Abstract
The increasing prevalence of multidrug-resistant (MDR) bacteria is a serious global challenge. Here, we studied prospectively whether bacterial whole-genome sequencing (WGS) for real-time MDR surveillance is technical feasible, returns actionable results, and is cost-beneficial. WGS was applied to all MDR isolates of four species (methicillin-resistant Staphylococcus aureus [MRSA], vancomycin-resistant Enterococcus faecium, MDR Escherichia coli, and MDR Pseudomonas aeruginosa) at the University Hospital Muenster, Muenster, Germany, a tertiary care hospital with 1,450 beds, during two 6-month intervals. Turnaround times (TAT) were measured, and total costs for sequencing per isolate were calculated. After cancelling prior policies of preemptive isolation of patients harboring certain Gram-negative MDR bacteria in risk areas, the second interval was conducted. During interval I, 645 bacterial isolates were sequenced. From culture, TATs ranged from 4.4 to 5.3 days, and costs were €202.49 per isolate. During interval II, 550 bacterial isolates were sequenced. Hospital-wide transmission rates of the two most common species (MRSA and MDR E. coli) were low during interval I (5.8% and 2.3%, respectively) and interval II (4.3% and 5.0%, respectively). Cancellation of isolation of patients infected with non-pan-resistant MDR E. coli in risk wards did not increase transmission. Comparing sequencing costs with avoided costs mostly due to fewer blocked beds during interval II, we saved in excess of €200,000. Real-time microbial WGS in our institution was feasible, produced precise actionable results, helped us to monitor transmission rates that remained low following a modification in isolation procedures, and ultimately saved costs.
INTRODUCTION
The increasing prevalence of multidrug-resistant (MDR) bacteria, e.g., methicillin-resistant Staphylococcus aureus (MRSA), vancomycin-resistant enterococci (VRE), or multidrug-resistant Gram-negative bacilli, poses serious challenges to global health (1). Infections caused by MDR bacteria present limited or even no therapeutic options, especially in nosocomial settings. To trace intrainstitutional spread, besides epidemiological investigations that take place, time, and patient information into consideration, molecular typing adds a fourth dimension, i.e., the genotype, to confirm similarity or the absence of similarity among such pathogens more precisely. However, the resolution of classical typing methods is limited. This changed dramatically with the introduction of next-generation whole-genome sequencing (WGS), which can elucidate the origin and spread of bacterial pathogens in a rapid manner due to the availability of benchtop next-generation sequencers since the large Shiga toxin-producing Escherichia coli (STEC) O104:H4 outbreak in 2011 (2, 3). Currently, WGS not only provides highly discriminatory typing data (4–7) but also enables a richer profiling of virulence traits and antibiotic resistance genes in pathogens (8, 9). Furthermore, in contrast to classical typing methods, WGS is a universal method that does not require species-specific protocols. Together, these advantages have generated an abundance of data in support of the theoretical value of molecular typing (10, 11), but these studies have been either retrospective molecular reconstructions of transmission events or used under dedicated study conditions (4, 12). Here, we prospectively applied WGS over two 6-month intervals in a clinical diagnostic microbiology laboratory and used these data to inform infection control measures that were applied in the second interval. In interval I, we sequenced the four most common MDR bacterial pathogens at our hospital (MRSA, VRE, Escherichia coli, and Pseudomonas aeruginosa) to determine if prospective and daily real-time WGS is technically feasible and whether results can be made available in a clinically actionable time frame for infection control-relevant decisions. After scaling back isolation procedures during interval II, we compared transmission rates in the two intervals. Moreover, we investigated the extent to which WGS genotyping efforts are economically justifiable.
MATERIALS AND METHODS
Experimental design.
The study was conducted at the University Hospital Muenster, Muenster, Germany, a tertiary care hospital with 1,450 beds, between 15 October 2013 and 15 April 2014 (interval I) and between 15 October 2014 and 15 April 2015 (interval II). We prospectively subjected all MRSA, VRE, MDR E. coli, and MDR P. aeruginosa isolates from patients (one phenotypic variant per patient/case) to WGS. These four species account for >90% of all MDR isolates at our hospital. In addition to bacteria from routine clinical testing, isolates from screening efforts, e.g., to determine the nasal carriage of MRSA, were analyzed. At our hospital, all patients receive a screening (combined nasal/pharyngeal swab) for MRSA at admission. For VRE and Gram-negative MDR bacteria, no general screening procedures are implemented. Patients harboring Gram-positive MDR bacteria, i.e., MRSA and VRE, were treated under strict isolation procedures. Patients carrying Gram-negative bacteria with resistance to the four substances/groups piperacillin, cefotaxime/ceftazidime, ciprofloxacin, and imipenem/meropenem (4MDR-GN) were similarly treated. In interval I, patients infected or colonized with Gram-negative bacteria that were resistant to the three substances/groups piperacillin, cefotaxime/ceftazidime, and ciprofloxacin but susceptible to imipenem/meropenem (3MDR-GN) were isolated only in dedicated risk areas, such as intensive care units (ICUs) and cancer wards, in accordance with national guidelines (13). Before interval II, starting from 11 July 2014, we stopped isolation of patients harboring 3MDR-GN in all risk wards except the neonatal ICU and the bone marrow transplantation unit. WGS results were not used for individual patient treatment. The study was approved by the Ethical Committee of the Aerztekammer Westfalen-Lippe and of the Medical Faculty, University of Muenster (vote no. 2013-302-f-S).
WGS and data analysis.
MDR bacterial isolates were delivered to our laboratory on working days immediately after detection, and WGS was performed on a single MiSeq instrument (Illumina, San Diego, CA, USA). Two or three runs were completed weekly, depending on specimen availability. For subsequent WGS, a single colony was inoculated into nutrient broth (Heipha, Eppelheim, Germany) and incubated overnight (37°C). Genomic DNA was purified using a MagAttract HMW DNA kit (Qiagen, Hilden, Germany) following the manufacturer's instructions with the addition of 120 U Lysostaphin (Sigma, Taufkirchen, Germany) to lyse MRSA. Subsequently, 1 ng of genomic DNA was introduced into library preparation with a Nextera XT DNA sample preparation kit (Illumina) and paired-end sequenced with a MiSeq Reagent kit v2 250 bp (Illumina) with an average insertion size of 300 bp. Libraries were scaled to reach 100-fold sequencing coverage for an average genome size of 5 MB. Only sequencing runs that fulfilled the manufacturers' specifications (Illumina) with respect to cluster density and Q30 were further analyzed. The resulting sequence files (fastq file format) were de novo assembled using CLC bio genomics workbench version 6.5 (Qiagen) during the first period as described previously (14). The resulting sequence assembly files (ACE file format) were analyzed in Ridom SeqSphere+ software version 1.0 (Ridom GmbH, Muenster, Germany) (15). During the second period, the automated quality trimming and de novo assembly pipeline of SeqSphere+ software version 2.2 was used. Core genome target definition, de novo assembly, and allele calling parameters were exactly the same as described previously (16).
To analyze the genomic data with respect to the molecular epidemiology and to enable continuous surveillance, we applied the core genome multilocus sequence typing (cgMLST) approach (2, 17). This approach relies on species-specific schemes with a fixed number of chromosomal target genes (see Tables S1 and S2 in the supplemental material) as the basis for genome-wide gene-by-gene comparison on an allelic level. This strategy is analogous to classical multilocus sequence typing (MLST), where for each gene—independently of the number of single nucleotide polymorphisms (SNP)—and for each unique sequence a new allele is assigned to mitigate the effects of recombination (18). For MRSA, we applied the published and public cgMLST scheme (8). For the other three species, we created ad hoc local schemes using the SeqSphere+ target definer and all available NCBI RefSeq genomes (see Tables S1 and S2). For backwards compatibility with conventional typing efforts, we also extracted the MLST sequence types (ST) and spa types (14) from the genome sequences.
After quality control of the sequencing run, we checked the sequence quality of every sample by assessing the percentage of successfully extracted cgMLST targets, which required sequence similarities of 90% and 100% overlap. Genomes containing ≥95% of the successfully extracted cgMLST targets of the respective gene set (see Tables S1 and S2 in the supplemental material) were accepted; otherwise, genome sequencing was repeated starting from the library preparation step. After passing the quality control, numerical allele designations were assigned to the draft genome sequences by the SeqSphere+ software. The combination of all alleles resulted in an allelic profile, i.e., the typing result. In parallel, clinical and epidemiological data of all patients harboring MDR bacteria were compiled. To illustrate the clonal relationships between different isolates, minimum-spanning tree analyzes were used based on the determined allelic profiles. Thresholds of differences of >6, >8, >10, and >14 alleles in a pairwise comparison of genotypes were applied to exclude transmission for MRSA, VRE, MDR E. coli, and MDR P. aeruginosa, respectively. For the remaining isolates, we used epidemiological data such as clinic/ward and sampling date to determine whether or not a transmission event was likely (see Table S3). In all situations where isolates were detected within 1 month and where the corresponding patients were located at the same or related wards, genotypic clusters were rated as probable transmission events. During interval II, genotyping results were immediately used to inform infection control personnel and target control procedures. Fisher's exact test was used to access statistical significance. A P value of <0.05 was deemed statistically significant.
Cost calculation.
During the study, we captured all costs for sequencing consumables. For the MiSeq instrument and the analysis computer, we calculated full depreciation over a period of 3 years while assuming a throughput of 1,500 isolates/year. For labor, we used the labor costs to the employers of an experienced laboratory technician according to the national labor agreement in Germany. All costs include the value-added tax (VAT), which is 19% in Germany, and are provided in euros (€).
Accession number(s).
Raw reads are deposited at European Nucleotide Archive (ENA) under study accession numbers PRJEB7089 and PRJEB8084.
RESULTS
During interval I, we analyzed a total 645 MDR bacterial isolates (412 MRSA, 102 MDR E. coli [all were 3MDR-GN, i.e., Gram-negative bacteria that were resistant to the three substances/groups piperacillin, cefotaxime/ceftazidime, and ciprofloxacin but susceptible to imipenem/meropenem], 79 VRE, and 52 MDR P. aeruginosa [28 were 4MDR-GN, i.e., Gram-negative bacteria with an additional resistance to imipenem/meropenem]). The 645 genome sequences were determined in 58 runs on the MiSeq instrument with a mean of 13.0 samples per run. Only once, because of overclustering and a resulting low Q30 value (59%), the MiSeq instrument failed, necessitating one repeat run. After quality control of each single sample, 561 (87%) isolates were immediately successfully sequenced with a mean measured turnaround time (TAT) of 4.4 days (Table 1). Among the remaining 84 isolates (13.0%) requiring a second sequencing run, the most common cause for repeating this analysis was failure to achieve at least 95% successfully extracted cgMLST targets because the level of sequencing coverage was too low (n = 56, 66.7%). This problem was more common with P. aeruginosa, which has the largest genome of the pathogens in this study. Repeated sequencing increased the turnaround time for all isolates overall to 5.3 days. In total, depending on the species, 97.2% to 99.2% of the cgMLST targets on average were successfully extracted (Table 1; see also Tables S2 and S3 in the supplemental material).
TABLE 1.
Summary statistics of sequencing results, turnaround times, and reasons for sequencing failures during interval I
| Organism | No. of isolates | Mean % of successfully extracted cgMLST targets | No. (%) of isolates that required repeated sequencing | Mean (SD)a turnaround time (in days) for all samples without repeaters | Mean (SD) turnaround time (in days) for all samples, including failed samples | Reasons for sequencing failure (no. of samples)b |
|---|---|---|---|---|---|---|
| S. aureus | 412 | 98.5 | 38 (9.2) | 4.4 (1.6) | 5.0 (2.6) | Low coverage (22), sequencing run failure (12), primary base-calling failure (4) |
| E. coli | 102 | 99.2 | 11 (10.8) | 4.4 (1.4) | 5.3 (3.0) | Low coverage (10), sequencing run failure (1) |
| E. faecium | 79 | 97.2 | 20 (25.3) | 4.1 (1.5) | 6.2 (4.6) | Low coverage (14), mixed culture (5), sequencing run failure (1) |
| P. aeruginosa | 52 | 97.8 | 15 (28.8) | 4.8 (1.8) | 6.8 (4.0) | Low coverage (10), sequencing run failure (5) |
| Total | 645 | 98.4 | 84 (13.0) | 4.4 (1.5) | 5.3 (3.2) | Low coverage (56), sequencing run failure (19), mixed culture (5), primary base-calling failure (4) |
SD, standard deviation.
The low coverage led to a failure to achieve at least 95% successfully extracted cgMLST targets.
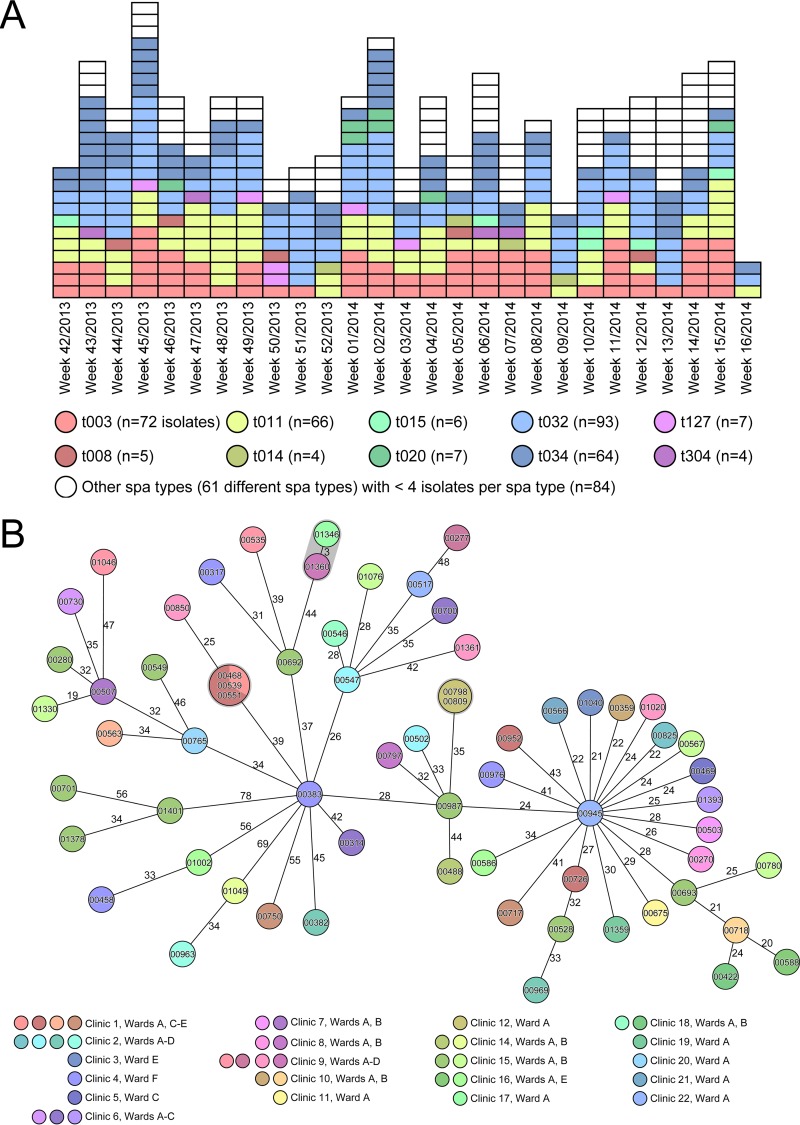
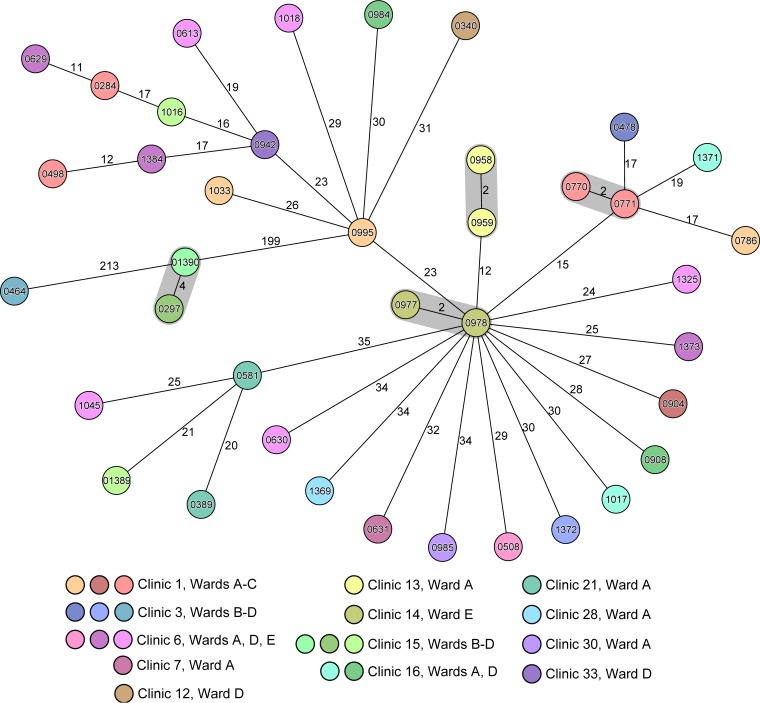
Figure 1A displays the epidemiological curve of all 412 MRSA isolates and their spa type distribution. Because only a few predominant spa type clones were putatively circulating in our institution in this interval, differentiation between sporadic cases and nosocomial transmissions based on classical typing applied to epidemiological data is very difficult. Therefore, we based our analysis on the precedent of the enhanced discriminatory power of WGS. Most notably, we illustrate the power of WGS to differentiate the 66 livestock-associated (LA)-MRSA isolates of spa type t011 (MLST ST398) (19). As they are considered to represent a very recently emergent clone and thus to have had little time to diverge (20), it was interesting to see if WGS could more precisely differentiate among these isolates. Indeed, the addition of WGS-based cgMLST analysis identified a bona fide cluster, where the identical LA-MRSA isolate (0468) was transmitted to two other patients at the same ward (one patient [isolate 0539] had already been transferred to another ward at the time of sampling) (Fig. 1B). A second pair of isolates (0798 and 0809) also exhibited identical genotypes; further investigations demonstrated that the two originated from the same specimen exhibiting different colony morphologies. The ability to detect their commonality attests to the high reproducibility of the method. All other LA-MRSA isolates differed by 3 to 78 alleles (Fig. 1B). Applying a threshold of a >6-allele difference in a pairwise comparison for a secure exclusion of transmission, which is a threshold based on our previous experience (8), we confidently excluded the intrainstitutional spread of 59 of the 66 LA-MRSA isolates. Focused epidemiological investigation refuted a nosocomial transmission of the two isolates (01346 and 01360) that differed in only 3 alleles. We also observed a similar situation in MDR E. coli of ST131, the globally most common MDR E. coli clone (21), where nosocomial transmission could be disproved for most isolates by the use of a threshold of >10 differing alleles for a secure exclusion of a nosocomial transmission (Fig. 2). Here, only four pairs of isolates (shaded in gray in Fig. 2) warranted further investigations; however, either these bacterial isolates (0297 and 01390) originated from the same patient during different hospital stays or there were no plausible epidemiological links between the hosts that harbored them.
FIG 1.
Prospective real-time WGS-based typing of all MRSA isolates exhibiting 71 different spa types. (A) Epidemic curve over interval I for all MRSA isolates detected. Each box represents a single isolate; the 10 most common spa types (≥4 isolates per spa type) are color coded. (B) Clonal relationship of all 66 livestock-associated (LA) MRSA indistinguishable by MLST (all ST398) and spa typing (spa type t011) in a minimum-spanning tree based on whole-genome sequencing. Each circle represents a single genotype, i.e., an allelic profile based on up to 1,861 target genes present in the isolates with the “pairwise ignoring missing values” option turned on in the SeqSphere+ software during comparison. The circles are named with the isolate identifiers (IDs) and colored according to ward, and the sizes are proportional to the number of isolates with identical genotypes. The number on connecting lines represents the number of alleles that differ between the connected genotypes.
FIG 2.
Minimum-spanning tree of all MDR E. coli isolates of ST131. The figure illustrates the clonal relationship of all 39 MDR E. coli isolates of MLST ST131 of interval I in a minimum-spanning tree based on whole-genome sequencing. Each circle represents a single genotype, i.e., an allelic profile based on up to 2,325 target genes present in the isolates with the “pairwise ignoring missing values” option turned on in the SeqSphere+ software during comparisons. The circles are named with the isolate IDs and colored according to the ward, and the sizes are proportional to the number of isolates with identical genotypes. The number on connecting lines represents the number of alleles that differ between the connected genotypes.
Due to overall low transmission rates, only the rates for our two most commonly identified species are summarized in Table 2. Here, we counted all genotypically determined strain clusters and retrospectively performed in-depth epidemiological investigations. This resulted in an assessment of whether or not each cluster was part of probable transmission events. Finally, the number of patient cases involved in these clusters was counted. Overall, transmission rates of MRSA and MDR E. coli were low, i.e., 5.8% and 2.3%, respectively.
TABLE 2.
Comparison of transmission rates of MRSA and 3MDR-GN E. coli during the two study intervals
| Study interval | Pathogen (no. of isolates/total no. of patient cases/no. of cases at risk wards) | No. of genotypic clusters (maximal distance for cluster recognition) | Epidemiological assessment of genotypic clusters | Total no. of cases involved in probable transmissions (%) | No. of cases in risk wards with changed infection control procedures for 3MDR-GN (%) during interval II |
|---|---|---|---|---|---|
| I | MRSA (412/397/68) | 32 (≤6 alleles) | 8 clusters with probable transmissions, 16 clusters with unlikely transmissions, isolation 8 times from same patient but different colony morphology/phenotype results | 23 (5.8) | 15 (22.1) |
| E. coli (102/86/51) | 13 (≤10 alleles) | 1 cluster with probable transmission, 1 cluster with unlikely transmissions, isolation 11 times from same patient but different cases/colony morphology/phenotype results | 2 (2.3) | 2 (3.9) | |
| II | MRSA (325/325/57) | 15 (≤6 alleles) | 6 clusters with probable transmissions, 9 clusters with unlikely transmissions | 14 (4.3) | 6 (10.5)a |
| E. coli (120/120/45) | 8 (≤10 alleles) | 1 cluster with probable transmissions, 7 clusters with unlikely transmissions | 6 (5.0) | 0 (0)a |
Results of comparisons of transmission rates between interval I and II were not statistically significant for MRSA (P = 0.0980) or MDR E. coli (P = 0.4967).
These results prompted us, in agreement with our managing board, to cease the policies of isolating patients harboring 3MDR-GN in all risk wards with the exception of the neonatal ICU and the bone marrow transplantation unit. For MRSA, VRE, and 4MDR-GN, the infection control procedures remained unchanged. After the staff members adapted to the reduced infection control measures implemented on 11 July 2014, we started interval II. In total, we analyzed 550 MDR bacterial isolates (325 MRSA, 120 MDR E. coli [all 3MDR-GN], 56 VRE, and 49 MDR P. aeruginosa [18 4MDR-GN]) (see Table S3 in the supplemental material). In addition, 48 isolates of 11 other MDR species were detected during this interval (see Table S4).
Again, we determined the number of probable transmissions of MRSA and MDR E. coli (Table 2, interval II). The overall MRSA transmission rate slightly decreased from 5.8% to 4.3%. On risk wards where the intervention took place, the same trend was noted (Table 2). Therefore, negative effects of the intervention on general hygiene procedures could be excluded. For MDR E. coli, the number of transmission events was low. Overall, an increase of transmissions (from 2.3% to 5.0%) in interval II was noted, but on risk wards with discontinued policies of isolating patients harboring 3MDR-GN, transmission rates even decreased (Table 2). Overall, there were no statistically significant changes of risk ward transmission rates between interval I and II for MRSA (P = 0.0980) and MDR E. coli (P = 0.4967), respectively.
Finally, we calculated the aggregate costs per isolate associated with the sequencing. Overall, including all repetitions, the costs were €202.49 per patient isolate. These costs consisted of 70.4% sequencing consumables (€142.45), 19.9% hardware depreciation and software costs (€40.27), and 9.7% labor expense (€19.69). To put this into context, we calculated that the less discriminatory and sometimes misleading pulsed-field gel electrophoresis (PFGE) procedure (22) cost approximately €100 for consumables and labor expenses per isolate with a similar TAT. In total, we spent €130,608.84 and €111,371.88 for WGS in the first and second intervals, respectively. To test the cost-benefit of WGS-based surveillance of MDR bacteria, during the second interval, we compared the overall sequencing costs with cost reductions that arose mainly from the decreased number of blocked beds due to the reduction of isolation measures for 3MDR-GN at risk wards (45 patients harbored E. coli and 11 P. aeruginosa, respectively). These indirect costs were calculated together with direct costs related to the extra workload reported by Herr et al. for MRSA precautions on a German surgical non-ICU ward in 2000 as being €305.74 and €66.21 per day and bed, respectively (23). These costs (in total, €371.95), a mean bed occupancy of 85.3% in our hospital in 2014 and 2015, and the assumption of one blocked bed per isolated MDR patient were used to calculate the avoided costs of isolation of patients with 3MDR-GN on risk wards. For the affected 56 patients, which had a mean residence time after MDR detection of 17.9 days, the avoided costs were €317,180.37.
DISCUSSION
Here we demonstrate that continuous real-time WGS-based bacterial gene-by-gene allele (cgMLST) typing is a powerful adjunct to classical epidemiological information and should be considered when implementing infection control measures. To facilitate interlaboratory communication for public cgMLST schemes, cgMLST allelic profiles of very closely related genomes are “lumped” together in a numerical cluster type (8, 16). The technology is sufficiently rapid and accurate, the costs for sequencing are diminishing, and we are now at a point where the data and the speed can be applied to real-world settings and result in cost savings. On top of this continuous genomic surveillance, in outbreak situations genomic data can be immediately used to design an outbreak-specific PCR for rapid screening of a large number of patients within 24 h (24).
There is a broad global consensus that strict isolation procedures, frequently represented by isolation in a single room, are required for all MRSA-, VRE-, and 4MDR-GN-positive patients. Nevertheless, we purposely included them in our study, although we could not immediately realize any cost savings, to monitor the general efficacy of our hygiene measures. For example, WGS-based typing of MRSA enabled us to immediately exclude the majority of isolates (Table 2) as being transmitted nosocomially, which would have necessitated further investigations with less-discriminatory methodologies. The few detected transmission events allowed us to improve measures in a focused manner to prevent further transmission. Moreover, the continuous genomic monitoring of the epidemiology of MDR bacteria enhanced also the surveillance of 3MDR-GN in non-risk wards, where patients with these pathogens have never been isolated.
During interval II, we wanted to determine if we could reduce extensive and costly hygiene measures, namely, isolation procedures in rooms with single occupancy, for patients colonized or infected with 3MDR-GN. These pathogens are increasing in prevalence, and their control measures are not well validated (25–28). The German “Protection against Infection Act” of 2011 and its related guidelines for infection control procedures recommend isolation of patients harboring 3MDR-GN in ICUs and in other high-risk areas that have to be defined by the managing board of each hospital (13). During the first interval, patients positive for such pathogens were isolated in a manner consistent with these guidelines. However, our data from interval I suggested that most patients infected with 3MDR-GN, as exemplarily shown for MDR E. coli ST131 (Fig. 2), were not members of identifiable institutional clusters. Therefore, we stopped the general preemptive isolation of 3MDR-GN on most risk wards during the second interval and restricted isolation to confirmed outbreak situations only. Indeed, typing and epidemiological data did not show an increase of nosocomial transmissions (Table 2). Interestingly, recent data suggest high levels of asymptomatic gut colonization with these organisms in the community (29).
Before we reduced isolation procedures for 3MDR-GN at our hospital, current national recommendations (13) had the unintentional consequence of blocked beds in our setting, as all intensive care unit (ICU) rooms and the vast majority of our other ward rooms are multibed rooms. We therefore compared our overall sequencing costs in interval II (€111,372 for WGS-based typing of 550 isolates) with avoided direct costs related to the extra workload and indirect costs that would have resulted from these blocked beds. We conservatively calculated a cost saving of €205,808 based on the data of a German surgical ward in 2000 (23). However, we believe that, 15 years later and for risk wards such as ICUs, costs for isolation have at least doubled. As a positive side effect, we could increase the quality of patient care by avoiding the negative effects of isolation (30, 31). Overall, these results convinced our managing board after the study to retain and expand WGS to all MDR bacterial species and to cost with institutional resources, i.e., these efforts are financed by the hospital and are not reimbursed by the patients' health insurance providers.
To reduce turnaround time (TAT), during interval II, we implemented an analysis pipeline within the SeqSphere+ software, which continuously monitors the MiSeq output (and as soon as sequences are generated, the reads are automatically quality-trimmed, de novo assembled, and subsequently uploaded into the SeqSphere+ database for allele assignment). After the study was completed, we further reduced TAT and the costs of the laboratory processes: we have adapted a published rapid method for WGS directly from a single colony without the need of an overnight broth culture prior to DNA extraction (32) and have reduced the costs of library preparation by halving the sample volume. Another point of action is a modification of the sequencing protocol, currently the most time-consuming process (40 h in our setting). We have chosen the 2-by-250-bp sequencing protocol to balance throughput, costs per sample, and turnaround time. However, new sequencing technologies, e.g., single-molecule sequencing, will further reduce instrument time, increase plasmid sequence reconstitution, and ultimately enable point-of-care testing (POCT) (33).
There are some limitations in our study that warrant further comments. As there is no general screening for Gram-negative MDR in our hospital, we detect only transmission events that are associated with an infection. The first transmission emanating from an unrecognized colonized patient is impossible for us to detect. Thus, we overlook these transmission events, resulting in a potential underestimation of our Gram-negative MDR transmission rate. However, as soon as a second patient is affected by an infection, WGS genotyping would at least reveal precisely a pair of highly related isolates, i.e., a genotypic cluster, although we would still miss the index patient. This cluster triggers focused and intensified infection control measures in our setting to prevent further transmissions and to identify the infection source. Moreover, transmission rates in our setting are of course also dependent on factors such as quantity and quality of staff members, general infection control procedures, and other infrastructure variables. Also, our cost-benefit analysis is biased by institutional characteristics such as the nearly exclusive presence of multibed rooms and the mean bed occupancy rate. Furthermore, we did not perform any resistance prediction from WGS data, although, for example, knowing whether a resistance phenotype is plasmid mediated would have helped us to make more risk-based decisions. Finally, use of WGS in a hospital with higher transmission rates would very likely not lead to dropping of isolation precautions. As demonstrated here in interval I, a baseline of transmission rates needs to be determined anyway before considering changes in infection control procedures.
It is conceivable that this technology could provide virulence and antimicrobial resistance potential data from WGS (8, 9, 34) to further characterize isolates independently of culture and to develop microbiologically informed individual risk assessment for colonized or infected patients (35). However, this personalized WGS usage is still hampered in many countries due to missing test accreditation and reimbursement regulations. For example, in the United States, current accreditation test regulations, including those promulgated by the Medical & Medicaid Services (CMS) under the mandate of the Clinical Laboratory Improvement Amendments (CLIA), the College of American Pathologists (CAP), and the U.S. Centers for Disease Control and Prevention, focus on human genetics and have not included microbial genomics (36). This problem is compounded in using laboratory-developed tests (LDT), in view of complex jurisdictional considerations (37, 38). In contrast, in Europe, oversight of LDT is not envisioned to be done by a national agency like the FDA and the regulation is still within the responsibility of each single laboratory according to EN ISO 15189. Therefore, we completed recently the EN ISO accreditation and the first audit of our microbial genomic genotyping workflow. Finally, compensation for these tests will need to be addressed. However, our data suggest that WGS can provide cost savings to health care systems by increasing the ability to implement epidemiologically appropriate infection control policies, rather than blanket interventions, and that investment in WGS could easily be recouped.
In summary, in our setting, prospective real-time microbial WGS is feasible, clinically actionable, and most likely cost-beneficial and helps to improve patient safety.
Supplementary Material
ACKNOWLEDGMENTS
We thank Phillip I. Tarr (Washington University School of Medicine, St. Louis, MO, USA) for fruitful discussion and critical reading of the manuscript.
This study was funded by the European Community's Seventh Framework Program (grant FP7/2007-2013 to D.H. and A.M.) under grant 278864 in the framework of the European Union Patho-NGen-Trace project, by the German Research Foundation (grant no. Me3205/2-1 to A.M.), by the German Federal Ministry of Education and Research (grant no. 01KI1301A to K.B.), and by the Medical Faculty of the University of Muenster (grant BD9817044 to A.M.). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
D.H. is one of the developers of the Ridom SeqSphere+ software mentioned in the article, which is a development of the company Ridom GmbH (Muenster, Germany) that is partially owned by him. The other authors have declared no conflict of interests.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.00790-16.
For a commentary on this article, see doi:10.1128/JCM.01714-16.
REFERENCES
- 1.World Health Organization. 2014. Antimicrobial resistance: global report on surveillance. World Health Organization, Geneva, Switzerland: http://apps.who.int/iris/bitstream/10665/112642/1/9789241564748_eng.pdf Accessed 2 January 2015. [Google Scholar]
- 2.Mellmann A, Harmsen D, Cummings CA, Zentz EB, Leopold SR, Rico A, Prior K, Szczepanowski R, Ji Y, Zhang W, McLaughlin SF, Henkhaus JK, Leopold B, Bielaszewska M, Prager R, Brzoska PM, Moore RL, Guenther S, Rothberg JM, Karch H. 2011. Prospective genomic characterization of the German enterohemorrhagic Escherichia coli O104:H4 outbreak by rapid next generation sequencing technology. PLoS One 6:e22751. doi: 10.1371/journal.pone.0022751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rohde H, Qin J, Cui Y, Li D, Loman NJ, Hentschke M, Chen W, Pu F, Peng Y, Li J, Xi F, Li S, Li Y, Zhang Z, Yang X, Zhao M, Wang P, Guan Y, Cen Z, Zhao X, Christner M, Kobbe R, Loos S, Oh J, Yang L, Danchin A, Gao GF, Song Y, Li Y, Yang H, Wang J, Xu J, Pallen MJ, Wang J, Aepfelbacher M, Yang R; E. coli O104:H4 Genome Analysis Crowd-Sourcing Consortium. 2011. Open-source genomic analysis of Shiga-toxin-producing E. coli O104:H4. N Engl J Med 365:718–724. doi: 10.1056/NEJMoa1107643. [DOI] [PubMed] [Google Scholar]
- 4.Eyre DW, Golubchik T, Gordon NC, Bowden R, Piazza P, Batty EM, Ip CL, Wilson DJ, Didelot X, O'Connor L, Lay R, Buck D, Kearns AM, Shaw A, Paul J, Wilcox MH, Donnelly PJ, Peto TE, Walker AS, Crook DW. 2012. A pilot study of rapid benchtop sequencing of Staphylococcus aureus and Clostridium difficile for outbreak detection and surveillance. BMJ Open 2:e001124. doi: 10.1136/bmjopen-2012-001124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harris SR, Feil EJ, Holden MT, Quail MA, Nickerson EK, Chantratita N, Gardete S, Tavares A, Day N, Lindsay JA, Edgeworth JD, de Lencastre H, Parkhill J, Peacock SJ, Bentley SD. 2010. Evolution of MRSA during hospital transmission and intercontinental spread. Science 327:469–474. doi: 10.1126/science.1182395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Köser CU, Holden MT, Ellington MJ, Cartwright EJ, Brown NM, Ogilvy-Stuart AL, Hsu LY, Chewapreecha C, Croucher NJ, Harris SR, Sanders M, Enright MC, Dougan G, Bentley SD, Parkhill J, Fraser LJ, Betley JR, Schulz-Trieglaff OB, Smith GP, Peacock SJ. 2012. Rapid whole-genome sequencing for investigation of a neonatal MRSA outbreak. N Engl J Med 366:2267–2275. doi: 10.1056/NEJMoa1109910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turabelidze G, Lawrence SJ, Gao H, Sodergren E, Weinstock GM, Abubucker S, Wylie T, Mitreva M, Shaikh N, Gautom R, Tarr PI. 2013. Precise dissection of an Escherichia coli O157:H7 outbreak by single nucleotide polymorphism analysis. J Clin Microbiol 51:3950–3954. doi: 10.1128/JCM.01930-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leopold SR, Goering RV, Witten A, Harmsen D, Mellmann A. 2014. Bacterial whole-genome sequencing revisited: portable, scalable, and standardized analysis for typing and detection of virulence and antibiotic resistance genes. J Clin Microbiol 52:2365–2370. doi: 10.1128/JCM.00262-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Priest NK, Rudkin JK, Feil EJ, van den Elsen JM, Cheung A, Peacock SJ, Laabei M, Lucks DA, Recker M, Massey RC. 2012. From genotype to phenotype: can systems biology be used to predict Staphylococcus aureus virulence? Nat Rev Microbiol 10:791–797. doi: 10.1038/nrmicro2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Didelot X, Bowden R, Wilson DJ, Peto TE, Crook DW. 2012. Transforming clinical microbiology with bacterial genome sequencing. Nat Rev Genet 13:601–612. doi: 10.1038/nrg3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fricke WF, Rasko DA. 2014. Bacterial genome sequencing in the clinic: bioinformatic challenges and solutions. Nat Rev Genet 15:49–55. [DOI] [PubMed] [Google Scholar]
- 12.Roach DJ, Burton JN, Lee C, Stackhouse B, Butler-Wu SM, Cookson BT, Shendure J, Salipante SJ. 2015. A year of infection in the intensive care unit: prospective whole genome sequencing of bacterial clinical isolates reveals cryptic transmissions and novel microbiota. PLoS Genet 11:e1005413. doi: 10.1371/journal.pgen.1005413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kommission fur Krankenhaushygiene und Infektionspravention (KRINKO) beim Robert Koch-Institut (RKI). 2012. Hygiene measures for infection or colonization with multidrug-resistant gram-negative bacilli. Commission recommendation for hospital hygiene and infection prevention (KRINKO) at the Robert Koch Institute (RKI). Bundesgesundheitsblatt Gesundheitsforschung Gesundheitsschutz 55:1311–1354. (In German.) [DOI] [PubMed] [Google Scholar]
- 14.Bletz S, Mellmann A, Rothgänger J, Harmsen D. 2015. Ensuring backwards compatibility: traditional genotyping efforts in the era of whole genome sequencing. Clin Microbiol Infect 21:347.e1–347.e4. doi: 10.1016/j.cmi.2014.11.005. [DOI] [PubMed] [Google Scholar]
- 15.Jünemann S, Sedlazeck FJ, Prior K, Albersmeier A, John U, Kalinowski J, Mellmann A, Goesmann A, von Haeseler A, Stoye J, Harmsen D. 2013. Updating benchtop sequencing performance comparison. Nat Biotechnol 31:294–296. doi: 10.1038/nbt.2522. [DOI] [PubMed] [Google Scholar]
- 16.Ruppitsch W, Pietzka A, Prior K, Bletz S, Fernandez HL, Allerberger F, Harmsen D, Mellmann A. 2015. Defining and evaluating a core genome multilocus sequence typing scheme for whole-genome sequence-based typing of Listeria monocytogenes. J Clin Microbiol 53:2869–2876. doi: 10.1128/JCM.01193-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maiden MC, van Rensburg MJ, Bray JE, Earle SG, Ford SA, Jolley KA, McCarthy ND. 2013. MLST revisited: the gene-by-gene approach to bacterial genomics. Nat Rev Microbiol 11:728–736. doi: 10.1038/nrmicro3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maiden MC, Bygraves JA, Feil E, Morelli G, Russell JE, Urwin R, Zhang Q, Zhou J, Zurth K, Caugant DA, Feavers IM, Achtman M, Spratt BG. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci U S A 95:3140–3145. doi: 10.1073/pnas.95.6.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Witte W, Strommenger B, Stanek C, Cuny C. 2007. Methicillin-resistant Staphylococcus aureus ST398 in humans and animals, Central Europe. Emerg Infect Dis 13:255–258. doi: 10.3201/eid1302.060924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schijffelen MJ, Boel CH, van Strijp JA, Fluit AC. 2010. Whole genome analysis of a livestock-associated methicillin-resistant Staphylococcus aureus ST398 isolate from a case of human endocarditis. BMC Genomics 11:376. doi: 10.1186/1471-2164-11-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nicolas-Chanoine MH, Blanco J, Leflon-Guibout V, Demarty R, Alonso MP, Canica MM, Park YJ, Lavigne JP, Pitout J, Johnson JR. 2008. Intercontinental emergence of Escherichia coli clone O25:H4-ST131 producing CTX-M-15. J Antimicrob Chemother 61:273–281. [DOI] [PubMed] [Google Scholar]
- 22.Jackson BR, Tarr C, Strain E, Jackson KA, Conrad A, Carleton H, Katz LS, Stroika S, Gould LH, Mody RK, Silk BJ, Beal J, Chen Y, Timme R, Doyle M, Fields A, Wise M, Tillman G, Defibaugh-Chavez S, Kucerova Z, Sabol A, Roache K, Trees E, Simmons M, Wasilenko J, Kubota K, Pouseele H, Klimke W, Besser J, Brown E, Allard M, Gerner-Smidt P. 18 April 2016. Implementation of nationwide real-time whole-genome sequencing to enhance Listeriosis outbreak detection and investigation. Clin Infect Dis doi: 10.1093/cid/ciw242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Herr CEW, Heckrodt TH, Hofmann FA, Schnettler R, Eikmann TF. 2003. Additional costs for preventing the spread of methicillin-resistant Staphylococcus aureus and a strategy for reducing these costs on a surgical ward. Infect Control Hosp Epidemiol 24:673–678. doi: 10.1086/502274. [DOI] [PubMed] [Google Scholar]
- 24.Kupferschmidt K. 2011. Epidemiology. Outbreak detectives embrace the genome era. Science 333:1818–1819. [DOI] [PubMed] [Google Scholar]
- 25.Centers for Disease Control and Prevention: NNIS System. 2003. National Nosocomial Infections Surveillance (NNIS) System Report, data summary from January 1992 through June 2003, issued August 2003. Am J Infect Control 31:481–498. doi: 10.1016/j.ajic.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 26.European Centre for Disease Prevention and Control. 2014. Antimicrobial resistance surveillance in Europe 2013. Annual Report of the European Antimicrobial Resistance Surveillance Network (EARS-Net). ECDC, Stockholm, Sweden: http://www.ecdc.europa.eu/en/publications/Publications/antimicrobial-resistance-surveillance-europe-2013.pdf Accessed 2 January 2015. [Google Scholar]
- 27.Siegel JD, Rhinehart E, Jackson M, Chiarello L; Healthcare Infection Control Practices Advisory Committee. 2007. Management of multidrug-resistant organisms in health care settings, 2006. Am J Infect Control 35:S165–193. doi: 10.1016/j.ajic.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 28.Tacconelli E, Cataldo MA, Dancer SJ, De Angelis G, Falcone M, Frank U, Kahlmeter G, Pan A, Petrosillo N, Rodríguez-Baño J, Singh N, Venditti M, Yokoe DS, Cookson B; European Society of Clinical Microbiology. 2014. ESCMID guidelines for the management of the infection control measures to reduce transmission of multidrug-resistant Gram-negative bacteria in hospitalized patients. Clin Microbiol Infect 20(Suppl 1):1–55. doi: 10.1111/1469-0691.12427. [DOI] [PubMed] [Google Scholar]
- 29.Gurnee EA, Ndao IM, Johnson JR, Johnston BD, Gonzalez MD, Burnham CA, Hall-Moore CM, McGhee JE, Mellmann A, Warner BB, Tarr PI. 2015. Gut colonization of healthy children and their mothers with pathogenic ciprofloxacin-resistant Escherichia coli. J Infect Dis 212:1862–1868. doi: 10.1093/infdis/jiv278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saint S, Higgins LA, Nallamothu BK, Chenoweth C. 2003. Do physicians examine patients in contact isolation less frequently? A brief report. Am J Infect Control 31:354–356. doi: 10.1016/S0196-6553(02)48250-8. [DOI] [PubMed] [Google Scholar]
- 31.Tarzi S, Kennedy P, Stone S, Evans M. 2001. Methicillin-resistant Staphylococcus aureus: psychological impact of hospitalization and isolation in an older adult population. J Hosp Infect 49:250–254. doi: 10.1053/jhin.2001.1098. [DOI] [PubMed] [Google Scholar]
- 32.Köser CU, Fraser LJ, Ioannou A, Becq J, Ellington MJ, Holden MT, Reuter S, Torok ME, Bentley SD, Parkhill J, Gormley NA, Smith GP, Peacock SJ. 2014. Rapid single-colony whole-genome sequencing of bacterial pathogens. J Antimicrob Chemother 69:1275–1281. doi: 10.1093/jac/dkt494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Quick J, Ashton P, Calus S, Chatt C, Gossain S, Hawker J, Nair S, Neal K, Nye K, Peters T, De Pinna E, Robinson E, Struthers K, Webber M, Catto A, Dallman TJ, Hawkey P, Loman NJ. 2015. Rapid draft sequencing and real-time nanopore sequencing in a hospital outbreak of Salmonella. Genome Biol 16:114. doi: 10.1186/s13059-015-0677-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Köser CU, Bryant JM, Becq J, Torok ME, Ellington MJ, Marti-Renom MA, Carmichael AJ, Parkhill J, Smith GP, Peacock SJ. 2013. Whole-genome sequencing for rapid susceptibility testing of M. tuberculosis. N Engl J Med 369:290–292. doi: 10.1056/NEJMc1215305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bletz S, Bielaszewska M, Leopold SR, Köck R, Witten A, Schuldes J, Zhang W, Karch H, Mellmann A. 2013. Evolution of enterohemorrhagic Escherichia coli O26 based on single-nucleotide polymorphisms. Genome Biol Evol 5:1807–1816. doi: 10.1093/gbe/evt136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gargis AS, Kalman L, Berry MW, Bick DP, Dimmock DP, Hambuch T, Lu F, Lyon E, Voelkerding KV, Zehnbauer BA, Agarwala R, Bennett SF, Chen B, Chin EL, Compton JG, Das S, Farkas DH, Ferber MJ, Funke BH, Furtado MR, Ganova-Raeva LM, Geigenmuller U, Gunselman SJ, Hegde MR, Johnson PL, Kasarskis A, Kulkarni S, Lenk T, Liu CS, Manion M, Manolio TA, Mardis ER, Merker JD, Rajeevan MS, Reese MG, Rehm HL, Simen BB, Yeakley JM, Zook JM, Lubin IM. 2012. Assuring the quality of next-generation sequencing in clinical laboratory practice. Nat Biotechnol 30:1033–1036. doi: 10.1038/nbt.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Food and Drug Administration. 2014. Optimizing FDA's regulatory oversight of Next Generation Sequencing diagnostic tests—preliminary discussion paper. http://www.fda.gov/downloads/MedicalDevices/NewsEvents/WorkshopsConferences/UCM427869.pdf Accessed 2 January 2015.
- 38.Evans JP, Watson MS. 2015. Genetic testing and FDA regulation: overregulation threatens the emergence of genomic medicine. JAMA 313:669–670. doi: 10.1001/jama.2014.18145. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.