Abstract
Mutations in the insulin receptor gene cause the inherited insulin resistant syndromes Leprechaunism and Rabson–Mendenhall syndrome. These recessive conditions are characterized by intrauterine and post-natal growth restrictions, dysmorphic features, altered glucose homeostasis, and early demise. The insulin receptor gene (INSR) maps to the short arm of chromosome 19 and is composed of 22 exons. Here we optimize the conditions for sequencing this gene and report novel mutations in patients with severe insulin resistance.
Methods
PCR amplification of the 22 coding exons of the INSR gene was performed using M13-tailed primers. Bidirectional DNA sequencing was performed with BigDye Terminator chemistry and M13 primers and the product was analyzed on the ABI 3100 genetic analyzer. Data analysis was performed using Mutation Surveyor software comparing the sequence to a reference INSR sequence (Genbank NC_000019).
Results
We sequenced four patients with Leprechaunism or Rabson–Mendenhall syndromes as well as seven samples from normal individuals and confirmed previously identified mutations in the affected patients. Three of the four mutations identified in this group caused premature insertion of a stop codon. In addition, the INSR gene was sequenced in 14 clinical samples from patients with suspected insulin resistance and one novel mutation was found in an infant with a suspected diagnosis of Leprechaunism.
Discussion
Leprechaunism and Rabson–Mendenhall syndrome are very rare and difficult to diagnose. Diagnosis is currently based mostly on clinical criteria. Clinical availability of DNA sequencing can provide an objective way of confirming or excluding the diagnosis.
Keywords: Insulin receptor, Leprechaunism, Donohue syndrome, Rabson–Mendenhall syndrome, Insulin resistance, Sequencing
1. Introduction
The insulin receptor is a membrane protein composed of two extracellular α subunits that bind insulin and two β subunits which span the plasma membrane and have an intracellular tyrosine kinase domain [1], [2]. Insulin binding to the α-subunits causes a conformational change that results in the activation of the kinase activity of the β-subunits with subsequent autophosphorylation and activation of kinase activity toward intracellular substrates [1], [2]. A single gene codes for both subunits. The resulting preprotein is post-translationally cleaved into mature alpha and beta subunits that assemble together as a heterotetramer to generate the mature insulin receptor [1], [2], [3]. The INSR gene maps to the short arm of chromosome 19 and is composed of 22 exons. Alternative splicing of the 36 base pair exon 11 results in two isoforms which differ in sequence at the C-terminal end of the insulin-binding alpha-subunit [3].
Mutations in INSR cause the insulin-resistant syndromes Leprechaunism, also known as Donohue syndrome [4], Rabson–Mendenhall syndrome and type A insulin resistance [5], [6]. Leprechaunism, (OMIM 246200), the most severe of the insulin resistant syndromes, is characterized by intrauterine growth restriction (IUGR), loss of glucose homeostasis, hyperinsulinemia, and dysmorphic features, with prominent eyes, thick lips, upturned nostrils, low-set posteriorly rotated ears, thick skin with lack of subcutaneous fat, distended abdomen, and enlarged genitalia in the male and cystic ovaries in the female [7], [8], [9]. Cells from most patients with Leprechaunism have markedly reduced insulin binding, although exceptions were reported [10], [11].
The slightly less severe Rabson–Mendenhall syndrome (OMIM 262190) was first described in three siblings with dental and skin abnormalities, abdominal distension, phallic enlargement, early dentition, coarse senile-looking facies, striking hirsutism, intellectual disability, prognathism, thick fingernails and acanthosis nigricans. Insulin-resistant diabetes mellitus, ketoacidosis, intercurrent infections, pineal hyperplasia and ovarian tumor [12]. Children have initial postprandial hyperglycemia and fasting hypoglycemia, caused by inappropriately elevated insulin levels at the time of fasting [6], [13]. Patients with Rabson–Mendenhall syndrome can survive beyond 1 year of age and, with time, develop constant hyperglycemia followed by diabetic ketoacidosis and death. This is accompanied by a progressive decline of insulin levels, which become insufficient to prevent liver glucose synthesis and release of fatty acids by adipocytes [13].
Mutations in the insulin receptor can cause disease with a dominant pattern of inheritance as well. For example, a mutation (p.Gly996Val) in a conserved Gly-X-Gly-X-X-Gly motif impairs tyrosine kinase activity of the insulin receptor and is associated with insulin-resistant diabetes mellitus and acanthosis nigricans, suggesting a dominant-negative pathogenesis [14], [15], [16]. A different mutation (p.Arg1174Gln) with unknown functional effects in INSR is implicated in familial hyperinsulinemic hypoglycemia type 5 in a few patients (HHF5) [17].
Leprechaunism and Rabson–Mendenhall syndrome are inherited as autosomal recessive traits. There is some correlation between genotype and phenotype, with mutations that markedly impair insulin binding resulting in the most severe phenotypes, while the presence of at least one mutation leaving residual insulin binding activity is associated with longer survival [6], [18]. Definitive genotype–phenotype correlation for INSR defects is difficult to establish primarily due to the rarity of these syndromes [6], a paucity of functional studies to determine the effect of mutations on insulin binding or signaling, and difficulty in establishing a precise molecular diagnosis due to the lack of clinically validated INSR gene sequencing [6], [19].
Herein we develop a clinically validated sequencing method to discover mutations in the INSR gene. Bidirectional sequencing with BigDye terminator and M13 primers was used to examine mutations in the coding regions and exon–intron boundaries of the INSR gene. A combination of the biochemical and DNA tests can provide accurate diagnosis for the insulin receptor deficiency.
2. Materials and methods
2.1. Patients/samples
DNA from 11 unrelated individuals (7 controls and 4 patients with Leprechaunism) was used to determine performance characteristics of this INSR full gene sequencing assay. Of these four patients with Leprechaunism, three of them, referred to here as 452, NY1, and 5880, had previously been described [6], [7], [23] Fibroblasts from each of these patients were received and DNA was extracted by MagNA Pure. The fourth patient with Leprechaunism, SLC, was not previously described but fit the clinical criteria. The diagnosis of Leprechaunism for all four patients was established from clinical presentation (failure to thrive, growth retardation, markedly elevated insulin levels, hirsutism, and acanthosis nigricans) and markedly reduced insulin binding to patients' fibroblasts. The samples were de-identified following an Institutional Review Board (IRB)-approved protocol. Fourteen additional samples referred to the ARUP Sequencing Laboratory by the patients' clinicians for INSR mutation detection were sequenced and analyzed.
2.2. DNA sequencing of the INSR gene
DNA was extracted from leukocytes in blood using MagNAPure Compact instrument (Roche Applied Science, Indianapolis, IN). Nucleic acid sequencing for the INSR gene coding region was performed by standard dideoxy termination. PCR primers were developed for the 22 exons of the INSR isoform containing exon 11 (NM_002082). Eighteen sets of PCR primers used in this validation were previously published [20], however, in the current study four sets of primers were re-designed to optimize PCR and sequencing results (see Table 1). We added another internal primer set to exon three interior to the homopolymer region to obtain cleaner sequence. In addition, exons 18 and 19 were consolidated into one amplicon. polymerase chain reaction of the 22 coding exons of the INSR gene was performed using M13-tailed primers Premix D (Epicentre, Madison, WI), and Platinum Taq (Invitrogen, Carlsbad, CA) using PCR conditions shown in Table 2 below. Unused PCR primers and unincorporated nucleotides were inactivated by incubation with ExoSAP (USB Corporation, Cleveland, OH). Bidirectional DNA sequencing was performed with BigDye Terminator chemistry (ABI, Foster City, CA) and M13 primers (IDT, Coralville, IA) and the product was analyzed on the ABI 3730. Data analysis was performed using Mutation Surveyor software (SoftGenetics, State College, PA) and GenBank reference sequence NG_008852.1.
Table 1.
Sequence of INSR primers used in the current study. Lower case letters represent the M13 tail sequence.
| Primer name | Sequence |
|---|---|
| IR E1F | tgtaaaacgacggccagtCGCGCTCTGATCCGAGGAGA |
| IR E1R | caggaaacagctatgaccAGGGTTCTCAGTCCACAAGC |
| IR E2F #2 | tgtaaaacgacggccagtTCTTGCTTTCTGTTCATTTTC |
| IR E2R #2 | caggaaacagctatgaccACGAGACACTGCTTAGAACC |
| IR E3F #2 | tgtaaaacgacggccagtCAGACAGGAATTGGACAAA |
| IR E3F Int | tgtaaaacgacggccagtGACCATCTGTAAGTCACACG |
| IR E3R | caggaaacagctatgaccAGCAGAGACCTCACTCATAGCCAA |
| IR E4F | tgtaaaacgacggccagtGCCTGAGATGTCTGAAGGAC |
| IR E4R | caggaaacagctatgaccGCCACTGAACGACCATCCTA |
| IR E5F | tgtaaaacgacggccagtCTCACCATGGAGAATCATGA |
| IR E5R | caggaaacagctatgaccCTAATACACGAACTTCCTAG |
| IR E6F #2 | tgtaaaacgacggccagtCACACCATCTTGGAGTTGTA |
| IR E6R | caggaaacagctatgaccTGTAATGCACTTGAATCATGCTG |
| IR E7F #2 | tgtaaaacgacggccagtTTGGTCTGAAACTACACTGAAA |
| IR E7R | caggaaacagctatgaccAAACGTAGCAAGCACAGAGC |
| IR E8F | tgtaaaacgacggccagtCGGTCTTGTAAGGGTAACTG |
| IR E8R #2 | caggaaacagctatgaccGCCAATAACCATATCAAGGA |
| IR E9F | tgtaaaacgacggccagtGCACACTGTTTCTCATGATG |
| IR E9R | caggaaacagctatgaccAGAGGTGAAGCAAAGTGCAT |
| IR E10F | tgtaaaacgacggccagtTGTTCAGCCGCAGAGACTTG |
| IR E10R | caggaaacagctatgaccCGGTCCCTAAGTAATGACCT |
| IR E11F | tgtaaaacgacggccagtGTGGTCTGTCTAATGAAGTT |
| IR E11R | caggaaacagctatgaccGAATTGGTGAAGCATCTGCT |
| IR E12F | tgtaaaacgacggccagtTGATGGTGATGGTGTCATCATA |
| IR E12R | caggaaacagctatgaccTGTCCTTGGTCAGCCTTGATGT |
| IR E13F #2 | tgtaaaacgacggccagtCAATCTTGTGGGATGAGTTT |
| IR E13R | caggaaacagctatgaccTACTAATAGCACAGTACCTG |
| IR E14F | tgtaaaacgacggccagtTGGACACTCCCAGATGTGCA |
| IR E14R | caggaaacagctatgaccACCATGCTCAGTGCTAAGCA |
| IR E15F | tgtaaaacgacggccagtGTGAACTTTGTTGGAAACACATTG |
| IR E15R | caggaaacagctatgaccCCTATACCTATATCAAGGCATG |
| IR E16F | tgtaaaacgacggccagtTCTGCTGGTAAGGGCTGCCA |
| IR E16R | caggaaacagctatgaccCTCACTCAATGGTGAAGGCA |
| IR E17F | tgtaaaacgacggccagtCCAAGGATGCTGTGTAGATAAG |
| IR E17R | caggaaacagctatgaccTCAGGAAAGCCAGCCCATGTC |
| IR E18–19F | tgtaaaacgacggccagtGGAGAACCCTGGTGAGTC |
| IR E18–19R | caggaaacagctatgaccTCCTTCTGAAATCAAACCTG |
| IR E20F | tgtaaaacgacggccagtAGGTTAAGAGCGTGTGAACCT |
| IR E20R | caggaaacagctatgaccGAATTCAAGCCCAGCGTCCAT |
| IR E21F | tgtaaaacgacggccagtTGTTACTACTATCAACTGTC |
| IR E21R | caggaaacagctatgaccACCTGTAACATACAGCATGC |
| IR E22F | tgtaaaacgacggccagtACTCACCCAGGACGTGTCCTTCT |
| IR E22R | caggaaacagctatgaccACCAGAGGAAAGCGAAAATG |
Table 2.
PCR conditions used in this study.
| Temp (°C) |
Rate (Δ°/cycle) |
Time |
Cycles |
|---|---|---|---|
| 95 | 5 m | ||
| 94 | 30 s | ||
| 62 | −0.5 | 45 s | 10 |
| 72 | 1 m | ||
| 94 | 30 s | ||
| 57 | 45 s | 25 | |
| 72 | 5 m | ||
3. Results
3.1. INSR mutation update
The INSR gene product contains 120 kilobases and is composed of 22 exons. There are three transcription initiation sites located at 276, 282 and 283 base pairs upstream of the translation initiation site. The alpha subunit is encoded by exons one through 11 (and part of exon 12) whereas the beta subunit is encoded by exons 12–22 [1], [2]. The insulin receptor is synthesized as a single protein that is post-translationally cleaved at a four amino acid site (p.759_762, RKRR, encoded by exon 12) to generate the mature alpha and beta subunit.
The INSR gene product contains a leader sequence of 27 amino acids. Cleavage of these amino acids results in the mature active protein. As a result of this cleavage, the nomenclature of reported variations differs depending on the author, time of publication, and source of their reference DNA sequence. For this reason, we reviewed the published literature for all known INSR variations to determine the consistent amino acid position using the current nomenclature, in both the immature and the mature protein. Absolute nucleotide positions were kept consistent with the beginning of the cDNA regardless of protein cleavage (recommendations of the Human Genome Variation Society, http://www.hgvs.org/rec.html). In addition, the INSR gene has two isoforms that differ only by the 12 amino acids encoded by the alternatively spliced exon 11. These isoforms have slightly different reported biological activity and different abundance in different tissues with the isoform containing exon 11 being predominant in the liver; the other in leukocytes; with similar expression levels in most other tissues such as skeletal muscle, placenta and adipose tissue [21].
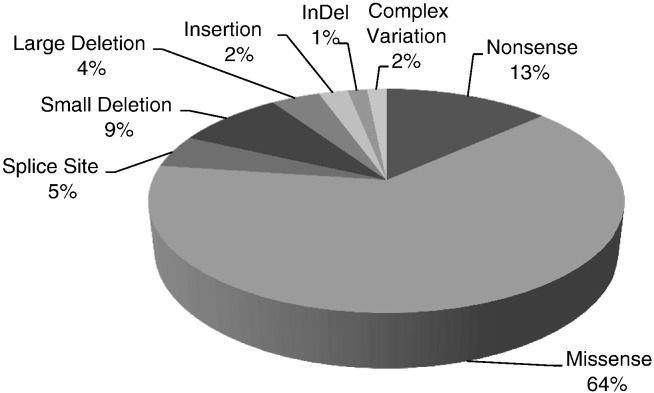
To date, there are 132 reports of disease causing mutations in the INSR gene in the literature (Table 4). The majority of the mutations (64%, or 85 of 132) are missense mutations, 13% (17 of 132) are nonsense mutation, 4.7% are splice site mutations, 8.3% are deletions (11/132), 2.3% are insertions (3/132), 1.5% are insertions and deletions (indel, 2/132), 5.3% are gross deletions or complex gene rearrangements (7/132) (Fig. 1). Most of the mutations located in the first 11 exons result in Leprechaunism while the mutations in the beta subunit are found more frequently in patients with Rabson–Mendenhall syndrome.
Table 4.
Compilation of reported INSR mutations.
| A. Missense/nonsense mutations | ||||||
|---|---|---|---|---|---|---|
| Location | Mutation type | Nucleotide change | Amino acid change (HGVS nomenclature) | Amino acid change (legacy, mature protein) | Phenotype | Reference |
| Exon 1 | Nonsense | c.90C > A | p.Tyr30Term | Tyr3Term | Rabson–Mendenhall syndrome | [30] |
| Exon 2 | Missense | c.121C > T | p.Arg41Trp | Arg14Trp | Rabson–Mendenhall syndrome | [31] |
| Exon 2 | Missense | c. 126C > A | p.Asn42Lys | Asn15Lys | Leprechaunism | [32] |
| Exon 2 | Missense | c.164T > C | p.Val55Ala | Val28Ala | Leprechaunism | [33] |
| Exon 2 | Missense | c.172G > A | p.Gly58Arg | Gly31Arg | Leprechaunism | [34] |
| Exon 2 | Missense | c.257A > G | p.Asp86Gly | Asp59Gly | Insulin resistance | [35] |
| Exon 2 | Missense | c.266T > C | p.Leu89Pro | Leu62Pro | Insulin resistance | [35] |
| Exon 2 | Missense | c.338G > C | p.Arg113Pro | Arg86Pro | Leprechaunism | [8] |
| Exon 2 | Nonsense | c.337C > T | p.Arg113Term | Arg86Term | Insulin resistance | [36] |
| Exon 2 | Missense | c.356C > T | p.Ala119Val | Ala92Val | Leprechaunism | [6] |
| Exon 2 | Missense | c.359T > A | p.Leu120Gln | Leu93Gln | Insulin resistance | [37] |
| Exon 2 | Missense | c.425G > T | p.Gly142Val | Gly115Val | Leprechaunism | This report |
| Exon 2 | Missense | c.433C > T | p.Arg145Cys | Arg118Cys | Insulin resistance A | [38] |
| Exon 2 | Missense | c.438C > G | p.Ile146Met | Ile119Met | Insulin resistance | [39] |
| Exon 2 | Nonsense | c.442A > T | p.Lys148Term | Lys121Term | Leprechaunism | [40] |
| Exon 2 | Nonsense | c.451G > T | p.Glu151Term | Glu124Term | Leprechaunism | [6] |
| Exon 2 | Nonsense | c.479G > A | p.Trp160Term | Trp133Term | Insulin resistance | [32] |
| Exon 2 | Missense | c.499G > T | p.Val167Leu | Val140Leu | Insulin resistance A | [41] |
| Exon 2 | Missense | c.511T > A | p.Tyr171Asn | Tyr144Asn | Diabetes, NIDDM | [42] |
| Exon 2 | Missense | c.515T > G | p.Ile172Ser | Ile145Ser | Diabetes, NIDDM | [42] |
| Exon 2 | Missense | c.557G > T | p.Cys186Phe | Cys159Phe | Rabson–Mendenhall syndrome | [19] |
| Exon 2 | Missense | c.586T > A | p.Cys196Ser | Cys169Ser | Diabetes, NIDDM | [42] |
| Exon 2 | Missense | c.628T > A | p.Trp210Arg | Trp183Arg | Diabetes, NIDDM | [42] |
| Exon 3 | Missense | c.659C > T | p.Pro220Leu | Pro193Leu | Leprechaunism | [43] |
| Exon 3 | Missense | c.679G > A | p.Gly227Ser | Gly200Ser | Diabetes, NIDDM | [42] |
| Exon 3 | Missense | c.694G > A | p.Gly232Ser | Gly205Ser | Diabetes, NIDDM | [42] |
| Exon 3 | Missense | c.707A > G | p.His236Arg | His209Arg | Leprechaunism | [32] |
| Exon 3 | Missense | c.712G > A | p.Glu238Lys | Glu211Lys | Rabson–Mendenhall syndrome | [30] |
| Exon 3 | Missense | c.766C > T | p.Arg256Cys | Arg229Cys | Rabson–Mendenhall syndrome | [19] |
| Exon 3 | Missense | c.779T > C | p.Leu260Pro | Leu233Pro | Insulin resistance | [44] |
| Exon 3 | Missense | c.835C > T | p.Arg279Cys | Arg252Cys | Insulin resistance | [45] |
| Exon 3 | Missense | c.836G > A | p.Arg279His | Arg252His | Insulin resistance | [37] |
| Exon 3 | Missense | c.839G > A | p.Cys280Tyr | Cys253Tyr | Insulin resistance A | [46] |
| Exon 3 | Nonsense | c.895C > T | p.Gln299Term | Gln272Term | Leprechaunism | [47] |
| Exon 3 | Missense | c.902G > A | p.Cys301Tyr | Cys274Tyr | Leprechaunism | [48] |
| Exon 3 | Missense | c.932G > A | p.Cys311Tyr | Cys284Tyr | Rabson–Mendenhall syndrome | [49] |
| Exon 4 | Missense | c.1049C > T | p.Ser350Leu | Ser323Leu | Insulin resistance | [50] |
| Exon 4 | Nonsense | c.1072C > T | p.Arg358Term | Arg331Term | Insulin resistance | [51] |
| Exon 4 | Nonsense | c.1114C > T | p.Arg372Term | Arg345Term | Insulin resistance A | [52] |
| Exon 5 | Missense | c.1156G > A | p.Gly386Ser | Gly359Ser | Rabson–Mendenhall syndrome | [53] |
| Exon 5 | Missense | c.1177G > A | p.Gly393Arg | Gly366Arg | Leprechaunism | [54] |
| Exon 5 | Nonsense | c.1195C > T | p.Arg399Term | Arg372Term | Insulin resistance | [55] |
| Exon 5 | Missense | c.1225T > G | p.Phe409Val | Phe382Val | Insulin resistance | [56] |
| Exon 5 | Nonsense | c.1246C > T | p.Arg416Term | Arg389Term | Leprechaunism | [57] |
| Exon 6 | Missense | c.1316G > C | p.Trp439Ser | Trp412Ser | Leprechaunism | [58] |
| Exon 6 | Missense | c.1372A > G | p.Asn458Asp | Asn431Asp | Insulin resistance | [37] |
| Exon 6 | Missense | c.1459A > G | p.Lys487Glu | Lys460Glu | Leprechaunism | [59] |
| Exon 6 | Missense | c.1466A > G | p.Asn489Ser | Asn462Ser | Insulin resistance | [32] |
| Exon 8 | Missense | c.1627A > T | p.Thr543Ser | Thr516Ser | Diabetes, NIDDM | [42] |
| Exon 8 | Missense | c.1650G > A | p.Ala550Ala | Ala523Ala | Association with reduced diastolic blood pressure | [60] |
| Exon 9 | Missense | c.1975T > C | p.Trp659Arg | Trp632Arg | Leprechaunism | [61] |
| Exon 10 | Nonsense | c.2095C > T | p.Gln699Term | Gln672Term | Leprechaunism | [59] |
| Exon 10 | Missense | c.2201A > C | p.Asp734Ala | Asp707Ala | Leprechaunism | [62] |
| Exon 12 | Missense | c.2286G > T | p.Arg762Ser | Arg735Ser | Insulin resistance | [63] |
| Exon 12 | Nonsense | c.2437C > T | p.Arg813Term | Arg786Term | Leprechaunism | [64] |
| Exon 12 | Missense | c.2453A > C | p.Tyr818Cys | Tyr791Cys | Leprechaunism | [65] |
| Exon 13 | Missense | c.2572A > G | p.Thr858Ala | Thr831Ala | Diabetes, NIDDM | [66] |
| Exon 13 | Missense | c.2621C > T | p.Pro874Leu | Pro847Leu | Leprechaunism/Rabson–Mendenhall syndrome | [31] |
| Exon 13 | Nonsense | c.2668C > T | p.Arg890Term | Arg863Term | Leprechaunism | [65] |
| Exon 13 | Missense | c.2669G > C | p.Arg890Pro | Arg863Pro | Diabetes, NIDDM | [42] |
| Exon 13 | Nonsense | c.2673T > A | p.Tyr891Term | Tyr864Term | Insulin resistance A | [46] |
| Exon 14 | Missense | c.2717C > G | p.Ala906Gly | Ala879Gly | Diabetes, NIDDM | [42] |
| Exon 14 | Nonsense | c.2770C > T | p.Arg924Term | Arg897Term | Leprechaunism | [23] |
| Exon 14 | Missense | c.2774T > C | p.Ile925Thr | Ile898Thr | Leprechaunism | [6] |
| Exon 14 | Missense | c.2776C > T | p.Arg926Trp | Arg899Trp | Leprechaunism | [6] |
| Exon 14 | Missense | c.2810C > T | p.Thr937Met | Thr910Met | Leprechaunism | [67] |
| Exon 16 | Missense | c.2971C > A | p.Leu991Ile | Leu964Ile | Leprechaunism | This report |
| Exon 16 | Missense | c.2989C > A | p.Pro997Thr | Pro970Thr | Rabson–Mendenhall syndrome | [6] |
| Exon 17 | Missense | c.3034G > A | p.Val1012Met | Val985Met | Diabetes, NIDDM | [68] |
| Exon 17 | Missense | c.3059G > A | p.Arg1020Gln | Arg993Gln | Insulin resistance | [69] |
| Exon 17 | Missense | c.3067A > T | p.Ile1023Phe | Ile996Phe | Insulin resistance | [70] |
| Exon 17 | Nonsense | c.3079C > T | p.Arg1027Term | Arg1000Term | Insulin resistance | [32] |
| Exon 17 | Missense | c.3104G > T | p.Gly1035Val | Gly1008Val | Diabetes, NIDDM | [14] |
| Exon 17 | Missense | c.3143G > A | p.Gly1048Asp | Gly1021Asp | Insulin resistance | [71] |
| Exon 17 | Missense | c.3160G > A | p.Val1054Met | Val1027Met | Leprechaunism | [61] |
| Exon 17 | Missense | c.3164C > T | p.Ala1055Val | Ala1028Val | Insulin resistance A | [41] |
| Exon 17 | Missense | c.3224C > A | p.Ala1075Asp | Ala1048Asp | Insulin resistance | [72] |
| Exon 17 | Missense | c.3255C > T | p.His1085His | His1058His | Association with polycystic ovary syndrome in lean women | [29] |
| Exon 17 | Missense | c.3257T > A | p.Val1086Glu | Val1059Glu | Diabetes, NIDDM | [42] |
| Exon 18 | Missense | c.3283A > G | p.Lys1095Glu | Lys1068Glu | Diabetes, NIDDM | [68] |
| Exon 18 | Missense | c.3220G > C | p.Glu1074Gln | Glu1047Gln | Rabson–Mendenhall syndrome | [31] |
| Exon 18 | Missense | c.3356G > A | p.Arg1119Gln | Arg1092Gln | Leprechaunism | [11] |
| Exon 18 | Missense | c.3355C > T | p.Arg1119Trp | Arg1092Trp | Leprechaunism | [49] |
| Exon 19 | Missense | c.3428T > C | p.Ile1143Thr | Ile1116Thr | Rabson–Mendenhall syndrome | [13] |
| Exon 19 | Missense | c.3436G > C | p.Gly1146Arg | Gly1119Arg | Insulin resistance | [71] |
| Exon 19 | Missense | c.3439A > T | p.Met1147Leu | Met1120Leu | Insulin resistance A | [73] |
| Exon 19 | Nonsense | c.3447C > A | p.Tyr1149Term | Tyr1122Term | Insulin resistance | [37] |
| Exon 19 | Missense | c.3470A > G | p.His1157Arg | His1130Arg | Insulin resistance | [74] |
| Exon 19 | Missense | c.3471T > A | p.His1157Gln | His1130Gln | Diabetes, NIDDM | [42] |
| Exon 19 | Missense | c.3473G > A | p.Arg1158Gln | Arg1131Gln | Insulin resistance | [75] |
| Exon 19 | Missense | c.3472C > T | p.Arg1158Trp | Arg1131Trp | Rabson–Mendenhall syndrome | [13] |
| Exon 19 | Missense | c.3481G > A | p.Ala1161Thr | Ala1134Thr | Insulin resistance | [76] |
| Exon 19 | Missense | c.3485C > A | p.Ala1162Glu | Ala1135Glu | Insulin resistance | [77] |
| Exon 20 | Missense | c.3540G > A | p.Met1180Ile | Met1153Ile | Insulin resistance | [78] |
| Exon 20 | Missense | c.3572G > A | p.Arg1191Gln | Arg1164Gln | Diabetes, NIDDM | [79] |
| Exon 20 | Missense | c.3602G > A | p.Arg1201Gln | Arg1174Gln | Insulin resistance | [80] |
| Exon 20 | Missense | c.3601C > T | p.Arg1201Trp | Arg1174Trp | Leprechaunism | [81] |
| Exon 20 | Missense | c.3614C > T | p.Pro1205Leu | Pro1178Leu | Insulin resistance | [82] |
| Exon 20 | Missense | c.3618G > C | p.Glu1206Asp | Glu1179Asp | Insulin resistance | [83] |
| Exon 20 | Missense | c.3616G > A | p.Glu1206Lys | Glu1179Lys | Leprechaunism | [49] |
| Exon 20 | Missense | c.3659G > T | p.Trp1220Leu | Trp1193Leu | Insulin resistance | [83] |
| Exon 21 | Missense | c.3680G > C | p.Trp1227Ser | Trp1200Ser | Insulin resistance | [76] |
| Exon 21 | Nonsense | c.3769C > T | p.Gln1257Term | Gln1230Term | Insulin resistance A | [73] |
| Exon 22 | Missense | c.4082A > G | p.Tyr1361Cys | Tyr1334Cys | Diabetes, NIDDM | [66] |
| Exon 22 | Missense | c.4133G > A | p.Arg1378Gln | Arg1351Gln | Insulin resistance | [50] |
| B. Splice site, insertion/deletion, and large gene rearrangement mutations | ||||
|---|---|---|---|---|
| Location | Mutation type | Nucleotide change | Phenotype | Reference |
| Intron 2 | Splice site | c.1124-2A > G | Insulin resistance | [67] |
| Intron 5 | Splice site | c.1268 + 2T > C | Rabson–Mendenhall syndrome | [31] |
| Intron 6 | Splice site | c.1483 + 43G > T | Diabetes, type 2, association with | [84] |
| Intron 13 | Splice site | c.2682 + 1G > A | Leprechaunism | [9] |
| Intron 14 | Splice site | c.2842 + 1G > A | Insulin resistance A | [85] |
| Exon 17 | Splice site | c.3258G > A | Fiber-type disproportion myopathy, congenital | [86] |
| Intron 21 | Splice site | c.3794 + 1G > T | Leprechaunism | [31] |
| Intron 21 | Splice site | c.3795-1G > A | Insulin resistance A | [41] |
| Exon 1 | Deletion | c.22_31del10 | Insulin resistance | [87] |
| Exon 2 | Deletion | c.404delA | Leprechaunism | [47] |
| Exon 2 | Deletion | c.444_446delGAA | Leprechaunism | [88] |
| Exon 3 | Deletion | c.927_929delCAA | Leprechaunism | [9] |
| Exon 4 | Deletion | c.1084_1086delGTC | Leprechaunism | [89] |
| Exon 9 | Deletion | c.1998_2001delTGAG | Leprechaunism | [6] |
| Exon 12 | Deletion | c.2480_2487del8 | Insulin resistance | [67] |
| Exon 15 | Deletion | c.2944_2945delAG | Leprechaunism | [90] |
| Exon 17 | Deletion | c.3077_3079delTTC | Insulin resistance | [91] |
| Exon 19 | Deletion | c.3408delG | Leprechaunism | [67] |
| Intron 20 | Deletion | c.3659 + 1_3659 + 3delGTG | Insulin resistance | [92] |
| Exon 3 | Insertion | c.866_867ins12 | Rabson–Mendenhall syndrome | [36] |
| Exon 10 | Insertion | c.2050_2051insG | Leprechaunism | [6] |
| Exon 10 | Insertion | c.2125_2126insA | Leprechaunism | [6] |
| Exon 2 | Large deletion | ex. 2 (c.101-652) | Insulin resistance | [70] |
| Exon 3 | Large deletion | ex. 3 (c.653-974) | Insulin resistance | [93] |
| Exons 10–13 | Large deletion | > 12 kb incl. ex. 10-13 | Leprechaunism | [49] |
| Exon 14 | Large deletion | 1.2 kb incl. ex. 14 | Acanthosis nigricans | [94] |
| Full gene | Large deletion | Entire gene | Leprechaunism | [26] |
| Exon 13 | Indel | c.2630_2642delins5 | Leprechaunism | [95] |
| Exon 14 | Indel | c.2752_2753delinsTG | Diabetes, NIDDM | [42] |
| Complex | Rec. INSR/Alu | Acanthosis nigricans, insulin related | [96] | |
| Complex | Translocation t(7;19)(p15.2;p13.2) | Insulin resistance | [97] | |
Fig. 1.
Summary of types of mutations found in the INSR gene.
3.2. Sequencing INSR
Four patients with known mutations were verified by the above sequencing protocol. The first patient, NY1, with clinically-confirmed Leprechaunism [6], [22], had a homozygous G to T variation at nucleotide 451 converting Glu 151 to a premature stop codon (c.451G > T,p.Glu151term). The second patient, 452, was a female infant with symptoms including repeated transient hypoglycemic episodes, prominent female genitalia, marked hirsutism, breast hyperplasia, loose and pachydermatous skin, decreased adipose tissue, acanthosis nigricans, and abdominal distention [7]. Sequencing results showed a heterozygous C to T nucleotide change at position 1195 coding for a premature stop codon at amino acid position 399 (c.1195C > T, p.Arg399term). A second mutation could not be detected by the assay as in the initial publication [7]. The third patient, 5880, had physical features of Leprechaunism and his lymphoblasts had a 90% decrease in the number of insulin receptors. This patient had a heterozygous C to T nucleotide change at position 2734 resulting in a change of arginine 924 to a premature stop codon (c.2770C > T, p.Arg924term) [23]. A second mutation could not be found even in this patient as in the original manuscript [24].
The forth patient, SLC, died before one year of age and had physical features of Leprechaunism. Insulin binding was reduced to about 4% of normal in fibroblasts from this patient. A novel G to T missense mutation was identified at nucleotide position 425 resulting in a change of glycine 142 to a valine (c.425G > T, p.Gly142Val). Computational prediction with the program Polyphen 2 (Harvard) predicts that a glycine to valine amino acid change at this position is “possibly damaging” with a score of 0.814 while SIFT (J Craig Venter Institute) predicts that the substitution is “damaging”. A second mutation could not be identified in this patient either.
Seven additional samples from normal, healthy individuals displayed no INSR variants. However all samples (as well as the clinical samples above) were found to have a benign polymorphism at nucleotide position 5 changing alanine 2 to glycine (c.5C > G, p.Ala2Gly).
An additional fourteen clinical samples (one sample from cultured amniocytes, four samples from pediatric patients and nine from adult patients) were referred to our lab for INSR sequencing. The clinical phenotype and laboratory results are summarized in Table 3. According to patient history received by ARUP with the amniotic sample, it was previously tested for deletions and duplications using a SNP array at another laboratory and was found to have a 63 kb deletion at 19p13.2 (7,143,507-7,206,857), including deletion of several exons of the INSR gene. Sequencing analysis detected no additional mutations. The 13 pediatric and adult patients presented with anomalies including, intra-uterine growth restriction (IUGR), failure to thrive (FTT), dysmorphic features, distended abdomen, and acanthosis nigricans. Although the major symptom was insulin resistance, the nine oldest patients tested were disproportionately female (7:2) with gynecological symptoms including menstrual irregularities and cystic ovaries. One 16 year old male patient had a history of IUGR, FTT, dysmorphic features, and poor response to exogenous insulin. Thirteen samples had no mutation detected by Sanger sequencing in the coding regions and exon/intron boundaries. An eleven week old boy with suspected Leprechaunism was homozygous for a variant of unknown clinical significance, c.2971C > A, p. Leu991Ile. For this patient, no positions of heterozygosity were observed in INSR, therefore we cannot rule out a partial or complete gene deletion. The infant was in intensive care and presented with IUGR, bilateral club feet, congenital hydrocephalus and dysmorphic features. Patient had only the right kidney and renal tubular acidosis. The patient had sporadic hypoglycemia and was noted to have glucose levels decreasing to 40 mg/dL range after 4–5 h of fasting, but given the age and size of the patient, this is of uncertain clinical significance. This patient also had elevated beta-hydroxybutyric acid of 14.1 mg/dL (reference range: 0.0–3.0) and a random insulin level of 1 μU/mL (reference range: 3–19 μU/mL). This variant (rs150114699) has been seen in the general population with a frequency of 0.4% in 1000 genomes and 0.6% in 6500 exomes in African Americans. The homozygous variant, c.2971C > A, p. Leu991Ile, has never been reported in the literature; sequence prediction programs give conflicting results about whether this substitution is likely to be deleterious (SIFT: deleterious; PolyPhen2: benign at score: 0.442). The next residue, Y992 is a conserved phosphorylation site in a highly conserved region (DGPLGPLyASSNPEY, http://www.phosphosite.org/siteAction.do?id=13426). The putative amino acid change at position L991 to the branched amino acid isoleucine may result in steric hindrance and decreased transporter activity. In light of the fact that the patient has only one kidney and is hypoglycemic, sequencing of HNF1B in this patient may be appropriate.
Table 3.
Clinical information and laboratory results for patient samples sequenced in this study.
| Patient | Age | Ethnicity | Gender | Clinical and other findings |
|---|---|---|---|---|
| 1 | Fetus | Asian | NA | Reported advanced maternal age. A SNP array detected a 63 kb deletion involving deletion of exons 3–11 of the INSR gene: 19p13.2(7,143,507-7,206,857)x1; GRCh37/hg 19 sequencing of the coding exons ruled out second mutation |
| 2 | 11 weeks | African-American | M | Possible IUGR, dysmorphic features, distended abdomen, can fast for only 4–5 h after which the glucose levels drop to 40 s, insulin 1 μU/mL (refa 3–19), renal tubular acidosis type 4, only one kidney, bilateral club feet, congenital hydrocephalus not requiring shunt; c.2971C > A, p.Leu991Ile |
| 3 | 1 yr | b | F | IUGR, low glucose fasting (35–67 mg/dL), seizures; previous testing found “regions of homozygosity” in INSR region by SNP array |
| 4 | 5 yr | Multi-ethnicity | M | Delivered at 27 weeks and has complications of prematurity, holoprosencephaly, absence of corpus; loss of white matter on both occipital lobes, FTT, insulin 379.6 μU/mL (ref 3–17) |
| 5 | 11 yr | c | F | Hypertriglyceridemia, low HDL cholesterol, high LDL cholesterol, nonalcoholic steatohepatitis, acanthosis nigricans, glucose fasting normal, insulin 70.5 μU/mL (ref 3–12) |
| 6 | 15 yr | NA | M | Extreme insulin resistance type A |
| 7 | 15 yr | African-American | F | Acanthosis nigricans, amenorrhea, insulin 21 μU/mL (ref 3–19) |
| 8 | 16 yr | African-American | M | IUGR, FTT, dysmorphic features, lack of subcutaneous fat, poor response to exogenous insulin or hyperforin |
| 9 | 21 yr | African-American | F | Acanthosis nigricans, cystic ovaries, insulin 65.8 μU/mL (ref 2.6–24.9), severe insulin resistance, cystic ovaries. Medications: Trajenta, metformin, Depo–Provera therapy |
| 10 | 28 yr | Caucasian | F | Cystic ovaries, glucose fasting 89 mg/dL, hx of heavy irregular periods, mental health symptoms, cystic ovaries |
| 11 | 29 yr | Asian Indian | F | Amenorrhea, cystic ovaries |
| 12 | 30 yr | Caucasian | F | Cystic ovaries |
| 13 | 50 yr | NA | F | Glucose fasting, 277 mg/dL (ref 70–99) unknown if fasting, insulin antibody 1.9 U/mL(< 0.4), triglyceride 1302 mg/dL (ref 40–149); cholesterol 231 mg/dL (ref 120–199) |
| 14 | 66 yr | Caucasian | F | Aggression, hyper-androgenism, gingival hyperplasia, thick skin, amenorrhea, distended abdomen, reported high fasting glucose fasting, high postprandial glucose 1501 mg/dL (ref < 180 mg/dL) |
Within normal reference range.
Patient from Haiti, ethnicity unknown.
Patient from Puerto Rico.
4. Discussion
Mutations in INSR can cause the insulin-resistant syndromes Leprechaunism, Rabson–Mendenhall syndrome, and type A insulin resistance [5], [6]. Diagnosis is established on clinical examination as well as laboratory diagnostic tests with markedly elevated insulin levels being a constant feature. Functional studies (insulin binding to cultured fibroblasts) and DNA analysis can be used for definitive confirmation, keeping in mind that certain mutations do not decrease insulin binding and that DNA analysis is still not identifying all putative mutations. Although there is no straightforward genotype–phenotype correlation, mutations affecting the alpha subunit of the receptor are associated with a more severe phenotype than the mutations affecting the beta subunit [25].
Due to the lack of a central repository of INSR mutations, we compiled a list of the published mutations, using currently accepted standards (Table 4). Our literature search of INSR mutations identified 132 causative variations. The vast majority of these variations are missense and nonsense mutations (78%) (Fig. 1). Interestingly, different missense mutations in the same codon have been reported to produce different phenotypes (Table 4). This highlights the need to expand the currently available databases to allow better understanding of the genotype–phenotype correlation.
There are five reports of large deletions within the INSR gene including an entire gene deletion [26]. Gross deletions and gene rearrangements account for about 5% of the mutations [26]. Large deletions, as in one of our patients, can be detected by CGH/SNP arrays. For this reason, development of a commercial test to detect single exon and whole gene deletions may be attractive. No commercial deletion/duplication testing is currently available in the US; however, deletion and duplication testing is offered at laboratories in the United Kingdom and Germany. A multiplex ligation dependent probe amplification (MLPA) assay could be used to detect single exon deletions in the INSR gene.
DNA sequencing can identify novel sequence variants of unknown clinical significance. In our study, we detected a novel c.425G > T, p. Gly142Val affecting the insulin binding alpha subunit of the insulin receptor. The evolutionary conservation analysis by Polyphen and SIFT predicts that a glycine to valine amino acid change at this position is “possibly damaging” or “damaging” to the function of the protein. Cells from this patient (TGB) failed to bind insulin, supporting a damaging role of the identified mutation. A second mutation in this patient could not be detected indicating the limitations of the current test in detecting mutations in the deep intronic or promoter regions or deletions, duplications, and rearrangements of the gene. In fact, sequencing failed to identify the second mutation in three patients with markedly reduced insulin binding in which previous studies also failed to detect the second pathogenic change [6], [13].
An additional sample of a pediatric patient referred for possible Leprechaunism was an apparent homozygous for c.2971C > A, p. Leu991Ile. A review of clinical data indicated normal to low insulin levels, a finding inconsistent with severe insulin resistance and indicating that the amino acid change is of unknown significance. As no positions of heterozygosity were observed in INSR, we cannot rule out a partial or complete gene deletion. The effect of a deletion of one copy of INSR in conjunction with this variant is yet to be studied.
It should be noted that only one mutation was detected in the 14 samples sent for clinical testing. This may be related to the poor clinical selection of patients whose phenotypes were inconsistent with insulin resistance but were nevertheless referred for this INSR mutation detection assay. More specific selection of candidate patients may enhance the utility of the assay.
Association studies show a strong correlation between single nucleotide polymorphism (SNP) in the INSR gene and a predisposition to type 2 diabetes [27]. An alternative isoform of exon 8 in the INSR gene in the Han population confers increased risk for central obesity, hypertension, glucose intolerance, hyperinsulinemia and type 2 diabetes [28], whereas variation in exon 17 is associated with insulin resistance, hyperandrogenemism and polycystic ovarian syndrome (PCOS) [29].
In conclusion, we report the development of a sequencing assay to detect mutations within the coding region and intron/exon boundaries of the INSR gene. Further development of deletion/duplication analysis is needed to detect deletions, duplications and large gene rearrangement of the INSR gene. A compilation of all the mutations reported to date using current terminology (Table 4) is the first step toward development of a publicly available online mutation database for the INSR gene.
Acknowledgments
Funding for this study was provided by the ARUP Institute for Clinical and Experimental Pathology.
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
References
- 1.Ebina Y., Ellis L., Jarnagin K., Edery M., Graf L., Clauser E., Ou J.H., Masiarz F., Kan Y.W., Goldfine I.D. The human insulin receptor cDNA: the structural basis for hormone-activated transmembrane signalling. Cell. 1985;40:747–758. doi: 10.1016/0092-8674(85)90334-4. [DOI] [PubMed] [Google Scholar]
- 2.Ullrich A., Bell J.R., Chen E.Y., Herrera R., Petruzzelli L.M., Dull T.J., Gray A., Coussens L., Liao Y.C., Tsubokawa M. Human insulin receptor and its relationship to the tyrosine kinase family of oncogenes. Nature. 1985;313:756–761. doi: 10.1038/313756a0. [DOI] [PubMed] [Google Scholar]
- 3.Seino S., Bell G.I. Alternative splicing of human insulin receptor messenger RNA. Biochem. Biophys. Res. Commun. 1989;159:312–316. doi: 10.1016/0006-291x(89)92439-x. [DOI] [PubMed] [Google Scholar]
- 4.Donohue W.L., Uchida I. Leprechaunism: a euphemism for a rare familial disorder. J. Pediatr. 1954;45:505–519. doi: 10.1016/s0022-3476(54)80113-2. [DOI] [PubMed] [Google Scholar]
- 5.Longo N., Singh R., Griffin L.D., Langley S.D., Parks J.S., Elsas L.J. Impaired growth in Rabson–Mendenhall syndrome: lack of effect of growth hormone and insulin-like growth factor-I. J. Clin. Endocrinol. Metab. 1994;79:799–805. doi: 10.1210/jcem.79.3.8077364. [DOI] [PubMed] [Google Scholar]
- 6.Longo N., Wang Y., Smith S.A., Langley S.D., DiMeglio L.A., Giannella-Neto D. Genotype–phenotype correlation in inherited severe insulin resistance. Hum. Mol. Genet. 2002;11:1465–1475. doi: 10.1093/hmg/11.12.1465. [DOI] [PubMed] [Google Scholar]
- 7.Longo N., Langley S.D., Griffin L.D., Elsas L.J. Mutations in the insulin receptor and their effect on glucose transport. Trans. Assoc. Am. Phys. 1992;105:204–213. [PubMed] [Google Scholar]
- 8.Longo N., Langley S.D., Griffin L.D., Elsas L.J. Activation of glucose transport by a natural mutation in the human insulin receptor. Proc. Natl. Acad. Sci. U. S. A. 1993;90:60–64. doi: 10.1073/pnas.90.1.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Longo N., Langley S.D., Griffin L.D., Elsas L.J. Two mutations in the insulin receptor gene of a patient with leprechaunism: application to prenatal diagnosis. J. Clin. Endocrinol. Metab. 1995;80:1496–1501. doi: 10.1210/jcem.80.5.7538143. [DOI] [PubMed] [Google Scholar]
- 10.Desbois-Mouthon C., Danan C., Amselem S., Blivet-Van Eggelpoel M.J., Sert-Langeron C., Goossens M., Besmond C., Capeau J., Caron M. Severe resistance to insulin and insulin-like growth factor-I in cells from a patient with leprechaunism as a result of two mutations in the tyrosine kinase domain of the insulin receptor. Metabolism. 1996;45:1493–1500. doi: 10.1016/s0026-0495(96)90178-x. [DOI] [PubMed] [Google Scholar]
- 11.Takahashi Y., Kadowaki H., Momomura K., Fukushima Y., Orban T., Okai T., Taketani Y., Akanuma Y., Yazaki Y., Kadowaki T. A homozygous kinase-defective mutation in the insulin receptor gene in a patient with leprechaunism. Diabetologia. 1997;40:412–420. doi: 10.1007/s001250050695. [DOI] [PubMed] [Google Scholar]
- 12.Rabson S.M., Mendenhall E.N. Familial hypertrophy of pineal body, hyperplasia of adrenal cortex and diabetes mellitus; report of 3 cases. Am. J. Clin. Pathol. 1956;26:283–290. doi: 10.1093/ajcp/26.3.283. [DOI] [PubMed] [Google Scholar]
- 13.Longo N., Wang Y., Pasquali M. Progressive decline in insulin levels in Rabson–Mendenhall syndrome. J. Clin. Endocrinol. Metab. 1999;84:2623–2629. doi: 10.1210/jcem.84.8.5902. [DOI] [PubMed] [Google Scholar]
- 14.Odawara M., Kadowaki T., Yamamoto R., Shibasaki Y., Tobe K., Accili D., Bevins C., Mikami Y., Matsuura N., Akanuma Y. Human diabetes associated with a mutation in the tyrosine kinase domain of the insulin receptor. Science. 1989;245:66–68. doi: 10.1126/science.2544998. [DOI] [PubMed] [Google Scholar]
- 15.Kahn C.R., Goldstein B.J. Molecular defects in insulin action. Science. 1989;245:13. doi: 10.1126/science.2662406. [DOI] [PubMed] [Google Scholar]
- 16.Yamamoto R., Shiba T., Tobe K., Shibasaki Y., Koshio O., Izumi T., Odawara M., Mikami Y., Matsuura N., Akanuma Y. Defect in tyrosine kinase activity of the insulin receptor from a patient with insulin resistance and acanthosis nigricans. J. Clin. Endocrinol. Metab. 1990;70:869–878. doi: 10.1210/jcem-70-4-869. [DOI] [PubMed] [Google Scholar]
- 17.Hojlund K., Hansen T., Lajer M., Henriksen J.E., Levin K., Lindholm J., Pedersen O., Beck-Nielsen H. A novel syndrome of autosomal-dominant hyperinsulinemic hypoglycemia linked to a mutation in the human insulin receptor gene. Diabetes. 2004;53:1592–1598. doi: 10.2337/diabetes.53.6.1592. [DOI] [PubMed] [Google Scholar]
- 18.Taylor S.I. Insulin action, insulin resistance, and type 2 diabetes mellitus. In: Scriver C.R., Beaudet A.L., Sly W.S., Valle D., editors. The Metabolic and Molecular Bases of Inherited Disease. McGraw-Hill; New York: 2000. pp. 1433–1470. [Google Scholar]
- 19.Thiel C.T., Knebel B., Knerr I., Sticht H., Müller-Wieland D., Zenker M., Reis A., Dörr H.-G., Rauch A. Two novel mutations in the insulin binding subunit of the insulin receptor gene without insulin binding impairment in a patient with Rabson–Mendenhall syndrome. Mol. Genet. Metab. 2008;94:356–362. doi: 10.1016/j.ymgme.2008.02.013. [DOI] [PubMed] [Google Scholar]
- 20.Seino S., Seino M., Bell G.I. Human insulin-receptor gene. Partial sequence and amplification of exons by polymerase chain reaction. Diabetes. 1990;39:123–128. doi: 10.2337/diacare.39.1.123. [DOI] [PubMed] [Google Scholar]
- 21.Benecke H., Flier J.S., Moller D.E. Alternatively spliced variants of the insulin receptor protein. Expression in normal and diabetic human tissues. J. Clin. Invest. 1992;89:2066–2070. doi: 10.1172/JCI115819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Melis R., Pruett P.B., Wang Y., Longo N. Gene expression in human cells with mutant insulin receptors. Biochem. Biophys. Res. Commun. 2003;307:1013–1020. doi: 10.1016/s0006-291x(03)01293-2. [DOI] [PubMed] [Google Scholar]
- 23.Kadowaki T., Kadowaki H., Taylor S.I. A nonsense mutation causing decreased levels of insulin receptor mRNA: detection by a simplified technique for direct sequencing of genomic DNA amplified by the polymerase chain reaction. Proc. Natl. Acad. Sci. U. S. A. 1990;87:658–662. doi: 10.1073/pnas.87.2.658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taylor S.I., Samuels B., Roth J., Kasuga M., Hedo J.A., Gorden P., Brasel D.E., Pokora T., Engel R.R. Decreased insulin binding in cultured lymphocytes from two patients with extreme insulin resistance. J. Clin. Endocrinol. Metab. 1982;54:919–930. doi: 10.1210/jcem-54-5-919. [DOI] [PubMed] [Google Scholar]
- 25.Longo N., Singh R., Elsas L.J. Decreased half-life of insulin-like growth factor I in Rabson–Mendenhall syndrome. J. Inherit. Metab. Dis. 2001;24:546–550. doi: 10.1023/a:1012411709972. [DOI] [PubMed] [Google Scholar]
- 26.Wertheimer E., Lu S.P., Backeljauw P.F., Davenport M.L., Taylor S.I. Homozygous deletion of the human insulin receptor gene results in leprechaunism. Nat. Genet. 1993;5:71–73. doi: 10.1038/ng0993-71. [DOI] [PubMed] [Google Scholar]
- 27.Barroso I., Luan J., Middelberg R.P., Harding A.H., Franks P.W., Jakes R.W., Clayton D., Schafer A.J., O'Rahilly S., Wareham N.J. Candidate gene association study in type 2 diabetes indicates a role for genes involved in beta-cell function as well as insulin action. PLoS Biol. 2003;1:E20. doi: 10.1371/journal.pbio.0000020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang C., Wang B., He H., Li X., Wei D., Zhang J., Ma M., Pan L., Yu T., Xue F., Li L., Shan G. Association between insulin receptor gene polymorphism and the metabolic syndrome in Han and Yi Chinese. Asia Pac. J. Clin. Nutr. 2012;21:457–463. [PubMed] [Google Scholar]
- 29.Mukherjee S., Shaikh N., Khavale S., Shinde G., Meherji P., Shah N., Maitra A. Genetic variation in exon 17 of INSR is associated with insulin resistance and hyperandrogenemia among lean Indian women with polycystic ovary syndrome. Eur. J. Endocrinol. 2009;160:855–862. doi: 10.1530/EJE-08-0932. [DOI] [PubMed] [Google Scholar]
- 30.Kim D., Cho S.Y., Yeau S.H., Park S.W., Sohn Y.B., Kwon M.J., Kim J.Y., Ki C.S., Jin D.K. Two novel insulin receptor gene mutations in a patient with Rabson–Mendenhall syndrome: the first Korean case confirmed by biochemical, and molecular evidence. J. Korean Med. Sci. 2012;27:565–568. doi: 10.3346/jkms.2012.27.5.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grasso V., Colombo C., Favalli V., Galderisi A., Rabbone I., Gombos S., Bonora E., Massa O., Meschi F., Cerutti F., Iafusco D., Bonfanti R., Monciotti C., Barbetti F. Six cases with severe insulin resistance (SIR) associated with mutations of insulin receptor: is a Bartter-like syndrome a feature of congenital SIR? Acta Diabetol. 2013;50:951–957. doi: 10.1007/s00592-013-0490-x. [DOI] [PubMed] [Google Scholar]
- 32.Barbetti F., Raben N., Kadowaki T., Cama A., Accili D., Gabbay K.H., Merenich J.A., Taylor S.I., Roth J. Two unrelated patients with familial hyperproinsulinemia due to a mutation substituting histidine for arginine at position 65 in the proinsulin molecule: identification of the mutation by direct sequencing of genomic deoxyribonucleic acid amplified by polymerase chain reaction. J. Clin. Endocrinol. Metab. 1990;71:164–169. doi: 10.1210/jcem-71-1-164. [DOI] [PubMed] [Google Scholar]
- 33.Accili D., Barbetti F., Cama A., Kadowaki H., Kadowaki T., Imano E., Levy-Toledano R., Taylor S.I. Mutations in the insulin receptor gene in patients with genetic syndromes of insulin resistance and acanthosis nigricans. J. Invest. Dermatol. 1992;98:77S–81S. doi: 10.1111/1523-1747.ep12462281. [DOI] [PubMed] [Google Scholar]
- 34.van der Vorm E.R., van der Zon G.C., Moller W., Krans H.M., Lindhout D., Maassen J.A. An Arg for Gly substitution at position 31 in the insulin receptor, linked to insulin resistance, inhibits receptor processing and transport. J. Biol. Chem. 1992;267:66–71. [PubMed] [Google Scholar]
- 35.Rouard M., Macari F., Bouix O., Lautier C., Brun J.F., Lefebvre P., Renard E., Bringer J., Jaffiol C., Grigorescu F. Identification of two novel insulin receptor mutations, Asp59Gly and Leu62Pro, in type A syndrome of extreme insulin resistance. Biochem. Biophys. Res. Commun. 1997;234:764–768. doi: 10.1006/bbrc.1997.6695. [DOI] [PubMed] [Google Scholar]
- 36.Muller-Wieland D., van der Vorm E.R., Streicher R., Krone W., Seemanova E., Dreyer M., Rudiger H.W., Rosipal S.R., Maassen J.A. An in-frame insertion in exon 3 and a nonsense mutation in exon 2 of the insulin receptor gene associated with severe insulin resistance in a patient with Rabson–Mendenhall syndrome. Diabetologia. 1993;36:1168–1174. doi: 10.1007/BF00401062. [DOI] [PubMed] [Google Scholar]
- 37.Maassen J.A., Tobias E.S., Kayserilli H., Tukel T., Yuksel-Apak M., D'Haens E., Kleijer W.J., Fery F., van der Zon G.C. Identification and functional assessment of novel and known insulin receptor mutations in five patients with syndromes of severe insulin resistance. J. Clin. Endocrinol. Metab. 2003;88:4251–4257. doi: 10.1210/jc.2003-030034. [DOI] [PubMed] [Google Scholar]
- 38.Alzahrani A.S., Zou M., Baitei E.Y., Parhar R.S., Al-Kahtani N., Raef H., Almahfouz A., Amartey J.K., Al-Rijjal R., Hammami R., Meyer B.F., Al-Mohanna F.A., Shi Y. Molecular characterization of a novel p.R118C mutation in the insulin receptor gene from patients with severe insulin resistance. Clin. Endocrinol. (Oxf.) 2012;76:540–547. doi: 10.1111/j.1365-2265.2011.04258.x. [DOI] [PubMed] [Google Scholar]
- 39.Hone J., Accili D., al-Gazali L.I., Lestringant G., Orban T., Taylor S.I. Homozygosity for a new mutation (Ile119→Met) in the insulin receptor gene in five sibs with familial insulin resistance. J. Med. Genet. 1994;31:715–716. doi: 10.1136/jmg.31.9.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krook A., Brueton L., O'Rahilly S. Homozygous nonsense mutation in the insulin receptor gene in infant with leprechaunism. Lancet. 1993;342:277–278. doi: 10.1016/0140-6736(93)91820-c. [DOI] [PubMed] [Google Scholar]
- 41.Rique S., Nogues C., Ibanez L., Marcos M.V., Ferragut J., Carrascosa A., Potau N. Identification of three novel mutations in the insulin receptor gene in type A insulin resistant patients. Clin. Genet. 2000;57:67–69. doi: 10.1034/j.1399-0004.2000.570110.x. [DOI] [PubMed] [Google Scholar]
- 42.Kazemi B., Seyed N., Moslemi E., Bandehpour M., Bikhof Torbati M., Saadat N., Eidi A., Ghayoor E., Azizi F. Insulin receptor gene mutations in Iranian patients with type II diabetes mellitus. Iran. Biomed. J. 2009;13:161–168. [PubMed] [Google Scholar]
- 43.Carrera P., Cordera R., Ferrari M., Cremonesi L., Taramelli R., Andraghetti G., Carducci C., Dozio N., Pozza G., Taylor S.I. Substitution of Leu for Pro-193 in the insulin receptor in a patient with a genetic form of severe insulin resistance. Hum. Mol. Genet. 1993;2:1437–1441. doi: 10.1093/hmg/2.9.1437. [DOI] [PubMed] [Google Scholar]
- 44.Klinkhamer M.P., Groen N.A., van der Zon G.C., Lindhout D., Sandkuyl L.A., Krans H.M., Moller W., Maassen J.A. A leucine-to-proline mutation in the insulin receptor in a family with insulin resistance. EMBO J. 1989;8:2503–2507. doi: 10.1002/j.1460-2075.1989.tb08387.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hamer I., Foti M., Emkey R., Cordier-Bussat M., Philippe J., De Meyts P., Maeder C., Kahn C.R., Carpentier J.L. An arginine to cysteine(252) mutation in insulin receptors from a patient with severe insulin resistance inhibits receptor internalisation but preserves signalling events. Diabetologia. 2002;45:657–667. doi: 10.1007/s00125-002-0798-5. [DOI] [PubMed] [Google Scholar]
- 46.Osawa H., Nishimiya T., Ochi M., Niiya T., Onuma H., Kitamuro F., Kaino Y., Kida K., Makino H. Identification of novel C253Y missense and Y864X nonsense mutations in the insulin receptor gene in type A insulin-resistant patients. Clin. Genet. 2001;59:194–197. doi: 10.1034/j.1399-0004.2001.590309.x. [DOI] [PubMed] [Google Scholar]
- 47.Ogilvy-Stuart A.L., Soos M.A., Hands S.J., Anthony M.Y., Dunger D.B., O'Rahilly S. Hypoglycemia and resistance to ketoacidosis in a subject without functional insulin receptors. J. Clin. Endocrinol. Metab. 2001;86:3319–3326. doi: 10.1210/jcem.86.7.7631. [DOI] [PubMed] [Google Scholar]
- 48.Dib K., Whitehead J.P., Humphreys P.J., Soos M.A., Baynes K.C., Kumar S., Harvey T., O'Rahilly S. Impaired activation of phosphoinositide 3-kinase by insulin in fibroblasts from patients with severe insulin resistance and pseudoacromegaly. A disorder characterized by selective postreceptor insulin resistance. J. Clin. Invest. 1998;101:1111–1120. doi: 10.1172/JCI119884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Desbois-Mouthon C., Magre J., Duprey J., Caron M., Blivet-Van Eggelpoel M.J., Daubas C., Gourmelen M., Chevallier B., Rizkalla S., Robert J.J., Capeau J. Major circadian variations of glucose homeostasis in a patient with Rabson-Mendenhall syndrome and primary insulin resistance due to a mutation (Cys284→Tyr) in the insulin receptor alpha-subunit. Pediatr. Res. 1997;42:72–77. doi: 10.1203/00006450-199707000-00012. [DOI] [PubMed] [Google Scholar]
- 50.Krook A., Kumar S., Laing I., Boulton A.J., Wass J.A., O'Rahilly S. Molecular scanning of the insulin receptor gene in syndromes of insulin resistance. Diabetes. 1994;43:357–368. doi: 10.2337/diab.43.3.357. [DOI] [PubMed] [Google Scholar]
- 51.Takahashi I., Yamada Y., Kadowaki H., Horikoshi M., Kadowaki T., Narita T., Tsuchida S., Noguchi A., Koizumi A., Takahashi T. Phenotypical variety of insulin resistance in a family with a novel mutation of the insulin receptor gene. Endocr. J. 2010;57:509–516. doi: 10.1507/endocrj.k09e-339. [DOI] [PubMed] [Google Scholar]
- 52.Hashiramoto M., Osawa H., Ando M., Murakami A., Nishimiya T., Nakano M., Nishida W., Onuma H., Makino H. A nonsense mutation in the Arg345 of the insulin receptor gene in a Japanese type A insulin-resistant patient. Endocr. J. 2005;52:499–504. doi: 10.1507/endocrj.52.499. [DOI] [PubMed] [Google Scholar]
- 53.Tuthill A., Semple R.K., Day R., Soos M.A., Sweeney E., Seymour P.J., Didi M., O'Rahilly S. Functional characterization of a novel insulin receptor mutation contributing to Rabson–Mendenhall syndrome. Clin. Endocrinol. (Oxf.) 2007;66:21–26. doi: 10.1111/j.1365-2265.2006.02678.x. [DOI] [PubMed] [Google Scholar]
- 54.Barbetti F., Gejman P.V., Taylor S.I., Raben N., Cama A., Bonora E., Pizzo P., Moghetti P., Muggeo M., Roth J. Detection of mutations in insulin receptor gene by denaturing gradient gel electrophoresis. Diabetes. 1992;41:408–415. doi: 10.2337/diab.41.4.408. [DOI] [PubMed] [Google Scholar]
- 55.Longo N., Langley S.D., Griffin L.D., Elsas L.J., II Reduced mRNA and a nonsense mutation in the insulin-receptor gene produce heritable severe insulin resistance. Am. J. Hum. Genet. 1992;50:998–1007. [PMC free article] [PubMed] [Google Scholar]
- 56.Accili D., Frapier C., Mosthaf L., McKeon C., Elbein S.C., Permutt M.A., Ramos E., Lander E., Ullrich A., Taylor S.I. A mutation in the insulin receptor gene that impairs transport of the receptor to the plasma membrane and causes insulin-resistant diabetes. EMBO J. 1989;8:2509–2517. doi: 10.1002/j.1460-2075.1989.tb08388.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Semple R.K., Soos M.A., Luan J., Mitchell C.S., Wilson J.C., Gurnell M., Cochran E.K., Gorden P., Chatterjee V.K., Wareham N.J., O'Rahilly S. Elevated plasma adiponectin in humans with genetically defective insulin receptors. J. Clin. Endocrinol. Metab. 2006;91:3219–3223. doi: 10.1210/jc.2006-0166. [DOI] [PubMed] [Google Scholar]
- 58.van der Vorm E.R., Kuipers A., Kielkopf-Renner S., Krans H.M., Moller W., Maassen J.A. A mutation in the insulin receptor that impairs proreceptor processing but not insulin binding. J. Biol. Chem. 1994;269:14297–14302. [PubMed] [Google Scholar]
- 59.Kadowaki T., Bevins C.L., Cama A., Ojamaa K., Marcus-Samuels B., Kadowaki H., Beitz L., McKeon C., Taylor S.I. Two mutant alleles of the insulin receptor gene in a patient with extreme insulin resistance. Science. 1988;240:787–790. doi: 10.1126/science.2834824. [DOI] [PubMed] [Google Scholar]
- 60.Thomas G.N., Tomlinson B., Chan J.C., Lee Z.S., Cockran C.S., Critchley J.A. An insulin receptor gene polymorphism is associated with diastolic blood pressure in Chinese subjects with components of the metabolic syndrome. Am. J. Hypertens. 2000;13:745–752. doi: 10.1016/s0895-7061(00)00265-x. [DOI] [PubMed] [Google Scholar]
- 61.Jiang L., Liu C., Wang W.Q., Ye L., Zhu N., Zhou W.W., Su T.W., Li X.Y., Ning G. Leprechaunism: an inherited insulin resistance syndrome caused by the defect of insulin receptor. Zhonghua Nei Ke Za Zhi. 2006;45:730–733. [PubMed] [Google Scholar]
- 62.Hart L.M., Stolk R.P., Heine R.J., Grobbee D.E., van der Does F.E., Maassen J.A. Association of the insulin-receptor variant Met-985 with hyperglycemia and non-insulin-dependent diabetes mellitus in the Netherlands: a population-based study. Am. J. Hum. Genet. 1996;59:1119–1125. [PMC free article] [PubMed] [Google Scholar]
- 63.Yoshimasa Y., Seino S., Whittaker J., Kakehi T., Kosaki A., Kuzuya H., Imura H., Bell G.I., Steiner D.F. Insulin-resistant diabetes due to a point mutation that prevents insulin proreceptor processing. Science. 1988;240:784–787. doi: 10.1126/science.3283938. [DOI] [PubMed] [Google Scholar]
- 64.Jospe N., Kaplowitz P.B., Furlanetto R.W. Homozygous nonsense mutation in the insulin receptor gene of a patient with severe congenital insulin resistance: leprechaunism and the role of the insulin-like growth factor receptor. Clin. Endocrinol. (Oxf.) 1996;45:229–235. doi: 10.1046/j.1365-2265.1996.d01-1548.x. [DOI] [PubMed] [Google Scholar]
- 65.Nobile S., Semple R.K., Carnielli V.P. A novel mutation of the insulin receptor gene in a preterm infant with Donohue syndrome and heart failure. J. Pediatr. Endocrinol. Metab. 2012;25:363–366. doi: 10.1515/jpem-2011-0448. [DOI] [PubMed] [Google Scholar]
- 66.Kan M., Kanai F., Iida M., Jinnouchi H., Todaka M., Imanaka T., Ito K., Nishioka Y., Ohnishi T., Kamohara S. Frequency of mutations of insulin receptor gene in Japanese patients with NIDDM. Diabetes. 1995;44:1081–1086. doi: 10.2337/diab.44.9.1081. [DOI] [PubMed] [Google Scholar]
- 67.Kadowaki H., Takahashi Y., Ando A., Momomura K., Kaburagi Y., Quin J.D., MacCuish A.C., Koda N., Fukushima Y., Taylor S.I., Akanuma Y., Yazaki Y., Kadowaki T. Four mutant alleles of the insulin receptor gene associated with genetic syndromes of extreme insulin resistance. Biochem. Biophys. Res. Commun. 1997;237:516–520. doi: 10.1006/bbrc.1997.7181. [DOI] [PubMed] [Google Scholar]
- 68.O'Rahilly S., Choi W.H., Patel P., Turner R.C., Flier J.S., Moller D.E. Detection of mutations in insulin-receptor gene in NIDDM patients by analysis of single-stranded conformation polymorphisms. Diabetes. 1991;40:777–782. doi: 10.2337/diab.40.6.777. [DOI] [PubMed] [Google Scholar]
- 69.Kusari J., Takata Y., Hatada E., Freidenberg G., Kolterman O., Olefsky J.M. Insulin resistance and diabetes due to different mutations in the tyrosine kinase domain of both insulin receptor gene alleles. J. Biol. Chem. 1991;266:5260–5267. [PubMed] [Google Scholar]
- 70.Moritz W., Boni-Schnetzler M., Stevens W., Froesch E.R., Levy J.R. In-frame exon 2 deletion in insulin receptor RNA in a family with extreme insulin resistance in association with defective insulin binding: a case report. Eur. J. Endocrinol. 1996;135:357–363. doi: 10.1530/eje.0.1350357. [DOI] [PubMed] [Google Scholar]
- 71.Ogawa W., Iwamoto K., Mori H., Hashiramoto M., Miyake K., Sakaguchi K., Kasuga M. Two related cases of type A insulin resistance with compound heterozygous mutations of the insulin receptor gene. Diabetes Res. Clin. Pract. 2009;83:e75–e77. doi: 10.1016/j.diabres.2008.12.002. [DOI] [PubMed] [Google Scholar]
- 72.Haruta T., Takata Y., Iwanishi M., Maegawa H., Imamura T., Egawa K., Itazu T., Kobayashi M. Ala1048→Asp mutation in the kinase domain of insulin receptor causes defective kinase activity and insulin resistance. Diabetes. 1993;42:1837–1844. doi: 10.2337/diab.42.12.1837. [DOI] [PubMed] [Google Scholar]
- 73.Yang G.Q., Wang B.A., Zhao W.R., Gu W.J., Lui Z.H., Dou J.T., Mu Y.M., Lu J.M. Clinical and genetic analysis of the insulin receptor gene in a Chinese patient with extreme insulin resistance. Diabetes Res. Clin. Pract. 2010;89:e56–e58. doi: 10.1016/j.diabres.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 74.Vambergue A., Lautier C., Valat A.S., Cortet-Rudelli C., Grigorescu F., Dewailly D. Follow-up study of two sisters with type A syndrome of severe insulin resistance gives a new insight into PCOS pathogenesis in relation to puberty and pregnancy outcome: a case report. Hum. Reprod. 2006;21:1274–1278. doi: 10.1093/humrep/dei455. [DOI] [PubMed] [Google Scholar]
- 75.Kasuga M., Kishimoto M., Hashiramoto M., Yonezawa K., Kazumi T., Hagino H., Shii K. Insulin receptor Arg1131→Gln: a novel mutation in the catalytic loop of insulin receptor observed in insulin resistant diabetes. Nihon Geka Gakkai Zasshi. 1992;93:968–971. [PubMed] [Google Scholar]
- 76.Moller D.E., Yokota A., White M.F., Pazianos A.G., Flier J.S. A naturally occurring mutation of insulin receptor alanine 1134 impairs tyrosine kinase function and is associated with dominantly inherited insulin resistance. J. Biol. Chem. 1990;265:14979–14985. [PubMed] [Google Scholar]
- 77.Cama A., de la Luz Sierra M., Quon M.J., Ottini L., Gorden P., Taylor S.I. Substitution of glutamic acid for alanine 1135 in the putative “catalytic loop” of the tyrosine kinase domain of the human insulin receptor. A mutation that impairs proteolytic processing into subunits and inhibits receptor tyrosine kinase activity. J. Biol. Chem. 1993;268:8060–8069. [PubMed] [Google Scholar]
- 78.Cama A., de la Luz Sierra M., Ottini L., Kadowaki T., Gorden P., Imperato-McGinley J., Taylor S.I. A mutation in the tyrosine kinase domain of the insulin receptor associated with insulin resistance in an obese woman. J. Clin. Endocrinol. Metab. 1991;73:894–901. doi: 10.1210/jcem-73-4-894. [DOI] [PubMed] [Google Scholar]
- 79.Cocozza S., Porcellini A., Riccardi G., Monticelli A., Condorelli G., Ferrara A., Pianese L., Miele C., Capaldo B., Beguinot F. NIDDM associated with mutation in tyrosine kinase domain of insulin receptor gene. Diabetes. 1992;41:521–526. doi: 10.2337/diab.41.4.521. [DOI] [PubMed] [Google Scholar]
- 80.Moritz W., Froesch E.R., Boni-Schnetzler M. Functional properties of a heterozygous mutation (Arg1174→Gln) in the tyrosine kinase domain of the insulin receptor from a type A insulin resistant patient. FEBS Lett. 1994;351:276–280. doi: 10.1016/0014-5793(94)00876-0. [DOI] [PubMed] [Google Scholar]
- 81.Whitehead J.P., Soos M.A., Jackson R., Tasic V., Kocova M., O'Rahilly S. Multiple molecular mechanisms of insulin receptor dysfunction in a patient with Donohue syndrome. Diabetes. 1998;47:1362–1364. doi: 10.2337/diab.47.8.1362. [DOI] [PubMed] [Google Scholar]
- 82.Kim H., Kadowaki H., Sakura H., Odawara M., Momomura K., Takahashi Y., Miyazaki Y., Ohtani T., Akanuma Y., Yazaki Y. Detection of mutations in the insulin receptor gene in patients with insulin resistance by analysis of single-stranded conformational polymorphisms. Diabetologia. 1992;35:261–266. doi: 10.1007/BF00400927. [DOI] [PubMed] [Google Scholar]
- 83.Imamura T., Takata Y., Sasaoka T., Takada Y., Morioka H., Haruta T., Sawa T., Iwanishi M., Hu Y.G., Suzuki Y. Two naturally occurring mutations in the kinase domain of insulin receptor accelerate degradation of the insulin receptor and impair the kinase activity. J. Biol. Chem. 1994;269:31019–31027. [PubMed] [Google Scholar]
- 84.Barroso W.K., Jardim P.C., Jardim T.S., Souza C., Magalhes A.L., Ibrahim F.M., Couto P.V., Silveira A., Monego E.T. Hypertensive diabetic patients: guidelines for conduct and their difficulties. Arq. Bras. Cardiol. 2003;81:143–147. doi: 10.1590/s0066-782x2003001000003. [DOI] [PubMed] [Google Scholar]
- 85.Magre J., Karayanni C., Hadjiathanasiou C.G., Desbois-Mouthon C., Meier M., Vigouroux C., Stavrinadis C., Sinaniotis C., Caron M., Capeau J. Dominant transmission of insulin resistance in a type A family resulting from a heterozygous nonsense mutation in the insulin receptor gene and associated with decreased mRNA level and insulin binding sites. Diabetes. 1997;46:1901–1903. doi: 10.2337/diab.46.11.1901. [DOI] [PubMed] [Google Scholar]
- 86.Vorwerk P., Christoffersen C.T., Muller J., Vestergaard H., Pedersen O., De Meyts P. Alternative splicing of exon 17 and a missense mutation in exon 20 of the insulin receptor gene in two brothers with a novel syndrome of insulin resistance (congenital fiber-type disproportion myopathy) Horm. Res. 1999;52:211–220. doi: 10.1159/000023464. [DOI] [PubMed] [Google Scholar]
- 87.Cama A., Sierra M.L., Kadowaki T., Kadowaki H., Quon M.J., Rudiger H.W., Dreyer M., Taylor S.I. Two mutant alleles of the insulin receptor gene in a family with a genetic form of insulin resistance: a 10 base pair deletion in exon 1 and a mutation substituting serine for asparagine-462. Hum. Genet. 1995;95:174–182. doi: 10.1007/BF00209397. [DOI] [PubMed] [Google Scholar]
- 88.Jospe N., Zhu J., Liu R., Livingston J.N., Furlanetto R.W. Deletion of 3 basepairs resulting in the loss of lysine-121 in the insulin receptor alpha-subunit in a patient with leprechaunism: binding, phosphorylation, and biological activity. J. Clin. Endocrinol. Metab. 1994;79:1294–1302. doi: 10.1210/jcem.79.5.7962321. [DOI] [PubMed] [Google Scholar]
- 89.George S., Johansen A., Soos M.A., Mortensen H., Gammeltoft S., Saudek V., Siddle K., Hansen L., O'Rahilly S. Deletion of V335 from the L2 domain of the insulin receptor results in a conformationally abnormal receptor that is unable to bind insulin and causes Donohue's syndrome in a human subject. Endocrinology. 2003;144:631–637. doi: 10.1210/en.2002-220815. [DOI] [PubMed] [Google Scholar]
- 90.Najjar S.M., Philippe N., Suzuki Y., Ignacio G.A., Formisano P., Accili D., Taylor S.I. Insulin-stimulated phosphorylation of recombinant pp 120/HA4, an endogenous substrate of the insulin receptor tyrosine kinase. Biochemistry. 1995;34:9341–9349. doi: 10.1021/bi00029a009. [DOI] [PubMed] [Google Scholar]
- 91.Awata T., Matsumoto C., Momomura K., Takahashi Y., Odawara M., Kasuga M., Kadowaki T., Iwamoto Y. A 3-basepair in-frame deletion (delta Leu999) in exon 17 of the insulin receptor gene in a family with insulin resistance. J. Clin. Endocrinol. Metab. 1994;79:1840–1844. doi: 10.1210/jcem.79.6.7989492. [DOI] [PubMed] [Google Scholar]
- 92.Kirk J., Porter K.M., Parker V., Barroso I., O'Rahilly S., Hendriksz C., Semple R.K. Loss of NPC1 function in a patient with a co-inherited novel insulin receptor mutation does not grossly modify the severity of the associated insulin resistance. J. Inherit. Metab. Dis. 2010;33(Suppl. 3):227–232. doi: 10.1007/s10545-010-9107-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wertheimer E., Litvin Y., Ebstein R.P., Bennet E.R., Barbetti F., Accili D., Taylor S.I. Deletion of exon 3 of the insulin receptor gene in a kindred with a familial form of insulin resistance. J. Clin. Endocrinol. Metab. 1994;78:1153–1158. doi: 10.1210/jcem.78.5.8175972. [DOI] [PubMed] [Google Scholar]
- 94.Shimada F., Taira M., Suzuki Y., Hashimoto N., Nozaki O., Tatibana M., Ebina Y., Tawata M., Onaya T. Insulin-resistant diabetes associated with partial deletion of insulin-receptor gene. Lancet. 1990;335:1179–1181. doi: 10.1016/0140-6736(90)92695-e. [DOI] [PubMed] [Google Scholar]
- 95.Hone J., Accili D., Psiachou H., Alghband-Zadeh J., Mitton S., Wertheimer E., Sinclair L., Taylor S.I. Homozygosity for a null allele of the insulin receptor gene in a patient with leprechaunism. Hum. Mutat. 1995;6:17–22. doi: 10.1002/humu.1380060105. [DOI] [PubMed] [Google Scholar]
- 96.Taira M., Hashimoto N., Shimada F., Suzuki Y., Kanatsuka A., Nakamura F., Ebina Y., Tatibana M., Makino H. Human diabetes associated with a deletion of the tyrosine kinase domain of the insulin receptor. Science. 1989;245:63–66. doi: 10.1126/science.2544997. [DOI] [PubMed] [Google Scholar]
- 97.Suliman S.G., Stanik J., McCulloch L.J., Wilson N., Edghill E.L., Misovicova N., Gasperikova D., Sandrikova V., Elliott K.S., Barak L., Ellard S., Volpi E.V., Klimes I., Gloyn A.L. Severe insulin resistance and intrauterine growth deficiency associated with haploinsufficiency for INSR and CHN2: new insights into synergistic pathways involved in growth and metabolism. Diabetes. 2009;58:2954–2961. doi: 10.2337/db09-0787. [DOI] [PMC free article] [PubMed] [Google Scholar]