Abstract
Tay–Sachs disease is an autosomal recessive neurodegenerative disorder occurring due to impaired activity of β-hexosaminidase-A (EC 3.2.1.52), resulting from the mutation in HEXA gene. Very little is known about the molecular pathology of TSD in Indian children except for a few mutations identified by us. The present study is aimed to determine additional mutations leading to Tay–Sachs disease in nine patients confirmed by the deficiency of β-hexosaminidase-A (< 2% of total hexosaminidase activity for infantile patients) in leucocytes. The enzyme activity was assessed by using substrates 4-methylumbelliferyl-N-acetyl-β-d-glucosamine and 4-methylumbelliferyl-N-acetyl-β-d-glucosamine-6-sulfate for total-hexosaminidase and hexosaminidase-A respectively, and heat inactivation method for carrier detection. The exons and exon–intron boundaries of the HEXA gene were bi-directionally sequenced on an automated sequencer. ‘In silico’ analyses for novel mutations were carried out using SIFT, Polyphen2 and MutationT@ster software programs. The structural study was carried out by UCSF Chimera software using the crystallographic structure of β-hexosaminidase-A (PDB-ID: 2GJX) as the template. Our study identified four novel mutations in three cases. These include a compound heterozygous missense mutation c.524A>C (D175A) and c.805G>C (p.G269R) in one case; and one small 1 bp deletion c.426delT (p.F142LfsX57) and one splice site mutation c.459+4A>C in the other two cases respectively. None of these mutations were detected in 100 chromosomes from healthy individuals of the same ethnic group. Three previously reported missense mutations, (i) c.532C>T (p.R178C), (ii) c.964G>T (p.D322Y), and (iii) c.1385A>T (p.E462V); two nonsense mutations (i) c.709C>T (p.Q237X) and (ii) c.1528C>T (p.R510X), one 4 bp insertion c.1277_1278insTATC (p.Y427IfsX5) and one splice site mutation c.459+5G>A were also identified in six cases. We observe from this study that novel mutations are more frequently observed in Indian patients with Tay–Sachs disease with clustering of ~ 73% of disease causing mutations in exons 5 to 12. This database can be used for a carrier rate screening in the larger population of the country.
Abbreviations: TSD, Tay–Sachs disease; SD, Sandhoff disease; Hex-A, β-Hexosaminidase A; LSDs, Lysosomal storage disorders; MUG, 4-Methylumbelliferyl-N-acetyl-β-d-glucosamine; MUGS, 4-Methylumbelliferyl-N-acetyl-β-d-glucosamine-6-sulfate
Keywords: Tay–Sachs disease, HEXA gene, β-Hexosaminidase-A, Lysosomal enzyme
1. Introduction
Tay–Sachs disease (TSD) (MIM# 272800) is the second most common lipid storage disorder among the majority of lysosomal storage disorders (LSDs) studied in India [1]. It is an autosomal recessive neurodegenerative disorder caused due to β-hexosaminidase-A (Hex-A) deficiency owing to a mutation in the HEXA gene (MIM* 606869). The gene encodes α-subunit of β-hexosaminidase-A, a lysosomal enzyme composed of α- and β-polypeptides [2]. The resulting phenotype of TSD can be variable with an acute infantile form of early onset leading to rapid neuroregression and early death to a progressive later onset form compatible with a longer survival. Clinical features include the presence of neuroregression, generalized hypotonia, exaggerated startle response, and a cherry-red spot on the fundus. Affected patients have a deficient enzyme activity of β-hexosaminidase-A (Hex-A) in leukocytes/plasma or fibroblasts [3]. The human HEXA gene is located on chromosomes 15q23–q24 encompassing 14 exons. As per HGMD (human gene mutation database), nearly 134 mutations have been reported so far in the gene to cause TSD (http://www.hgmd.cf.ac.uk/). Majority of these mutations were private mutations present in a single or few families. Only few mutations have been commonly reported for TSD in particular ethnic communities or geographically isolated populations. In Ashkenazi Jews, one of the three common mutations i.e. c.1277_1278insTATC, c.1421+1G>C and c.805G>A (p.G269S) has been reported in 94–98% affected patients [4], [5]. The c.1277_1278insTATC and the c.805G>A (p.G269S) mutations are also commonly found in non-Ashkenazi Jewish populations, along with the intron 9 splice site mutation (c.1073+1G>A), 7.6 kb French Canadian deletion [6], and c.571-1G>T in Japanese patients [2]. About 35% non-Jewish individuals also carry one of the two pseudo-deficiency alleles: c.739C>T (p.R247W) and c.745C>T (p.R249W), which are associated with no clinical phenotypes [7]. Although the mutation spectrum for Tay–Sachs disease in India has been partly addressed [1], [8], our research continues to understand the molecular pathology of the disease. The present study was planned to identify additional novel and/or known mutations that will provide an insight into the molecular mechanism of the HEXA gene in Indian subjects.
2. Material and methods
2.1. Subject and sample collection
Nine families were included in the study. The Institutional ethics committee approved the study. For blood collection, clinical details were filled up in the case record forms after an informed written consent was obtained from the guardian of the study subjects. The clinical inclusion criteria were the presence of seizures, cherry red spots, neuroregression, exaggerated startle, and hypotonia followed by the confirmation of enzyme deficiency of Hex-A and mutation identification. Though neuroimaging criteria were not strictly followed up, it mainly included the presence of gray matter disease as shown by hypointensities in T2W images of the brain MRI.
2.2. Enzyme assay
Six milliliter peripheral blood was collected in an EDTA vacutainer from all the patients for leukocyte enzyme assay and DNA extraction. The enzyme activity was determined by the fluorimetric method using a specific synthetic substrate. The total-Hex activity was measured from the hydrolysis of the synthetic substrate 4-methylumbelliferyl-N-acetyl-β-d-glucosamine (MUG) that releases fluorescent 4-methylumbelliferone when acted upon by β-hexosaminidase. Hexosaminidase B (Hex-B) was determined as the activity left over after incubating the sample for 3 h at 50 °C. This procedure led to the loss of heat-labile Hex-A but not Hex-B or intermediate isoenzyme. Thus Hex-A activity was obtained after subtracting Hex-B activity from the total-Hex activity. Hex-A was also assayed with a more specific sulfated substrate 4-methylumbelliferyl-N-acetyl-β-d-glucosamine-6-sulfate (MUGS) [9]. Hex-A activity (obtained from MUGS lysis) was expressed as the percentage of total-Hex activity (obtained from MUG lysis).
2.3. Molecular analysis of HEXA gene by sequencing
Genomic DNA was extracted by the standard salting out method [10]. The exonic and intronic flanking sequences of the HEXA gene were PCR amplified in 14 fragments using the primer pairs as described earlier [11]. Amplification included 5-min denaturation at 94 °C followed by 30 cycles each consisting of 1 min denaturation at 94 °C for 45 s of annealing at temperature ranging from 60 to 65 °C, depending on the characteristic of each exon (melting temperature), and 45 sec extension at 72 °C. Final extension was carried out at 72 °C for 10 min. The negative control contained all the above ingredients except DNA. PCR products were electrophoresed on 2% agarose along with the appropriate negative controls and 100 base-pair DNA ladder. Products that passed this quality check were purified by treatment with Exo-SAP-IT™ (USB Corporation, OH, USA) or Column purification and then sequenced using BigDye Terminator v3.1. Capillary electrophoresis was performed on an automated sequencer ABI-3130 (Applied Biosystems, CA, USA) and nucleotide numbers are derived from HEXA cDNA sequences (RefSeq NM_000520.4). Putative mutations were confirmed in two separate PCR products from the patient's DNA. Heterozygosity for these mutations was confirmed in parents. The mutations identified were then looked up in public databases such as The Human Gene Mutation Database (http://www.hgmd.cf.ac.uk), SNP database (http://www.ncbi.nlm.nih.gov/SNP/index.html) and McGill University database (http://www.hexdb.mcgill.ca). Novel variants were further confirmed as pathogenic by studying 50 control individuals with normal leukocyte enzyme results.
2.4. In silico analysis
Prediction of functional effects of non-synonymous single nucleotide substitutions (nsSNPs) was done using software SIFT (Sorting Intolerant From Tolerant) (available at http://sift.jcvi.org/), Polyphen2 (Polymorphism Phenotyping v2) (available at http://genetics.bwh.harvard.edu/pph2/) and MutationT@ster (available at http://www.mutationtaster.org/) [12], [13], [14]. HumVar-trained prediction model of Polyphen2 was used to distinguish mutations with drastic effects from all the remaining human variations, including many mildly deleterious alleles. Evolutionary conservation of the amino acid residues of Hex A was analyzed using ClustalW program available online at (http://www.uniprot.org/help/sequence-alignments).
2.5. Structural studies
Structural study of two missense mutant alleles was carried out by UCSF Chimera software using the crystallographic structure of Hex-A (PDB ID: 2GJX) as the template. The Root Mean Square Deviation (RMSD) of the mutant structures with respect to the wild-type structure was calculated. RMSD values of more than 0.15 were considered as significant structural perturbations that could have functional implications for the protein [15].
3. Results
Consanguinity was present in 4/9 (44.4%) patients. The mean age at presentation was 12.3 months (± 5.3). All the cases were classified as infantile as they had a history of neuroregression since infancy. All the cases were presented with seizures, neuroregression, cherry red spot on fundus, exaggerated startle, hypotonia, and brisk deep tendon reflexes. However, none of the cases had hepato-splenomegaly. The CT/MRI brain was available in 4/9 cases and showed characteristic findings of decrease in thalami and decreased attenuation of basal ganglia isodense with white matter, and few cases had dysmyelination.
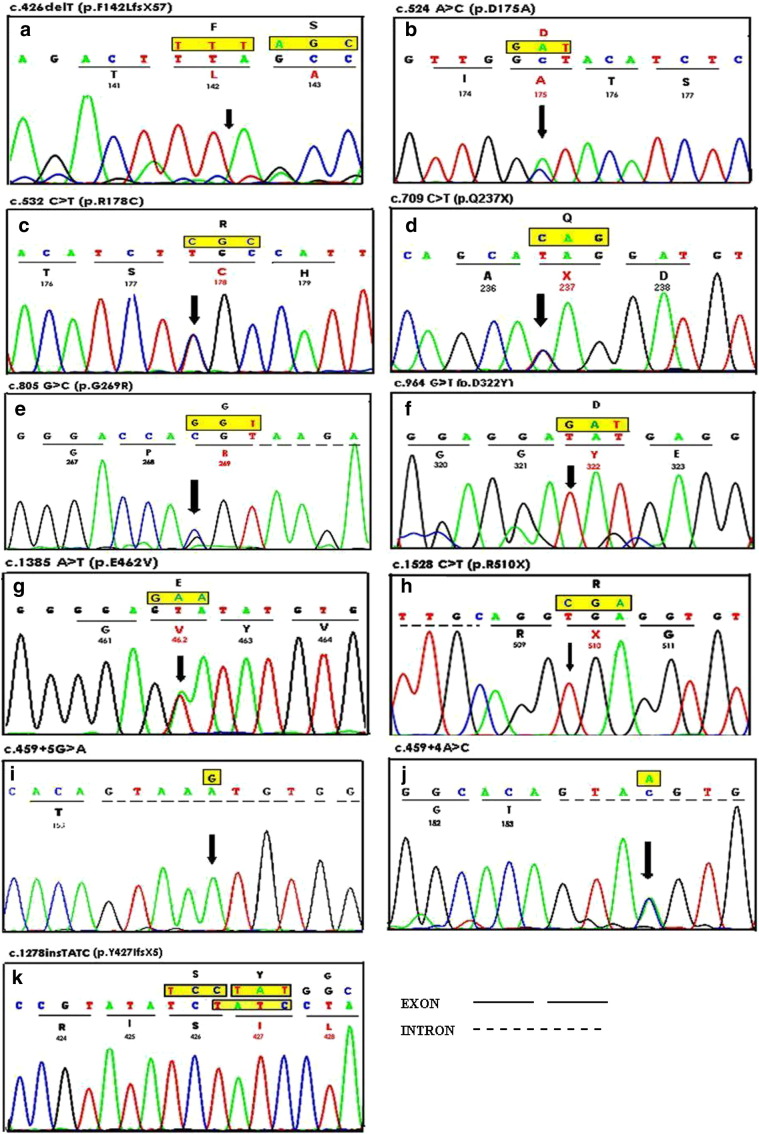
We confirmed 7/9 cases with TSD [deficient activity of β-hexosaminidase-A (Hex-A) in leucocytes], with carrier detection (% Hex-A) in 2 parents where proband was not alive but was earlier diagnosed as a TSD by enzyme study (Table 1). Molecular analysis was carried out in all the 9 affected families with TSD followed by bi-directional sequencing for common mutation screening. Exon sequencing analysis revealed 11 mutations in 9 patients, 4 of which were novel including compound heterozygous missense mutations c.524A>C (p.D175A) and c.805G>C (p.G269R) in exons 5 and 7 respectively, one small 1 bp deletion c.426delT (p.F142LfsX57) in exon 4 and one patient was a compound heterozygote for novel splice site mutation c.459+4A>C and known missense mutation c.532C>T (p.R178C). In addition, previously known missense mutations c.964G>T (p.D322Y) in exon 9, two nonsense mutations c.709C>T (p.Q237X) and c.1528C>T (p.R510X) in exons 7 and 14 respectively and one insertion mutation c.1277_1278insTATC (p.Y427IfsX5) were detected. One patient was found to be heterozygous for c.1385A>T (p.E462V) with unknown second allele and another patient with splice site homozygous mutation c.459+5G>A in intron 4 as shown in Table 1 and Fig. 1. The novel mutations were not found in 100 control individuals from the same ethnic background.
Table 1.
Clinical, biochemical and molecular details of the Indian patients with Tay–Sachs disease.
| Patient ID | Age at diagnosis | Consanguinity | Hex-A activity (MUGS) (nmol/h/mg) = (x) | Total Hex activity (MUG) = (y) | Hex A % = (x / y) × 100 | MUG/MUGS ratiob | Genotypes |
|
|---|---|---|---|---|---|---|---|---|
| Mutations at DNA level | Mutations at protein level | |||||||
| 1. | 1.3 yr | Yes | 9.3 | 2250.3 | 0.4 | 1.50 | c.1277_1278insTATC/c.1277_1278insTATC | p.Y47IfsX5/p.Y47IfsX5 |
| 2. | 1.5 months | No | 8.9 | 2254.3 | 0.39 | 1.44 | c.964G>T/c.964G>T | p.D322Y/p.D322Y |
| 3. | 11 months | Yes | 10.8 | 1273.2 | 0.84 | 3.1 | c.1528C>T/c.1528C>T | p.R510X/p.R510X |
| 4. | 1.1 yr | No | 7.0 | 2602.7 | 0.27 | 1.0 | c.524A>C/c.805G>C | p.D175A/p.G269R |
| 5. | 1.5 yr | No | 0.00 | 1781.0 | 0.0 | 0.0 | c.459+5G>A/c.459+5G>A | –/– |
| 6. | 1.5 yr | Yes | 6.2 | 2090.0 | 0.29 | 1.07 | c.426delT/c.426delT | p.F142Lfsx57/p.F142Lfsx57 |
| 7. | 1 yr | No | 7.9 | 2185.7 | 0.36 | 1.33 | c.1385A>T/? | p.E462V/? |
| 8.a | 1 yr | Yes | 0.9 Mother HLC — 53.8% Father HLC — 58.8% |
761 | 0.11 | 0.41 | –/– Mother — [c.532C>T/normal] Father — [c.459+4A>C/normal] |
[–/–] Mother — [p.R178C/normal] Father — [–/–] |
| 9.a | 1.3 yr | No | 1.5 Mother HLC — 50.5% Father HLC — 48.8% |
1526.3 | 0.098 | 0.36 | –/– Mother — [c.709>T/normal] Father — [c.709C>T/normal] |
[–/–] Mother — [p.Q237X/normal] Father — [p.Q237X/normal] |
Normal total-hexosaminidase values using MUG substrate in our controls — 723 to 2700 nmol/h/mg protein, normal hexosaminidase A levels — 62 to 77%; and normal MUGS activity 80 to 410 nmol/h/mg.
DNA of index case is not available, HLC—heat labile activity in carrier parents.
The MUG/MUGS ratio for Hex A is 3.7:1 [19].
Fig. 1.
Sequence chromatogram of HEXA gene mutations. (a): c.426delT (p.F142LFsX57) (homozygous); (b): c.524A>C (p.D175A) (heterozygous); (c): c.532C>T (p.R178C) (heterozygous); (d): c.709C>T (p.Q237X) (heterozygous); (e): c.805G>C (p.G269R) (heterozygous); (f): c.964G>T (p.D322Y) (homozygous); (g): c.1385A>T (p.E462V) (heterozygous); (h): c.1528C>T (p.R510X); (i): c.459+5G>A (homozygous); (j): c.459+4A>C (heterozygous); (k): c.1277_1278insTATC (p.Y47IfsX5) (homozygous).
The SIFT index, Polyphen2 scores and MutationT@ster scores for the non-synonymous single nucleotide substitutions are presented in Table 2. The predictions of the SIFT, Polyphen2 and MutationT@ster algorithms were in concordance with the observed pathogenicity in the case of mutations p.D175A, p.G269R, p.D322Y and p.E462V as described in Table 2. The RMSD (Root Mean Square Deviation) values for the modeled mutants were significant for the pathogenicity of all missense mutations (Table 2).
Table 2.
Details of HEXA missense mutations detected in infantile TSD patients and in silico analysis.
| Location | Codon number | Codon change | Amino acid change | MutationT@ster score | SIFT score | Polyphen2 score (sensitivity, specificity) | RMSD between native and mutant structures | Amino acid change |
|---|---|---|---|---|---|---|---|---|
| Exon 5 | 175 | GAT–GCT | Asp175Ala | 3.44 (DC) | 0.00 (IT) | 1.00 (0, 1) (PD) | 0.268, (PD) | Acidic to nonpolar |
| Exon 7 | 269 | GGT–GCT | Gly269Arg | 3.41 (DC) | 0.00 (IT) | 1.00 (0,1)(PD) | 0.225, (PD) | Hydrophilic to charged |
| Exon 8 | 322 | GAT–TAT | Asp322Tyr | 4.36 (DC) | 0.00 (IT) | 1.00 (0,1) (PD) | 0.178, (PD) | Non- cyclic to cyclic |
| Exon 12 | 462 | GAA–GTA | Glu462Val | 3.3 (DC) | 0.00 (IT) | 1.00 (0,1) (PD) | 0.182, (PD) | Acidic to nonpolar |
DC = disease causing, P = polymorphism, IT = intolerant, PD = probably damaging.
The MutationT@ster score is taken from an amino acid substitution matrix (Grantham Matrix) which takes into account the physico-chemical characteristics of amino acids and scores substitutions according to the degree of difference between the original and the new amino acid which scores may range from 0.0 to 6.0.
The SIFT score is the normalized the probability that the amino acid change is tolerated and ranges from 0 to 1. The amino acid substitution is predicted damaging if the score id ≤ 0.05, and tolerated if the score is > 0.05.
The Polyphen2 score is the naïve Bayes posterior to probability that this mutation is damaging and thus ranges from 0 to 1.
4. Discussion
The clinical features of infantile Tay–Sachs disease seen in our set of patients were consistent with the defined phenotype, except for the presence of prominent Mongolian spots on the back even beyond infancy in two patients. However, Mongolian spots are also reported in association with many of the patients with other LSDs [1], [16] and in our case it could be an accidental finding. The enzyme activity was in accordance with the previous observations of infantile TSD cases [17], [18]. However, MUG substrate is cleaved by all three isozymes and therefore is used to measure total-Hex activity. MUGS substrate is cleaved primarily by the α-subunit containing isozymes (Hex-A and S). However, the β active site can slowly hydrolyze this substrate with MUG/MUGS ratios of ~ 300:1 for Hex-B and 3.7:1 for Hex-A [19]. Thus each of the %-residual Hex-A values reported in Table 1 is multiplied by 3.7 and is varying from 0% to 3.1%, which is in accordance with the previously reported value.
Among 11 mutations demonstrated here, six have been previously reported in different populations that include missense mutations p.E462V and p.D322Y from the Indian population [1], [8], p.R178C from Czechoslovakia [20], c.1277_1278insTATC from Indian, Ashkenazi Jews and non-Jewish populations [8], [20], nonsense mutation p.R510X from Saudi Arabia [21] and Iranian population [22], and splice site mutation c.459+5G>A from Argentinean [23] and Spanish populations [24]. The nonsense mutation p.Q237X was previously reported as benign in accordance with the NCBI SNP database. However, a subunit lacking its last 293 C-terminal residues is highly likely to make the protein non-functional. In the present study this mutation was observed in both parents with the history of an affected child with TSD. Though, mutation study in the affected child could not be carried out due to sudden death and unavailability of a sample. The National Center for Biotechnology Information's (NCBI) single nucleotide polymorphism database (dbSNP) (http://www.ncbi.nlm.nih.gov/SNP/snp_ref.cgi?searchType=adhoc_search&type=rs&rs=rs150675340) reveals that the minor allele count and pathogenic potential of this polymorphism (rs150675340) are not available. Generally, nonsense and frame-shift mutations result in the reduction of mRNA in the HEXA gene. Diminished amounts of mRNA have been reported in several mutations in the HEXA gene [25]. The presence of one mutant and one normal copy of this allele in both the parents suggests that the child would have had the mutation in the homozygous state, thus being responsible for causing of the disease.
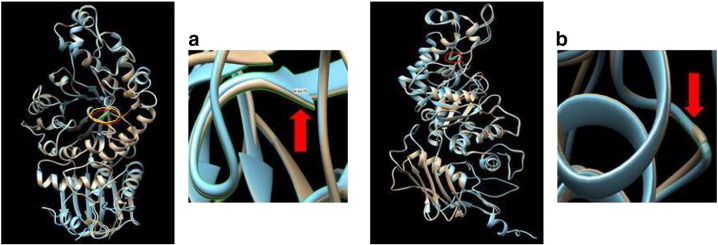
In the present study, novel mutations p.D175A and p.G269R were found as compound heterozygous in infantile TSD. The missense mutation p.D175A disrupts the β-sheet due to an amino acid change from a positively charged aspartic acid to a hydrophobic alanine residue at codon 175 of exon 5. At the same time, p.G269R mutation creates over-packing and backbone disruption due to the replacement of a small hydrophilic group (glycine) with a positively charged residue (Alanine) at codon 269 of exon-7 [Fig. 2(a)–(b)]. Therefore, disruption of the backbone from αGly269Arg may result in the movement of αHis262, with the likelihood of disrupting the coordination of residues in the active site. The RMSD of the modeled mutant is also significantly higher as compared to the wild-type protein (Table 2). Additionally, p.G269 is also the site of the mutation in late onset Tay–Sachs (LOTS) (p.G269S) with high frequency in the Ashkenazi Jews and results in an unstable alpha subunit precursor. This fails to associate with the beta subunit, suggesting that the mutation reported here, found in a patient with the infantile acute disease, has a more deleterious impact on the enzyme or its expression [15]. Protein sequence alignment demonstrated that p.D175A and p.G269R are highly conserved residues among different species. All these novel mutations were found to be pathogenic by SIFT, Polyphen2 and MutationT@aster softwares.
Fig. 2.
Superimposed native structures (brown) and mutant structure (blue) produced using USCF Chimera software: (a) mutation p.D175A disrupts the β-sheet and (b) p.G269R creates overpacking and backbone disruption.
Another novel mutation (1 bp deletion c.426delT) was observed in exon 4 in an infantile TSD patient that changes the reading frame at codon 142 and introduces a premature stop at codon 199 (p.F142LfsX57), leading to a truncated protein or a nonsense-mediated mRNA decay (NMD). This mutation was also evaluated for pathogenicity using MutationT@ster and was found to be disease causing. Nonetheless, in a non-Jewish French child, a 2 bp deletion in the same codon (codon-142) is reported by Akli et al. [26].
In one case, the novel intronic mutation c.459+4A>C was found with previously reported mutation p.R178C in a compound heterozygous state. This mutation is in close proximity to the reported splice mutation c.459+5G>A in Argentinean and Spanish populations [23], [24]. It was evaluated for pathogenicity using MutationT@ster and was found to be disease causing; and was predicted to lead to a loss of aberrant donor splice site at this position (score 0.87). The patient was not available for further functional study of the mutation. As per our knowledge, these novel mutations have never been identified in TSD patients from other ethnic groups.
A large number of gene mutations causing TSD have been previously reported in the HEXA mutation database (http://www.hexdb.mcgill.ca/). Among them, mutations resulting in gross alterations in the Hex α-subunit sequence are generally found in the severe infantile form. However, missense mutations causing amino acid substitutions have been found in both the infantile and late onset phenotypes [27]. The detrimental effect of most mutations is on the overall folding and/or transport of the protein rather than on the functional sites, as reported by Mahuran [28]. Some of the mutations identified in our series affect crucial active residues or cause significant structural aberrations leading to the infantile form of the disease.
5. Conclusion
Identification of four novel mutations provides additional insight into the molecular pathology of TSD in Indian subjects. Combining this with our previous study of 28 cases, mutations responsible for TSD in Indian patients are mostly novel with nearly 73% mutations clustering in between exons 5 to 12. This database can be used as a tool for the carrier screening of a larger Indian population.
Conflict of interest
The authors have declared that no competing interests exist.
Author contributions
JS was involved in the designing of the study, standardization of technical procedure, and preparation of manuscript and will also act as a guarantor. MM was involved in processing the sample, standardization of molecular techniques and preparation of the manuscript. FS have critically evaluated the manuscript. CD, MK, SP, HS, and UK were involved in clinical data collection. SG helped in technical support. All the authors have read and approved the manuscript.
Acknowledgments
We are grateful to all the referring clinicians and patients for their support; without their consent this study would not have been possible. We acknowledge the Indian Council of Medical Research (ICMR) for their financial support to conduct this study.
References
- 1.Sheth J., Mistri M., Sheth F., Shah R., Bavdekar A., Godbole K., Nanavaty N., Datar C., Kamate M., Oza N., Ankleshwaria C., Mehta S., Jackson M. JIMD Rep. 2013. Burden of lysosomal storage disorders in India: experience of 387 affected children from a single diagnostic facility. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tanaka A., Fujimaru M., Choeh K., Isshiki G. Novel mutations, including the second most common in Japan, in the B-hexosaminidase A subunit gene, a simple screening of Japanese patients with Tay–Sachs disease. J. Hum. Genet. 1999;44:91–95. doi: 10.1007/s100380050116. [DOI] [PubMed] [Google Scholar]
- 3.Fernades Filho J.A., Shapiro B.E. Tay–Sachs disease. Arch. Neurol. 2004;61:1466–1468. doi: 10.1001/archneur.61.9.1466. [DOI] [PubMed] [Google Scholar]
- 4.Myerowitz R., Costigan F.C. The major defect in Ashkenazi Jews with Tay–Sachs disease is an insertion in the gene for alpha-chain of beta-hexosaminidase. J. Biol. Chem. 1988;263:18587–18589. [PubMed] [Google Scholar]
- 5.Kaback M.M., Lim-steele J.S., Dabholkar D., Brown D., Levy N., Zeiger K. Tay–Sachs disease carrier screening, prenatal diagnosis and the molecular era. An international perspective, 1970–1993. The International TSD data collection Network. JAMA. 1993;270:2307–2315. [PubMed] [Google Scholar]
- 6.Myerowitz R., Hogikyan N.D. Different mutations in Ashkenazi Jewish and non-Jewish French Canadians with Tay–Sachs disease. Science. 1986;232(4758):1646–1648. doi: 10.1126/science.3754980. [DOI] [PubMed] [Google Scholar]
- 7.Cao Z., Natowicz M.R., Kaback M.M., Lim-steele J.S., Prence E.M., Brown D., Chabot T., Triggs-Raine B.L. A second mutation associated with, apparent beta-hexosaminidase A pseudodeficiency: identification and frequency estimation. Am. J. Hum. Genet. 1993;53:1198–1205. [PMC free article] [PubMed] [Google Scholar]
- 8.Mistri M., Tamhankar P.M., Sheth F., Sanghavi D., Kondurkar P., Patil S., Idicula-Thomas S., Gupta S., Sheth J. Identification of novel mutations in HEXA Gene in children affected with Tay Sachs disease from India. PLoS ONE. 2012;7(6):e39122. doi: 10.1371/journal.pone.0039122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wendeler M., Sandhoff K. Hexosaminidase assays. Glycoconj. J. 2009;26(8):945–952. doi: 10.1007/s10719-008-9137-5. [DOI] [PubMed] [Google Scholar]
- 10.Miller S.A., Dykes D.D., Polesky H.F. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16(3):1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Trigss Raine B.L., Akerman B.R., Clarke J.T., Gravel R.A. Sequencing of DNA flanking the exons of the HEXA gene, and identification of mutation in Tay–Sachs disease. Am. J. Hum. Genet. 1991;49:1041–1054. [PMC free article] [PubMed] [Google Scholar]
- 12.Kumar P., Henikoff S., Ng P.C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 2009;4(7):1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 13.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7(4):248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schwarz J.M., Rödelsperger C., Schuelke M., Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat. Methods. 2010;7(8):575–576. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- 15.Ohno K., Saito S., Sugawara K., Sakuraba H. Structural consequences of amino acid substitutions causing Tay–Sachs disease. Mol. Genet. Metab. 2008;94:462–468. doi: 10.1016/j.ymgme.2008.04.006. [DOI] [PubMed] [Google Scholar]
- 16.Staretz-Chacham O., Lang T.C., LaMarca M.E., Krasnewich D., Sidransky E. Lysosomal storage disorders in the newborn. Pediatrics. 2009;123(4):1191–1207. doi: 10.1542/peds.2008-0635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaback M.M. Population-based genetic screening for reproductive counseling: the Tay–Sachs disease model. Eur. J. Pediatr. 2000;159(Suppl. 3):S192–S195. doi: 10.1007/pl00014401. [DOI] [PubMed] [Google Scholar]
- 18.Nalini A., Christopher R. Cerebral glycolipidoses: clinical characteristics of 41 pediatric patients. J. Child Neurol. 2004;19(6):447–452. doi: 10.1177/088307380401900610. [DOI] [PubMed] [Google Scholar]
- 19.Hou Y., Tse R., Mahuran D.J. The direct determination of the substrate specificity of the α-active site in heterodimeric β-hexosaminidase A. Biochemistry. 1996;35:3963–3969. doi: 10.1021/bi9524575. [DOI] [PubMed] [Google Scholar]
- 20.Tanaka A., Ohno K., Sandhoff K., Maire I., Kolodny E.H., Brown A., Suzuki K. GM2-gangliosidosis B1 variant: analysis of beta-hexosaminidase alpha gene abnormalities in seven patients. Am. J. Hum. Genet. 1990;46(2):329–339. [PMC free article] [PubMed] [Google Scholar]
- 21.Kaya N., Al- Owain M., AbuDheim N., Al-Zahrani J., Colak D., Al-Sayed M., Milanlioglu A., Ozand P.T., Alkuraya F.S. GM2 gangliosidosis in Saudi Arabia: multiple mutations and considerations for future carrier screening. Am. J. Hum. Genet. A. 2011:1–4. doi: 10.1002/ajmg.a.33932. [DOI] [PubMed] [Google Scholar]
- 22.Jamali S., Eskandari N., Aryani O., Salehpour S., Zaman T., Kamalidehghan B., Houshmand M. Three novel mutations in Iranian patients with Tay–Sachs disease. Iran. Biomed. J. 2014;18:114–119. doi: 10.6091/ibj.1137.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zampieri S., Montalvo A., Blanco M., Zanin I., Amartino H., Vlahovicek K., Szlago M., Schenone A., Pittis G., Bembi B., Dardis A. Molecular analysis of HEXA gene in Argentinean patients affected with Tay–Sachs disease: possible common origin of the prevalent c.459+5A>G mutation. Gene. 2012;499(2):262–265. doi: 10.1016/j.gene.2012.03.022. [DOI] [PubMed] [Google Scholar]
- 24.Gort L., de Olano N., Macías-Vidal J., Coll M.A. Spanish GM2 Working Group, GM2 gangliosidoses in Spain: analysis of the HEXA and HEXB genes in 34 Tay–Sachs and 14 Sandhoff patients. Gene. 2012;506(1):25–30. doi: 10.1016/j.gene.2012.06.080. [DOI] [PubMed] [Google Scholar]
- 25.Drucker L., Proia R., Navon R. Identification and rapid detection of three Tay–Sachs mutations in the Moroccan Jewish population. Am. J. Hum. Genet. 1992;51:371–377. [PMC free article] [PubMed] [Google Scholar]
- 26.Akli S., Chomel J.C., Lacorte J.M., Bachner L., Kahn A., Poenaru L. Ten novel mutations in the HEXA gene in non-Jewish Tay–Sachs patients. Hum. Mol. Genet. 1993;2:61–67. doi: 10.1093/hmg/2.1.61. [DOI] [PubMed] [Google Scholar]
- 27.Gravel R.A., Kaback M.M., Proia R.L., Sandhoff K., Suzuki K. The GM2 gangliosidosis. In: Scriver C.R., Beaudet A.L., Sly W.S., Valle D., editors. The Metabolic and Molecular Bases of Inherited Disease. McGraw-Hill; New York: 1995. pp. 3827–3876. [Google Scholar]
- 28.Mahuran D.J. Biochemical consequences of mutations causing the GM2 gangliosidosis. Biochem. Biophys. Acta. 1999;1455:105–138. doi: 10.1016/s0925-4439(99)00074-5. [DOI] [PubMed] [Google Scholar]