Abstract
Human Gingival Mesenchymal Stem Cells (hGMSCs) are multipotential cells that can expand and differentiate in culture under specific and standardized conditions. In the present study, we have investigated whether in vitro pre-treatment of hGMSCs with Cannabidiol (CBD) can influence their expression profile, improving the therapeutic potential of this cell culture. Following CBD treatment (5 μM) for 24 h, gene expression analysis through Next Generation Sequencing (NGS) has revealed several genes differentially expressed between CBD-treated hGMSCs (CBD-hGMSCs) and control cells (CTR-hGMSCs) that were linked to inflammation and apoptosis. In particular, we have demonstrated that CBD treatment in hGMSCs prevented the activation of the NALP3-inflammasome pathway by suppressing the levels of NALP3, CASP1, and IL18, and in parallel, inhibited apoptosis, as demonstrated by the suppression of Bax. CBD treatment was also able to modulate the expression of the well-known mesenchymal stem cell markers (CD13, CD29, CD73, CD44, CD90, and CD166), and other surface antigens. Specifically, CBD led to the downregulation of genes codifying for antigens involved in the activation of the immune system (CD109, CD151, CD40, CD46, CD59, CD68, CD81, CD82, CD99), while it led to the upregulation of those implicated in the inhibition of the immune responses (CD47, CD55, CD276). In conclusion, the present study will provide a new simple and reproducible method for preconditioning hGMSCs with CBD, before transplantation, as an interesting strategy for improving the hGMSCs molecular phenotype, reducing the risk of immune or inflammatory reactions in the host, and in parallel, for increasing their survival and thus, their long-term therapeutic efficacy.
Keywords: cannabidiol, human gingival mesenchymal stem cells, immunophenotype, next generation sequencing, inflammasome
Introduction
During the last decades, Mesenchymal Stem Cells (MSCs) have attracted much attention for their ability to transdifferentiate into several tissues, under specific conditions, and therefore, to potentially reconstitute the damaged organs and tissues (Zhang et al., 2009; Patel et al., 2013). They have already been isolated from different tissues, including bone marrow, adipose tissue, umbilical cord blood and oral cavity (Hass et al., 2011). In 2006, The International Society for Cellular Therapy has established the minimal criteria for defining MSCs: they must be plastic-adherent in standard culture conditions, they must express specific surface antigens like the cluster of differentiation (CD) CD105, CD73, and CD90 in flow cytometry, and they must lack expression of CD45, CD34, CD14, or CD11b, CD79a, or CD19 and HLA class II (Dominici et al., 2006). Moreover, they must show a multipotent ability to differentiate into several mesenchymal lineages such as osteoblasts, adipocytes and chondroblasts under standard in vitro differentiating conditions (Dominici et al., 2006).
In a recent work, our group has demonstrated the beneficial effect of the human periodontal ligament stem cells, an oral-derived MSC lineage, in experimental autoimmune encephalomyelitis models (Trubiani et al., 2016a). Among oral-derived MSCs, the human Gingival Mesenchymal Stem Cells (hGMSCs) have been recently explored since they have shared many common features to other MSCs such as a spindle-like cell morphology, plastic adherence, the expression profile of specific cell surface markers, multipotent differentiation, and even immunomodulatory functions (Xiao and Nasu, 2014; Jin et al., 2015; Fawzy El-Sayed and Dörfer, 2016). One of the major advantage of hGMSCs compared to MSCs isolated from other tissue, is their abundance and easy isolation from gingiva through a minimally invasive dental procedure (Du et al., 2016; Fawzy El-Sayed and Dörfer, 2016). Therefore, following expansion and pre-differentiation into a desired tissue type, hGMSCs potentially could be re-implanted for tissue repair in the patients.
Despite the therapeutic potential of MSCs in stem cell-based therapies, certain issues need to be addressed, such as the risk of immune rejection which can affect MSCs survival and thus their long-term fate once transplanted in vivo. Current approaches to prevent immune rejections are based on immunosuppressive drugs which induce an immunological tolerance in the host. However, the prolonged use of these therapies has been associated with increased risk to develop other pathological conditions, such as diabetes, hypertension, and nephrotoxicity (Merville, 2005). As a consequence, new strategies that prevent immune rejections affecting stem cell survival in vivo and that assure a successful long-term engraftment, are needed. Here, we propose an alternative strategy to overcome these issues which consists in preconditioning hGMSCs in vitro with a cannabinoid compound, the Cannabidiol (CBD). Thus far, scientists have found that cannabinoids by interacting with several receptors, including the endocannabinoid receptors type 1 and type 2 (CB1R and CB2R), can modulate several pathways, such as stem cells self-renewal, proliferation and differentiation (Galve-Roperh et al., 2013). Therefore, it is not surprising that many studies are aimed at evaluating the therapeutic potential of different exogenous cannabinoids, among which CBD has received much attention since it is devoid of psychotropic effects compared to other cannabinoids.
In this study, we have performed a Next Generation Sequencing (NGS)-based gene expression profiling in order to evaluate whether CBD treatment can modify the molecular phenotype of hGMSCs, enhancing their therapeutic efficacy.
Materials and methods
Extraction and isolation of CBD
Pure CBD (>99%) was isolated from Carmagnola, an Italian variety of industrial C. sativa and obtained from greenhouse cultivation at CREA-CIN, Rovigo (Italy), according to a standardized method of the cannabinoid purification reported by Taglialatela-Scafati et al. (2010), in order to avoid any trace of THC that could interfere in the trial or cause legal limitation. The isolation and purification of cannabinoids were done in accordance with Italian legal status (Authorization SP/106 23/05/2013 of the Ministry of Health, Rome, Italy).
Isolation and immunophenotyping of hGMSCs
hGMSCs culture establishment
Gingival tissue biopsies were obtained from five healthy adult volunteers with no gingival inflammation. The gingival specimens were completely de-epithelialized with a scalpel, for the exclusion of the most of the keratinocytes resident in the gingival. In brief, the connective tissues were grinded and then washed several time with PBS (LiStarFish, Milan, Italy) and subsequently cultured using TheraPEAK™MSCGM-CD™ BulletKit serum free, chemically defined (MSCGM-CD) medium for the growth of human MSCs (Lonza, Basel, Switzerland). The medium was changed twice a week, and cells spontaneously migrating from the explant fragments after reaching about 80% of confluence, were trypsinized using Triple Select (LiStar Fish) (Diomede et al., 2014). On day 6, colonies of 50 or more cells were scored as colony-forming unit fibroblasts (CFU).
Cytofluorimetric evaluation
Samples were stained for surface or intracellular antigens, as previously described (Cianci et al., 2016). For surface staining, samples were resuspended in 100 μl washing buffer (0.1% sodium azide and 0.5% bovine serum albumine in PBS) containing the appropriate amount of surface antibodies; samples were incubated for 30 min at 4°C in the dark. Cells were washed (3 ml of washing buffer), centrifuged (4°C, 400 X g, 8 min), resuspended with 1ml PBS 0.5% paraformaldehyde, incubated for 5 min at RT, washed, centrifuged (4°C, 400 X g, 8min) and stored at 4°C in the dark until their acquisition. For intracellular staining, cells were resuspended in 1 ml of FACS Lysing solution (BD), vortexed and incubated at room temperature (RT) in the dark for 10 min. Samples were centrifuged (4°C, 400 X g, 8 min); 1 ml of Perm 2 (BD) was added to each tube and cells were incubated at RT in the dark for 10 min. Samples were washed and centrifuged (4°C, 400 X g, 8 min). Cells were resuspended in 100 μl of washing buffer containing the appropriate amount of intracellular antibodies and incubated for 30 min at 4°C in the dark. Cells were centrifuged (4°C, 400 X g, 8 min), resuspended with 1 ml PBS 0.5 % paraformaldehyde for 5 min at RT, washed, centrifuged (4°C, 400 X g, 8 min) and stored at 4°C in the dark until the acquisition. Cells were analyzed on a FACS Calibur flow cytometer (BD), using CellQuest™ software (BD). Quality control included regular check-ups with Rainbow Calibration Particles (BD Biosciences). Debris was excluded from the analysis by gating on morphological parameters; 20,000 non-debris events in the morphological gate were recorded for each sample. To assess non-specific fluorescence we used isotype controls. All antibodies were titrated under assay conditions and optimal photomultiplier voltages (PMT) were established for each channel. Data were analyzed using FlowJo™ software (TreeStar, Ashland, OR). Mean Fluorescence Intensity Ratio (MFI Ratio) was calculated dividing the MFI of positive events by the MFI of negative events (Trubiani et al., 2016b).
Multipotent differentiation of hGMSCs
Osteogenic differentiation
7.5 × 103 cells /cm2 CTR-hGMSCs and CBD-hGMSCs, were seeded at 2nd passage in a 6-well cell culture plate (Falcon) and incubated overnight at 37°C, in a humidified 5% CO2 atmosphere. The culture medium was replaced with osteogenic induction medium represented of MSCGM-CD medium supplemented with 100 nM dexamethasone (Applichem GmbH, Darmstadt, Germany), 10 nM β-glycerol-phosphate (Applichem) and 0.05 mM 2-phosphate-ascorbic acid (Sigma-Aldrich). The medium was replaced every 3 days. At days 21 Alizarin Red S staining (Sigma-Aldrich Co.) was performed to detect calcium formation (Trubiani et al., 2015).
Adipogenic differentiation
CTR-hGMSCs and CBD-hGMSCs were seeded in a 6-well cell culture plate (Falcon) at a density of 2 7.5 × 103 cells /cm2 and incubated overnight. The medium was replaced with adipogenic induction medium (Lonza) composed by induction and maintenance medium. The medium After 28 days of treatment samples were fixed and stained with Oil Red O working solution (Sigma-Aldrich) and counterstained with hematoxylin to detect the oil globules. Images were collected using light microscopy Leica DMIL (Leica Microsystem, Milan, Italy) (Trubiani et al., 2016b).
Cell culture conditions and drug treatment
hGMSCs were cultured in monolayer using DMEM high-glucose medium (CARLO ERBA, Italy) containing 10% fetal bovine serum (FBS) (Sigma-Aldrich Co. Ltd., USA). The cells were grown in logarithmic phase at 37°C in a moisturized atmosphere of 5% CO2 and 95% air. For drug treatment, cells at passage 10 were grown until 70–80% confluence and then, they were treated with CBD (5 μM) for 24 h (CBD-hGMSCs). Untreated cells were used as control (CTR-hGMSCs) and cells treated with vehicle (0.1% DMSO) were also included as a control (DMSO-hGMSCs). For antagonists study, cells were incubated for 2 h with different combinations of SR141716A (CB1R antagonist; 1 μM) or AM630 (CB2R antagonist;100 nM) before CBD administration for 24 h. All antagonists were purchased from Tocris Bioscience, UK. All the experiments were made in triplicates and repeated for three independent times.
Preparation of RNA libraries for deep sequencing
Total RNA was isolated with a Reliaprep RNA Cell Miniprep System (Promega, Milano, Italy). RNA sequencing libraries were prepared using TruSeq RNA Access library kit (Illumina, Inc., San Diego, CA, USA) according to the manufacturer's protocol. Thereafter, RNA samples (50 ng) were fragmented at 94°C for 8 min on a thermal cycler. First strand cDNA syntheses were performed at 25°C for 10 min, 42°C for 15 min, and 70°C for 15 min, using random hexameres and the SuperScript II Reverse Transcriptase (Invitrogen, Milan, Italy). In a second strand cDNA synthesis, the RNA templates were removed and a second replacement strand was generated by incorporation dUTP (in place of dTTP, to keep strand information) to generate double strands cDNA. The AMPure XP beads (Beckman Coulter) were used to clean up the blunt-ended cDNA from the second strand reaction mix. The 3′ends of the cDNA were then adenylated to facilitate the adaptor ligation in the next step. After ligation of indexing adaptors, the AMPure XP beads were used to clean up the libraries. A first Polymerase Chain Reaction (PCR) amplification step (15 cycles of 98°C for 10 s, 60°C for 30 s, and 72°C for 30 s) was performed to selectively enrich those DNA fragments that have adapter molecules on both ends and also to amplify the amount of DNA in the library. After validation of the libraries, the first hybridization step was performed using exome capture probes. Before hybridization a 2-plex pool of libraries was made, by combining 200 ng of each DNA library. The hybridization was performed by 18 cycles of 1 min incubation, starting at 94°C, and then decreasing 2°C for cycle. Then streptavidin-coated magnetic beads were used to capture probes hybridized to the target regions. The enriched libraries were then eluted from the beads and prepared for a second round of hybridization. This second hybridization (18 cycles of 1 min incubation, starting at 94°C, and then decreasing 2°C per cycle) was required to ensure high specificity of the capture regions. A second capture with streptavidin-coated beads was performed, followed by two heated wash procedures to remove the non-specific binding from the beads. The enriched libraries were then eluted from the beads and cleaned up by the AMPure XP beads, prior to a second PCR amplification. The amplification step was performed by 10 cycles (98°C for 10 s, 60°C for 30 s, and 72°C for 30 s) followed by a second PCR clean up. Finally, the libraries were quantitated by quantitative PCR using the KAPA Library Quantification Kit-Illumina/ABI Prism® (Kapa Biosystems, Inc., Wilmington, MA, USA) and validated using the Agilent High Sensitivity DNA Kit on a Bioanalyzer. The size range of the DNA fragments was measured to be in the range of 200–650 bp and peaked around 250 bp.
Libraries were normalized to 12 pM and subjected to cluster and the single read sequencing was performed for 150 cycles on a MiSeq instrument (Illumina, Inc. San Diego, CA, USA), according to the manufacturer's instructions. Finally, the libraries generated were loaded for clustering on a MiSeq Flow Cell v3 and sequenced through the MiSeq Instrument.
Gene expression data processing
CASAVA (version 1.8.2, Illumina) software was used to demultiplex reads into specific sample and groups of the indexes. Each sample was mapped against its reference sequences “Homo sapiens UCSC hg19” using RNA-Seq Alignment version 1.0.0 with its default parameters, in particular for the Read mapping was used the TopHat 2 (Bowtie 1). The Fragments Per Kilobase Of Exon Per Million Fragments Mapped (FPKM) values were calculated for each sample using the normalized reads counts for each annotated gene [(1000 × read count) ÷ (number of gene covered bases × number of mapped fragments in million)]. Unmapped reads were removed, retaining only read pairs with both reads aligned to the reference sequences “Homo sapiens UCSC hg19.” The comparison between two different samples was visualized by a scatter plot of the LOG2 of the FPKM.
Bioinformatics analysis of genes differentially expressed
Once identified the genes differentially expressed between CTR-hGMSCs and CBD-hGMSCs, the enrichment analyses of the Gene Ontology processes (GO) and the pathway analyses were conducted using the online free databases such as bioDBnet (Mudunuri et al., 2009), STRING (Szklarczyk et al., 2015) and KEEG (Kanehisa et al., 2016). This strategy, based on testing multiple tools, is recommended in order to obtain the most satisfactory results (Rhee et al., 2008). We considered only the GO categories or KEGG pathways reported to be significantly enriched.
Immunocytochemistry
Cells on coverslips (10 mm, Thermo SCIENTIFIC, Germany) were fixed with 4% paraformaldehyde at room temperature for 20 min, followed by phosphate buffered saline (PBS, pH 7.5) washes. Then, cells were incubated with 3% hydrogen peroxide (H2O2) at room temperature for 15 min to suppress the endogenous peroxidase activity. Following three washes with PBS, cells were blocked with horse serum + 0.1% Triton X-100 for 20 min followed by incubation for overnight at 4°C with primary antibodies against examined proteins: Caspase-1 (CASP1, 1:100, Abcam), Interleukin-18 (IL-18, 1:100, Abcam), NLR family pyrin domain containing 3 (NRLP3/NALP3, 1:100, R&D Systems), Bcl2 associated X (Bax, 1:100 Santa Cruz Biotechnology Inc) and Bcl-2 (1:100 Santa Cruz Biotechnology Inc). After PBS wash, cells were incubated with biotinylated secondary antibody (1:200, Vector Laboratories, Burlingame, CA) and streptavidin ABComplex-HRP (ABC-kit from Dako, Glostrup, Denmark). The immunostaining was developed with peroxidase substrate kit DAB (Vector Laboratories, Burlingame, CA) (brown color; positive staining) and counterstaining with nuclear fast red (Vector Laboratories, Burlingame, CA) (pink background; negative staining). Microscopy was performed using light microscopy (LEICA DM 2000 combined with LEICA ICC50 HD camera). Immunocytochemistry images were assessed for densitometry analysis using LEICA Application Suite V4.2.0 software to calculate the percentage of positive staining of the cells. All images are representative of three independent experiments.
Protein extraction and Western blot analysis
In order to extract proteins, cells were washed with ice-cold PBS and lysed using Buffer A [320 mM Sucrose, 10 mM, 1 mM EGTA, 2 mM EDTA, 5 mM NaN3, 50 mM NaF, β-mercaptoethanol, and protease/phosphatase inhibitor cocktail (Roche, USA)] in ice for 15 min, followed by centrifugation at 1000 g for 10 min at 4°C. The supernatant was served as cytosolic extract. The pellet was further lysed using Buffer B [150 mM NaCl, 10 mM Tris-HCl (pH 7.4), 1 mM EGTA, 1 mM EDTA, Triton X-100, and protease/phosphatase inhibitor cocktail (Roche, USA)] in ice for 15 min, followed by centrifugation at 15,000 g for 30 min at 4°C. The supernatant was collected and used as nuclear extract. Then, the concentrations of proteins were assessed by using the Bio-Rad Protein Assay (Bio-Rad, Segrate, Italy) using BSA as standard, and 30 μg of total extracts from each sample were analyzed. Total proteins were separated on sodium dodecyl sulfate-polyacrylamide mini-gels and then transferred onto polyvinylidene fluoride membranes (Immobilon-P Transfer membrane, Millipore), blocked with PBS containing 5% nonfat dried milk (PM) for 45 min at room temperature, and subsequently incubated at 4°C overnight with a specific antibody for the cannabinoid receptor 1 (CB1R, 1:200; ThermoFisher, Scientific), for caspase-1 (CASP1, 1:1000, Abcam) and for pro-caspase-1 (pro-CASP1, 1:1000, Abcam).
The membranes were incubated with secondary antibodies, HRP-conjugated goat anti-mouse IgG or HRP-conjugated goat anti-rabbit IgG (1:2000; Santa Cruz Biotechnology, Inc.) for 1 h at room temperature. To ascertain that blots were loaded with equal amounts of protein lysates, they were also incubated with antibody for GAPDH HRP Conjugated (1:1000; Cell Signaling Technology). The relative expression of protein bands was visualized using an enhanced chemiluminescence system (Luminata Western HRP Substrates, Millipore) and protein bands were acquired and quantified with ChemiDoc™ MP System (Bio-Rad) and analyzed by using the software Image J.
Statistical data analysis
The statistical analyses to determine the proportion of differentially expressed genes between two samples on the read counts were performed with the Cufflinks Assembly & DE package version 2.0.
Immunocytochemical and Western blot data were analyzed by using the software GraphPad Prism 6.0 (GraphPad Software, La Jolla, CA) and applying the one-way ANOVA statistic test, followed by a Bonferroni post-hoc test for multiple comparisons. A p-value of < 0.05 was considered statistically significant.
Results
Cytofluorimetric characterization of hGMSCs
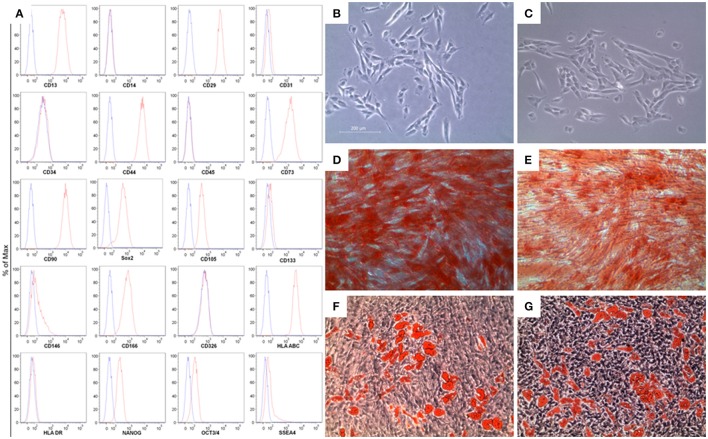
Flow cytometry was used to assess the MSC immunoprofile. In Figure 1, section A, we have shown that hGMSCs expressed surface molecules such as CD13, CD29, CD44, CD73, CD90, CD105, CD 146, CD166, and the human leukocyte antigen (HLA)-ABC, while they were negative for the subsequent markers: CD14, CD31, CD34, CD45, CD117, CD133, CD326, HLA-DR, according to the minimal criteria for defining MSCs (Dominici et al., 2006). Moreover, hGMSCs expressed pluripotency associated markers like OCT3/4, NANOG, SOX2, SSEA4.
Figure 1.
Characterization of gingiva-derived mesenchymal stem cells (hGMSCs). (A) Flow cytometry phenotype of hGMSCs at the second passage during in vitro cultures of surface (CD13, CD14, CD29, CD31, CD34, CD44, CD45, CD73, CD90, CD105, CD117, CD133, CD146, CD166, CD326, HLA-ABC, and HLA-DR) and intracellular (SSEA4, Oct3/4, Sox2, and NANOG) marker expression levels were detected. Red histograms show the distribution of each antigen expression, whereas Blue histograms represent the distribution of the respective background control. (B) The gingival connective tissue-derived mesenchymal stem cells showed colony-forming potency and plastic-adherent characteristics at day 6. (C) CBD-hGMSCs and CTR-hGMSCs showed spindle-shaped, fibroblast-like morphology under light microscope. (D) Osteogenic differentiation at day 21 after Alizarin Red S staining highlighted calcium deposits in CTR-hGMSCs and in (E) CBD-hGMSCs. (F) CTR-hGMSCs and (G) CBD-hGMSCs induced to a dipogenic differentiation after 28 days showed many oil droplets at cytoplasmic level stained with Oil Red O solution. Mag: 10X; bar: 200 μm.
Morphological and functional characterization of hGMSCs
The first CFU appeared after 6 days of the initiation of the primary culture. Under optical microscopy, primary cells showed spindle-shaped with fibroblastic-like morphology (Figure 1B). No differences in morphological features have been evidenced after CBD treatment for 24 h (Figure 1C). Moreover, we have proved the multipotent potential of hGMSCs to differentiate in osteoblasts and adipocytes. During osteogenic differentiation, at day 21, after Alizarin Red S staining, mineralized extracellular deposits were observed in CTR-hGMSCs and in CBD-hGMSCs (Figures 1D,E). For adipogenic differentiation, Oil Red O solution stained oil globules containing many lipid-rich vacuoles at cytoplasmic level in CTR-hGMSCs and in CBD-hGMSCs 28 days of culture in adipogenic medium (Figures 1E,F).
Global gene expression
To explore the effect of CBD treatment on the expression profile of the hGMSCs, we performed a high-throughput sequencing through the NGS platform. This technique allowed us to map about 14.320 for the CTR-hGMSCs group and 10.158 for CBD-hGMSCs group. Statistical analysis revealed 5843 genes differentially expressed following CBD treatment, among which 2798 were down-regulated, while 3045 were up-regulated.
CBD treatment modulated the expression of genes related to inflammation, immune response and apoptosis
Interestingly, here we found that CBD totally inhibited the expression of 25 genes in CBD-hGMSCs (False Discovery Rate (FDR), q-value < 0.05). According to GO processes, these suppressed genes belonging to the following GO categories: apoptotic process and regulation of apoptotic process (H1FX, CASP6, PDCL3, SIVA1, ING4, AQP1, MIEN1, CAPN3,CASP1, WWOX, IFI6, GAS1, PTN, PPP3CC, CARD16, PYCARD), inflammatory response (FLT3LG, DDT, NLRP3, BRCC36, LY96, ATAT1, PLGRKT) and innate immune response (HLA-H) and signal transduction (GNG10). In Table 1A have been reported the GO process categories, the expression levels of CTR-hGMSCs and CBD-hGMSCs (Exp_Value), the Fold Change (FC) and the False Discovery Rate (FDR) for each aforementioned genes.
Table 1A.
CBD treatment suppressed genes linked to apoptosis, inflammatory and innate immune responses.
| Gene | Description | Go processes | CTR-hGMSCs Exp_Value | CBD-hGMSCs Exp_Value | LOG10FC | FDR Q_VALUE |
|---|---|---|---|---|---|---|
| AQP1 | Aquaporin 1 | Apoptotic process, regulation of apoptotic process extrinsic apoptotic signaling pathway | 6.01 | 0.00 | −5.78 | 9.79e-05 |
| CAPN3 | Calpain 3 | Apoptotic process, regulation of apoptotic process, inflammatory response | 5.16 | 0.00 | −5.71 | 9.79e-05 |
| CARD16 | Caspase recruitment domain family member 16 | Regulation of apoptotic process, inflammatory response | 6.60 | 0.00 | −5.82 | 9.79e-05 |
| CASP1 | Caspase 1 | Apoptotic process, regulation of apoptotic process, inflammatory response | 8.22 | 0.00 | −5.92 | 9.79e-05 |
| CASP6 | Caspase 6 | Apoptotic process, regulation of apoptotic process, | 2.19 | 0.00 | −5.3 | 9.79e-05 |
| GAS1 | Growth arrest specific 1 | Regulation of apoptotic process, regulation of response to stimulus | 14.63 | 0.00 | −6.17 | 2.96e-02 |
| H1FX | H1 histone family member X | Apoptotic DNA fragmentation, nucleosome assembly | 6.41 | 0.00 | −5.81 | 9.79e-05 |
| IFI6 | Interferon alpha inducible protein 6 | Apoptotic process, innate immune response | 5.07 | 0.00 | −5.71 | 9.79e-05 |
| ING4 | Inhibitor of growth family member 4 | Apoptotic process, regulation of apoptotic process | 5.88 | 0.00 | −5.77 | 9.79e-05 |
| MIEN1 | Migration and invasion enhancer 1 | Apoptotic process, regulation of apoptotic process, cell redox homeostasis | 16.44 | 0.00 | −6.22 | 9.79e-05 |
| PDCL3 | Phosducin like 3 | Apoptotic process, protein folding | 11.78 | 0.00 | −6.07 | 9.79e-05 |
| PPP3CC | Protein phosphatase 3 catalytic subunit gamma | Apoptotic process, positive regulation of apoptotic process | 8.82 | 0.00 | −5.95 | 9.79e-05 |
| PTN | Pleiotrophin | Positive regulation of apoptotic process | 6.62 | 0.00 | −5.82 | 9.79e-05 |
| PYCARD | PYD and CARD domain containing | Regulation of apoptotic process, inflammatory response, innate immune response | 8.12 | 0.00 | −5.91 | 9.79e-05 |
| SIVA1 | SIVA1 apoptosis inducing factor | Apoptotic process, regulation of apoptotic process | 21.16 | 0.00 | −6.33 | 9.79e-05 |
| WWOX | WW domain containing oxidoreductase | Apoptotic process, regulation of apoptotic process | 8.16 | 0.00 | −5.91 | 2.14e-02 |
| FLT3LG | fms related tyrosine kinase 3 ligand | Regulation of apoptotic process, inflammatory response | 11.22 | 0.00 | −6.05 | 9.79e-05 |
| DDT | D-dopachrome tautomerase | Inflammatory response | 16.71 | 0.00 | −6.22 | 9.79e-05 |
| NLRP3 | NLR family, pyrin domain containing 3 | Inflammatory response, apoptotic process | 3.17 | 0.00 | −5.34 | 9.79e-05 |
| BRCC36 | BRCA1/BRCA2-containing complex subunit 3 | Inflammatory response, G2 DNA damage checkpoint | 5.02 | 0.00 | −5.70 | 9.79e-05 |
| LY96 | Lymphocyte antigen 96 | Inflammatory response, innate immune response, regulation of apoptotic process | 14.95 | 0.00 | −6.17 | 9.79e-05 |
| ATAT1 | Alpha tubulin acetyltransferase 1 | Inflammatory response | 4.88 | 0.00 | −5.69 | 9.79e-05 |
| PLGRKT | Plasminogen receptor with a C-terminal lysine | Inflammatory response | 5.68 | 0.00 | −5.75 | 9.79e-05 |
| HLA-H | Major histocompatibility complex, class I, H | Antigen processing and presentation, regulation of immune response | 8.08 | 0.00 | −5.91 | 9.79e-05 |
| GNG10 | G protein subunit gamma 10 | G-protein coupled receptor signaling pathway, signal transduction | 6.37 | 0.00 | −5.80 | 9.79e-05 |
The differential expression between CBD-hGMSCs and CTR-hGMSCs is given in Fold Change (log10 FC). GO processes indicate their classification in different biological processes. Instead, the statistical significance is indicated by False Discovery Rate (FDR), p-values ≤ 0.05 were considered statistically significant.
However, by interrogating the KEGG database for pathway analysis, we found only a pathway predicted to be statistically significant for this class of genes: the NOD-like receptor signaling (FDR = 0.006; Table 1B), an intracellular platform for the assembly of protein complexes known as inflammasomes, which includes NLRP3, CASP1, and PYCARD (Table 1A) (Shaw et al., 2011). NALP3 is a NOD-like receptor and together with the adaptor protein PYCARD forms a CASP1 activating complex, in response to a dangerous stimulus, known as the NALP3 inflammasome, which is the best characterized inflammasome complex (Schroder and Tschopp, 2010). Active CASP1 can eventually process IL-1 and IL-18 precursors, enhancing multiple pro-inflammatory pathways, including NF-κB, the mitogen-activated protein kinase (MAPK) and Interferon (IFN) (Fuentes-Antrás et al., 2014). Our gene expression results indicated a downregulation of the genes implicated in the NALP3-inflammasone pathway in CBD-HGMSCs.
Table 1B.
CBD suppressed the expression of genes belonging to the NOD-like receptor signaling pathway.
| KEGG pathway | Gene symbol | FDR |
|---|---|---|
| NOD-like receptor signaling pathway | NLRP3, CASP1, PYCARD | 0.006 |
Although for the other genes suppressed by CBD and reported in Table 1A, none statistical pathways were predicted by the KEEG software, these preliminary results have encouraged us to further investigate the potential of CBD to modulate genes implicated in immune response, inflammation and apoptosis.
CBD treatment attenuated the expression of pro-inflammatory genes
Gene expression profiling has showed that CBD treatment in hGMSCs downregulated the expression of genes codifying for pro-inflammatory cytokines (IL6ST, IL-1β, and IL-18), Interleukin receptors or subunits (IL1R1, IL11RA, IL13RA), the Toll-like receptor adaptor (MYD88), Interferon Gamma Receptors (IFNGR1 and IFNGR2), Mitogen-Activated Protein Kinases (MAPK1, MAPK12, and MAPK14), transcription factors (STAT3 and STAT6), the complex of NFκB (NFKB2, NFKB3/RELA), and the Matrix Metallopeptidase 3 (MMP3) (Table 2A). Specifically, these genes can be classified in the following GO terms: response to cytokine, regulation of immune response, regulation of defense response. etc. (Table 2A). The GO annotations, the expression levels (Exp-Value), the fold changes (FC) and the False Discovery Rates (FDR) of these genes differentially expressed between CBD-hGMSCs and CTR-hGMSCs have been reported in Table 2A.
Table 2A.
CBD treatment reduced the expression of pro-inflammatory genes.
| Gene | Description | Go processes | CTR-hGMSCs Exp_Value | CBD-hGMSCs Exp_Value | LOG10 FC | FDR Q_VALUE |
|---|---|---|---|---|---|---|
| CARD8 | Caspase Recruitment Domain Family Member 8 | Regulation of inflammatory response | 2.87 | 0.91 | −0.49 | 3.52e-04 |
| CTSB | Cathepsin B | Response to cytokine, regulation of inflammatory response, regulation of defense response | 150.13 | 104.14 | −0.15 | 9.79e-05 |
| HSP90AA1 | Heat shock protein 90 alpha family class A member 1 | Regulation of apoptotic process, negative regulation of apoptotic process, regulation of apoptotic signaling pathway | 423.83 | 724.1 | 0.23 | 9.79E-05 |
| IFNGR1 | Interferon Gamma Receptor 1 | Response to cytokine, regulation of immune response, regulation of defense response | 14.43 | 7.32 | −0.29 | 9.79e-05 |
| IFNGR2 | Interferon Gamma Receptor 2 | Response to cytokine, regulation of immune response, regulation of defense response | 40.42 | 35.86 | −0.05 | 4.5e-02 |
| IL11RA | Interleukin 11 Receptor Subunit Alpha | Response to cytokine | 23.77 | 12.22 | −0.28 | 9.79e-05 |
| IL13RA1 | Interleukin 13 Receptor Subunit Alpha 1 | Response to cytokine, inflammatory response, cell surface receptor signaling pathway, | 6.01 | 0.60 | −0.99 | 9.79e-05 |
| IL1B | Interleukin 1 Beta | Response to cytokine, inflammatory response, regulation of inflammatory response | 6.48 | 4.71 | −0.13 | 1.91 e-03 |
| IL1R1 | IL1R1 Interleukin 1 Receptor Type 1 | Response to cytokine, regulation of defense response, regulation of inflammatory response | 6.48 | 4.71 | −0.13 | 1.90e-03 |
| IL6ST | Interleukin 6 | Response to cytokine, regulation of immune response, regulation of defense response, regulation of inflammatory response | 24.86 | 22.91 | −0.03 | 0.021783 |
| IL18 | Interleukin 18 | Response to cytokine, inflammatory response, regulation of inflammatory response | 0.89 | −1.06 | −0.31 | 2.58e-05 |
| MAPK1 | Mitogen-Activated Protein Kinase 1 | Response to cytokine, inflammatory response, regulation of inflammatory response, MAPK cascade | 35.18 | 27.59 | −0.10 | 2.82e-03 |
| MAPK12 | Mitogen-Activated Protein Kinase 12 | Inflammatory response, regulation of inflammatory response, MAPK cascade | 7.16 | 4.35 | −0.21 | 8.23e-03 |
| MAPK14 | Mitogen-Activated Protein Kinase 14 | Inflammatory response, regulation of immune response, regulation of defense response | 14.06 | 5.83 | −0.38 | 9.79e-05 |
| MMP3 | Matrix Metallopeptidase 3 | Inflammatory response, regulation of inflammatory response, extracellular matrix disassembly | 85.78 | 55.05 | −0.19 | 9.79e-05 |
| MYD88 | Myeloid Differentiation Primary Response Gene 88 | Response to cytokine, regulation of immune response, regulation of defense response, regulation of inflammatory response | 12.55 | 2.62 | −0.67 | 9.79e-05 |
| NFKB2 | Nuclear Factor Kappa B Subunit 2 | Response to cytokine, regulation of immune response | 8.64 | 5.53 | −0.19 | 4.50e-05 |
| NFKBIA | NFKB Inhibitor Alpha | Response to cytokine, inflammatory response, negative regulation of NF-kB transcription factor activity | 28.48 | 34.80 | 0.08 | 1.34e-02 |
| RELA | RELA Proto-Oncogene, NF-κB Subunit | Response to cytokine, regulation of immune response, regulation of defense response | 17.21 | 11.29 | −0.18 | 9.79e-05 |
| STAT3 | Signal Transducer And Activator Of Transcription 3 | Response to cytokine, inflammatory response, regulation of inflammatory response | 45.10 | 20.27 | −0.34 | 9.79e-05 |
| STAT6 | Signal Transducer And Activator Of Transcription 6 | Response to cytokine, regulation of immune response, inflammatory response, regulation of inflammatory response | 47.66 | 37.66 | −0.10 | 9.79e-05 |
Gene expression levels of CTR-hGMSCs and CBD-hGMSCs are given in log10, as well as the Fold Changes (FC). GO processes indicate their classification in different biological processes. Instead, the statistical significance is indicated by the False Discovery Rate (FDR), p-values ≤ 0.05 were considered statistically significant.
According to KEEG pathway predictions, this group of genes can be clustered in the most following significant pathways: the NOD-like receptor signaling pathway, the TNF signaling pathway, the Jak-STAT signaling pathway and the NF-kB signaling pathway (Table 2B). These results altogether indicated that CBD negatively regulates differential pathways involved in inflammatory responses, but in particular the NOD-like receptor signaling pathway.
Table 2B.
CBD negatively modulated the transcription of genes involved in NOD-like receptor signaling pathway and other inflammatory pathways.
| KEGG pathway | Gene symbol | FDR |
|---|---|---|
| NOD-like receptor signaling pathway | CARD8, IL1B, IL18, MAPK1, MAPK12, MAPK14, NFKBIA, RELA | 2.3e-14 |
| TNF signaling pathway | MMP3, IL1B, MAPK1, MAPK12, MAPK14, NFKBIA, RELA | 1.43e-09 |
| Jak-STAT signaling pathway | STAT6, IFNGR1, IFNGR2, IL13RA1, IL6ST, STAT3 | 1.43e-08 |
| NF-kB signaling pathway | MYD88, IL1B, IL1R1, NFKBIA, NFKB2, RELA | 2.19e-08 |
The statistical significance is indicated by FDR, p-values of ≤ 0.05 were considered statistically significant.
CBD prevented the activation of the NALP3-inflammasome in hGMSCs
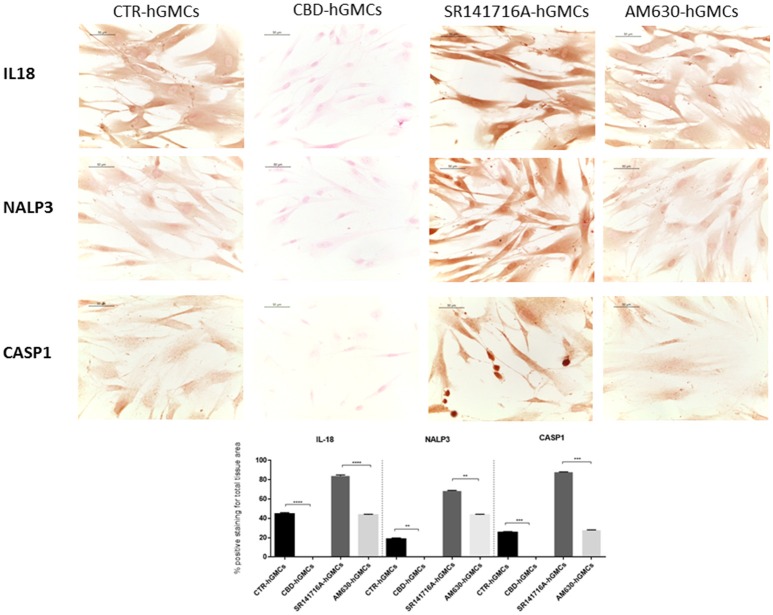
The NALP3-inflammasome has been correlated with several inflammatory conditions and increasing evidences have indicated that the inhibition of the inflammasome could be a promising strategy for treating inflammatory disorders and also for preventing allograft-rejections in cell-based therapies (Foley, 2013; Coll et al., 2015; Shah et al., 2015). Our gene expression data have revealed a downregulation of several genes codifying for key players of the NALP3-inflammasome in CBD-hGMSCs. In order to confirm the ability of CBD to modulate the NALP3-inflammasome, we have performed immunocytochemistry and western blot analyses. Achieved immune-localization data have shown that NALP3, IL-18, and CASP1 were completely negative in CBD-hGMSCs, in a significant manner compared to CTR-hGMSCs (Figure 2). A representative negative control of immunocytochemistry is showed in Supplementary Material. The expression levels of CASP1 were also investigated by Western blotting and normalized on pro-CASP1, showing a reduced CASP1 expression in CBD-hGMSCs compared to CTR-hGMSCs (Figure 3).
Figure 2.
Immunocytochemical staining for IL18, NALP3 and CASP1. CBD-hGMSCs showed a negative staining for IL-18, NALP3, and CASP1 compared to CTR-hGMSCs. Instead, hGMSCs-SR141716A and hGMSCs-AM630 showed a significant positive staining for IL-18, NALP3, and CASP1 compared to CBD-hGMSCs. The graph represented the densytometric quantitative analysis. For IL-18 CTR-hGMSCs vs. CBD-hGMSCs ***p < 0.001; SR141716A-hGMSCs vs. AM630-hGMSCs ****p < 0.0001. For NALP3 CTR-hGMSCs vs. CBD-hGMSCs **p < 0.01; SR141716A-hGMSCs vs. AM630-hGMSCs **p < 0.01. For CASP-1 CTR-hGMSCs vs. CBD-hGMSCs ***p < 0.001; SR141716A-hGMSCs vs. AM630-hGMSCs ***p < 0.001.
Figure 3.
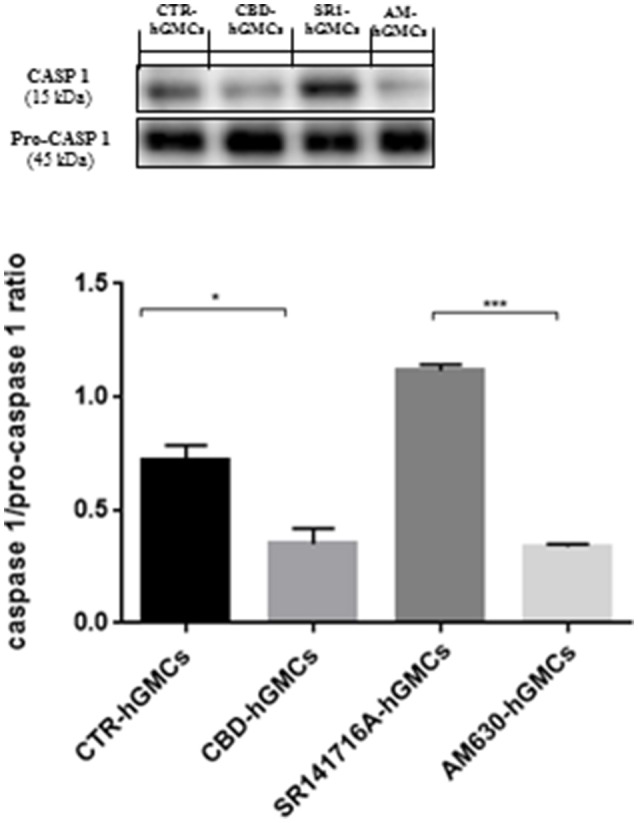
Western blot analysis for CASP1. CASP1 expression is decreased in CBD-hGMSCs compared to CTR-DMSO). The levels of CASP1 are significant increase in SR141716A-hGMSCs compared with CBD-hGMSCs. CTR-hGMSCs vs. CBD-hGMSCs *p < 0.05; SR141716A-hGMSCs vs. AM630-hGMSCs ***p < 0.001.
Moreover, in order to understand whether the inhibition of the inflammasome by CBD could be mediated by the cannabinoid receptors (CB1R and CB2R), we have pre-treated hGMSCs with selective receptor antagonists for CB1R (SR141716A) and CB2R (AM630), before CBD treatment for 24 h. Immunochemistry has showed that SR141716A reverted completely the downregulation of NALP3, IL-18, and CASP1 induced by CBD, instead AM630 was also able to antagonize only partially the CBD effects. Western blotting has confirmed that blocking CB1R reverted the CBD-mediated reduction of CASP1 (Figure 3).
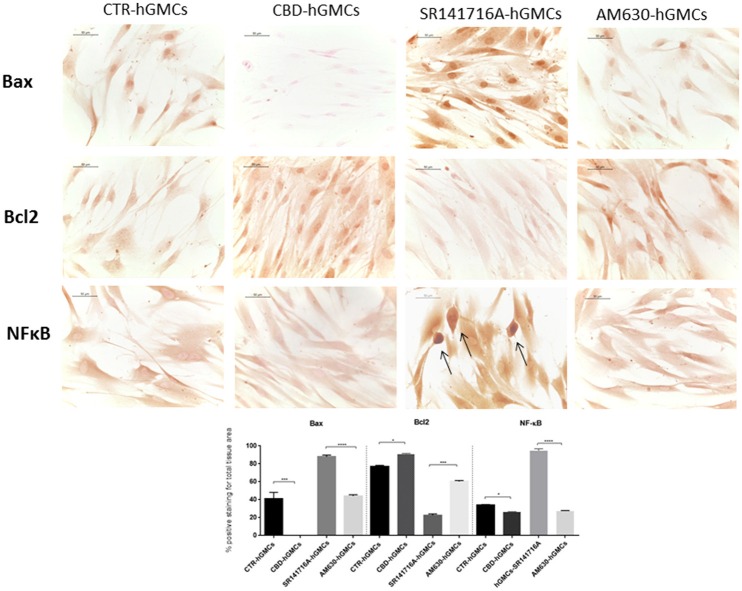
Moreover, we evaluated the expression of NF-κB since it is considered as a regulator of NALP3 activity (Bauernfeind et al., 2009), finding that NF-κB levels were statically reduced by CBD treatment, as demonstrated by immunocytochemistry analyses (Figure 4). These results suggested that CBD promotes the inactive state of the NALP3-inflammasome pathway.
Figure 4.
Immunocytochemical staining for Bax, Bcl2 and NFκB. CBD-hGMSCs showed a negative staining for Bax, a reduced expression for NF-KB and a positive staining for Bcl2, compared to CTR-hGMSCs. hGMSCs-SR141716A showed a marked positive cytoplasmatic/nuclear staining for NF-kB, as indicated by the arrows. Moreover, hGMSCs-SR141716A showed an increased nuclear expression for Bax and reduced expression for Bcl2 at both cytoplasmatic and nuclear compartment, compared to CBD-hGMSCs. Instead, hGMSCs-AM630 showed a reduced expression for Bax and Bcl2 compared to CBD-hGMSCs. Whereas, no differences statistically differences were found for the NF-kB expression between CBD-hGMSCs and hGMSCs-AM630. The graph represented the densytometric quantitative analysis. For Bax CTR-hGMSCs vs. CBD-hGMSCs ***p < 0.001; SR141716A-hGMSCs vs. AM630-hGMSCs ****p < 0.0001. For Bcl2 CTR-hGMSCs vs. CBD-hGMSCs *p < 0.05; SR141716A-hGMSCs vs. AM630-hGMSCs ***p < 0.001. For NF-kB CTR-hGMSCs vs. CBD-hGMSCs *p < 0.05; SR141716A-hGMSCs vs. AM630-hGMSCs ****p < 0.0001.
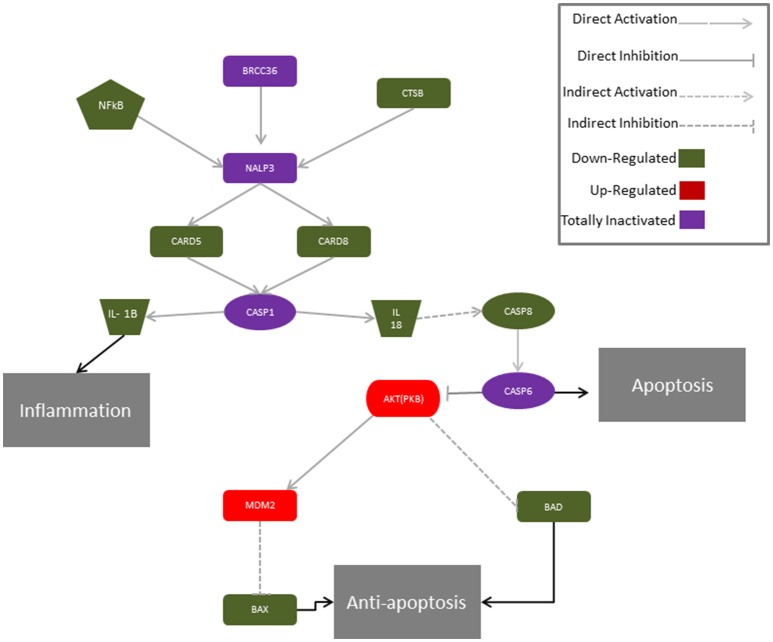
Moreover, only the CB1 agonist has shown to antagonize the CBD-mediated reduction of NF-kB, indicating that CBD may act in a CB1R dependent manner, while CB2R is likely to be not involved in this signaling. These results might suggest that CBD can interact with both CB1R and CB2R, although it acts preferably via a CB1R mechanism, showing a selectivity for the CB1R respect to the CB2R in hGMSCs. Additionally, Western blot results (Figure 5) have showed an increased CB1R expression in CBD-hGMSCs compared to control groups (DMSO-hGMSCs and CTR-hGMSCs). A schematic representation of the NALP3-inflammasome pathway in CBD-hGMSCs is illustrated in the Figure 6.
Figure 5.
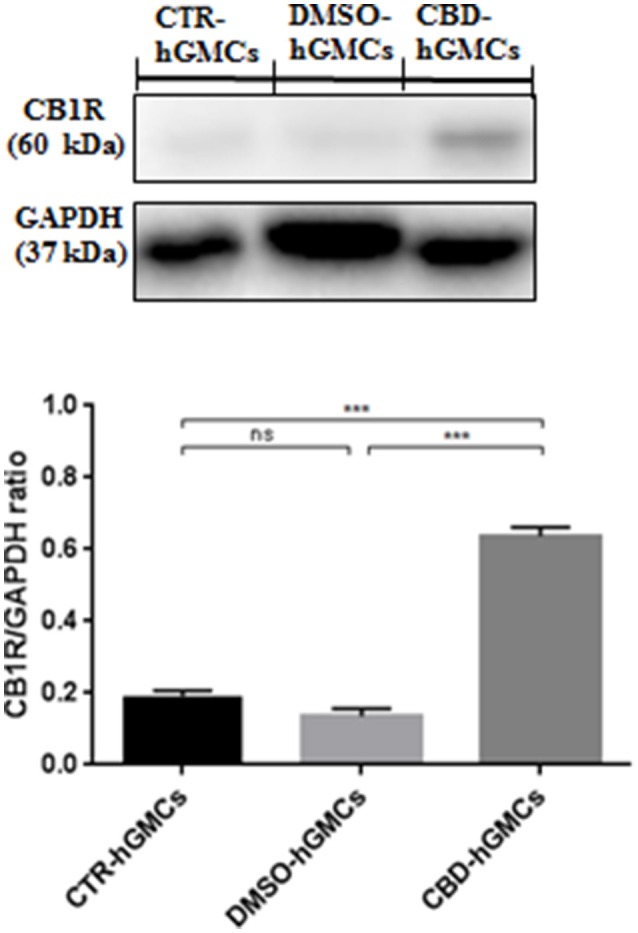
Western blot analysis for CB1R. CB1R expression is increased in CBD-hGMSCs compared to control groups (DMSO-hGMSCs and CTR-hGMSCs). ***p < 0.001. ns, no statistical differences.
Figure 6.
Proposed molecular mechanism for CBD modulation of the genes of the NALP3-inflammasome in hGMSCs. The activation of the NALP3-inflammasome is regulated by NF-kB which promotes NALP3 deubiquitination via activation of BRCC36. NALP3 interacts with CASP1 through CARD5 and CARD8. Activated CASP1 cleaves pro-IL-1 beta and pro-IL-18, leading to subsequent release of mature cytokines which trigger inflammation. In parallel, IL-18 indirectly via CASP8 and CASP6 can stimulate apoptosis. In hGMSCs, BRCC6, NALP3 and CASP1 transcripts are suppressed (violet) and the expression of the downstream mediators (CARD5, CARD8, IL1β, IL18, CASP8) is downregulated (green), while CASP8 is totally suppressed (violet). Instead, the pro-survival AKT1 and MDM2 were upregulated (red), leading to the downregulation of BAX and BAD (green).
CBD reduced the expression of pro-apoptotic genes
Following infusion of MSCs, the hostile environment could induce apoptosis few hours after transplantation. Thus, researchers had tried to develop new strategies to improve survival of MSCs, such as preconditioning of MSCs by cultivation under peroxide hydrogen, hypoxic and serum deprivation (Farzaneh Taban et al., 2016).
Here, we have previously showed that CBD treatment in hGMSCs inhibited the expression of several genes correlated to the apoptosis, according to the GO biological processes. In this study, we have also found that CBD downregulated the expression of additional genes related to apoptosis, among which we detected genes codifying for the TNF receptor superfamily members (TNFRSF10B, TNFRSF11B, TNFRSF12A, and TNFRSF19), the initiator caspases (CASP4 and CASP8), the pro-apoptotic mediators (BAX, BAD, BID, BCL7B, BCL2L13, CYCS), and the apoptotic peptidase APAF-1 (Table 3A). Some of these genes were predicted by KEGG to be involved in the apoptotic pathway in a significant manner, as reported in Table 3B.
Table 3A.
CBD treatment reduced the expression of pro-apoptotic genes.
| Gene | Description | Go processes | CTR-hGMSCs Exp_Value | CBD-hGMSCs Exp_Value | LOG10FC | FDR Q_VALUE |
|---|---|---|---|---|---|---|
| AKT1 | AKT serine/threonine kinase 1 | Regulation of apoptotic process, negative regulation of apoptotic process, regulation of apoptotic signaling pathway | 47.16 | 59.52 | 0.10 | 9.79e-05 |
| APAF1 | Apoptotic peptidase activating factor 1 | Regulation of apoptotic process, positive regulation of cysteine-type endopeptidase activity involved in apoptotic process, regulation of apoptotic signaling pathway | 7.14 | 5.34 | −0.12 | 2.77e-03 |
| BAD | BCL2 associated agonist of cell death | Positive regulation of cysteine-type endopeptidase activity involved in apoptotic process, regulation of apoptotic signaling pathway | 12.89 | 11.18 | −0.06 | 4.34e-05 |
| BAX | BCL2 associated X, apoptosis regulator | Regulation of apoptotic process, extrinsic apoptotic signaling pathway | 37.48 | 13.20 | −0.45 | 9.79e-05 |
| BCL2L13 | BCL2 like 13 | Regulation of apoptotic process, positive regulation of cysteine-type endopeptidase activity involved in apoptotic process | 9.96 | 4.33 | −0.36 | 9.79e-05 |
| BCL7B | BCL tumor suppressor 7B | Regulation of apoptotic process, regulation of apoptotic signaling pathway | 8.40 | 1.58 | −0.72 | 9.79e-05 |
| BID | BH3 interacting domain death agonist | Positive regulation of cysteine-type endopeptidase activity involved in apoptotic process, regulation of apoptotic process, extrinsic apoptotic signaling pathway | 5.33 | 1.37 | −0.60 | 9.79e-05 |
| CASP4 | Caspase 4 | Regulation of apoptotic process, negative regulation of apoptotic process, regulation of apoptotic signaling pathway | 74.8 | 23.66 | −0.50 | 9.79e-05 |
| CASP8 | Caspase 8 | Positive regulation of cysteine-type endopeptidase activity involved in apoptotic process, regulation of apoptotic process, extrinsic apoptotic signaling pathway | 10.07 | 3.79 | −0.42 | 9.79e-05 |
| CYCS | Cytochrome c, somatic | Positive regulation of cysteine-type endopeptidase activity involved in apoptotic process, negative regulation of apoptotic process, regulation of apoptotic signaling pathway | 9.07 | 3.42 | −0.42 | 9.79e-05 |
| HSP90AA1 | Heat shock protein 90 alpha family class A member 1 | Regulation of apoptotic process, negative regulation of apoptotic process, regulation of apoptotic signaling pathway | 423.83 | 724.1 | 0.23 | 9.79e-05 |
| MDM2 | MDM2 proto-oncogene | Regulation of apoptotic process, negative regulation of apoptotic process, regulation of apoptotic signaling pathway | 15.7341 | 17.81 | 0.05 | 9.728e-02 |
| PIK3CA | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha | Regulation of apoptotic process, negative regulation of apoptotic process, regulation of apoptotic signaling pathway | 14.77 | 18.38 | 0.095 | 1.35e-03 |
| PIK3CB | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit beta | Regulation of apoptotic process, negative regulation of apoptotic process, regulation of apoptotic signaling pathway | 3.33 | 5.60 | 0.22 | 9.79e-05 |
| TNFRSF10B | TNF receptor superfamily member 10B | Positive regulation of cysteine-type endopeptidase activity involved in apoptotic process, regulation of apoptotic process, extrinsic apoptotic signaling pathway | 63.87 | 38.70 | −0.21 | 9.79e-05 |
| TNFRSF11B | TNF receptor superfamily member 11B | Regulation of apoptotic process, regulation of apoptotic signaling pathway | 136.65 | 97.87 | −0.14 | 9.79e-05 |
| TNFRSF12A | TNF receptor superfamily member 12A | Regulation of apoptotic process, extrinsic apoptotic signaling pathway | 8.54 | 2.97 | −0.47 | 9.07e-3 |
| TNFRSF19 | TNF receptor superfamily member 12A | Regulation of apoptotic process, regulation of apoptotic signaling pathway | 25.96 | 9.10 | −0.41 | 9.79e-05 |
Both the expression levels (Exp_Value) and the fold changes (FC) are expressed in log10. GO terms indicate their classification in different biological processes. Instead, the statistical significance is indicated by False Discovery Rate (FDR), p-values ≤ 0.05 were considered statistically significant.
Table 3B.
CBD modulated the apoptotic pathway in hGMSCs.
| KEGG pathway | Gene symbol | FDR |
|---|---|---|
| Apoptosis | APAF-1, AKT-1, BAX, BAD, BID, CYCS, CASP8, MDM2, PIK3CA PIK3CB, TNFRSF10B | 4.01e-17 |
The statistical significance is indicated by the False Discovery Rate (zFDR), p-values ≤ 0.05 were considered statistically significant.
In parallel, CBD enhanced the transcription of the phosphoinositid-3 kinase (PIK3) subunits (PIK3CA and PIK3CB) and AKT1 serine/threonine kinase 1 (AKT1). The upregulation of the PI3K/Akt signaling has been associated with increased stem cell survival (Hossini et al., 2016).
Moreover, we have observed a negative staining for the pro-apoptotic mediator Bax in CBD-hGMSCs and positive staining for the anti-apoptotic protein Bcl-2, by immunocytochemistry. These data confirmed that CBD treatment inhibited apoptosis in hGMSCs (Figure 4). By pre-treating cells with the cannabinoid receptor agonists, we found that this inhibition seems to be mainly mediated by the CB1R (Figure 4). Our results suggested that CBD treatment may improve stem cell survival through two complementary strategies: by reducing the expression of pro-apoptotic mediators and by promoting the anti-apoptotic ones.
CBD modified the immunophenotype of hGMSCs
MSCs have been reported to exert immunosuppressive properties in the host (Schu et al., 2012). However, evidence also suggests that they can also elicit a allogeneic immune response (Nauta et al., 2006). Thus, we investigated whether CBD was able to modulate the cell surface expression of immunogenic markers. We detected a differential expression of the well-known mesenchymal markers: specifically CD13, CD29, and CD73 were upregulated in CBD-hGMSCs (Table 4), while CD44, CD90, and CD166 were downregulated (Table 4).
Table 4.
Mesenchymal markers differentially expressed between CTR-hGMSCs and CBD-hGMSCs.
| Gene | Description | Go processes | CBD-HGMSCS Exp_Value | CBD-HGMSCS Exp_Value | LOG10FC | FDR Q_VALUE |
|---|---|---|---|---|---|---|
| CD13 (ANPEP) | Alanyl Aminopeptidase | Single organismal cell-cell adhesion | 114.68 | 120.90 | 0.02 | 4.36e-02 |
| CD29 (ITGB1) | Integrin Subunit Beta 1 | Cell adhesion Cell-matrix adhesion Single organismal cell-cell adhesion | 511.60 | 666.69 | 0.11 | 9.79e-05 |
| CD73 (NT5E) | Nt5e 5'-Nucleotidase Ecto | Cell adhesion Single organismal cell-cell adhesion | 314.08 | 360.84 | 0.06 | 9.79e-05 |
| CD44 | CD44 Molecule | Cell adhesion Cell-matrix adhesion Single organismal cell-cell adhesion | 306.34 | 295.16 | −0.01 | 2.96e-02 |
| CD90 (THY1) | Thy-1 Cell Surface Antigen | Cell adhesion Cell-matrix adhesion Single organismal cell-cell adhesion | 75.20 | 58.37 | −0.11 | 9.79e-05 |
| CD166 (ALCAM) | Activated Leukocyte Cell Adhesion Molecule | Cell adhesion, single organismal cell-cell adhesion | 58.64 | 53.18 | −0.04 | 1.30e-02 |
The expression levels (Exp_Value) and the fold changes (FC) are expressed in log10. The statistical significance is indicated by the False Discovery Rate (FDR) value, p-values ≤ 0.05 were considered statistically significant.
Furthermore, we found that following CBD treatment, other genes codifying for surface antigens were differentially expressed, in particular those involved in the inhibition of the immune system activation such as CD47, CD55, and CD276 were upregulated whereas, the expression levels of the costimulatory molecules CD40, CD46, CD81, and CD82, as well as the levels of the T-cells and macrophage activators CD59, CD68, and CD99 and, the immune surface markers CD109 and CD151 were decreased by CBD (Table 5). The GO process categories, the expression levels (Exp_Value), the fold change (FC) and the False Discovery Rate (FDR) have been given in Table 5. None pathway statistically significant was found for this group of genes, according to KEEG database.
Table 5.
CBD modulated the transcription of genes codifying for the antigenic repertoire of hGMSCs.
| Gene | Description | Go processes | CBD-HGMSCS Exp_Value | CBD-HGMSCS Exp_Value | LOG10FC | FDR Q_VALUE |
|---|---|---|---|---|---|---|
| CD276 | CD276 molecule | Regulation of immune system process, negative regulation of T cell proliferation, negative regulation of inflammatory response | 20.86 | 25.95 | 0.09 | 3.64e-4 |
| CD47 | CD47 molecule | Regulation of immune system process, leukocyte migration, integrin-mediated signaling pathway | 13.11 | 16.64 | 0.10 | 2.77e-03 |
| CD55 | CD55 molecule | Regulation of immune system process, Complement and coagulation cascades | 12.29 | 15.22 | 0.09 | 0.45e-02 |
| CD40 | CD40 molecule | Immune system process, immune response, regulation of immune system process | 5.60 | 1.73 | −0.50 | 9.79e-05 |
| CD46 | CD46 molecule | Regulation of immune system process, T cell mediated immunity | 54.35 | 42.68 | −0.10 | 9.79e-05 |
| CD59 | CD59 molecule | Immune system process, immune response, regulation of immune system process | 130.87 | 78.57 | −0.22 | 9.79e-05 |
| CD68 | CD68 molecule | Immune system process | 134.37 | 116.70 | −0.06 | 9.79e-05 |
| CD81 | CD81 molecule | Regulation of immune system process, cell surface receptor signaling pathway | 404.95 | 270.42 | −0.17 | 9.79e-05 |
| CD82 | CD82 molecule | Cell surface receptor signaling pathway, regulation of immune system process | 24.53 | 15.88 | −0.18 | 9.79e-05 |
| CD99 | CD99 molecule | Immune system process, cell adhesion | 685.80 | 638.84 | −0.03 | 3.64e-03 |
Both the expression levels (Exp_Value) and the fold changes (FC) are expressed in log10. GO processes indicate their classification in different biological processes. Instead, the statistical significance is indicated by False Discovery Rate (FDR), p-values ≤ 0.05 were considered statistically significant.
Discussion
The ability of hGMSCs to self-renew, to differentiate into different tissues, together with their easy accessibility and abundance, renders them a promising alternative source of MSCs for regenerative medicine. However, prior to their clinical applications, further investigations are required to ensure that they do not elicit immune or inflammatory responses that could affect their efficacy and survival in vivo.
In this work, we asked whether preconditioning donor cells (hGMSCs) with CBD, before transplantation in the host, could modify their gene expression profile, improving their therapeutic efficacy.
Global gene expression data have showed that CBD modulated the expression of 5843 genes in hGMSCs, among which, we found that CBD treatment downregulated the expression of genes correlated to inflammation and apoptosis. Specifically, we have detected a reduced expression of several key genes involved in the NOD-like receptor signaling and more specifically, belonging to the NALP3 inflammasome complex. The NALP3-inflammasome could be considered as an integrated signaling system which converges multiple signaling pathways such as cell death and innate immune responses (Nakanishi et al., 2001). Indeed, following a dangerous stimulus, NALP3 binds its adaptor protein PYCARD to assembly the inflammasome complex which recruits CASP1 (Latz et al., 2013). CASP1 in turn promotes the cleavage of the pro-inflammatory cytokines such as IL-1β and IL-18 (Lalor et al., 2011), which promote the inflammatory and immune responses (Nakanishi et al., 2001; Netea et al., 2010). Here, we have found that CBD promotes the inactive state of the NALP3-inflammasome pathway in hGMSCs by inhibiting some key members of this signaling, including NALP3, CASP1, and IL18, via a preferably CB1R mechanism, which does not exclude completely the CB2R involvement. Moreover, we have found that CBD led to increased CB1R expression in hGMSCs. These findings are in according with previous studies which have showed an increased CB1R expression following in vitro exposure to exogenous or endogenous cannabinoids (Börner et al., 2007; Laprairie et al., 2013).
Additionally, we have observed a reduced expression of NF-κB, a crucial transcription factor which promotes the transcription of several pro-inflammatory mediators, including NALP3, both at transcriptional and protein levels. Moreover, the blockade of CB1R, through the administration of a specific antagonist, led to increased NF-kB expression, while no statistically changes were observed after the exposure to CB2R antagonist, indicating a direct involvement of CB1R in this signaling.
Overall, our results have suggested that CBD negatively regulated the NALP3-inflammasome activation, mainly via a CB1R mechanism. Although a direct modulation of the inflammasome complex by the cannabinoid receptors CB1R and CB2R, has been already investigated by previous studies, it role remains still controversial. Indeed, it has been reported that CB2R activation, by inhibiting the NLRP3-inflammasome pathway, played a protective role in experimental models of inflammatory and autoimmune disorders (Shao et al., 2014; Ke et al., 2016). Conversely, Jourdan et al. (2013) have showed that CB1R mediated the NALP3-inflammasome activation and, thus inflammation in macrophages derived from rat model of diabetes.
However, accumulating evidences have highlighted the NALP3 inflammasome as potential target for treating inflammation and for preventing graft failures (Foley, 2013; Jankovic et al., 2013; Coll et al., 2015), thus we proposed that CBD treatment, by antagonizing the NALP3 inflammasome activation in hGMSCs, renders them less predispose to trigger inflammation, thus more resistant to the hostile microenvironment, once transplanted in the host. All these evidences suggested that preconditioning hGMSCs with CBD might be a strategy to improve cell engraftment in the context of stem cell-based therapy.
Following MSCs infusion, the hostile environment could induce apoptosis few hours after transplantation, as consequence, new strategies to counteract apoptosis or to improve survival of MSCs are needed. In this study, we have showed that CBD pre-treatment in hGMSCs correlated with the downregulation of several genes involved in apoptosis. Moreover, we have demonstrated that Bax expression was totally suppressed in CBD-hGMSCs, while Bcl2 levels were increased, confirming an anti-anti-apoptotic role for CBD in these cells. We have also found that this pro-survival effect of CBD could be mediated by the CB1R. These results are corroborated by other studies which have showing that CB1R activation correlated with reduced apoptosis (Gómez del Pulgar et al., 2000) and increased mesenchymal stem cell survival (Gowran et al., 2013).
Immune rejection is one of the major complication that may occur with stem cell transplantation. The main targets for the immune response to transplanted grafts are major histocompatibility complex (MHC)-encoded molecules (Ayala García et al., 2012). Despite, the hGMSCs have showed to express the human leukocyte antigen (HLA) major histocompatibility complex (MHC) class I and to be negative for MHC II (Castro-Manrreza et al., 2015), they have still not been adequately characterized with respect to their immunogenic properties. Several studies reported that CBD could have a role in the inhibition of the graft rejection T-cells mediated (Kaplan et al., 2008; Nagarkatti et al., 2010; Robinson et al., 2013), exerting immunosuppressive properties.
Here, we have found that CBD treatment modified the expression of genes codifying for surface antigens in hGMSCs. Specifically, in CBD-hGMSCs we detected upregulated levels of the well-known MSCs markers CD13, CD29, CD73, CD13, CD29, CD73, involved in promoting MSCs adhesion and migration. The overexpression of these markers has been associated with improved MSCs therapeutic efficacy (Ode et al., 2011; Rahman et al., 2014). Moreover, CBD treatment led to the down-regulation of the MSCs markers CD44, CD90, CD166. Interestingly, the downregulation of CD90, CD44, and CD166 has been linked to a more differentiated stem cell phenotype and to a less tumorigenic potential (Moraes et al., 2016). Additionally, we detected others surface markers involved the activation of the immune system which expression was downregulated by CBD such as CD109, CD151, CD40, CD46, CD59, CD68, CD81, CD82, CD99. CD109, and CD151 are surface markers expressed by different cell types of the immune system (Zhang et al., 2015; Seu et al., 2016). CD59 and CD99 and play a role in of T-cells activation and migration (Kimberley et al., 2007), whereas CD68 promotes macrophage activation (Ji, 2012). CD40, CD46, CD81, and CD82 are immune-costimulatory antigens which promote the activation of different cells of the immune system (Levy et al., 1998; Shibagaki et al., 1998; Zaffran et al., 2001; Elgueta et al., 2009; Johnson and Ruffell, 2009; Saleh et al., 2013). In parallel, CBD treatment led to the upregulation of antigens which negatively regulate T cell activation via the disruption of APC/T interactions as well as of antigen processing and presentation, such as CD47, CD55, and CD276. In particular, CD47 is expressed on most innate immune cells able to dampen the inflammatory immune response (Latour et al., 2001). Instead, CD276 (B7-H3) is a negative regulator of T cell proliferation, as well as CD55 (DAF), a complement inhibitor protein that suppresses T cells activation in vivo. Of note, transgenic overexpression of human DAF is a strategy to reduce the immune rejection complement-mediated in xenotransplants (Liu et al., 2005). Based on our results, we suggested that CBD treatment by modifying the molecular immunophenotype of hGMSCs, make them less immunogenic. A less immunogenic phenotype can potentially prevent the allogenic immune responses without requiring a systemic immune suppression in the host.
In conclusion, we found that CBD negatively regulated the NALP3-inflammasome activation and cell death, via the inhibition of apoptosis, through a CB1R-dependent. Moreover, CBD treatment reduced the immunogenicity of hGMSCs.
In the present study, we will provide a new simple and reproducible method for preconditioning hGMSCs with CBD before transplantation. This method could represent an interesting strategy for reducing the risk of immune reactions following cell transplantation, and in parallel, for increasing their survival and thus, their long-term therapeutic efficacy.
Ethics statement
The protocol and informed consent from human gingival biopsies were carried out in accordance with the approved guidelines of Medical Ethics Committee at the Medical School, “G. d'Annunzio” University, Chieti, Italy (n°266/17.04.14).
Author contributions
RL wrote the manuscript. DS provided technical support for NGS experiments and data analysis. FD and MM were involved in acquisition, analysis and interpretation of data and helped in revising the manuscript critically for important intellectual content. GG and FP have performed CBD isolation and purification. AP has performed the recruitment of patients for hGMSCs explanation. PB, EM, and OT conceived and designed the experiments, and were involved in revising the manuscript.
Funding
This study was supported by current research funds 2016 of Health Ministry to IRCCS Centro Neurolesi “Bonino-Pulejo.”
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fphys.2016.00559/full#supplementary-material
References
- Ayala García M. A., González Yebra B., López Flores A. L., Guaní Guerra E. (2012). The major histocompatibility complex in transplantation. J. Transplant. 2012, 1–7. 10.1155/2012/842141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauernfeind F. G., Horvath G., Stutz A., Alnemri E. S., MacDonald K., Speert D., et al. (2009). Cutting edge: NF-kappaB activating pattern recognition and cytokine receptors license NLRP3 inflammasome activation by regulating NLRP3 expression. J. Immunol. 183, 787–791. 10.4049/jimmunol.0901363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Börner C., Höllt V., Sebald W., Kraus J. (2007). Transcriptional regulation of the cannabinoid receptor type 1 gene in T cells by cannabinoids. J. Leukoc. Biol. 81, 336–343. 10.1189/jlb.0306224 [DOI] [PubMed] [Google Scholar]
- Castro-Manrreza M. E., Montesinos J. J., Castro-Manrreza M. E., Montesinos J. J. (2015). Immunoregulation by mesenchymal stem cells: biological aspects and clinical applications. J. Immunol. Res. 2015, 1–20. 10.1155/2015/394917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galve-Roperh I., Chiurchiù V., Díaz-Alonso J., Bari M., Guzmán M., Maccarrone M. (2013). Cannabinoid receptor signaling in progenitor/stem cell proliferation and differentiation. Prog. Lipid Res. 52, 633–650. 10.1016/j.plipres.2013.05.004 [DOI] [PubMed] [Google Scholar]
- Cianci E., Recchiuti A., Trubiani O., Diomede F., Marchisio M., Miscia S., et al. (2016). Human periodontal stem cells release specialized proresolving mediators and carry immunomodulatory and prohealing properties regulated by lipoxins. Stem Cells Transl. Med. 5, 20–32. 10.5966/sctm.2015-0163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coll R. C., Robertson A. A. B., Chae J. J., Higgins S. C., Muñoz-Planillo R., Inserra M. C., et al. (2015). A small-molecule inhibitor of the NLRP3 inflammasome for the treatment of inflammatory diseases. Nat. Med. 21:248. 10.1038/nm.3806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diomede F., Caputi S., Merciaro I., Frisone S., D'Arcangelo C., Piattelli A., et al. (2014). Pro-inflammatory cytokine release and cell growth inhibition in primary human oral cells after exposure to endodontic sealer. Int. Endod. J. 47, 864–872. 10.1111/iej.12230 [DOI] [PubMed] [Google Scholar]
- Dominici M., Le Blanc K., Mueller I., Slaper-Cortenbach I., Marini F., Krause D., et al. (2006). Minimal criteria for defining multipotent mesenchymal stromal cells. The international society for cellular therapy position statement. Cytotherapy 8, 315–317. 10.1080/14653240600855905 [DOI] [PubMed] [Google Scholar]
- Du L., Yang P., Ge S. (2016). Isolation and characterization of human gingiva-derived mesenchymal stem cells using limiting dilution method. J. Dent. Sci. 11, 304–314. 10.1016/j.jds.2016.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgueta R., Benson M. J., de Vries V. C., Wasiuk A., Guo Y., Noelle R. J. (2009). Molecular mechanism and function of CD40/CD40L engagement in the immune system. Immunol. Rev. 229, 152–172. 10.1111/j.1600-065X.2009.00782.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farzaneh Taban Z., Khatibi S., Halabian R., Mohammadi Roushandeh A. (2016). The effects of preconditioning on survival of mesenchymal stem cells in vitro. Gene Cell Tissue. 10.17795/gct-40229 [DOI] [Google Scholar]
- Fawzy El-Sayed K. M., Dörfer C. E. (2016). Gingival mesenchymal stem/progenitor cells: a unique tissue engineering gem. Stem Cells Int. 2016:7154327. 10.1155/2016/7154327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley J. F. (2013). The inflammasome and transplantation. Sci. Signal. 6:ec240 10.1126/scisignal.2004791 [DOI] [Google Scholar]
- Fuentes-Antrás J., Ioan A. M., Tuñón J., Egido J., Lorenzo O. (2014). Activation of toll-like receptors and inflammasome complexes in the diabetic cardiomyopathy-associated inflammation. Int. J. Endocrinol. 2014:847827. 10.1155/2014/847827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez del Pulgar T., Velasco G., Guzma M. (2000). The CB 1 cannabinoid receptor is coupled to the activation of protein kinase B/Akt. Biochem. J. 347, 369–373. 10.1042/bj3470369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowran A., McKayed K., Campbell V. A. (2013). The cannabinoid receptor type 1 is essential for mesenchymal stem cell survival and differentiation: implications for bone health. Stem Cells Int. 2013:796715. 10.1155/2013/796715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hass R., Kasper C., Böhm S., Jacobs R. (2011). Different populations and sources of human mesenchymal stem cells (MSC): a comparison of adult and neonatal tissue-derived MSC. Cell Commun. Signal. 9:12. 10.1186/1478-811X-9-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossini A. M., Quast A. S., Plötz M., Grauel K., Exner T., Küchler J., et al. (2016). PI3K/AKT signaling pathway is essential for survival of induced pluripotent stem cells. PLoS ONE 11:e0154770. 10.1371/journal.pone.0154770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankovic D., Ganesan J., Bscheider M., Stickel N., Weber F. C., Guarda G., et al. (2013). The Nlrp3 inflammasome regulates acute graft-versus-host disease. J. Exp. Med. 210, 1899–1910. 10.1084/jem.20130084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji R.-C. (2012). Macrophages are important mediators of either tumor- or inflammation-induced lymphangiogenesis. Cell. Mol. Life Sci. 69, 897–914. 10.1007/s00018-011-0848-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S. H., Lee J. E., Yun J.-H., Kim I., Ko Y., Park J. B. (2015). Isolation and characterization of human mesenchymal stem cells from gingival connective tissue. J. Periodontal Res. 50, 461–467. 10.1111/jre.12228 [DOI] [PubMed] [Google Scholar]
- Johnson P., Ruffell B. (2009). CD44 and its role in inflammation and inflammatory diseases. Inflamm. Allergy Drug Targets 8, 208–220. 10.2174/187152809788680994 [DOI] [PubMed] [Google Scholar]
- Jourdan T., Godlewski G., Cinar R., Bertola A., Szanda G., Liu J., et al. (2013). Activation of the Nlrp3 inflammasome in infiltrating macrophages by endocannabinoids mediates beta cell loss in type 2 diabetes. Nat. Med. 19, 1132–1140. 10.1038/nm.3265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Sato Y., Kawashima M., Furumichi M., Tanabe M. (2016). KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 44, D457–D462. 10.1093/nar/gkv1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan B. L. F., Springs A. E. B., Kaminski N. E. (2008). The profile of immune modulation by cannabidiol (CBD) involves deregulation of nuclear factor of activated T cells (NFAT). Biochem. Pharmacol. 76, 726–737. 10.1016/j.bcp.2008.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke P., Shao B.-Z., Xu Z.-Q., Wei W., Han B.-Z., Chen X.-W., et al. (2016). Activation of cannabinoid receptor 2 ameliorates DSS-induced colitis through inhibiting NLRP3 inflammasome in macrophages. PLoS ONE 11:e0155076. 10.1371/journal.pone.0155076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimberley F. C., Sivasankar B., Paul Morgan B. (2007). Alternative roles for CD59. Mol. Immunol. 44, 73–81. 10.1016/j.molimm.2006.06.019 [DOI] [PubMed] [Google Scholar]
- Lalor S. J., Dungan L. S., Sutton C. E., Basdeo S. A., Fletcher J. M., Mills K. H. G. (2011). Caspase-1-processed cytokines IL-1beta and IL-18 promote IL-17 production by gammadelta and CD4 T cells that mediate autoimmunity. J. Immunol. 186, 5738–5748. 10.4049/jimmunol.1003597 [DOI] [PubMed] [Google Scholar]
- Laprairie R. B., Kelly M. E. M., Denovan-Wright E. M. (2013). Cannabinoids increase type 1 cannabinoid receptor expression in a cell culture model of striatal neurons: implications for Huntington's disease. Neuropharmacology 72, 47–57. 10.1016/j.neuropharm.2013.04.006 [DOI] [PubMed] [Google Scholar]
- Latour S., Tanaka H., Demeure C., Mateo V., Rubio M., Brown E. J., et al. (2001). Bidirectional negative regulation of human T and dendritic cells by CD47 and its cognate receptor signal-regulator protein-alpha: down-regulation of IL-12 responsiveness and inhibition of dendritic cell activation. J. Immunol. 167, 2547–2554. 10.4049/jimmunol.167.5.2547 [DOI] [PubMed] [Google Scholar]
- Latz E., Xiao T. S., Stutz A. (2013). Activation and regulation of the inflammasomes. Nat. Rev. Immunol. 13, 397–411. 10.1038/nri3452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy S., Todd S. C., Maecker H. T. (1998). CD81 (TAPA-1): a molecule involved in signal transduction and cell adhesion in the immune system. Annu. Rev. Immunol. 16, 89–109. 10.1146/annurev.immunol.16.1.89 [DOI] [PubMed] [Google Scholar]
- Liu J., Miwa T., Hilliard B., Chen Y., Lambris J. D., Wells A. D., et al. (2005). The complement inhibitory protein DAF (CD55) suppresses T cell immunity in vivo. J. Exp. Med. 201, 567–577. 10.1084/jem.20040863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merville P. (2005). Combating chronic renal allograft dysfunction : optimal immunosuppressive regimens. Drugs 65, 615–631. 10.2165/00003495-200565050-00004 [DOI] [PubMed] [Google Scholar]
- Moraes D. A., Sibov T. T., Pavon L. F., Alvim P. Q., Bonadio R. S., Da Silva J. R., et al. (2016). A reduction in CD90 (THY-1) expression results in increased differentiation of mesenchymal stromal cells. Stem Cell Res. Ther. 7:97. 10.1186/s13287-016-0359-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudunuri U., Che A., Yi M., Stephens R. M. (2009). bioDBnet: the biological database network. Bioinformatics 25, 555–556. 10.1093/bioinformatics/btn654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagarkatti M., Rieder S. A., Hegde V. L., Kanada S., Nagarkatti P. (2010). Do cannabinoids have a therapeutic role in transplantation? Trends Pharmacol. Sci. 31, 345–350. 10.1016/j.tips.2010.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi K., Yoshimoto T., Tsutsui H., Okamura H. (2001). Interleukin-18 regulates both Th1 and Th2 responses. Annu. Rev. Immunol. 19, 423–474. 10.1146/annurev.immunol.19.1.423 [DOI] [PubMed] [Google Scholar]
- Nauta A. J., Westerhuis G., Kruisselbrink A. B., Lurvink E. G. A., Willemze R., Fibbe W. E. (2006). Donor-derived mesenchymal stem cells are immunogenic in an allogeneic host and stimulate donor graft rejection in a nonmyeloablative setting. Blood 108, 2114–2120. 10.1182/blood-2005-11-011650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netea M. G., Simon A., van de Veerdonk F., Kullberg B.-J., Van der Meer J. W. M., Joosten L. A. B. (2010). IL-1β processing in host defense: beyond the inflammasomes. PLoS Pathog. 6:e1000661. 10.1371/journal.ppat.1000661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ode A., Kopf J., Kurtz A., Schmidt-Bleek K., Schrade P., Kolar P., et al. (2011). CD73 and CD29 concurrently mediate the mechanically induced decrease of migratory capacity of mesenchymal stromal cells. Eur. Cell. Mater. 22, 26–42. [DOI] [PubMed] [Google Scholar]
- Patel D. M., Shah J., Srivastava A. S., Patel D. M., Shah J., Srivastava A. S. (2013). Therapeutic potential of mesenchymal stem cells in regenerative medicine. Stem Cells Int. 2013, 1–15. 10.1155/2013/496218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman M. M., Subramani J., Ghosh M., Denninger J. K., Takeda K., Fong G.-H., et al. (2014). CD13 promotes mesenchymal stem cell-mediated regeneration of ischemic muscle. Front. Physiol. 4:402. 10.3389/fphys.2013.00402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee S. Y., Wood V., Dolinski K., Draghici S. (2008). Use and misuse of the gene ontology annotations. Nat. Rev. Genet. 9, 509–515. 10.1038/nrg2363 [DOI] [PubMed] [Google Scholar]
- Robinson R. H., Meissler J. J., Breslow-Deckman J. M., Gaughan J., Adler M. W., Eisenstein T. K. (2013). Cannabinoids inhibit T-cells via cannabinoid receptor 2 in an in vitro assay for graft rejection, the mixed lymphocyte reaction. J. Neuroimmune Pharmacol. 8, 1239–1250. 10.1007/s11481-013-9485-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh S. M., Parhar R. S., Al-Hejailan R. S., Bakheet R. H., Khaleel H. S., Khalak H. G., et al. (2013). Identification of the tetraspanin CD82 as a new barrier to xenotransplantation. J. Immunol. 191, 2796–2805. 10.4049/jimmunol.1300601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder K., Tschopp J. (2010). The inflammasomes. Cell 140, 821–832. 10.1016/j.cell.2010.01.040 [DOI] [PubMed] [Google Scholar]
- Schu S., Nosov M., O'Flynn L., Shaw G., Treacy O., Barry F., et al. (2012). Immunogenicity of allogeneic mesenchymal stem cells. J. Cell. Mol. Med. 16, 2094–2103. 10.1111/j.1582-4934.2011.01509.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seu L., Sun J. J., Schaaf K. R., Duverger A. E., Kutsch O., Goepfert P. A. (2016). The tetraspanin CD151 is an activation molecule that characterizes M. tuberculosis- specific effector CD4+ T cells. J. Immunol. 196:65.9. [Google Scholar]
- Shah K. B., Mauro A. G., Flattery M., Toldo S., Abbate A. (2015). Formation of the inflammasome during cardiac allograft rejection. Int. J. Cardiol. 201, 328–330. 10.1016/j.ijcard.2015.08.070 [DOI] [PubMed] [Google Scholar]
- Shao B.-Z., Wei W., Ke P., Xu Z.-Q., Zhou J.-X., Liu C. (2014). Activating cannabinoid receptor 2 alleviates pathogenesis of experimental autoimmune encephalomyelitis via activation of autophagy and inhibiting NLRP3 inflammasome. CNS Neurosci. Ther. 20, 1021–1028. 10.1111/cns.12349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw P. J., McDermott M. F., Kanneganti T.-D. (2011). Inflammasomes and autoimmunity. Trends Mol. Med. 17, 57–64. 10.1016/j.molmed.2010.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibagaki N., Hanada K., Yamaguchi S., Yamashita H., Shimada S., Hamada H. (1998). Functional analysis of CD82 in the early phase of T cell activation: roles in cell adhesion and signal transduction. Eur. J. Immunol. 28, 1125–1133. [DOI] [PubMed] [Google Scholar]
- Szklarczyk D., Franceschini A., Wyder S., Forslund K., Heller D., Huerta-Cepas J., et al. (2015). STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 43, D447–D452. 10.1093/nar/gku1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taglialatela-Scafati O., Pagani A., Scala F., De Petrocellis L., Di Marzo V., Grassi G., et al. (2010). Cannabimovone, a cannabinoid with a rearranged terpenoid skeleton from hemp. European J. Org. Chem. 2010, 2067–2072. 10.1002/ejoc.200901464 [DOI] [Google Scholar]
- Trubiani O., Giacoppo S., Ballerini P., Diomede F., Piattelli A., Bramanti P., et al. (2016a). Alternative source of stem cells derived from human periodontal ligament: a new treatment for experimental autoimmune encephalomyelitis. Stem Cell Res. Ther. 7, 1. 10.1186/s13287-015-0253-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trubiani O., Guarnieri S., Diomede F., Mariggiò M. A., Merciaro I., Morabito C., et al. (2016b). Nuclear translocation of PKCα isoenzyme is involved in neurogenic commitment of human neural crest-derived periodontal ligament stem cells. Cell. Signal. 28, 1631–1641. 10.1016/j.cellsig.2016.07.012 [DOI] [PubMed] [Google Scholar]
- Trubiani O., Piattelli A., Gatta V., Marchisio M., Diomede F., D'Aurora M., et al. (2015). Assessment of an efficient xeno-free culture system of human periodontal ligament stem cells. Tissue Eng. Part C. Methods 21, 52–64. 10.1089/ten.TEC.2014.0024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao L., Nasu M. (2014). From regenerative dentistry to regenerative medicine: progress, challenges, and potential applications of oral stem cells. Stem Cells Cloning 7, 89–99. 10.2147/sccaa.s51009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaffran Y., Destaing O., Roux A., Ory S., Nheu T., Jurdic P., et al. (2001). CD46/CD3 costimulation induces morphological changes of human T cells and activation of Vav, Rac, and extracellular signal-regulated kinase mitogen-activated protein kinase. J. Immunol. 167, 6780–6785. 10.4049/jimmunol.167.12.6780 [DOI] [PubMed] [Google Scholar]
- Zhang J.-M., Murakumo Y., Hagiwara S., Jiang P., Mii S., Kalyoncu E., et al. (2015). CD109 attenuates TGF-β1 signaling and enhances EGF signaling in SK-MG-1 human glioblastoma cells. Biochem. Biophys. Res. Commun. 459, 252–258. 10.1016/j.bbrc.2015.02.093 [DOI] [PubMed] [Google Scholar]
- Zhang Q., Shi S., Liu Y., Uyanne J., Shi Y., Shi S., et al. (2009). Mesenchymal stem cells derived from human gingiva are capable of immunomodulatory functions and ameliorate inflammation-related tissue destruction in experimental colitis. J. Immunol. 183, 7787–7798. 10.4049/jimmunol.0902318 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.