Abstract
Objective
To establish the relative importance of Salmonella enterica serovar Typhi with non-classical quinolone resistance.
Methods
Eight hundred and ninety-one isolates of S. Typhi, isolated between 2004 and 2011, were tested for antibiotic susceptibility determination using disc diffusion and E-test. The mechanisms of fluoroquinolone resistance were studied in a sub-set of the NALS (nalidixic acid susceptible) isolates by wave nucleic acid fragment analysis of PCR products from gyrA, gyrB, parC and parE and from the plasmid borne determinants: qnrA,B,S; aac(6′)-Ib-cr and qepA. To assess genetic relatedness multi-locus variable number tandem repeat analysis was carried out using five loci.
Results
Eighty isolates with a nalidixic acid MIC of <32 mg/L (NALS) and a ciprofloxacin MIC of >0.064 mg/L CIPI (ciprofloxacin reduced susceptibility) were found. In 36 NALS CIPI isolates two distinct genotypes were identified when compared with 16 susceptible controls: Group B (n = 34), mutation in gyrB at codon 464, NAL MIC of 3–12 mg/L and CIP MIC of 0.064–0.5 mg/L.; and Group C, mutation in gyrA at codon 83 (n = 2) NAL MIC of 16 mg/L and CIP MIC of 0.25–0.38 mg/L. Group B isolates were found in different strain backgrounds as defined by MLVA.
Conclusion
The use of nalidixic acid to screen for reduced susceptibility to fluoroquinolones in S. Typhi misses CIPI-NALS isolates, an established phenotype in India.
Keywords: Salmonella Typhi, Decreased ciprofloxacin susceptibility, DHPLC, gyrB mutation, VNTR
1. Introduction
Typhoid fever, caused by Salmonella enterica serovar Typhi (S. Typhi), remains a global public health problem centred in developing countries [1]. The emergence of multi-drug resistant (MDR) S. Typhi, with resistance to all first line drugs: (chloramphenicol, co-trimoxazole and ampicillin) led to the fluoroquinolones becoming the treatment of choice for enteric fever [2]. There is a clear relationship between increasing fluoroquinolone MIC and outcome during fluoroquinolone treatment of typhoid fever. Therefore for clinical management, any increase in fluoroquinolone MIC needs to be detected [3]. No zone around a nalidixic acid 30 μg disc has been used as a screening test for isolates with a clinically relevant ciprofloxacin resistance; MIC of ≥0.125 mg/L with a sensitivity of 92.9% (248/267); and a specificity of 98.4% (540/549) [4]. The most common mutation associated with reduced susceptibility to fluoroquinolones in S. Typhi is in the quinolone resistance determining region of the GyrA sub-unit of DNA gyrase. This mutation, at amino acid 83, also confers resistance to nalidixic acid (NALR) and so nalidixic acid can be a marker for fluoroquinolone resistance where this mutation is the sole cause of resistance. Strains of S. Typhi with decreased susceptibility to fluoroquinolones (e.g. ciprofloxacin (CIPI) MIC > 0.064 mg/L are now very common in India [3]. The reports of alternative mechanisms of fluoroquinolone resistance are increasing [5], [6]. In response to this the CLSI has redefined the breakpoints for ciprofloxacin as ≤0.064 mg/L and ≥1 mg/L and ≥31 mm and ≤20 mm for susceptibility and resistance using MIC and disc diffusion [7], and strains that are >0.064 mg/L and <1 mg/L as intermediate for Salmonella enterica. Many laboratories in India however continue to test nalidixic acid as a marker for decreased susceptibility to fluoroquinolones (CIPI) and so a formal report on the utility of the nalidixic acid screening test in India is of great importance to guide clinical management. Furthermore there is conflicting evidence on the role of fluoroquinolones mutations in clinical isolates; two mutations in gyrA and/or gyrB can be associated with full resistance to fluoroquinolones (MIC ≥ 4 mg/L) [8] but genetic manipulation of isogenic mutants shows that this is not the full story – other mutations must also be present [9]. It is therefore important to characterize circulating strain.
The Indian sub-continent reports more typhoid fever than any other world region and so monitoring of antibiotic resistance in S. Typhi in India can be used to detect any trend towards increasing ciprofloxacin resistance. Our centre is one of the busiest in India and surveillance of ciprofloxacin resistance has been carried out since 2001. In a previous study from 2003 the majority (282/285) of isolates with CIPI exhibited the classical quinolone resistance phenotype, NALR-CIPI and three isolates with NALS-CIPI (CIP-MIC 0.125 mg/L) were observed [10]. Here we present data on the genetic and phenotypic characterization of CIPI isolates of S. Typhi from 2004 to 2011.
2. Methods
All isolates were from patients admitted to the Safdarjung Hospital during 2004–2011 and were from blood or stool cultures, deduplication was carried out as far as possible using the clinical data available retrospectively. The Minimum Inhibitory Concentration (MIC) for nalidixic acid and ciprofloxacin against 891 S. Typhi was determined by E-test (AB Biodisk, Solna, Sweden). Results were interpreted by two microbiologists and discrepant results were repeated. Susceptibilities to nalidixic acid (30 μg), ciprofloxacin (5 μg) chloramphenicol (30 μg), co-trimoxazole (25 μg) and ampicillin (10 μg) were determined by disc diffusion (DD) test according to CLSI guidelines [7]. The breakpoints used for CIPI were 0.064–1.0 mg/L and 21–30 mm for MIC and DD respectively. MDR was defined as resistance to chloramphenicol, co-trimoxazole, and ampicillin. The data was analyzed using WHONET 5.6. software.
From the NALS S. Typhi (MIC < 32 mg/L) 52 strains were selected to represent all years of isolation and all antibiograms to characterize the mechanism of FQ resistance. PCR was used to amplify the QRDRs of gyrA, gyrB, parC and parE genes [3]. Denaturing high-pressure liquid chromatography (DHPLC: Wave Nucleic Acid Fragment Analysis System; Transgenomic Inc.) was used to study mutations [11]. Mutations in the genes studied were identified by comparison with DNA sequences from S. Typhi strains Ty2 (GenBank accession no. AE014613). Screening for the plasmid determinants (qnrA,B,S; aac(6′)-Ib-cr and qepA) was carried out by PCR (primers in Table 1).
Table 1.
Primer sequences of plasmid mediated resistance (PMQR) genes: qnrA, qnrB, qnrS, aac(6′)-Ib-cr and qepA.
| S. No. | Primer of PMQR genes | Amplicon size |
|---|---|---|
| 1. |
qnrA FP: 5′-ATT TCT CAC GCC AGG ATT TG-3′ RP: 5′-GAT CGG CAA AGG TTA GGT CA-3′ |
516 bp |
| 2. |
qnrB FP: 5′-GAT CGT GAA AGC CAG AAA GG-3′ RP: 5′-ACG ATG CCT GGT AGT TGT CC-3′ |
469 bp |
| 3. |
qnrS FP: 5′-ACG ACA TTC GTC AAC TGC AA-3′ RP: 5′-TAA ATT GGC ACC CTG TAG GC-3′ |
417 bp |
| 4. |
aac(6′)-Ib-cr FP: 5′-TTG CGA TGC TCT ATG AGT GGC TA-3′ RP: 5′-CTC GAA TGC CTG GCG TGT TT-3′ |
489 bp |
| 5. |
qepA FP: 5′-GCA GGT CCA GCA GCG GGT AG-3′ RP: 5′-CTT CCT GCC CGA GTA TCG TG-3′ |
1100 bp |
Multi-locus variable number tandem repeat (VNTR) analysis (MLVA) was carried out to assess genetic relatedness using five VNTR loci (TR1,TR2, TR4699, Sal02 and Sal16) [12]. Primers were tagged with 6′FAM (blue) fluorescent dye. Fragment length analysis was done by capillary electrophoresis (ABI3130 Genetic analyzer, Applied Biosystems). Fragment sizes were binned into alleles. Copy number was confirmed by sequencing. A cluster dendogram was constructed using R-software.
3. Results and discussion
Among 891 S. Typhi isolates the numbers of each antibacterial phenotype isolated were as follows: of wild type (NALS-CIPS) 46 (5.2%), classical fluoroquinolone decreased susceptibility (NALR-CIPI) 675 (75.8%), non-classical fluoroquinolone decreased susceptibility (NALS-CIPI) 80 (9%) and fluoroquinolone resistance 90 (10%).
The proportion of NALR increased from 76.1% in 2004 to 97.0% in 2011. Of interest is that among the NALS-isolates CIPI was isolated every year although the number varied (Fig. 1). In 2011 only 1/104 isolates was fully susceptible to ciprofloxacin and nalidixic acid. The NALS-CIPI isolates appear to be established within the population. In order to assess the reasons for this stability we decided to investigate the genetic variation present within the population of isolates with the NALS-CIPI phenotype.
Fig. 1.
The relative numbers of isolates of Salmonella Typhi at Safdarjung hospital. (A) NALS and NALR-CIPI. (B) NALS and NALS-CIPI. Note: NALS = nalidixic acid susceptible (MIC ≤ 16 mg/L). NALR = nalidixic acid resistance (MIC ≥ 32 mg/L). CIPI = decreased ciprofloxacin susceptible (MIC > 0.064 mg/L).
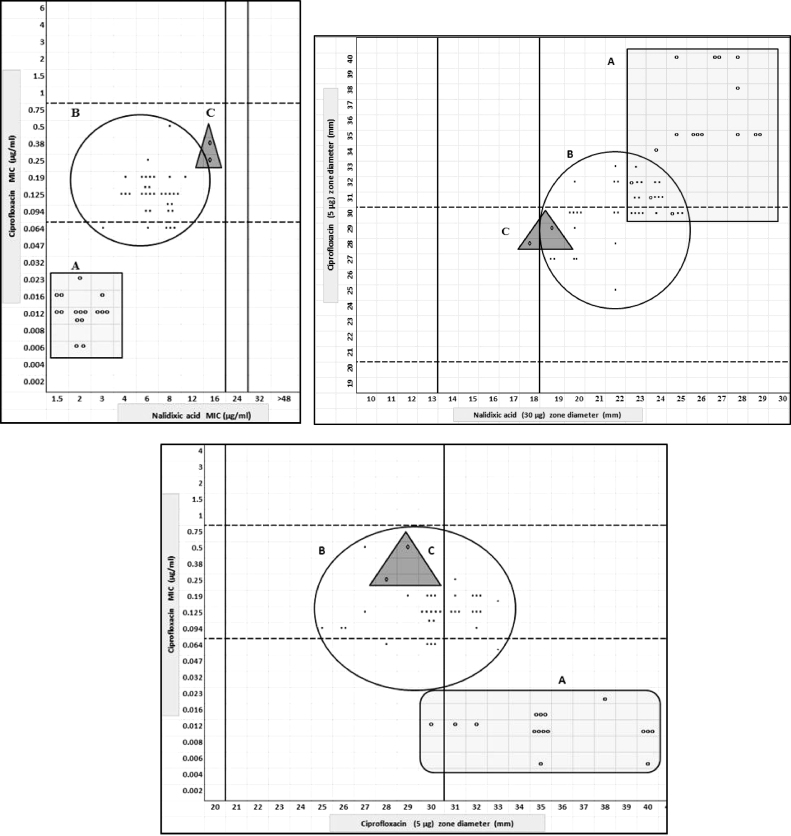
No plasmid-encoded determinants were detected. No mutations were observed in the QRDR of parC and parE. Mutations were found in the QRDR of gyrA and gyrB. Results are shown in Table 2. From the mutations detected we split all isolates into three groups. Group A (n = 16): isolates with wild type gyrA and gyrB gene; Group B (n = 34): gyrB mutation only; and Group C (n = 2) gyrA mutation only. Scatter plots correlating the zone diameters and MIC of CIP vs NAL and CIP MIC vs CIP zone diameters of these isolates are shown in Fig. 2. Although there is some heterogeneity in this group of 52 isolates it is clear that each group: A, B and C, could be distinctly identified based on the MIC data for CIP and NAL (Table 2). It is also clear that the mutations on gyrB (Group B) are a common cause of the NALS-CIPI phenotype in S. Typhi in India. Group B isolates (mutation in gyrB) were distinct, with NAL (3–12 mg/L) and CIP (0.064–0.5 mg/L) MIC being 3- and 15-fold higher respectively than the wild type group A isolates but where difficult to identify by a single phenotypic test; only 16/34 group B isolates were CIPI both by disc diffusion and MIC. Thus using the revised CLSI breakpoints the group B isolates would be very difficult to classify: the ciprofloxacin zone diameter breakpoint and MICs for this group of isolates cut across the defined breakpoints. This is in agreement with reports from France (n = 12) [13] and UK (n = 8) where reported CIP-MIC range among S. Typhi NALS-CIPI with gyrB mutation were 0.032–0.19 mg/L and 0.03–0.05 mg/L respectively [14]. The predominant gyrB mutation among isolates of Indian origin in these studies was Ser-464→Phe (accession no. KF993966). Our findings corroborate this. The gyrB mutations at Asp 466, Leu 465 and Glu 468 reported earlier [6], [13] were not observed in our study. Other mutations observed among our isolates were Ser-464→Tyro (n = 2) (accession no. KF993965) and a novel mutation not reported so far was observed at Ser-464→Thre (n = 1) (accession no. KF993964).
Table 2.
Characteristics of the 52 Salmonella enterica serovar Typhi isolates belonging to Groups A, B and C, India 2004–2011.
| Group | No. of isolates | NAL MIC (μg/ml) |
CIP MIC (μg/ml) |
QRDR mutation | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| MIC50 | MIC90 | GM | MIC range | MIC50 | MIC90 | GM | MIC range | |||
| A | 16 | 2 | 3 | 2.06 | 1.5–3 | 0.012 | 0.016 | 0.012 | 0.006–0.023 | No mutation |
| B | 34a | 6 | 8 | 6.44 | 3–12 | 0.125 | 0.19 | 0.13 | 0.064–0.5 |
gyrB 464 Ser→Tyr (n = 2) Ser→Phe (n = 31) Ser→Thre (n = 1) |
| C | 2 | 16 | 16 | 16 | 16 | 0.25 | 0.38 | 0.38 | 0.25–0.38 |
gyrA 83 Ser→Tyro |
NAL, nalidixic acid; CIP, ciprofloxacin; GM, geometric mean; QRDR, quinolone resistance-determining region of the gyrA, gyrB, parC and parE genes.
13 isolates were additionally resistant to chloramphenicol, co-trimoxazole and ampicillin.
Fig. 2.
Scatter plot showing the relationship between nalidixic acid and ciprofloxacin MIC, and zone size diameter, for 52 S. Typhi isolates with (A) no mutation in topoisomerase genes; (B) a mutation in gyrB only; and (C) a mutation in gyrA only.
One explanation for the phenotypic variation could be strain background and their genetic relatedness was assessed by MLVA. Among group B isolates tested, 17 distinct VNTR profiles were observed (data not shown). This suggests that the selective advantage of the resistance phenotype associated with gyrB Ser-464→Phe is not associated with a biological cost. Were compensatory mutations necessary the primary mutation would be present in a limited portion of the population–this is not the case, gyrB Ser-464→Phe has become fixed in several branches of the population. There were also two isolates which were MDR as well as NALSCIPI with a mutation at gyrB Ser-464→Phe and two NALS-CIPI isolates with different MDR phenotype also showed an identical VNTR profile suggesting that there was no linkage between the MDR phenotype and gyrB Ser-464→Phe. This data means that losing the mutations, and by association the CIPI phenotype, from the population by reducing selection from the use of fluoroquinolones is very unlikely.
Two isolates Group C, selected as NALS CIPI (MIC 16 mg/L and zone diameter 18 and 19 mm for nalidixic acid) had gyrA mutation (Ser-83→Tyr) which would normally encode resistance to nalidixic acid, these strains must have an unusual background which potentiated the action of nalidixic acid but not the fluoroquinolones. We did not have the resource to investigate these interesting strains further.
4. Conclusions
It is important to know the clinical significance of NALS-CIPI among S. Typhi. The impact of the NALS-CIPI on fluoroquinolone treatment in typhoid fever is unknown as there are no studies that document outcome of this infection following treatment. Even in our study, patients with infection due to these strains could not be followed as most of them presented in the outpatient department. However, the CIP MIC is likely to be the most important determinant of clinical response to therapy, irrespective of resistance mechanism and therefore CIPI must be considered as likely to fail ciprofloxacin therapy. Furthermore the emergence of NALS-CIPI in several different background strains for S. Typhi, as evidenced by the MLVA data, suggests that these CIPI mutants are biologically fit. We therefore have significant concern that under-reporting of CIPI by a failure to detect CIPI-NALS isolates (by conventional NAL resistance) may facilitate the subsequent emergence of high-level FQ resistance. Our results show that screening with nalidixic acid only will not detect 14/34 and 5/34 of the CIPI-NALS isolates supporting the CLSI guidelines to use CIP testing to detect CIPI Salmonella. Alternative screening tests need to be explored to identify these isolates for optimal treatment. Co-existence of the MDR phenotype in 38.2% of CIPI-NALS may further restrict therapeutic options. There has been a rapid evolution of NALR-CIPI that has been maintained through selective pressure and these strains are now endemic in India subcontinent. It is thus important to study the spread of NALS-CIPI strains. These isolates represented 11.6% (49/421) of the S. Typhi isolated in England, Scotland, and Wales (1999–2003) and 4.6% (36/770) of S. Typhi isolates in the United States (1999–2006). In France among 685 S. Typhi (1997–2009) the incidence of these isolates was rare (1.0%). In our study (2004–2011) overall prevalence of these isolates was 9.0%. Among the NAL susceptible population during 2004–2011 the ratio of isolates CIPI-NALS to wild type QRDR shifted from 1:1.3 to 2:1, but their isolation has decreased from 10.3 to 1.9%. Although the overall prevalence of these strains is declining, they have arisen and spread on more than one occasion and so will likely reemerge in the future.
Funding
This work was supported by Indian Council of Medical Research, Govt. of India (Grant No. 5/8-1(8) 2010-11/ECD-II IRIS cell, ID-2010-04020) to Dr. Rajni Gaind.
Acknowledgments
We would like to thank the staff of the Safdarjung Hospital and the editors and reviewers of the BDQ journal.
Contributor Information
Ruchi Gupta, Email: ruchigupta_08@yahoo.com.
Rajni Gaind, Email: rgaind5@hotmail.com.
Seemi Farhat Basir, Email: seemifb@gmail.com.
References
- 1.Buckle G.C., Walker C.L., Black R.E. Typhoid fever and paratyphoid fever: systematic review to estimate global morbidity and mortality for 2010. J Glob Health. 2012;2(1):10401. doi: 10.7189/jogh.02.010401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wain J., Kidgell C. The emergence of multidrug resistance to antimicrobial agents for the treatment of typhoid fever. Trans R Soc Trop Med Hyg. 2004;98(7):423–430. doi: 10.1016/j.trstmh.2003.10.015. [DOI] [PubMed] [Google Scholar]
- 3.Wain J., Hoa N.T., Chinh N.T., Vinh H., Everett M.J., Diep T.S. Quinolone-resistant Salmonella typhi in Viet Nam: molecular basis of resistance and clinical response to treatment. Clin Infect Dis. 1997;25(6):1404–1410. doi: 10.1086/516128. [DOI] [PubMed] [Google Scholar]
- 4.Parry C.M., Thuy C.T., Dongol S., Karkey A., Vinh H., Chinh N.T. Suitable disk antimicrobial susceptibility breakpoints defining Salmonella enterica serovar Typhi isolates with reduced susceptibility to fluoroquinolones. Antimicrob Agents Chemother. 2010;54(12):5201–5208. doi: 10.1128/AAC.00963-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cooke F.J., Day M., Wain J., Ward L.R., Threlfall E.J. Cases of typhoid fever imported into England: Scotland and Wales (2000–2003) Trans R Soc Trop Med Hyg. 2007;101(4):398–404. doi: 10.1016/j.trstmh.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 6.Song Y., Roumagnac P., Weill F.X., Wain J., Dolecek C., Mazzoni C.J. A multiplex single nucleotide polymorphism typing assay for detecting mutations that result in decreased fluoroquinolone susceptibility in Salmonella enterica serovars Typhi and Paratyphi A. J Antimicrob Chemother. 2010;65(8):1631–1641. doi: 10.1093/jac/dkq175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.CLSI . vol. 33 (1) 2013. (Performance Standards for Antimicrobial Susceptibility Testing: 23rd Informational Supplement Clinical Laboratory Standards Institute Document M100-S23). [Google Scholar]
- 8.Chau T.T., Campbell J.I., Galindo C.M., Van Minh Hoang N., Diep T.S., Nga T.T. Antimicrobial drug resistance of Salmonella enterica serovar Typhi in Asia and molecular mechanism of reduced susceptibility to the fluoroquinolones. Antimicrob Agents Chemother. 2007;51(12):4315–4323. doi: 10.1128/AAC.00294-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Turner A.K., Nair S., Wain J. The acquisition of full fluoroquinolone resistance in Salmonella Typhi by accumulation of point mutations in the topoisomerase targets. J Antimicrob Chemother. 2006;58(4):733–740. doi: 10.1093/jac/dkl333. [DOI] [PubMed] [Google Scholar]
- 10.Walia M., Gaind R., Mehta R., Paul P., Aggarwal P., Kalaivani M. Current perspectives of enteric fever: a hospital-based study from India. Ann Trop Paediatr. 2005;25(3):161–174. doi: 10.1179/146532805X58085. [DOI] [PubMed] [Google Scholar]
- 11.Randall L.P., Coldham N.G., Woodward M.J. Detection of mutations in Salmonella enterica gyrA: gyrB, parC and parE genes by denaturing high performance liquid chromatography (DHPLC) using standard HPLC instrumentation. J Antimicrob Chemother. 2005;56(4):619–623. doi: 10.1093/jac/dki293. [DOI] [PubMed] [Google Scholar]
- 12.Octavia S., Lan R. Multiple-locus variable-number tandem-repeat analysis of Salmonella enterica serovar Typhi. J Clin Microbiol. 2009;47(8):2369–2376. doi: 10.1128/JCM.00223-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Accou-Demartin M., Gaborieau V., Song Y., Roumagnac P., Marchou B., Achtman M. Salmonella enterica serotype Typhi with nonclassical quinolone resistance phenotype. Emerg Infect Dis. 2011;17(6):1091–1094. doi: 10.3201/eid1706.101242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cooke F.J., Wain J., Threlfall E.J. Fluoroquinolone resistance in Salmonella Typhi. BMJ. 2006;333(7563):353–354. doi: 10.1136/bmj.333.7563.353-b. [DOI] [PMC free article] [PubMed] [Google Scholar]