Abstract
Clinical microbiology and public health laboratories are beginning to utilize next-generation sequencing (NGS) for a range of applications. This technology has the potential to transform the field by providing approaches that will complement, or even replace, many conventional laboratory tests. While the benefits of NGS are significant, the complexities of these assays require an evolving set of standards to ensure testing quality. Regulatory and accreditation requirements, professional guidelines, and best practices that help ensure the quality of NGS-based tests are emerging. This review highlights currently available standards and guidelines for the implementation of NGS in the clinical and public health laboratory setting, and it includes considerations for NGS test validation, quality control procedures, proficiency testing, and reference materials.
INTRODUCTION
Next-generation sequencing (NGS) is transforming the landscape of clinical microbiology and public health laboratories. The applications of NGS are wide-ranging and include whole-genome sequencing, microbiome analysis/metagenomics, transcriptome profiling, infectious disease diagnosis, pathogen discovery, and public health surveillance. For example, NGS has recently been used to better understand hospital outbreaks and inform infection control practices (1), and it can be used in the clinical microbiology laboratory to identify unknown organisms, predict antimicrobial resistance, assess virulence gene content, and inform molecular epidemiology efforts (2). Metagenomic “unbiased” NGS applications, coupled with recently developed bioinformatics solutions (3–5) that enable the identification of all pathogens directly from a clinical sample based on sequence homology, have the potential to complement or even replace current standard clinical laboratory tests. For example, the use of metagenomics combined with a rapid bioinformatics pipeline recently facilitated a clinically actionable diagnosis of neuroleptospirosis when conventional testing was initially unable to identify the causative organism (6). A number of agencies are working to bring NGS into the public health laboratory setting. For example, through the U.S. Centers for Disease Control and Prevention (CDC) Advanced Molecular Detection (AMD) Initiative, national, state, and local partners are beginning to incorporate NGS-based methods into disease surveillance systems. AMD initiatives include broad applications of NGS to address public health problems, including vaccine improvement, identification of emerging threats, and tracking diseases and outbreaks (http://www.cdc.gov/amd/). The CDC, Food and Drug Administration (FDA), National Institutes of Health (NIH), National Center for Biotechnology Information (NCBI), National Library of Medicine, and the U.S. Department of Agriculture/Food Safety and Inspection Service (USDA/FSIS) have established an Interagency Collaboration on Genomics and Food Safety (Gen-FS), with the goal of fostering timely access to genomic data for foodborne pathogen surveillance and outbreak response (http://www.cdc.gov/oid/docs/bsc_oid_fsma_surv_wg_2015_annual_report.pdf). Both the emergence of affordable and user-friendly benchtop sequencers and the resources and funding made available through federal initiatives have helped transition NGS into public health laboratories. A 2014 survey conducted by the Association of Public Health Laboratories (APHL) revealed that public health laboratories are embracing the adoption of NGS technologies, especially for foodborne pathogen surveillance activities (7). The APHL survey also indicated that the use of NGS in public health laboratories is expected to expand and be applied to an increasing diversity of public health investigations and applications (7). For example, the New York State Department of Health is now using whole-genome sequencing to track the emergence of drug resistance for influenza virus (8).
Several issues and capacity gaps were identified by the 2014 APHL survey, including the need to identify public health laboratory NGS applications beyond sequencing of foodborne pathogens, the development and support of information technology (IT) infrastructure, and the need for training of public health laboratorians (7). Additional issues have been identified that should be addressed to fully realize the integration of NGS into the clinical and public health laboratory setting. These include reducing the cost and turnaround time of sequencing, the development of fully automated user-friendly sequencing and data analysis pipelines, the creation of comprehensive and well-curated reference genome databases, curation of genotype-phenotype correlations for clinically relevant microorganisms (for example, when making predictions about antimicrobial resistance), establishment of proficiency testing (PT) and quality control (QC) measures, and the development of practice guidelines to ensure the quality of NGS-based tests (7, 9–12). This review highlights currently available standards and guidelines for the implementation of NGS in the clinical and public health laboratory setting, and it includes considerations for NGS test validation, QC procedures, PT, and reference materials.
NGS WORKFLOW
NGS is a term used to represent different technologies that enable massively parallel sequencing of clonally amplified or single DNA molecules. High-throughput sequencing approaches have been commercially available for over a decade, and the technologies continue to evolve and improve (13). The various commercially available platforms differ in the chemistries used, read lengths, and throughput capabilities and can be divided into short-read and long-read sequencing technologies (9). While short-read technologies (e.g., read lengths in the hundreds of bases) offer a lower per-base cost of sequencing, they are challenged with creating finished high-quality genomes because longer reads are needed to fill in sequence gaps. The emergence of long-read sequencing platforms, which can produce reads tens of kilobases in length, allows for the finishing of microbial genomes for well under $1,000 per genome (14). Short-read technologies are sufficient for microbial genomic analyses, including strain typing, outbreak tracing, and pangenome surveys. However, studies that investigate structural variants (e.g., genome rearrangements, duplications, or deletions) or interspersed repeats (particularly insertion sequences) are limited when short-read technologies are used, because these large gaps or repeated regions can be difficult or impossible to resolve without the use of a long-read technology.
Despite the differences in NGS technologies, the sequencing workflows of most NGS platforms are conceptually similar and are made up of both wet-lab (i.e., sample processing steps) and dry-lab (i.e., the data analysis performed using a bioinformatics pipeline) steps (Fig. 1). The wet-lab process steps may include DNA extraction and quantification (or for RNA viral sequencing or microbial transcriptome profiling studies, RNA-to-cDNA conversion by reverse transcription), followed by a library preparation step, where the DNA is fragmented, and adaptors are added to each fragment and amplified prior to sequence generation (15). The dry-lab steps are composed of commercial and/or laboratory-developed custom software tools and scripts that are assembled to create a bioinformatics pipeline used to perform the sequence analysis steps.
FIG 1.
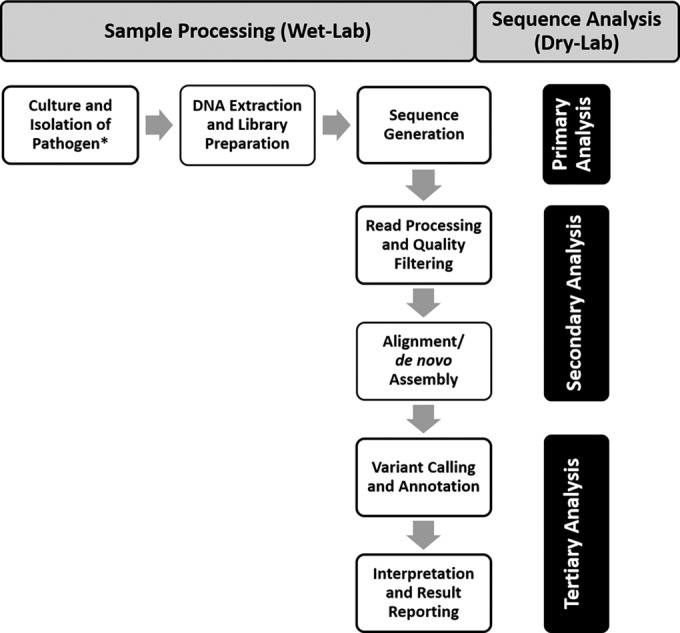
General NGS workflow. NGS workflows contain both wet-lab (and sample processing steps) and dry-lab (bioinformatics pipeline) steps. Sequence generation (primary analysis) occurs on the instrument and is the process of taking images or signals from the instrument and converting them into base calls that are assigned quality scores. During secondary analysis, primary sequence data are further processed and assessed for quality before either alignment to a reference sequence or de novo assembly is performed. During tertiary analysis, results are interpreted, clinically significant findings are identified, and a final report is generated. These workflow steps will vary depending on platform and application-specific requirements. Asterisk indicates metagenomics, or unbiased sequencing applications, do not require culture or isolation steps.
The bioinformatics workflow can be considered in terms of primary, secondary, and tertiary analyses (Fig. 1) (16). Primary analysis is the process of converting the images or signals from the instrument into base calls that are assigned quality scores. These quality scores describe the probability that a base has been correctly assigned. During secondary analysis, primary sequence data are further processed and assessed for quality, trimmed, and filtered based on laboratory-established quality thresholds. The sequence reads are either aligned to a reference sequence, or in the absence of a reference, assembled to create a full-length sequence using a process referred to as de novo assembly. Tertiary analysis is the stage when results are interpreted, clinically significant findings are identified, and a final report is generated. The tertiary analyses steps may include pathogen identification, variant calling, functional annotation, taxonomic classification, etc.
The primary, secondary, and tertiary analysis steps involve substantial automated informatics components, which is a significant change in operations for many clinical and public health laboratories. For example, the majority of laboratories do not routinely establish or maintain large computational servers and databases that are necessary for NGS applications (11). Similarly, the use of bioinformatics tools for the analysis and interpretation of NGS data is not a routine skill set of most clinical and public health laboratorians. Education of the workforce will be critical for the successful adoption of NGS into the clinical and public health laboratory setting (11, 12). There are a variety of commercially available software packages that contain complete bioinformatics workflows that are optimized for particular NGS applications, have a user-friendly graphical interface, and are designed for use by biologists without requiring knowledge of programming and scripting languages (e.g., CLC Genomics Workbench, Geneious, Bionumerics, Galaxy [https://galaxyproject.org/], and Illumina BaseSpace). These resources will help make NGS techniques more accessible to laboratory personnel lacking bioinformatics expertise.
STANDARDS AND GUIDELINES FOR IMPLEMENTATION OF CLINICAL NGS
Standards to ensure the reliability of NGS-based test results, guidance for the application of regulatory requirements, and professional standards for human genetic analysis have been introduced. In 2012, the CDC's Next-Generation Sequencing: Standardization of Clinical Testing (Nex-StoCT) working group published the first consensus guidance document that presented recommendations for NGS test system validation, QC procedures, reference material, and PT mainly for human genetic testing. That same year, the College of American Pathologists (CAP) published a checklist specific to NGS as part of the molecular pathology checklist for accrediting clinical laboratories for human testing (17). The CAP NGS checklist outlines requirements for documentation, validation, QC, and quality monitoring for both the sequencing (wet-lab) work and bioinformatics (dry-lab) steps. This checklist will provide the basis for a new checklist that is currently in development for infectious disease applications of NGS (9). In 2013, the American College for Medical Genetics and Genomics (ACMG) published detailed clinical laboratory standards for NGS as applied to human genetic testing (18). Good laboratory practice guidelines that focus on considerations for the development and implementation of a clinical bioinformatics pipeline for human testing were also published by the CDC's Nex-StoCT II working group (16). While the currently available guidelines cover various aspects and applications of NGS and may target different audiences, there is a general consensus on the recommendations presented across these publications. This is largely due to the fact that there was intentional overlap in the participants involved in developing the guidance documents.
Although the aforementioned initiatives were primarily directed to applications of NGS for human genetic testing of heritable diseases, many of the resulting recommendations and CAP checklist items are applicable to clinical microbiology and public health NGS applications. This commonality is largely due to the shared process steps in the NGS workflow and common certification and accreditation requirements. As such, these early guidance documents have been seminal in the development of an overall validation framework for clinical NGS and in the establishment of good laboratory practices for meeting regulatory and professional requirements. However, there are key differences between human genome sequencing and NGS-based approaches to infectious disease testing. One fundamental difference is the critical role of comprehensive reference databases for reference-based microbial sequencing methods, which is arguably less important for human NGS applications, given the availability of the human genome reference assembly. In addition, variant calling is essential for human NGS-based tests and is required in comparative genomic infectious disease applications (e.g., use of genomic variants for phylogenetic analysis, comparative genomics, or outbreak investigations); however, variant calling is not necessarily important for all infectious disease methods, e.g., pathogen detection. With the large variety of NGS-based infectious disease applications, assay-specific considerations will be required to ensure the quality of these diverse testing approaches. There have been a number of reports and reviews that highlight the diversity of NGS-based approaches that have the potential for use in clinical and public health laboratory settings (9, 11, 12, 19, 20). Recently, guidance documents that include specific considerations for the use of NGS for infectious disease testing applications have become available, including the 2014 update of the Clinical and Laboratory Standards Institute (CLSI) MM-09 document “Nucleic acid sequencing methods in diagnostic laboratory medicine” (21), as well as the publication of a validation framework for NGS and microbial forensics applications (20). In the United Kingdom, the PHG Foundation released a 2015 report that presents a “road map” with over 30 recommendations to help achieve patient and population benefits from pathogen genomics (22). The American Academy of Microbiology recently published the outcomes of a colloquium composed of subject matter experts tasked with defining the specific challenges and establishing recommendations for the transition of NGS from research to the clinical and public health laboratory setting (12). In 2016, the FDA issued a draft guidance with recommendations for the establishment of analytical and clinical performance characteristics for NGS-based diagnostic devices for microbial identification and the detection of antimicrobial resistance and virulence markers (23). The following sections will review the current paradigm of NGS test validation, including considerations for QC procedures, proficiency testing, and reference materials.
CONSIDERATIONS FOR NGS ASSAY VALIDATION AND QUALITY CONTROL
The majority of NGS-based assays currently used in the clinical setting for patient testing are laboratory-developed tests (LDTs). LDTs are defined as in vitro diagnostic tests that are developed by, manufactured by, and used within a single laboratory (http://www.fda.gov/MedicalDevices/ProductsandMedicalProcedures/InVitroDiagnostics/ucm407296.htm). The FDA released its draft guidance on the regulation of LDTs in October of 2014 (24); however, a final decision and specific details regarding the FDA's approach to the regulation of LDTs has yet to be established. In the United States, clinical laboratory tests, including those that make use of LDTs, are subject to the Clinical Laboratory Improvement Amendments (CLIA) regulations, which require laboratories to establish analytical performance specifications for certain performance characteristics to ensure the analytical validity of test results prior to patient testing (25). This process is commonly referred to as assay validation. Currently, there are three NGS instruments that are FDA cleared, including the class II exempt MiSeqDx, along with its associated reagent kit (Illumina, San Diego, CA), the Ion PGM Dx platform (Life Technologies, Carlsbad, CA), and the Sentosa SQ301 (Vela Diagnostics, Fairfield, NJ), which were registered, listed, and can now be marketed under the same regulation (http://www.fda.gov/NewsEvents/Newsroom/PressAnnouncements/ucm375742.htm, http://www.accessdata.fda.gov/scripts/cdrh/cfdocs/cfRL/rl.cfm?lid=427645&lpcd=PFF, and http://www.accessdata.fda.gov/scripts/cdrh/cfdocs/cfRL/rl.cfm?lid=430009&lpcd=PFF, respectively). The use of these instruments still requires the establishment of laboratory-developed components (e.g., development of the bioinformatics pipeline) specific for the test's intended clinical application. Therefore, a test developed using the cleared instrument would still be considered an LDT and would still require validation to establish performance under CLIA. In 2013, two NGS-based diagnostic tests for cystic fibrosis, in which no component is laboratory developed, were FDA cleared. For these tests, under CLIA, clinical laboratories are not required to perform a validation but must verify the ability to meet the performance specifications established by the manufacturer.
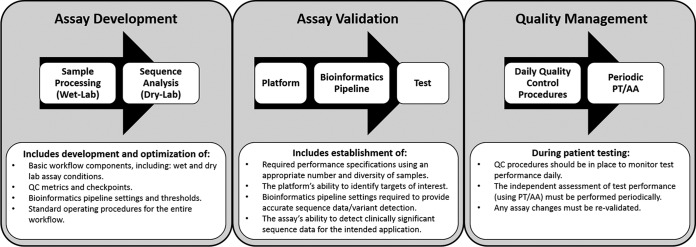
During the validation process, the laboratory must establish the following performance characteristics: accuracy, precision, analytic sensitivity, analytic specificity, reportable range, reference range, and any other characteristics that are necessary to define test performance (26). The definitions traditionally used for these performance characteristics, as described in the CLIA regulations, were originally developed for quantitative single-analyte tests, such as quantitation of blood glucose. Establishment of these characteristics for an NGS assay that is capable of detecting a potentially unlimited number of targets can be challenging. Instead, a method-based validation that can identify and reduce potential sources of error has been suggested as an alternative approach (11). Several groups have recently provided updated definitions and guidance for establishment of these analytical performance characteristics as applied to NGS (9, 20, 21, 26, 27); these are summarized in Table 1. These and other proposed frameworks for clinical NGS implementation divide the analytical validation process into three phases: test development, assay validation, and quality management (18, 26) (Fig. 2).
TABLE 1.
Definitions and guidance for establishment of analytical performance characteristics as applied to NGS
| Performance characteristic | Definition(s) as applied to NGS | Guidance for implementation of NGSa |
|---|---|---|
| Accuracy | Degree or closeness of agreement between material measured (e.g., nucleic acid sequences derived from the assay), and material's true value (e.g., a reference sequence) (20, 21, 26). | Reference sequences used can be derived from samples with well-characterized genomic DNA, synthetic DNA, or reference data sets (26); accuracy may be calculated by: (TP + TN)/(TP + FP + FN + TN) (20). |
| Precision | Degree to which a repeated measurement (e.g., sequence analyses) gives same result: repeatability (within-run precision) and reproducibility (between-run precision) (21, 26). | Repeatability (within-run precision): degree to which the same result(s) is obtained in sequencing the same sample many times under the same conditions (i.e., sequencing of same samples by same operator and/or detection instrument in replicates within a run) (9, 20, 21). |
| Precision is typically expressed numerically by measures of imprecision, including standard deviation, variance, or coefficient of variation under specified measurement conditions (21). | Reproducibility (between-run precision): degree to which the same result(s) is obtained for a sample when sequencing is performed by multiple operators, with multiple lots of reagents, on more than one instrument, and if applicable, at multiple sites/testing locations (i.e., sequencing same samples between/among different operators, on different runs, and/or detection instruments) (9, 20, 21). | |
| Additional considerations: if only a limited no. of samples can be sequenced and compared, other parameters (e.g., avg depth of coverage) may be useful for establishing repeatability and reproducibility (21); precision can also be assessed for variances that may occur during the library preparation process (i.e., between-library precision) by sequencing different library preparations of the same samples on the same sequencing run (9). | ||
| Analytic sensitivity | Likelihood that the assay will detect a target (e.g., variant[s], targeted regions, functional elements, etc.), if present; can include target attribution when defined as strain- or isolate-level detection (20, 26). The true-positive rate is a useful measurement for the sensitivity of sequencing assays (i.e., dividing the no. of true positives by the sum of true positive and false negatives: TP/[TP + FN]) (20, 27). | Assay's LOD is associated with analytical sensitivity. Establishment of the LOD is critical for sequencing assays used to detect the presence of low-level variants or sequences (e.g., viral quasispecies, mixed populations, metagenomics approaches, etc.) (27). For NGS, the LOD can be defined as the minimum amount of input material proportional to the total material available for which all replicates are consistently positive for a defined sequence target (20). |
| Recommendation for microbial variant detection: use of mixtures of strains with known variants and wild-type strains at different percentages and at low, medium, and high levels (e.g., viral loads) (9). | ||
| Recommendation for microbial identification: use of serial dilutions of a known pathogen(s) in a clinically relevant matrix to establish the minimum coverage needed to detect the pathogen(s) (9). | ||
| Analytic specificity | Probability that the assay will not detect a target (e.g., targeted sequence region or variant) when that target is not present in the sample (20, 21, 26); the false-positive rate is a useful measurement for the specificity of sequencing assays (i.e., dividing true negatives by the sum of true negatives plus false positives: TN/[TN + FP]) (20, 21, 26). | Establishment of specificity should include considerations for interfering substances that may be found in the sample (21, 27). |
| It may be impractical to calculate specificity for sequencing approaches designed to detect any and all potential pathogens present in a sample (e.g., unbiased sequencing/metagenomics) (20). | ||
| Recommendation for microbial variant detection and microbial identification: estimation of the false-positive rate should be evaluated at various read depths (20). | ||
| Reportable range | Region(s) of the sequenced genome(s) for which sequence of an acceptable quality can be derived by the laboratory test (21, 26). | Reportable range is not traditionally applicable to qualitative assays; however, this parameter can be interpreted to describe the regions of the genome (e.g., genes, and/or targeted regions) that are sequenced and included in the analysis (27) or from which information is drawn for comparison or attribution. |
| Reference range/intervals | Reportable sequence variants or targeted regions that the assay can detect and are expected to occur in a reference population (normal values) (21, 26). | Reference range is not traditionally applicable to qualitative assays; however, this parameter can be interpreted to describe the types of sequence variants that can occur at a genomic region/position in a reference population (e.g., single-nucleotide variants, insertions or deletions, or other structural variant) (21) and can also describe the reference sequence(s) used for analysis and interpretation of results (27). |
TP, true positive; TN, true negative; FP, false positive; FN, false negative; LOD, limit of detection.
FIG 2.
Assay validation framework. Proposed frameworks for clinical NGS implementation divide the analytical validation process into three phases: test development, assay validation, and quality management (18, 26). This figure was adapted in part from frameworks that were previously described (18, 26). SOP, standard operating procedure; QC, quality control; PT, proficiency testing; AA, alternate assessment.
The test development phase involves iterative cycles of testing until all assay conditions and bioinformatics pipeline settings are optimized and a standard operating procedure for the entire workflow is established. The formal assay validation is the phase when required assay performance specifications (e.g., accuracy, precision, etc.) are established using an appropriate number and diversity of sample types (e.g., representative pathogen types in clinical matrices of interest) and assay conditions (e.g., different operators) to demonstrate that the assay can accurately identify the sequence information the test is designed to detect (e.g., identification of a pathogen, variant calling, etc.). During validation, it is important to establish appropriate QC procedures for the entire testing process, including both the wet-lab and the dry-lab components (18, 26) (Fig. 1).
Quality control procedures monitor whether each component of an assay functions properly and delivers accurate results. The QC procedures should be designed to confirm that the previously established performance specifications are met for each run of a patient sample, and if a change occurs, it may be an indication of an error in the testing process. Examples of QC metrics useful for monitoring NGS tests performance include DNA quality and quantity, quality scores, depth and uniformity of read coverage, GC bias, strand bias, along with a variety of other application-specific metrics for the data processing and analysis steps (18–20, 26). Use of these QC parameters can help ensure that no sample or sequence data move forward in the testing process without meeting the laboratory-established minimum quality standards. Quality assurance procedures, such as use of confirmatory testing with a separate clinically validated method (e.g., an orthogonal or gold standard method) may be necessary to reduce the risk of errors or to exclude the possibility of contamination (11, 18, 26). This is of particular importance when the assay's analytic false-positive rate is high or not yet well established and for assays intended for pathogen discovery or clinical detection of unusual or unexpected agents (11, 18). In some cases, the high discriminatory power of NGS can result in assays that are more sensitive than other tests, and there may be no orthogonal or gold standard method to confirm the results. In these instances, other methods may be used, such as seeking independent replicates across different CLIA laboratories using similar or different NGS technologies. CLIA precludes the use of research-based PCR or other types of testing as the orthogonal method.
Following validation, the test should be considered “locked down” and cannot be changed. Any changes to the assay, such as changes in instrumentation, specimen types, reagents, and/or sample preparation kit, software updates, or other modifications, require that performance specifications be reestablished or otherwise shown to be unchanged by a validation study (26). The extent of revalidation will depend on the extent of the change. For example, changes that do not affect the test process, such as replacement of a depleted reagent, will likely not require a revalidation, only confirmation that the established performance specifications are not altered by the change. For a more extensive change, such as the inclusion of new targets to an existing gene panel or an update to the bioinformatics pipeline, a broader revalidation will be necessary to ensure the capability to detect new sequence targets without compromising the quality of the original assay.
NGS platforms, software, reference databases, and bioinformatics pipelines are continuously evolving and updated frequently. These changes will present a challenge for clinical laboratories that are required to maintain a validated assay. In some instances, it may only be necessary to reestablish performance specifications at or after certain steps in the process, depending on what the change has affected. For example, if only the bioinformatics pipeline is altered, it may not be necessary to revalidate the wet-lab process steps. A particular challenge when using Web-based software tools and databases for sequence analysis is that these resources are frequently updated, and these updates are not always announced or obvious. If using Web-based tools that are not archived and versioned online, it has been recommended that clinical laboratories consider bringing the software tools in-house so that modifications can be versioned, documented, and referenced for each test that is performed, as well as to ensure that clinical laboratories can reproduce results (16).
CONSIDERATIONS FOR NGS REFERENCE MATERIALS AND PROFICIENCY TESTING
Reference materials (RMs) represent a variety of material types, including certified or standard reference materials, quality control materials, and calibrators (28). RMs are essential for the evaluation of both the wet- and dry-lab NGS process steps. They are also used for test development and validation, as QC materials, and for PT. It is recommended that RMs resemble patient specimens as closely as possible (29). For example, well-characterized biological reference organisms (e.g., bacterial strains) can be spiked into appropriate clinical matrices to assess each step in the NGS testing process from DNA extraction to data analysis. Extracted genomic DNA, along with corresponding well-characterized reference sequence data, is also a useful RM, as it can be incorporated into QC procedures; however, this material would not measure the success of DNA extraction. There is a need to develop RMs for the variety of pathogenic organisms that are relevant to the public health and the clinical laboratory setting. To address these needs, the National Institute of Standards and Technology (NIST) is currently working to develop RMs for bacterial genomic sequencing. The strains chosen are relevant to food safety and clinical microbiology NGS applications and represent diverse genome sizes, plasmid contents, and GC contents (10). Other RMs that may be useful for NGS for validation and evaluation of the bioinformatics pipeline include synthetic DNA samples, such as plasmids containing known variants that can be engineered to represent a broad range of sequences and variant types or synthetic “armored RNA” capsids that can be used to simulate infectious viral particles spiked into clinical matrices; and electronic or digital reference materials, such as curated benchmark data sets (i.e., well-characterized and complete genome data sets derived from a variety of organisms relevant to clinical and public health microbiology) (10, 11, 16, 30).
Clinical laboratories are required to demonstrate the independent assessment of test performance through proficiency testing. This can be achieved by participation in formal PT programs, in which blinded samples are provided to a laboratory that performs testing and the results are used for the assessment of interlaboratory performance. If a formal PT program is not available for a particular test, alternate assessment activities, for example, a sample exchange with a laboratory performing similar tests, can be utilized (31, 32). Due to the extremely large variety of possible target sequences, use of a methods-based evaluation of interlaboratory performance, rather than traditional analyte-specific PT, has been recommended for clinical NGS applications (26, 31, 33). The CAP currently offers methods-based PT challenges for germ line and somatic variants for human molecular genetic testing. These CAP PT challenges were designed to be applicable to laboratories that use a variety of sequencing platforms and test applications, with plans to further emphasize bioinformatics-based challenges moving forward (33). It is anticipated that the lessons learned from these PT challenges will help inform the development of CAP PT challenges for NGS-based assays for infectious diseases (http://www.captodayonline.com/3-new-ngs-surveys-cap-2016-pt-launchpad/). The European Molecular Genetics Quality Network also offers a NGS pilot sequencing and dry-lab schemes for human genetic testing (http://www.emqn.org/emqn/Home). Following a survey to guide the development of PT for bacterial whole-genome sequencing (34), the Global Microbial Identifier (GMI) launched their 2015 PT for NGS (http://www.globalmicrobialidentifier.org/Workgroups/About-the-GMI-Proficiency-Test-2015). The test, for which enrollment closed in November 2015, focused on Salmonella enterica, Escherichia coli, and Staphylococcus aureus. Three types of testing material were provided to participants for analysis: 6 bacterial cultures (two Salmonella enterica, two Escherichia coli, and two Staphylococcus aureus strains), prepared DNA from the same bacterial strains, and whole-genome sequence data sets from each of the strains. The PT contained three optional components that included an assessment of the laboratory's DNA preparation and sequencing procedures, the sequencing output, and the procedures used to identify variants and distinguish samples based on those variants. The overarching goals of this PT challenge are to better understand how to quantify differences among laboratories that perform whole-genome sequencing, assess the reliability of results, and improve NGS data that are uploaded to databases. A report summarizing the results of the GMI PT challenge will be published following completion of the challenge. The GMI PT challenge is limited to bacterial whole-genome sequencing, and the results will not directly apply to other applications (e.g., viral sequencing, metagenomics, infectious disease diagnostics, etc.); however, the challenge is an important step toward the development of PT for NGS-based testing in clinical and public health laboratories.
NEED FOR ADDITIONAL STANDARDS AND GUIDANCE FOR CLINICAL MICROBIOLOGY AND PUBLIC HEALTH NGS
There have been a variety of activities focused on the development of resources, reference materials, and guidance for the transition of NGS into the clinical laboratory (17, 18, 26, 33). While the majority of these efforts have focused on human molecular genetic testing applications, many of the principles and guidelines that have been developed are also applicable to clinical microbiology and public health laboratories. One example of this overlap is the variant-calling process. Comparative microbial genomic applications (e.g., identification of genomic variants for phylogenetic analysis, comparative genomics, or outbreak investigations), like human genetic testing, rely on variant calling for the elucidation of nucleotide-level organismal differences (10).
Standardized methods for performance evaluation and reporting of variants are critical due to the various potential sources for error in the sequencing and variant-calling processes, as well as the need for consistency between laboratories. Best practice guidelines for variant-calling methods for human and microbial genomics are available (10, 16). Many of the recommendations and best practices to optimize the quality of the data used to generate variant calls are shared between human and microbial genomic NGS assays, including the recommendation to minimize amplification steps in library preparation (when applicable), use of paired-end sequencing, removal of duplicate reads, realignment around insertions and deletions, and recalibration of base quality scores (10, 16).
Despite similarities to human testing for inherited disorders or cancer, there are unique challenges for the application of NGS to clinical infectious disease testing. For example, the use of de novo genome assembly, which is commonly used for microbial analysis, does not require a reference sequence or knowledge of the sample's sequence prior to NGS. However, similar to human testing, microbial identification, gene prediction, and variant analysis still require the use of a reference database. The development of public curated reference databases is required for successful pathogen identification and discovery of novel variants and genes (11, 12). The database should include accurate annotations and high-quality reference sequences from relevant organisms (e.g., bacteria, viruses, fungi, yeasts, and parasites) that provide a true diversity of strains, including both current circulating organisms and older strains (11, 12). Reference databases, by their very nature, are heavily biased toward commonly sequenced organisms, and smaller databases with more limited entries are even more so. Biases and incompleteness in reference databases are challenges for moving the field forward. For example, there are limited databases containing sequences for clinically important fungi, yeast, and parasite species (12). It can be difficult to avoid bias, given that many organisms are rare or uncommon, or there is a public health focus on certain high-priority agents (e.g., Ebola virus, Zika virus, Listeria, Salmonella, influenza virus, etc.). Efforts are under way to address these issues, for example, the Reference Sequence (RefSeq) database (http://www.ncbi.nlm.nih.gov/refseq/), maintained and curated at the NCBI, is a collection of taxonomically diverse, nonredundant, and well-annotated genomic, transcript, and protein sequence records constructed from sequence data from the International Nucleotide Sequence Database Collaboration (INSDC). The RefSeq project represents sequences from more than 55,000 organisms, including viruses, prokaryotes, and eukaryotes, with efforts to further expand the taxonomic diversity of the collection (35). For infectious disease NGS-based diagnostic devices, the FDA, in collaboration with various federal agencies, has established the publically available FDA-ARGOS database (FDA dAtabase for Regulatory-Grade micrObial Sequences) that contains a set of validated regulatory-grade microbial genomic sequences that are intended to cover the diversity of circulating strains, including clinically and environmentally important microbes (http://www.ncbi.nlm.nih.gov/bioproject/231221). The database, which is still growing, is anticipated to be used by both diagnostic test manufacturers for assay development and by the FDA to support the regulatory review of NGS-based diagnostic devices (e.g., the use of the FDA-ARGOS sequences as an alternative comparator for clinical evaluation).
CONCLUSIONS
In 2015, the American Academy of Microbiology held a colloquium to begin defining the specific challenges and establishing recommendations for the transition of NGS from research to the clinical and public health laboratory setting. One of the recommendations from the colloquium report is that clinical microbiologists and other relevant stakeholders should work with representatives from organizations, including the FDA, Centers for Medicare and Medicaid Services (CMS), NIH, CDC, and CAP, to develop specific guidelines for the validation of NGS-based diagnostic assays (12, 30). There is a need for pathogen-specific guidance for the validation and QC procedures unique to the variety of etiological agents that can be detected using NGS (e.g., bacteria, viruses, fungi, yeasts, and parasites). Likewise, the diversity of NGS applications in the clinical and public health microbiology laboratory (e.g., surveillance, genotypic antimicrobial resistance prediction, direct detection of unknown disease-associated pathogens in clinical specimens, investigation of microbial population diversity in the human host, and comparative genomics approaches, like strain typing) require unique considerations for assuring the quality of sequence results (9–11).
NGS technologies have been commercially available for over a decade, and microbial genomes can be fully sequenced in hours for pennies per base; however, a widespread clinical diagnostic role for NGS has yet to be realized. No clinical microbiology NGS tests have been approved by the FDA, and the limited number of clinical infectious disease NGS-based assays currently offered are being performed as LDTs. The FDA is beginning to evaluate the critical components for the clearance/approval of infectious disease NGS-based assays, particularly for pathogen identification and detection of antimicrobial resistance markers, and it has published both a discussion paper and draft guidance describing the current considerations for the approaches for approval/clearance of NGS diagnostic devices for clinical microbiology (23, 30, 36).
In the absence of FDA-approved/cleared assays, clinical laboratories must validate their assays in-house. Performing a CLIA validation for microbial NGS-based tests is complex, and the establishment of validation standards and guidelines will help with the transition of NGS tests into clinical laboratories. Clinical microbiology laboratories need user-friendly bioinformatics tools, sufficient training, comprehensive curated microbial databases, and standard reference materials, like those being developed by the NIST, for test development, validation, and for QC and PT procedures used to establish and monitor test quality. The development of these standards and tools will require the collaboration of a multidisciplinary team, including laboratories, clinicians, manufacturers of platforms and reagents, software developers, professional organizations, and state and government agencies.
ACKNOWLEDGMENTS
The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention/The Agency for Toxic Substances and Disease Registry. The use of trade names is for identification only and does not imply endorsement by the Centers for Disease Control and Prevention, the Agency for Toxic Substances and Disease Registry, the Public Health Service, or the U.S. Department of Health and Human Services.
Biography

Amy S. Gargis, Ph.D., is a Microbiologist at the U.S. Centers for Disease Control and Prevention (CDC), Atlanta, GA. Dr. Gargis earned her M.S. and Ph.D. in Microbiology from The University of Alabama. After receiving her Ph.D. in 2010, she joined the Genetics Team within the Division of Laboratory Programs, Standards, and Services at the CDC, where she focused on efforts to improve the quality of genetic testing in the clinical and public health laboratory setting. Dr. Gargis was colead in organizing two national workgroups of experts to review and establish consensus guidelines for scientific principles, clinical laboratory practices, regulatory requirements, and professional standards for NGS (16, 26). In 2014, Dr. Gargis joined the BioDefense Research and Development Laboratory within the Division of Preparedness and Emerging Infections at the CDC. In this position, she works to develop and optimize rapid molecular-based assays, including NGS-based approaches, to characterize biological threat agents, with an emphasis on antibiotic resistance.
Funding Statement
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
REFERENCES
- 1.Snitkin ES, Zelazny AM, Thomas PJ, Stock F, NISC Comparative Sequencing Program Group, Henderson DK, Palmore TN, Segre JA. 2012. Tracking a hospital outbreak of carbapenem-resistant Klebsiella pneumoniae with whole-genome sequencing. Sci Transl Med 4:148ra116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Long SW, Williams D, Valson C, Cantu CC, Cernoch P, Musser JM, Olsen RJ. 2013. A genomic day in the life of a clinical microbiology laboratory. J Clin Microbiol 51:1272–1277. doi: 10.1128/JCM.03237-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Naccache SN, Federman S, Veeraraghavan N, Zaharia M, Lee D, Samayoa E, Bouquet J, Greninger AL, Luk KC, Enge B, Wadford DA, Messenger SL, Genrich GL, Pellegrino K, Grard G, Leroy E, Schneider BS, Fair JN, Martinez MA, Isa P, Crump JA, DeRisi JL, Sittler T, Hackett J Jr, Miller S, Chiu CY. 2014. A cloud-compatible bioinformatics pipeline for ultrarapid pathogen identification from next-generation sequencing of clinical samples. Genome Res 24:1180–1192. doi: 10.1101/gr.171934.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wood DE, Salzberg SL. 2014. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol 15:R46. doi: 10.1186/gb-2014-15-3-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Freitas TA, Li PE, Scholz MB, Chain PS. 2015. Accurate read-based metagenome characterization using a hierarchical suite of unique signatures. Nucleic Acids Res 43:e69. doi: 10.1093/nar/gkv180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wilson MR, Naccache SN, Samayoa E, Biagtan M, Bashir H, Yu G, Salamat SM, Somasekar S, Federman S, Miller S, Sokolic R, Garabedian E, Candotti F, Buckley RH, Reed KD, Meyer TL, Seroogy CM, Galloway R, Henderson SL, Gern JE, DeRisi JL, Chiu CY. 2014. Actionable diagnosis of neuroleptospirosis by next-generation sequencing. N Engl J Med 370:2408–2417. doi: 10.1056/NEJMoa1401268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.APHL. 2015. Next generation sequencing in public health laboratories, 2014 survey results. Association of Public Health Laboratories, Silver Spring, MD: http://www.aphl.org/AboutAPHL/publications/Documents/ID_NGSSurveyReport_52015.pdf. [Google Scholar]
- 8.McGinnis J, Laplante J, Shudt M, George KS. 2016. Next generation sequencing for whole genome analysis and surveillance of influenza A viruses. J Clin Virol 79:44–50. doi: 10.1016/j.jcv.2016.03.005. [DOI] [PubMed] [Google Scholar]
- 9.Lefterova MI, Suarez CJ, Banaei N, Pinsky BA. 2015. Next-generation sequencing for infectious disease diagnosis and management: a report of the Association for Molecular Pathology. J Mol Diagn 17:623–634. doi: 10.1016/j.jmoldx.2015.07.004. [DOI] [PubMed] [Google Scholar]
- 10.Olson ND, Lund SP, Colman RE, Foster JT, Sahl JW, Schupp JM, Keim P, Morrow JB, Salit ML, Zook JM. 2015. Best practices for evaluating single nucleotide variant calling methods for microbial genomics. Front Genet 6:235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chiu C, Miller S. 2016. Next-generation sequencing. In Persing D, Tenover F, Hayden R, Ieven M, Miller M, Nolte F, Tang Y, van Belkum A (ed), Molecular microbiology: diagnostic principles and practice, 3rd ed ASM Press, Washington, DC. [Google Scholar]
- 12.AAM. 2016. Applications of clinical microbial next-generation sequencing. A report on an American Academy of Microbiology colloquium held in Washington, DC, in April 2015. American Academy of Microbiology, American Society for Microbiology, Washington, DC: http://academy.asm.org/images/Colloquia-report/NGS_Report.pdf. [PubMed] [Google Scholar]
- 13.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, Dewell SB, Du L, Fierro JM, Gomes XV, Godwin BC, He W, Helgesen S, Ho CH, Irzyk GP, Jando SC, Alenquer ML, Jarvie TP, Jirage KB, Kim JB, Knight JR, Lanza JR, Leamon JH, Lefkowitz SM, Lei M, Li J, Lohman KL, Lu H, Makhijani VB, McDade KE, McKenna MP, Myers EW, Nickerson E, Nobile JR, Plant R, Puc BP, Ronan MT, Roth GT, Sarkis GJ, Simons JF, Simpson JW, Srinivasan M, Tartaro KR, Tomasz A, Vogt KA, Volkmer GA, et al. 2005. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437:376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koren S, Phillippy AM. 2015. One chromosome, one contig: complete microbial genomes from long-read sequencing and assembly. Curr Opin Microbiol 23:110–120. doi: 10.1016/j.mib.2014.11.014. [DOI] [PubMed] [Google Scholar]
- 15.Loman NJ, Constantinidou C, Chan JZ, Halachev M, Sergeant M, Penn CW, Robinson ER, Pallen MJ. 2012. High-throughput bacterial genome sequencing: an embarrassment of choice, a world of opportunity. Nat Rev Microbiol 10:599–606. doi: 10.1038/nrmicro2850. [DOI] [PubMed] [Google Scholar]
- 16.Gargis AS, Kalman L, Bick DP, da Silva C, Dimmock DP, Funke BH, Gowrisankar S, Hegde MR, Kulkarni S, Mason CE, Nagarajan R, Voelkerding KV, Worthey EA, Aziz N, Barnes J, Bennett SF, Bisht H, Church DM, Dimitrova Z, Gargis SR, Hafez N, Hambuch T, Hyland FC, Luna RA, MacCannell D, Mann T, McCluskey MR, McDaniel TK, Ganova-Raeva LM, Rehm HL, Reid J, Campo DS, Resnick RB, Ridge PG, Salit ML, Skums P, Wong LJ, Zehnbauer BA, Zook JM, Lubin IM. 2015. Good laboratory practice for clinical next-generation sequencing informatics pipelines. Nat Biotechnol 33:689–693. doi: 10.1038/nbt.3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aziz N, Zhao Q, Bry L, Driscoll DK, Funke B, Gibson JS, Grody WW, Hegde MR, Hoeltge GA, Leonard DG, Merker JD, Nagarajan R, Palicki LA, Robetorye RS, Schrijver I, Weck KE, Voelkerding KV. 2015. College of American Pathologists' laboratory standards for next-generation sequencing clinical tests. Arch Pathol Lab Med 139:481–493. doi: 10.5858/arpa.2014-0250-CP. [DOI] [PubMed] [Google Scholar]
- 18.Rehm HL, Bale SJ, Bayrak-Toydemir P, Berg JS, Brown KK, Deignan JL, Friez MJ, Funke BH, Hegde MR, Lyon E, Working Group of the American College of Medical Genetics, Genomics Laboratory Quality Assurance Committee. 2013. ACMG clinical laboratory standards for next-generation sequencing. Genet Med 15:733–747. doi: 10.1038/gim.2013.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kwong JC, McCallum N, Sintchenko V, Howden BP. 2015. Whole genome sequencing in clinical and public health microbiology. Pathology 47:199–210. doi: 10.1097/PAT.0000000000000235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Budowle B, Connell ND, Bielecka-Oder A, Colwell RR, Corbett CR, Fletcher J, Forsman M, Kadavy DR, Markotic A, Morse SA, Murch RS, Sajantila A, Schmedes SE, Ternus KL, Turner SD, Minot S. 2014. Validation of high throughput sequencing and microbial forensics applications. Invest Genet 5:9. doi: 10.1186/2041-2223-5-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.CLSI. 2014. Nucleic acid sequencing methods in diagnostic laboratory medicine; approved guideline. CLSI document MM09-A2. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 22.Luheshi L, Raza S, Moorthie S, Hall A, Blackburn L, Rands C, Sagoo G, Chowdhury S, Kroese M, Burton H. 2015. Pathogen genomics into practice. PHG Foundation, Cambridge, United Kingdom: http://www.phgfoundation.org/file/16848/. [Google Scholar]
- 23.U.S. FDA. 2016. Infectious disease next generation sequencing based diagnostic devices: microbial identification and detection of antimicrobial resistance and virulence markers: draft guidance for industry and Food and Drug Administration staff. U.S. Food and Drug Administration, Silver Spring, MD: http://www.fda.gov/downloads/MedicalDevices/DeviceRegulationandGuidance/GuidanceDocuments/UCM500441.pdf. [Google Scholar]
- 24.U.S. FDA. 2016. Draft guidance for industry, Food and Drug Administration staff, and clinical laboratories: framework for regulatory oversight of laboratory developed tests (LDTs). U.S. Food and Drug Administration, Silver Spring, MD: http://www.fda.gov/downloads/medicaldevices/deviceregulationandguidance/guidancedocuments/ucm416685.pdf. [Google Scholar]
- 25.CMS. 1988. Centers for Medicare and Medicaid Services. Clinical laboratory improvement amendments of 1988 (part 493). U.S. Department of Health and Human Services, Rockville, MD. [Google Scholar]
- 26.Gargis AS, Kalman L, Berry MW, Bick DP, Dimmock DP, Hambuch T, Lu F, Lyon E, Voelkerding KV, Zehnbauer BA, Agarwala R, Bennett SF, Chen B, Chin EL, Compton JG, Das S, Farkas DH, Ferber MJ, Funke BH, Furtado MR, Ganova-Raeva LM, Geigenmuller U, Gunselman SJ, Hegde MR, Johnson PL, Kasarskis A, Kulkarni S, Lenk T, Liu CS, Manion M, Manolio TA, Mardis ER, Merker JD, Rajeevan MS, Reese MG, Rehm HL, Simen BB, Yeakley JM, Zook JM, Lubin IM. 2012. Assuring the quality of next-generation sequencing in clinical laboratory practice. Nat Biotechnol 30:1033–1036. doi: 10.1038/nbt.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pont-Kingdon G, Gedge F, Wooderchak-Donahue W, Schrijver I, Weck KE, Kant JA, Oglesbee D, Bayrak-Toydemir P, Lyon E, Biochemical and Molecular Genetic Resource Committee of the College of American Pathologists. 2012. Design and analytical validation of clinical DNA sequencing assays. Arch Pathol Lab Med 136:41–46. doi: 10.5858/arpa.2010-0623-OA. [DOI] [PubMed] [Google Scholar]
- 28.Emons H, Fajgelj A, van der Veen AMH, Watters R. 2006. New definitions on reference materials. Accred Qual Assur 10:576–578. doi: 10.1007/s00769-006-0089-9. [DOI] [Google Scholar]
- 29.CLSI. 2008. Verification and validation of multiplex nucleic acid assays; approved guideline. CLSI document MM17-A. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 30.Goldberg B, Sichtig H, Geyer C, Ledeboer N, Weinstock GM. 2015. Making the leap from research laboratory to clinic: challenges and opportunities for next-generation sequencing in infectious disease diagnostics. mBio 6(6):e01888-15. doi: 10.1128/mBio.01888-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kalman LV, Lubin IM, Barker S, du Sart D, Elles R, Grody WW, Pazzagli M, Richards S, Schrijver I, Zehnbauer B. 2013. Current landscape and new paradigms of proficiency testing and external quality assessment for molecular genetics. Arch Pathol Lab Med 137:983–988. doi: 10.5858/arpa.2012-0311-RA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.CLSI. 2008. Assessment of laboratory tests when proficiency testing is not available; approved guideline. CLSI document GP-29. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 33.Schrijver I, Aziz N, Jennings LJ, Richards CS, Voelkerding KV, Weck KE. 2014. Methods-based proficiency testing in molecular genetic pathology. J Mol Diagn 16:283–287. doi: 10.1016/j.jmoldx.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 34.Moran-Gilad J, Sintchenko V, Pedersen SK, Wolfgang WJ, Pettengill J, Strain E, Hendriksen RS, Global Microbial Identifier Initiative's Working Group 4 (GMI-WG4). 2015. Proficiency testing for bacterial whole genome sequencing: an end-user survey of current capabilities, requirements and priorities. BMC Infect Dis 15:174. doi: 10.1186/s12879-015-0902-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O'Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, McVeigh R, Rajput B, Robbertse B, Smith-White B, Ako-Adjei D, Astashyn A, Badretdin A, Bao Y, Blinkova O, Brover V, Chetvernin V, Choi J, Cox E, Ermolaeva O, Farrell CM, Goldfarb T, Gupta T, Haft D, Hatcher E, Hlavina W, Joardar VS, Kodali VK, Li W, Maglott D, Masterson P, McGarvey KM, Murphy MR, O'Neill K, Pujar S, Rangwala SH, Rausch D, Riddick LD, Schoch C, Shkeda A, Storz SS, Sun H, Thibaud-Nissen F, Tolstoy I, Tully RE, Vatsan AR, Wallin C, Webb D, Wu W, Landrum MJ, Kimchi A, et al. 2016. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res 44:D733–D745. doi: 10.1093/nar/gkv1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sichtig H. 2014. High-throughput sequencing technologies for microbial identification and detection of antimicrobial resistance markers. U.S. Food and Drug Administration, Silver Spring, MD: http://www.fda.gov/downloads/MedicalDevices/NewsEvents/WorkshopsConferences/UCM390380.pdf. [Google Scholar]