Abstract
Extensive phenotypic variability is commonly observed in individuals with Mendelian disorders, even among those with identical genotypes in the disease-causing gene. To determine whether variants within and surrounding CFTR contribute to phenotypic variability in cystic fibrosis (CF), we performed deep sequencing of CFTR in 762 patients homozygous for the common CF-causing variant, F508del. In phase 1, ~200 kb encompassing CFTR and extending 10 kb 5′ and 5 kb 3′ of the gene was sequenced in 486 F508del homozygotes selected from the extremes of sweat chloride concentration. In phase 2, a 510 kb region, which included the entire topologically associated domain of CFTR, was sequenced in 276 F508del homozygotes drawn from extremes of lung function. An additional 163 individuals who carried F508del and a different CF-causing variant were sequenced to inform haplotype construction. Region-based burden testing of both common and rare variants revealed seven regions of significance (α=0.01), five of which overlapped known regulatory elements or chromatin interactions. Notably, the −80 kb locus known to interact with the CFTR promoter was associated with variation in both CF traits. Haplotype analysis revealed a single rare recombination event (1.9% frequency) in intron 15 of CFTR bearing the F508del variant. Otherwise, the majority of F508del chromosomes were markedly similar, consistent with a single origin of the F508del allele. Together, these high-resolution variant analyses of the CFTR locus suggest a role for non-coding regulatory motifs in trait variation among individuals carrying the common CF allele.
Introduction
Cystic fibrosis (CF) is an excellent example of a highly variable Mendelian disease. The condition affects ~70,000 patients worldwide and the primary traits exhibited in affected individuals include elevated sweat chloride levels, lung disease, and pancreatic insufficiency. CFTR, the protein defective in CF, facilitates chloride, and bicarbonate movement across the apical membranes of epithelial tissues is abnormal.1 Aberrant ion transport leads to unusually viscous secretions in the lung and pancreas, resulting in damage to both organ systems. The severity of these traits in each individual is primarily determined by the CF-causing variants they have inherited.2 The most common disease-causing variant leads to the loss of phenylalanine at amino acid position 508, commonly referred to as F508del (ΔF508, rs113993960 or rs199826652, which yields the same mutant DNA sequence), often erroneously annotated rs121909001, which yields the same amino acid deletion but a different and uncommon mutant DNA sequence. This variant is present in the homozygous state in ~50% and in the heterozygous state in a further 40% of CF patients,3 where it results in the improper folding and eventual degradation of the final protein product.4
Significant phenotypic heterogeneity is observed in the F508del homozygous CF patient population. Genome-wide association studies have identified loci that may modify lung function, neonatal intestinal obstruction and diabetes risk.5 However, there have been no comprehensive studies to determine whether variation within the CFTR locus itself contributes to the variance observed in disease traits. In this study, we have sought to determine whether previously untyped genetic variation within or near CFTR is underlying the observed distribution of phenotypes in a select patient population. These variants have the potential to alter not only the disease presentation, but also a patient’s response to CFTR-targeted therapeutics.
Treatment of CF has been revolutionized by the advent of small-molecule drugs that target defective CFTR.6 Due to the commonness of F508del, there has been intense effort to develop drugs that recover the function of CFTR bearing F508del.7 These efforts have resulted in the development of CFTR-targeted treatment (Orkambi) that combines a ‘corrector’ compound VX-809 that rectifies misfolding and a ‘potentiator’, VX-770, that activates the chloride channel of CFTR bearing the F508del variant.6,8 However, Orkambi produces only modest improvement in lung function in F508del homozygotes and no significant improvement in patients that have only one copy of F508del.6 Consequently, the research community is seeking to identify molecules with higher effectiveness for CFTR-F508del by empirical screening.9
To inform the search for the cause of disease variation and the development of small molecules that target CFTR-F508del, we have systematically investigated genetic variation in CFTR alleles bearing F508del. Our overarching goal was to identify any variants or combination of variants that modify disease severity associated with the F508del mutation. Studies of F508del homozygotes have revealed a broad range of both sweat chloride values and lung function.2,10 Affected twin and sibling studies indicate that the CFTR locus is the primary determinant of variation in sweat chloride concentration, accounting for 56% of the total variance.11 Variation elsewhere in the genome (i.e., genetic modifiers) appears to play a minor role in sweat variability. Thus, F508del homozygotes at the extremes of the distribution of CF traits, particularly sweat chloride concentration, provide an opportunity to find variants that modulate the effect of F508del upon CFTR function. Consequently we have performed deep re-sequencing of intragenic and extragenic regions surrounding CFTR in 762 F508del homozygotes. Region-based burden testing was used to test whether genetic variation within and near CFTR is associated with sweat chloride concentration or lung function. High-resolution haplotypes and linkage disequilibrium (LD) patterns were used to map recombination events on the F508del-bearing chromosomes.
Materials and methods
Please see Supplementary Text (S1) for information regarding cohort selection, sample preparation, re-sequencing capture design, variant and haplotype calling, and association testing.
Results
Variation in CFTR chromosomes bearing F508del
A total of 925 individuals with CF (762 F508del homozygotes and 163 F508del heterozygotes) were sequenced in two phases. To increase the power to detect associations with modifying variants in the CFTR locus, all available F508del homozygous individuals in the Johns Hopkins CF Twin and Sibling Study (TSS) and the Genetic Modifier Study (GMS) with extremes of sweat chloride levels (Supplementary Figure 1A) were selected for analysis. A 210 kb region encompassing CFTR and extending 10 kb 5′ and 5 kb 3′ of the gene was sequenced in 583 subjects (486 F508del homozygotes and 97 heterozygotes: Supplementary Table 1). Advances in capture technology following completion of the first phase enabled us to expand coverage of the CFTR locus in a second phase. It has been shown previously that the three-dimensional structure of genomic DNA plays a key role in regulating gene expression. These regions of increased DNA interactions and resultant chromosome looping are often referred to as topologically associated domains (TADs), and have been well characterized for the CFTR locus.12,13 Consequently, in phase 2, a 305 kb region fully encompassing the TAD of CFTR12,13 plus an additional 300 kb flanking this TAD was sequenced in 342 subjects (276 F508del homozygotes and 66 heterozygotes). The expanded region includes the neighboring genes WNT2, ASZ1 and CTTNBP2. In the second phase, we selected the F508del homozygotes in the TSS from the extremes of lung function (Supplementary Figure 1B). By combining the two phases, we obtained F508del homozygous individuals drawn from the entire phenotype spectrum for sweat chloride function and lung function (Supplementary Figure 2). The F508del heterozygous samples were used only to inform haplotype studies. Otherwise, the following results are specific to the sequence-verified F508del homozygous population (n=762).
A total of 652 variants were observed within CFTR in the F508del homozygous subjects. Twenty-four variants were observed within CFTR exons (n=13) (Table 1) or in the 5′ and 3′ UTRs (n=11), while the remaining 628 were intronic. Two of the 13 exonic variants had minor allele frequencies (MAF)>1% (p.I1027T and p.Q1463Q), while a third variant, L467F (rs1800089), was detected at a frequency of 0.3% in CFTR chromosomes bearing F508del. Of note from a clinical diagnostic perspective, L467F has been reported in individuals also carrying one copy of F508del diagnosed with CF-related metabolic disorder.14–17 Our results indicate that L467F is in cis with the F508del allele, indicating that it is not the trans CF variant leading to disease in these patients. However, due to low frequency in this population, we were not able to resolve the revertant potential of L467F or three other variants that cause an amino-acid substitution (p.Q1330Q, p.R1438Y and p.V1475M) or five synonymous variants (p.L130L, p.I203I, p.T854T, p.I1404I and p.Q1463Q). None of these variants were predicted to activate cryptic RNA splicing. Functional testing will be required to assess whether the amino acid substitutions affect the function of CFTR bearing F508del. Of note, the p.V1475M allele detected here was present in one of the original CFTR cDNAs that was widely distributed, and it appears to have no functional effect.18 Two frameshifting mutations were also observed (p.His1402Glyfs*2 and p.Arg1403Serfs*60), each resulting in a premature stop codon. While one would expect these frameshifts to result in a completely null protein, further functional testing may be warranted for p.Arg1403Seqfs*60, which truncates near the end of the protein. Finally, five variants initially mapped to the CFTR gene were discovered to be variants in regions of high homology to CFTR’s exon 1019,20 (Supplementary Table 2). Together, these results indicate that there is limited variation in the coding regions of CFTR bearing the F508del mutation.
Table 1. Exonic CFTR variants in 762 F508del homozygotes.
| Chr7 bp (hg19) | Variant type | Transcript annotation | No. of chromosomes | Frequency |
Samples in extremes of sweat Cl− levels |
Samples in extremes of lung function |
rsID | ||
|---|---|---|---|---|---|---|---|---|---|
| −1 s.d. | +1 s.d. | −1 s.d. | +1 s.d. | ||||||
| 117171069 | Synonymous | p.L130L | 1 | 0.001 | — | — | — | 1 | |
| 117175331 | Synonymous | p.I203I | 1 | 0.001 | 1 | — | — | — | rs1800081 |
| 117199524 | Nonsynonymous | p.L467F | 4 | 0.003 | — | 2 | 1 | 1 | rs1800089 |
| 117199533 | Nonsynonymous | p.V470M | 1524 | 1 | — | — | — | — | rs213950 |
| 117235055 | Synonymous | p.T854T | 5 | 0.003 | 2 | 1 | 1 | 1 | rs1042077 |
| 117250664 | Nonsynonymous | p.I1027T | 42 | 0.028 | 11 | 14 | 7 | 14 | rs1800112 |
| 117304766 | Nonsynonymous | p.Q1330E | 1 | 0.001 | — | — | 1 | — | rs375661578 |
| 117305579 | Frameshift deletion | c.4204_4207del:p.His1402Glyfs*2 | 1 | 0.001 | — | — | 1 | — | |
| 117305584 | Frameshift insertion | c.4208_4209insCTGC:p.Arg1403Serfs*60 | 1 | 0.001 | — | — | 1 | — | |
| 117305588 | Synonymous | p.I1404I | 1 | 0.001 | — | — | — | 1 | |
| 117307031 | Nonsynonymous | p.R1438Y | 1 | 0.001 | 1 | — | — | — | |
| 117307108 | Synonymous | p.Q1463Q | 35 | 0.023 | 12 | 13 | 8 | 4 | rs1800136 |
| 117307142 | Nonsynonymous | p.V1475M | 1 | 0.001 | — | — | 1 | — | |
*Association beta values provided for variants with >1 chromosomes only.
Single-variant association analysis reveals no significant association with sweat chloride concentration or lung function
Common variants with MAF>1% (602 total variants; 235 in phase 1; 598 in phase 2) were assayed for association with either sweat chloride levels (Supplementary Table 3A) or lung function as measured by the age and survival-adjusted phenotype SaKnorm (Supplementary Table 3B). Linear regressions between number of minor alleles and the phenotypes were conducted on the Phase 1 and Phase 2 study samples separately and, for regions sequenced in both captures (the 210 kb encompassing CFTR), in combination. All P-values were calculated by permutation because of possibly non-normally distributed phenotypes (Supplementary Figure 1C). Forty-three variants showed some evidence of association with point-wise permutation P-values (P<0.05) and 15 variants had β values in the same direction when observed in both phase 1 and phase 2 cohorts (Supplementary Tables 3A and 3B). None of these variants was statistically significant after multiple test correction using either max(T) permutation (Supplementary Table 3) or Bonferroni correction (not shown).
One coding variant (p. I1027T) showed weak evidence of association with lung function (uncorrected point-wise permutation P=0.013). Recognizing the limited power to detect associations of single rare variants with CF traits (57 and 18% power at MAF 0.01 and an effect size of 1 s.d. for sweat chloride and lung function, respectively), we tested I1027T for association in a second, unrelated group of 748 F508del homozygotes. However, the association of I1027T with lung function did not replicate (P=0.8239). These results indicate no single common variant in cis with F508del was associated with either sweat chloride concentration or lung function in this study.
Clusters of variants 5′ of and within CFTR correlate with sweat chloride concentration and lung function in F508del homozygotes
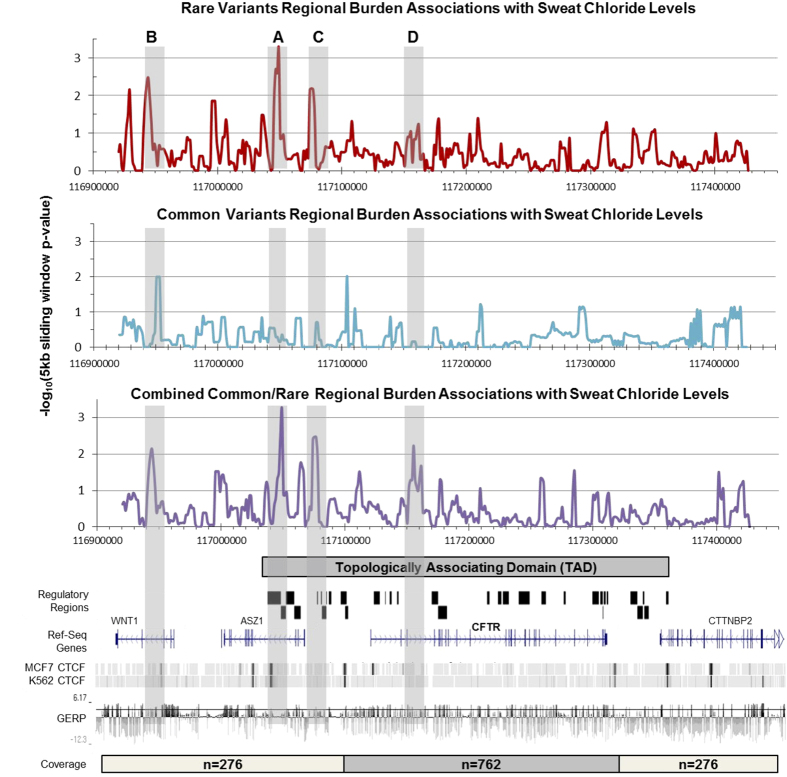
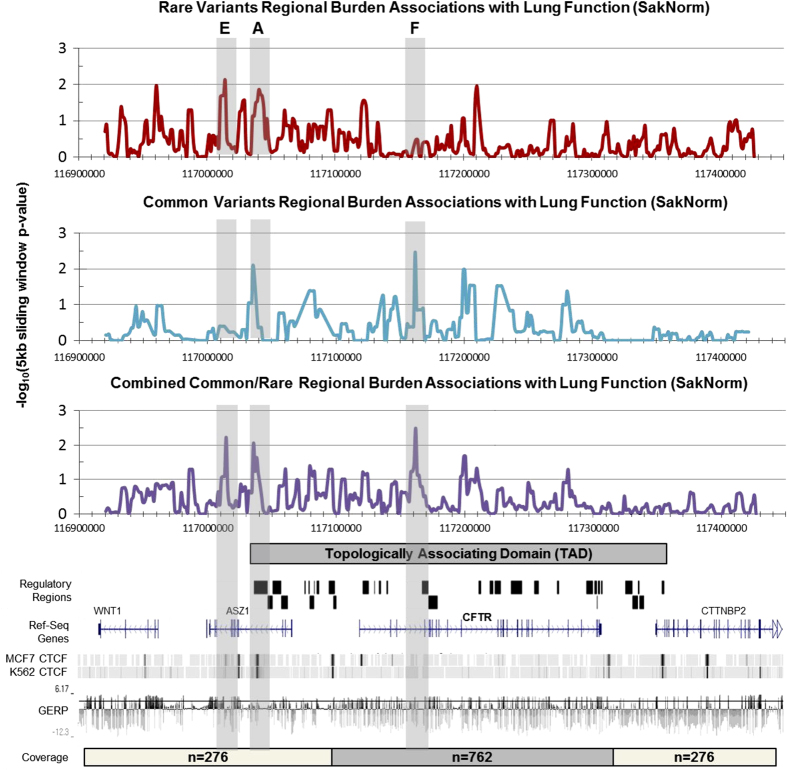
A region-based burden assay was performed to identify groupings of variants that are associated with trait variation. Variants were tested for association with sweat chloride or lung function in groups defined by a series of overlapping 5 kb windows (offset in 1,250 bp increments), with each window generating a test P-value. The P-value for each window was Bonferroni corrected for multiple testing based on the total number of unique windows assayed (see Methods). Regions of significance (hg19 coordinates, study-wide P<0.01) were highlighted for the combined test of common and rare variants associated with either sweat chloride concentration (Figure 1) and/or lung function (Figure 2). Regions that coincided with known regulatory and boundary elements are discussed below (Supplementary Table 4).13,21,22
Figure 1.
Burden testing of common and rare variants associating with sweat Cl− levels. All variants within each 5 kb window, moved across the entire region in increments of 1,250 bp, were tested for a combined association with sweat chloride levels (mM) via SKAT-O test. The x axis denotes chr7 hg19 bp position, y axis is −log10 of the regional P-value. Association values were plotted at the center of each 5 kb window. Top (red): rare variants with minor allele frequency (MAF)<1% only. Middle (blue): common variants with MAF>1% only. Bottom (purple): combined test of common and rare variants with variants weighted inversely proportional to their frequency. Vertical shaded boxes: regions of significant association in the combined analysis (α=0.01). Genome browser style tracks: Top, packed view of known CFTR regulatory regions of interest and TAD as previously reported (see Supplementary File). Middle, view of genes with exonic/intronic structure. Bottom, CCCTC-binding factor (CTFC) binding signals in two cell types, and mammalian conservation as assayed by genomic evolutionary rate profiling (GERP) (horizontal bar indicating a GERP score of 4). CFTR, the protein defective in cystic fibrosis; TAD, topologically associated domain.
Figure 2.
Burden testing of common and rare variants associating with lung function (SaKnorm). All variants within each 5 kb window, moving across the entire region in increments of 1,250 bp, were tested for a combined association with lung function (SaKnorm) via SKAT-O test. The x axis denotes chr7 hg19 bp position, y axis is −log10 of the regional P-value. Association values were plotted at the center of each 5 kb window. Top (red): rare variants with minor allele frequency (MAF)<1% only. Middle (blue): common variants with MAF>1% only. Bottom (purple): combined test of common and rare variants with variants weighted inversely proportional to their frequency. Vertical shaded boxes: regions of significant association in the combined analysis (α=0.01). Genome browser style tracks: Top: packed view of known CFTR regions of interest and TAD as previously reported (see Supplementary File). Middle: view of genes with exonic/intronic structure. Bottom: CCCTC-binding factor (CTFC) binding signals in two cell types, and mammalian conservation as assayed by genomic evolutionary rate profiling (GERP) (horizontal bar indicating GERP score of 4). CFTR, the protein defective in cystic fibrosis; TAD, topologically associated domain.
A regulatory locus at −80 kb is associated with both sweat chloride levels and lung function
Variation in a locus denoted ‘Region A’ was associated with both sweat chloride concentration (study-wide corrected P=5.9e−4; region of statistical significance: chr7: 117, 039,250–117,053,000, Figure 1) and lung function (study-wide corrected P=8.9e−3; chr7: 117,030,500–117,050,000, Figure 2). Region A is located approximately 80 kb 5′ of the CFTR transcription start site and within intron 4 of ASZ1. Chromatin conformation capture assays have shown that this region interacts with sequences in the CFTR promoter.13,21 It also contains CCCTC-binding factor (CTCF) binding sites (Figure 1), which may assist in looping of distant regulatory elements to CFTR’s transcriptional start site.13,21,22 A total of 12 common and 9 rare variants are observed under the sweat chloride peak (Supplementary Table 5). Of note are two variations in the length of a poly A tract, both of which may be associated with higher sweat chloride concentration (7:117047463:TA>T and 7:117047463:TAA>T). A total of 13 common and 3 rare variants are observed under the lung function peak (Supplementary Table 6). One of these variants (rs4730780, 7:117041448:T:A) that results in a decrease in length of a poly T tract, and an increase in length of the adjacent poly A tract is located ~100 bp from a known CTCF binding site.23 All the four individuals with this variant had above-average lung function (β=+0.81 SaKnorm).
Three loci associated with sweat chloride levels
Variation in three regions was associated with sweat chloride concentration (Figure 1). Region B (chr7: 116,941,750–116,951,750, P=7.1e−3, phase 2 coverage only, n=276) is located within intron 3 of WNT2 and contains 21 common and 9 rare variants (Supplementary Table 5). This region lies outside CFTR’s TAD, and is adjacent to known CTCF binding sites in MCF7 and K562 cells (Figure 1). Three individuals harbored what appears to be a haplotype of four rare variants (116942115:G>A, 116942433:G>T, 116943135:C>T, 116944283:T>A, 3/552 chromosomes) and had lower mean sweat chloride concentration (~17 mM Cl−). These variants lie within or near a region of open chromatin and CEBPB binding site in fetal lung fibroblasts cells (IMR90).23 A common variant in the same region also showed evidence of association (P=0.02 uncorrected) with decreased sweat chloride levels (7:116943793:A>T, −8.63 mM Cl−). Region C (chr7: 117,074,250–117,078,000, P=3.3e−3, phase 1 and phase 2 coverage, n=762) contains six rare variants and a known regulatory locus ~44 kb upstream of CFTR.13 The variant with the most significant uncorrected P-value (117076029:G>A, P=1.6e−3, Supplementary Table 5) lies within MTA3 and PML binding regions in GM12878 cells.23 The final region significantly enriched for variants associated with sweat chloride levels (Region D, chr7: 117,153,000–117,156,750, P=5.8e-3, phase 1 and phase 2 coverage, n=762) is located within intron 3 of CFTR, and contains two common and seven rare variants. Region D contains no known functional elements and has various repetitive sequences.
Two distinct loci associated with lung function
Variation in two regions was associated with SaKnorm (Figure 2). Region E (chr7: 117,010,500–117,014,250, P=5.8e−3, phase 2 coverage only, n=276, ASZ1 intron 10) lies just outside of the proposed TAD containing CFTR (Figure 2), and contains six common and one rare variants (Supplementary Table 6). Most of the variants are 2–4 kb 3′ of known CTCF binding sites, but may influence interactions with regions outside of the CFTR TAD. Region F (chr7: 117,159,250–117,164,250, P=3.3e−3, n=762) is located within intron 3 of CFTR, slightly 3′ to region B associated with sweat chloride (Figure 1). The association here appears primarily due to common variation, specifically variations within a poly T tract (chr7: 11,716,0319, 17T, 18T or 16T). Increasing the length of this tract is marginally associated with improved lung function (uncorrected P=3.6e−3), and, conversely, decreasing the length of this tract is associated with poorer lung function (uncorrected P=3.3e−3) (Supplementary Tables 3A and 3B, Supplementary Table 6). The region has a large number of repetitive elements and no known functional elements.
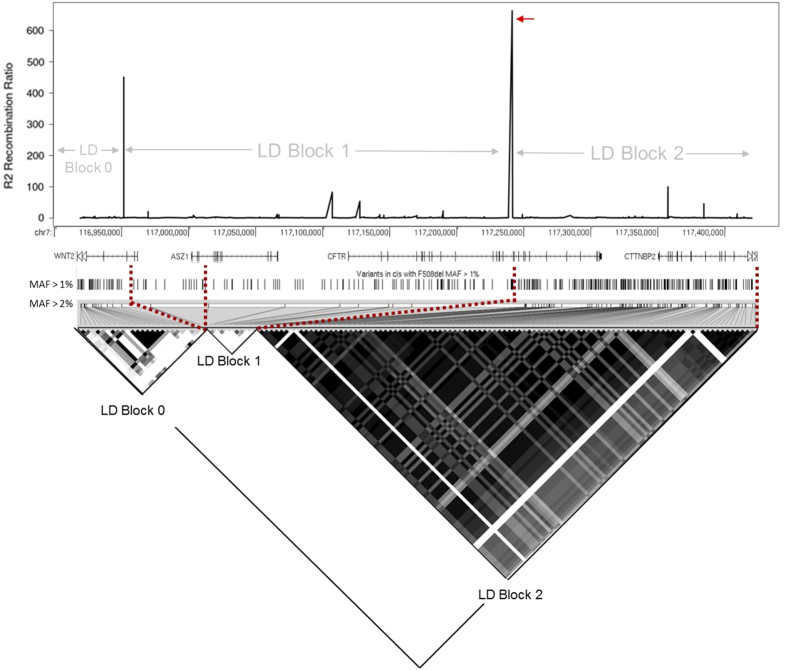
One recombination event defines two blocks of LD within CFTR bearing F508del
In addition to association testing, we also sought to systematically determine the genetic architecture at this locus in the F508del homozygous population. Haplotypes are combinations of variants that tend to be inherited together (in cis). Their borders are often delineated by meiotic recombination events. Derivation of haplotypes is useful for establishing the degree of genetic diversity in a locus, associating functional variants with background variation, and inferring ancestral origins of disease-causing variants. To assemble haplotypes, single-nucleotide variants with MAF>1% within the 510 kb region surrounding CFTR were phased using SHAPEIT2.24 Samples bearing non-F508del chromosomes were used to deduce the locations of alternative recombination events and additional diversity of the CFTR locus. LD among variants with MAF>2% in 762 F508del homozygotes revealed three primary regions of high LD, two of which encompassed CFTR (Figure 3). A recombination event was observed within intron 15 of CFTR, resulting in an alternative haplotype in LD block 2. This recombination event is not present in the 206 non-F508del chromosomes that were sequenced using the same capture design. The intron 15 recombination event in F508del homozygotes is unique to this population, and is not apparent in HapMap populations (which contain a diversity of CFTR haplotypes only ~5% of which is the F508del ancestral haplotype), where a distinct recombination event within intron 22 is observed.25
Figure 3.
Recombination ratio and linkage disequilibrium observed in 762 F508del homozygous samples (1,524 chromosomes) across a 506 kb re-sequencing region surrounding CFTR. Top: recombination ratio plotted by genomic location. A recombination event occurring within intron 15 of CFTR is indicated by the red arrow. Below is an intronic and exonic map of known RefSeq genes, and re-sequencing study variants with minor allele frequency (MAF)>1% (hg19 coordinates). Bottom: LD heat map of variants with MAF>2% in the F508del population. Dashed red lines indicate projection of variants from their genomic positions to the heat-map of r2 values below. Within the heatmap, black boxes indicate an r2 value of 1 or complete LD, while white boxes indicate an r2 of 0 or linkage equilibrium. Three proposed LD blocks are outlined (triangles). The first extends from the start of the sequencing capture to intron 3 of WNT2. LD block 1 then extends from the WNT2 locus to intron 15 of CFTR. Finally, LD block 2 extends from intron 15 of CFTR to the end of the sequencing capture (mid-CTTNBP2). LD blocks 0 and 2 likely extend far beyond the capture design. CFTR, the protein defective in cystic fibrosis; LD, linkage disequilibrium.
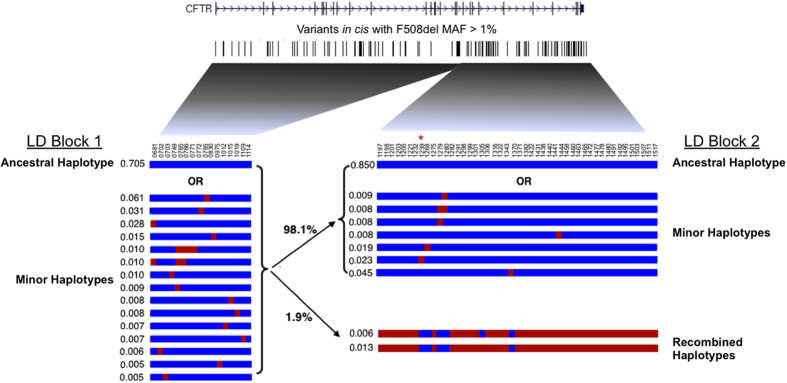
Using common variants with a MAF>1%, we found that 16 haplotypes occurred at a frequency of 0.5% or higher in LD block 1 (CFTR exons 1–15) on chromosomes bearing F508del (Figure 4, Supplementary Table 7). The second LD block encompasses exons 16–27 of CFTR, and 10 haplotypes above 0.5% frequency are observed in the F508del homozygous cohort. Additional diversity is seen via inclusion of INDELs, but due to the large number of such sites observed in this region and their poly-allelic state they were not included in the analysis presented.
Figure 4.
Haplotypes observed in 762 F508del homozygous samples (1,524 chromosomes) across the CFTR locus. Top: intronic and exonic map of CFTR, containing re-sequencing study variants with minor allele frequency (MAF)>1%. Bottom: representation of CFTR SNP haplotypes with MAF>1% and MHF>0.5%. Each numbered variant is represented vertically by blue (reference allele) or red (alternate allele) squares. Each haplotype is represented horizontally as a row, with respective minor haplotype frequencies to the left. Two LD blocks are shown, with primary recombination events indicated by bold connecting lines. LD block 1 shows one ancestral haplotype with minor haplotypes below, while LD block 2 shows an additional recombined haplotype. CFTR, the protein defective in cystic fibrosis; LD, linkage disequilibrium; MHF, minor haplotype frequency.
LD block 1 of CFTR encompassing the F508del allele displays primarily rare variation
Limited common variation would be expected in the region of high LD surrounding the F508del variant if it were inherited from a common founder ancestor of European descent.26 Indeed, the autozygosity of this region is confirmed by the dearth of common variants in this haplotype (2/17 SNPs with MAF>5%). The majority of variation within this locus is very low frequency (15/17 SNPs have MAF between 1–5%). Overall, ~70% of F508del homozygotes carry the same ancestral haplotype in LD block 1, with minor variations of this haplotype occurring in another 30% of samples (Figure 4).
LD block 2 of CFTR contains two distinct haplotypes
Due to the historical recombination event in intron 15 that occurred on an F508del-containing chromosome, a second distinct haplotype is observed in LD block 2 that represents 1.9% of F508del chromosomes (Figure 4, bottom right). The alternative haplotype is observed in two forms (0.6% and 1.3% minor haplotype frequency), which vary by one marker (rs138427389) (Figure 4, variant #1305). Of note, this alternative haplotype contains the synonymous variant Q1463Q (rs1800136) (Figure 4, variant #1507).
When samples bearing this alternative haplotype are removed from the F508del homozygous population, the recombination event within intron 15 is not observed (Supplementary Figure 3). Most variation observed in LD block 2 is rare (46/47 SNPs with MAF between 1–5%), and thus the majority of haplotypes in LD block 2 deviate from the ancestral haplotype by only one marker. One such haplotype contains I1027T (rs1800112), a variant known to be observed in cis with F508del.27 Our data, as well as data from the CFTR2 database (K Raraigh and P Sosnay, personal correspondence), indicate this allele and its associated haplotype are present on ~2.5% of F508del chromosomes (Figure 4, variant #1239, red asterisk). Overall, 85% of F508del chromosomes carry the ancestral haplotype in LD block 2, with the alternative haplotype as well as minor variations representing another 15% of chromosomes. Finally, when considering only SNPs with MAF>1% within the CFTR locus, ~55% of F508del chromosomes are completely identical across both regions of LD.
Thirteen single-nucleotide variants capture all common (>1% frequency) haplotypes found on CFTR chromosomes bearing F508del
By definition, haplotypes of a given LD block contain SNPs in high LD. As such, these haplotypes can be simplified by tagging variants. In this process, all variants above a particular LD threshold are grouped and represented by a single variant, or ‘tag’. All haplotypes composed of SNPs with a MAF>1% within the ~200 kb surrounding CFTR are detailed in Supplementary Table 7, where a subset of 31 tagged SNPs (Supplementary Table 8) captured 99.2% of the total ‘common’ variation at an r2 correlation value greater than 0.9. Of these tagged SNPs, 13 were capable of representing all haplotypes with a minor haplotype frequency >1% (Supplementary Table 7, gray highlights). Haplotype-based studies using 13 tagged SNPs did not reveal any significant association with either sweat chloride concentration or lung function (data not shown).
Mapping of restriction fragment length polymorphisms at the CFTR locus
CFTR was originally mapped using restriction fragment length polymorphisms (RFLPs).28,29 We mapped eight of these RFLPs to within 2 kb of their hg19 genomic coordinates, and have determined the coordinate and rsID of an additional three RFLPs (Supplementary Figure 4, Supplementary Table 9). These RFLPs primarily lie within LD block 1 (Intron 3 WNT2 to Intron 15 CFTR), which includes the F508del allele. Even RFLPs lying beyond this linkage block show residual LD with the F508del allele. An example of this is H2.3A (XV-2C, rs3779549), which lies just beyond the recombination event in WNT2, but displays residual LD with the F508del locus. This allele likely was on the same haplotype as F508del, but, over time, recombination events decreased the LD between these two markers. For this reason, the reference allele of H2.3A is enriched in the F508del population. As the majority of CF patients carry at least one copy of the F508del allele, the high LD of these markers with this variant facilitated the localization of CFTR.30
Discussion
This study’s initial goal was to determine if a coding region variant might moderate the deleterious effect of the F508del allele. We found that neither a single common variant nor a combination of variants (i.e., haplotype) within this region is associated with CF trait variation. However, we were unable to exclude four rare amino-acid substitutions (p.L467F, p.Q1330E, p.R1438Y and p.V1475M) as these variants were not frequent enough to allow for statistically valid association testing in this population. Functional studies will be required to assess if these rare variants have any effect on CFTR bearing the F508del allele.
The basal transcription of CFTR is primarily driven by binding of factors at the 5′ promoter element.31 However, recent studies have shown that additional cis regulatory elements are required for tissue specificity, abundance and temporal expression.32,33 These cis regulatory elements have been shown to interact with the CFTR promoter, likely through a chromatin looping mechanism in part facilitated by CTCF binding.34 Multiple chromatin interaction studies have now shown that these regulatory loci are encompassed within a TAD,13,22,35 which is defined by boundary elements at −80 and +49 kb from CFTR. While much progress has been made regarding the chromatin structure in this region, resolving the function of each of these regulatory elements continues to be an active area of research.
The results presented here posit that a burden of both rare and common variants at these key loci may modulate the CF phenotype by alteration in the level and/or timing of expression of CFTR bearing F508del. Our findings may inform future functional studies of the cis-regulatory elements identified in chromatin studies. The shared burden of rare and common variants associating with both CF traits at the −80 kb regulatory motif is possibly the most striking finding we report. We hypothesize that variants here may affect CTCF binding, or increase inherent enhancer activity. This could lead to altered expression of the F508del transcript, which has some residual processing and function.36–41 Presence of even small amounts of partially functional CFTR over the lifetime of an individual might be sufficient to moderate CF traits such as sweat chloride concentration and lung function.38 The concept that natural variation in the expression level of mutated genes may underlie differences in the severity of inherited diseases is supported by recent studies of loss-of-function C. elegans phenotypes.42 Additionally, we posit that the intragenic and extragenic variation present in the F508del population may confer increased or decreased response to Orkambi or future CFTR-specific drugs.
The most 5′ regions of interest (Regions B and E, Figures 1 and 2) were located within the introns of WNT2 and ASZ1. The region in WNT2 is located in an adjacent TAD to CFTR. In a recent study, there was no report of this region interacting with the CFTR locus.22 However, previous studies in epididymis cells indicate there are weak long-range chromatin interactions with this region that may be cell type specific.13 It is possible that some of the rare variants associating with sweat chloride in this region modify overall chromatin organization in certain cell types, such as the sweat gland. Another distant region of interest was found within ASZ1, and is located just outside of the proposed CFTR TAD.22 These regions are often enriched for TAD-TAD interactions. Variants here could alter CTCF binding, TAD architecture or inter-TAD interactions. Assaying both of these possible inter-TAD interactions could lead to additional insight into distant regulatory elements in ASZ1 and WNT2. Of note, the 5′ TAD boundary proposed by Smith and Dekker22 closely follows the recombination event in intron 10 of ASZ1 in this study, suggesting a possible link between recombination events and chromatin structure in this region.
Interestingly, adjacent regions within intron 3 of CFTR were found to associate with both sweat chloride levels and lung function (albeit, there is no distinct overlap given the coordinates identified here). To our knowledge, this region has not previously been shown to have regulatory function. While the intron 3 signal for sweat chloride was primarily composed of rare variation, common variation in the length of a poly T tract resulted in the lung function association. Interestingly, while not achieving significance, the 18T and 16T alleles at this locus trended toward association with sweat chloride levels as well (P=0.06, beta=−3.58 mM Cl− and P=0.08, β=+3.49 mM Cl−, respectively). We do note that this poly T tract still modulates lung function when the cohorts are considered independently (18T allele: P=0.051, β=+0.18, n=486 and P=0.12, β=+0.19, n=276). Given the lack of functional elements and low conservation in this region, it is challenging to imagine a mechanism by which this alteration could modulate CF traits. However, poly T tracts may regulate gene expression by acting as matrix attachment regions,43 or may participate in RNA triplex formation.44
A recurring theme throughout the variation observed in this study was variable lengths of repetitive elements associating with disease severity. These INDELs may represent a mode of phenotype modification that is not well characterized,45 but has been previously observed to modify the phenotype of other CF-causing alleles (i.e., R117H and polyT tract).46 This type of variation is observed in five of the seven regions of interest. A limitation of the current study is that insertion/deletion variants may be inadequately characterized because of limitations of current sequencing methods. However, all INDELs reported here were of both high mapping and variant call qualities, and variant frequencies did not deviate from Hardy-Weinberg equilibrium. It is possible that these small variants may be partially marking larger repetitive sequences that could not be typed in this study due to read length (or high homology). Additional studies of common and rare INDELs at these loci could reveal a mechanism of phenotype modification. Finally, we recognize that some of the associations employed here have limited power, especially at low minor allele frequencies given the cohort size (which ranges from 276 to 762, depending on the region). Power is additionally limited when assaying sweat chloride associations in the phase 2 cohort, as this cohort was selected for extremes of lung function, and thus contains intermediate sweat chloride values. Given these limitations, the study presented here likely contains false negatives, which could only be resolved using larger cohorts.
Some sequencing studies fail to consider regions of known homology with the region of interest. In this study, we opted to allow for a higher frequency of false positives in regions of the capture with high homology to pseudogenes (specifically intron 9 and exon 10 of CFTR, Supplementary Table 2).20 This was to allow for more consistent tiling of baits, better detection of large structural variants and a more complete capture overall. Clinical labs should be aware of these regions when designing assays in order to minimize erroneous calls. For example, the nonsynonymous mutation A455E is a high-frequency CF-causing allele in exon 10. This variant is also present in a pseudogene present on chromosome 20. While this variant can be correctly typed using a longer read length, short amplifications cannot distinguish between these two forms.47 The variants reported in Supplementary Table 2 could be assigned to either the chromosome 9 or chromosome 20 pseudogenes due to their reoccurrence in a small subset of samples (n=5); however, alternative methods would be required in a clinical setting.
Using the rich dataset produced by sequencing the entire CFTR locus, we were able to resolve the genetic architecture surrounding the common CF-causing variant to an unprecedented level of detail. We have now made available a detailed map of common variation and population-based haplotypes for the F508del locus (Supplementary Table 7). Specifically, we describe 13 haplotype-tagging SNPs that represent the vast majority of the genetic variation surrounding F508del. These SNPs could be used to parse F508del homozygotes into subpopulations to test whether variation at the CFTR locus underlies differences in responses to molecular-targeted treatments. Furthermore, they could be used to infer F508del carrier status in non-CF genome-wide association studies.
Overall variation was rare within the LD block containing F508del, consistent with a single ancestral origin of this allele in the population. When considering only common SNPs, the majority of F508del chromosomes (~55%) are completely identical. These results indicate the F508del homozygous population is highly homogenous, with the majority of variation being private or due to a low-frequency recombination event within intron 15. Because this event is not observed on non-F508del chromosomes, it likely occurred after F508del arose. Previously, a recombination event was reported to have occurred within intron 22.25 However, this recombination event was based on population-level data provided by the HapMap project, which used wild-type CFTR and had significantly reduced marker density compared to this study.48 Newer HapMap releases suggest two possible primary recombination events in the general population: intron 11 and intron 15-intron 16. A recombination event at intron 15-intron 16 event appears to have occurred more than once, both in F508del-containing and in wild-type chromosomes. The previously reported intron 22 event may have some limited evidence in Mexican and Italian Hapmap cohorts.
In summary, this study has methodically characterized variation in cis with the F508del allele and the genetic architecture of this locus in great depth. Collectively, our findings suggest a combination of rare and common variation within suspect and known regulatory regions at the CFTR locus may contribute to the phenotypic heterogeneity observed in F508del homozygous CF patients. The identified variation may modify CFTR expression levels and/or timing of expression, and should inform future regulatory studies of this locus.
Acknowledgments
We would like to acknowledge David Mohr and CIDR for their thoughtful and methodical design of the resequencing capture. We would also like to thank the Cystic Fibrosis Foundation (CUTTIN13A2) and the National Institutes of Health (DK44003) for funding this work.
The authors declare no conflict of interest.
Footnotes
Supplementary Information for this article can be found on the Human Genome Variation website (http://www.nature.com/hgv).
References
- Gadsby DC, Vergani P, Csanady L. The ABC protein turned chloride channel whose failure causes cystic fibrosis. Nature 2006; 440: 477–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerem E, Corey M, Kerem B-S, Rommens J, Markiewicz D, Levison H et al. The relation between genotype and phenotype in cystic fibrosis--analysis of the most common mutation (deltaF508). N Engl J Med 1990; 323: 1517–1522. [DOI] [PubMed] [Google Scholar]
- Bobadilla JL, Macek M, Fine JP, Farrell PM. Cystic fibrosis: a worldwide analysis of CFTR mutations - correlation with incidence data and application to screening. Hum Mutat 2002; 19: 575–606. [DOI] [PubMed] [Google Scholar]
- Qu BH, Strickland E, Thomas PJ. Cystic fibrosis: a disease of altered protein folding. J Bioenerg Biomembr 1997; 29: 483–490. [DOI] [PubMed] [Google Scholar]
- Cutting GR. Modifier genes in Mendelian disorders: the example of cystic fibrosis. Ann NY Acad Sci 2010; 1214: 57–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wainwright CE, Elborn JS, Ramsey BW, Marigowda G, Huang X, Cipolli M et al. Lumacaftor-Ivacaftor in patients with cystic fibrosis homozygous for Phe508del CFTR. N Engl J Med 2015; 373: 220–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukacs GL, Verkman AS. CFTR: folding, misfolding and correcting the DeltaF508 conformational defect. Trends Mol Med 2012; 18: 81–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Goor F, Hadida S, Grootenhuis PD, Burton B, Stack JH, Straley KS et al. Correction of the F508del-CFTR protein processing defect in vitro by the investigational drug VX-809. Proc Natl Acad Sci USA 2011; 108: 18843–18848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phuan PW, Veit G, Tan J, Roldan A, Finkbeiner WE, Lukacs GL et al. Synergy-based small-molecule screen using a human lung epithelial cell line yields DeltaF508-CFTR correctors that augment VX-809 maximal efficacy. Mol Pharmacol 2014; 86: 42–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamosh A, Corey M. Correlation between genotype and phenotype in patients with cystic fibrosis. The Cystic Fibrosis Genotype-Phenotype Consortium. N Engl J Med 1993; 329: 1308–1313. [DOI] [PubMed] [Google Scholar]
- Collaco JM, Blackman SM, Raraigh KS, Corvol H, Rommens JM, Pace RG et al. Sources of Variation in Sweat Chloride Measurements in Cystic Fibrosis. Am J Respir Crit Care Med (e-pub ahead of print 3 June 2016; doi:10.1164/rccm.201603-0459OC). [DOI] [PMC free article] [PubMed]
- Blackledge NP, Ott CJ, Gillen AE, Harris A. An insulator element 3′ to the CFTR gene binds CTCF and reveals an active chromatin hub in primary cells. Nucleic Acids Res 2009; 37: 1086–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R, Kerschner JL, Gosalia N, Neems D, Gorsic LK, Safi A et al. Differential contribution of cis-regulatory elements to higher order chromatin structure and expression of the CFTR locus. Nucleic Acids Res 2015; 44: 3082–3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobczyńska-Tomaszewska A, Ołtarzewski M, Czerska K, Wertheim-Tysarowska K, Sands D, Walkowiak J et al. Newborn screening for cystic fibrosis: Polish 4 years' experience with CFTR sequencing strategy. Eur J Hum Genet 2013; 21: 391–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolesár P, Minárik G, Baldovic M, Ficek A, Kovács L, Kádasi L. Mutation analysis of the CFTR gene in Slovak cystic fibrosis patients by DHPLC and subsequent sequencing: identification of four novel mutations. Gen Physiol Biophys 2008; 27: 299–305. [PubMed] [Google Scholar]
- Amato F, Bellia C, Cardillo G, Castaldo G, Ciaccio M, Elce A et al. Extensive molecular analysis of patients bearing CFTR-related disorders. J Mol Diagn 2012; 14: 81–89. [DOI] [PubMed] [Google Scholar]
- Elahi E, Khodadad A, Kupershmidt I, Ghasemi F, Alinasab B, Naghizadeh R et al. A haplotype framework for cystic fibrosis mutations in Iran. J Mol Diagn 2006; 8: 119–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit LS, Wilkinson DJ, Mansoura MK, Collins FS, Dawson DC. Functional roles of the nucleotide-binding folds in the activation of the cystic fibrosis transmembrane conductance regulator. Proc Natl Acad Sci USA 1993; 90: 9963–9967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Seedy A, Dudognon T, Bilan F, Pasquet MC, Reboul MP, Iron A et al. Influence of the duplication of CFTR exon 9 and its flanking sequences on diagnosis of cystic fibrosis mutations. J Mol Diagn 2009; 11: 488–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozmahel R, Heng HH, Duncan AM, Shi XM, Rommens JM, Tsui LC. Amplification of CFTR exon 9 sequences to multiple locations in the human genome. Genomics 1997; 45: 554–561. [DOI] [PubMed] [Google Scholar]
- Gheldof N, Smith EM, Tabuchi TM, Koch CM, Dunham I, Stamatoyannopoulos JA et al. Cell-type-specific long-range looping interactions identify distant regulatory elements of the CFTR gene. Nucleic Acids Res 2010; 38: 4325–4336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith EM, Lajoie BR, Jain G, Dekker J. Invariant TAD boundaries constrain cell-type-specific looping interactions between promoters and distal elements around the CFTR locus. Am J Hum Genet 2016; 98: 185–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium, E. A user's guide to the Encyclopedia of DNA Elements (ENCODE). PLoS Biol 2011; 9: e1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaneau O, Zagury JF, Marchini J. Improved whole-chromosome phasing for disease and population genetic studies. Nat Methods 2013; 10: 5–6. [DOI] [PubMed] [Google Scholar]
- Cordovado S, Hendrix M, Greene C, Mochal S, Earley M, Farrell P et al. CFTR mutation analysis and haplotype associations in CF patients. Mol Genet Metab 2012; 105: 249–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morral N, Bertranpetit J, Estivill X, Nunes V, Casals T, Giménez J et al. The origin of the major cystic fibrosis mutation (deltaF508) in European populations. Nature Genet 1994; 7: 169–175. [DOI] [PubMed] [Google Scholar]
- Fichou Y, Genin E, Le MC, Audrezet MP, Scotet V, Ferec C. Estimating the age of CFTR mutations predominantly found in Brittany (Western France). J Cyst Fibros 2008; 7: 168–173. [DOI] [PubMed] [Google Scholar]
- Rommens JM, Zengerling S, Burns J, Melmer G, Kerem BS, Plavsic N et al. Identification and regional localization of DNA markers on chromosome 7 for the cloning of the cystic fibrosis gene. Am J Hum Genet 1988; 43: 645–663. [PMC free article] [PubMed] [Google Scholar]
- Kerem B, Rommens JM, Buchanan JA, Markiewicz D, Cox TK, Chakravarti A et al. Identification of the cystic fibrosis gene: genetic analysis. Science 1989; 245: 1073–1080. [DOI] [PubMed] [Google Scholar]
- Rommens JM, Iannuzzi MC, Kerem B, Drumm ML, Melmer G, Dean M et al. Identification of the cystic fibrosis gene: chromosome walking and jumping. Science 1989; 245: 1059–1065. [DOI] [PubMed] [Google Scholar]
- McCarthy VA, Harris A. The CFTR gene and regulation of its expression. Pediatr Pulmonol 2005; 40: 1–8. [DOI] [PubMed] [Google Scholar]
- Ott CJ, Suszko M, Blackledge NP, Wright JE, Crawford GE, Harris A. A complex intronic enhancer regulates expression of the CFTR gene by direct interaction with the promoter. J Cell Mol Med 2009; 13: 680–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broackes-Carter FC, Mouchel N, Gill D, Hyde S, Bassett J, Harris A. Temporal regulation of CFTR expression during ovine lung development: implications for CF gene therapy. Hum Mol Genet 2002; 11: 125–131. [DOI] [PubMed] [Google Scholar]
- Gosalia N, Harris A. Chromatin dynamics in the regulation of CFTR expression. Genes 2015; 6: 543–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moisan S, Berlivet S, Ka C, Le Gac G, Dostie J, Férec C. Analysis of long-range interactions in primary human cells identifies cooperative CFTR regulatory elements. Nucl Acids Res 2015; 44: 2564–2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke F, van Barneveld A, Hedtfeld S, Wolfl S, Becker T, Tummler B. The CF-modifying gene EHF promotes p. Phe508del-CFTR residual function by altering protein glycosylation and trafficking in epithelial cells. Eur J Hum Genet 2014; 22: 660–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mall M, Wissner A, Seydewitz HH, Hübner M, Kuehr J, Brandis M et al. Effect of genistein on native epithelial tissue from normal individuals and CF patients and on ion channels expressed in Xenopus oocytes. Br J Pharmacol 2000; 130: 1884–1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veeze HJ, Halley JJ, deJongste JC, deJonge HR, Sinaasappel M. Determinants of mild clinical symptoms in cystic fibrosis patients. J Clin Invest 1994; 93: 461–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penque D, Mendes F, Beck S, Farinha C, Pacheco P, Nogueira P et al. Cystic fibrosis F508del patients have apically localized CFTR in a reduced number of airway cells. Lab Invest 2000; 80: 857–868. [DOI] [PubMed] [Google Scholar]
- Dekkers JF, van der Ent CK, Beekman JM. Novel opportunities for CFTR-targeting drug development using organoids. Rare Dis 2013; 1: 939–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Barneveld A, Stanke F, Tamm S, Siebert B, Brandes G, Derichs N et al. Functional analysis of F508del CFTR in native human colon. Biochim Biophys Acta 2010; 1802: 1062–1069. [DOI] [PubMed] [Google Scholar]
- Vu V, Verster AJ, Schertzberg M, Chuluunbaatar T, Spensley M, Pajkic D et al. Natural Variation in Gene Expression Modulates the Severity of Mutant Phenotypes. Cell 2015; 162: 391–402. [DOI] [PubMed] [Google Scholar]
- Boulikas T. Chromatin domains and prediction of MAR sequences. Int Rev Cytol 1996; 162: 279–388. [DOI] [PubMed] [Google Scholar]
- Devi G, Zhou Y, Zhong Z, Toh DFK, Chen G. RNA triplexes: from structural principles to biological and biotech applications. Wiley Interdiscipl Rev 2015; 6: 111–128. [DOI] [PubMed] [Google Scholar]
- Gymrek M, Willems T, Guilmatre A, Zeng H, Markus B, Georgiev S et al. Abundant contribution of short tandem repeats to gene expression variation in humans. Nat Genet 2016; 48: 22–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiesewetter S, Macek M Jr., Curristin S, Chu C, Graham C, Shrimpton AE et al. The CFTR mutation R117H produces different phenotypes depending on genetic background. Am J Med Genet 1993; 53: #86. [DOI] [PubMed] [Google Scholar]
- Deeb KK, Metcalf JD, Sesock KM, Shen J, Wensel CA, Rippel LI et al. The c.1364C>A (p.A455E) mutation in the CFTR pseudogene results in an incorrectly assigned carrier status by a commonly used screening platform. J Mol Diagn 2015; 17: 360–365. [DOI] [PubMed] [Google Scholar]
- Consortium, TIH. The International HapMap Project. Nature 2003; 426: 789–796. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.