Abstract
Melioidosis is a potentially fatal infection caused by the bacterium Burkholderia pseudomallei. Clinical diagnosis of melioidosis can be challenging since there is no pathognomonic clinical syndrome, and the organism is often misidentified by methods used routinely in clinical laboratories. Although the disease is more prevalent in Thailand and northern Australia, sporadic cases may be encountered in areas where it is not endemic, including the United States. Since the organism is considered a tier 1 select agent according to the Centers for Disease Control and Prevention and the U.S. Department of Agriculture Animal and Plant Health Inspection Service, clinical laboratories must be proficient at rapidly recognizing isolates suspicious for B. pseudomallei, be able to safely perform necessary rule-out tests, and to refer suspect isolates to Laboratory Response Network reference laboratories. In this minireview, we report a case of melioidosis encountered at our institution and discuss the laboratory challenges encountered when dealing with clinical isolates suspicious for B. pseudomallei or clinical specimens from suspected melioidosis cases.
CASE
A 67-year-old Filipino woman with a previous history of treated tuberculosis, hypertension, type 2 diabetes, coronary artery disease, and complete heart block requiring a pacemaker and drug-eluting stent that was placed 4 months earlier presented to an outside hospital with 2 weeks of progressive left lower quadrant abdominal pain, chills, and subjective fever. She was evaluated in different emergency departments for similar complaints on two occasions in the preceding week, but was discharged home. She was admitted to an outside hospital on this visit and computed tomography (CT) of the abdomen and pelvis with intravenous contrast revealed an 8 by 8 by 8 mm suprarenal saccular aneurysm arising from the posterior aortic wall with surrounding inflammation. Blood cultures drawn in the emergency department grew a Gram-negative rod, which was identified by the Vitek 2 (bioMérieux, Durham NC) as Burkholderia pseudomallei. This identification was confirmed several weeks later by the Centers for Disease Control and Prevention (CDC). Transthoracic echocardiography showed no vegetation. The patient was treated with meropenem 1 g intravenously (i.v.) every 8 hours (q8h). Follow-up CT scans performed 9 days after presentation showed that the mycotic aneurysm had enlarged to 13 by 24 by 20 mm and revealed a second aneurysm of the lateral wall of the aorta measuring 4 by 4 mm.
The patient was transferred to our institution for surgical evaluation. On arrival, two sets of blood cultures (BD Bactec FX system; Becton, Dickinson and Company, NJ) were collected. Within 48 h of transfer, the patient underwent surgical excision of the mycotic aneurysm and reconstruction of her aorta using bioprosthetic homografts. During the procedure, the aneurysm was found to have ruptured, with the resulting pseudoaneurysm encased in inflammatory material and purulent fluid. Intraoperative samples from the aorta, the para-aortic tissue, and tissue from the aortic aneurysm were submitted to the laboratory. Oxidase-positive, indole-negative, lactose nonfermenting Gram-negative rods were isolated from one out of two blood cultures and the surgical specimens at our institution. At 24 h, both isolates had good growth on blood agar plates (BAP), MacConkey agar plates (MAC), and triple iron sugar (TSI) agar with no change in slant or butt. Per laboratory protocol, the blood isolate was subjected to identification by Vitek MS (bioMérieux, Durham NC), which resulted in identification of Burkholderia multivorans with 94.9% confidence. The isolate from the aortic tissue was identified by the Vitek 2 GN ID card as B. pseudomallei (with 86% confidence and a note to confirm by an alternative method). The laboratory at the time had only implemented Vitek mass spectrometry (MS) for the routine identification of blood isolates. The laboratory director and Infectious Diseases service were consulted, and it was revealed that the patient was known to be infected with B. pseudomallei.
Select agent protocols were immediately engaged, including submitting the isolate to the local Laboratory Response Network (LRN) Los Angeles County Public Health reference laboratory, which confirmed the identification of B. pseudomallei. Nine employees had worked with the organism on an open bench prior to engagement of select agent protocols, including procedures concerning aerosol generation, such as the mixing of saline suspensions for Vitek 2 identification, subculturing, and catalase testing. All exposed employees were evaluated by the occupational health department and prescribed either trimethoprim-sulfamethoxazole (n = 7) or doxycycline (n = 2) as prophylaxis. Baseline serology (IgM/IgG) results were submitted to the CDC, and repeated 1, 2, 4, and 6 weeks after exposure. Staff were counseled on symptoms of melioidosis and instructed to report to the occupational health department should they experience any symptoms. Antimicrobial susceptibility testing results, which had been performed by the Vitek 2 at the outside hospital, were requested, and they revealed resistance to trimethoprim-sulfamethoxazole and tetracycline and susceptibility to meropenem, imipenem, and amoxicillin-clavulanic acid. Out of concern that the prophylaxis might not be effective, reference broth microdilution susceptibility testing was performed in a class II biological safety cabinet (BSC). Note that such testing should not be attempted by routine clinical laboratories. Test results revealed the isolate was susceptible to trimethoprim-sulfamethoxazole (MIC, ≤1 μg/ml), doxycycline (MIC, 4 μg/ml), amoxicillin-clavulanic acid (MIC, 4 μg/ml), ceftazidime (MIC, 2 μg/ml), imipenem (MIC, ≤0.25 μg/ml), and meropenem (MIC, 0.5 μg/ml). These MICs were also found with testing by the CDC, with results available 1 month later. The staff was instructed to continue with the originally prescribed prophylaxis. Two members had adverse reactions to the trimethoprim-sulfamethoxazole (vomiting) and were transitioned to doxycycline. None of the staff developed an infection with B. pseudomallei or had positive antibody tests. Despite the laboratory precautions, including labeling all patient cultures with “select agent precaution required” stickers, one additional technologist was exposed when they violated the laboratory standard operating procedures and opened the culture plates outside the biological safety cabinet. This employee was also started on prophylaxis, had serological testing performed, and did not become ill or develop antibodies to B. pseudomallei.
The patient continued to be intermittently febrile and had recurrence of bacteremia on hospital day 15 (2 weeks after surgery). Eventually, the patient's condition stabilized, and she was discharged home on hospital day 20, 1 month after her initial presentation to the outside hospital, with a plan to complete a prolonged course of intravenous imipenem (1 g, q8h). During her clinic visit 1 week after discharge from the hospital, she was feeling well, tolerating her intravenous antibiotics, and stable based on physical and laboratory examinations. However, at her 1-month follow-up visit, a surveillance CT scan of the abdomen revealed a large rim-enhancing fluid collection in the left hemiabdomen, which prompted her referral back to the emergency room. Within hours of presentation to the hospital, she developed acute-onset dyspnea, followed by hemodynamic instability. She was intubated and admitted to the surgical intensive care unit where she was found to have rapidly declining hemoglobin levels. CT angiography demonstrated extensive bleeding into the soft tissues of the neck, chest, back, and abdomen, and mediastinal and left retroperitoneal hematomas. The integrity of the aortic graft could not be assessed definitively. Despite a transfusion of multiple blood units and the initiation of vasopressors, her condition rapidly declined and she died shortly after arrival in the intensive care unit, only 13 h after returning to the hospital.
BACKGROUND
Melioidosis is a disease that occurs in both humans and animals, and causes a wide range of clinical presentations, including asymptomatic infection, localized skin ulcers, abscesses, chronic pneumonia mimicking tuberculosis, and fulminant septic shock (1). Over the past 3 decades, the disease has emerged as an important cause of mortality in Southeast Asia and northern Australia (2), and has been reported in the Indian subcontinent and Central and South Americas (3–5). Recognition of melioidosis based on clinical presentation can be challenging, and a delay in diagnosis can result in fatality (6). The etiological agent of melioidosis, B. pseudomallei, has been designated a tier 1 overlap select agent by the U.S. CDC and U.S. Department of Agriculture and Animal and Plant Health Inspection Service (USDA/APHIS). Identification of B. pseudomallei and all occupational exposures must be reported to the Federal Select Agent Program within 24 h (7). As demonstrated in this case, B. pseudomallei appears similar in culture to many other non-glucose-fermenting Gram-negative rods, and identification of the bacterium using commercially available automated systems is unreliable (8). This minireview focuses on the laboratory challenges involved in dealing with B. pseudomallei or clinical specimens from suspected melioidosis cases.
MICROBIOLOGY
The genus Burkholderia, first described by Walter H. Burkholder at Cornell University, is comprised of >60 species of obligatory aerobic, non-spore-forming, straight or slightly curved Gram-negative rods that are ubiquitous in nature (6, 9, 10). Historically classified in the genus Pseudomonas, Burkholderia was separated into its own genus based on heterogeneity in rRNA (3). While the majority of Burkholderia species are not considered to be pathogenic, B. pseudomallei is the etiological agent of melioidosis, Burkholderia mallei is the etiological agent of glanders, and the Burkholderia cepacia complex, comprising at least 17 species, causes opportunistic pulmonary infections, predominantly in cystic fibrosis patients (5, 6, 11). Burkholderia fangorum, Burkholderia glumae, and Burkholderia thailandensis also have rarely been associated with human infections (3).
The genome of B. pseudomallei is made up of two chromosomes (4.04 and 3.17 megabase pairs in size), the larger of which carries genetic elements involved in core physiological functions, while the smaller chromosome carries genes associated with accessory functions, such as adaptation to different environmental niches (12). These relatively large chromosomes encode several putative virulence factors, including quorum sensing, type III secretion systems, and the capsular polysaccharide (13). The chromosomes are thought to be highly dynamic, evidenced by the presence of several recently acquired mobile genetic elements in the chromosome (6, 12) and the extensive genetic diversification observed over time in a single patient (14, 15). The portion of the genome with high variability between strains may contribute to virulence and antibiotic resistance (16).
EPIDEMIOLOGY
Melioidosis is endemic in northern Australia, Papua New Guinea, southeast Asia, in most of the Indian subcontinent, and in southern China, Hong Kong, and Taiwan, and it is considered “highly endemic” in northeast Thailand, northern Australia, Singapore, and in parts of Malaysia (1, 17). In these regions, B. pseudomallei is present in surface water and soil. Humans are typically infected via percutaneous inoculation, inhalation, and ingestion (3, 6). One study of children residing in northeast Thailand found that 80% of the children tested had developed antibodies to B. pseudomallei by the age of 4 years (18). A minority of exposed patients will develop melioidosis, and risk factors for disease include diabetes, heavy alcohol use, chronic pulmonary disease, chronic kidney disease, thalassemia, treatment with glucocorticoid therapy, and cancer (6). Nonetheless, the incidence of melioidosis is high in these regions (2) with ≥21.3 cases/100,000 people per year reported in Thailand in 2006 and 41.7 cases/100,000 people per year reported in Australia in 1998 (6, 19). The majority of the cases occur during the rainy season (6). In northeast Thailand, melioidosis is the second most common cause of community-acquired bacteremia (20). Sporadic cases of melioidosis have been reported in areas outside the regions where it is endemic, including the Middle East, Africa, the Caribbean, and the Americas (1, 5), and among individuals with no history of travel to regions where it is endemic (5). A recent comprehensive review identified 120 human cases of melioidosis that were reported in the Americas (5). Among these, 95 (79%) cases were likely acquired in the Americas (12 cases from North America, 41 cases from Central America and the Caribbean, and 42 cases from South America) (5). Similarly, the CDC recently reported 37 laboratory-confirmed cases of melioidosis in the United States between 2008 and 2013 (21). Among these, 3 cases occurred in U.S. residents with no history of travel outside the United States (21). The sources of infections in the Americas remain to be determined, but may be associated with transmission from returning travelers, animals, or environmentally contaminated materials from regions where it is endemic (5), although zoonotic and person-to-person transmission is thought to be rare (3). Importantly, latent infections have been described and may manifest into disease decades after infection. In one case, a patient developed cutaneous melioidosis 62 years after his internment in Thailand as a Japanese prisoner of war (22).
CLINICAL PRESENTATION
Melioidosis has been dubbed “the Great Imitator” due to the absence of a pathognomonic clinical syndrome and the ability to exhibit clinical manifestations that mimic other diseases, such as cancer or tuberculosis (5, 23). It is important to include melioidosis in the differential diagnosis for any patient from, or has a history of travel to, an area where it is endemic and who presents with community-acquired sepsis, pneumonia, or abscesses (5). The incubation period varies between 1 and 21 days with a mean of 9 days (24). Latent infection with subsequent reactivation has been reported (3), notably among veterans from the Korean and Vietnam wars (hence the moniker “Vietnamese Time Bomb”) and immigrants from regions where it is endemic (25). In these instances, reactivation can occur decades after the first exposure. Clinical manifestations of the disease vary from acute septicemia to chronic infection (88% and 12% of cases in Australia, respectively) (24). Intracellular survival and cell-to-cell spread may have contributed to the organism's ability to evade the immune response, causing a persistent infection (26). The Darwin prospective melioidosis study examined 540 cases of culture-confirmed melioidosis from the Top End of Australia between 1989 and 2009 and identified pneumonia as the most common clinical manifestation (51%, 278 cases), followed by genitourinary infection (14%), skin infection (13%), bacteremia without evident focus (11%), septic arthritis or osteomyelitis (4%), and neurological melioidosis (meningoencephalitis, myelitis, and cerebral abscesses, 3%) (27). Fifty-five percent of the cases were bacteremic upon presentation, and about one-fifth of all patients developed septic shock, with a fatality rate of 50% even when their care was supervised by infectious diseases specialists (27). Most primary pneumonia cases exhibit acute or subacute manifestations, with chronic disease occurring less commonly (28). Pneumonia may occur in 20% of cases with other primary manifestations (28). In Thailand, suppurative parotitis was reported in 29% of melioidosis cases in children (29), while in Australia, this clinical manifestation is very rare (6).
LABORATORY DIAGNOSIS AND RULE-OUT GUIDELINES
Culture remains the gold standard method for the diagnosis of melioidosis (5). While it is not required that human cases be reported to the CDC's Special Pathogens Branch, physicians must notify the clinical laboratory of suspected cases prior to submitting diagnostic specimens to ensure the implementation of proper biosafety precautions to prevent accidental laboratory exposure (7). The CDC recommends that blood, throat, and urine cultures be performed on all patients with suspected melioidosis, regardless of their symptoms (5). Specimens from localized disease, such as sputum, surface swabs, and aspirates from abscesses, should also be collected (5). Standard techniques for the collection, transport, and storage of specimens for microbiological testing are sufficient to ensure viability and recovery of B. pseudomallei (3). Blood and bone marrow should be inoculated into blood culture bottles or processed using a lysis-centrifugation method to prepare the inoculum for plating. Standard laboratory media (e.g., 5% sheep blood and chocolate agar) support the growth of B. pseudomallei. However, the use of selective media, such as MacConkey agar, Ashdown's agar, B. pseudomallei selective agar (BPSA), or B. cepacia selective agar (BCSA), is recommended, particularly for tissues or any specimen expected to be contaminated with normal flora, such as respiratory secretions (3, 5). In the absence of these specialized media, a colistin or polymyxin B disk may also be placed in the first quadrants of blood agar plates to help select for Burkholderia spp., as these are inherently resistant to polymyxins. Urine specimens should be inoculated on blood and MacConkey agar plates using the standard technique for quantitative urine culture. All culture media should be incubated at 37°C for at least 4 days before being finalized as negative (3).
Of challenge to the laboratory, B. pseudomallei displays few “early” clues to allow its prompt recognition as a potential select agent. In our case, the organism appeared similar to any of the other non-Pseudomonas aeruginosa nonfermenters isolated from blood samples of highly immunocompromised transplant patients served by our laboratory. On sheep blood agar, B. pseudomallei is typically small, smooth, cream-colored with a metallic sheen, and may develop a dry or wrinkled appearance upon incubation beyond 24 to 48 h, although in our experience, this took 5 days of incubation to manifest (3, 5). On MacConkey agar, colonies are lactose, nonfermenting, and colorless, and may develop a metallic sheen with a pinkish, rugose appearance after ≥48 h (5). On Ashdown agar, colonies are typically pinpoint in size at 18 h and develop into purple, flat, wrinkled colonies at 48 h (3).
Upon Gram staining, the organism appears as a small Gram-negative rod and may exhibit bipolar staining (3). The organism is motile, indole negative, oxidase positive, inherently resistant to the polymyxins and gentamicin (3), and may produce a musty or earthy odor, although plates should not be sniffed due to the risk of laboratory exposure (30). Another rarely encountered pathogen, B. thailandensis, may resemble B. pseudomallei based on routine biochemical profiles. However, B. thailandensis is able to utilize l-arabinose as a sole carbon source, while B. pseudomallei is not (3). The etiological agent of glanders, B. mallei, differs from B. pseudomallei by its nonmotility, susceptibility to gentamicin, and much slower growth (3).
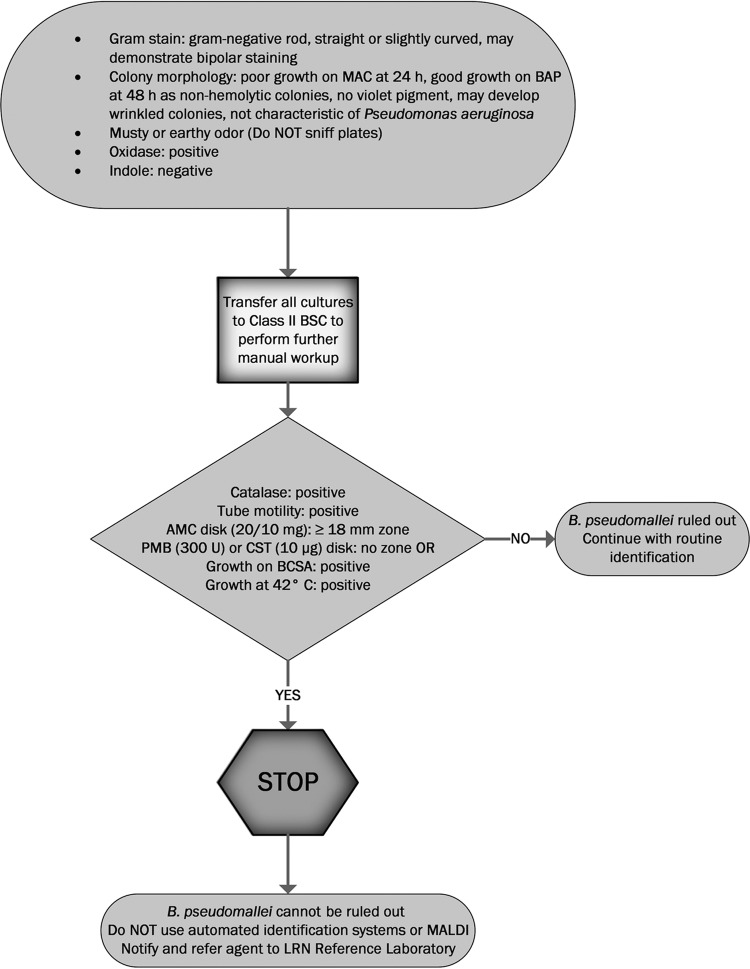
Early recognition of a potential B. pseudomallei isolate and immediate referral to a Laboratory Response Network (LRN) reference laboratory are key in minimizing laboratory exposure to the organism. Any oxidase-positive, indole-negative, Gram-negative rod that does not morphologically resemble Pseudomonas aeruginosa should be suspected as B. pseudomallei until this identification is ruled out (5). Biosafety level 2 laboratories that perform routine bacteriological workups on an open bench (such as clinical laboratories) should transfer such cultures to a class II BSC or higher, especially when performing potentially aerosol-generating procedures (7, 31). The American Society for Microbiology (ASM) provides guidelines for LRN sentinel level clinical laboratories to aid in the identification of such potential B. pseudomallei isolates (32). Key features, in addition to oxidase-positive, indole-negative reactions, include growth on MacConkey agar (may be poor at 24 h), no hemolysis on blood agar, no violet pigment, susceptibility to amoxicillin-clavulanic acid (zone of inhibition ≥18 mm surrounding a 20/10 mg amoxicillin-clavulanic acid disk), and resistance to colistin or polymyxin B (showing no zone of inhibition surrounding a disk with 10 μg colistin or 300 U polymyxin B). Such isolates should be referred immediately to an LRN reference laboratory. All work surfaces should be disinfected immediately using phenol or 10% bleach (32), and all inoculated plates must be sealed. Culture workup may continue only after B. pseudomallei and B. mallei have been ruled out by the reference LRN laboratory. A suggested workflow for clinical laboratories when encountering a possible B. pseudomallei isolate from clinical specimens based on ASM and Association for Public Health Laboratories (APHL) guidelines (32, 33) is demonstrated in Fig. 1.
FIG 1.
Suggested workflow for clinical laboratories when encountering a possible B. pseudomallei isolate from clinical specimens based on ASM and Association for Public Health Laboratories (APHL) guidelines (30, 31). MAC, MacConkey agar plate; BAP, sheep blood agar plate; BSC, biological safety cabinet; AMC, amoxicillin-clavulanic acid; PMB, polymyxin B; CST, colistin; BCSA, B. cepacia selective agar; MALDI, matrix-assisted laser desorption ionization; LRN, laboratory response network.
The performances of commercially available identification systems, such as API 20NE, Phoenix, and Vitek 2, vary greatly for identifying B. pseudomallei (5). The API 20NE system performs generally well with an accuracy of 80 to 99%; an exception was in a study using Australian isolates where the test only identified 37% of B. pseudomallei isolates (n = 37) in a challenge panel (34). API 20NE has been reported to misidentify B. pseudomallei isolates as P. aeruginosa, Pseudomonas fluorescens, B. cepacia, Chromobacterium violaceum, and Comamonas testosteroni (34, 35). Moreover, B. thailandensis has been misidentified as B. pseudomallei by the API 20NE (35). Although, in our case, the automated Vitek 2 system at the outside hospital laboratory correctly identified the isolate as B. pseudomallei, the automated Vitek 2 system with a newer-generation identification card has been reported to perform variably, with an accuracy of between 63% and 81% (8), and to misidentify B. pseudomallei as B. cepacia (8). The Phoenix and MicroScan WalkAway 96 systems have reportedly performed poorly in the identification of B. pseudomallei (8). With these potential identification errors, it is not advisable for a clinical laboratory to use any of these systems to rule out an isolate as B. pseudomallei. Additionally, these systems may generate aerosols, generating a biosafety risk (36). Identification of clinical isolates using matrix-assisted laser desorption ionization (MALDI) mass spectrometry is increasingly common in clinical laboratories and has shown benefit in identifying rare human pathogens (37). However, the accuracy of a MALDI result relies largely on the spectral database used. Misidentification of B. pseudomallei as B. thailandensis has been reported with the MALDI Biotyper system (Bruker Daltonik GmbH, Bremen, Germany) utilizing a standard Biotyper reference library (38, 39). The Biotyper instrument has been shown to correctly identify B. pseudomallei and B. thailandensis only if using a database extended with reference strains for both species (40). Bruker has developed a spectral database containing 6 bioterrorism agents (including B. pseudomallei), but the availability of this database is limited (39). For the Vitek MS system, only the research-use-only (RUO) database (and not the in vitro diagnostic [IVD] database) includes B. pseudomallei (39). The lack of spectral data on B. pseudomallei in standard IVD databases for both MALDI platforms has been shown in two cases to date, including our case (39), to lead to the misidentification of the organism as B. thailandensis and B. multivorans and to accidental exposure to the pathogen among laboratory personnel, especially in laboratories that have adopted MALDI in a routine bacteriology workflow.
Other methods to aid in identifying B. pseudomallei are available to LRN reference laboratories and laboratories that serve where it is endemic. These include nucleic acid amplification (real-time PCR or loop-mediated isothermal amplification) and polyclonal/monoclonal antibody-based immunofluorescence microscopy, which have been used successfully to identify B. pseudomallei directly from clinical specimens (41). A latex agglutination test has also been developed to aid in identifying B. pseudomallei in culture or blood culture broth (42). In the United States, the LRN-developed real-time PCR assay for B. pseudomallei is used to presumptively identify the organism (5). Serological tests, such as the indirect hemagglutination and enzyme-linked immunosorbent assay (ELISA), are available in reference laboratories and could be useful in determining if laboratory workers, military personnel, and other returning travelers have been exposed to B. pseudomallei (5). A recently developed ELISA based on purified O-polysaccharide (OPS) antigen demonstrated a high specificity (>95%) among specimens from healthy Thai and U.S. donors, suggesting the potential usefulness of the assay in areas where the pathogen is endemic and in those that are not endemic (43).
BIOSAFETY CONCERNS AND ACCIDENTAL LABORATORY EXPOSURE
B. pseudomallei is a tier 1 overlap select agent, which can affect both humans and animals (7, 21). Physicians and clinical laboratories should be aware of federal and state regulations regarding melioidosis, which include the reporting of cases, laboratory results, and special handling of specimens and cultures. When the organism is identified, it must be reported to the Federal Select Agent Program within 24 h. Clinical laboratories must also submit a report via APHIS/CDC form 4A within 7 days (44). The organism must be destroyed or transferred to an entity eligible to receive such agents within 7 days after identification (44). Some states have additional special reporting requirements. The California Department of Public Health (CDPH) requires reporting via telephone within 1 h when encountering patients with laboratory findings that satisfy their communicable disease surveillance case definitions for melioidosis (45, 46). These definitions include the culture and identification of possible B. pseudomallei from any clinical specimen or evidence of a 4-fold or greater rise in B. pseudomallei antibody titers by indirect hemagglutination (IHA) between acute- and convalescent-phase serum specimens obtained at least 2 weeks apart (46).
Laboratory workers may be exposed to B. pseudomallei during culture workups, which as noted, may be prolonged due to the many challenges associated with identifying this organism. To date, two reported cases of laboratory-acquired melioidosis have been reported, with symptoms occurring 3 to 4 days after presumed inhalational exposure to B. pseudomallei cultures (7). The first patient was supposedly exposed while cleaning a centrifuge spill using their bare hands, and the second patient performed susceptibility testing at an open bench on an isolate that was previously identified biochemically as B. cepacia (47). Between 2008 and 2013, at least 159 individuals were at risk for occupational exposures to B. pseudomallei while performing laboratory diagnostics in the United States, although none of them developed a clinical infection (21). Although the risk of occupational exposure to B. pseudomallei leading to a laboratory-associated infection is low, clinical laboratories should perform a risk assessment and develop a plan to prevent accidental exposure to B. pseudomallei. This includes a strict enforcement of standard laboratory precautions, conducting culture workups for suspicious isolates in biological safety cabinets, and wearing appropriate personal protective equipment (PPE) when working with cultures, as described in Biosafety in Microbiological and Biomedical Research Laboratories (48).
If an accidental exposure occurs, the exposure risk level for each laboratory worker should be determined based on the guidelines described by Peacock et al. (7). Low-risk exposures include plate sniffing, opening of the lid of an agar plate growing B. pseudomallei outside a biological safety cabinet, visible contact with the organism with intact skin or protected body and hands without evidence of aerosols, and spillage of small volumes of liquid culture (<1 ml) within a biological safety cabinet. High-risk exposures include a penetrating injury (including needlestick) with contaminated equipment, a bite or scratch by an infected animal, any splash causing contamination of the mouth or eyes, and aerosol-generating activities (sonication or centrifuging) performed outside the biological safety cabinet. Exposure of laboratory workers with certain health conditions, such as diabetes mellitus and chronic liver or kidney disease, in the absence of proper PPE is also considered high risk. The complete list of predisposing health conditions that are at risk is stated in the guidelines by Peacock et al. (7). Serum specimens should be obtained from at-risk individuals on the day of exposure, and weeks 1, 2, 4, and 6 postexposure for serological testing (7). Three weeks of postexposure prophylaxis (PEP) with trimethoprim-sulfamethoxazole should be initiated as soon as possible in individuals involved in high- and low-risk exposure incidents. For individuals allergic to trimethoprim-sulfamethoxazole or with isolates that are resistant, oral doxycycline or amoxicillin-clavulanate may be considered (7). However, amoxicillin-clavulanate was shown to be ineffective as a postexposure prophylaxis in a murine model of inhalational B. pseudomallei infection, as opposed to treatment with trimethoprim-sulfamethoxazole and doxycycline (49). All personnel at risk should be monitored for signs and symptoms of melioidosis and should be instructed to seek medical assistance if they start feeling ill (7).
THERAPEUTIC APPROACHES, ANTIMICROBIAL RESISTANCE, AND PREVENTION
Melioidosis has a prolonged course of illness and usually requires a lengthy course of antimicrobial treatment (6), typically consisting of 10 to 14 days of ceftazidime, meropenem, or imipenem intravenously followed with oral trimethoprim-sulfamethoxazole for 3 to 6 months (6). Monitoring of treatment compliance is crucial as it has been proposed that adherence may be the most important factor in determining recurrence, which is the most serious complication of melioidosis (2). Recurrent melioidosis occurs in 5% to 25% of cases and has a high mortality rate of 25% (2).
B. pseudomallei is intrinsically resistant to many antibiotics, including penicillin, first- and second-generation cephalosporins, macrolides, rifamycins, polymyxins, and aminoglycosides (50). Many mechanisms of acquired antimicrobial resistance in B. pseudomallei, especially by efflux pumps (51), have been characterized (6). The Clinical and Laboratory Standards Institute (CLSI) provides interpretative guidelines for susceptibility testing of B. pseudomallei in the M45 guideline (52). Resistance of B. pseudomallei to first-line agents is relatively uncommon. Resistance to either ceftazidime or amoxicillin-clavulanic acid among 4,012 cases from Thailand was only 0.6% (n = 24) (53). Another large-scale study from Thailand reported a trimethoprim-sulfamethoxazole resistance rate of 0.33% (n = 10) among a collection of 3,038 clinical isolates (54). The trimethoprim-sulfamethoxazole resistance rate in Thailand was previously reported as 13% (55), but the majority of these isolates were misclassified as resistant due to erroneous determination of MICs by Etest (54). Of note, the performance of automated commercial AST systems is poorly documented, but as seen in our case, it can give erroneous results. Further, laboratories should not attempt to determine MICs on these isolates due to the concern for inadvertent laboratory exposure. If there is a concern for resistance to first-line antimicrobial agents, clinical laboratories should immediately inform their LRN reference laboratory to determine the necessity for antimicrobial susceptibility testing at a facility that is capable of handling B. pseudomallei.
Since there is currently no vaccine available, avoidance of contact with potential sources, such as soil and water by using full-length boots and gloves, and agricultural mechanization may be needed to prevent infection (2). Development of vaccines to prevent naturally acquired infections and to protect from use of the organism as a biological weapon is under way (56–58). These include live-attenuated, whole-cell killed, subunit, glycoconjugate, outer-membrane vesicle, DNA, and dendritic cell vaccines (56). Several animal models, including those of mouse and nonhuman primates, are being used to develop and study the efficacy of experimental vaccines (56) with aims to immunize individuals in high-risk groups, such as persons with diabetes living in areas where the organism is highly endemic, such as in northeast Thailand, and to use as a countermeasure against biological warfare (56, 58).
CONCLUSION
Clinical and laboratory diagnoses of melioidosis are challenging. The disease is increasingly being reported worldwide and is gaining the status of an emerging infectious disease. Timely diagnosis of the disease and prompt initiation of treatment play important roles in determining the treatment outcome. Although B. pseudomallei is not considered endemic in most areas of the world, physicians and clinical laboratories must be aware that sporadic cases may occur at any time. To prevent any accidental exposure, clinical laboratories must be able to recognize suspicious organisms and safely perform the necessary rule-out tests before forwarding the isolate to LRN reference laboratories.
REFERENCES
- 1.Currie BJ. 2010. Burkholderia pseudomallei and Burkholderia mallei: melioidosis and glanders, p 2869–2885. Mandell, Douglas, and Bennett's principles and practice of infectious diseases, 7th ed, Elsevier, Churchill Livingstone, London, United Kingdom. [Google Scholar]
- 2.Limmathurotsakul D, Peacock SJ. 2011. Melioidosis: a clinical overview. Br Med Bull 99:125–139. doi: 10.1093/bmb/ldr007. [DOI] [PubMed] [Google Scholar]
- 3.Versalovic J; American Society for Microbiology. 2011. Burkholderia, Stenotrophomonas, Ralstonia, Cupriavidus, Pandoraea, Brevundimonas, Comamonas, Delftia and Acidovorax, p 692–713, Manual of clinical microbiology, 10th ed, ASM Press, Washington, DC. [Google Scholar]
- 4.Gopalakrishnan R, Sureshkumar D, Thirunarayan MA, Ramasubramanian V. 2013. Melioidosis: an emerging infection in India. J Assoc Physicians India 61:612–614. [PubMed] [Google Scholar]
- 5.Benoit TJ, Blaney DD, Doker TJ, Gee JE, Elrod MG, Rolim DB, Inglis TJ, Hoffmaster AR, Bower WA, Walke HT. 2015. A review of melioidosis cases in the Americas. Am J Trop Med Hyg 93:1134–1139. doi: 10.4269/ajtmh.15-0405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wiersinga WJ, Currie BJ, Peacock SJ. 2012. Melioidosis. N Engl J Med 367:1035–1044. doi: 10.1056/NEJMra1204699. [DOI] [PubMed] [Google Scholar]
- 7.Peacock SJ, Schweizer HP, Dance DA, Smith TL, Gee JE, Wuthiekanun V, DeShazer D, Steinmetz I, Tan P, Currie BJ. 2008. Management of accidental laboratory exposure to Burkholderia pseudomallei and B. mallei. Emerg Infect Dis 14:e2. doi: 10.3201/eid1407.071501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zong Z, Wang X, Deng Y, Zhou T. 2012. Misidentification of Burkholderia pseudomallei as Burkholderia cepacia by the VITEK 2 system. J Med Microbiol 61:1483–1484. doi: 10.1099/jmm.0.041525-0. [DOI] [PubMed] [Google Scholar]
- 9.Weissert C, Dollenmaier G, Rafeiner P, Riehm J, Schultze D. 2009. Burkholderia pseudomallei misidentified by automated system. Emerg Infect Dis 15:1799–1801. doi: 10.3201/eid1511.081719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Estrada-de los Santos P, Vinuesa P, Martinez-Aguilar L, Hirsch AM, Caballero-Mellado J. 2013. Phylogenetic analysis of Burkholderia species by multilocus sequence analysis. Curr Microbiol 67:51–60. doi: 10.1007/s00284-013-0330-9. [DOI] [PubMed] [Google Scholar]
- 11.Ramsay KA, Butler CA, Paynter S, Ware RS, Kidd TJ, Wainwright CE, Bell SC. 2013. Factors influencing acquisition of Burkholderia cepacia complex organisms in patients with cystic fibrosis. J Clin Microbiol 51:3975–3980. doi: 10.1128/JCM.01360-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holden MT, Titball RW, Peacock SJ, Cerdeno-Tarraga AM, Atkins T, Crossman LC, Pitt T, Churcher C, Mungall K, Bentley SD, Sebaihia M, Thomson NR, Bason N, Beacham IR, Brooks K, Brown KA, Brown NF, Challis GL, Cherevach I, Chillingworth T, Cronin A, Crossett B, Davis P, DeShazer D, Feltwell T, Fraser A, Hance Z, Hauser H, Holroyd S, Jagels K, Keith KE, Maddison M, Moule S, Price C, Quail MA, Rabbinowitsch E, Rutherford K, Sanders M, Simmonds M, Songsivilai S, Stevens K, Tumapa S, Vesaratchavest M, Whitehead S, Yeats C, Barrell BG, Oyston PC, Parkhill J. 2004. Genomic plasticity of the causative agent of melioidosis, Burkholderia pseudomallei. Proc Natl Acad Sci U S A 101:14240–14245. doi: 10.1073/pnas.0403302101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wiersinga WJ, van der Poll T, White NJ, Day NP, Peacock SJ. 2006. Melioidosis: insights into the pathogenicity of Burkholderia pseudomallei. Nat Rev Microbiol 4:272–282. doi: 10.1038/nrmicro1385. [DOI] [PubMed] [Google Scholar]
- 14.Price EP, Sarovich DS, Mayo M, Tuanyok A, Drees KP, Kaestli M, Beckstrom-Sternberg SM, Babic-Sternberg JS, Kidd TJ, Bell SC, Keim P, Pearson T, Currie BJ. 2013. Within-host evolution of Burkholderia pseudomallei over a twelve-year chronic carriage infection. mBio 4:pii: e00388-13. doi: 10.1128/mBio.00388-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Price EP, Hornstra HM, Limmathurotsakul D, Max TL, Sarovich DS, Vogler AJ, Dale JL, Ginther JL, Leadem B, Colman RE, Foster JT, Tuanyok A, Wagner DM, Peacock SJ, Pearson T, Keim P. 2010. Within-host evolution of Burkholderia pseudomallei in four cases of acute melioidosis. PLoS Pathog 6:e1000725. doi: 10.1371/journal.ppat.1000725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sim SH, Yu Y, Lin CH, Karuturi RK, Wuthiekanun V, Tuanyok A, Chua HH, Ong C, Paramalingam SS, Tan G, Tang L, Lau G, Ooi EE, Woods D, Feil E, Peacock SJ, Tan P. 2008. The core and accessory genomes of Burkholderia pseudomallei: implications for human melioidosis. PLoS Pathog 4:e1000178. doi: 10.1371/journal.ppat.1000178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Currie BJ, Dance DA, Cheng AC. 2008. The global distribution of Burkholderia pseudomallei and melioidosis: an update. Trans R Soc Trop Med Hyg 102 Suppl 1:S1–S4. doi: 10.1016/S0035-9203(08)70002-6. [DOI] [PubMed] [Google Scholar]
- 18.Wuthiekanun V, Chierakul W, Langa S, Chaowagul W, Panpitpat C, Saipan P, Thoujaikong T, Day NP, Peacock SJ. 2006. Development of antibodies to Burkholderia pseudomallei during childhood in melioidosis-endemic northeast Thailand. Am J Trop Med Hyg 74:1074–1075. [PubMed] [Google Scholar]
- 19.Cheng AC, Currie BJ. 2005. Melioidosis: epidemiology, pathophysiology, and management. Clin Microbiol Rev 18:383–416. doi: 10.1128/CMR.18.2.383-416.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Limmathurotsakul D, Wongratanacheewin S, Teerawattanasook N, Wongsuvan G, Chaisuksant S, Chetchotisakd P, Chaowagul W, Day NP, Peacock SJ. 2010. Increasing incidence of human melioidosis in northeast Thailand. Am J Trop Med Hyg 82:1113–1117. doi: 10.4269/ajtmh.2010.10-0038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Benoit TJ, Blaney DD, Gee JE, Elrod MG, Hoffmaster AR, Doker TJ, Bower WA, Walke HT; Centers for Disease Control and Prevention (CDC). 2015. Melioidosis cases and selected reports of occupational exposures to Burkholderia pseudomallei—United States, 2008-2013. MMWR Surveill Summ 64:1–9. [PubMed] [Google Scholar]
- 22.Ngauy V, Lemeshev Y, Sadkowski L, Crawford G. 2005. Cutaneous melioidosis in a man who was taken as a prisoner of war by the Japanese during World War II. J Clin Microbiol 43:970–972. doi: 10.1128/JCM.43.2.970-972.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vishnu Prasad NR, Balasubramaniam G, Karthikeyan VS, Ramesh CK, Srinivasan K. 2012. Melioidosis of chest wall masquerading as a tubercular cold abscess. J Surg Tech Case Rep 4:115–117. doi: 10.4103/2006-8808.110254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Currie BJ, Fisher DA, Howard DM, Burrow JN, Selvanayagam S, Snelling PL, Anstey NM, Mayo MJ. 2000. The epidemiology of melioidosis in Australia and Papua New Guinea. Acta Trop 74:121–127. doi: 10.1016/S0001-706X(99)00060-1. [DOI] [PubMed] [Google Scholar]
- 25.Dance DA. 1991. Melioidosis: the tip of the iceberg? Clin Microbiol Rev 4:52–60. doi: 10.1128/CMR.4.1.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Welkos SL, Klimko CP, Kern SJ, Bearss JJ, Bozue JA, Bernhards RC, Trevino SR, Waag DM, Amemiya K, Worsham PL, Cote CK. 2015. Characterization of Burkholderia pseudomallei strains using a murine intraperitoneal infection model and in vitro macrophage assays. PLoS One 10:e0124667. doi: 10.1371/journal.pone.0124667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Currie BJ, Ward L, Cheng AC. 2010. The epidemiology and clinical spectrum of melioidosis: 540 cases from the 20 year Darwin prospective study. PLoS Negl Trop Dis 4:e900. doi: 10.1371/journal.pntd.0000900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meumann EM, Cheng AC, Ward L, Currie BJ. 2012. Clinical features and epidemiology of melioidosis pneumonia: results from a 21-year study and review of the literature. Clin Infect Dis 54:362–369. doi: 10.1093/cid/cir808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lumbiganon P, Chotechuangnirun N, Kosalaraksa P, Teeratakulpisarn J. 2011. Localized melioidosis in children in Thailand: treatment and long-term outcome. J Trop Pediatr 57:185–191. doi: 10.1093/tropej/fmq078. [DOI] [PubMed] [Google Scholar]
- 30.Ashdown LR. 1992. Melioidosis and safety in the clinical laboratory. J Hosp Infect 21:301–306. doi: 10.1016/0195-6701(92)90140-H. [DOI] [PubMed] [Google Scholar]
- 31.Centers for Disease Control and Prevention (CDC). 2004. Laboratory exposure to Burkholderia pseudomallei—Los Angeles, California, 2003. MMWR Morb Mortal Wkly Rep 53:988–990. [PubMed] [Google Scholar]
- 32.American Society for Microbiology. 2015. Sentinel level clinical laboratory guidelines for suspected agents of bioterrorism and emerging infectious diseases. Glanders: Burkholderia mallei and melioidosis: Burkholderia pseudomallei. https://www.asm.org/images/PSAB/LRN/Burkholderia316.pdf.
- 33.Association for Public Health Laboratories. 2016. Biothreat agent bench cards for the sentinel clinical laboratory. https://www.aphl.org/programs/preparedness/Documents/APHL-Sentinel-Laboratory-Biothreat-Bench-Cards.pdf.
- 34.Inglis TJ, Merritt A, Chidlow G, Aravena-Roman M, Harnett G. 2005. Comparison of diagnostic laboratory methods for identification of Burkholderia pseudomallei. J Clin Microbiol 43:2201–2206. doi: 10.1128/JCM.43.5.2201-2206.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Amornchai P, Chierakul W, Wuthiekanun V, Mahakhunkijcharoen Y, Phetsouvanh R, Currie BJ, Newton PN, van Vinh Chau N, Wongratanacheewin S, Day NP, Peacock SJ. 2007. Accuracy of Burkholderia pseudomallei identification using the API 20NE system and a latex agglutination test. J Clin Microbiol 45:3774–3776. doi: 10.1128/JCM.00935-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller JM, Astles R, Baszler T, Chapin K, Carey R, Garcia L, Gray L, Larone D, Pentella M, Pollock A, Shapiro DS, Weirich E, Wiedbrauk D; Biosafety Blue Ribbon Panel; Centers for Disease Control and Prevention (CDC). 2012. Guidelines for safe work practices in human and animal medical diagnostic laboratories. Recommendations of a CDC-convened, Biosafety Blue Ribbon Panel. MMWR Suppl 61:1–102. [PubMed] [Google Scholar]
- 37.Seng P, Abat C, Rolain JM, Colson P, Lagier JC, Gouriet F, Fournier PE, Drancourt M, La Scola B, Raoult D. 2013. Identification of rare pathogenic bacteria in a clinical microbiology laboratory: impact of matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol 51:2182–2194. doi: 10.1128/JCM.00492-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cunningham SA, Patel R. 2013. Importance of using Bruker's security-relevant library for Biotyper identification of Burkholderia pseudomallei, Brucella species, and Francisella tularensis. J Clin Microbiol 51:1639–1640. doi: 10.1128/JCM.00267-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dingle TC, Butler-Wu SM, Abbott AN. 2014. Accidental exposure to Burkholderia pseudomallei in the laboratory in the era of matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol 52:3490–3491. doi: 10.1128/JCM.01238-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lau SK, Tang BS, Curreem SO, Chan TM, Martelli P, Tse CW, Wu AK, Yuen KY, Woo PC. 2012. Matrix-assisted laser desorption ionization-time of flight mass spectrometry for rapid identification of Burkholderia pseudomallei: importance of expanding databases with pathogens endemic to different localities. J Clin Microbiol 50:3142–3143. doi: 10.1128/JCM.01349-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tandhavanant S, Wongsuvan G, Wuthiekanun V, Teerawattanasook N, Day NP, Limmathurotsakul D, Peacock SJ, Chantratita N. 2013. Monoclonal antibody-based immunofluorescence microscopy for the rapid identification of Burkholderia pseudomallei in clinical specimens. Am J Trop Med Hyg 89:165–168. doi: 10.4269/ajtmh.13-0066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Anuntagool N, Naigowit P, Petkanchanapong V, Aramsri P, Panichakul T, Sirisinha S. 2000. Monoclonal antibody-based rapid identification of Burkholderia pseudomallei in blood culture fluid from patients with community-acquired septicaemia. J Med Microbiol 49:1075–1078. doi: 10.1099/0022-1317-49-12-1075. [DOI] [PubMed] [Google Scholar]
- 43.Suttisunhakul V, Wuthiekanun V, Brett PJ, Khusmith S, Day NP, Burtnick MN, Limmathurotsakul D, Chantratita N. 2016. Development of rapid enzyme-linked immunosorbent assays for detection of antibodies to Burkholderia pseudomallei. J Clin Microbiol 54:1259–1268. doi: 10.1128/JCM.02856-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Federal Select Agent Program. 2014. Report of identification of select agent of toxin FAQs. http://www.selectagents.gov/faq-reporting.html Accessed 6 October 2016.
- 45.State of California. 2015. Barclays official California code of regulations, Title 17, Section 2505. Notification by Laboratories. https://www.cdph.ca.gov/HealthInfo/Documents/Title17Section2505List.pdf.
- 46.California Department of Public Health. January 2014. Division of Communicable Disease Control. Guidelines for laboratories on reporting select agent test requests and results. https://www.cdph.ca.gov/programs/lfs/Documents/AB186_Guidance%20on%20Reporting%20Select%20Agents.pdf.
- 47.Schlech WF III, Turchik JB, Westlake RE Jr, Klein GC, Band JD, Weaver RE. 1981. Laboratory-acquired infection with Pseudomonas pseudomallei (melioidosis). N Engl J Med 305:1133–1135. doi: 10.1056/NEJM198111053051907. [DOI] [PubMed] [Google Scholar]
- 48.Chosewood LC, Wilson DE (ed). 2009. Biosafety in microbiological and biomedical laboratories, 5th ed, U.S. Department of Health and Human Services, Public Health Service, Centers for Disease Control and Prevention, National Institutes of Health, Washington, DC. [Google Scholar]
- 49.Sivalingam SP, Sim SH, Jasper LC, Wang D, Liu Y, Ooi EE. 2008. Pre- and post-exposure prophylaxis of experimental Burkholderia pseudomallei infection with doxycycline, amoxicillin/clavulanic acid and co-trimoxazole. J Antimicrob Chemother 61:674–678. doi: 10.1093/jac/dkm527. [DOI] [PubMed] [Google Scholar]
- 50.Khosravi Y, Vellasamy KM, Mariappan V, Ng SL, Vadivelu J. Antimicrobial susceptibility and genetic characterisation of Burkholderia pseudomallei isolated from Malaysian patients. ScientificWorldJournal 2014:132971. doi: 10.1155/2014/132971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Podnecky NL, Rhodes KA, Schweizer HP. 2015. Efflux pump-mediated drug resistance in Burkholderia. Front Microbiol 6:305. doi: 10.3389/fmicb.2015.00305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.CLSI. 2010. Methods for antimicrobial dilution and disk susceptibility testing of infrequently isolated or fastidious bacteria; approved guideline, 2nd ed CLSI Document M45-A2. CLSI, Wayne, PA. [Google Scholar]
- 53.Wuthiekanun V, Amornchai P, Saiprom N, Chantratita N, Chierakul W, Koh GC, Chaowagul W, Day NP, Limmathurotsakul D, Peacock SJ. 2011. Survey of antimicrobial resistance in clinical Burkholderia pseudomallei isolates over two decades in northeast Thailand. Antimicrob Agents Chemother 55:5388–5391. doi: 10.1128/AAC.05517-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Saiprom N, Amornchai P, Wuthiekanun V, Day NP, Limmathurotsakul D, Peacock SJ, Chantratita N. 2015. Trimethoprim/sulfamethoxazole resistance in clinical isolates of Burkholderia pseudomallei from Thailand. Int J Antimicrob Agents 45:557–559. doi: 10.1016/j.ijantimicag.2015.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wuthiekanun V, Cheng AC, Chierakul W, Amornchai P, Limmathurotsakul D, Chaowagul W, Simpson AJ, Short JM, Wongsuvan G, Maharjan B, White NJ, Peacock SJ. 2005. Trimethoprim/sulfamethoxazole resistance in clinical isolates of Burkholderia pseudomallei. J Antimicrob Chemother 55:1029–1031. doi: 10.1093/jac/dki151. [DOI] [PubMed] [Google Scholar]
- 56.Limmathurotsakul D, Funnell SG, Torres AG, Morici LA, Brett PJ, Dunachie S, Atkins T, Altmann DM, Bancroft G, Peacock SJ; Steering Group on Melioidosis Vaccine Development. 2015. Consensus on the development of vaccines against naturally acquired melioidosis. Emerg Infect Dis 21:. doi: 10.3201/eid2106.141480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Patel N, Conejero L, De Reynal M, Easton A, Bancroft GJ, Titball RW. 2011. Development of vaccines against Burkholderia pseudomallei. Front Microbiol 2:198. doi: 10.3389/fmicb.2011.00198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Easton A, Haque A, Chu K, Patel N, Lukaszewski RA, Krieg AM, Titball RW, Bancroft GJ. 2011. Combining vaccination and postexposure CpG therapy provides optimal protection against lethal sepsis in a biodefense model of human melioidosis. J Infect Dis 204:636–644. doi: 10.1093/infdis/jir301. [DOI] [PMC free article] [PubMed] [Google Scholar]