Abstract
3-Hydroxyisobutyryl-CoA hydrolase (HIBCH) deficiency is an autosomal recessive disorder characterized by episodes of ketoacidosis and a Leigh-like basal ganglia disease, without high concentrations of pyruvate and lactate in the cerebrospinal fluid. Only 4 cases of HIBCH deficiency have been reported. However, clinical–biochemical correlation in HIBCH deficiency by determining the detailed residual enzyme activities has not yet been elucidated. Here, we report a case of two Japanese siblings with HIBCH deficiency carrying a new homozygous missense mutation (c.287C > A, [p.A96D]) at the substrate-binding site. A transfection study using HIBCH expression vectors harboring wild type or 4 reported mutations, including the newly identified mutation (p.A96D, p.Y122C, p.G317E, and p.K74Lfs*13), revealed a correlation between residual HIBCH activities and the severity of the disease. All HIBCH mutants, except p.K74Lfs*13, showed residual enzyme activity and only the patient with p.K74Lfs*13 had congenital anomalies. p.G317E showed only low enzyme activity (~ 3%) of that of wild-type HIBCH. Although p.A96D had approximately 7 times higher enzyme activity than p.G317E, patients with p.A96D died during childhood. These findings are essential for clinical management, genetic counseling, and specific meal and concomitant drug considerations as part of the treatment for patients with HIBCH deficiency.
Keywords: HIBCH, Valine catabolic pathway, Leigh-like disease, Ketoacidosis
1. Introduction
Essential amino acids, leucine, isoleucine, and valine, are commonly termed as branched-chain amino acids (BCAAs). In humans, BCAAs are found at high levels in skeletal muscle, where they contribute to energy production by catabolism after the activation of branched-chain α-keto acid dehydrogenase complex (BCKDC) during energy-compromised states such as, exercise or fever [1]. 3-Hydroxyisobutyryl-CoA hydrolase (HIBCH) localizes in the mitochondrial matrix and is crucial to prevent the accumulation of toxic methacrylyl-CoA by removing CoA from 3-hydroxyisobutyryl-CoA in the valine catabolic pathway [2]. HIBCH deficiency is an autosomal recessive disorder characterized by episodes of ketoacidosis and a Leigh-like basal ganglia disease. Presently, only 4 mutations from 4 cases of HIBCH deficiency have been reported, but the clinical–biochemical correlation has not yet been evaluated in terms of detailed residual enzyme activities [3], [4], [5]. Here, we report a newly identified case of two Japanese siblings with HIBCH deficiency and discuss the clinical–biochemical correlation of all 6 known cases of HIBCH deficiency by determining the relative residual enzyme activity of the 4 mutant HIBCH proteins. We also describe, in detail, the clinical features of both patients and brain magnetic resonance imaging (MRI) findings for one patient in response to treatment with frequent carbohydrate-rich meals.
2. Materials and methods
2.1. Bioethics approval
The experiments were conducted after approval by the institutional review board at the Institute for Developmental Research, Aichi Human Service Center. Written informed consent was obtained from the patients, other members of the patients' family, and the normal controls who participated in this study. Lymphoblastoid cell lines were established by the Institute for Developmental Research, Aichi Human Service Center, through Epstein–Barr virus transformation of peripheral blood samples obtained from the patients and the normal controls.
2.2. Patients
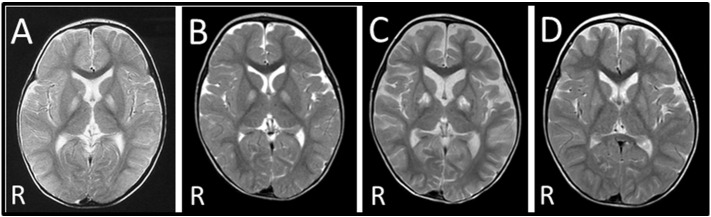
Patient II-2 was the second child of healthy, nonconsanguineous, Japanese parents. She was born at full term gestation after an uncomplicated pregnancy. Her birth weight was 2714 g, and she measured 44 cm (− 2.6 SD) in length, with an occipitofrontal circumference of 32.5 cm (− 0.5 SD). She could hold her head up at 5 months and roll over at 10 months. At 12 months, she was referred to the hospital due to generalized hypotonia and a developmental delay after viral infection. Brain MRI revealed bilateral signal abnormalities in the globi pallidi. A muscle biopsy from the right biceps brachii showed no abnormalities. Lactate and pyruvate levels in arterial blood (16.5 mg/dL and 1.48 mg/dL, respectively; normal range: 5–20 mg/dL and 0.3–0.9 mg/dL, respectively) and cerebrospinal liquor (13.5 mg/dL and 1.12 mg/dL, respectively; normal range: 9.1–18.8 mg/dL and 0.26–1.32 mg/dL, respectively) measured once in a non-acute phase were almost normal [6]. At 15 months, she had febrile illness and impaired consciousness with severe metabolic acidosis. Arterial blood ketone bodies, pH, pCO2, and base excess were (8700 μmol/L; normal range: < 130 μmol/L), 7.051, 6.7 mm Hg, and − 26.9 mEq/L, respectively. She required ventilator support and frequent infusions of bicarbonate. A brain MRI showed bilateral signal abnormalities in the globi pallidi and right caudate nucleus (Fig. 1A). Consequently, she developed subarachnoid hemorrhage, had no spontaneous respirations, and demonstrated no pupillary response to light and corneal reflex. At 17 months, electroencephalography (EEG) revealed an absence of brain activity, and auditory brainstem response audiometry showed no response. She used a home mechanical ventilation system until she died at 4 years of age with acute heart failure and severe metabolic acidosis (pH; 6.87, pCO2; 13.5 mm Hg, and base excess; − 29.4 mEq/L).
Fig. 1.
Brain MRI of the patients. (A) Brain MRI of patient II-2 at 14 months revealed bilateral signal abnormalities in the globi pallidi and right caudate nucleus. (B) Brain MRI of patient II-4 at 14 months showed bilateral signal abnormalities in the globi pallidi. (C) Brain MRI of patient II-4 at 18 months revealed progression of the pathological lesions, with atrophy of the cortex and enlargement of the ventricles. (D) The brain MRI scan of patient II-4 at 24 months shows improvement in signal abnormalities in the basal ganglia when compared to scans at 14 (B) and 18 (C) months.
Patient II-4 was the younger sister of patient II-2, and was delivered by normal vaginal delivery at 39 weeks gestation. Her birth weight was 3245 g and she measured 51.5 cm (0.9 SD) in length, with an occipitofrontal circumference of 33.5 cm (0.3 SD). She acquired head control and rolled over at 4 months. However, she could not sit independently even at 9 months and was referred to the hospital. She exhibited generalized hypotonia and poor body-weight gain. Deep tendon reflexes were mildly increased in the lower limbs and pathological reflexes were not detected. Cranial MRI revealed no abnormalities at 11 months. She developed bilateral signal abnormalities in the globi pallidi (Fig. 1B) at 14 months without any episodes. She also showed dystonia. Cardiac ultrasonography performed at 16 months showed no abnormalities. At 18 months, she contracted influenza and developed impaired consciousness with severe metabolic acidosis. Arterial blood pH, pCO2, and base excess were 7.286, 20.8 mm Hg, and − 14.8 mEq/L, respectively. Plasma levels of ketone bodies, lactate, and pyruvic acid were 5804 μmol/L, 15.3 mg/dL, and 1.2 mg/dL, respectively. A brain MRI revealed the progression of the pathological lesions (Fig. 1C). EEG showed continuous, generalized, high-voltage slow waves. She required frequent infusions of bicarbonate and lost head control and eye pursuit. Since the patient was found to be homozygous for a pathogenic HIBCH mutation, she was treated with frequent carbohydrate-rich meals to avoid the onset of fasting as suggested in a previous report [5] and coenzyme Q10 (30 mg/day), vitamin C (375 mg/day), and vitamin E (75 mg/day), which are known to have a positive antioxidant effect [7]. Consequently, she steadily recovered and exhibited a social smile again. It was noted that the abnormal brain regions improved with recovery of the physical condition (Fig. 1D). However, at 3 years and 10 months, she suddenly acquired an infection, developed impaired consciousness, and was hospitalized. At that time, she showed severe metabolic acidosis: arterial blood pH, pCO2, and base excess were 7.198, 21.4 mm Hg, and − 18 mEq/L, respectively. Her family did not consent to intensive treatment with a ventilator and she died 2 days after admission. Enzyme activities of multiple respiratory chain (RC) enzymes and the pyruvate dehydrogenase complex (PDHc) of both patients were not determined. The clinical features of the presented patients are summarized in Table 1.
Table 1.
Summary of 6 Cases with HIBCH deficiency.
| S1 | S2 | S3 | S4 | II-2 | II-4 | |
|---|---|---|---|---|---|---|
| Mutations | Lys74Leufs*13 | Tyr122Cys/IVS2-3C > G | Gly317Glu | Gly317Glu | Ala96Asp | Ala96Asp |
| Gender | Male | Male | Male | Male | Female | Female |
| Family history | Consanguinity (+) parents are first cousins |
Consanguinity (–) | Consanguinity (+) distantly related parents |
Consanguinity (+) brother of S3 |
Consanguinity (–) | Consanguinity (–) sister of II-2 |
| Initial present | Dysmorphism at birth | Head bobbing at 4 m | Developmental regression at 3 m | Poor feeding at birth | Poor head control at 4 m | Sitting with support at 9 m |
| Hypotonia | + | + | ++ | ++ | + | + |
| Seizures | ND | Absences of eye rolling at 9 m | Myoclonus at 8 m generalized at 10 m | Infantile spasm at 7 m | – | – |
| Dystonia | ND | + | + | + | – | + |
| Others | Dysmorphism, tetralogy of Fallot, vertebral abnormalities | Cerebellar ataxia intention tremor | Vomiting, irritability sleep disturbance | Vomiting, irritability sleep disturbance | – | – |
| Age at death | 3 m | 8 y (alive) | 3 y | 2 y 8 m | 4 y 3 m | 3 y 10 m |
| Abnormal signals in (brain MRI) | ND | Globi pallidi | Globi pallidi | Dentate nuclei globi pallidi |
Globi pallidi | Globi pallidi |
| CSF lactate (< 2.0 mmol/L) | ND | 1.3 | 3.5, 2.2 | 2.1, 2.6 | 1.5 | ND |
| Relative HIBCH activity (WT = 100%) | 0 | > 27.5 | 3 | 3 | 22 | 22 |
| References | [3], [5] | [5] | [4] | [4] | This study | This study |
+, present; −, not present; ND, not described; m, month; y, year; WT, Wild Type.
2.3. Mutation analysis
Whole-exome analysis was performed on 3 family members [patient II-2, the father (I-1), and the mother (I-2)]. Genomic DNA was isolated from peripheral blood leukocytes by standard phenol/chloroform extraction, sheared into approximately 300 bp fragments, and used to create a library for multiplexed, paired-end sequencing (Illumina, San Diego, CA, USA). Targeted sequences were purified, amplified, and sequenced using an Illumina HiSeq2000 platform with paired-end 100 bp configuration. The mapping result was corrected using Picard (ver. 1.73) to remove duplicates and GATK (ver. 1.6-13) for local alignment and quality score recalibration. Annotations of single nucleotide variants (SNVs) and indels were based on dbSNP135, CCDS (NCBI, Nov 2011), RefSeq (UCSC Genome Browser, Nov 2011), Encode (UCSC Genome Browser, ver. 7), and 1000Genomes (Oct 2011). We filtered the raw data based on the assumption of autosomal recessive inheritance, and that penetrance was 1.0. Variants were further filtered according to the following criteria: with predicted functions of frameshift, nonsense, read-through, missense, deletion, insertion, or insertion-deletion.
To confirm the mutation identified in exon 4 of HIBCH by whole-exome analysis, a DNA segment was amplified from genomic DNA by PCR using the HIBCH-specific primers S1 (sense primer; 5′-gtactgtcacaaacacatatgtat-3′) and A1 (antisense primer; 5′-tactaactgcagaaagtaagtct-3′). The resulting PCR product was purified and then sequenced using the GenomeLab™ GeXP Genetic Analysis System (Beckman Coulter, Fullerton, CA, USA) and the GenomeLab™ Dye Terminator Cycle Sequencing with Quick Start Kit (Beckman Coulter). DNA fragments amplified with S1 and A2 (5′-ctttacctctgatatcacacgcg-3′) from family members and controls were subjected to restriction fragment length polymorphism (RFLP) analysis using MluI. The gcg sequence (underlined) in A2 was introduced to generate an MluI recognition site (acgcgt) in the amplified fragments from the mutant allele.
2.4. Construction of wild-type and mutant HIBCH cDNA expression vectors
A96D HIBCH and wild-type cDNA were amplified from first-strand cDNA of lymphoblastoid cells obtained from patient II-4 and normal controls, respectively, using primers S2 (exon 1; 5′-ttcggaattcttggcgatggggca-3′) and A3 (exon 14; 5′-gcctggatcctcaaaatttcaaatcac-3′). An EcoRI recognition site (gaattc) or a BamHI recognition site (ggatcc) was introduced into S2 or A3, respectively. After the sequences of the PCR products were confirmed, EcoRI/BamHI fragments of wild-type and A96D HIBCH cDNA were subcloned into the EcoRI/BamHI site of the mammalian expression vector p3 × FLAG-CMV (Sigma-Aldrich, St. Louis, MO, USA) to yield pHIBCH and pHIBCH-A96D, respectively. Both pHIBCH and pHIBCH-A96D contained a termination codon (TGA) upstream of the 3 × FLAG-tag sequence. Three mutant HIBCH expression vectors, pHIBCH-Y122C, pHIBCH-G317E, and pHIBCH-K74Lfs*13, which contained p.Y122C, p.G317E, and p.K74Lfs*13 IVS3-9T>G mutation at the intron 3–exon 4 boundary, a frameshift) mutations, respectively,were generated using the in vitro mutagenesis method according to Yamada et al. [8].
2.5. Determination of HIBCH activity in patient lymphoblastoid cells and HEK293 cells transiently expressing mutant HIBCH
HIBCH activity was measured via a coupled enzyme assay with crotonase using methacrylyl-CoA as a substrate, according to the method described by Shimomura et al. [9]. Briefly, cells were solubilized with a solution containing 50 mM potassium phosphate, pH 7.5, 1 mM EDTA, 0.5% (w/v) Triton X-100, 0.1 mM phenylmethanesulfonyl fluoride (PMSF), 10 μM N-tosyl-l-phenylalanine chloromethyl ketone (TPCK), and, 10 μg leupeptin/mL. The assay cocktail (1 mL) contained 100 mM Tris–HCl, pH 8.0, 1 mM EDTA, 0.1% (w/v) Triton X-100, and 0.1 mM 5,5′-dithiobis (2-nitrobenzoic acid) (Sigma-Aldrich), and the reaction was started by mixing the enzyme solution, 0.2 mM methacrylyl-CoA, and crotonase (10 units/mL, Sigma-Aldrich) in a cuvette at 30 °C, and changes in absorbance at 412 nm were monitored for 2 min. One unit of HIBCH activity catalyzes the formation of 1 μmol of CoA per minute. Protein concentration was determined by the Bradford method, using IgG as the standard. The Km values of HIBCH in the cells of patient II-4 and in 3 control lymphoblastoid cells were determined using Lineweaver–Burk plot by measuring HIBCH activities with different substrate concentrations.
HEK293 cells were maintained in Dulbecco's modified Eagle's medium (Sigma-Aldrich) supplemented with 10% heat-inactivated fetal calf serum. The cells were grown to 80–90% confluence in 24-well dishes at 37 °C with 5% carbon dioxide and then cotransfected with 10 ng pCMV-β-gal (an Escherichia coli β-galactosidase expression vector) and 0.8 μg of either a human HIBCH expression vector (pHIBCH, pHIBCH-A96D, pHIBCH-Y122C, pHIBCH-G317E, or pHIBCH-K74Lfs*13) or the control vector (p3 × FLAG-CMV), using the Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA). After 48 h the cells were harvested, washed twice with PBS, and solubilized in 200 μL of ice-cold 50 mM potassium phosphate buffer (pH 7.5) containing 1 mM EDTA, 0.5% (w/v) Triton X-100, 0.1 mM PMSF, 10 μM TPCK, and 10 μg leupeptin/mL.
The efficiency of the DNA transfection was verified by measuring β-galactosidase activity using o-nitrophenyl-β-d-galactopyranoside as the substrate [10]. Lysed cell extracts (containing 15 μg protein each) were electrophoresed on 10% SDS-polyacrylamide gels, transferred onto PVDF membranes (Immobilon-P, Millipore, Billerica, MA, USA), and incubated with a polyclonal antibody specific for HIBCH (at a dilution of 1:1,000), which was raised in rabbit by injection of purified HIBCH of rat liver, at 4 °C overnight. After incubation with a secondary horseradish peroxidase-conjugated anti-rabbit IgG antibody (at a dilution of 1:20,000; Promega, Madison, WI, USA), the signals were visualized with an enhanced chemiluminescence western blotting detection system (GE Healthcare, Tokyo, Japan).
2.6. Quantitative acylcarnitine analysis
Acylcarnitines (ACs) in serum were analyzed using electrospray tandem mass spectrometry (LCMS/MS) (LCMS-8040, Shimadzu, Kyoto, Japan) [11]. Briefly, 150 μL methanol-extracted sample was dried under a nitrogen stream and butyl-derivatized with 50 μL of 3 N n-butanol-HCl at 65 °C for 15 min. The dried butylated sample was dissolved in 100 μL of 80% acetonitrile:water (4:1 v/v). The ACs in 1 μL of the resultant aliquots were analyzed using LCMS/MS and evaluated using LabSolutions LCMS (Shimadzu) and Neonatal software (Shimadzu).
3. Results
3.1. Identification of a novel missense mutation (p.A96D) in HIBCH from the 2 patients
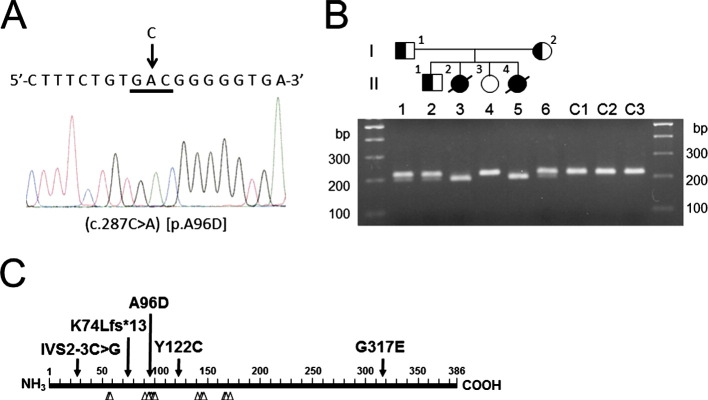
Whole-exome analysis of genomic DNA from a family member revealed a novel missense mutation (c.287C > A, [p.A96D]) in exon 4 of HIBCH on chromosome 2q32.3 (Fig. 2A). A96 is one of the substrate-binding residues in HIBCH and is highly conserved in eukaryotes (e.g., Arabidopsis thaliana, Caenorhabditis elegans, and other higher eukaryotes whose sequence information is available in the NCBI reference protein database; Fig. 2C). The mutation is absent from the NCBI reference assembly SNP database and could not be located using the Human Genetic Variation Browser. The mutation has a maximum predicted pathogenicity score, as determined by Polyphen-2 [12]. Direct sequencing and PCR-RFLP analysis demonstrated that both patients were homozygous for the mutation and that their parents were heterozygous (Fig. 2B). We could not trace the relationship beyond the grandparents of the parents. However, ancestors of the grandparents of the parents lived in the same prefecture of Japan, raising the possibility that the patients and their parents in the present study have inherited the same mutation from a common ancestor.
Fig. 2.
Identification of the mutation in HIBCH. (A) Direct sequence analysis of DNA from patient II-2 revealed a C to A substitution at nucleotide position 287 in exon 4 of HIBCH, resulting in the substitution of alanine (GCC) with aspartic acid (GAC) at codon 96 (c.287C > A, [p.Ala96Asp]), as indicated by the arrow. (B) The pedigree of the family with HIBCH deficiency. Affected individuals are indicated by filled symbols, unaffected individuals by hollow symbols, and carrier individuals by half-filled symbols. PCR-RFLP analysis using MluI-digested PCR products from family members and 3 normal controls (C1, C2, and C3) were run on a 1.5% low-melting agarose gel. The size of the DNA markers is indicated on both sides. (C) Schematic representation of HIBCH. The positions of the mutations identified in the previously reported cases and the patients from this study are indicated by arrows. Predicted substrate-binding residues are indicated by arrowheads. A96 is a substrate-binding residue.
3.2. HIBCH activity was decreased in lymphoblastoid cells from patient II-4 and HEK293 cells transiently expressing mutant HIBCH
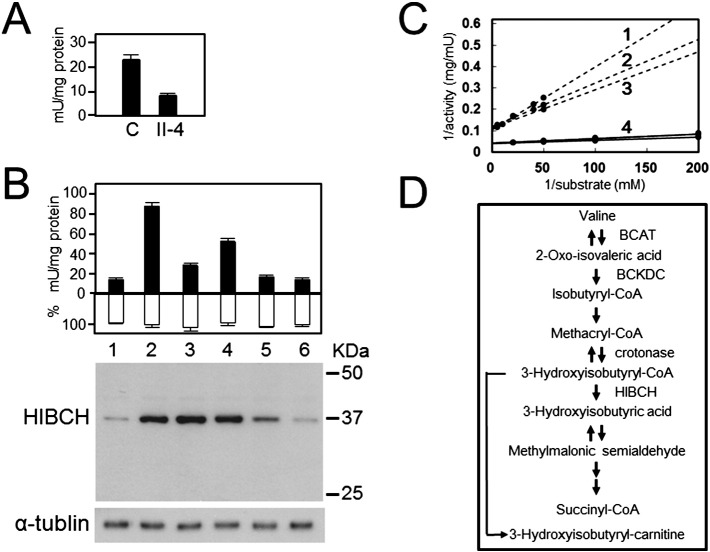
The HIBCH activity in patient II-4 lymphoblastoid cells was decreased to 35% of that of the healthy controls' cells (Fig. 3A). A transfection study showed that transient expression of pHIBCH increased the level of HIBCH activity approximately 5-fold when compared with that in control cells mock transfected with p3 × FLAG-CMV. HIBCH activity in cells transiently expressing the HIBCH mutants p.A96D, p.Y122C, p.G317E, or p.K74Lfs*13 were 22%, 55%, 3%, and 0% of the wild-type HIBCH activity, respectively (Fig. 3B). Western blot analysis demonstrated that the amounts of HIBCH harboring p.A96D and p.Y122C mutations were the same as that of wild type. However, the amount of HIBCH harboring the p.G317E mutation decreased, while HIBCH harboring the frameshift mutation (p.K74Lfs*13) was not detected (Fig. 3B). The mean Km of HIBCH in the cells of patient II-4 and in 3 control lymphoblastoid cells was determined to be 20.1 μM and 3.7 μM, respectively (Fig. 3C).
Fig. 3.
Residual activity and western blot of HIBCH mutants. (A) HIBCH activity in lymphoblastoid cells from patient II-4 and 3 controls (1 U = 1 μmol/min). (B) The black columns indicate mean HIBCH activity, the white columns indicate relative β-gal activity, and vertical bars indicate standard error of the mean (n = 3). Western blot analysis of HEK293 cells transfected with each of the HIBCH-expressing vectors, using antibodies specific for HIBCH (upper panel) or α-tubulin (lower panel). Lane 1, p3 × FLAG; lane 2, wild type; lane 3, p.A96D; lane 4, p.Y122C; lane 5, p.G317E; lane 6, p.K74Lfs*13. (C) Lineweaver–Burk plot. Triplicate measurement of Km of p.A96D HIBCH (No. 1–3) in patient II-4's lymphoblastoid cells gave the following results: 27.7 μM, 17.1 μM, and 15.5 μM (mean ± SD; 20.1 ± 6.6 μM). The Km of HIBCH from 3 control lymphoblastoid cells was obtained as 3.7 ± 0.7 μM (No. 4). (D) Schematic illustration of the valine catabolic pathway.
3.3. Serum hydroxy-C4-carnitine (C4OH) was increased in patient II-2
Serum C4OH levels of patient II-2 in the non-acute phase and the 6 controls (mean ± SD) were 0.17 and 0.023 ± 0.010 nmol/mL, respectively. Thus, the serum concentration of C4OH in II-2 was approximately 7 times higher than those of normal controls.
4. Discussion
HIBCH deficiency is quite a rare disease, and prior to this study, only 4 cases had been reported [3], [4], [5]. In the initial case report by Brown et al. [3], the authors noted that specific characteristics of this disease were caused by the accumulation of methacrylyl-CoA in multiple tissues as the result of an enzyme deficiency in the valine catabolic pathway. It is hypothesized that intramitochondrial methacrylyl-CoA then reacts with thiol compounds (e.g., cysteamine, cysteine and reduced glutathione (GSH)) to form cysteine, cysteamine and glutathione conjugates of methacrylyl-CoA [3] and essential cysteine residues of mitochondrial enzymes, including multiple RC enzymes and PDHc, and reduces their activities [4]. This leads to cell damage, especially in the basal ganglia, by dramatically decreasing the cellular reduction state and reducing ATP production [4].
As the residual HIBCH activities in cultured fibroblasts derived from previously reported patients were below detectable limits, they could not be precisely determined [4], [5]. In this study, we measured HIBCH activity in lymphoblastoid cells, as cultured fibroblasts from patient II-4 were not available. Thus, it was not possible to compare residual HIBCH activities in the 6 reported patients [3], [4], [5]. Therefore, to investigate the clinical–biochemical correlation in this disease, we constructed wild-type and mutant HIBCH expression vectors, transiently expressed them in HEK293 cells, and then determined HIBCH activities (Fig. 3B). The cells expressing p.K74Lfs*13 had no HIBCH activity in the absence of the mature protein (Fig. 3B). Thus, HIBCH harboring p.K74Lfs*13 is nonfunctional. However, only low enzyme activity (~ 3%) was detected in p.G317E when compared with cells expressing wild-type HIBCH (Fig. 3B). p.G317E HIBCH was detected in small amounts probably since the mutation affected the protein structure and stability (Fig. 3B). Patients homozygous for the p.G317E mutations died within 3 years of their birth [4]. The patient who was homozygous for p.K74Lfs*13 died at 3 months of age. He presented dysmorphic features, tetralogy of Fallot, and vertebral abnormalities in addition to features commonly found in HIBCH deficiency (i.e., hypotonia, neurological regression, developmental delay in infancy, episodes of ketoacidosis, and abnormal MRI findings in the basal ganglia) (Table 1) [3]. In contrast, HEK293 cells expressing the p.Y122C HIBCH had relatively high (55%) enzyme activity when compared to cells expressing wild-type HIBCH. The patient harboring the Y122C mutation also had a splicing mutation (IVS2-3C > G) in another allele, and was still alive at 8 years of age [5]. Therefore, HIBCH activity caused by heterozygous Y122C mutation was estimated to be 27.5%, from results of the transfection study. The presented patients (harboring the p.A96D mutation) also showed a relatively high HIBCH activity (22%) and did not present with symptoms of infantile spasms or generalized seizures before 10 months of age, which were observed in the sibling patients with homozygous G317E mutation. Thus, our results suggest a correlation between the level of residual HIBCH activity and severity of the disease, except the acute phase (e.g. infection). The high (approximately 35%) residual enzyme activity in the lymphoblastoid cells of patient II-4 is not common in autosomal recessive disease. The Km of p.A96D HIBCH in the patient's lymphoblastoid cells is approximately 5.4-fold greater than that of wild-type HIBCH (20.1 μM vs 3.7 μM, Fig. 3C). Since we measured the maximal enzyme activity detected under standard conditions (0.2 mM substrate) in vitro, the enzyme activity indicated by the “35% and 22% values” would be much more reduced with low substrate concentration in vivo thus affecting cell metabolism. This suggestion is supported by the results of LCMS/MS analysis, which demonstrated that increased serum hydroxy-C4-carnitine levels in patient II-2 reflected an accumulation of the substrate (3-hydroxyisobutyryl-CoA) and methacrylyl-CoA. The slow and steady accumulation of toxic methacrylyl-CoA in the mitochondria during normal activities of daily life and its rapid and intensive accumulation in energy-compromised states such as exercise or fever could explain the pathogenesis of the presented patients.
Loupatty et al. suggested treating HIBCH deficiency with frequent, carbohydrate-rich meals to avoid the onset of fasting [5]. This treatment may prevent excess ATP production from the catabolism of BCAAs including valine, since the main energy source for the patient would now be carbohydrates. We started administering this therapy with antioxidants (coenzyme Q10, vitamin C and vitamin E) to patient II-4 at 18 months, after the first episode of infection accompanied by a high fever (38 °C). Although the therapy was effective where the regions with abnormal basal ganglia improved concurrently with physical recovery (Fig. 1), the patient died after a second episode of infection. This showed that therapy with frequent, carbohydrate-rich meals and the prescribed antioxidants cannot prevent progression of the disease. The first and rate-controlling step in the catabolism of valine is catalyzed by BCAA aminotransferase (BCAT, Fig. 3D). The enzyme has a very high Km value (2.96 mM) for valine as substrate [13]. Thus, a low-valine diet may result in therapeutically low plasma valine concentrations and the administration of carnitine (to activate the excretion of 3-hydroxyisobutyryl-CoA as 3-hydroxyisobutyryl-carnitine in urine) [5] could reduce the production of methacrylyl-CoA in neuronal cells. N-acetyl-cysteine (NAC), a cell membrane-permeable derivative of cysteine, can cross the blood–brain barrier and replenish depleted neuronal glutathione content [14], [15], [16]. NAC is used for treatment of many diseases related to oxidative stress including neurodegenerative diseases [17]. Therefore, administration of NAC to patients with HIBCH deficiency to increase intramitochondrial GSH should also be considered for suppressing the consumption of GSH caused by increased methacrylyl-CoA, since the pathogenesis of HIBCH deficiency is considered to be via the binding of thiol compounds and essential cysteine residues of mitochondrial enzymes to accumulated methacrylyl-CoA.
Conflicts of interest statements
The authors declare no conflict of interests.
Acknowledgments
We are grateful to the patients and their families for agreeing to participate in this study. This work was supported by the Takeda Science Foundation to N.W.
References
- 1.Rennie M.J. Handbook of Physiology, Rowell LB, Shepherd JT (Eds.), Section 12. Exercise. Regulation and Integration of Multiple Systems. Oxford University Press; New York: 1996. Influence of exercise on protein and amino acid metabolism; pp. 995–1035. [Google Scholar]
- 2.Shimomura Y., Murakami T., Fujitsuka N., Nakai N., Sato Y., Sugiyama S., Shimomura N., Irwin J., Hawes J.W., Harris R.A. Purification and partial characterization of 3-hydroxyisobutyryl-coenzyme A hydrolase of rat liver. J. Biol. Chem. 1994;269:14248–14253. [PubMed] [Google Scholar]
- 3.Brown G.K., Hunt S.M., Scholem R., Fowler K., Grimes A., Mercer J.F., Truscott R.M., Cotton R.G., Rogers J.G., Danks D.M. Beta-hydroxyisobutyryl coenzyme A deacylase deficiency: a defect in valine metabolism associated with physical malformations. Pediatrics. 1982;70:532–538. [PubMed] [Google Scholar]
- 4.Ferdinandusse S., Waterham H.R., Heales S.J., Brown G.K., Hargreaves I.P., Taanman J.W., Gunny R., Abulhoul L., Wanders R.J., Clayton P.T., Leonard J.V., Rahman S. HIBCH mutations can cause Leigh-like disease with combined deficiency of multiple mitochondrial respiratory chain enzymes and pyruvate dehydrogenase. Orphanet J. Rare Dis. 2013;8:188. doi: 10.1186/1750-1172-8-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Loupatty F.J., Clayton P.T., Ruiter J.P., Ofman R., Ijlst L., Brown G.K., Thorburn D.R., Harris R.A., Duran M., Desousa C., Krywawych S., Heales S.J., Wanders R.J. Mutations in the gene encoding 3-hydroxyisobutyryl-CoA hydrolase results in progressive infantile neurodegeneration. Am. J. Hum. Genet. 2007;80:195–199. doi: 10.1086/510725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang W.M., Natowicz M.R. Cerebrospinal fluid lactate and pyruvate concentrations and their ratio. Clin. Biochem. 2013;46:694–697. doi: 10.1016/j.clinbiochem.2012.11.008. [DOI] [PubMed] [Google Scholar]
- 7.Shargorodsky M., Debby O., Matas Z., Zimlichman R. Effect of long-term treatment with antioxidants (vitamin C, vitamin E, coenzyme Q10 and selenium) on arterial compliance, humoral factors and inflammatory markers in patients with multiple cardiovascular risk factors. Nutr. Metab. (Lond.) 2010;7:55. doi: 10.1186/1743-7075-7-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamada K., Takado Y., Kato Y.S., Yamada Y., Ishiguro H., Wakamatsu N. Characterization of the mutant β-subunit of β-hexosaminidase for dimer formation responsible for the adult form of Sandhoff disease with the motor neuron disease phenotype. J. Biochem. 2013;153:111–119. doi: 10.1093/jb/mvs131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shimomura Y., Murakami T., Nakai N., Huang B., Hawes J.W., Harris R.A. 3-Hydroxyisobutyryl-CoA hydrolase. Methods Enzymol. 2000;324:229–240. doi: 10.1016/s0076-6879(00)24235-3. [DOI] [PubMed] [Google Scholar]
- 10.Gotoda Y., Wakamatsu N., Kawai H., Nishida Y., Matsumoto T. Missense and nonsense mutations in the lysosomal α-mannosidase gene (MANB) in severe and mild forms of α-mannosidosis. Am. J. Hum. Genet. 1998;63:1015–1024. doi: 10.1086/302048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Azzari C., la Marca G., Resti M. Neonatal screening for severe combined immunodeficiency caused by an adenosine deaminase defect: a reliable and inexpensive method using tandem mass spectrometry. J. Allergy Clin. Immunol. 2011;127:1394–1399. doi: 10.1016/j.jaci.2011.03.040. [DOI] [PubMed] [Google Scholar]
- 12.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schadewaldt P. Analysis of (S)- and (R)-3-methyl-2-oxopentanoate enantiomorphs in body fluids. Methods Enzymol. 2000;324:33–39. doi: 10.1016/s0076-6879(00)24216-x. [DOI] [PubMed] [Google Scholar]
- 14.Aoyama K., Suh S.W., Hamby A.M., Liu L., Chan W.Y., Chen Y., Swanson R.A. Neuronal glutathione deficiency and age-dependent neurodegeneration in the EAAC1 deficient mouse. Nat. Neurosci. 2006;9:119–126. doi: 10.1038/nn1609. [DOI] [PubMed] [Google Scholar]
- 15.Farr S.A., Poon H.F., Dogrukol-Ak D., Drake J., Banks W.A., Eyerman E., Butterfield D.A., Morley J.E. The antioxidants alpha-lipoic acid and N-acetylcysteine reverse memory impairment and brain oxidative stress in aged SAMP8 mice. J. Neurochem. 2003;84:1173–1183. doi: 10.1046/j.1471-4159.2003.01580.x. [DOI] [PubMed] [Google Scholar]
- 16.Dringen R., Hamprecht B. N-acetylcysteine, but not methionine or 2-oxothiazolidine-4-carboxylate, serves as cysteine donor for the synthesis of glutathione in cultured neurons derived from embryonal rat brain. Neurosci. Lett. 1999;259:79–82. doi: 10.1016/s0304-3940(98)00894-5. [DOI] [PubMed] [Google Scholar]
- 17.Dodd S., Dean O., Copolov D.L., Malhi G.S., Berk M. N-acetylcysteine for antioxidant therapy: pharmacology and clinical utility. Expert. Opin. Biol. Ther. 2008;8:1955–1962. doi: 10.1517/14728220802517901. [DOI] [PubMed] [Google Scholar]