Abstract
Single amino acid substitutions in Fibroblast Growth Factor Receptor 1 (FGFR1) destabilize protein and have been implicated in several genetic disorders like various forms of cancer, Kallamann syndrome, Pfeiffer syndrome, Jackson Weiss syndrome, etc. In order to gain functional insight into mutation caused by amino acid substitution to protein function and expression, special emphasis was laid on molecular dynamics simulation techniques in combination with in silico tools such as SIFT, PolyPhen 2.0, I-Mutant 3.0 and SNAP. It has been estimated that 68% nsSNPs were predicted to be deleterious by I-Mutant, slightly higher than SIFT (37%), PolyPhen 2.0 (61%) and SNAP (58%). From the observed results, P722S mutation was found to be most deleterious by comparing results of all in silico tools. By molecular dynamics approach, we have shown that P722S mutation leads to increase in flexibility, and deviated more from the native structure which was supported by the decrease in the number of hydrogen bonds. In addition, biophysical analysis revealed a clear insight of stability loss due to P722S mutation in FGFR1 protein. Majority of mutations predicted by these in silico tools were in good concordance with the experimental results.
Abbreviations: SNPs, Single Nucleotide Polymorphisms; FGFR1, Fibroblast growth factor type 1; SIFT, Sorting Intolerant From Tolerant; PolyPhen 2.0, Polymorphism Phenotyping; OMIM, Online Mendelian Inheritance in Man; NCBI, National Center for Biological Information; SNAP, Screening for Non acceptable Polymorphisms; MSA, Multiple Sequence Alignments; RI, Reliability Index; RMSD, Root Mean Square Deviation; RMSF, Root Mean Square Fluctuation; SPC, Simple Point Charge; GV, Grantham Variance; GD, Grantham Deviation
Keywords: SNPs, Molecular dynamics simulation, FGFR1
Highlights
► Predicting the significance of novel genetic variants associated with FGFR1 gene using in silico tools. ► Biophysical validation of nsSNPs in FGFR1 gene. ► Analysis of structural and functional impact due to mutation by molecular dynamics approach. ► Stability loss of P722S mutation was observed in RMSD, RMSF and Hydrogen bond analysis.
1. Introduction
The number of identified amino acid variants in the human genome has grown rapidly owing to the application of high-throughput sequencing methods, but identification of variants responsible for specific phenotypes is understood poorly. Hence, the use of computational based tools with different algorithms significantly helps to overcome the difficulty of selection and prioritizing pathogenic variants from a pool of data. Amino acid substitutions may disrupt protein binding sites or ligand-binding pockets that are critical in protein function and may leads to alterations in the protein structure, folding or stability. In recent years, there has been considerable interest in understanding the genetic basis of FGFR1 associated with human disorder (Jiao et al., 2011, Rodriguez-Otero et al., 2011, Hitosugi et al., 2011). FGFR1 is one of the most commonly amplified gene involved in cancer which regulates cell proliferation, migration and differentiation (Ford et al., 2001). FGFR1 comprises of an extracellular region contacting three Ig-G like domains, single transmembrane helix and intracellular region containing tyrosine kinase domain.
Molecular dynamics (MD) simulation study may be useful to gain insight to the impact of non-synonymous polymorphisms (nsSNPs) on structural changes that may affect the activity of FGFR1. In particular, the effect of amino acid substitution that disrupts protein–protein interaction has been investigated for selected nsSNPs in our study. A number of algorithms based on sequence and structure based approach have been developed to predict the impact of missense mutations on protein function. To increase the confidence in prediction of functional and deleterious nsSNPs in this analysis, we have incorporated most commonly used computational methods like sorting intolerant from tolerant (SIFT) (Ng and Henikoff, 2003), polymorphism phenotyping (PolyPhen 2.0) (Adzhubei et al., 2010), I-Mutant 3.0 (Capriotti et al., 2008), and screening for non acceptable polymorphisms (SNAP) (Bromberg et al., 2008). Based on the results obtained from these methods, we proposed a model structure for the mutant proteins and compared this with the native protein in the three dimensional (3D) modeled structure of the FGFR1. In order to quantify the structural changes resulting from the SNPs, the native and mutant modeled proteins were evaluated using a range of structure assessment software. The ProSA-web z-score (Wiederstein and Sippl, 2007) was used to determine any change in the quality of the structure as a result of the mutation. Verify 3D (Luthy et al., 1992) was used to check improperly built segments based on the range of score between native and mutated residues. In order to biophysically validate the proposed impact of mutation on protein structure and function, align GVDV (Tavtigian et al., 2006) and what if web service (WIWS) (Hekkelman et al., 2010) were used. By analyzing the structural environment of substituted amino acids, we were able to develop a physiochemical hypothesis on the effect of the substitution in FGFR1. Furthermore, we suggest future experimental work that could be undertaken to confirm these findings and thus improve our knowledge in understanding the molecular basis of FGFR1 functionality. To the best of our knowledge this is the first study that incorporates the results of polymorphism analysis in conjunction with molecular dynamics approach for predicting disease causing mutation in FGFR1 gene.
2. Materials and methods
2.1. SNP dataset
Human FGFR1 gene data were collected from Online Mendelian Inheritance in Man (OMIM) (Amberger and Bocchin, 2009) and Entrez Gene on National Center for Biological Information (NCBI) web site. The SNP information (protein accession number (NP), mRNA accession number (NM) and SNP ID) of FGFR1 was retrieved from the NCBI dbSNP (http://www.ncbi.nlm.nih.gov/snp/) (Sherry et al., 2001), and SWISS-Prot databases (http://expasy.org/) (Amos and Rolf, 1996). Protein 3D structure was obtained from protein data bank (PDB) (Berman et al., 2002).
2.2. Predicting functional context of missense mutation
The functional context of nsSNPs was predicted using SIFT, PolyPhen 2.0, I-Mutant 3.0 and SNAP. SIFT is a sequence homology-based tool that predicts variants as neutral or deleterious using normalized probability score. Variants at position with normalized probability score less than 0.05 are predicted to be deleterious and score greater than 0.05 is predicted to be neutral (Ng and Henikoff, 2006). PolyPhen 2.0 utilizes a combination of sequence and structure based attributes and uses naive Bayesian classifier for the identification of an amino acid substitution and the effect of mutation. The output levels of probably damaging and possibly damaging were classified as deleterious (≤ 0.5) and the benign level being classified as tolerated (≥ 0.51). I-Mutant 3.0 (http://gpcr2.biocomp.unibo.it/cgi/predictors/I-Mutant3.0/I-Mutant3.0.cgi) is a support vector machine (SVM)-based tool. We used the sequence-based version of I-Mutant 3.0 that classifies the prediction in three classes: neutral mutation (− 0.5 ≤ DDG ≤ 0.5 kcal/mol), large decrease (≤− 0.5 kcal/mol) and large increase (> 0.5 kcal/mol). The output file shows the predicted free energy change (DDG) which is calculated from the unfolding Gibbs free energy change of the mutated protein minus the unfolding free energy value of the native protein (kcal/mol). SNAP is used for the prediction of impact of missense mutation based on neural network and improved machine-learning methodologies. For each mutant, SNAP returns three values: the binary prediction (neutral/non-neutral), the reliability index (RI, range 0–9) and the expected accuracy that estimates accuracy on a large dataset at the given RI.
2.3. Modeling of the mutant protein structure
For understanding the significance of a single amino acid substitution on protein function, knowledge about 3D structure of protein is very important. We used the dbSNP to identify the protein coded by FGFR1 (PDB ID 3RHX). We also confirmed the mutation positions and residues from this server. These mutation positions and residues were in complete agreement with the results obtained with SIFT, PolyPhen 2.0, I-Mutant 3.0 and SNAP. The mutation analysis was performed using SWISSPDB viewer, and energy minimization for three-dimensional structures was performed using NOMAD-Ref server (Lindahl et al., 2006). NOMAD-Ref use Gromacs as default force field for energy minimization based on methods of steepest descent, conjugate gradient and L-BFGS methods. In order to quantify the structural changes resulting from the SNPs, the wild and native type structures were evaluated using a range of structure assessment software.
2.4. Model verification
The quality of 3D models was assessed by ProSA-web and Verify 3D. ProSA-web calculates energy profiles (z-score) for modeled structure by using molecular mechanics force field. The z-score predicts overall model quality and measures the total energy deviation of the structure using random conformations. The modeled structure is predicted to be erroneous if the z-scores range outside the characteristic for native proteins. z-Score plot can be used for better interpretation of the z-score of the specified protein, which displays z-scores of all experimentally determined protein chains in current PDB. This plot can be used to check whether the z-score of the protein is within the range of scores typically found for proteins of similar groups. Verify 3D is used to identify unreliable regions in protein that have been improperly modeled and constructs a 3D model profile in which each amino acid residue position is characterized by its environmental score. Scores of mutated amino acid residues were compared with wild type residue to identify any structural problems arising from the mutation. For experimentally verified high resolution structure, Verify 3D score is positive and highly consistent.
2.5. Molecular dynamics simulation
All the molecular dynamics simulations were carried out using the program package GROMACS 4.0.5 (Hess et al., 2008) along with GROMOS9643a1 force field (van Gunsteren et al., 1996). Initially all models were solvated with the 0.9 nm simple point charge (SPC) water embedded in the simulation boxes. In order to neutralize the systems, one chlorine ion was added to replace one SPC water molecule (Jorgensen et al., 1983). Subsequently, all the systems investigated were subjected to a steepest descent energy minimization until reaching a tolerance of 100 kJ/mol. After the solvent molecules were equilibrated with the fixed protein at 300 K for a while, the entire system was gradually relaxed and heated up to 300 K. Finally, 6 ns MD simulations were performed under the normal temperature and pressure with coupling time constant 1.0 ps. The particle mesh Ewald method (Essmann et al., 1995) was used to treat long-range Coulombic interactions and the simulations performed using the SANDER module. The SHAKE algorithm was used to constrain bond lengths involving hydrogen, permitting a time step of 2 fs. Van der Waals force was maintained at 1.4 nm, and coulomb interactions were truncated at 0.9 nm.
2.6. Analysis of molecular dynamics trajectory
The trajectory files were analyzed by using g_rmsd and g_rmsf GROMACS utilities in order to obtain the root-mean-square deviation (RMSD), root-mean square fluctuation (RMSF). Number of distinct intermolecular hydrogen bonds formed between during the simulation was calculated using g_hbond utility. Number of hydrogen bond is prominent when donor–acceptor distance is smaller than 3.9 nm and donor–hydrogen–acceptor angle is larger than 90 nm.
2.7. Biophysical validation of nsSNPs
Align-GVGD (http://agvgd.iarc.fr/) combines the biophysical characteristics such as side chain composition, polarity and volume of amino acids and protein multiple sequence alignments (Grantham Variation (GV) and Grantham Deviation (GD) scores) to predict where amino acid substitutions fall in a spectrum of deleterious to neutral. The prediction is based on GV and GD scores (0 to > 200) and graded classifiers (C0 to C65). WIWS predicts accessible surfaces and the contact surfaces for a water probe with a radius of 1.4 Å. The default parameters of all programs were applied, and only the protein sequence and missense variant were given as input information for each program.
3. Results
3.1. Analysis of deleterious mutation
The functional impact of nsSNPs can be assessed by evaluating the importance of the amino acids they affect. A total of 38 nsSNPs was retrieved for our analysis. Protein sequence with mutational position and amino acid residue variants were submitted as input in SIFT. Out of 38 nsSNPs, 8 nsSNPs were predicted to be highly deleterious with score ranging from 0.00, 6 nsSNPs were predicted to be deleterious with a score range of 0.01–0.05 and, the remaining 14 nsSNPs were categorized as benign. All protein sequences submitted to SIFT were submitted to PolyPhen 2.0 server. PolyPhen 2.0 reports a score ranging from 0 (neutral) to 1 (damaging), which represents the confidence of its internal classifier. A total of 15 nsSNPs were predicted to be probably damaging with score ranging from 0.99 to 1.00, 8 nsSNPs were predicted to be possibly damaging with a score range of 0.5–0.9 and the remaining 15 nsSNPs were categorized as benign. The protein stability change due to a single point mutation was predicted using support vector machine-based tool I-Mutant 3.0. All the nsSNPs submitted to SIFT and PolyPhen 2.0 was submitted as input to I-Mutant 3.0. A total of 26 nsSNPs were predicted to cause stabilizing mutation (ΔΔG ≤ − 0.5 kcal/mol) and, the remaining 12 nsSNPs were found to be neutral mutations (− 0.5 ≤ ΔΔG ≤ 0.5 kcal/mol). SNAP was used to predict the overall severity of the missense mutations based on neural network and improved machine-learning methodologies. Out of 38 nsSNPs, SNAP predicted 22 nsSNPs as non neutral which could bring about changes in protein function and, the remaining 16 nsSNPs were predicted as neutral (Table 1).
Table 1.
Summary of nsSNPs (tolerated/deleterious) that were analyzed by computational methods SIFT, PolyPhen 2.0, I-Mutant 3.0 and SNAP.
| SNP ID | Allele | Variant | SIFT | PolyPhen 2.0 | I-Mutant 3.0 | SNAP | References |
|---|---|---|---|---|---|---|---|
| rs143341876 | C/T | P23L | 0.2 | 0.994 | − 0.12 | N | |
| rs149206728 | C/T | P25L | 0.39 | 0.00 | − 0.02 | N | |
| rs145434725 | C/T | P28L | 0.42 | 0.111 | 0.06 | N | |
| rs121909640 | C/T | G48S | 0.01 | 0.999 | − 0.96 | N | (Trarbach et al., 2006) |
| rs145315779 | C/A | R54H | 0.13 | 0.962 | − 1.26 | N | |
| rs150042321 | A/T | D59V | 0.16 | 0.383 | − 0.18 | N | |
| rs140254426 | G/A | G70R | 0.13 | 0.871 | − 0.63 | N.N | |
| rs143241978 | C/T | A74V | 0.29 | 0.00 | − 0.15 | N | |
| rs139867599 | G/T | V88L | 0.37 | 0.074 | − 0.62 | N | |
| rs150973404 | C/A | A94E | 1.00 | 0.00 | − 0.23 | N | |
| rs55642501 | G/A | V102I | 0.47 | 0.016 | − 0.55 | N | (Albuisson et al., 2005) |
| rs140382957 | C/T | S107L | 0.68 | 0.001 | − 0.19 | N | |
| rs121913473 | C/T | S125L | 0.37 | 0.018 | − 0.49 | N | (Greenman et al., 2007) |
| rs77734798 | A/C | D128A | 0.63 | 0.998 | − 0.68 | N | |
| rs121909630 | G/T | A167S | 0.19 | 1.00 | − 0.77 | N.N | (Dode et al., 2003) |
| rs17851623 | T/G | W213G | 0.03 | 1.00 | − 2.29 | N.N | (Gerhard et al., 2004) |
| rs121909635 | G/A | G237S | 0.01 | 1.00 | − 1.26 | N.N | (Pitteloud et al., 2006) |
| rs186746130 | G/A | V248M | 0.12 | 0.95 | − 1.11 | N.N | |
| rs121909645 | G/A | R250Q | 0.03 | 0.967 | − 0.95 | N.N | (Trarbach et al., 2006) |
| rs121913472 | C/A | P252T | 0.52 | 0.575 | − 1.3 | N | (Muenke et al., 1994) |
| rs121909627 | C/G | P252R | 0.00 | 0.999 | − 0.96 | N.N | (Greenman et al., 2007) |
| rs4647901 | G/C | L261F | 0.02 | 1.00 | − 1.1 | N | |
| rs121909633 | T/C | I300T | 0.39 | 0.01 | − 2.33 | N.N | (Pitteloud et al., 2006) |
| rs121909632 | A/T | N330I | 0.00 | 1.00 | − 0.97 | N.N | (Muenke et al., 1994) |
| rs121909638 | T/C | L342S | 0.13 | 0.976 | − 1.02 | N.N | |
| rs121909641 | C/T | P366L | 0.34 | 0.002 | − 0.38 | N.N | (Trarbach et al., 2006) |
| rs121909631 | A/G | Y374C | 0.23 | 0.997 | − 1.11 | N.N | (Muenke et al., 1994) |
| rs121909634 | T/C | C381R | 0.29 | 0.99 | − 0.16 | N.N | (Muenke et al., 1994) |
| rs183376882 | G/A | R424H | 0.55 | 0.032 | − 1.45 | N.N | |
| rs121909637 | G/T | R470L | 0.00 | 0.002 | − 0.45 | N.N | |
| rs77988343 | T/G | V513G | 0.00 | 0.999 | − 2.28 | N.N | |
| rs121909629 | G/A | V607M | 0.00 | 0.998 | − 1.81 | N.N | (Albuisson et al., 2005) |
| rs121909642 | C/T | P722S | 0.00 | 1.00 | − 2.69 | N.N | (Trarbach et al., 2006) |
| rs121909643 | G/T | Q764H | 0.01 | 0.841 | − 0.93 | N.N | |
| rs149979921 | T/G | L767R | 0.00 | 0.987 | − 1.71 | N.N | |
| rs2956723 | C/G | L769V | 0.32 | 0.046 | − 1.39 | N.N | |
| rs56234888 | C/T | P772S | 0.15 | 0.017 | − 1.68 | N.N | (Kress et al., 2009) |
| rs17182463 | C/T | R822C | 0.00 | 0.999 | − 0.84 | N | (Kress et al., 2009) |
AA — Amino Acid; NA — Not Available, N.N — Non neutral, and N — Neutral. SNP ID highlighted in bold is predicted to be deleterious by all 5 tools.
3.2. Concordance analysis of predicted results using in silico tools
The accuracy of deleterious nsSNPs predicted can be increased by combining different computational methods. Out of 38 nsSNPs, 14 nsSNPs were predicted to be deleterious by SIFT, 23 nsSNPs were predicted to be damaging by PolyPhen 2.0, 26 nsSNPs were predicted to be deleterious by I-Mutant 3.0, and 22 nsSNPs were predicted to be non neutral by SNAP server. For the results we could infer that, I-Mutant 3.0 predict 68% deleterious nsSNPs, slightly higher than SIFT (37%), PolyPhen 2.0 (61%) and SNAP (58%). Most of the nsSNPs predicted to be deleterious were in very well concordance with the experimentally derived data, highlighting the accuracy of our prediction method (Trarbach et al., 2006, Albuisson et al., 2005, Greenman et al., 2007, Dode et al., 2003, Gerhard et al., 2004, Pitteloud et al., 2006, Muenke et al., 1994, Kress et al., 2009, White et al., 2005, Dode et al., 2007).
3.3. Modeling deleterious nsSNPs
Single amino acid mutations can significantly alter protein structure thereby disturbs stability. In this context, knowledge of a protein's 3D structure is essential for better understanding the functionality of protein. Mutation analysis was performed based on the results obtained from various in silico tools. SWISS-PDB viewer was used to perform mutations at their respective coordinates and energy minimizations were done by NOMAD-Ref server for the native protein and mutant modeled structures. The crystal structure of human FGFR1 [3RHX] at 2.01 Å resolution was obtained from protein data bank (PDB) for structural analysis. By visualizing the position of the mutated amino acid residues, it is possible to suggest a physiochemical rationale for the effect on protein activity. The quality of 3D structure was assessed two programs: Verify 3D and ProSA-web. Furthermore, the results of each nsSNPs examined are reported in detail.
3.3.1. V513G variant
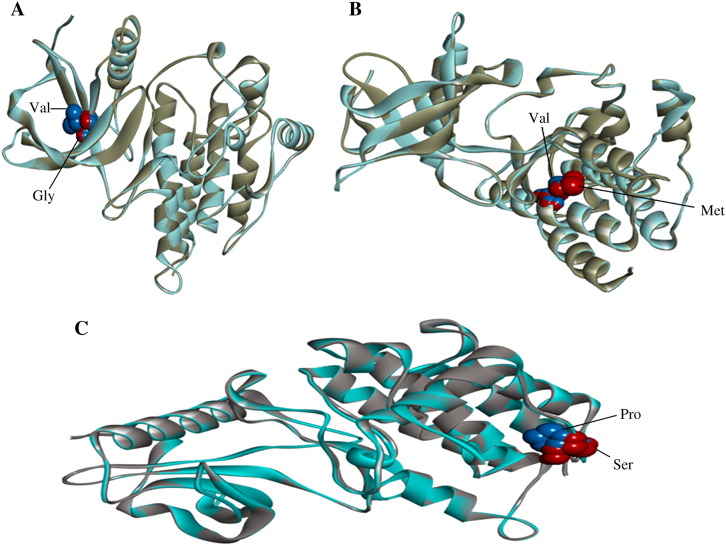
Each amino acid has unique size, charge and hydrophobicity value. SNP with ID rs77988343 results in the mutation of valine to glycine at position 513. The mutant residue is smaller than the wild type residue which leads to an empty space in the core of the protein. This mutation might cause loss of hydrophobic interactions in the core of the protein. Substitution of valine to glycine results in a slight worsening of ProSA-web z-score, from − 9.13 to − 9.05, while there was no change in Verify 3D score (0.81). The total energy of native protein after energy minimization using NOMAD-Ref was − 890,123.34 kcal/mol, and for the mutant protein was found to be − 889,931.09 kcal/mol. The RMSD value between native and mutant modeled protein was 1.01 Å. The superimposed structures of the native protein 3RHX (chain A) with the mutant model is shown in Fig. 1A.
Fig. 1.
Superimposition of native and mutant modeled structures (cartoon shape) of FGFR1.
A. Superimposed structure of native amino acid valine in sphere shape (blue color) with mutant amino acid glycine (red color) at position 513 in PDB ID 3RHX of FGFR1.
B. Superimposed structure of native amino acid valine in sphere shape (blue color) with mutant amino acid methionine (red color) at position 607 in PDB ID 3RHX of FGFR1.
C. Superimposed structure of native amino acid proline in sphere shape (blue color) with mutant amino acid serine (red color) at position 722 in PDB ID 3RHX of FGFR1.
3.3.2. V607M variant
SNP with ID rs121909629 resulted in the mutation of valine to methionine at position 607. The wild type residue is buried in the core of the protein, while the mutant residue being larger probably does not fit. Substitution of valine to methionine results in a slight worsening of ProSA-web z-score, from − 9.13 to − 9.10, while there was no change in Verify 3D score (0.81). The total energy of native protein after energy minimization using NOMAD-Ref was − 890,123.34 kcal/mol whereas, for the mutant model, it was found to be − 889,755.16 kcal/mol. The RMSD value between native and mutant modeled proteins was 1.12 Å. The superimposed structures of the native protein 3RHX (chain A) with the mutant model is shown in Fig 1B.
3.3.3. P722S variant
SNP with ID rs121909642 resulted in the mutation of proline to serine at position 722. The mutant residue is smaller than the wild type residue. The mutation will cause empty space in the core of the protein. Proline is in a cis conformation, and its side chain is engaged in numerous hydrophobic contacts with residues from neighboring α helices of the kinase domain. The P722S substitution could weaken these hydrophobic contacts and induce structural perturbations, which at the active site of kinase domain lead to a reduction in tyrosine kinase activity of FGFR1. Proline is known to have a very rigid structure, sometimes forcing the backbone in a specific conformation. Therefore, mutation of proline to uncharged polar serine may disturb the local structure of protein thereby altering protein function. Substitution of proline to serine results in a slight increasing of ProSA-web z-score from − 9.13 to − 9.14 and Verify 3D structure score from 0.81 to 0.88. The total energy of native protein after energy minimization using NOMAD-Ref was − 890,123.34 kcal/mol, whereas for the mutant model total energy was found to be − 889,012.14 kcal/mol. The RMSD value between native and mutant modeled protein was 1.21 Å. The superimposed structures of the native protein 3RHX (chain A) with the mutant model is shown in Fig. 1C.
3.4. Molecular dynamics conformational flexibility and stability analysis
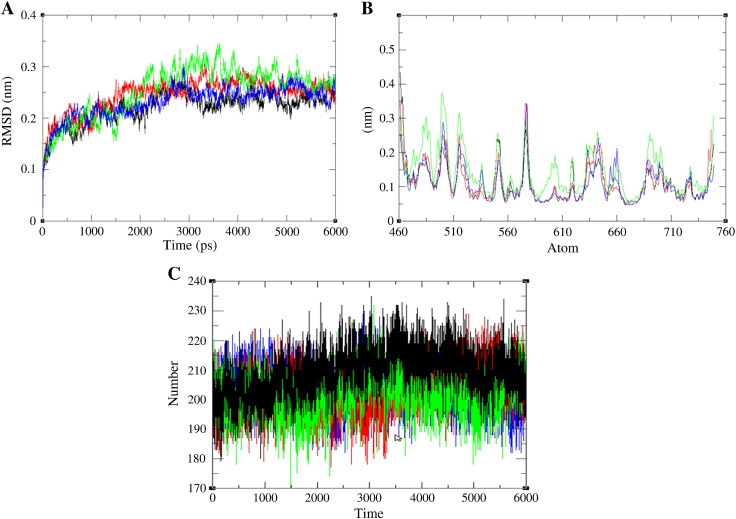
To examine the extent to which mutation effects protein structure, RMSD values were determined for native and mutant protein structure. We calculated the RMSD for all the atoms from the initial structure, which were considered as a central criterion to measure the convergence of the protein system concerned. It is evident that the native (3RHX) and mutant structures (V513G, V607M, P7222S) remain close to its starting conformation till 200 ps resulting in a backbone RMSD of about 0.14 nm (Fig. 2A). Between ranges of 500–2000 ps, wild type structure attained a maximum RMSD value of about 0.25 nm and among mutants 607 attained a maximum deviation of about 0.28 nm. From 2000 ps till end, mutant P722S retained a large deviation from other structure attaining a maximum RMSD of about 0.35 nm around 3600 ps. Throughout the analysis, mutant model P722S showed maximum deviation, while mutant model V607M exhibited intermediated deviated and native and mutant model V513G showed least deviation. A small variation in the average RMSD values of native and mutants after the relaxation period (~ 0.14 nm) lead to the conclusion that the mutations could affect the dynamic behavior of mutant protein, thus providing a suitable basis for further analyses. For determining the mutation affects dynamic behavior of residues, RMSF values of mutant and native structure were calculated. RMSF value of native residues fluctuates from a range of 0.08–0.28 nm in the entire simulation period. Moreover, mutant model V513G and V607M exhibited flexibility of ~ 0.35 nm and ~ 0.36 nm, while mutant P722S showed a maximum flexibility of about 0.38 nm (Fig. 2B). Analysis of the fluctuations revealed that the greatest degree of flexibility was shown by mutant model P722S. The reason for deviation in flexibility of residues was further validated by hydrogen bond analysis. Native protein exhibited maximum number of hydrogen bond 178–235, while the mutant model V513G and V607M showed an intermediate number of hydrogen bonds in the range of 180–235 (Fig. 2C). P722S exhibited least number of hydrogen bond ranging from ~ 170 to 213, which was in agreement with the stability of mutant models observed from the RMSD and RMSF analyses. These results imply that mutations might destroy the ability of hydrogen bond formation.
Fig. 2.
Molecular dynamics simulation of native and mutant model protein at 6000 ps.
A. Time evolution of backbone RMSDs is shown as a function of time of the wild and mutant structures at 6000 ps. The symbol coding scheme is as follows: wild (black color), mutant P722S (Green color), V607M (red color) and V513G (blue color).
B. RMSF of the backbone carbon alpha over the entire simulation. The ordinate is RMSF (nm), and the abscissa is atom. The symbol coding scheme is as follows: wild (black color), mutant P722S (green color), V607M (red color) and V513G (blue color).
C. Average number of intermolecular hydrogen bond in native and mutant versus time. The symbol coding scheme is as follows: wild (black color), mutant P722S (green color), V607M (red color) and V513G (blue color).
3.5. Biophysical analysis of missense mutation
We used Align-GVGD to assess the functional effect of missense variants, with alignment to 95 similar sequences down to human beings (BlastP). 21 nsSNPs occurred at strongly conserved residues (GV = 0) and had a GD ≥ 65. Thus, these were inferred to belong to the class (C65) of substitutions most likely to interfere with function. Two FGFR1 variants were defined as interfering with function (A-GVGD class C55), and the additional 12 nsSNPs had either a low GV or high GD score which lifted them above class C0 and the remaining 3 amino acid substitutions were less likely to compromise function (C0) (Table 2). In order to compare the biophysical property of native and mutant amino acids, solvent accessibility for surface was calculated. The location and type of a mutated residue affects the stability changes induced by mutations. In particular, as the solvent accessibility of a residue decreases, stability of protein due to mutation decreases. Based on WIWS, the solvent accessibility of V513G increases from 0.00 (native) to 0.873 (mutant), contrary there was a decrease in solvent accessibility value for V607M and P722S. A huge drift in solvent accessible surface area was observed in P722S (native 3.42 and mutant 0.873).
Table 2.
Prediction of functional effect of missense variants in FGFR1 using the Align-GVGD program.
| Variant | A-GVDV |
||
|---|---|---|---|
| GV | GD | Prediction class | |
| P23L | 73.35 | 97.78 | Class C15 |
| P25L | 102.71 | 56.87 | Class C0 |
| P28L | 0.00 | 97.78 | Class C65 |
| G48S | 0.00 | 55.27 | Class C55 |
| R54H | 0.00 | 28.82 | Class C25 |
| D59V | 0.00 | 152.22 | Class C65 |
| G70R | 0.00 | 125.13 | Class C65 |
| A74V | 65.28 | 0.00 | Class C0 |
| V88L | 0.00 | 30.92 | Class C25 |
| A94E | 0.00 | 106.71 | Class C65 |
| V102I | 0.00 | 28.68 | Class C25 |
| S107L | 144.08 | 0.00 | Class C0 |
| S125L | 0.00 | 144.08 | Class C65 |
| D128A | 0.00 | 125.75 | Class C65 |
| A167S | 0.00 | 99.13 | Class C65 |
| W213G | 0.00 | 183.79 | Class C65 |
| G237S | 0.00 | 55.27 | Class C55 |
| V248M | 0.00 | 20.52 | Class C15 |
| R250Q | 0.00 | 42.81 | Class C35 |
| P252T | 0.00 | 37.56 | Class C35 |
| P252R | 0.00 | 102.71 | Class C65 |
| L261F | 0.00 | 21.82 | Class C15 |
| I300T | 0.00 | 89.28 | Class C65 |
| N330I | 0.00 | 148.91 | Class C65 |
| L342S | 0.00 | 144.08 | Class C65 |
| P366L | 0.00 | 97.78 | Class C65 |
| Y374C | 0.00 | 193.72 | Class C65 |
| C381R | 0.00 | 179.53 | Class C65 |
| R424H | 0.00 | 28.82 | Class C25 |
| R470L | 0.00 | 101.88 | Class C65 |
| V513G | 0.00 | 189.55 | Class C65 |
| V607M | 0.00 | 12.52 | Class C15 |
| P722S | 0.00 | 193.35 | Class C65 |
| Q764H | 0.00 | 24.08 | Class C15 |
| L767R | 0.00 | 101.88 | Class C65 |
| L769V | 0.00 | 30.92 | Class C25 |
| P772S | 0.00 | 73.35 | Class C65 |
| R822C | 0.00 | 179.53 | Class C65 |
- GD >= 65 + tan(10) × (GV^2.5) = > Class C65 < = > most likely.
- GD >= 55 + tan(10) × (GV^2.0) = > Class C55.
- GD >= 35 + tan(50) × (GV^1.1) = > Class C35.
- GD >= 25 + tan(55) × (GV^0.95) = > Class C25.
- GD >= 15 + tan(75) × (GV^0.6) = > Class C15.
- Else (GD < 15 + tan(75) × (GV^0.6)) = > Class C0 < = > less likely.
4. Discussion
Predicting the phenotypic effect of nsSNPs using in silico methods may provide a greater understanding of genetic differences in susceptibility to disease. Our previous studies on polymorphisms screening using in silico analysis helped in predicting the functional nsSNPs associated with genes such as G6PD and ATM (George and Rajith, 2012, Rajith and George, 2011). Our findings also revealed that combination of different algorithms often serves as powerful tools for prioritizing candidate functional nsSNPs. Recent work by Thusberg and Vihinen (2009) compared several in silico tools, out of which SIFT, PolyPhen 2.0 and SNAP were reported to have better performance in identifying functional nsSNPs. The accuracy of SIFT and PolyPhen 2.0 was further validated by Hicks et al. (2011), which makes these tools more appropriate for the prediction. I-Mutant 3.0 was ranked as the one of the most reliable predictor based on the work performed by Khan and Vihinen (2010). Based on these in silico studies, we choose SIFT, PolyPhen, I-Mutant and SNAP for the prediction of functional and deleterious nsSNPs in FGFR1. It has been estimated that 68% nsSNPs were predicted to be deleterious by I-Mutant, slightly higher than SIFT (37%), PolyPhen 2.0 (61%) and SNAP (58%). In addition, we choose highly deleterious nsSNPs namely rs77988343 (V513G), rs121909629 (V607M) and rs121909642 (P722S) for further structural analysis. Out of this, V607M and P722S exhibited transition (2761 G → A, 3106 C → T) while V513G exhibited transversion (2480 T → G). Several groups have studied the relationships between nsSNPs and their location in protein structure (Capriotti and Altman, 2011, Yue and Moult, 2006). As a result, 3D model of native protein (PDB ID 3RHX) was compared with mutated modeled protein using SWISS PDB viewer (Fig. 1). Calculating the total energy difference between native and mutant model proteins gives the information about the protein structure stability. We compared RMSD value and total energy values (kcal/mol) of native and mutated modeled structure (V513G, V607M and P722S). Mutant model P722S showed an increase in total energy level (less favorable change) and increase in RMSD value deviation in comparison with native structure. Divergence in mutant structure with native structure is due to mutation, deletions, and insertions (Han et al., 2006) and the deviation between the two structures is evaluated by their RMSD values which could affect stability and functional activity (Varfolomeev et al., 2002). To better understand how these mutations affect the structural behavior of FGFR1, we incorporated molecular dynamic approach using GROMACS force field 43a1. Wang and Moult (2001) in his analysis revealed the key atomic events that allow substrate access and kinase activation due to mutation using molecular dynamics approach. The results that we have presented highlight the difficulty of unambiguously distinguishing native and mutant trajectories. The precise difference in the RMSD trajectories of P722S mutation, indicate the differences in the path of transition of structures from the starting conformation to their final states despite the initial structures being identical (except at the mutation sites). This information clearly speaks of the influence of amino acid substitutions on the dynamics of the protein. The RMSF data indicate that mutations are characterized by a subtle, but significant increase in the flexibility of the molecule. A clear insight of stability loss was observed in the RMSD and RMSF, which was further accompanied by decreased number of intermolecular hydrogen bonds in P722S mutant structure. This might eventually disrupt FGFR1 domain function which in turn alters the interaction with its protein partner there by affecting the signalling pathway. A more comprehensive characterization of disease causing and benign variants based on biophysical property were performed using Align GVDV and WIWS. Both relative entropy (Grantham parameters) and solvent accessibility (WIWS score) exclusively characterize the mutation site in a protein. Tokuriki et al. (2007) in his work argued that as the solvent accessibility of a residue decreases, the destabilizing ΔΔG values of its mutation increases. We observed a good concordance between stability of the protein (I-Mutant 3.0) and solvent accessibility (WIWS), in which P722S showed a huge drift in solvent accessibility followed by decrease in protein stability. Our analysis strongly indicates that amino acid substitution P722S is highly deleterious mutation which has been experimentally verified by Trarbach et al. (2006).
5. Conclusion
Impact of single amino acid substitution on protein stability remains one of the most promising setbacks in protein science. But its illumination by experiments that take advantage of large numbers, both experimentally and computationally, offers new hope for a solution in the years ahead. In our analysis, we identified the most deleterious mutation in FGFR1 based on various in silico tools. The following mutations V513G, V607M and P722S were screened for its deleterious impact on protein function based on these tools. To examine the structural consequences of these mutations, molecular dynamics simulations were carried out. A clear insight of stability loss of P722S mutation was observed in RMSD, RMSF and number of hydrogen bond when compared to other mutations. Impact of P722S mutation on protein biophysical property was further validated based on solvent accessibility analysis and Grantham parameters. In conclusion, our study shows that SNP analysis could be an ideal platform for identifying both somatic and germline genetic variants that leads to various disease. Hence the in silico analysis we performed proved to be both practical and valuable for a posteriori comprehension of human disorder, thereby greatly facilitating valuable resource for the pharmacogenomics approach.
Author disclosure statement
No competing financial interests exist.
Acknowledgments
The authors take this opportunity to thank the management of VIT University for providing the facilities and encouragement to carry out this work.
References
- Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. A method and server for predicting damaging missense mutations. Nature Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albuisson J., Pêcheux C., Carel J.C., Lacombe D., Leheup B., Lapuzina P., Bouchard P., Legius E., Matthijs G., Wasniewska M., Delpech M., Young J., Hardelin J.P., Dodé C. Kallmann syndrome: 14 novel mutations in KAL1 and FGFR1 (KAL2) Human Mutation. 2005;25:98–99. doi: 10.1002/humu.9298. [DOI] [PubMed] [Google Scholar]
- Amberger J., Bocchin C.A., Scott A.F., Hamosh A. Online Mendelian inheritance in man (OMIM) Nucleic Acids Research. 2009;37:D793–D796. doi: 10.1093/nar/gkn665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amos B., Rolf A. The SWISS-PROT protein sequence data bank and its new supplement TREMBL. Nucleic Acids Research. 1996;24:21–25. doi: 10.1093/nar/24.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The protein data bank. Nucleic Acids Research. 2002;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromberg Y., Yachdav G., Rost B. SNAP predicts effect of mutations on protein function. Bioinformatics. 2008;20:2397–2398. doi: 10.1093/bioinformatics/btn435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capriotti E., Altman R.B. Improving the prediction of disease-related variants using protein three-dimensional structure. BMC Bioinformatics. 2011;12:S3. doi: 10.1186/1471-2105-12-S4-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capriotti E., Fariselli P., Rossi I., Casadio R. A three-state prediction of single point mutations on protein stability changes. BMC Bioinformatics. 2008;9(Suppl. 2-S6) doi: 10.1186/1471-2105-9-S2-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dode C., Levilliers J., Dupont J.M. Loss-of-function mutations in FGFR1 cause autosomal dominant Kallmann syndrome. Nature Genetics. 2003;33:463–465. doi: 10.1038/ng1122. [DOI] [PubMed] [Google Scholar]
- Dode C., Fouveaut C., Mortier G., Janssens S., Bertherat J., Mahoudeau J., Kottler M.L., Chabrolle C., Gancel A., Francois I., Devriendt K., Wolczynski S., Pugeat M., Pineiro-Garcia A., Murat A., Bouchard P., Young J., Delpech M., Hardelin J.P. Novel FGFR1 sequence variants in Kallmann syndrome, and genetic evidence that the FGFR1c isoform is required in olfactory bulb and palate morphogenesis. Human Mutation. 2007;28:97–98. doi: 10.1002/humu.9470. [DOI] [PubMed] [Google Scholar]
- Essmann U., Perera L., Berkowitz M.L., Darden T., Lee H., Peterson L.G. A smooth particle meshes Ewald method. Journal of Chemical Physics. 1995;103:8577–8593. [Google Scholar]
- Ford P.M., Abud H., Murphy M. Fibroblast growth factors in the developing central nervous system. Clinical and Experimental Pharmacology and Physiology. 2001;28:493–503. doi: 10.1046/j.1440-1681.2001.03477.x. [DOI] [PubMed] [Google Scholar]
- George P.D.C., Rajith B. Computational refinement of functional single nucleotide polymorphisms associated with ATM gene. PLoS One. 2012;7(4):e34573. doi: 10.1371/journal.pone.0034573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhard D.S., Wagner L., Feingol E.A. The status, quality, and expansion of the NIH full-length cDNA project: the Mammalian Gene Collection (MGC) Genome Research. 2004;14:2121–2127. doi: 10.1101/gr.2596504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenman C., Stephens P., Smith R., Dalgilesh G.L., Hunter C., Bignell G. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J.H., Kerrison N., Chothia C., Teichmann S.A. Divergence of interdomain geometry in two-domain proteins. Structure. 2006;14:935–945. doi: 10.1016/j.str.2006.01.016. [DOI] [PubMed] [Google Scholar]
- Hekkelman M.L., Te Beek T.A., Pettifer S.R., Thorne D., Attwood T.K., Vriend G. WIWS: a protein structure bioinformatics Web service collection. Nucleic Acids Research. 2010;38:W719–W723. doi: 10.1093/nar/gkq453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess B., Kutzner C., van der Spoel D. GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. Journal of Chemical Theory and Computation. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- Hicks S., Wheeler D.A., Plon S.E., Kimmel M. Prediction of missense mutation functionality depends on both the algorithm and sequence alignment employed. Human Mutation. 2011;6:661–668. doi: 10.1002/humu.21490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitosugi T., Fan J., Chung T.W., Lythgoe K., Wang X., Xie J., Ge Q., Gu T.L., Polakiewicz R.D., Roesel J.L., Chen G.Z., Boggon T.J., Lonial S., Fu H., Khuri F.R., Kang S., Chen J. Tyrosine phosphorylation of mitochondrial pyruvate dehydrogenase kinase 1 is important for cancer metabolism. Molecular Cell. 2011;44:864–877. doi: 10.1016/j.molcel.2011.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao H., Arner P., Dickson S.L., Vidal H., Mejhert N., Henegar C., Taube M., Hansson C., Hinney A., Galan P., Simon C., Silveira A., Benrick A., Jansson J.O., Bouloumié A., Langin D., Laville M., Debard C., Axelsson T., Rydén M., Kere J., Dahlman-Wright K., Hamsten A., Clement K., Dahlman I. Genetic association and gene expression analysis identify FGFR1 as a new susceptibility gene for human obesity. Journal of Clinical Endocrinology and Metabolism. 2011;96:E962–E966. doi: 10.1210/jc.2010-2639. [DOI] [PubMed] [Google Scholar]
- Jorgensen W.L., Chandrasekhar J., Madura J.D., Impey R.W., Klein M.L. Comparison of simple potential functions for simulating liquid water. Journal of Chemical Physics. 1983;79:926. [Google Scholar]
- Khan S., Vihinen M. Performance of protein stability predictors. Human Mutation. 2010;31:675–678. doi: 10.1002/humu.21242. [DOI] [PubMed] [Google Scholar]
- Kress W., Petersen B., Collmann H. An unusual FGFR1 mutation (fibroblast growth factor receptor 1 mutation) in a girl with non-syndromic trigonocephaly. Cytogenetics and Cell Genetics. 2009;91:138–140. doi: 10.1159/000056834. [DOI] [PubMed] [Google Scholar]
- Lindahl E., Azuara C., Koehl P., Delarue M. NOMAD-Ref: visualization, deformation and refinement of macromolecular structures based on all-atom normal mode analysis. Nucleic Acids Research. 2006;34:52–56. doi: 10.1093/nar/gkl082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luthy R., Bowie J.U., Eisenberg D. Assessment of protein models with three dimensional profiles. Nature. 1992;356:83–85. doi: 10.1038/356083a0. [DOI] [PubMed] [Google Scholar]
- Muenke M., Schell U., Hehr A. A common mutation in the fibroblast growth factor receptor 1 gene in Pfeiffer syndrome. Nature Genetics. 1994;8:269–274. doi: 10.1038/ng1194-269. [DOI] [PubMed] [Google Scholar]
- Ng P.C., Henikoff S. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng P.C., Henikoff S. Predicting the effects of amino acid substitutions on protein function. Annual Review of Genomics and Human Genetics. 2006;7:61–80. doi: 10.1146/annurev.genom.7.080505.115630. [DOI] [PubMed] [Google Scholar]
- Pitteloud N., Acierno J.S., Meysing A., Eliseenkova A.V., Ma J., Ibrahimi O.A. Mutations in fibroblast growth factor receptor 1 cause both Kallmann syndrome and normosmic idiopathic hypogonadotropic hypogonadism. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:6281–6286. doi: 10.1073/pnas.0600962103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajith B., George P.D.C. Path to facilitate the prediction of functional amino acid substitutions in red blood cell disorders — a computational approach. PLoS One. 2011;6(9):e24607. doi: 10.1371/journal.pone.0024607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Otero P., Román-Gómez J., Vilas-Zornoza A., José-Eneriz E.S., Martín-Palanco V., Rifón J., Torres A., Calasanz M.J., Agirre X., Prosper F. Deregulation of FGFR1 and CDK6 oncogenic pathways in acute lymphoblastic leukaemia harbouring epigenetic modifications of the MIR9 family. British Journal of Haematology. 2011;155:73–83. doi: 10.1111/j.1365-2141.2011.08812.x. [DOI] [PubMed] [Google Scholar]
- Sherry S.T., Ward M., Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Research. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavtigian S.V., Deffenbaugh A.M., Yin L., Judkins T., Scholl T., Samollow P.B., de Silva D., Zharkikh A., Thomas A. Comprehensive statistical study of 452 BRCA1 missense substitutions with classification of eight recurrent substitutions as neutral. Journal of Medical Genetics. 2006;43:295–305. doi: 10.1136/jmg.2005.033878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thusberg J., Vihinen M. Pathogenic or not? And if so, then how? Studying the effects of missense mutations using bioinformatics methods. Human Mutation. 2009;30:703–714. doi: 10.1002/humu.20938. [DOI] [PubMed] [Google Scholar]
- Tokuriki N., Stricher F., Schymkowitz J., Serrano L., Tawfik D.S. The stability effects of protein mutations appear to be universally distributed. Journal of Molecular Biology. 2007;369:1318–1332. doi: 10.1016/j.jmb.2007.03.069. [DOI] [PubMed] [Google Scholar]
- Trarbach E.B., Costa E.M., Versiani B., de Castro M., Baptista M.T., Garmes H.M., de Mendonca B.B., Latronico A.C. Novel fibroblast growth factor receptor 1 mutations in patients with congenital hypogonadotropic hypogonadism with and without anosmia. Journal of Clinical Endocrinology and Metabolism. 2006;91:4006–4012. doi: 10.1210/jc.2005-2793. [DOI] [PubMed] [Google Scholar]
- van Gunsteren W.F., Billeter S.R., Eising A.A., Hünenberger P.H., Krüger P., Mark A.E., Scott W.R.P., Tironi I.G. Vdf Hochschulverlag AG an der ETH Zürich; Zürich, Switzerland: 1996. Biomolecular Simulation: The GROMOS96 Manual and User Guide. pp. 1–1042. [Google Scholar]
- Varfolomeev S.D., Uporov I.V., Fedorov E.V. Bioinformatics and molecular modeling in chemical enzymology: active sites of hydrolases. Biochemistry. 2002;67:1099–1108. doi: 10.1023/a:1020907122341. [DOI] [PubMed] [Google Scholar]
- Wang Z., Moult J. SNPs, protein structure, and disease. Human Mutation. 2001;17:263–270. doi: 10.1002/humu.22. [DOI] [PubMed] [Google Scholar]
- White K.E., Cabral J.M., Davis S.I. Mutations that cause osteoglophonic dysplasia define novel roles for FGFR1 in bone elongation. American Journal of Human Genetics. 2005;6:361–367. doi: 10.1086/427956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiederstein M., Sippl M.J. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Research. 2007;35:W407–W410. doi: 10.1093/nar/gkm290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue P., Moult J. Identification and analysis of deleterious human SNPs. Journal of Molecular Biology. 2006;356:1263–1274. doi: 10.1016/j.jmb.2005.12.025. [DOI] [PubMed] [Google Scholar]