell division the progeny of each blastemal cell will undergo, instructions that scale appendage size without impacting the spatial patterns of cell contributions that create a bi-lobed pattern (Figure 6G).
Conclusions
In conclusion, we describe here what is, to our knowledge, the first clonal analysis of a progenitor compartment within a regenerating vertebrate appendage. Our findings reveal prominent heterogeneity among connective tissue progenitors that comprise a major compartment of the regenerating fin blastema. This heterogeneity is explained in part by the emergence of a pre-pattern in which blastemal cells acquire PD preferences by 2-3 dpa (Figure 6G), although there is some plasticity for non-conformance to the pre-pattern. Our data argue against a model in which cells within a lineage compartment of the blastema have similar PD fates that progressively change as regeneration occurs, which would be analogous to the proposed progress zone for embryonic limb development [27, 28]. Interestingly, a recent assessment of blastemal organization using gene expression assays and tissue transplantation in axolotls indicated that PD positional information is progressively acquired during limb regeneration [12]. One potential molecular indicator of these species-dependent mechanisms is the expression kinetics of HoxA genes. In regenerating zebrafish fins, HoxA9, HoxA11, and HoxA13 are each induced early after amputation (data not shown and [29]), whereas induction of HoxA13 in regenerating axolotl upper arms occurs several days after induction of HoxA9 and HoxA11 [12]. We note that we cannot formally exclude the possibility that a cloaked subpopulation of zebrafish fin blastemal cells might escape labeling by the tph1:CreER reagent and make contributions that are distinct from what we describe in our study.
Precisely when blastemal heterogeneity and the pre-pattern emerge during zebrafish fin regeneration, and the underlying molecular factors, remain to be determined. Our data indicate that Calcineurin activity, while having a profound influence on the amount of proliferation and on the ultimate size of the regenerate, has minimal effects on these variables. It will be important to monitor the proliferation dynamics of other fin cell lineages in zebrafish to assess similar principles, and to apply clonal analysis to models like salamander limb regeneration, which is now amenable to genetic fate-mapping [30]. Understanding how intrinsic and extrinsic molecular effectors organize the blastema in highly regenerative contexts will provide insights for engineering limb regeneration in mammals.
EXPERIMENTAL PROCEDURES
Zebrafish Maintenance and Procedures
Wild-type or transgenic zebrafish of the outbred Ekkwill (EK) strain of both sexes were used for all experiments, ranging in age from 4 to 12 months. Water temperature for animals was maintained at 26°C. Zebrafish were anae sthetized, and caudal fins were amputated using razor blades at 50% of their original length except where noted. For FK506 treatment, caudal fins were amputated halfway and treated with 0.1 μg/ml FK506 or 0.004% DMSO from 3 dpa until 21 dpa. In these experiments, fish were fed and aquarium water was exchanged every other day, with water temperature at 24°C. Experiments with animals were approved by the Institutional Animal Care and Use Committee at Duke University.
Construction of tph1b:CreER Zebrafish
Approximately 5 kb of sequence upstream of the tph1b coding start site was subcloned into a Gateway plasmid upstream of the CreER coding sequence using primers tph1b-f, TCTAAGGTGAATCTGTCACATTC and tph1b-r, GGATGGATGGTCTTGTTTTATAG. The vector contains a α-crystallin:EGFP minigene [31] for transgene identification and GAB and 5’HS4 insulator sequences [32]. The resulting plasmid was purified, linearized by I-SceI digestion, and injected into one-cell zebrafish eggs. Stable transgenic lines were identified by lens fluorescence and test for tamoxifen-inducible Cre recombination at embryonic and adult stages by crosses to priZm [24], β-act2:RSG [33], and ubi:Switch [18]. One line was selected for experiments in this study, the full name of which is Tg(tph1b:CreER)pd250.
Histology
Immunohistochemistry on cryosections of 4% paraformaldehyde-fixed fins was performed as previously described [19], using a antibody against DsRed (rabbit, 632496, Clontech) at 1:500 or Zns-5 (mouse, ZIRC) at 1:200. Imaging of 12 μm tissue sections was performed using a Leica DM6000B compound microscope.
Clonal Analysis
tph1b:CreER; ubi:switch adult fish were genotyped for Cre using DNA from dorsal fin clips. Adult caudal fins were amputated at 50% and allowed to regenerate normally. For heavy labeling of blastemal cells, fish were treated with 4 μM tamoxifen from 24-36 hpa. For clonal analysis in tph1b:CreER; priZm fins, animals were treated with 2 μM tamoxifen from 24-36 hpa. For analysis of spontaneous clone formation in tph1b:CreER; ubi:switch fish, zebrafish were anaesthetized using phenoxyethanol and regenerating fins were imaged using a Zeiss AxioZoom microscope twice daily from 36 hpa until 7 dpa, then every 4-5 days until 25-29 dpa. To allow for transgene expression, recombination, and fluorescence detection, we identified distinct labeling events occurring between 36-72 hpa. No animal or sample was excluded unless the animal died during experiments, over half the rays (more than 7 rays) contained labeled cells (3 fish), there were too many labeled cells in a blastema to be defined as a clone (13 blastemas), the blastema developed abnormally (11 blastemas), or label was not maintained (66 of 282 events). Clutchmates were randomized into different groups of animals maintained at equal aquarium densities for experiments. For FK506 treatment, fins were imaged at 3, 4, 5, 7, 14, and 21 dpa for clone tracking. In re-amputation experiments, caudal fins with labeled connective tissue clones were amputated after regeneration had completed (> 30 dpa) slightly distal to isolated, small clusters of labeled cells. Fins were imaged at 6 hours post re-amputation, daily from 1-4 dpra, and then at 6, 8, 10, 14, 21, and 28 dpra.
Data Analysis
All raw images were processed using FIJI software, and images were organized by individual fish identifiers to track respective clones over time. Clones for tph1b:CreER; priZm analysis were identified at 7 dpa and retrospectively traced back to initial labeling events occurring by 3 dpa. Clones for tph1b:CreER; ubi:switch analysis were identified, tagged, and categorized at the last imaged timepoint (25-29 dpa), then retrospectively traced back to initial labeling events occurring between 36-72 hpa. Raw images were organized by clone, and combined into stacks using custom FIJI scripts. Images were manually segmented using FIJI software to identify the position of individual labeled cells, amputation plane, distal plane, and lateral planes at each timepoint. Higher magnification images were used to better quantify cells and their positions in larger clones. To confirm cell counting accuracy from AxioZoom images, 11 clones were first imaged at 10 dpa and manually segmented for cell counts. These same fins were then collected, fixed in 4% paraformaldehyde, washed, and mounted with DAPI for optical sectioning using a Zeiss confocal 700 microscope. These images were then segmented at each Z plane, and cells quantified for 11 representative clones of varying lengths, densities, and PD positions. Cell numbers determined from AxioZoom images were 87.5 – 100% similar to those from confocal projections.
Cell numbers, physical positions in the ray, position of amputation, distal, lateral planes, and timestamps for all cells in each clone were extracted at each timepoint by means of a custom written FIJI script. The calculation of distances of each cell from the corresponding amputation and distal planes was calculated using MATLAB. Each cell’s relative position in the regenerating fin was defined as its distance to the amputation plane, divided by the regenerate’s mean amputation-plane-to-distal-tip distance and multiplied by 100. Segmentation from the first detectable image of each labeled clone was used to generate initial blastema plots, with rescaled PD and DV axes as described above. A triples test for symmetry was used to assess spatial distribution of labeling across the PD and DV axes.
Prospective analyses were performed by binning rescaled initial PD blastema coordinates. To collapse all clone coordinates on top of a virtual fin, a fin image was manually drawn to scale, and the relative lengths of each of the different rays were measured. Relative coordinates of the positions of cells in each clone were then rotated and dilated to match relative length of each of the rays in the virtual fin and to preserve the relative position along the ray. Statistical testing was performed using Mann-Whitney one- or two-tailed tests in order to avoid the assumption of normality of the data. All data manipulations and statistical analyses were performed in MATLAB.
Supplementary Material
Highlights.
-
-
Large-scale, longitudinal clonal analysis of a regenerating vertebrate appendage
-
-
Connective tissue progenitors assemble a pre-pattern in the zebrafish fin blastema
-
-
Quantitative imaging of cell recruitment dynamics that create the blastema
-
-
Calcineurin scales regeneration by regulating blastemal progeny division
ACKNOWLEDGMENTS
We thank J. Burris, T. Thoren, T. Laffredo, and B. Thomas for zebrafish care; A. Dickson for technical assistance and artwork; B. Carlson and A. Srivastava for help with image processing; J. A. Goldman for help in generating the tph1b:CreER plasmid; C. Mosimann and L. Zon for transgenic animals; and D. Fox, M. Bagnat, and B. Hogan for comments on the manuscript. V.A.T. and L.A.S. were supported by Graduate Research Fellowships from the National Science Foundation. M.K. is an INSERM Researcher. This work was supported by Finalized Research and Funding for Investments in Basic Research (RBAP11BYNP-Newton; A.F. and L.P.), a Whitehead Faculty Scholar Award to S.D., and a grant (R01 GM074057) from NIH to K.D.P.
Footnotes
Supplemental Information includes 6 Supplemental Figures, One Supplemental Table, and 3 Supplemental Movies
AUTHOR CONTRIBUTIONS
V.A.T. and K.D.P. designed the experimental strategy, analyzed data, and prepared the manuscript. V.A.T. generated transgenic animals and performed regeneration experiments. A.P. designed and performed quantitative image and data analysis. L.P. and S.D. contributed quantitative analysis support. L.S. generated the transgenic construct. J.T. performed regeneration experiments and analyzed data. G.N. generated and analyzed transcriptome data. A.K. and M.K. contributed unpublished reagents and transgenic animals. All authors commented on the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Tornini VA, Poss KD. Keeping at arm's length during regeneration. Developmental Cell. 2014;29:139–145. doi: 10.1016/j.devcel.2014.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Simon A, Tanaka EM. Limb regeneration. Wiley Interdiscip Rev Dev Biol. 2013;2:291–300. doi: 10.1002/wdev.73. [DOI] [PubMed] [Google Scholar]
- 3.Kragl M, Knapp D, Nacu E, Khattak S, Maden M, Epperlein HH, Tanaka EM. Cells keep a memory of their tissue origin during axolotl limb regeneration. Nature. 2009;460:60–65. doi: 10.1038/nature08152. [DOI] [PubMed] [Google Scholar]
- 4.Rinkevich Y, Lindau P, Ueno H, Longaker MT, Weissman IL. Germ-layer and lineage-restricted stem/progenitors regenerate the mouse digit tip. Nature. 2011;476:409–413. doi: 10.1038/nature10346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Knopf F, Hammond C, Chekuru A, Kurth T, Hans S, Weber CW, Mahatma G, Fisher S, Brand M, Schulte-Merker S, et al. Bone regenerates via dedifferentiation of osteoblasts in the zebrafish fin. Developmental Cell. 2011;20:713–724. doi: 10.1016/j.devcel.2011.04.014. [DOI] [PubMed] [Google Scholar]
- 6.Singh SP, Holdway JE, Poss KD. Regeneration of amputated zebrafish fin rays from de novo osteoblasts. Developmental Cell. 2012;22:879–886. doi: 10.1016/j.devcel.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lehoczky JA, Robert B, Tabin CJ. Mouse digit tip regeneration is mediated by fate-restricted progenitor cells. Proc Natl Acad Sci U S A. 2011;108:20609–20614. doi: 10.1073/pnas.1118017108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Echeverri K, Tanaka EM. Proximodistal patterning during limb regeneration. Developmental Biology. 2005;279:391–401. doi: 10.1016/j.ydbio.2004.12.029. [DOI] [PubMed] [Google Scholar]
- 9.Nacu E, Glausch M, Le HQ, Damanik FF, Schuez M, Knapp D, Khattak S, Richter T, Tanaka EM. Connective tissue cells, but not muscle cells, are involved in establishing the proximo-distal outcome of limb regeneration in the axolotl. Development. 2013;140:513–518. doi: 10.1242/dev.081752. [DOI] [PubMed] [Google Scholar]
- 10.Pescitelli MJ, Jr., Stocum DL. The origin of skeletal structures during intercalary regeneration of larval Ambystoma limbs. Developmental Biology. 1980;79:255–275. doi: 10.1016/0012-1606(80)90115-3. [DOI] [PubMed] [Google Scholar]
- 11.Stocum DL, Melton DA. Self-organizational capacity of distally transplanted limb regeneration blastemas in larval salamanders. J Exp Zool. 1977;201:451–461. doi: 10.1002/jez.1402010312. [DOI] [PubMed] [Google Scholar]
- 12.Roensch K, Tazaki A, Chara O, Tanaka EM. Progressive specification rather than intercalation of segments during limb regeneration. Science. 2013;342:1375–1379. doi: 10.1126/science.1241796. [DOI] [PubMed] [Google Scholar]
- 13.Blanpain C, Simons BD. Unravelling stem cell dynamics by lineage tracing. Nat Rev Mol Cell Biol. 2013;14:489–502. doi: 10.1038/nrm3625. [DOI] [PubMed] [Google Scholar]
- 14.Buckingham ME, Meilhac SM. Tracing Cells for Tracking Cell Lineage and Clonal Behavior. Developmental Cell. 2011;21:394–409. doi: 10.1016/j.devcel.2011.07.019. [DOI] [PubMed] [Google Scholar]
- 15.Gemberling M, Bailey TJ, Hyde DR, Poss KD. The zebrafish as a model for complex tissue regeneration. Trends Genet. 2013;29:611–620. doi: 10.1016/j.tig.2013.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kang J, Hu J, Karra R, Dickson AL, Tornini VA, Nachtrab G, Gemberling M, Goldman JA, Black BL, Poss KD. Modulation of tissue repair by regeneration enhancer elements. Nature. 2016;532:201–206. doi: 10.1038/nature17644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Soulika M, Kaushik AL, Mathieu B, Lourenco R, Komiarczuk AZ, Romano SA, Jourary A, Lardennois A, Tissot N, Okada S, et al. Diversity in cell motility reveals the dynamic nature of the formation of zebrafish taste sensory organs. Development. 2016;143:2012–2024. doi: 10.1242/dev.134817. [DOI] [PubMed] [Google Scholar]
- 18.Mosimann C, Kaufman CK, Li P, Pugach EK, Tamplin OJ, Zon LI. Ubiquitous transgene expression and Cre-based recombination driven by the ubiquitin promoter in zebrafish. Development. 2011;138:169–177. doi: 10.1242/dev.059345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nachtrab G, Kikuchi K, Tornini VA, Poss KD. Transcriptional components of anteroposterior positional information during zebrafish fin regeneration. Development. 2013;140:3754–3764. doi: 10.1242/dev.098798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tu S, Johnson SL. Fate restriction in the growing and regenerating zebrafish fin. Developmental Cell. 2011;20:725–732. doi: 10.1016/j.devcel.2011.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poss KD, Nechiporuk A, Hillam AM, Johnson SL, Keating MT. Mps1 defines a proximal blastemal proliferative compartment essential for zebrafish fin regeneration. Development. 2002;129:5141–5149. doi: 10.1242/dev.129.22.5141. [DOI] [PubMed] [Google Scholar]
- 22.Nechiporuk A, Keating MT. A proliferation gradient between proximal and msxb-expressing distal blastema directs zebrafish fin regeneration. Development. 2002;129:2607–2617. doi: 10.1242/dev.129.11.2607. [DOI] [PubMed] [Google Scholar]
- 23.Kang J, Nachtrab G, Poss KD. Local Dkk1 crosstalk from breeding ornaments impedes regeneration of injured male zebrafish fins. Developmental Cell. 2013;27:19–31. doi: 10.1016/j.devcel.2013.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gupta V, Poss KD. Clonally dominant cardiomyocytes direct heart morphogenesis. Nature. 2012;484:479–484. doi: 10.1038/nature11045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Azevedo AS, Grotek B, Jacinto A, Weidinger G, Saude L. The regenerative capacity of the zebrafish caudal fin is not affected by repeated amputations. PLoS One. 2011;6:e22820. doi: 10.1371/journal.pone.0022820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kujawski S, Lin W, Kitte F, Bormel M, Fuchs S, Arulmozhivarman G, Vogt S, Theil D, Zhang Y, Antos CL. Calcineurin regulates coordinated outgrowth of zebrafish regenerating fins. Developmental Cell. 2014;28:573–587. doi: 10.1016/j.devcel.2014.01.019. [DOI] [PubMed] [Google Scholar]
- 27.Summerbell D, Lewis JH, Wolpert L. Positional information in chick limb morphogenesis. Nature. 1973;244:492–496. doi: 10.1038/244492a0. [DOI] [PubMed] [Google Scholar]
- 28.Towers M, Tickle C. Growing models of vertebrate limb development. Development. 2009;136:179–190. doi: 10.1242/dev.024158. [DOI] [PubMed] [Google Scholar]
- 29.Geraudie J, Borday Birraux V. Posterior hoxa genes expression during zebrafish bony fin ray development and regeneration suggests their involvement in scleroblast differentiation. Dev Genes Evol. 2003;213:182–186. doi: 10.1007/s00427-003-0307-y. [DOI] [PubMed] [Google Scholar]
- 30.Sandoval-Guzman T, Wang H, Khattak S, Schuez M, Roensch K, Nacu E, Tazaki A, Joven A, Tanaka EM, Simon A. Fundamental differences in dedifferentiation and stem cell recruitment during skeletal muscle regeneration in two salamander species. Cell Stem Cell. 2014;14:174–187. doi: 10.1016/j.stem.2013.11.007. [DOI] [PubMed] [Google Scholar]
- 31.Waxman JS, Keegan BR, Roberts RW, Poss KD, Yelon D. Hoxb5b acts downstream of retinoic acid signaling in the forelimb field to restrict heart field potential in zebrafish. Developmental Cell. 2008;15:923–934. doi: 10.1016/j.devcel.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bessa J, Tena JJ, de la Calle-Mustienes E, Fernandez-Minan A, Naranjo S, Fernandez A, Montoliu L, Akalin A, Lenhard B, Casares F, et al. Zebrafish enhancer detection (ZED) vector: a new tool to facilitate transgenesis and the functional analysis of cis-regulatory regions in zebrafish. Dev Dyn. 2009;238:2409–2417. doi: 10.1002/dvdy.22051. [DOI] [PubMed] [Google Scholar]
- 33.Kikuchi K, Holdway JE, Werdich AA, Anderson RM, Fang Y, Egnaczyk GF, Evans T, Macrae CA, Stainier DY, Poss KD. Primary contribution to zebrafish heart regeneration by gata4(+) cardiomyocytes. Nature. 2010;464:601–605. doi: 10.1038/nature08804. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.