Abstract
Lipid transfer proteins (LTPs) are a class of cysteine-rich soluble proteins having small molecular weights. LTPs participate in flower and seed development, cuticular wax deposition, also play important roles in pathogen and abiotic stress responses. A non-specific LTP gene (SiLTP) was isolated from a foxtail millet (Setaria italica) suppression subtractive hybridization library enriched for differentially expressed genes after abiotic stress treatments. A semi-quantitative reverse transcriptase PCR analysis showed that SiLTP was expressed in all foxtail millet tissues. Additionally, the SiLTP promoter drove GUS expression in root tips, stems, leaves, flowers, and siliques of transgenic Arabidopsis. Quantitative real-time PCR indicated that the SiLTP expression was induced by NaCl, polyethylene glycol, and abscisic acid (ABA). SiLTP was localized in the cytoplasm of tobacco leaf epidermal cells and maize protoplasts. The ectopic expression of SiLTP in tobacco resulted in higher levels of salt and drought tolerance than in the wild type (WT). To further assess the function of SiLTP, SiLTP overexpression (OE) and RNA interference (RNAi)-based transgenic foxtail millet were obtained. SiLTP-OE lines performed better under salt and drought stresses compared with WT plants. In contrast, the RNAi lines were much more sensitive to salt and drought compared than WT. Electrophoretic mobility shift assays and yeast one-hybrids indicated that the transcription factor ABA-responsive DRE-binding protein (SiARDP) could bind to the dehydration-responsive element of SiLTP promoter in vitro and in vivo, respectively. Moreover, the SiLTP expression levels were higher in SiARDP-OE plants compared than the WT. These results confirmed that SiLTP plays important roles in improving salt and drought stress tolerance of foxtail millet, and may partly be upregulated by SiARDP. SiLTP may provide an effective genetic resource for molecular breeding in crops to enhance salt and drought tolerance levels.
Keywords: foxtail millet, lipid transfer protein, salt stress, drought tolerance, transcription factor
Introduction
Presently, environmental problems are becoming more severe. Among the major factors, drought, salinity, and cold stresses adversely affect the growth and yield of plants (Mahajan and Tuteja, 2005). In response to abiotic stresses, plants have developed diverse signaling pathways. Various genes, including those of enzymes, transcription factors (TFs), and functional proteins that participate in mitigating the effects of stress, adjust to the cellular milieu and increase plant tolerance.
Foxtail millet (Setaria italica) is an important millet crop worldwide and has been cultivated in China for more than 7,000 years. Foxtail millet has a greater tolerance to drought and adverse soil conditions than maize and sorghum (Panaud, 2006). In India, China, and Japan, foxtail millet is normally cultivated in the salinity- and drought-prone regions. With remarkable drought tolerance, extensive adaptability, rich genetic diversity, and a small diploid genome about 490 Mb (Doust et al., 2009), foxtail millet provides a rich resource of stress resistance genes for study and application. Recently, considerable progress has been made on the whole-genome sequencing of foxtail millet (Bennetzen et al., 2012; Zhang et al., 2012; Jia et al., 2013), meanwhile, foxtail millet were recommended as an excellent C4 crop model (Doust et al., 2009; Bennetzen et al., 2012; Kumar et al., 2013; Muthamilarasan and Prasad, 2015).
Approximately 40 years ago, lipid transfer proteins (LTPs) were discovered (Kader, 1975) and defined by their ability to facilitate the transfer of phospholipids between membranes in vitro (Kader, 1996). LTPs are small peptides, each with eight highly conserved cysteine residues, which form the internal hydrophobic cavity of the three-dimensional structure, and an N-terminal hydrophobic signal peptide. When the N-terminal hydrophobic signal peptide is excised, the mature LTP protein targets the cell secretory pathway (Kader, 1996). Based on the molecular masses, LTPs have traditionally been classified into two families, including ∼9 kDa (LTP I) and ∼7 kDa (LTP II), respectively (Arondel and Kader, 1990; Castagnaro and García-Olmedo, 1994). Recently, based on the occurrence and distribution of non-specific LTPs (nsLTPs) in different plant species, additional subfamilies, including C, D, E, F, G, H, J, and K, were proposed (Edstam et al., 2011). The locations of the LTPs are varied. LTPs occur in the plasma membrane (Debono et al., 2009; Kim et al., 2012), cell wall (Thoma et al., 1993), or cytoplasm (Guo et al., 2013; Edstam et al., 2014). LTPs are reported to probable participated in cutin synthesis (Pyee et al., 1994; Han et al., 2001; Debono et al., 2009; Kim et al., 2012), pathogen defense responses (Maldonado et al., 2002; Silverstein et al., 2007; Guo et al., 2013; Yu et al., 2013), reproductive development (Chae et al., 2009; Zhang D. et al., 2010; Zhang Y. et al., 2010), and adaption to abiotic stresses (Guo et al., 2013; Pitzschke et al., 2014), even though their functions remain unclear.
Since the LTP gene induced by abscisic acid (ABA) and low temperature was discovered (Hughes et al., 1992), more LTPs responsive to abiotic stress have been found and studied. Moreover, the expression of LTP genes was also induced by signal molecules which involved in the signaling pathway. Cryoprotectin is induced by cold (Hincha, 2002), LpLtp1 and LpLtp2 are induced by drought (Trevino and O’Connell, 1998), BG-14 is induced by abiotic stresses like drought, cold, and heat shock duration. It is regulated by signal molecules including ABA, anisomycin, and sphingosine, as well (Wu et al., 2004). AZI1 from Arabidopsis thaliana is induced by cold and salt stresses, as well as by ethylene (Xu et al., 2011; Atkinson et al., 2013; Pitzschke et al., 2014).
In plants, the main abiotic stress responses include ABA-independent and ABA-dependent signal transduction pathways (Yamaguchi-Shinozaki and Shinozaki, 2005, 2006). Different TFs are activated in response to cold, salinity, and drought pathways, and TFs, such as DREB2A, DREB2B, bZip, MYC, and MYB, are important in responding to drought and salt stresses in several plant species (Mahajan and Tuteja, 2005; Abuqamar et al., 2009; Prasad et al., 2011; Sham et al., 2014, 2015). As functional proteins, several LTPs are regulated by upstream proteins, like TFs or kinases/phosphatases. In Arabidopsis, LTP3 is positively regulated by MYB96 through direct binding to the LTP3 promoter, and it is involved in plant tolerance levels to freezing and drought stress (Guo et al., 2013). AZI1, a LTP-related hybrid proline-rich protein, and a novel target of mitogen-activated protein kinase 3, is positively regulated by the latter, and plays a role in salt tolerance in plants (Pitzschke et al., 2014).
In this study, we isolated a novel nsLTP gene (SiLTP) from a S. italica suppression subtractive hybridization (SSH) cDNA library, and investigated its biological functions. The transcription levels of SiLTP were induced by NaCl, polyethylene glycol (PEG) and ABA. The expression of SiLTP enhanced the salt and drought tolerance levels of transgenic tobacco and foxtail millet. SiARDP which plays important role in the abiotic stress response (Li et al., 2014) could bind to the DRE element of SiLTP promoter region. Moreover, SiLTP transcription level was upregulated in SiARDP-OE (overexpression) foxtail millet. SiLTP plays important roles in response to salt and drought stresses in foxtail millet, and may be a candidate of SiARDP downstream genes.
Materials and Methods
Plant Materials and Treatments
Foxtail millet (S. italica cv. Jigu11) was cultivated in the greenhouse or growth chamber. The condition is that the temperature is 25°C and the photoperiod is 16 h/8 h (light/dark). For SSH library construction, 21-day-old seedlings were pre-cultured for 3 days in 1/3 Hoagland liquid medium with bubbling. Then, seedlings were transferred to the same medium applied with 20% (m/v) PEG 6000 (Sigma-Aldrich, Shanghai, China) or 250 mM NaCl, respectively. Meanwhile, plants cultured in the medium without any addition as control. After treatment with PEG or NaCl for 0, 1, 3, 6, 12, and 24 h, the roots and shoots were separately stored at -80°C for RNA extraction. To analyze the LTP expression pattern, foxtail millet roots, stems, leaves, inflorescences, and seeds of different developmental stage: 5, 10, and 15 days after pollination, were collected and stored at -80°C until use. To confirm SiLTP expression levels under stress, 14-day-old foxtail millet seedlings were subjected to 1/4 Hoagland liquid medium containing 100 mM NaCl, 20% (m/v) PEG 6000 or 10 μM ABA, independently, for the indicated time. Harvested seedlings were immediately frozen and stored at -80°C until use.
Arabidopsis thaliana (Col-0) were grown in the growth chamber at 21–22°C under 16 h/8 h (light/dark) conditions for transformation or GUS staining.
Suppression Subtractive Hybridization
Total RNA was extracted from foxtail millet seedlings using a hot phenol method (Longeman et al., 1987). mRNA was purified from total RNA using a PolyATtract mRNA Isolation System (Promega, Madison, WI, USA). Then, 4 μg mRNA mixed with equal amounts of roots and shoots mRNA of the seedlings at indicated times after independent NaCl and PEG treatments, as well as from non-treated seedlings. These samples were the testers and drivers in the construction of the SSH cDNA library. A Clontech PCR-Select cDNA Subtraction Kit (Clontech, Palo Alto, CA, USA) was used to construct the library. After first round PCR, the secondary round PCR was carried out and the products were ligated to the vector of pGEM T-easy (Promega). The ligation product was transferred into the competent cells of Escherichia coli DH5α. Colonies growing on the medium were collected and cultured at 37°C for 12 h. Then, plasmids were extracted from the E. coli and digested by NotI. The resulting fragments between 800 and 2,000 bp were recovered, and inserted into the plasmid 19-T (Promega). The resultant cDNA libraries were transformed to E. coli DH5 and sequenced.
RNA Extraction, Semi-qRT-PCR, and qPCR
Total RNA extracted by TRIzol reagent (Invitrogen, USA) was digested with RNase-free DNaseI (Takara, Japan), and revised transcribed into first-strand cDNA using MLV-Reverse Transcriptase (Promega). For quantitative real-time PCR (qPCR), a 20-μL reaction system, which containing 10 μL 2 × SYBR mix (CoWin Biotech, Beijing, China) and 100 ng of cDNA template, was adopted. Primers of the SiLTP gene (GenBank: LOC101782694) and the reference Actin gene (GenBank: AF288226) are shown in Supplementary Table S1. qPCR was conducted by the Lightcycler 480 Real-Time PCR System (Roche, Indianapolis, IN, USA). Thermal Cycler as we described before (Pan et al., 2015). Each qPCR was run in three biological replicates. The ΔΔCT method was adopted for the calculation of relevant genes expression levels (Bustin et al., 2009). Data represent the means and standard deviations (SD) of three replicates. To analyze LTP expression patterns, semi-quantitative reverse transcriptase PCR (semi-qRT-PCR) was conducted, with the following cycling parameters: 94°C for 1 min, followed by 25 cycles of 94°C for 1 min, 60°C for 30 s and 72°C for 30 s (Pan et al., 2015). Total RNA was extracted from 30 plants at the indicated times after the treatments.
Subcellular Localization of SiLTP
SiLTP coding region was amplified with the primers (Supplementary Table S1) without the terminating codon. The PCR products were digested with XbaI/KpnI and inserted into the corresponding sites of pSuper1300-GFP. The constructed pSuper1300-LTP-GFP was used for Agrobacterium-mediated transient transformation of tobacco epidermal cells (Zhang Y. et al., 2010). pSuper1300-GFP was used as the control. Maize protoplasts were prepared and transformed as described by Wang et al. (2014). Green fluorescent protein (GFP) signals were observed using a confocal laser scanning microscopy (LSM 510, Carl Zeiss MicroImaging GmbH, Jena, Germany).
Generation of Transgenic Plants
The coding region of SiLTP was amplified and ligated into the XbaI/KpnI sites of pCAMBIA2300 and the SalI/KpnI sites of pCOU to generate the constructs pCAMBIA2300-SiLTP and pCOU-SiLTP, respectively. The forward and reverse sequences of SiLTP were connected with the 400 bp GUS DNA fragment spacer, and then inserted into the BamHI/SacI sites of pCOU to generate the pCOU-SiLTP-RNAi (RNA interference) vector. pCAMBIA2300-SiLTP was introduced into the Agrobacterium tumefaciens strain LBA4404 and transformed into tobacco. The constructs pCOU-SiLTP and pCOU-SiLTP-RNAi were transformed into foxtail millet mediated by A. tumefaciens strain LBA4404 as described previously (Qin et al., 2008; Wang et al., 2011; Pan et al., 2015). The transgenic tobacco and foxtail millet plants were confirmed by PCR. The expression of SiLTP in transgenic plants was determined by semi-qRT-PCR and qRT-PCR using SiLTP-specific primers (Supplementary Table S1). Actin was used as the endogenous reference. In order to analyze the SiLTP promoter, we amplified the putative SiLTP promoter with the primers (Supplementary Table S1) and inserted into the pCAMBIA1391-GUS. The construct pCAMBIA1391-proLTP::GUS was then introduced into A. tumefaciens strain GV3101, and then transformed into Arabidopsis (Clough and Bent, 1998). Seeds were obtained following self-pollination.
Seed Germination Assay of SiLTP Transgenic Tobacco and Foxtail Millet under Abiotic Stress
Seeds of wild type (WT) and T1 transgenic tobacco plants were sterilized by chlorine gas (Clough and Bent, 1998) and spread on 1/2 Murashige and Skoog (MS) medium supplemented with 0, 200, or 250 mM mannitol, or 100 mM NaCl, independently, and were grown for 10 days in a growth chamber under the conditions of 25°C and 16 h/8 h (light/dark) photoperiod. The percentage of seeds germinated from each sample by the 10th day was calculated, and the lengths of seedling roots and shoots were measured.
Both the WT and T2 transgenic foxtail millet seeds were germinated on filter papers moistened with water containing 0, 100, 150, or 250 mM NaCl, or -0.5 MPa PEG at 25°C in the darkness, independently, and the following formulae were used to calculate the germination stress index (GSI). The promptness index (PI) = nd2 + 3/4 (nd4) + 1/2 (nd6) + 1/4(nd8), the nd2, 4, 6, and 8 indicate the percentage of seeds observed to germinate after 2, 4, 6, and 8 days, separately, and the GSI = (PI of stressed seeds/PI of control seeds) × 100 (Bouslama and Schapaugh, 1984). The shoot and root lengths of seedlings were measured on the 8th day after sowing.
Abiotic Stress Tolerance of SiLTP Transgenic Tobacco and Foxtail Millet
Four-week-old seedlings of WT and T1 transgenic tobacco were irrigated with water supplemented with 250 mM NaCl every 4 days, or not irrigated for 21 days, and then re-watered and grown for 3 days, the phenotypes of transgenic lines and WT were investigated and surviving plants were counted. Two-week-old foxtail millet seedlings were irrigated with water supplemented with 100 mM NaCl every 4 days, or not irrigated for 10 days, and then re-watered and grown for 3 days. The phenotypes were investigated and surviving plants were counted. The above-ground parts of seedlings at the indicated times of stress treatments were collected and used in subsequent proline and sugar measurements.
For the heading stage drought treatment, foxtail millet was grown in the greenhouse. After the 8th or 9th leaf appeared, the plants were not irrigated for 28 days, then re-watered and grown for 3 days, the survival rates were calculated.
Proline Content Measurement
The free proline contents were measured as described by Bates et al. (1973) and Wang et al. (2014). 0.05 g dry leaf tissue was used. Free proline was extracted with 3% sulphosalicylic acid at 95°C for 15 min. Two milliliters of supernatant was transferred to a new tube, 2 mL of acetic acid and 2 mL of acidified ninhydrin reagent were added and mixed well. The reaction was carried out at 95°C for 30 min. After that, 5 mL of toluene was added under continuously shaking. The toluene layer were fetched and measured at 520 nm for absorbance.
Soluble Sugar Content Measurement
Soluble sugar contents were examined according to Yemm and Willis (1954) and Wang et al. (2014). 0.1 g dry leaf tissue was used for the extraction of soluble sugar. Ten milliliters of 80% ethanol was added and the reaction was conducted at 80°C for 30 min with constant stirring. After filtration, 1 mL extracting solution was transferred to a new tube, and incubated in a boiling water bath to evaporate ethanol. Next, water was added to make the final volume of this solution up to 1 mL. Then 5 mL 0.15% anthrone solution were added, mixed well, and incubated for 15 min at 95°C. The reaction solution were cooled to room temperature and measured the absorbance at 620 nm. Glucose was used for making standard curve.
Statistical Analysis
For stress tolerance analysis, the experiment was repeated three times, and each time with three replicates. For Proline and sugar content measure, each data point had three replicates. Data represent the means and SD of three replicates, respectively. Student’s t-test were analyzed by the GraphPad Prism 5 software.
Histochemical GUS Staining
In transgenic Arabidopsis, GUS expression were detected by histochemical GUS staining (Jefferson et al., 1987; Pan et al., 2015). Chlorophyll was removed from tissues in 70% ethanol after the GUS staining, then the tissues were photographed in a dissecting microscope (Olympus SEX16, Tokyo, Japan).
Electrophoretic Mobility Shift Assay
SiARDP-GST and SiAREB1-GST vectors were constructed and transferred into E. coli (BL21) by Li et al. (2014). After induction, the fusion proteins were purified trough Glutathione Sepharose 4B column (GE, USA). Oligonucleotides (P1-R, P2-R, and P3-R) and their reverse complementary oligonucleotides (P1-F, P2-F, and P3-F, respectively), which were labeled with biotin, were synthesized. Meanwhile, the oligonucleotides (competitor2-1R, competitor2-1F, competitor2-2R, and competitor2-2F) for competitors were synthesized. All of these sequences are shown in Supplementary Table S1. Double-stranded DNA was obtained by heating oligonucleotides at 92°C for 30 s and annealing at 30°C overnight. The gel-shift assay was performed following the manufacturer’s protocol for the LightShift Chemiluminescent Electrophoretic Mobility Shift Assay (EMSA) Kit (Thermo, USA).
Yeast One-Hybrid Assay
The bait oligonucleotides of DRE2 (DRE2-F and DRE2-R) and the mutant bait oligonucleotides (mDRE2-F and mDRE2-R) were synthesized (Supplementary Table S1). All of the bait and mutant bait sequences were inserted into the pAbAi vector at the HindIII and XhoI sites to create the bait vectors. The bait vectors were transformed into yeast strain Y1HGold following the protocol of the Yeastmaker Yeast Transformation System 2 (Clontech, USA), and the transformed yeast was cultivated on synthetic medium without uracil but supplied with various contents of aureobasidin A. Then, SiARDP prey vector constructed by Li et al. (2014) was transformed into bait strains. The medium without leucine and with 500 ng mL-1 aureobasidin A was used. The transformed bait strain was cultivated at 30°C for 3 days.
Results
Characterization of SiLTP and its Expression Pattern in Foxtail Millet
SSH libraries was constructed using foxtail millet “Jigu 11” seedlings independently treated with PEG and NaCl. The SiLTP was cloned from this library. It contains 282 nucleotides, and the deduced protein contained 93 amino acid residues. The tertiary structure of SiLTP was predicted using SWISSMODEL (Arnold et al., 2006), and nsLTP2 from rice (Oryza sativa) is the most appropriate structural template for SiLTP. Between nsLTP2 and SiLTP, 64.7% of positional sequence identity and 53% similarity reflected the high degree of matching between the nsLTP2 model and SiLTP. SignalP 3.0 (Bendtsen et al., 2004) indicated that there is a signal peptide containing 25 amino acid residues in SiLTP which is located at N-terminal and hydrophobic. After cleavage of the signal peptide, a mature protein of 68 amino acids was produced. The predicted molecular weight is 6.9 kDa. This indicated that SiLTP belongs to the LTP II subfamily.
To analyze the SiLTP expression pattern, total RNAs were extracted from seeds of different developmental stage, roots, stems, leaves, inflorescences which collected before, and used for semi-qRT-PCR. SiLTP was detected in all of the tested tissues. However, the transcription level of SiLTP was lower in roots compared with other tissues (Figure 1A). Additionally, the SiLTP promoter was cloned and fused with GUS. The construct p1391-proSiLTP::GUS was transformed into Arabidopsis, 13 transgenic lines were obtained, and three T2 transgenic lines were selected for GUS staining. The GUS signals were detected in the root tips, stems, leaves, anthers, and both ends of siliques (Figures 1B–D). qPCR was carried out to determine the expression levels of SiLTP under stress conditions. The maximum SiLTP transcript level was detected 6 h after the salt treatment began, and then gradually decreased (Figure 2A). The PEG treatment resulted in the maximum SiLTP accumulation level, which was 12-fold 9 h after the treatment began (Figure 2B). The SiLTP transcript level increased significantly 1 h after the ABA treatment began, reached its highest level (ninefold) after 3 h, and then decreased (Figure 2C). These results imply that SiLTP is involved in plant responses to abiotic stresses through an ABA-dependent pathway.
FIGURE 1.
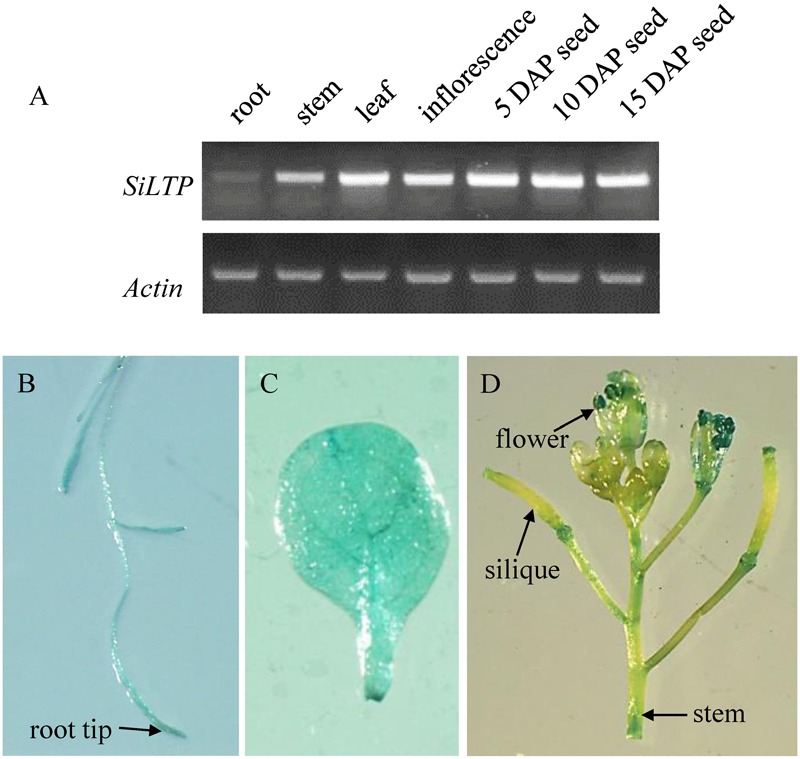
Expression pattern of SiLTP. (A) Semi-qRT-PCR analysis of SiLTP in various foxtail millet organs. (B–D) Histochemical analysis of the SiLTP promoter::GUS expression in a transgenic Arabidopsis root (B), 10-day-old leaf (C), and inflorescence (D).
FIGURE 2.
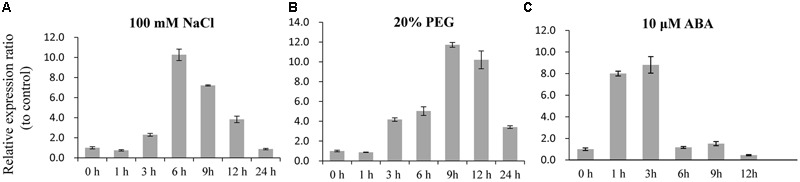
SiLTP transcript accumulation in response to various treatments. Two-week-old foxtail millet seedlings were treated independently with 100 mM NaCl (A), 20% (m/v) PEG 6000 (B), and 10 μM ABA (C) for the indicated times.
Subcellular Localization of SiLTP
To determine the subcellular localization of SiLTP, the pSuper::SiLTP-GFP construct was generated and transformed into Nicotiana benthamiana epidermal cells. The GFP fluorescence of pSuper::SiLTP-GFP was distributed in the cytoplasm, especially the cytoplasm near the cell membrane, and the fluorescence of control cells containing pSuper::GFP was ubiquitously localized (Figure 3A). To further confirm these results, maize protoplasts were subsequently used. The GFP fluorescence was observed throughout the cytoplasm (Figure 3B). Thus, SiLTP was localized in the cytoplasm.
FIGURE 3.
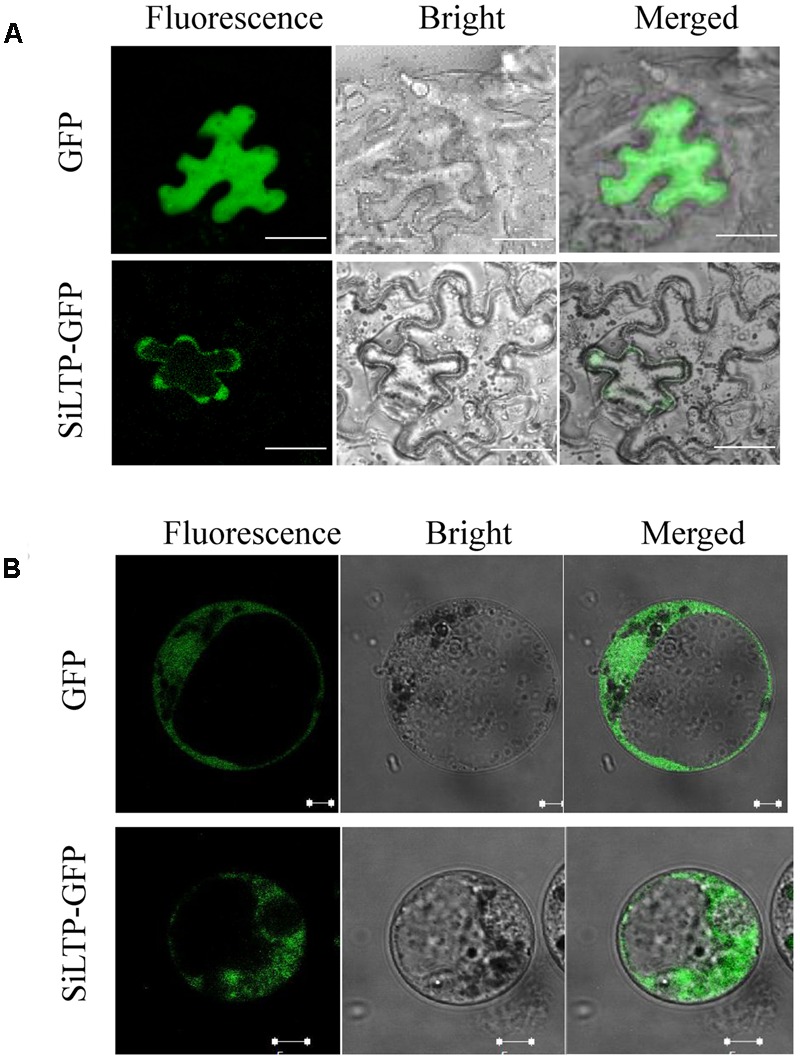
Subcellular localization of SiLTP in tobacco epidermal cells (A) and maize protoplasts (B). The green fluorescence signals, bright-field, and an overlay of the green fluorescence and bright-field images are shown from left to right. (A) Bar = 50 μm; (B) bar = 5 μm.
Ectopic Expression of SiLTP Improves the Abiotic Stress Tolerance of Transgenic Tobacco
Transgenic tobacco plants were generated using the construct pCOMBIA2300-SiLTP, and three independent T1 transgenic lines (LTP-11, LTP-14, and LTP-17) with high expression levels were chosen for further analysis (Figure 4A). To investigate the influence of salt and drought stress on seed germination and growth, the WT and transgenic tobacco seeds were spread on 1/2 MS medium supplied with various contents of mannitol or NaCl. The root and shoot lengths were measured, and no obvious differences were observed between the transgenic and WT plants on 1/2 MS medium. However, on medium with 100 mM NaCl, 200 mM mannitol, and 250 mM mannitol, SiLTP transgenic lines had significantly higher germination ratios than those of the WT (Figures 4B,C), and the SiLTP transgenic plants had longer roots and shoots on the media supplemented with either 100 mM NaCl or 200 mM mannitol compared than the WT seedlings (Figures 4D,E). To further determine the effect of abiotic stress on SiLTP transgenic tobacco, 4-week-old plants growing in soil were not irrigated with water, simulating drought stress (Figure 5A) or were irrigated with water supplemented with 250 mM NaCl (Figure 6A). After 3 weeks of the no irrigation drought treatment or 4 weeks of the salt treatment, the survival rates of these three transgenic lines were both significantly higher than those of WT plants (Figures 5B and 6B). Accordingly, transgenic lines also had significantly higher proline and soluble sugar contents than those of the WT under drought and salt treatments (Figures 5C,D and 6C,D). These results indicated that SiLTP improves the salt and drought tolerance of transgenic tobacco.
FIGURE 4.
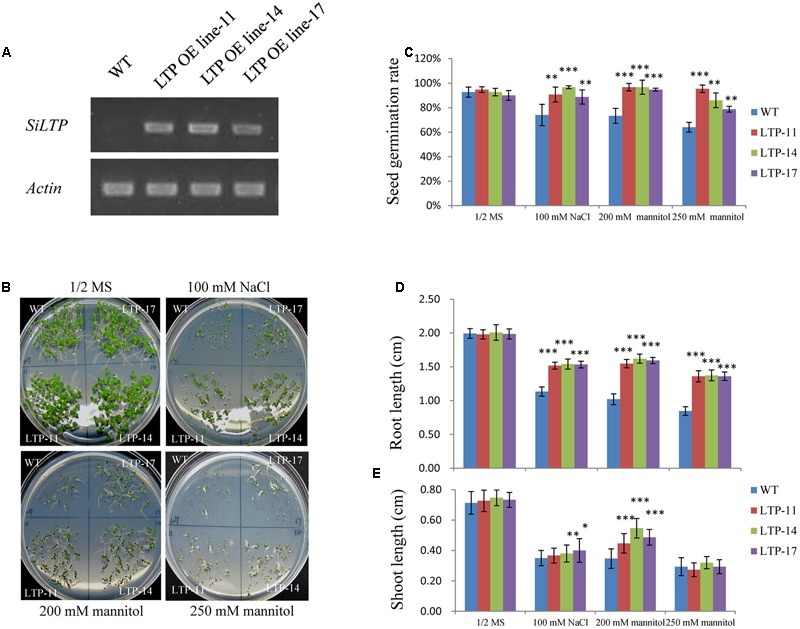
Effects of NaCl and mannitol stresses on SiLTP transgenic tobacco seed germination. Expression levels of SiLTP in transgenic tobacco and WT plants (A). NaCl or mannitol sensitivity of WT and transgenic plants (B). Seeds of WT and transgenic lines were germinated and then grown for 8 days on medium independently supplemented with 0 (control), 100 mM NaCl, 250 mM mannitol, and 300 mM mannitol. The corresponding seed germination rates (C), root lengths (D), and shoot lengths (E) are compared. ∗, ∗∗, and ∗∗∗ indicate statistically significant differences at P < 0.05, P < 0.01, and P < 0.001 (Student’s t-test), respectively.
FIGURE 5.
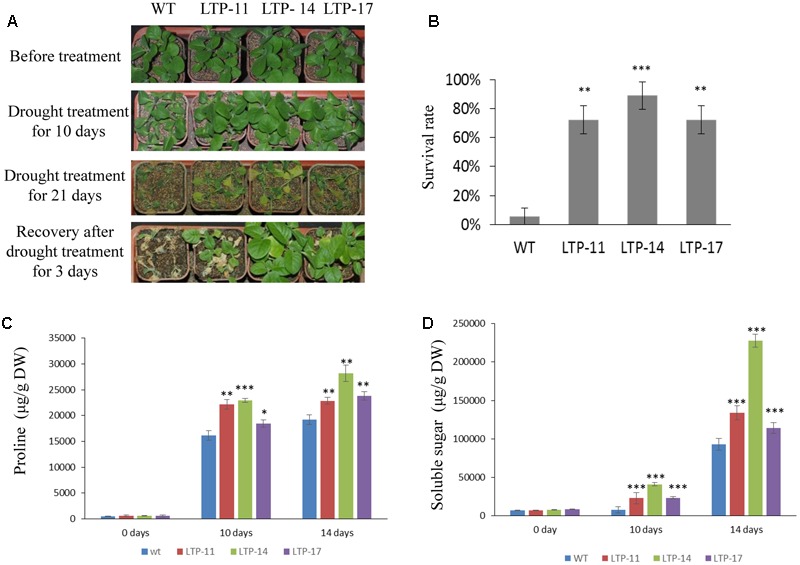
Drought resistance analysis of T1 transgenic and WT tobacco seedlings. Four-week-old seedlings of transgenic lines and WT were not watered for 0, 10, and 21 days, and were then re-watered (A); the survival rates of transgenic and WT seedlings after re-watering (B); the proline content (C), and soluble sugar content (D) were analyzed in transgenic and WT plants after exposure to drought for 10 and 14 days, respectively. Six seedlings of each transgenic line and WT were planted in each small basin for the drought treatment and corresponding analysis; 18 plants were used in each experiment. ∗, ∗∗, and ∗∗∗ indicate statistically significant differences at P < 0.05, P < 0.01, and P < 0.001 (Student’s t-test), respectively.
FIGURE 6.
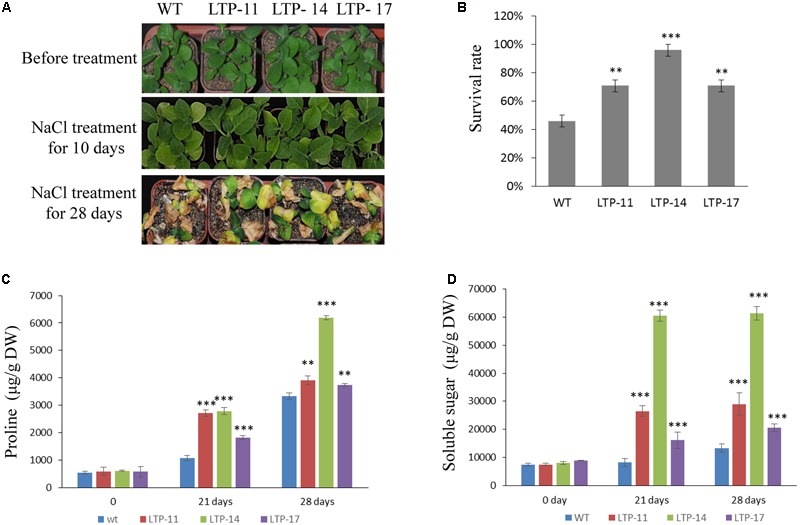
Salt resistance analysis of T1 transgenic and WT tobacco seedlings. Four-week-old seedlings of the transgenic lines and WT were irrigated for 0, 10, and 28 days with 250 mM NaCl (A); the survival rates of transgenic and WT seedlings after watering for 28 days with NaCl (B); the proline content (C); and soluble sugar content (D) were analyzed in transgenic and WT plants after watering for 21 and 28 days with NaCl, respectively. Six seedlings of each transgenic line and WT were planted in each small basin for the NaCl treatment and corresponding analysis; 18 plants were used in each experiment. ∗∗ and ∗∗∗ indicate statistically significant differences at P < 0.01 and P < 0.001 (Student’s t-test), respectively.
SiLTP Participates in Salt and Drought Tolerance of Foxtail Millet
For further analyzing SiLTP function in foxtail millet, OE and RNAi transgenic plants were generated. According to the result of qPCR, two OE lines (OE14 and OE69) and two RNAi lines (Ri21 and Ri27) were selected for further analysis (Figure 7A). The T2 transgenic seeds were used to analyze germination under normal conditions, and after salinity and drought treatments (Figure 7B). No significant germination stress indices changed among the OE, RNAi and WT lines (Figure 7C). However, after the independent 100 mM NaCl and -0.5 MPa PEG treatments, the roots and shoots of the OE lines were significantly longer than those of the WT and RNAi lines. Additionally, after the 150 mM NaCl treatment, the OE lines’ roots, but not the shoots, were significantly longer than in WT and RNAi lines (Figures 7D,E), the result was likely with that of SiLTP transgenic tobacco seeds on 250 mM mannitol treatment (Figures 4D,E).
FIGURE 7.
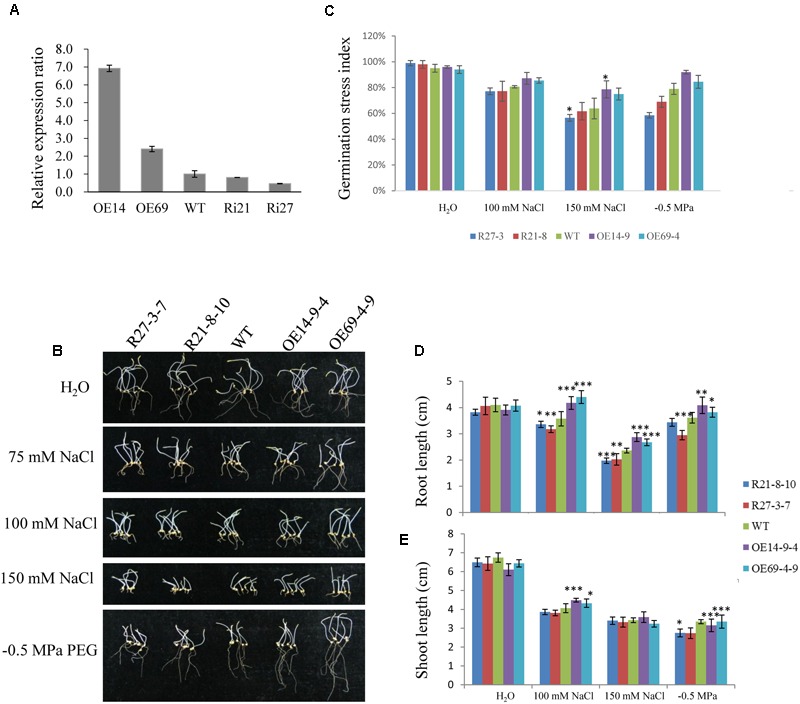
Effects of NaCl and PEG stresses on foxtail millet germination rates. Relative expression levels of SiLTP in T1 transgenic foxtail millet and WT plants as determined by qPCR (A). Seeds of WT and transgenic lines were germinated and grown for 8 days in water independently supplemented with 0 (control), 100 mM, 150 mM NaCl, or -0.5 MPa PEG (B). The corresponding seed germination rates (C), root lengths (D), and shoot lengths (E) were compared. ∗, ∗∗, and ∗∗∗ indicate statistically significant differences at P < 0.05, P < 0.01, and P < 0.001 (Student’s t-test), respectively.
Furthermore, 2-week-old seedlings of WT and transgenic plants were not irrigated for 10 days, and were then re-irrigated and grown under normal conditions for 3 days. The survival rates of the OE lines were higher than that of the WT, and the RNAi lines had much lower survival rates compared with the WT (Figures 8A,B). The proline and soluble sugar contents of the OE lines (especially the OE14 line) were higher than those of WT and RNAi lines after 10 days of drought treatment, and the RNAi lines had significantly lower soluble sugar contents than those of the WT and OE lines (Figures 8C,D). Moreover, the drought treatment was implemented during the reproductive stage of foxtail millet grown in a greenhouse. After 22 days of drought treatment, RNAi lines had visually retarded growth rates than WT and OE lines (Figure 8E). After 28 days of the drought treatment, the survival rate of RNAi lines significantly decreased compared with those of OE lines and the WT, and the OE line survival rates were much higher than those of WT and RNAi lines (Figures 8E,F). When 2-week-old seedlings of WT and transgenic plants were irrigated with water supplemented with 100 mM NaCl, none of the seedlings showed any significant differences after 14 days of treatment (Figure 9A). SiLTP-OE seedlings showed more tolerance compared with those of WT and RNAi lines after 21 days and 28 days of treatments (Figure 9A). After 21 days of treatment, the proline and soluble sugar contents of SiLTP-OE line 14 were significantly greater than those of the WT and RNAi lines (Figures 9B,C). The SiLTP-OE 69 line had a significantly greater proline content compared with the WT and RNAi lines, and its soluble sugar content was also greater, but not significantly (Figures 9B,C).
FIGURE 8.
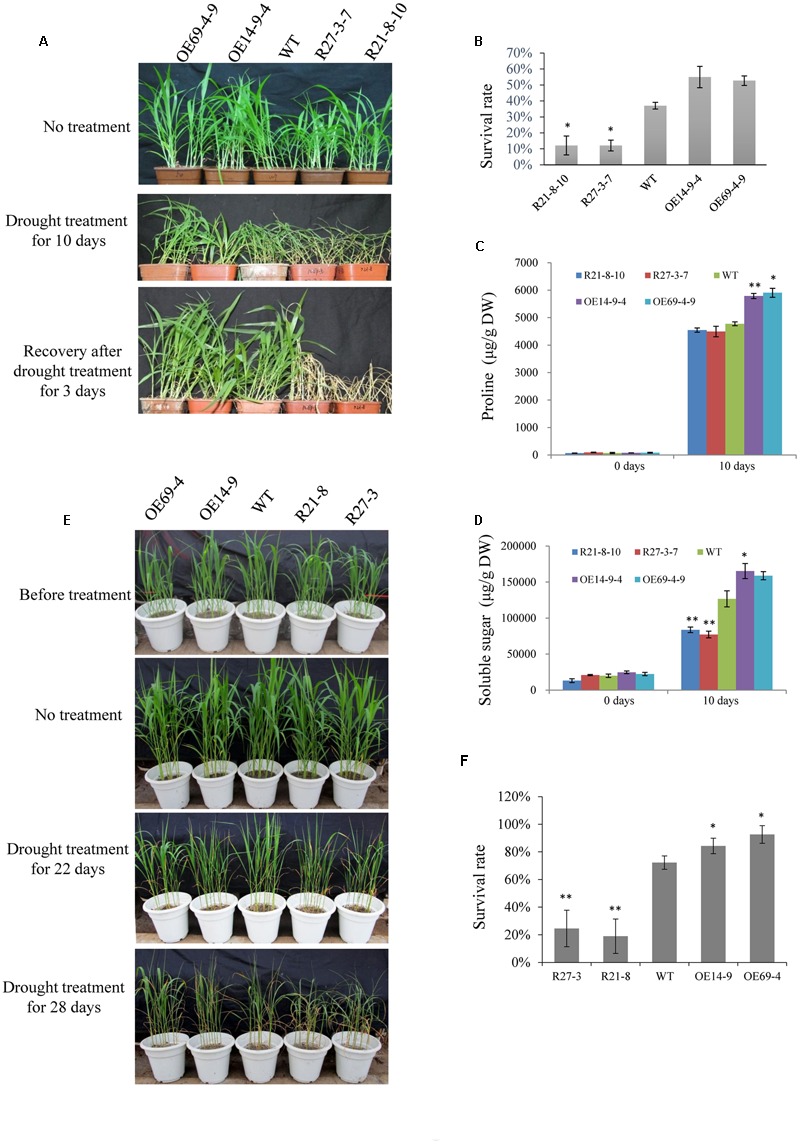
Drought tolerance of SiLTP-OE and RNAi foxtail millet. The phenotype of the 2-week-old transgenic foxtail millet seedlings under drought treatment (A). The survival rates of transgenic foxtail millet and WT plants (B). Proline contents (C) and soluble sugar contents (D) in WT and transgenic plants after drought stress. The drought tolerance phenotypes of SiLTP-OE and RNAi foxtail millet at the reproductive stage (E) and the survival rates of transgenic foxtail millet (F). ∗ and ∗∗ indicate statistically significant differences at P < 0.05 and P < 0.01 (Student’s t-test), respectively.
FIGURE 9.
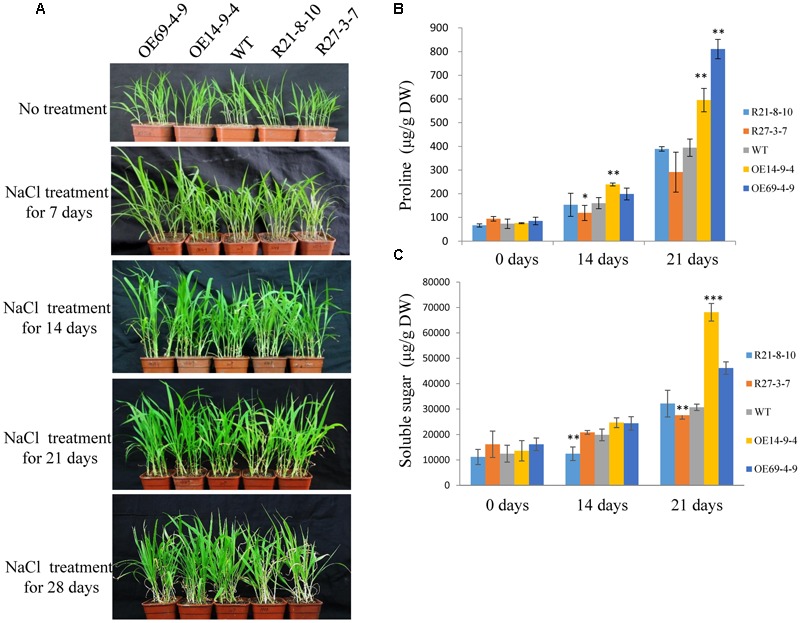
Salt tolerance of SiLTP-OE and RNAi foxtail millet seedlings. The phenotypes of transgenic foxtail millet under a 100 mM NaCl treatment (A). Proline contents (B) and soluble sugar contents (C) were analyzed in WT and transgenic plants after watering for 14 and 21 days with NaCl, respectively. ∗, ∗∗, and ∗∗∗ indicate statistically significant differences at P < 0.05, P < 0.01, and P < 0.001 (Student’s t-test), respectively.
Thus, the OE of SiLTP in foxtail millet enhanced salt and drought stress-related tolerance, while the down regulation of SiLTP led to an increased sensitivity to salt and drought stresses.
SiARDP Regulates SiLTP Expression by Directly Binding the DRE Element
PlantCARE was used to analyze the promoter region of SiLTP, three cis-elements including two DRE and one AREB, were discovered (Supplementary Table S2) (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/). EMSA was carried out and the result indicated that SiARDP bound to the SiLTP promoter through the DRE elements in vitro, but SiABRE1 could not bind to the AREB element (Figure 10A). Meanwhile, yeast one-hybrid results showed that SiARDP could bind to the SiLTP DRE elements in vivo (Figure 10B). The SiLTP transcription levels in SiARDP-OE lines were measured as well. The qPCR results showed that SiLTP was upregulated in SiARDP-OE lines (Li et al., 2014) by ∼1.5- to 2.0-fold compared with the WT (Figure 10C). Thus, SiLTP may be a downstream gene of SiARDP TF.
FIGURE 10.
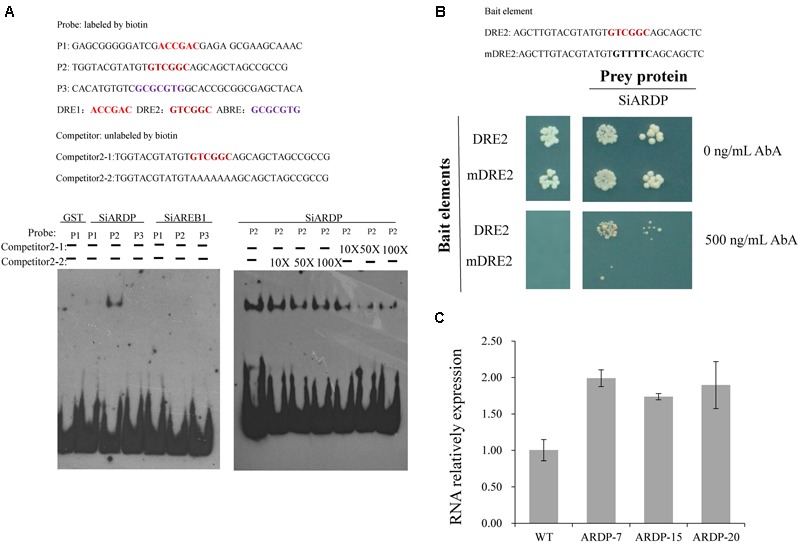
SiARDP regulates SiLTP expression by directly binding to the DRE element. EMSA for SiARDP and SiAREB1 binding to the promoter of SiLTP (A); yeast one-hybrid assay for SiARDP binding to the DRE element of the SiLTP promoter (B) and the SiLTP expression levels in SiARDP transgenic foxtail millet (C). Total RNA was extracted from 30 plants at the indicated times after the treatments. Data represent means and standard deviations for three biological replicates.
Discussion
The nsLTPs are known for their ability to transfer different lipids in vitro, but their functions have not yet been elucidated in vivo. LTPs were subdivided into two families based on size, LTP I and LTP II. So far, functional studies of the LTP II family are still very limited. Most found that LTP II-type genes are associated with seed development (GarcćA-Garrido et al., 1998; Monnet et al., 2001; Castro et al., 2003). LTP2 is an aleurone-specific expressed gene, and the LTP2 promoter drove GUS specific expression in the aleurone layer of rice immature seeds (Kalla et al., 1994). A 7-kDa nsLTP2 isolated from rice (O. sativa) seeds (Liu et al., 2002) has an unclear function. Only two type II LTP genes that respond to drought stress have been cloned, and the expression was detected in leaves, stems, and crowns (Jang et al., 2002). Here, we reported the identification and characterization of SiLTP, a novel gene belonging to the LTP II family in foxtail millet, and this will help further the understanding of LTP II functions in abiotic stress responses.
Drought stress affects plant growth and development. In the early stages of drought stress, leaf and stem growth is suppressed, while root growth is increased (Sharp et al., 1988; Nonami and Boyer, 1990). The leaves and stems stop growing to preserve the limited carbohydrate content and maintain their own metabolism. Additionally, it enriches the insoluble substances in the cell to maintain osmotic balance (Osorio et al., 1998). Root elongation allows plants to absorb water from deeper soils to maintain their physiological activities (Munns et al., 2000). In this study, when seeds of SiLTP transgenic and WT tobacco were germinated on 1/2 MS medium containing 200 mM mannitol, the SiLTP transgenic plants had significantly longer roots and shoots compared than the WT (Figure 4D). As the mannitol content increased to 250 mM, both WT and transgenic plants were more seriously restrained with little changes in shoot length and significant difference of root length (Figures 4D,E). When 5-day-old tobacco seedlings were placed on the 1/2 MS medium that contained 200, 250, or 300 mM mannitol, the transgenic seedlings had longer root length than the WT, even though the roots and shoots of WT and transgenic seedlings were restrained (Supplementary Figure S1). In foxtail millet, the same trends in root and shoot elongation were observed under -0.5 MPa PEG treatments (Figures 7D,E). All these results implied that SiLTP may function in drought stress resistance through enhancing root elongation. Osmotic adjustment is an important way in which plants resist environmental stresses. Under salt and drought stresses, a large number of organic osmolytes, such as sugars, alcohols, amino acids, and betaine, accumulate in plant cells. These substances maintain inflation pressure and the cell’s structural stability (Yancey et al., 1982). Increases in the proline and sugar contents of cells can enhance the abiotic stress resistance of plants (Gilmour et al., 2000; Streeter et al., 2001; Roosens et al., 2002; Ashraf and Foolad, 2007). We found the similar results in SiLTP transgenic plants. The proline and soluble sugar contents in SiLTP transgenic tobacco plants were significantly greater than those of WT when treated with salt and drought stress (Figures 5 and 6). The proline contents of SiLTP-OE lines were also significant greater than that of WT and RNAi lines after 10 days drought treatment, while the soluble sugar contents of RNAi lines were significant lower than that of WT and OE lines (Figures 8C,D).
The plant hormone ABA has vital roles in the plant signaling pathways of abiotic stress responses (Danquah et al., 2013). SiARDP, a DREB-type TF, is upregulated by the AREB-type TFs, SiAREB1 and SiAREB2. SiAREB1, SiAREB2, and SiARDP are involved in the ABA-dependent signal transduction pathways, and can improve the salt and drought stress resistance of transgenic foxtail millet (Li et al., 2014). In this study, the SiLTP mRNA level was induced by the ABA treatment (Figure 2C), and the SiLTP promoter sequence contained two DRE elements. SiARDP and SiLTP have similar expression patterns under drought and ABA treatments. SiLTP expression was upregulated by ∼1.5- to 2.0-fold in the SiARDP-OE foxtail millet. In addition, an EMSA and a yeast one-hybrid assay showed interactions between SiARDP and the SiLTP promoter in vitro and in vivo (Figures 10A,B). All these implied that SiLTP may be a direct target of SiARDP during the positive regulation of plant responses to salt and drought stresses. MYB and MYC TFs are also important in responses to drought and salt stress (Agarwal and Jha, 2010). In Arabidopsis, LTP3 expressed in various tissues and its expression is induced by cold, drought, and ABA. The LTP3 is localized in the cytoplasm. MYB96 directly binds to the LTP3 promoter, regulating LTP3 expression, and participating in the plant’s tolerance to freezing and drought stresses (Guo et al., 2013). In this study, SiLTP was induced by NaCl, drought, and ABA. SiLTP was expressed in all of the tested tissues, and SiLTP localized to the cytoplasm. The expression pattern and localization of SiLTP are similar to LTP3, and their regulation patterns may also be similar. In the SiLTP promoter region, we found five MYB and two MYC recognition sites (Supplementary Table S2). Therefore, it could be speculated that the MYB and MYC TFs take part in the regulation of SiLTP transcript levels except DREB type TFs. Further research is needed to investigate the regulation of SiLTP.
In transgenic tobacco and foxtail millet, SiLTP was proven to take part in the response to salt and drought stress, however, how the protein work in cells is unknown. When response to abiotic stress, cells would reduce lipid fluidity or decrease the membrane solute permeability to keep its integrity (Liu et al., 2015). Cryoprotectin, a LTP from cabbage, was proved to posse the cryoprotective activity without lipid transfer activity in cold treatment (Hincha et al., 2001). EARLI1 from Arabidopsis, can reduce electrolyte leakage during freezing damage, and was speculated to participate in the cell membrane or cell wall modification (Bubier and Schläppi, 2004). The Ca2+ sensor, calmodulin (CaM) is a mediator of intracellular Ca2+ signal transduction pathways (Golldack et al., 2014). Some evidence showed that LTPs can bind to CaM by their CaM-binding site (Wang et al., 2005, 2008; Gao et al., 2009), imply LTPs may through Ca2+ signal transduction pathways to participate in abiotic stress. Besides abiotic stress tolerance, some LTPs were found to possess biotic resistance or effects on plant growth (Sabine and Jutta, 2015; Safi et al., 2015). In our study, SiLTP RNAi lines showed slight morphological retardation in growth at the earlier stage under drought and salt treatments, we inferred that SiLTP may also participate in plant growth. Further research is needed to study the biological function of SiLTP.
In summary, SiLTP induced by NaCl, PEG, and ABA treatments and plays important roles in salt and drought stresses resistance in foxtail millet. SiLTP may be involved in ABA-dependent signaling pathway as a candidate of SiARDP downstream genes. This study provides insights into SiLTP and its regulator in plants response to abiotic stress, and a valid approach for improving abiotic stress resistance of crop.
Author Contributions
YP and JY designed the experiments; YP, JL, and LJ did the experiments; YP completed the data analysis and wrote the original manuscript; CL constructed vectors and SiARDP transgenic plants; DZ planted the foxtail millet; JY modified the manuscript and finalize the manuscript. All authors consent to the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Prof. Xianmin Diao (Chinese Academy of Agricultural Sciences) provided the seeds of “Jigu 11” foxtail millet, Prof. Chuanqing Sun (China Agricultural University) offered the pCOU vector, and Prof. Dongtao Ren (China Agricultural University) provided the pSuper1300-GFP vector. We thanks here.
Footnotes
Funding. This work was funded by the financial support (National Basic Research Programme of China, Project No. 2012CB215301).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.01752/full#supplementary-material
References
- Abuqamar S., Luo H., Laluk K., Mickelbart M. V., Mengiste T. (2009). Crosstalk between biotic and abiotic stress responses in tomato is mediated by the aim1 transcription factor. Plant J. 58 347–360. 10.1111/j.1365-313X.2008.03783.x [DOI] [PubMed] [Google Scholar]
- Agarwal P. K., Jha B. (2010). Transcription factors in plants and ABA dependent and independent abiotic stress signaling. Biol. Plant. 54 201–212. 10.1007/s10535-010-0038-7 [DOI] [Google Scholar]
- Arondel V., Kader J. C. (1990). Lipid transfer in plants. Experientia 46 579–585. 10.1007/BF01939696 [DOI] [PubMed] [Google Scholar]
- Arnold K., Bordoli L., Kopp J., Schwede T. (2006). The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22 195–201. 10.1093/bioinformatics/bti770 [DOI] [PubMed] [Google Scholar]
- Ashraf M., Foolad M. R. (2007). Roles of glycine betaine and proline in improving plant abiotic stress resistance. Environ. Exp. Bot. 59 206–216. 10.1016/j.envexpbot.2005.12.006 [DOI] [Google Scholar]
- Atkinson N. J., Lilley C. J., Urwin P. E. (2013). Identification of genes involved in the response of Arabidopsis to simultaneous biotic and abiotic stresses. Plant Physiol. 162 2028–2041. 10.1104/pp.113.222372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates L. S., Waldren R. P., Teare I. D. (1973). Rapid determination of free proline for water-stress studies. Plant Soil 39 205–207. 10.1016/j.dental.2010.07.006 [DOI] [Google Scholar]
- Bendtsen J. D., Nielsen H., von Heijne G., Brunak S. (2004). Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340 783–795. 10.1016/j.jmb.2004.05.028 [DOI] [PubMed] [Google Scholar]
- Bennetzen J. L., Schmutz J., Wang H., Percifield R., Hawkins J., Pontaroli A. C., et al. (2012). Reference genome sequence of the model plant Setaria. Nat. Biotechnol. 30 555–561. 10.1038/nbt.2196 [DOI] [PubMed] [Google Scholar]
- Bouslama M., Schapaugh W. T. (1984). Stress tolerance in soybeans. I. evaluation of three screening techniques for heat and drought tolerance. Crop Sci. 24 933–937. [Google Scholar]
- Bubier J., Schläppi M. (2004). Cold induction of EARLI1 a putative Arabidopsis lipid transfer protein, is light and calcium dependent. Plant Cell Environ. 27 929–936. 10.1111/j.1365-3040.2004.01198.x [DOI] [Google Scholar]
- Bustin S. A., Benes V., Garson J. A., Hellemans J., Huggett J., Kubista M., et al. (2009). The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55 611–622. 10.1373/clinchem.2008.112797 [DOI] [PubMed] [Google Scholar]
- Castagnaro A., García-Olmedo F. (1994). A fatty-acid-binding protein from wheat kernels. FEBS Lett. 349 117–119. 10.1016/0014-5793(94)00660-1 [DOI] [PubMed] [Google Scholar]
- Castro M. S., Gerhardt I. R., Orrù S., Pucci P., Bloch C. (2003). Purification and characterization of a small (7.3 kDa) putative lipid transfer protein from maize seeds. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 794 109–114. 10.1016/S1570-0232(03)00423-9 [DOI] [PubMed] [Google Scholar]
- Chae K., Kieslich C. A., Morikis D., Kim S. C., Lord E. M. (2009). A gain-of-function mutation of Arabidopsis lipid transfer protein 5 disturbs pollen tube tip growth and fertilization. Plant Cell 21 3902–3914. 10.1105/tpc.109.070854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S. J., Bent A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. 10.1046/j.1365-313x.1998.00343.x [DOI] [PubMed] [Google Scholar]
- Danquah A., Zelicourt A. D., Colcombet J., Hirt H. (2013). The role of ABA and MAPK signaling pathways in plant abiotic stress responses. Biotechnol. Adv. 32 40–52. 10.1016/j.biotechadv.2013.09.006 [DOI] [PubMed] [Google Scholar]
- Debono A., Yeats T. H., Rose J. K., Bird D., Jetter R., Kunst L., et al. (2009). Arabidopsis LTPG is a glycosylphosphatidylinositol-anchored lipid transfer protein required for export of lipids to the plant surface. Plant Cell 21 1230–1238. 10.1105/tpc.108.064451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doust A. N., Kellogg E. A., Devos K. M., Bennetzen J. L. (2009). Foxtail millet: a sequence-driven grass model system. Plant Physiol. 149 137–141. 10.1104/pp.108.129627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edstam M. M., Laurila M., Hoglund A., Raman A., Dahlstrom K. M., Salminen T. A., et al. (2014). Characterization of the GPI-anchored lipid transfer proteins in the moss Physcomitrella patens. Plant Physiol. Biochem. 75 55–69. 10.1016/j.plaphy.2013.12.001 [DOI] [PubMed] [Google Scholar]
- Edstam M. M., Viitanen L., Salminen T. A., Edqvist J. (2011). Evolutionary history of the non-specific lipid transfer proteins. Mol. Plant 4 947–964. 10.1093/mp/ssr019 [DOI] [PubMed] [Google Scholar]
- Gao G., Jin L. P., Xie K. Y., Qu D. Y. (2009). The potato StLTPa7 gene displays a complex Ca-associated pattern of expression during the early stage of potato-Ralstonia solanacearum interaction. Mol. Plant Pathol. 10 15–27. 10.1111/j.1364-3703.2008.00508.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- GarcćA-Garrido J. M., Menossi M., Puigdoménech P., MartıìNez-Izquierdo J. A., Delseny M. (1998). Corrigendum to: characterization of a gene encoding an abscisic acid-inducible type-2 lipid transfer protein from rice. FEBS Lett. 428 193–199. 10.1016/S0014-5793(98)00529-8 [DOI] [PubMed] [Google Scholar]
- Gilmour S. J., Sebolt A. M., Salazar M. P., Everard J. D., Thomashow M. F. (2000). Overexpression of the Arabidopsis CBF3 transcriptional activator mimics multiple biochemical changes associated with cold acclimation. Plant Physiol. 124 1854–1865. 10.1104/pp.124.4.1854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golldack D., Li C., Mohan H., Probst N. (2014). Tolerance to drought and salt stress in plants: unraveling the signaling networks. Front. Plant Sci. 5:151 10.3389/fpls.2014.00151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C., Ge X., Ma H. (2013). The rice OsDIL gene plays a role in drought tolerance at vegetative and reproductive stages. Plant Mol. Biol. 82 239–253. 10.1007/s11103-013-0057-9 [DOI] [PubMed] [Google Scholar]
- Han G. W., Lee J. Y., Song H. K., Chang C., Min K., Moon J., et al. (2001). Structural basis of non-specific lipid binding in maize lipid-transfer protein complexes revealed by high-resolution X-ray crystallography. J. Mol. Biol. 308 263–278. 10.1006/jmbi.2001.4559 [DOI] [PubMed] [Google Scholar]
- Hincha D. K. (2002). Cryoprotectin: a plant lipid-transfer protein homologue that stabilizes membranes during freezing. Philos. Trans. R. Soc. B Biol. Sci. 357 909–916. 10.1098/rstb.2002.1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hincha D. K., Neukamm B., Sror H. A., Sieg F., Weckwarth W., Schmitt J. M. (2001). Cabbage cryoprotectin is a member of the nonspecific plant lipid transfer protein gene family. Plant Physiol. 125 835–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes M. A., Dunn M. A., Pearce R. S., White A. J., Zhang L. (1992). An abscisic-acid-responsive, low temperature barley gene has homology with a maize phospholipid transfer protein. Plant Cell Environ. 15 861–865. 10.1111/j.1365-3040.1992.tb02155.x [DOI] [Google Scholar]
- Jang C., Kim D., Bu S., Kim J., Lee S., Kim J., et al. (2002). Isolation and characterization of lipid transfer protein (LTP) genes from a wheat-rye translocation line. Plant Cell Rep. 20 961–966. 10.1007/s00299-001-0424-x [DOI] [Google Scholar]
- Jefferson R. A., Kavanagh T. A., Bevan M. W. (1987). GUS fusions: b-Glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia G., Huang X., Zhi H., Zhao Y., Zhao Q., Li W., et al. (2013). A haplotype map of genomic variations and genome-wide association studies of agronomic traits in foxtail millet (Setaria italica). Nat. Genet. 45 957–961. 10.1038/ng.2673 [DOI] [PubMed] [Google Scholar]
- Kader J. C. (1975). Proteins and the intracellular exchange of lipids. I. Stimulation of phospholipid exchange between mitochondria and microsomal fractions by proteins isolated from potato tuber. Biochim. Biophys. Acta 380 31–44. [PubMed] [Google Scholar]
- Kader J. C. (1996). Lipid-transfer proteins in plants. Annu. Rev. Plant Biol. 47 627–654. 10.1146/annurev.arplant.47.1.627 [DOI] [PubMed] [Google Scholar]
- Kalla R., Shimamoto K., Potter R., Nielsen P. S., Linnestad C., Olsen O. A. (1994). The promoter of the barley aleurone-specific gene encoding a putative 7 kDa lipid transfer protein confers aleurone cell-specific expression in transgenic rice. Plant J. 6 849–860. 10.1046/j.1365-313X.1994.6060849.x [DOI] [PubMed] [Google Scholar]
- Kim H., Lee S. B., Kim H. J., Min M. K., Hwang I., Suh M. C. (2012). Characterization of glycosylphosphatidylinositol-anchored lipid transfer protein 2 (LTPG2) and overlapping function between LTPG/LTPG1 and LTPG2 in cuticular wax export or accumulation in Arabidopsis thaliana. Plant Cell Physiol. 53 1391–1403. 10.1093/pcp/pcs083 [DOI] [PubMed] [Google Scholar]
- Kumar K., Muthamilarasan M., Prasad M. (2013). Reference genes for quantitative real-time PCR analysis in the model plant foxtail millet (Setaria, italica, L.) subjected to abiotic stress conditions. Plant Cell Tissue Organ Cult. 115 13–22. 10.1007/s11240-013-0335-x [DOI] [Google Scholar]
- Li C., Yue J., Wu X., Xu C., Yu J. (2014). An ABA-responsive DRE-binding protein gene from Setaria italica, SiARDP, the target gene of SiAREB, plays a critical role under drought stress. J. Exp. Bot. 65 5415–5427. 10.1093/jxb/eru302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F., Zhang X., Lu C., Zeng X., Li Y., Fu D., et al. (2015). Non-specific lipid transfer proteins in plants: presenting new advances and an integrated functional analysis. J. Exp. Bot. 66 5663 10.1093/jxb/erv313 [DOI] [PubMed] [Google Scholar]
- Liu Y. J., Samuel D., Lin C. H., Lyu P. C. (2002). Purification and characterization of a novel 7-kDa non-specific lipid transfer protein-2 from rice (Oryza sativa). Biochem. Biophys. Res. Commun. 294 535–540. 10.1016/S0006-291X(02)00509-0 [DOI] [PubMed] [Google Scholar]
- Longeman J., Schell J., Willmitzer L. (1987). Improved method for the isolation of RNA from plant tissues. Anal. Biochem. 163 16–20. 10.1016/0003-2697(87)90086-8 [DOI] [PubMed] [Google Scholar]
- Mahajan S., Tuteja N. (2005). Cold, salinity and drought stresses: an overview. Arch. Biochem. Biophys. 444 139–158. 10.1016/j.abb.2005.10.018 [DOI] [PubMed] [Google Scholar]
- Maldonado A. M., Doerner P., Dixon R. A., Lamb C. J., Cameron R. K. (2002). A putative lipid transfer protein involved in systemic resistance signalling in Arabidopsis. Nature 419 399–403. 10.1038/nature00962 [DOI] [PubMed] [Google Scholar]
- Monnet F. P., Dieryck W., Boutrot F., Joudrier P., Gautier M. F. (2001). Purification, characterisation and cDNA cloning of a type 2 (7 kDa) lipid transfer protein from Triticumdurum. Plant Sci. 161 747–755. 10.1016/S0168-9452(01)00459-9 [DOI] [Google Scholar]
- Munns R., Passioura J. B., Guo J., Chazen O., Cramer G. R. (2000). Water relations and leaf expansion: importance of time scale. J. Exp. Bot. 51 1495–1504. 10.1093/jexbot/51.350.1495 [DOI] [PubMed] [Google Scholar]
- Muthamilarasan M., Prasad M. (2015). Advances in Setaria genomics for genetic improvement of cereals and bioenergy grasses. Theor. Appl. Genet. 128 1–14. 10.1007/s00122-014-2399-3 [DOI] [PubMed] [Google Scholar]
- Nonami H., Boyer J. S. (1990). Primary events regulating stem growth at low water potentials. Plant Physiol. 93 1601–1609. 10.1104/pp.93.4.1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osorio J., Osorio M. L., Chaves M. M., Pereira J. S. (1998). Water deficits are more important in delaying growth than in changing patterns of carbon allocation in Eucalyptus globulus. Tree Physiol. 18 363–373. 10.1093/treephys/18.6.363 [DOI] [PubMed] [Google Scholar]
- Pan Y., Ma X., Liang H., Zhao Q., Zhu D., Yu J. (2015). Spatial and temporal activity of the foxtail millet (Setaria italica) seed-specific promoter pF128. Planta 241 57–67. 10.1007/s00425-014-2164-5 [DOI] [PubMed] [Google Scholar]
- Panaud O. (2006). Foxtail Millet. In Cereals and Millets. Berlin: Springer; 325–332. [Google Scholar]
- Pitzschke A., Datta S., Persak H. (2014). Salt stress in Arabidopsis: lipid transfer protein AZI1 and its control by mitogen-activated protein kinase MPK3. Mol. Plant 7 722–738. 10.1093/mp/sst157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad M., Lata C., Yadav A. (2011). “Role of plant transcription factors in abiotic stress tolerance,” in Abiotic Stress Response in Plants – Physiological, Biochemical and Genetic Perspectives eds Shanker A. K., Venkateswarlu B. (Rijeka: InTech Press; ) 269–296. [Google Scholar]
- Pyee J., Yu H., Kolattukudy P. E. (1994). Identification of a lipid transfer protein as the major protein in the surface wax of broccoli (Brassica oleracea) leaves. Arch. Biochem. Biophys. 311 460–468. 10.1006/abbi.1994.1263 [DOI] [PubMed] [Google Scholar]
- Qin F., Zhao Q., Ao G., Yu J. (2008). Co-suppression of Si401 a maize pollen specific Zm401 homologous gene, results in aberrant anther development in foxtail millet. Euphytica 163 103–111. 10.1007/s10681-007-9610-4 [DOI] [Google Scholar]
- Roosens N. H., Bitar F., Loenders K., Angenon G., Jacobs M. (2002). Overexpression of ornithine-δ-aminotransferase increases proline biosynthesis and confers osmotolerance in transgenic plants. Mol. Breed. 9 73–80. 10.1023/A:1026791932238 [DOI] [Google Scholar]
- Sabine J., Jutta L. M. (2015). Response of Arabidopsis thaliana roots with altered lipid transfer protein (ltp) gene expression to the clubroot disease and salt stress. Plants 5:2 10.3390/plants5010002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safi H., Saibi W., Alaoui M. M., Hmyene A., Masmoudi K., Hanin M., et al. (2015). A wheat lipid transfer protein (TDLTP4) promotes tolerance to abiotic and biotic stress in Arabidopsis thaliana. Plant Physiol. Biochem. 89C 64–75. 10.1016/j.plaphy.2015.02.008 [DOI] [PubMed] [Google Scholar]
- Sham A., Alazzawi A., Alameri S., Almahmoud B., Awwad F., Alrawashdeh A., et al. (2014). Transcriptome analysis reveals genes commonly induced by botrytis cinerea infection, cold, drought and oxidative stresses in Arabidopsis. PLoS ONE 9:e113718 10.1371/journal.pone.0113718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sham A., Moustafa K., Al-Ameri S., Al-Azzawi A., Iratni R., Abuqamar S. (2015). Identification of Arabidopsis candidate genes in response to biotic and abiotic stresses using comparative microarrays. PLoS ONE 10:e0125666 10.1371/journal.pone.0125666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp R. E., Silk W. K., Hsiao T. C. (1988). Growth of the maize primary root at low water potentials: I. Spatial distribution of expansive growth. Plant Physiol. 87 50–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstein K. A., Moskal W. J., Wu H. C., Underwood B. A., Graham M. A., Town C. D., et al. (2007). Small cysteine-rich peptides resembling antimicrobial peptides have been under-predicted in plants. Plant J. 51 262–280. 10.1111/j.1365-313X.2007.03136.x [DOI] [PubMed] [Google Scholar]
- Streeter J. G., Lohnes D. G., Fioritto R. J. (2001). Patterns of pinitol accumulation in soybean plants and relationships to drought tolerance. Plant Cell Environ. 24 429–438. 10.1046/j.1365-3040.2001.00690.x [DOI] [Google Scholar]
- Thoma S., Kaneko Y., Somerville C. (1993). A non-specific lipid transfer protein from Arabidopsis is a cell wall protein. Plant J. 3 427–436. 10.1046/j.1365-313X.1993.t01-25-00999.x [DOI] [PubMed] [Google Scholar]
- Trevino M. B., O’Connell M. A. (1998). Three drought-responsive members of the nonspecific lipid-transfer protein gene family in Lycopersicon pennellii show different developmental patterns of expression. Plant Physiol. 116 1461–1468. 10.1104/pp.116.4.1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Xie W., Chi F., Hu W., Mao G., Sun D., et al. (2008). BcLTP, a novel lipid transfer protein in Brassica chinensis, may secrete and combine extracellular CaM. Plant Cell Rep. 27 159–169. 10.1007/s00299-007-0434-4 [DOI] [PubMed] [Google Scholar]
- Wang M., Li P., Li C., Pan Y., Jiang X., Zhu D., et al. (2014). SiLEA14 a novel atypical LEA protein, confers abiotic stress resistance in foxtail millet. BMC Plant Biol. 14:290 10.1186/s12870-014-0290-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Pan Y., Li C., Liu C., Zhao Q., Ao G., et al. (2011). Culturing of immature inflorescences and Agrobacterium-mediated transformation of foxtail millet (Setaria italica). Afr. J. Biotechnol. 10 16466–16479. [Google Scholar]
- Wang Z., Xie W., Chi F., Li C. (2005). Identification of non-specific lipid transfer protein-1 as a calmodulin-binding protein in Arabidopsis. FEBS Lett. 579 1683–1687. 10.1016/j.febslet.2005.02.024 [DOI] [PubMed] [Google Scholar]
- Wu G., Robertson A. J., Liu X., Zheng P., Wilen R. W., Nesbitt N. T., et al. (2004). A lipid transfer protein gene BG-14 is differentially regulated by abiotic stress, ABA, anisomycin, and sphingosine in bromegrass (Bromus inermis). J. Plant Physiol. 161 449–458. [DOI] [PubMed] [Google Scholar]
- Xu Z. Y., Zhang X., Schlappi M., Xu Z. Q. (2011). Cold-inducible expression of AZI1 and its function in improvement of freezing tolerance of Arabidopsis thaliana and Saccharomyces cerevisiae. J. Plant Physiol. 168 1576–1587. 10.1016/j.jplph.2011.01.023 [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K., Shinozaki K. (2005). Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters. Trends Plant Sci. 10 88–94. 10.1016/j.tplants.2004.12.012 [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K., Shinozaki K. (2006). Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 57 781–803. 10.1146/annurev.arplant.57.032905.105444 [DOI] [PubMed] [Google Scholar]
- Yancey P. H., Clark M. E., Hand S. C., Bowlus R. D., Somero G. N. (1982). Living with water stress: evolution of osmolyte systems. Science 217 1214–1222. 10.1126/science.7112124 [DOI] [PubMed] [Google Scholar]
- Yemm E. W., Willis A. J. (1954). The estimation of carbohydrates in plant extracts by anthrone. Biochem. J. 57 508–514. 10.1042/bj0570508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K., Soares J. M., Mandal M. K., Wang C., Chanda B., Gifford A. N., et al. (2013). A feedback regulatory loop between G3P and lipid transfer proteins DIR1 and AZI1 mediates azelaic-acid-induced systemic immunity. Cell Rep. 3 1266–1278. 10.1016/j.celrep.2013.03.030 [DOI] [PubMed] [Google Scholar]
- Zhang D., Liang W., Yin C., Zong J., Gu F., Zhang D. (2010). OsC6 encoding a lipid transfer protein, is required for postmeiotic anther development in rice. Plant Physiol. 154 149–162. 10.1104/pp.110.158865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G., Liu X., Quan Z., Cheng S., Xu X., Pan S., et al. (2012). Genome sequence of foxtail millet (Setaria italica) provides insights into grass evolution and biofuel potential. Nat. Biotechnol. 30 549–554. 10.1038/nbt.2195 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Zhang X., Niu S., Han C., Yu J., Li D. (2010). Nuclear localization of beet black scorch virus capsid protein and its interaction with importin α. Virus Res. 155 307–315. 10.1016/j.virusres.2010.10.029 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


