Abstract
In Manitoba, Canada, the overall incidence of Severe Combined Immunodeficiency (SCID) is three-fold higher than the national average, with SCID overrepresented in two population groups: Mennonites and First Nations of Northern Cree ancestries. T-cell receptor excision circle (TREC) assay is being used increasingly for neonatal screening for SCID in North America. However, the majority of SCID patients in Manitoba are T-cell-positive. Therefore it is likely that the TREC assay will not identify these infants. The goal of this study was to blindly and retrospectively perform TREC analysis in confirmed SCID patients using archived Guthrie cards. Thirteen SCID patients were tested: 5 T-negative SCID (3 with adenosine deaminase deficiency, 1 with CD3δ deficiency, and 1 unclassified) and 8 T-positive SCID (5 with zeta chain-associated protein kinase (ZAP70) deficiency and 3 with inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase beta (IKKβ) deficiency). As a non-SCID patient group, 5 Primary Immunodeficiency Disease (PID) patients were studied: 1 T-negative PID (cartilage-hair hypoplasia) and 4 T-positive PID (2 common immune deficiency (CID), 1 Wiskott–Aldrich syndrome, and 1 X-linked lymphoproliferative disease). Both patient groups required hematopoietic stem cell transplantation. In addition, randomly-selected de-identified controls (n = 982) were tested. Results: all T-negative SCID and PID had zero TRECs. Low-TRECs were identified in 2 ZAP70 siblings, 1 CID patient as well as 5 preterm, 1 twin, and 4 de-identified controls. Conclusions: TREC method will identify T-negative SCID and T-negative PID. To identify other SCID babies, newborn screening in Manitoba must include supplemental targeted screening for ethnic-specific mutations.
Abbreviations: ADA, adenosine deaminase deficiency; CHH, cartilage–hair hypoplasia; CPL, Cadham Provincial Laboratory; DBS, dried blood spots; CID, common immune deficiency; FNMI, First Nations, Metis, and Inuit; HSCT, hematopoietic stem cell transplant; IKKβ, inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase beta; NENSP, New England Newborn Screening Program, NICU, neonatal intensive care unit; PID, Primary Immunodeficiency Disease; SCID, Severe Combined Immunodeficiency; TREC, T-cell receptor excision circle; WAS, Wiskott–Aldrich syndrome; XLP, X-linked lymphoproliferative disease; ZAP70, zeta chain-associated protein kinase
Keywords: T-cell receptor excision circle, Severe Combined Immunodeficiency, T-cell positive primary immunodeficiency, Newborn screening, Archived Guthrie cards, Dried blood spots
1. Introduction
T-cell receptor excision circle (TREC) quantification assay is being increasingly used by newborn screening programs for detection of Severe Combined Immunodeficiency (SCID), which is the most profound form of the Primary Immunodeficiency Diseases (PID) [1], [2], [3], [4]. Babies born with SCID appear healthy at birth, making clinical diagnosis difficult, but are vulnerable to infections once maternal protection has diminished. SCID is fatal within the first year of life without treatment (usually, hematopoietic stem cell transplantation (HSCT)); early treatment is associated with better outcome [5]. Most SCID babies have very few or no functional T-cells and extremely low or absent TRECs that are normally produced as a by-product of the early stages of T-cell development in the thymus [1], [2], [3], [4]. Different versions of the assay that quantify TRECs in blood have been developed in several laboratories [6], [7]. The protocols were tested and validated to screen for TREC deficiency typically associated with SCID [1], [2], [3], [8]. The assay can be also used to detect other forms of T-cell lymphopenia regardless if there is a genetic or acquired cause [4], [6], [7]. Population-based newborn screening for SCID by quantification of TRECs on DNA isolated from dried blood spots (DBS) from standard newborn screening Guthrie cards was added to the newborn screening panel in numerous states throughout the United States [9], [10], [4] and, recently, in the province of Ontario, Canada [11].
In a Canadian SCID surveillance study, an almost three-fold higher incidence of SCID in First Nations, Metis, and Inuit children (FNMI, whose proportion in the pediatric population was estimated to be 6.3%) than in non-FNMI children was reported: 4.4 versus 1.4 per 100,000 live births, respectively [11]. However the TREC analysis was not available [11]. In the province of Manitoba, Canada, SCID is overrepresented in two population groups: First Nations of Cree ancestry and Mennonites. Overall an observed incidence of SCID in Manitoba appears to be three-fold higher than the average national rate: 4.8 per 100,000 live births (13 confirmed cases in 18 years with approximately 15,000 live births per year). The difficulty is that, in some of the conditions known to be present in Manitoba, such as inhibitor of kappa light polypeptide gene enhancer in B-cells kinase beta (IKKβ) deficiency and zeta chain-associated protein kinase (ZAP70) deficiency, the infants have T-cells; however, the T-cell function is abnormal [12], [13], [14], [15], [16]. It is therefore possible that the TREC method would not identify these babies unless their TRECs were significantly decreased as compared to normal controls. Thus, the purpose of this study was two-fold: 1) determine the normal distribution of TRECs in the Manitoba newborn population using a validated TREC assay [3], 2) retrospectively test confirmed SCID babies using archived Guthrie cards to determine the applicability of the TREC assay in the ethnically unique population with a higher proportion of infants affected by atypical forms of SCID.
2. Materials and methods
2.1. Patient selection
Eighteen Manitoba patients diagnosed with severe genetic immune deficiencies between 1992 and 2010 were included in the study. The cases were selected because they all received a stem cell transplant, knowing that both SCID infants and a number of children with other PID were included. The PID patients were included as a patient control group of non-SCID. They represent all the significant immune deficiency patients in our referral area. We are unaware of any patients being referred or transferred elsewhere for transplant although some infants may have died prior to referral.
Five categories of patients were included (Table 1): 1) typical T-cell deficient SCID (n = 5; this category included 3 patients with adenosine deaminase (ADA) deficiency, 1 with CD3δ deficiency, and 1 clinical SCID case with no molecular diagnosis (patient #17; 2) T-cell-positive SCID (n = 8; this heterogeneous group included 5 patients with ZAP70 deficiency and 3 patients with IKKβ deficiency; 3) T-cell deficient PID (n = 1, with a cartilage–hair hypoplasia (CHH) patient);T-cell positive PID (n = 4: 2 patients with CID, 1 with the Wiskott–Aldrich syndrome (WAS), and 1 with X-linked lymphoproliferative disease (XLP)), and 5) normal controls (n = 982) randomly selected from the archives of the Manitoba newborn screening facility at Cadham Provincial Laboratory (CPL) and anonymized and deidentified (meaning untraceable). The protocol was reviewed and approved by the University of Manitoba Health Research Ethics Board (REB).
Table 1.
Characteristics of SCID/PID patients.
| Clinical phenotype | Patient ID | Gender | Age at initial assessment, month | Phytohemagglutinin response | Treatment/outcome | TRECsa, per μl blood | Diagnosis |
|---|---|---|---|---|---|---|---|
| T− SCID | 2 | M | 4 | No response | BMT/Alive | 0 | ADAb |
| 11 | F | 2.5 | No response | BMT/Expired | 0 | ADA | |
| 12c | M | 0 | No response | BMT/Alive | 0 | ADA | |
| 15 | F | 6 | No response | BMT/Alive | 0 | CD3δd | |
| 17 | F | 5 | No response | Expired | 0 | Molecular test N/A | |
| T+ SCID | 1 | F | 13 | No response | BMTe/Alive | 71÷173 | ZAP70f |
| 3g | F | 0.5 | No response | BMT/Alive | 118÷216 | ZAP70 | |
| 5 | F | 4 | No response | BMT/Alive | 1393÷1763 | ZAP70 | |
| 7 | F | 19 | No response | BMT/Alive | 526÷697 | ZAP70 | |
| 8 | F | 3 | No response | BMT/Expired | 302÷706 | IKKβh | |
| 9 | F | 6 | No response | BMT/Alive | 286÷483 | ZAP70 | |
| 14 | F | 5 | Decreased response | BMT/Expired | 1043÷1792 | IKKβ | |
| 18 | F | 2 | Decreased response | BMT/Expired | 1305÷1619 | IKKβ | |
| T− PID | 4 | F | 7 | Decreased response | BMT/Alive | 0 | CHHi |
| T+ PID | 6 | M | 18 | Normal response | BMT/Alive | 920÷1136 | WASj |
| 10 | M | 14 | Decreased response | BMT/Alive | 163÷235 | CIDk | |
| 13 | F | 2 | Normal response | BMT/cured | 745÷1123 | CID | |
| 16 | M | 15 | Not tested | BMT/Alive | 780÷1485 | XLPl |
Range.
ADA, adenosine deaminase deficiency.
Younger sibling of patient #11.
CD3δ deficiency.
BMT, bone marrow transplant.
ZAP70, zeta chain-associated protein kinase deficiency.
Younger sibling of patient #1.
IKKβ, inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase beta deficiency.
CHH, cartilage–hair hypoplasia.
WAS, Wiskott–Aldrich syndrome.
CID: combined immune deficiency, not otherwise specified.
XLP, X-linked lymphoproliferative disease.
2.2. Sample coding
The names of the 18 known affected babies and their demographic identifiers were provided to CPL. Guthrie cards of the affected individuals as well as of control samples were retrieved from the archives by CPL staff. 3.2 mm samples were punched from the Guthrie cards. Each sample was de-identified, anonymized, coded and the information recorded by JRT in a secure confidential location at CPL. TREC testing was done in another laboratory by another researcher (OJ) in a “blind” fashion, i.e. the person performing the TREC assay knew only the unique identifier but did not know the name, immunologic status or any other information about the sample.
2.3. DNA extraction
A single 3.2-mm disk was punched from the DBS for DNA extraction according to the method developed by the New England Newborn Screening Program (NENSP) modified for local use [3]. Briefly, each 3.2-mm disk was placed in a 1.8 ml screw-cap tube and washed twice with 150 μl PBS-0.5% Triton X-100 buffer, once with PBS buffer, and once with Generation Solution 2 (S2, Qiagen), while being shaken in a Labnet Vortemp 56 Shaker/Incubator (Spectra Services, Inc.) at 800 rpm for 10 min at ambient temperature. Following the washes, DNA was eluted into 100 μl S2 while shaking at 800 rpm at 98.5 °C for 30 min. To account for the residual sample carry-over during the washing steps, a blank Guthrie card was punched at the end of each sample series and processed along with the DBS punches in the TREC assay; no TRECs were detected in the blank punches. Hemoglobin could not be completely removed from the archived DBS samples that were stored at ambient temperature, despite adding an extra wash. Thus typically DNA eluates contained a significant amount of hemoglobin.
2.4. TREC assay
A modified protocol developed by Comeau and colleagues [3] and described in detail elsewhere was used. Briefly, QRT-PCR multiplex reaction, multiplying two targets: TREC and RNaseP in the same reaction mixture, was used. To ensure amplification of DNA, the master mix resistant to hemoglobin contamination was used. The 20-microliter reaction mixture contained 5 μl TaqMan Fast Virus 1-Step MMix (FAM-MGB, Cat. No. 4444434, Applied Biosystems), 5 μl DNA eluate/calibrators, 1 μl 20 × TREC primer-probe mixture (prepared according to NENSP protocol), and 0.4 μl 20 × RNaseP primer/probe mixture (VIC-TAMRA, Cat. No. 4316844, Applied Biosystems) in RNAse and DNAse free water to a total of 20 μl. QRT-PCR was performed in 7500Fast Real-time PCR Thermo-cycler (Applied Biosystems).
TREC/RNaseP calibrators were used (in triplicate) to create eight standards of differing concentrations and used on each 96-well plate to generate standard curves. The standard curves were plotted in the range of 39–10,000 copies of TRECs and 625–40,000 copies of RNaseP, per reaction mixture. To ensure comparability of the data in different runs, Ct values for each copy number in the standard curves (for both targets, RNaseP and TRECs) were analyzed and compared against those obtained across runs as well as against common data points on average NENSP standard curves [3] to make sure that Ct values typically fall within ± 1 cycles of the “average” range, especially for the lower copy numbers.
For unknown samples, the copy number of TRECs and RNaseP per 5 μl DNA eluate was determined by interpolation on the graph. This number was converted into a copy number/μl of total blood by the following calculation:
A sample was found to be deficient in TRECs (i.e., designated screen-positive) if the copy number of TRECs measured in the sample was consistently lower than the lowest dilution point on a TREC standard curve (i.e., lower than 39 TREC copies/reaction mixture, corresponding to 252 TREC copies/μl of whole blood). Each DNA eluate was tested in duplicate in a 96-well plate thus providing two results. For each sample measured below the second lowest dilution point on the TREC standard curve (which was 78 TREC copies/reaction mixture or 503 TREC copies/μl of whole blood), a re-punch of the DBS and a new DNA eluate was re-tested in duplicate; thus, four TREC results were available. We used modified NENSP decision criteria [3]: a sample that had all four TREC results below 78 and at least three results below 39, copies/reaction mixture (corresponding to 501 and 252 TREC copies/μl of whole blood, respectively), was designated as screen-positive.
DNA eluted from a DBS prepared using adult whole blood was used as a SCID-like control on each plate. A total of 1000 samples were analyzed.
2.5. Statistics
Histograms were plotted using Microsoft Excel program. Data in the text are presented as mean ± standard deviation.
3. Results
3.1. Distribution of TRECs in the control archived samples
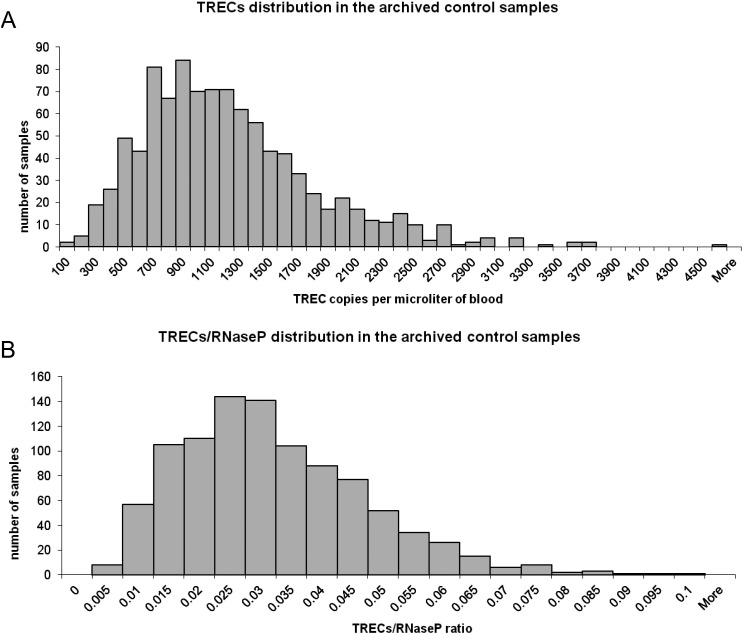
Satisfactory RNaseP amplification (above 4032 copies/μl of whole blood, according to the protocol by Gerstel-Thompson et al. [3]) was observed in all archived samples, thus no samples were excluded from the analysis. Distribution of TREC values and TREC/RNaseP ratio in the archived samples (known SCID/PID were excluded) are depicted in Fig. 1. A mean TREC value of 1162 ± 602 and mean RNaseP value of 42,439 ± 17,093 (in copies/μl blood) were determined in the archived samples. The mean ratio of TREC/RNaseP was 0.029 ± 0.015.
Fig. 1.
TREC distribution. TREC (panel A) and TREC/RNaseP ratio (panel B) distribution in the archived control samples.
3.2. Screening for TREC deficiency
Nineteen samples with TREC values below 252 copies/μl blood were identified: 9 were from known SCID and PID patients and 10 abnormal results were from newborn infants with no immunologic data available (Table 1, Table 2A). Six of the screen-positive confirmed SCID and PID patients had undetectable TRECs; they were: 3 patients with ADA deficiency, 1 patient with CD3δ deficiency, 1 patient, who was diagnosed with clinical SCID 16 years ago, but expired before HSCT and did not have a molecular diagnosis, and 1 patient with CHH (Table 1, Table 2A). Three samples with abnormally low TRECs were from patients with T-cell positive SCID and PID cases: 2 ZAP70 deficiencies and 1 CID, respectively. Notably, ZAP70 deficient samples that were identified by the TREC assay were from siblings (patients #1 and #3). The abnormal samples with no diagnosis available were: 5 preterm (average body weight of 905 g), 1 twin, and 4 unknown infants (Table 2A).
Table 2.
Results of screening 1000 patientsa for TREC deficiency.
| N (total) | TREC (copies per μl blood) | Diagnosis (n) | Mean age of sample(s) (years) | |
|---|---|---|---|---|
| A. Screen-positive samples | 6 | 0 | T− SCID: | |
| ADA (3) | 10 | |||
| CD3δ (1) | 2 | |||
| Unknown (1) | 16 | |||
| T− PID: | ||||
| CHH (1) | 14 | |||
| 3 | Low (< 252) | T+ SCID: | ||
| ZAP70 (2) | 18 | |||
| T+ PID: | ||||
| CID (1) | 11 | |||
| 10 | Low (< 252) | Abnormal, diagnosis N/A: | Unknown, up to 21 | |
| Preterm babies (5) | ||||
| Twin (1) | ||||
| Unknown (4) | ||||
| B. Screen-negative samples | 9 | Normal (> 252) | T+ SCID: | |
| ZAP70 (3) | 12 | |||
| IKKβ (3) | 7 | |||
| T+ PID: | ||||
| CID (1) | 20 | |||
| XLP (1) | 3 | |||
| WAS (1) | 19 | |||
| 972 | Normal (mean = 1162) | “True” negatives | Unknown, up to 21 |
Patient selection is described under “Materials and methods”, Section 2.1.
As expected, TREC assay did not identify most of the patients with T-cell positive SCID: 3 unrelated patients with ZAP70 deficiency and 3 unrelated patients with IKKβ deficiency, nor T-cell positive PIDs: 1 patient with CID, 1 WAS patient, and 1 patient with XLP (Table 2B).
3.3. In-vitro tests of T-cell function in SCID/PID patients
Mitogen response assays were done as part of the initial assessment to confirm the diagnosis of SCID. To determine how normal/abnormal TREC screen correlates with T-cell function assessed by mitogen stimulation, the results of T-cell stimulation tests were analyzed. In most patients, including patients with ZAP70 deficiency, upon stimulation with phytohemagglutinin, T-cell activation was severely impaired; thus demonstrating diminished immune response. Exceptions were patient #6 with WAS, patient #13 with CID, and two patients with IKKβ deficiency, who had moderately decreased response (Table 1). Patient #8 with IKKβ deficiency was critically ill at the time of testing, which may explain the “no response” result.
4. Discussion
Currently, over half of live births in the United States and all live births only in one of the 10 provinces and 2 territories in Canada, Ontario, are screened for SCID using the TREC assay [4], [11]. The novel aspect of our study was that the TREC screening was conducted, retrospectively, in a Canadian province with an overall incidence of SCID three-fold higher than the national average and with two distinct ethnic populations known to have a higher incidence of SCID: Mennonites and First Nations of Northern Cree ancestries [11], [12], [14], [16].
Several TREC protocols have been developed and validated [1], [2], [3], [8]. Depending on the assay parameters (such as DNA isolation method, plasmid, RT-PCR primers and probes, singleplex or multiplex mode, etc.), the absolute number of TRECs varies greatly, with the differences between laboratories being several-fold [4]. Therefore, positive screen results rather than actual numbers are used to compare data between laboratories [4]. From our analysis, mean TREC and RNaseP numbers were approximately 40% lower than those reported by Gerstel-Thompson et al. [3], whose protocol, with some modifications, we followed. Despite the differences in the numbers, the TREC/RNaseP ratio was similar, suggesting that the discrepancy was likely due to a lower efficiency of DNA elution from the archived samples and, possibly, hemoglobin contamination of the eluate. It is noteworthy that the assay was capable of working with DBS that were more than 20 years old.
TREC assay worked exceptionally well in correctly identifying all patients with T-cell-deficient forms of SCID (100% success rate). Importantly, not only SCID patients with confirmed molecular diagnoses, but also an archived SCID case (patient #17), who did not have a molecular diagnosis and succumbed to the disease before HSCT would have been identified soon after birth by the TREC assay. In addition, the assay established a lack of TRECs in the CHH patient (#4, Table 1) thus indicating that in this patient, CHH was associated with profound T-cell lymphopenia. The TREC assay also identified some, but not all, T-cell positive cases of SCID and PID who had TREC numbers below the assay cutoff. The assay was positive for two out of eight T-cell positive SCID (ZAP70 patients #1 and 3), but it did not identify the other T-cell positive SCID nor, as expected, the PID patients (with an exception of a borderline result for CID patient #10).
There are three prevalent SCID-causing mutations in our Mennonite population: ADA, CD3δ, and ZAP70 deficiencies [14]. It is known that patients with ADA and CD3δ deficiencies are T-cell negative [17]. Indeed, their TRECs were undetectable. However, infants with ZAP70 deficiency are T-cell-positive [12], [14]. Interestingly, the genetic background of the ZAP70 deficient patients apparently affects their TREC numbers: a variation from 71 to 1763 TREC numbers/μl blood was observed in ZAP70-deficient patients in this study, with TREC numbers below the assay cutoff detected in two siblings. This indicates that the degree of suppression of thymopoiesis in ZAP70-deficient patients may depend on unknown, possibly genetic, factors [14]. Out of these two siblings, the youngest (patient #3) came to medical attention very early because of the affected sibling (patient #1, Table 1). It has been demonstrated that earlier diagnosis is associated with better outcome [5]. Indeed, in a different pair of siblings, who had a T-cell-negative form of SCID (namely, ADA, patients #11 and 12), the younger sibling, who was diagnosed at birth because of the affected older sibling, survived; the older sibling, who was diagnosed at 2.5 months, did not survive (Table 1).
In a recent Canadian SCID surveillance study, it has been demonstrated that the incidence of SCID is almost three times higher in FNMI children; thus further necessitating screening for SCID in this particular group. Patients with IKKβ deficiency have a severe form of immunodeficiency that was recently described in Northern Cree in the Canadian provinces of Manitoba and Saskatchewan [16]. All patients carried a homozygous insertion c.1292dup (exon13) in IKBKB encoding IKKβ (IKK2), leading to a loss of IKKβ protein expression, a component of the IKK/NF-κB pathway. T-cells from these patients were almost exclusively of naïve phenotype and had differentiation and activation defects. Our findings that patients with IKKβ deficiency have normal numbers of TRECs are consistent with the data of Pannicke et al. [16] and indicate that the TREC assay will not capture these patients.
PID patients diagnosed with one of the following conditions: WAS, XLP, and one CID patient (#10) were older than 12 months when they were referred for HSCT (Table 1). They all had T-cell positive forms of PID [18], [19], [20], and were expected to have TRECs within the normal range. Indeed, the XLP and WAS patients had normal TREC values. CID is a heterogeneous group of immune deficiencies with several potentially affected genes [19]. The outcome of TREC testing was different for the two CID patients: the first CID patient (#10) had TREC values below the assay cutoff and could have been identified at birth, but the second CID patient (#13) had TRECs within the normal range.
The identity of control infants with abnormal TREC results, but no immunologic data could not be determined in this blinded retrospective study. It is not surprising that very low birth weight infants (i.e., preterm) have abnormally low TREC numbers (Table 2) as it has been reported previously that a higher proportion of infants in neonatal intensive care units (NICUs) had lower numbers of TRECs than non-NICU infants [3]. However, there were 4 “unknown abnormal” cases in this study (4/982, Table 2) who had very low TRECs. This finding is important because it suggests that we might be missing patients with a spectrum of conditions, accompanied by defects in thymopoiesis and TREC development, who would have been picked up by screening. Indeed, it has been reported recently that TREC screening done both prospectively and retrospectively identified infants, who appeared healthy at birth (i.e., “false-positive”), but whose whole genome sequencing identified deleterious mutations in the ataxia telangiectasia (ATM) gene [21]. An even more likely cause of neonatal T-cell lymphopenia could be 22q11.2 deletion (DiGeorge syndrome) [11]. Early detection of such cases would allow improvement in the early patient management (e.g., live vaccine avoidance and detection of hypocalcemia) and genetic counseling.
Overall the prevalence of SCID in Manitoba appears to be in the range of 1 in 15,000–20,000 live births. However, the incidence of the disease in Manitoba could still be underestimated because of undetected cases. Furthermore, individuals of Northern Cree and Mennonite ancestries are not confined to the province of Manitoba [12], [14], [16]; thus emphasizing the importance of broadening directed screening in other jurisdictions. All SCID and PID patients in this study were seen by the same clinician (MLS), their tests done in the same laboratory and all had their Guthrie cards stored under the same conditions, making the data analysis not only relevant, but also more comparable. The study analyzed a relatively large, but still limited number (n = 18) of SCID and PID cases, with some of the diverse conditions represented only once. Additional prospective screening will increase the power of the analysis and help further characterize atypical SCID and PID cases. However, our data demonstrate that the TREC test might not capture the majority of SCID cases in some ethnic populations and supplementary screening should be considered. Indeed, B-cell deficient immune deficiencies could be detected by using the κ-deleting recombination excision circle (KREC) assay [22]. However, KREC numbers were shown to be within the normal range for IKKβ patients [16]. A more direct albeit a more expensive approach in specific populations would be using DNA sequencing when a mutation is known.
5. Conclusions
TREC screening identified all (100%) T-cell-deficient forms of SCID and PID. As expected, the TREC assay did not identify the majority of T-cell positive forms of SCID and PID that are prevalent in the province of Manitoba. If population-based newborn screening for SCID using TREC assay is introduced in the future in our province, we recommend that screening includes supplemental targeted or multiplexed screening for ethnic-specific mutations to ensure that the highest proportion of affected babies is identified.
Conflict of interest statement
The authors declare that there are no conflict of interest.
Acknowledgments
TREC plasmid was developed by Dr. D. Douek and kindly provided by Dr. A.M. Comeau (New England Newborn Screening Program), who also provided TREC assay training. We acknowledge Cadham Provincial Laboratory for their participation in the project. We would like to thank Dr. C. Ellison for the helpful discussions, Dr. T. Zelinski for the insightful comments on the manuscript, Dr. G. Cuvelier for a critical reading of the manuscript, and Jessica Hartley for the helpful discussions regarding genetic counseling in SCID.
References
- 1.Chan K., Puck J.M. Development of population-based newborn screening for severe combined immunodeficiency. J. Allergy Clin. Immunol. 2005;115:391–398. doi: 10.1016/j.jaci.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 2.Baker M.W., Grossman W.J., Laessig R.H., Hoffman G.L., Brokopp C.D., Kurtycz D.F., Cogley M.F., Litsheim T.J., Katcher M.L., Routes J.M. Development of a routine newborn screening protocol for severe combined immunodeficiency. J. Allergy Clin. Immunol. 2009;124:522–527. doi: 10.1016/j.jaci.2009.04.007. [DOI] [PubMed] [Google Scholar]
- 3.Gerstel-Thompson J.L., Wilkey J.F., Baptiste J.C., Navas J.S., Pai S.Y., Pass K.A., Eaton R.B., Comeau A.M. High-throughput multiplexed T-cell-receptor excision circle quantitative PCR assay with internal controls for detection of severe combined immunodeficiency in population-based newborn screening. Clin. Chem. 2010;56:1466–1474. doi: 10.1373/clinchem.2010.144915. [DOI] [PubMed] [Google Scholar]
- 4.Kwan A., Church J.A., Cowan M.J., Agarwal R., Kapoor N., Kohn D.B., Lewis D.B., McGhee S.A., Moore T.B., Stiehm E.R., Porteus M., Aznar C.P., Currier R., Lorey F., Puck J.M. Newborn screening for severe combined immunodeficiency and T-cell lymphopenia in California: results of the first 2 years. J. Allergy Clin. Immunol. 2013;132:140–150. doi: 10.1016/j.jaci.2013.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Myers L.A., Patel D.D., Puck J.M., Buckley R.H. Hematopoietic stem cell transplantation for severe combined immunodeficiency in the neonatal period leads to superior thymic output and improved survival. Blood. 2002;99:872–878. doi: 10.1182/blood.v99.3.872. [DOI] [PubMed] [Google Scholar]
- 6.Douek D.C., Vescio R.A., Betts M.R., Brenchley J.M., Hill B.J., Zhang L., Berenson J.R., Collins R.H., Koup R.A. Assessment of thymic output in adults after haematopoietic stem-cell transplantation and prediction of T-cell reconstitution. Lancet. 2000;355:1875–1881. doi: 10.1016/S0140-6736(00)02293-5. [DOI] [PubMed] [Google Scholar]
- 7.Hazenberg M.D., Otto S.A., Cohen Stuart J.W., Verschuren M.C., Borleffs J.C., Boucher C.A., Coutinho R.A., Lange J.M., Rinke de Wit T.F., Tsegaye A., van Dongen J.J., Hamann D., de Boer R.J., Miedema F. Increased cell division but not thymic dysfunction rapidly affects the T-cell receptor excision circle content of the naive T cell population in HIV-1 infection. Nat. Med. 2000;6:1036–1042. doi: 10.1038/79549. [DOI] [PubMed] [Google Scholar]
- 8.Morinishi Y., Imai K., Nakagawa N., Sato H., Horiuchi K., Ohtsuka Y., Kaneda Y., Taga T., Hisakawa H., Miyaji R., Endo M., Oh-Ishi T., Kamachi Y., Akahane K., Kobayashi C., Tsuchida M., Morio T., Sasahara Y., Kumaki S., Ishigaki K., Yoshida M., Urabe T., Kobayashi N., Okimoto Y., Reichenbach J., Hashii Y., Tsuji Y., Kogawa K., Yamaguchi S., Kanegane H., Miyawaki T., Yamada M., Ariga T., Nonoyama S. Identification of severe combined immunodeficiency by T-cell receptor excision circles quantification using neonatal Guthrie cards. J. Pediatr. 2009;155:829–833. doi: 10.1016/j.jpeds.2009.05.026. [DOI] [PubMed] [Google Scholar]
- 9.Verbsky J.W., Baker M.W., Grossman W.J., Hintermeyer M., Dasu T., Bonacci B., Reddy S., Margolis D., Casper J., Gries M., Desantes K., Hoffman G.L., Brokopp C.D., Seroogy C.M., Routes J.M. Newborn screening for severe combined immunodeficiency; the Wisconsin experience (2008–2011) J. Clin. Immunol. 2012;32:82–88. doi: 10.1007/s10875-011-9609-4. [DOI] [PubMed] [Google Scholar]
- 10.Hale J.E., Bonilla F.A., Pai S.Y., Gerstel-Thompson J.L., Notarangelo L.D., Eaton R.B., Comeau A.M. Identification of an infant with severe combined immunodeficiency by newborn screening. J. Allergy Clin. Immunol. 2010;126:1073–1074. doi: 10.1016/j.jaci.2010.08.043. [DOI] [PubMed] [Google Scholar]
- 11.Rozmus J., Junker A., Thibodeau M.L., Grenier D., Turvey S.E., Yacoub W., Embree J., Haddad E., Langley J.M., Ramsingh R.M., Singh V.A., Long R., Schultz K.R. Severe Combined Immunodeficiency (SCID) in Canadian children: a national surveillance study. J. Clin. Immunol. 2013;33:1310–1316. doi: 10.1007/s10875-013-9952-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roifman C.M., Dadi H., Somech R., Nahum A., Sharfe N. Characterization of ζ-associated protein, 70 kD (ZAP70)-deficient human lymphocytes. J. Allergy Clin. Immunol. 2010;126:1226–1233. doi: 10.1016/j.jaci.2010.07.029. [DOI] [PubMed] [Google Scholar]
- 13.Puck J.M. Laboratory technology for population-based screening for severe combined immunodeficiency in neonates: the winner is T-cell receptor excision circles. J. Allergy Clin. 2012;129:607–616. doi: 10.1016/j.jaci.2012.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roifman C.M., Somech R., Kavadas F., Pires L., Nahum A., Dalal I., Grunebaum E. Defining combined immunodeficiency. J. Allergy Clin. Immunol. 2012;130:177–183. doi: 10.1016/j.jaci.2012.04.029. [DOI] [PubMed] [Google Scholar]
- 15.Guazzi V., Aiuti F., Mezzaroma I., Mazzetta F., Andolfi G., Mortellaro A., Pierdominici M., Fantini R., Marziali M., Aiuti A. Assessment of thymic output in common variable immunodeficiency patients by evaluation of T-cell receptor excision circles. Clin. Exp. Immunol. 2002;129:346–353. doi: 10.1046/j.1365-2249.2002.01893.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pannicke U., Baumann B., Fuchs S., Henneke P., Rensing-Ehl A., Rizzi M., Janda A., Hese K., Schlesier M., Holzmann K., Borte S., Laux C., Rump E.-M., Rosenberg A., Zelinski T., Schrezenmeier H., Wirth T., Ehl S., Schroeder M.L., Schwarz K. Deficiency of innate and acquired immunity caused by an IKBKB mutation. N. Engl. J. Med. 2013;369:2504–2514. doi: 10.1056/NEJMoa1309199. [DOI] [PubMed] [Google Scholar]
- 17.Serana F., Sottini A., Chiarini M., Zanotti C., Ghidini C., Lanfranchi A., Notarangelo L.D., Caimi L., Imberti L. The different extent of B and T-cell immune reconstitution after hematopoietic stem cell transplantation and enzyme replacement therapies in SCID patients with adenosine deaminase deficiency. J. Immunol. 2010;185:7713–7722. doi: 10.4049/jimmunol.1001770. [DOI] [PubMed] [Google Scholar]
- 18.Marangoni F., Trifari S., Scaramuzza S., Panaroni C., Martino S., Notarangelo L.D., Baz Z., Metin A., Cattaneo F., Villa A., Aiuti A., Battaglia M., Roncarolo M.G., Dupré L. WASP regulates suppressor activity of human and murine CD4(+)CD25(+)FOXP3(+) natural regulatory T cells. J. Exp. Med. 2007;204:369–380. doi: 10.1084/jem.20061334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schäffer A.A., Salzer U., Hammarström L., Grimbacher B. Deconstructing common variable immunodeficiency by genetic analysis. Curr. Opin. Genet. Dev. 2007;17:201–212. doi: 10.1016/j.gde.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 20.Rezaei N., Mahmoudi E., Aghamohammadi A., Das R., Nichols K.E. X-linked lymphoproliferative syndrome: a genetic condition typified by the triad of infection, immunodeficiency and lymphoma. Br. J. Haematol. 2011;152:13–30. doi: 10.1111/j.1365-2141.2010.08442.x. [DOI] [PubMed] [Google Scholar]
- 21.Mallott J., Kwan A., Church J., Gonzalez-Espinosa D., Lorey F., Tang L.F., Sunderam U., Rana S., Srinivasan R., Brenner S.E., Puck J. Newborn screening for SCID identifies patients with ataxia telangiectasia. J. Clin. Immunol. 2013;33:540–549. doi: 10.1007/s10875-012-9846-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Borte S., von Döbeln U., Fasth A., Wang N., Janzi M., Winiarski J., Sack U., Pan-Hammarström Q., Borte M., Hammarström L. Neonatal screening for severe primary immunodeficiency diseases using high-throughput triplex real-time PCR. Blood. 2012;119:2552–2555. doi: 10.1182/blood-2011-08-371021. [DOI] [PubMed] [Google Scholar]