Abstract
Molecular diagnostic measurements are currently underpinned by the polymerase chain reaction (PCR). There are also a number of alternative nucleic acid amplification technologies, which unlike PCR, work at a single temperature. These ‘isothermal’ methods, reportedly offer potential advantages over PCR such as simplicity, speed and resistance to inhibitors and could also be used for quantitative molecular analysis. However there are currently limited mechanisms to evaluate their quantitative performance, which would assist assay development and study comparisons. This study uses a sexually transmitted infection diagnostic model in combination with an adapted metric termed isothermal doubling time (IDT), akin to PCR efficiency, to compare quantitative PCR and quantitative loop-mediated isothermal amplification (qLAMP) assays, and to quantify the impact of matrix interference. The performance metric described here facilitates the comparison of qLAMP assays that could assist assay development and validation activities.
Abbreviations: Cq, quantification cycle; IDT, isothermal doubling time; MIQE, minimum information for the publication of quantitative real-time PCR experiments; NAA, nucleic acid amplification; qLAMP, quantitative loop-mediated amplification; qPCR, quantitative real-time polymerase chain reaction; td, doubling time; Tt, threshold time
Keywords: Diagnostics, Isothermal nucleic acid amplification, Quantitative LAMP, Quantitative real time PCR, Standardisation
1. Introduction
The use of molecular approaches for clinical diagnosis has increased over the past 30 years since the development of PCR [1] resulting in a wide variety of diagnostic applications [2], [3], [4]. Alternative nucleic acid amplification (NAA) technologies, reviewed by Craw and colleagues [5], [6], [7], utilising isothermal conditions offer a range of potential advantages over PCR, including speed and simplicity, and lend themselves to near patient and point of care diagnostic testing [8], [9]. Loop-mediated isothermal amplification (LAMP) [10] is an example of an isothermal NAA technology that is typically faster than PCR [11], [12], [13] and reportedly less susceptible to common biological inhibitors [14], [15], [16], [17]. LAMP has been successfully utilised in the development of a wide range of diagnostic assays [18], [19], [20].
The application of LAMP to measure template abundance, originally established by real time quantitative PCR (qPCR), has been explored by a variety of laboratories [17], [20], [21], [22], [23], [24], [25], [26]. However, real time quantitative LAMP (qLAMP) is a relatively immature technology compared with qPCR and demonstrates poor quantitation capabilities below 1000 target copies [22], [23], [26]. Threshold time (Tt) [26] is a real-time quantitative LAMP measurement analogous to quantification cycle (Cq) [27] and is central to quantifying template abundance using qLAMP. While Tt, allows quantification to be performed there is currently no agreed method for expressing assay performance or defining the magnitude of a difference between two results when conducting qLAMP; a role that ‘PCR efficiency’ has provided since the development of qPCR. Metrics for assay performance are essential to enable assay optimisation, evaluation of the impact of matrices [28], [29], [30], [31] and to facilitate laboratory comparison.
An isothermal performance metric, analogous to PCR efficiency, has been proposed for real-time helicase dependent amplification (HDA) [32] reactions by Goldmeyer and colleagues [33], termed doubling time (td) and calculated by comparing the time required by different starting quantities of template to reach a uniform threshold (2-point approach). We discuss the application of a more comprehensive metric termed isothermal doubling time (IDT) that utilised a standard curve-based approach to provide an equation (based on the slope of the Tt against concentration) to define the magnitude of a difference in results and estimate assay performance which also benefits from evaluating the linear dynamic range.
Effective mechanisms for comparing the performance of NAA technologies such as qLAMP to that of established PCR-based approaches is central to the uptake of these technologies and establishing confidence in their diagnostic potential. This paper describes a methodology for assessing quantitative isothermal nucleic acid amplification performance that, similar to PCR efficiency for qPCR [34], can assign a relative value to any qLAMP assay. We investigate the application of this metric to target pathogens and assess the impact of inhibition on assay performance.
2. Materials and methods
2.1. Artificial urine matrices
A panel of artificial urine matrices (Table S1) were developed from published work [35] that contained varying levels of urea concentration (500 or 1000 mM urea) which is a known PCR inhibitor [29]. For investigations into matrix impact, reactions were supplemented with 10% (v/v) of the respective artificial matrix.
2.2. Cultured DNA extracts
Chlamydia trachomatis, serovar E (ATCC VR-348BD) genomic DNA containing co-purified cryptic plasmid DNA and Mycoplasma genitalium G37 (ATCC 33530D) gDNA (LGC Standards, Teddington, United Kingdom) were used as genomic templates. Ten-fold serial dilutions (∼5 × 104 to ∼50 copies per reaction) of each DNA template (spectrophotometrically quantified) were prepared in 6.25 ng μl−1 sonicated salmon sperm DNA carrier (GE Healthcare, Chalfont St Giles, United Kingdom).
2.3. Clinical purified DNA samples
All initial clinical sample purification and molecular screening activities were performed at University College London Hospitals (UCLH), London, United Kingdom. 24 pre-screened DNA samples with defined M. genitalium and C. trachomatis content (mix of positives and negatives) were provided blind by UCLH for comparative analysis at the approval of the Chair of the Camden and Islington Community Research Ethics Committee.
C. trachomatis testing was performed as part of a routine clinical diagnostic protocol. Testing process overview: cervical swabs, self-taken vaginal swabs or urine samples were collected and transported in 3 or 4 ml of APTIMA transport medium (Gen-Probe Incorporated, San Diego, USA) mixed with urine 1:1 (v/v) for routine C. trachomatis testing. The test sample (400 μl) was analysed using the APTIMA CT assay on the TIGRIS® platform (Gen-Probe Incorporated, San Diego, USA).
For initial M. genitalium analysis, 200 μl samples were taken from materials previously tested for C. trachomatis, purified using a BioRobot 9604 automated workstation using the QIAamp® Virus BioRobot® 9604 Kit (QIAGEN, Hilden, Germany) and eluted in 50 μl elution buffer (QIAGEN, Hilden, Germany). The eluate (7 μl, equivalent to 28 μl of the original volume) was analysed by a qPCR assay adapted from Jensen et al. [36] incorporating a mouse CMV (mCMV) internal control system [37].
2.4. qPCR assay design and conditions
qPCR assays were designed to detect the C. trachomatis cryptic plasmid (GenBank Accession #X07547), and the M. genitalium partially sequenced MgPa gene (GenBank Accession #X91072) from strain M2300. The real-time PCR assays (Fig. 1, Table 1) were designed using Primer Express 2.0 (Life Technologies Ltd, Paisley, UK) using default design parameters and the sequences screened for homology using the BLASTn algorithm (http://blast.ncbi.nlm.nih.gov/) that revealed no database alignments likely to cause cross reactivity, other than the region of interest. HPLC purified oligonucleotide primers and FAM/TAMRA hydrolysis probes were synthesised by Eurofins Genomics (Ebersberg, Germany). HPLC purified FAM/NFQ TaqMan® MGB probes were provided by Life Technologies Ltd (Paisley, UK). Preliminary primer/probe optimisation was conducted to determine optimal PCR conditions.
Fig. 1.
Target sequences with corresponding amplicon regions for qLAMP (shaded sequence) and qPCR assays (underlined sequence). (a) Chlamydia trachomatis plasmid DNA for growth within mammalian cells (GenBank Acc#X07547) 1081–1560 bp target region for qPCR and qLAMP assay. (b) M. genitalium partial MgPa gene (strain M2300) (GenBank Acc#X91072) 161–480 bp target region for qPCR and qLAMP assay.
Table 1.
NAA assays – primer and probe details.
| Type | Target | Sequence names | Sequence details |
|---|---|---|---|
| qPCR | C. trachomatis | CRYPTIC_MGB_FWD1 | 5′-TCCGTAAAATGTCCTGATTAGTGAAAT-3′ |
| Cryptic plasmid | CRYPTIC_MGB_REV1 | 5′-TTCGGAACGAGTTTTCATGTTTATAT-3′ | |
| (#X07547) | CRYPTIC_MGB_PROBE1 | 5′-FAM-AGGATAGCACGCTCGGTA-MGB-NFQ-3′ | |
| M. genitalium | MGPA_TM_FWD2 | 5′-GGCGAGCCTATCTTTGATCCT-3′ | |
| MgPa gene | MGPA_TM_REV2 | 5′-AACTTTACCTTTGATCTCATTCCAATC-3′ | |
| (#X91072) | MGPA_TM_PROBE2 | 5′-FAM-AAGGCTTTGGTTTAACTGGTAATGCCCCT-TAMRA-3′ | |
| qLAMP | C. trachomatis | Cryptic F3(3) | 5′-ACTTAAAAGACAATGGATTACCT-3′ |
| Cryptic plasmid | Cryptic B3(3) | 5′-AATTACTCAAATTTTCTTCAGCG-3′ | |
| (#X07547) | Cryptic FIP(3) | 5′-CCGAGATACGATTTGTCCATATCTTATAACTGTAGACTCGGCTTG-3′ | |
| Cryptic BIP(3) | 5′-TGTTGCATGATGCTTTATCAAATGACACACGCTCAAATCATCGA-3′ | ||
| Cryptic LF(3) | 5′-CGCCGCAAAAGCTCTTCC-3′ | ||
| Cryptic LB(3) | 5′-CTTAGATCCGTTTCTCATACGGTTT-3′ | ||
| M. genitalium | PA F3(5) | 5′-GAAAACCCCTCAACGGTG-3′ | |
| MgPa gene | PA B3(5) | 5′-CTTGGTTATTCAGGTTGTGAT-3′ | |
| (#X91072) | PA FIP(5) | 5′-AGGGGCATTACCAGTTAAACCAAAAAAGGGGTTTAAATGGCGA-3′ | |
| PA BIP(5) | 5′-ATGAGATCAAAGGTAAAGTTCCAGTCACTAGTAACAGAAAATAGAGGTT-3′ | ||
| PA LF(5) | 5′-GCCTTTAAAAGGATCAAAGATAGGC-3′ | ||
| PA BF(5) | 5′-AGAAGTAGTTCAATCCCCCCAT-3′ | ||
# denotes GenBank accession number.
The qPCR reactions (20 μl volume) were performed comprising 1× TaqMan® Universal PCR Master Mix (Life Technologies Ltd, Paisley, UK), 900 nM forward and reverse primers, 200 nM 6-carboxyfluorescein (6-FAM) labelled hydrolysis probe, sample template (10 fold dilution series from ∼5 × 104 to ∼50 copies) and, where appropriate, 10% (v/v) synthetic matrices (Table S1). All oligonucleotide primers and hydrolysis probes were HPLC purified. Reactions were performed using the Applied Biosystems 7900HT Fast Real-Time PCR System (Life Technologies Ltd, Paisley, UK) and standard cycling conditions: 50 °C for 2 min, 95 °C for 10 min, 50 cycles at 95 °C for 15 s and 60 °C for 1 min. Data analysis was performed using Sequence Detection Software (SDS) version 2.4 (Life Technologies Ltd, Paisley, UK) with manual baseline/threshold settings to estimate quantification cycle (Cq). Water control assays were included in which template was omitted to assess the presence of contamination, all water control assays produced no detectable Cq signal.
2.5. qLAMP assay design and conditions
qLAMP assays were designed to detect a similar region of the C. trachomatis cryptic plasmid and partial M. genitalium MgPa gene as that designed for the qPCR assays, however, due to design constraints exactly the same genetic region could not be targeted for the cryptic plasmid target (Fig. 1 for qLAMP/qPCR amplicon comparisons). The qLAMP assays (Fig. 1, Table 1) comprising 4 core primers (F3/B3/FIP/BIP) and 2 loop primers (LF/LB) were designed using PrimerExplorer V4 (Eiken Chemical Co., Ltd., Tokyo, Japan) online software (http://primerexplorer.jp/elamp4.0.0/index.html) using default design parameters. The sequences were screened for homology using the same approach as for qPCR primers.
The qLAMP reactions (20 μl volume) were performed comprising 1× isothermal master mix (OptiGene Limited, Horsham, United Kingdom) containing proprietary intercalating fluorescent dye, 200 nM F3/B3/LF/LB primers, 800 nM FIP/BIP primers, sample template (∼5 × 104 to ∼50 copies per reaction) and, where appropriate, 10% (v/v) synthetic matrices (Table S1). All oligonucleotide primers were HPLC purified. Reactions were performed using the Applied Biosystems 7900HT Fast Real-Time PCR System (Life Technologies Ltd, Paisley, UK) under the following thermal cycling conditions: 65 °C/45 min (60 × 45 s cycles), and fluorescence monitored over the SYBR Green I spectral region. Data analysis was performed using Sequence Detection Software (SDS) version 2.4 (Life Technologies Ltd, Paisley, UK) with manual baseline/threshold settings to estimate Threshold Time (Tt). Water control assays were included in which template was omitted to assess the presence of contamination, all water control assays produced no detectable Tt signal.
2.6. Comparison studies
2.6.1. Matrix evaluation
Experiments were conducted to compare the performance of specific qLAMP assays with comparable qPCR assays in the presence of two different matrices (artificial urine matrix containing 50 or 100 mM urea final concentration, Table S1). Three independent experiments were performed per assay/NAA technology and each reaction performed in triplicate.
2.6.2. qLAMP and qPCR performance assessment
Standard curve analysis (log10 transformed input copy number plotted against Cq or Tt) was performed to evaluate the performance of the qPCR and qLAMP assays and enable calculation of PCR efficiencies [34] and isothermal doubling times (IDT = −0.301 m, whereby 0.301 corresponds on the base 10 logarithmic scale to a change in concentration of a factor of 2 and m = slope). The use of real time PCR instrument to undertake isothermal studies necessitates the conversion of the qPCR instrument output (Cq) to Tt through the application of a time multiplier (cycle time), which in this experiment consisted of 45s cycles.
2.6.3. Clinical comparison
The 24 pre-screened clinical DNA samples (pathogen status determined by UCLH) comprising 18 positives (7× M. genitalium, 7× C. trachomatis and 2× C. trachomatis and M. genitalium) and 8 negatives, were used to investigate the C. trachomatis and M. genitalium assays using both NAA platforms on real clinical samples. Due to sample limitations single replicates of each clinical sample, comprising 1 μl volumes, were interrogated using appropriate qPCR or qLAMP-based assays. This corresponded to 4 μl of the original sample volume and was less than the initial sample analysed by UCLH with the M. genitalium qPCR (28 μl) or the C. trachomatis Gen-Probe assay (400 μl). To simulate how the respective technologies performed with clinical extracts in the presence of inhibition, a repeat measurement was performed in which artificial urine matrix to include 100 mM urea (final reaction concentration) was also added.
3. Results
3.1. Establishing baseline performance
Standard curve-based analyses of the C. trachomatis and M. genitalium qPCR assays (Fig. 2a and b, Table S2a) show that the assays achieved good linearity and precision (mean R2 measurements of 1.00), displayed good levels of PCR sensitivity (Fig. 3a and b) and are capable of repeatedly detecting ∼5 copies of target material under experimental conditions.
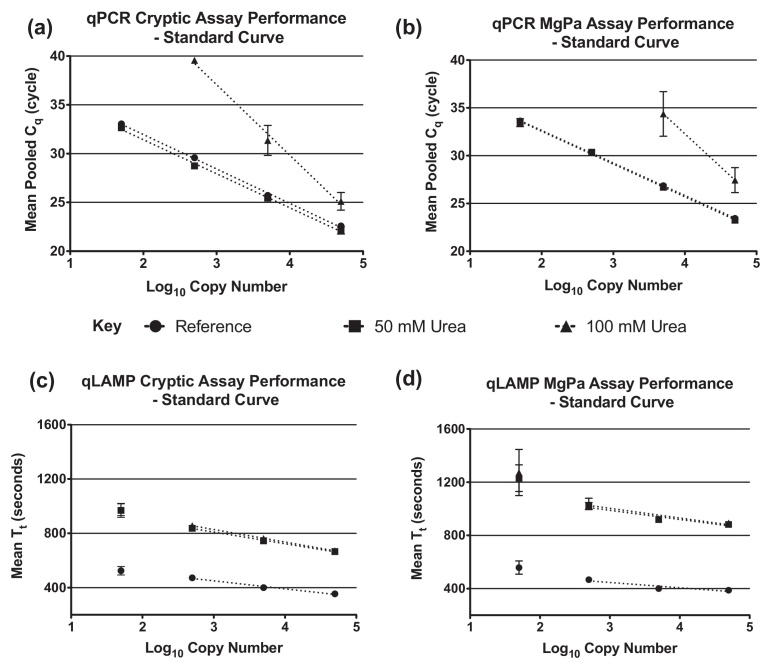
Fig. 2.
Standard curve based analysis of qPCR and qLAMP cryptic plasmid/MgPa assay performance. Plots displaying mean Cq [(a) & (b)] and Tt [(c) & (d)] from three separate reactions per three experiments when threshold set at the same respective fluorescence. Error bars illustrate standard deviations.
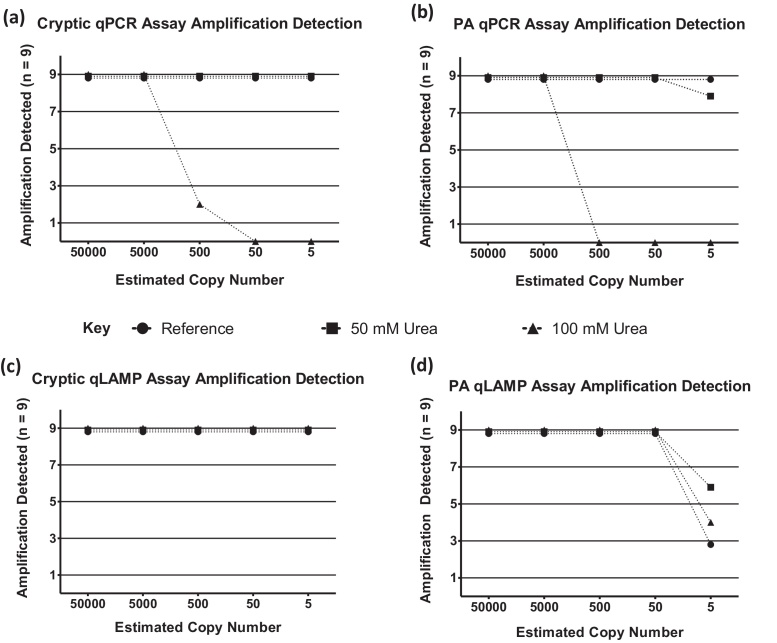
Fig. 3.
Comparative analysis of qPCR and qLAMP assay amplification success. Plots displaying qPCR [(a) & (b)] and qLAMP [(c) & (d)] successful amplification data from three separate reactions per 3 experiments (n = 9).
Comparable standard curve-based analyses of the qLAMP assays (Fig. 2c and d, Table S2b) displayed a reduced dynamic range as compared to the qPCR assays which was characterised by a loss of linear response below the 1000 target copies input level. Variable R2 measurements (0.82 and 0.97) and higher levels of error (Table S3) for both assays highlight the lower levels of precision associated with qLAMP compared to qPCR. Both assays display good lower levels of detection sensitivity (Fig. 3c and d) and are capable of detecting 5 theoretical copies of target material under ideal conditions, although this was not quantitative below 1000 copies.
3.2. Assessing the impact of urea containing matrices
We investigated the impact of artificial matrices containing two concentrations of urea, a known PCR inhibitor [29] and found that urea had a differential impact upon the performance of both NAA technologies typified by a right shift in Cq/Tt (Fig. 2). 100 mM urea affects the cryptic plasmid qPCR assay by causing a positive shift in Cq for the 50,000 copies sample from 23.50 ± 0.09 to 26.89 ±0.83 Cq while a comparable qLAMP reaction demonstrates a Tt shift from 387.24 ± 5.14 to 726.00 ± 12.95 s. While the presence of inhibitors produced an appreciable delay in qLAMP amplification, both assays were able to routinely detect ∼5 copies of template regardless of the inhibition levels (Fig. 3c and d). In comparison, the qPCR assays were only appreciably affected by the presence of 100 mM urea which reduces the detection capabilities of both assays to above 500 template copies (Fig. 3a and b).
3.3. Comparing NAA assay performance
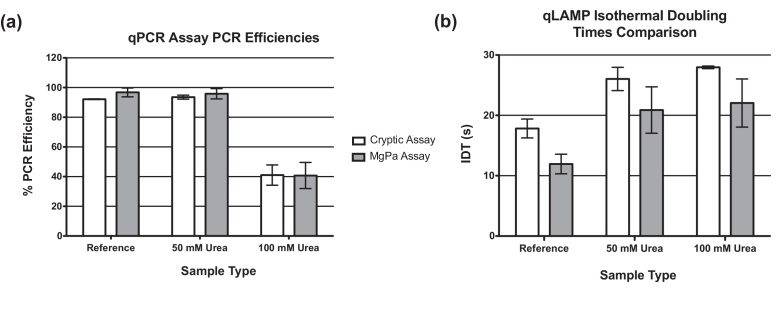
Initial evaluation work demonstrated that the NAA technologies displayed different performance characteristics and responses to matrices. PCR efficiency and IDT metrics were used to compare intra-assay performance and investigate the impact of an inhibitor on qLAMP and qPCR assay performance. The PCR efficiency and IDT data correlates well with the initial matrix assessment work utilising performance metrics as characterised by increased Ct/Tt and lower PCR efficiency/higher IDT in the presence of inhibitors. PCR efficiency and IDT data suggested that the qPCR assays (Fig. 4a) were less affected by the presence of 50 mM urea than the comparable qLAMP assays (Fig. 4b). Reductions in PCR efficiencies (below 50%) were only observed with the 100 mM urea matrix samples. Matrices containing 100 mM urea had a pronounced impact on the performance of both qPCR assays characterised by a loss of assay sensitivity (Fig. 3a and b), increased Cq and reduced efficiency (Fig. 4a).
Fig. 4.
Methodologies to compare general assay performance. (a) Percentage PCR efficiency, (b) Isothermal Doubling Times. Plots displaying IDT or PCR efficiency (n = 3 separate experiments, 3 technical replicates). Error bars denote inter-experimental standard deviations.
In comparison, qLAMP assays show a minimum ∼1.5× increase in IDT that is unaffected by urea concentration (Fig. 4b) and suggests that urea is not responsible for the observed impact on assay performance.
3.4. Clinical evaluation
The performance of the two NAA technologies was further evaluated using the M. genitalium and C. trachomatis qLAMP/qPCR assays with clinical samples. 24 Clinical DNA extracts with defined M. genitalium and C. trachomatis status were evaluated by all assays in the presence/absence of 100 mM urea. A detailed assessment of diagnostic efficacy for either technology could not be performed due to sample limitations, however this study clearly demonstrated that both qPCR and qLAMP were able to detect their respective pathogen targets from clinical sample extracts. We demonstrated that the MgPa qPCR assay was more sensitive than the corresponding qLAMP assay (Table 2a) while the cryptic plasmid qLAMP assay demonstrated comparable performance to the cryptic plasmid qPCR assay (Table 2b). With the exception of one sample, the qLAMP assay's detection response (Table 2) was unaffected by the presence of the artificial matrices, while both qPCR assays were always inhibited when 100 mM urea matrix was present.
Table 2.
Clinical samples screening results. 24 clinical DNA extracts screened against a panel of (a) Mycoplasma genitalium and (b) Chlamydia trachomatis qPCR and qLAMP assays in the presence or absence of urine type artificial matrix containing 100 mM urea (final concentration). Negative samples not displayed (100% concordance with both assays). Respective Cq and Tt (s) data displayed (rounded to nearest whole number), ‘ND’ denotes no detection and shaded regions highlight positive assay responses.
| Sample | qPCR (Cq) |
qLAMP (Tt) |
||
|---|---|---|---|---|
| Minus urine | Plus urine | Minus urine | Plus urine | |
| (a) Screening results for theMycoplasma genitaliumqPCR and qLAMP assays | ||||
| 1 | 38 | ND | ND | ND |
| 7 | 30 | ND | 796 | 1472 |
| 10 | 36 | ND | ND | ND |
| 12 | 38 | ND | ND | ND |
| 16 | 34 | ND | 922 | 2476 |
| 21 | ND | ND | ND | ND |
| 28 | 31 | ND | 848 | 1486 |
| 29 | 40 | ND | ND | ND |
| 34 | 30 | ND | 683 | 1292 |
| Total | 8 | 0 | 4 | 4 |
| (b) Screening results for theChlamydia trachomatisqPCR and qLAMP assays | ||||
| 5 | ND | ND | 985 | ND |
| 6 | ND | ND | ND | ND |
| 10 | 29 | ND | 694 | 1269 |
| 11 | 38 | ND | ND | 1565 |
| 16 | 31 | ND | 910 | 1267 |
| 22 | 29 | ND | 642 | 1069 |
| 23 | ND | ND | ND | ND |
| 24 | 32 | ND | 894 | 1316 |
| 26 | 20 | ND | 477 | 906 |
| 29 | ND | ND | ND | ND |
| Total | 6 | 0 | 6 | 6 |
4. Discussion
Current molecular diagnostic standardisation activities are driven by international organisations such as the Laboratory Standards Institute (CLSI) [38] and user driven initiatives such as ‘Minimum Information for Publication of Quantitative Real-Time PCR Experiments (MIQE) Guidelines’ which aim to improve measurement comparability and traceability [27]. Metrics such as PCR efficiency, aid qPCR measurement and ultimately reproducibility but comparable tools available to evaluate qLAMP, and other isothermal assays, are limited. To this end, this study developed a performance metric (IDT) and compared it to the application of equivalent qPCR metrics.
Differences in performance were observed between the two NAA technologies, with qPCR displaying better sensitivity and quantitative precision when compared to qLAMP. However, the qualitative potential of qLAMP was not affected by the presence of artificial urine-type matrices containing urea, which represents an inhibitor found in urine-based clinical samples/extracts. In addition, this finding supports the applicability of LAMP to the direct testing of clinical urine samples without the need for time-consuming extraction steps [12], [16].
While our findings support the general assumption that qualitative LAMP amplification may be less susceptible to inhibitors than qPCR, as others have observed [14], [39], we have demonstrated that quantification using qLAMP is susceptible to matrix interference. Our data suggests that LAMP-based assays are susceptible to alternative inhibitory processes than those observed for qPCR as characterised by an increase in Isothermal Doubling Time (IDT) and Tt shift that is not dependent upon urea concentration.
These results support our previous findings [17] which demonstrated that inhibitors can exhibit different levels of inhibition on different nucleic acid amplification methods and Tani et al. [14] who observed that urea (up to 400 mM) has little effect on the quantitative performance of qLAMP, indicating that other matrix components are responsible for the observed inhibitory effect. The observed gross change in IDT would affect any quantitative measurements and require appropriate correction mechanisms such as matched calibrator and test matrices.
Performance comparison was achievable through the application of the commonly used PCR efficiency metric and our adaptation of the doubling time (td) [33] approach that we have termed IDT. IDT provides a mechanism by which a fold difference can be attributed to multiple target dilution points rather than two points as utilised to calculate td. The standard curve-based approach provides an estimate of qLAMP assay performance that minimises biases associated with dual-point estimations. PCR is only able to perform a maximum of a doubling of all templates per cycle when it is at its optimum and thus PCR efficiency is reported as a percentage. qLAMP does not have the same cyclical constraint as PCR and thus IDT does not have a clearly defined physical maximum. IDT has the potential to be used for other quantitative isothermal technologies as long as there is a log linear relationship between Tt and concentration such as observed with real-time nucleic acid sequence-based amplification (NASBA) and recombinase polymerase amplification (RPA) approaches [40], [41].
IDT also provides users with a common baseline measurement to compare the performance of compatible isothermal NAA assays and support assay development activities such as optimisation or investigating the impact of inhibitors. Inhibition was demonstrated to affect the qLAMP and qPCR reactions through an increase in Tt or Cq respectively, as well as a reduction in PCR efficiency or increase in IDT.
IDT can be estimated using standard curves, as performed here, or to assess sample specific performance in the same way as qPCR when performing relative quantification [42]. This not only facilitates comparison between assays within the same laboratory, but also between laboratories and enables reporting of a key assay characteristic similar to qPCR efficiency; which is agreed to be an important factor that should be stated when performing qPCR [27].
This study highlighted the reduced precision associated with qLAMP when compared to qPCR (Table S3) and suggests further work on assay components such as enzymes and buffer systems is necessary to improve qLAMP performance. However, it should be remembered that qPCR has benefitted from over a decade of continuous development that contrasts with the relative immaturity of the LAMP.
LAMP has the potential to complement existing qPCR based techniques that are currently the gold standard within the molecular diagnostics field, but is limited by poor quantitation below 1000 target copies. This could be alleviated through the use of approaches similar to digital PCR, subject to sensitivity issues arising from current assayable sample volume limitations [17], or through improvements in LAMP chemistry, e.g. enzyme optimisation [43]. The metric proposed by this study provides a mechanism that can facilitate such research and development work and ultimately assist in the transfer of novel LAMP-based methods into diagnostic tests.
5. Conclusion
We demonstrate the application of a performance metric comparable to PCR efficiency in the evaluation of qLAMP assays which is potentially applicable to other isothermal NAA technologies. The isothermal doubling time metric described here facilitates the comparison of qLAMP assays that could assist assay development and validation activities within the developer and user communities.
Competing interests
The authors declare no competing interests.
Acknowledgements
We would like to thank Dr Simon Cowen for assistance with the development of the concept of isothermal doubling time. The work described in the paper was funded by the UK National Measurement System, UCL CBRC (project grant award reference 59) and the European Metrology Research Programme (EMRP). The EMRP is jointly funded by the EMRP participating countries within The European Association of National Metrology Institutes (EURAMET) and the European Union.
Footnotes
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.bdq.2014.11.001.
Appendix A. Supplementary data
The following are supplementary data to this article:
References
- 1.Mullis K., Faloona F., Scharf S., Saiki R., Horn G., Erlich H. Specific enzymatic amplification of DNA in vitro: the polymerase chain reaction. Cold Spring Harb Symp Quant Biol. 1986;51(Pt 1):263–273. doi: 10.1101/sqb.1986.051.01.032. [DOI] [PubMed] [Google Scholar]
- 2.Millar B.C., Xu J., Moore J.E. Molecular diagnostics of medically important bacterial infections. Curr Issues Mol Biol. 2007;9(1):21–39. [PubMed] [Google Scholar]
- 3.Josko D. Molecular diagnostics in the public health laboratories. Clin Lab Sci. 2010;23(4):242–247. [PubMed] [Google Scholar]
- 4.Espy M.J., Uhl J.R., Sloan L.M., Buckwalter S.P., Jones M.F., Vetter E.A. Real-time PCR in clinical microbiology: applications for routine laboratory testing. Clin Microbiol Rev. 2006;19(1):165–256. doi: 10.1128/CMR.19.1.165-256.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim J., Easley C.J. Isothermal DNA amplification in bioanalysis: strategies and applications. Bioanalysis. 2011;3(2):227–239. doi: 10.4155/bio.10.172. [DOI] [PubMed] [Google Scholar]
- 6.Gill P., Ghaemi A. Nucleic acid isothermal amplification technologies: a review. Nucleosides Nucleotides Nucleic Acids. 2008;27(3):224–243. doi: 10.1080/15257770701845204. [DOI] [PubMed] [Google Scholar]
- 7.Craw P., Balachandran W. Isothermal nucleic acid amplification technologies for point-of-care diagnostics: a critical review. Lab Chip. 2012;12(14):2469–2486. doi: 10.1039/c2lc40100b. [DOI] [PubMed] [Google Scholar]
- 8.Fang X., Liu Y., Kong J., Jiang X. Loop-mediated isothermal amplification integrated on microfluidic chips for point-of-care quantitative detection of pathogens. Anal Chem. 2010;82(7):3002–3006. doi: 10.1021/ac1000652. [DOI] [PubMed] [Google Scholar]
- 9.Bearinger J.P., Dugan L.C., Baker B.R., Hall S.B., Ebert K., Mioulet V. Development and initial results of a low cost, disposable, point-of-care testing device for pathogen detection. IEEE Trans Biomed Eng. 2011;58(3):805–808. doi: 10.1109/TBME.2010.2089054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe K., Amino N. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28(12):E63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Enomoto Y., Yoshikawa T., Ihira M., Akimoto S., Miyake F., Usui C. Rapid diagnosis of herpes simplex virus infection by a loop-mediated isothermal amplification method. J Clin Microbiol. 2005;43(2):951–955. doi: 10.1128/JCM.43.2.951-955.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hill J., Beriwal S., Chandra I., Paul V.K., Kapil A., Singh T. Loop-mediated isothermal amplification assay for rapid detection of common strains of Escherichia coli. J Clin Microbiol. 2008;46:2800–2804. doi: 10.1128/JCM.00152-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boehme C.C., Nabeta P., Henostroza G., Raqib R., Rahim Z., Gerhardt M. Operational feasibility of using loop-mediated isothermal amplification for diagnosis of pulmonary tuberculosis in microscopy centers of developing countries. J Clin Microbiol. 2007;45(6):1936–1940. doi: 10.1128/JCM.02352-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tani H., Teramura T., Adachi K., Tsuneda S., Kurata S., Nakamura K. Technique for quantitative detection of specific DNA sequences using alternately binding quenching probe competitive assay combined with loop-mediated isothermal amplification. Anal Chem. 2007;79(15):5608–5613. doi: 10.1021/ac070041e. [DOI] [PubMed] [Google Scholar]
- 15.Kaneko H., Kawana T., Fukushima E., Suzutani T. Tolerance of loop-mediated isothermal amplification to a culture medium and biological substances. J Biochem Biophys Methods. 2007;70(3):499–501. doi: 10.1016/j.jbbm.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 16.Bista B.R., Ishwad C., Wadowsky R.M., Manna P., Randhawa P.S., Gupta G. Development of a loop-mediated isothermal amplification assay for rapid detection of BK virus. J Clin Microbiol. 2007;45(5):1581–1587. doi: 10.1128/JCM.01024-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nixon G., Garson J.A., Grant P., Nastouli E., Foy C.A., Huggett J.F. Comparative study of sensitivity, linearity, and resistance to inhibition of digital and nondigital polymerase chain reaction and loop mediated isothermal amplification assays for quantification of human cytomegalovirus. Anal Chem. 2014;86(9):4387–4394. doi: 10.1021/ac500208w. [DOI] [PubMed] [Google Scholar]
- 18.Geojith G., Dhanasekaran S., Chandran S.P., Kenneth J. Efficacy of loop mediated isothermal amplification (LAMP) assay for the laboratory identification of Mycobacterium tuberculosis isolates in a resource limited setting. J Microbiol Methods. 2011;84(1):71–73. doi: 10.1016/j.mimet.2010.10.015. [DOI] [PubMed] [Google Scholar]
- 19.Wang G., Shang Y., Wang Y., Tian H., Liu X. Comparison of a loop-mediated isothermal amplification for orf virus with quantitative real-time PCR. Virol J. 2013;10:138. doi: 10.1186/1743-422X-10-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang G., Brown E.W., Gonzalez-Escalona N. Comparison of real-time PCR, reverse transcriptase real-time PCR, loop-mediated isothermal amplification, and the FDA conventional microbiological method for the detection of Salmonella spp. in produce. Appl Environ Microbiol. 2011;77(18):6495–6501. doi: 10.1128/AEM.00520-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kang Y., Deng R., Wang C., Deng T., Peng P., Cheng X. Etiologic diagnosis of lower respiratory tract bacterial infections using sputum samples and quantitative loop-mediated isothermal amplification. PLoS One. 2012;7(6):e38743. doi: 10.1371/journal.pone.0038743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang F., Jiang L., Ge B. Loop-mediated isothermal amplification assays for detecting shiga toxin-producing Escherichia coli in ground beef and human stools. J Clin Microbiol. 2012;50(1):91–97. doi: 10.1128/JCM.05612-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Francois P., Bento M., Hibbs J., Bonetti E.J., Boehme C.C., Notomi T. Robustness of loop-mediated isothermal amplification reaction for diagnostic applications. FEMS Immunol Med Microbiol. 2011;62(1):41–48. doi: 10.1111/j.1574-695X.2011.00785.x. [DOI] [PubMed] [Google Scholar]
- 24.Nakao R., Stromdahl E.Y., Magona J.W., Faburay B., Namangala B., Malele I. Development of loop-mediated isothermal amplification (LAMP) assays for rapid detection of Ehrlichia ruminantium. BMC Microbiol. 2010;10:296. doi: 10.1186/1471-2180-10-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen S., Ge B. Development of a toxR-based loop-mediated isothermal amplification assay for detecting Vibrio parahaemolyticus. BMC Microbiol. 2010;10:41. doi: 10.1186/1471-2180-10-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aoi Y., Hosogai M., Tsuneda S. Real-time quantitative LAMP (loop-mediated isothermal amplification of DNA) as a simple method for monitoring ammonia-oxidizing bacteria. J Biotechnol. 2006;125(4):484–491. doi: 10.1016/j.jbiotec.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 27.Bustin S.A., Benes V., Garson J.A., Hellemans J., Huggett J., Kubista M. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55(4):611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 28.Al Soud W.A., Radstrom P. Purification and characterization of PCR-inhibitory components in blood cells. J Clin Microbiol. 2001;39(2):485–493. doi: 10.1128/JCM.39.2.485-493.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khan G., Kangro H.O., Coates P.J., Heath R.B. Inhibitory effects of urine on the polymerase chain reaction for cytomegalovirus DNA. J Clin Pathol. 1991;44(5):360–365. doi: 10.1136/jcp.44.5.360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kontanis E.J., Reed F.A. Evaluation of real-time PCR amplification efficiencies to detect PCR inhibitors. J Forensic Sci. 2006;51(4):795–804. doi: 10.1111/j.1556-4029.2006.00182.x. [DOI] [PubMed] [Google Scholar]
- 31.Mahony J., Chong S., Jang D., Luinstra K., Faught M., Dalby D. Urine specimens from pregnant and nonpregnant women inhibitory to amplification of Chlamydia trachomatis nucleic acid by PCR, ligase chain reaction, and transcription-mediated amplification: identification of urinary substances associated with inhibition and removal of inhibitory activity. J Clin Microbiol. 1998;36(11):3122–3126. doi: 10.1128/jcm.36.11.3122-3126.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vincent M., Xu Y., Kong H. Helicase-dependent isothermal DNA amplification. EMBO Rep. 2004;5(8):795–800. doi: 10.1038/sj.embor.7400200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goldmeyer J., Kong H., Tang W. Development of a novel one-tube isothermal reverse transcription thermophilic helicase-dependent amplification platform for rapid RNA detection. J Mol Diagn. 2007;9(5):639–644. doi: 10.2353/jmoldx.2007.070012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rasmussen R. Quantification on the LightCycler. In: Meuer S., Wittwer C., Nakagawara K., editors. Rapid Cycle Real-time PCR, Methods and Applications. Springer Press; Heidelberg: 2001. pp. 21–34. [Google Scholar]
- 35.Brooks T., Keevil C.W. A simple artificial urine for the growth of urinary pathogens. Lett Appl Microbiol. 1997;24(3):203–206. doi: 10.1046/j.1472-765x.1997.00378.x. [DOI] [PubMed] [Google Scholar]
- 36.Jensen J.S., Bjornelius E., Dohn B., Lidbrink P. Use of TaqMan 5′ nuclease real-time PCR for quantitative detection of Mycoplasma genitalium DNA in males with and without urethritis who were attendees at a sexually transmitted disease clinic. J Clin Microbiol. 2004;42(2):683–692. doi: 10.1128/JCM.42.2.683-692.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garson J.A., Grant P.R., Ayliffe U., Ferns R.B., Tedder R.S. Real-time PCR quantitation of hepatitis B virus DNA using automated sample preparation and murine cytomegalovirus internal control. J Virol Methods. 2005;126(1–2):207–213. doi: 10.1016/j.jviromet.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 38.Holden M.J., Madej R.M., Minor P., Kalman L.V. Molecular diagnostics: harmonization through reference materials, documentary standards and proficiency testing. Expert Rev Mol Diagn. 2011;11(7):741–755. doi: 10.1586/erm.11.50. [DOI] [PubMed] [Google Scholar]
- 39.Kaneko H., Iida T., Aoki K., Ohno S., Suzutani T. Sensitive and rapid detection of herpes simplex virus and varicella-zoster virus DNA by loop-mediated isothermal amplification. J Clin Microbiol. 2005;43:3290–3296. doi: 10.1128/JCM.43.7.3290-3296.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Piepenburg O., Williams C.H., Stemple D.L., Armes N.A. DNA detection using recombination proteins. PLoS Biol. 2006;4(7):e204. doi: 10.1371/journal.pbio.0040204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yates S., Penning M., Goudsmit J., Frantzen I., van D.W.B., Van S.D. Quantitative detection of hepatitis B virus DNA by real-time nucleic acid sequence-based amplification with molecular beacon detection. J Clin Microbiol. 2001;39:3656–3665. doi: 10.1128/JCM.39.10.3656-3665.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pfaffl M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29(9):e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tanner N.A., Zhang Y., Evans T.C. Simultaneous multiple target detection in real-time loop-mediated isothermal amplification. Biotechniques. 2012;53(2):81–89. doi: 10.2144/0000113902. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.