Abstract
The aim of this study was to establish standardized drug susceptibility testing (DST) methodologies and reference MIC quality control (QC) ranges for bedaquiline, a diarylquinoline antimycobacterial, used in the treatment of adults with multidrug-resistant tuberculosis. Two tier-2 QC reproducibility studies of bedaquiline DST were conducted in eight laboratories using Clinical Laboratory and Standards Institute (CLSI) guidelines. Agar dilution and broth microdilution methods were evaluated. Mycobacterium tuberculosis H37Rv was used as the QC reference strain. Bedaquiline MIC frequency, mode, and geometric mean were calculated. When resulting data occurred outside predefined CLSI criteria, the entire laboratory data set was excluded. For the agar dilution MIC, a 4-dilution QC range (0.015 to 0.12 μg/ml) centered around the geometric mean included 95.8% (7H10 agar dilution; 204/213 observations with one data set excluded) or 95.9% (7H11 agar dilution; 232/242) of bedaquiline MICs. For the 7H9 broth microdilution MIC, a 3-dilution QC range (0.015 to 0.06 μg/ml) centered around the mode included 98.1% (207/211, with one data set excluded) of bedaquiline MICs. Microbiological equivalence was demonstrated for bedaquiline MICs determined using 7H10 agar and 7H11 agar but not for bedaquiline MICs determined using 7H9 broth and 7H10 agar or 7H9 broth and 7H11 agar. Bedaquiline DST methodologies and MIC QC ranges against the H37Rv M. tuberculosis reference strain have been established: 0.015 to 0.12 μg/ml for the 7H10 and 7H11 agar dilution MICs and 0.015 to 0.06 μg/ml for the 7H9 broth microdilution MIC. These methodologies and QC ranges will be submitted to CLSI and EUCAST to inform future research and provide guidance for routine clinical bedaquiline DST in laboratories worldwide.
INTRODUCTION
Newer drugs are being developed to counter the growing problem of multidrug-resistant (MDR) strains of Mycobacterium tuberculosis. Of the 9.6 million estimated new M. tuberculosis cases occurring globally in 2014, 3.3% were MDR, in addition to 20% of previously treated tuberculosis (TB) cases estimated to have MDR; this equates to an overall estimate of 480,000 people with MDR-TB annually. Furthermore, current treatment outcome data for patients started on MDR-TB treatment in 2015 suggest a success rate of only 50% (1). The approval of new antimycobacterials effective against MDR-TB strains has highlighted the need for validated and standardized drug susceptibility testing (DST) methods to enhance patient care and for facilitating drug resistance surveillance.
Bedaquiline, a diarylquinoline antimycobacterial (2), has received accelerated/conditional approval for use based on phase II trials in the United States (2012), the European Union (2014), and 7 countries with high MDR-TB burden (3–8). Interim policy guidance for the use of bedaquiline as part of combination therapy for adults who have pulmonary MDR-TB has been issued by the World Health Organization (WHO) and the U.S. Centers for Disease Control and Prevention (CDC) (9, 10).
Preliminary DST methodology for bedaquiline was previously piloted and then used in two phase II clinical studies (3–6). In these studies, bedaquiline DST was performed by 7H11 agar dilution and 7H9 broth microdilution methods using the resazurin microtiter assay (REMA) (11, 12). For bedaquiline DST, the standard quality control (QC) strain M. tuberculosis H37Rv should be used under the same conditions as the clinical M. tuberculosis isolates to ensure that the bedaquiline MIC of the reference falls within a predefined QC range. For QC purposes when testing clinical isolates prior to the present study, the provisional bedaquiline MIC ranges against strain H37Rv were 0.03 to 0.12 μg/ml for 7H11 agar and 0.03 to 0.12 μg/ml for REMA (7H9 broth) (7). These provisional DST methodologies and MIC QC ranges for bedaquiline required validation in a multilaboratory study. The validation of bedaquiline phenotypic DST methods is particularly important, since it is not foreseeable that a rapid bedaquiline molecular or genotypic DST method will be available in the near future for the following reasons. (i) Mutations in the bedaquiline target atpE gene that lead to high bedaquiline MICs have only been observed in vitro so far and not in M. tuberculosis isolates from MDR-TB patients. (ii) Mutations in Rv0678, a transcriptional repressor of the genes encoding the MmpS5-MmpL5 efflux pump, lead to 2- to 8-fold MIC increases and low-level resistance and have been observed in isolates obtained both in vitro and in the clinic (13, 14). These mutations include single-nucleotide insertions, deletions, and substitutions, as well as large deletions and random insertions of insertion sequence elements. (iii) Mutations in pepQ (Rv2435c), a putative Xaa-Pro aminopeptidase, produce a modest increase (up to 4-fold) in bedaquiline and clofazimine MICs but significantly reduce the efficacy of bedaquiline and clofazimine in the mouse model (15). Similar to atpE, the mutations in Rv0678 and pepQ are spread over the length of the gene. Additional resistance-associated variants (RAVs) carry mutations in the intergenic region between Rv0678 and Rv0677c (16). Thus, Rv0678-, pepQ-, and atpE-based rapid genotypic DST is not feasible using the current single-nucleotide polymorphism (SNP) molecular DST platform. A limited number of atpE mutations have been examined. (iv) As with most chromosome-borne antimicrobial resistance genetic tests, there is a lack of an algorithm to translate any known mutations in either atpE, Rv0678, or pepQ into bedaquiline MICs. Because of these shortcomings, reliable bedaquiline phenotypic DST methods are necessary.
Two multilaboratory studies were conducted to establish and validate standard DST methodologies for bedaquiline, as well as to determine reference bedaquiline MIC QC ranges on solid and liquid media. The validation methods were adapted from Clinical Laboratory and Standards Institute (CLSI) documents M07-A9 (17) and M24-A2 (18), and criteria for establishing DST QC ranges for new antimicrobial agents were sourced from CLSI document M23-A3 (19).
(Data contained in this article were presented during the 46th Union World Conference on Lung Health in Cape Town, South Africa, 2 to 6 December 2015 [20]).
MATERIALS AND METHODS
Study design.
Two concurrent multilaboratory, multicountry reproducibility studies were performed using bedaquiline phenotypic DST. One study was conducted using 7H10 and 7H11 agar dilution MIC methods (study TMC207-PMR-1988-003), and another study was performed using a 7H9 broth microdilution MIC method (study TMC207-PMR-1988-004). Both studies conformed to CLSI tier-2 criteria, as they sought to establish acceptable QC ranges for bedaquiline.
Eight independent laboratories participated (a minimum of seven laboratories is required under CLSI tier-2 criteria). Each laboratory was instructed to perform 10 replicate tests on separate days using three separate medium lots from different manufacturers and an internationally recognized M. tuberculosis reference strain according to CLSI document M23-A3 (19). The study protocol was developed by consensus among participating investigators. None of the laboratories had access to the results of the others.
Studies were performed according to CLSI document M07-A9 (17) with respect to preparation of the drug stock concentration and dilution process for the 7H10 and 7H11 agars, and they were based on the classical method for DST as outlined in M24-A2 (18) for the QC reference strain and the preparation of the inoculum.
The two studies evaluated the reproducibility of the DST methods within each participating laboratory, between laboratories, and between reagent lots. The MICs obtained from the agar DST method were also compared with those from the broth method.
Participating laboratories.
The eight participating laboratories were geographically diverse, being situated on three different continents. The investigators and their laboratories included CLSI members or advisors, the WHO Supranational Reference Laboratory Network (SRLN), the U.S. CDC, and other internationally recognized TB laboratories.
Materials. (i) Isolates.
The reference strain used was M. tuberculosis H37Rv, which was originally obtained from the American Type Culture Collection (ATCC number 27294). Each laboratory used its own stock of this H37Rv reference strain.
(ii) Antimicrobials.
Bedaquiline was provided to each laboratory by the manufacturer (Janssen Pharmaceutica NV, Beerse, Belgium).
(iii) Labware.
All regular plates and microtiter plates used to perform the DST were polystyrene, as polypropylene can lead to errors in MIC determination for bedaquiline (21).
(iv) Culture media.
For the agar dilution study, each laboratory was provided with three separate lots of Middlebrook 7H10 agar and three separate lots of Middlebrook 7H11 agar in powder form. Media were obtained from three different manufacturers: Becton Dickinson (lot numbers 3116410 for 7H10 agar and 3189213 for 7H11 agar; Franklin Lakes, NJ), Sigma-Aldrich (lot numbers BCBL7582V for 7H10 agar and BCBL4307V for 7H11 agar; St. Louis, MO), and Titan Biotech Ltd. (lot numbers M2B4KN01 for 7H10 agar and M7I6KN01 for 7H11 agar; Delhi, India). For the purpose of the analyses, each type of agar (7H10 or 7H11) was assigned a number from 1 to 3 depending on the manufacturer. Middlebrook oleic acid albumin dextrose catalase (OADC) growth supplement was also provided (Becton Dickinson, Franklin Lakes, NJ).
For the broth microdilution study, Middlebrook 7H9 broth base and OADC were obtained from Becton Dickinson (Franklin Lakes, NJ). Each laboratory was provided with three separate lots of custom-made frozen 96-well polystyrene microtiter plates (lot numbers 14181, 14202, and 14203; Thermo Fisher Scientific Inc., Waltham, MA). Laboratories were also provided with 12.5-ml sterile deionized water tubes and saline Tween broth to reconstitute the mycobacterial inoculum.
Lowenstein-Jensen medium is not recommended for bedaquiline DST due to its high protein content and therefore was not evaluated in this study (7, 21).
MIC determination by the agar dilution method. (i) Preparation of agar media.
The 7H10 and 7H11 agar media were prepared by the investigators according to CLSI document M07-A9 (17).
(ii) Preparation of bedaquiline dilutions.
Bedaquiline was dissolved in dimethyl sulfoxide (DMSO) to give a 200 μg/ml solution, and then 2-fold serial dilutions were further made from 200 μg/ml down to 0.8 μg/ml in DMSO (working solutions). Storage of aliquots of these solutions in DMSO was allowed for up to 3 months at −20°C, but once thawed, the solutions could not be stored further or refrozen. The working solutions in DMSO were diluted 1/100 in 7H10 or 7H11 agar medium to obtain the final desired concentrations of 2, 1, 0.5, 0.25, 0.12, 0.06, 0.03, 0.015, and 0.008 μg/ml in the polystyrene plates to be used for the DST.
(iii) M. tuberculosis culture and preparation of suspension.
An M. tuberculosis H37Rv suspension was prepared in biosafety level 3 (BSL3) laboratories according to the procedures in use for M. tuberculosis. M. tuberculosis isolates were grown on 7H11/7H10 media (or Lowenstein-Jensen medium). Several loops, 2 to 5 mg of mycobacterial growth, were harvested with the aim of selecting M. tuberculosis from each colony. Cultures older than 21 days were not considered acceptable, as they may yield unreliable DST results. The colonies were transferred to a 16- by 125-mm screw-cap tube containing 5 ml sterile saline Tween and 6 to 8 glass beads. The suspension was homogenized with a test tube mixer for 5 to 10 min and larger particles allowed to settle for 10 min. The supernatant suspension was then harvested and the density adjusted to that of a McFarland standard 1 suspension (∼5 × 107 CFU/ml) using sterile deionized water or saline (18, 22). There is good concordance between the McFarland scale and CFU per milliliter for M. tuberculosis (23). A new inoculum was prepared each time a set of agar plates was inoculated.
(iv) Inoculation of culture media.
Inoculum in all experiments was standardized. Working suspensions were made using a 10-fold dilution of the M. tuberculosis H37Rv suspension with sterile deionized water or saline. The undiluted (100 dilution) M. tuberculosis suspension, containing ∼5 × 107 CFU/ml, was mixed, and 0.1 ml was transferred to 0.9 ml of the first dilution tube (10−1 dilution) (∼5 × 106 CFU/ml). Care was taken to ensure all inocula were of the same standard size and fell within 0.5 log of the target in order for the resulting MIC values to be accepted as valid.
The preprepared bedaquiline-containing 7H10 or 7H11 agar plates were then inoculated with 0.1 ml of the 10−1 dilution, resulting in 5 × 105 CFU/ml plated.
To ascertain the accuracy of the inoculum plated, additional 10-fold serial dilutions of the suspension (10−3, 10−4, and 10−5 dilutions) were made from the 10−1-dilution tube. The 7H10 or 7H11 polystyrene plates containing no drugs were then inoculated with 0.1 ml of the 10−3, 10−4, and 10−5 dilutions, resulting in plates containing 5,000 CFU, 500 CFU, and 50 CFU, respectively. These plates also served as positive controls for growth.
(v) Incubation.
Inoculated polystyrene plates were incubated topside up for 1 to 2 days at 35 to 37°C until the inoculum was dry; they were then turned upside down. For incubators that were not humidified, plates were kept in plastic bags.
(vi) MIC assessment.
Results were reported at 21 days postinoculation. The MIC was defined as the lowest drug concentration (in micrograms per milliliter) that resulted in complete (100%) inhibition of visual growth (17). The positive control was checked to ensure that it showed mycobacterial growth.
MIC determination by the broth microdilution method.
Custom-made frozen microtiter plates designed for research use only were used and were prepared with bedaquiline serial dilutions in 2× supplemented 7H9 medium (7H9-S, which is 7H9 broth supplemented with 10% OADC; Becton Dickinson) at 2× final drug concentrations.
M. tuberculosis culture and preparation of suspension.
M. tuberculosis isolates were grown on 7H11 agar medium (or Lowenstein-Jensen medium) and colonies resuspended in 7H9-S. The turbidity of the M. tuberculosis H37Rv suspension was adjusted with phosphate-buffered saline to match that of a McFarland standard 1 suspension, corresponding to ∼5 × 107 CFU/ml.
Inoculation of the microtiter plates.
A 2× inoculum of M. tuberculosis H37Rv was prepared by adding (with a calibrated micropipetting device) 255 μl of the McFarland standard 1 suspension to 12.5 ml of sterile deionized water (50-fold dilution) in a polystyrene tube to give 1 × 106 CFU/ml. The 2× inoculum was poured into a disposable inoculum reservoir, and then 100 μl was transferred to the microtiter plate wells using an 8- or 12-channel micropipette and sterile tips with filters. Inoculum was added to all wells (including the growth control wells) except the negative-control wells, which received 100 μl of sterile deionized water. The final inoculum size in the plates was 5 × 105 CFU/ml, and the final bedaquiline concentrations were 4, 2, 1, 0.5, 0.25, 0.12, 0.06, 0.03, 0.015, 0.008, and 0 μg/ml.
Incubation.
After inoculation, the microtiter plates were sealed in plastic bags (10 plates per bag) and incubated at 35 to 37°C.
MIC assessment.
Results were reported at days 7, 10, and 14 postinoculation, with the microtiter plates read according to the usual laboratory procedure. The growth control was checked to ensure that it showed mycobacterial growth and the negative control to ensure it showed no growth. Any negative-control well with a turbid appearance was suspected of contamination and results discounted as invalid.
Data collection and statistical methods.
To ensure consistent data capture and reporting, and to allow compilation of the final analyses, investigators were presupplied with data collection forms. Each laboratory was assigned a unique identification number based on the order in which data were submitted. QC checks were performed on the data sets from all laboratories, and inconsistent data were queried with the investigator, who was then required to resubmit an updated file for the final analyses. Examples of inconsistencies included MIC values not within the specified dilution range, commas used as decimal separators, and erroneous dilutions. All finalized MIC data were consolidated in a master file and a final QC check performed.
Statistical analysis of MIC distribution frequencies, the modes (most frequently occurring MIC value), and the geometric mean MICs were performed using SAS software, version 9.2 (SAS Institute). For MIC values preceded by “<,” the sign was ignored, and the lower-end MIC value of the range was reported as “≤.” For MIC values preceded by “>,” the MIC value was reported as “≥” the next dilution (e.g., >1 was reported as ≥2).
CLSI document M23-A3 (19) instructs that whenever possible, the low end of the QC range for dilution testing should include concentrations that can be accurately prepared. In addition, dilutions should extend to no more than five dilutions below a drug's susceptibility breakpoint. For bedaquiline, the provisional EUCAST susceptibility breakpoint (≤0.25 μg/ml) (24) was used as a reference to select the lowest dilution. Initial QC ranges were determined by centering on the MIC mode ±1 dilution and doubling dilution over a 3-dilution QC range (19). When the mode occurred at the end of the proposed dilution range, the geometric mean was used instead. The proposed QC range was required to encompass at least 95% of the observed MIC values. In the event that a QC range over 3 dilutions could not be established, a 4-dilution QC range was analyzed. In this case, the initial proposed range was adjusted: (i) to include at least 95% of the observed MIC values to accommodate variability expected in routine testing, (ii) if a shoulder off the modal value with ≥60% data points of the mode was observed, and/or (iii) when a bimodal distribution was observed. If none of these criteria was met, it was concluded that the QC range could not be established. Following these initial analyses, if a laboratory produced outlying data, their entire data set (for all three medium lots) was discarded and the analyses repeated with the remaining laboratories. Similarly, if a medium lot produced outlying data, the lot from all laboratories would be excluded from the analyses (19). In addition, one laboratory performed 12 replicate tests for each lot and another performed 11 tests. Therefore, the number of observations did not always add up to the preplanned value of 240 (i.e., 8 laboratories times 30 tests).
A final analysis was performed to determine any potential correlations between the different DST media: 7H10 agar versus 7H11 agar, 7H10 agar versus 7H9 broth, and 7H11 agar versus 7H9 broth. First, the Pearson correlation coefficient (R values) and P values were calculated using regression analysis on a log2 scale. Second, a microbiologically meaningful intermethod correlation was performed to establish whether the two media were comparable (19). Correlation was assessed at a ±1 log2 dilution step and included ≥100 data points from each of the two media being compared. To be considered microbiologically significant, the target correlation between the two media was defined as ≥90% essential agreement (19).
RESULTS
Establishment of bedaquiline MIC QC ranges. (i) 7H10 agar dilution MIC.
In an initial analysis of data from all laboratories, comprising 243 observations (see Table S1 in the supplemental material), the mode MIC for bedaquiline was 0.03 μg/ml and the geometric mean was 0.041 μg/ml. A 3-dilution QC range centered around the mode was found to contain only 81.9% (199/243) of the observed bedaquiline MIC values. Moreover, a 4-dilution QC range covering 0.015 to 0.12 μg/ml and centered either around the mode or the geometric mean was found to contain only 90.9% (221/243) of bedaquiline MIC values (data not shown). However, the bedaquiline MIC was found to be unusually high, at 0.25 μg/ml, for one laboratory (Lab-5) for 7H10 agar Lot-1. In line with the protocol (19), all bedaquiline DST data from Lab-5 for all three lots of media were excluded from the bedaquiline MIC QC range assessment for the 7H10 agar dilution method.
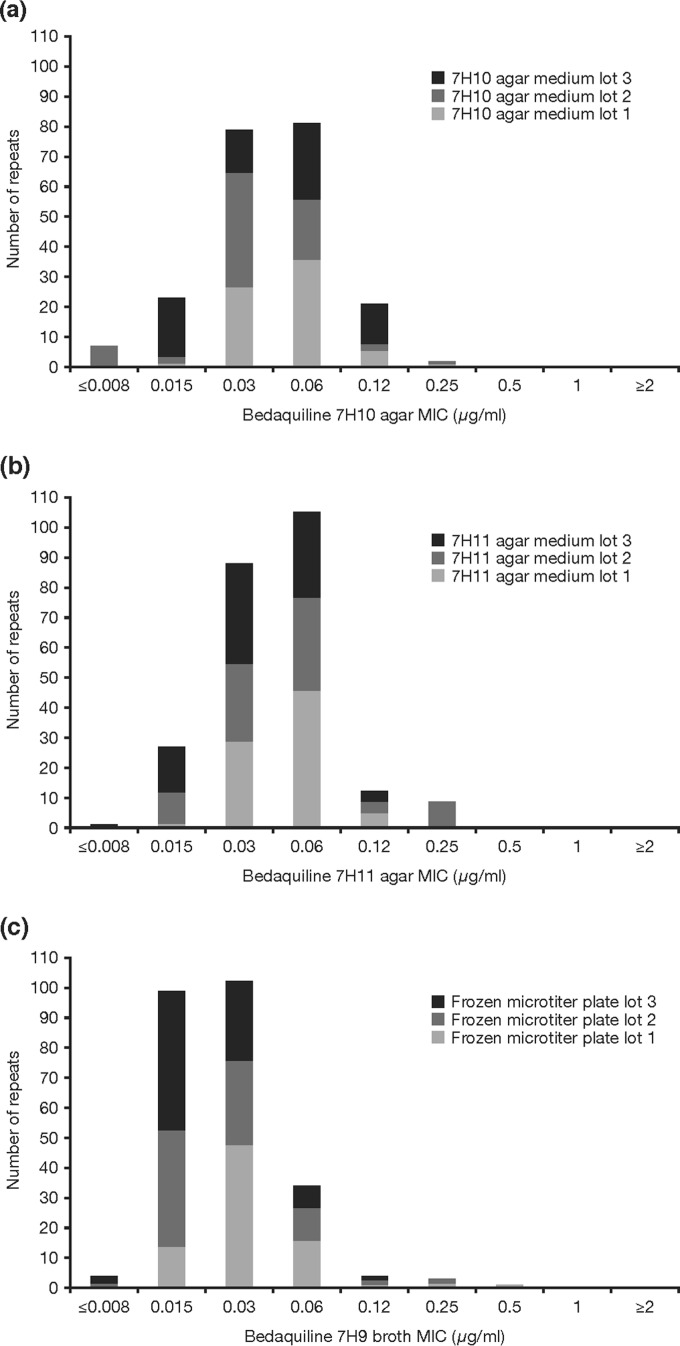
Following the exclusion of the Lab-5 data, a revised mode and geometric mean for bedaquiline MIC was calculated based on a total of 213 observations. The resulting values were 0.06 μg/ml and 0.041 μg/ml, respectively. Repeating the 4-dilution QC range (0.015 to 0.12 μg/ml) centered around the geometric mean resulted in 95.8% (204/213) of the bedaquiline MIC values being included. Therefore, the required CLSI criteria were met (19), and the bedaquiline MIC QC range for the 7H10 agar dilution method was set at 0.015 to 0.12 μg/ml (Fig. 1a).
FIG 1.
Bedaquiline MIC distributions against M. tuberculosis H37Rv and QC ranges. (a) 7H10 agar, including data using all medium lots but excluding data from Lab-5; (b) 7H11 agar, including data from all laboratories and using all medium lots; (c) 7H9 broth using all custom-made microtiter plates lots but excluding Lab-1 data.
The source of 7H10 agar medium did not appear to affect bedaquiline MIC determination, and a unimodal bedaquiline MIC distribution pattern was observed for each lot. The three different medium lots made similar contributions to the overall bedaquiline MIC distribution, with the exception of Lot-3, which showed a high proportion of bedaquiline MICs at 0.015 μg/ml compared with the other lots (Fig. 2a).
FIG 2.
Bedaquiline MIC distributions against M. tuberculosis H37Rv according to medium lot (a and b) or 96-well microtiter plate lot (c). (a) 7H10 agar excluding data from Lab-5; (b) 7H11 agar including data from all laboratories and using all medium lots; (c) 7H9 broth including data from all laboratories and using all microtiter plate lots.
(ii) 7H11 agar dilution MIC.
In the initial analysis of data from all laboratories (242 observations) (see Table S2 in the supplemental material), the mode MIC for bedaquiline was 0.06 μg/ml and the geometric mean was 0.043 μg/ml. A 3-dilution QC range centered around the mode was found to contain only 90.9% (220/242) of the observed bedaquiline MIC values. However, a 4-dilution QC range covering 0.015 to 0.12 μg/ml and centered either around the mode or the geometric mean was found to contain 95.9% (232/242) of the bedaquiline MIC values. Since the CLSI criteria were met (19), the bedaquiline MIC QC range for the 7H11 agar dilution method was set at 0.015 to 0.12 μg/ml (Fig. 1b). The source of 7H11 agar media did not appear to affect bedaquiline MIC determination, with all three lots contributing equally to the overall bedaquiline MIC distribution, and a unimodal bedaquiline MIC distribution pattern was observed for each lot (Fig. 2b).
(iii) 7H9 broth microdilution MIC.
In the initial analysis of the 7H9 broth microdilution method comprising a total of 247 observations (see Table S3 in the supplemental material), the bedaquiline MIC mode was calculated as 0.03 μg/ml and the geometric mean was 0.027 μg/ml. A 3-dilution QC range centered around the mode was found to include 95.1% (235/247) of the bedaquiline MIC values.
Overall, there were few unexpected MIC values between laboratories and between microtiter plate lots. Lab-1 reported 3 observations of bedaquiline MIC values of 0.25 μg/ml, of which 2 instances occurred in Lot-1 and 1 occurred in Lot-2 (see Text S1 in the supplemental material). The same laboratory also had 1 observation of a bedaquiline MIC value of 0.5 μg/ml in Lot-1. High MICs represented 11% (4/36) of the data generated by Lab-1. Such high MICs had not been seen previously for bedaquiline against a pansusceptible strain and disagreed with the results of the other study sites. Therefore, all bedaquiline MIC data from Lab-1 were excluded from the analyses according to the protocol (19).
Following the exclusion of the Lab-1 data, the new analysis, based on a total of 211 observations, showed a mode of 0.03 μg/ml and a geometric mean of 0.025 μg/ml. A 3-dilution QC range based on the mode ±1 dilution included 98.1% (207/211) of the total observations (Fig. 1c). A shoulder at 0.015 μg/ml with more than 60% of the mode was noticeable. However, the lower bound of the QC range was not extended to a 4-dilution range, as this would have included a concentration that cannot be accurately measured, consistent with CLSI document M23 (19).
Therefore, the bedaquiline MIC QC range for the 7H9 broth microdilution method was set at 0.015 to 0.06 μg/ml (Fig. 1c). Analysis of MIC values, including Lab-1 data, by manufactured lots of 96-well microtiter plates showed a balanced contribution of each lot to the bedaquiline MIC distribution (Fig. 2c).
(iv) Final bedaquiline MIC QC ranges for phenotypic DST.
Based on the overall data, bedaquiline MIC QC ranges against the M. tuberculosis reference strain H37Rv have been defined (Table 1). For all three media, the lower limit of the MIC QC range was 0.015 μg/ml; the upper limit was 0.12 μg/ml for 7H10 and 7H11 agar and 0.06 μg/ml for 7H9 broth.
TABLE 1.
Bedaquiline MIC QC ranges for Mycobacterium tuberculosis H37Rv
| Medium | Bedaquiline MIC (μg/ml) |
|---|---|
| 7H9 broth | 0.015–0.06 |
| 7H10 agar | 0.015–0.12 |
| 7H11 agar | 0.015–0.12 |
Comparison of bedaquiline MICs in different media.
For the correlation between 7H10 versus 7H11 agar, all data from Lab-5 were excluded. Similarly, all data from Lab-5 and Lab-1 were excluded in the correlations of 7H9 broth versus 7H10 agar and 7H9 broth versus 7H11 agar per the protocol.
Comparison of 7H10 and 7H11 agar dilution MICs.
Regression analysis revealed a statistically significant correlation between bedaquiline MICs obtained with 7H10 agar and 7H11 agar dilution (n = 212; Pearson correlation coefficient, 0.58550; P ≤ 0.0001) (see Fig. S1a in the supplemental material). The microbiologically meaningful intermethod correlation (with ±1 dilution) established that the 7H10 and 7H11 agar media were comparable, as there was 93.9% (199/212) essential agreement between the two.
Comparison of 7H10 agar dilution and 7H9 broth microdilution MICs.
The Pearson correlation coefficient was −0.15700 (n = 178) (see Fig. S1b in the supplemental material). There was no statistically significant correlation for bedaquiline MICs determined for 7H9 broth and 7H10 agar media. Furthermore, only 72.5% essential agreement between the two media was demonstrated, which is lower than the 90% target value used to define microbiologic equivalence (19). Therefore, when performing DST of bedaquiline, the 7H10 agar dilution and 7H9 broth microdilution MICs were deemed not equivalent.
Comparison of 7H11 agar dilution and 7H9 broth microdilution MICs.
Similar results were obtained when comparing the 7H11 agar dilution and 7H9 broth microdilution MICs. The Pearson correlation coefficient was −0.22055 (n = 177) (see Fig. S1c in the supplemental material). There was no statistically significant correlation for bedaquiline MICs determined for 7H9 broth and 7H11 agar media. Only 74.6% essential agreement was demonstrated between the two media. The 7H11 agar dilution and 7H9 broth microdilution MICs were deemed not equivalent for performing DST of bedaquiline.
DISCUSSION
These two concurrent, multicountry, tier-2 QC studies analyzed a total of 666 DST results (213 observations for the 7H10 agar dilution, 242 for the 7H11 agar dilution, and 211 for the 7H9 broth microdilution methods) generated by eight separate laboratories. Based on the overall data, bedaquiline MIC QC ranges for the M. tuberculosis H37Rv reference strain were established, and DST methodology was standardized for 7H10 and 7H11 agar dilution and 7H9 broth microdilution MICs. The two agar media were shown to be microbiologically equivalent, but bedaquiline DST using 7H9 broth microdilution MICs was not equivalent to the agar dilution MICs.
Strengths of the studies included the design, which adhered to strict CLSI criteria (19). The study protocols were developed by adapting the CLSI reference standard for DST methods for aerobic bacteria (17) and mycobacteria, nocardiae, and other aerobic actinomycetes (18). This adaptation was performed using feedback from the investigators; thus, the study findings have been produced through a consensus methodology. The robustness of the findings may have been aided by the participating laboratories being highly experienced in M. tuberculosis DST, with the investigators being members of the CLSI, U.S. CDC, and internationally recognized tuberculosis laboratories, such as members of the WHO SRLN. The considerable geographical diversity across the eight laboratories demonstrates that the findings were reproduced globally.
Phenotypic DST using agar-based methods is considered to be generally reliable for drugs such as isoniazid, rifampin, and, to a lesser extent, kanamycin but can be variable for others, such as ofloxacin, ethionamide, and para-aminosalicylic acid (25). Most clinical microbiology laboratories do not yet have experience in performing phenotypic DST with bedaquiline, as this antimycobacterial drug has only recently been approved. Therefore, documentation of the methodologies for bedaquiline phenotypic DST is important. 7H11 agar was originally developed to facilitate the growth of fastidious M. tuberculosis isolates, particularly MDR and extensively drug-resistant strains, and only differs from 7H10 in that it contains a pancreatic digest of casein. However, some researchers have suggested that the supplementation with casein pancreatic digest makes no difference to the growth of M. tuberculosis. Moreover, studies evaluating different media for M. tuberculosis detection in clinical samples have produced contradictory results regarding whether 7H10 or 7H11 agar medium is more reliable (26, 27). Hence, there is no consensus among laboratories for preference between these two agar media for growth of M. tuberculosis, with use largely dictated by historical preference.
On the basis of the observed results, bedaquiline phenotypic DST can be performed on 7H10 and 7H11 agar interchangeably. However, no equivalence was demonstrated for bedaquiline DST between the 7H9 broth microdilution MIC and agar dilution MIC using either 7H10 or 7H11 agar. Therefore, MIC values obtained by the agar dilution method cannot be used interchangeably with those obtained by the 7H9 broth microdilution method.
Both the agar and broth dilution methods can be used to accurately determine bedaquiline MIC, but various factors will dictate which method is selected in each laboratory. Expeditious reporting of DST results is critical to clinical decision-making, particularly regarding antimycobacterial agent selection for patients infected with MDR strains. Therefore, one significant advantage of the broth microdilution method over agar dilution is that it provides DST results in half the time, with an incubation period of 10 days or less, compared with 21 days or more for agar media with the current methodologies. Of note, this comparison does not take into account the 6 weeks required to obtain pure colonies for the preparation of the inoculum used in either method. The bedaquiline 7H9 broth microdilution DST using frozen microtiter plates can also be performed using manual dilution (see Text S2 in the supplemental material).
Limitations of the studies included the requirement to exclude from analysis the data from one laboratory (Lab-5) for the 7H10 agar dilution method and one laboratory (Lab-1) for the 7H9 broth due to unusually high bedaquiline MICs. Although some other specific issues were reported by individual laboratories, these did not affect the outcome for determination of bedaquiline MIC ranges (see Text S1 in the supplemental material). In addition, the bedaquiline MIC QC range using the broth microdilution method is not applicable to the widely used mycobacterial growth indicator tube (MGIT; Becton Dickinson) system (28), a commercial rapid liquid culture system for diagnosis and DST. Studies have been done to determine the mean bedaquiline MIC values (0.65 μg/ml) (29) or ranges (≤0.03 to 1.00 μg/ml) (30) and epidemiological cutoffs (1.6 μg/ml [29] and 1.0 μg/ml [30]) for M. tuberculosis using the MGIT 960 system. However, these studies were not tier-2, multilaboratory, reproducibility studies, and in one of the studies (30), crushed tablets were used instead of pure powder. We expect that the manufacturer of the MGIT system will perform further QC studies of bedaquiline DST in the MGIT 960 system under rigorous standards.
The methodologies established in these studies for 7H10/7H11 agar dilution MIC and 7H9 broth microdilution MIC, coupled with the definition of bedaquiline MIC QC range standards, will inform future research, as well as provide guidance for routine clinical bedaquiline phenotypic DST and M. tuberculosis drug resistance monitoring in laboratories worldwide.
Supplementary Material
ACKNOWLEDGMENTS
We thank Janssen team members, in particular Koen Andries, Brian Dannemann, Ross Underwood, Myriam Theeuwes, and Chrispin Kambili, for their input into this paper and Ben Van Baelen (independent contractor) for statistical analyses. We acknowledge Emanuele Borroni (TB Supranational Reference Laboratory, San Raffaele Scientific Institute, Milan, Italy), Maria Wijkander and Juan Carlos Toro (TB Supranational Reference Laboratory, The Public Health Agency of Sweden, Solna, Sweden), Shaheed Vally Omar (National TB Reference Laboratory, Center for Tuberculosis, National Institute of Communicable Diseases, Johannesburg, and Department of Medical Microbiology, University of Pretoria, Pretoria, South Africa), Luc Vranckx (Janssen Pharmaceutica NV, Beerse, Belgium), David Sikes (Reference Laboratory, Division of TB Elimination, U.S. Centers for Disease Control and Prevention, Atlanta, GA, USA), Franziska Wittwer (Department of Medical Microbiology, Center of Laboratory Medicine, Luzerner Kantonsspital LUKS, Luzern, Switzerland), Alex Sloutsky (Department of Public Health, Massachusetts State Laboratory Institute, Jamaica Plain, MA, USA), and Joni Mostert (TASK Applied Science, SA MRC Centre for TB Research, DST/NRF Center of Excellence for Biomedical TB Research, Division of Molecular Biology and Human Genetics, Faculty of Medicine and Health Sciences, Stellenbosch University, Tygerberg, South Africa) for their important contributions to these studies. Medical writing support was provided by Ian Woolveridge, of Zoetic Science, Macclesfield, UK, an Ashfield Company.
K.K. and N.L. are full-time employees of Janssen. We have no additional financial conflicts of interest to declare.
The findings and conclusions in this report are those of the author(s) and do not necessarily represent the views of the Centers for Disease Control and Prevention.
These studies were done in part to satisfy FDA postmarketing requirements.
All authors substantially contributed to the study's design and protocol and execution of the work described. All authors were involved in the development of the primary manuscript and interpretation of the data and have read and approved the final version and met the criteria for authorship as established by the ICMJE.
Funding Statement
All participating laboratories received funds for this study from Janssen Pharmaceuticals except the Reference Laboratory, Division of TB Elimination, U.S. Centers for Disease Control and Prevention, Atlanta, GA, USA. Support for medical writing assistance was provided by Janssen Pharmaceuticals.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.01123-16.
For a companion article on this topic, see doi:10.1128/JCM.01138-16.
REFERENCES
- 1.WHO. 2015. Global tuberculosis report 2015. WHO, Geneva, Switzerland: http://apps.who.int/iris/bitstream/10665/191102/1/9789241565059_eng.pdf?ua=1. [Google Scholar]
- 2.Andries K, Verhasselt P, Guillemont J, Göhlmann HW, Neefs JM, Winkler H, Van Gestel J, Timmerman P, Zhu M, Lee E, Williams P, de Chaffoy D, Huitric E, Hoffner S, Cambau E, Truffot-Pernot C, Lounis N, Jarlier V. 2005. A diarylquinoline drug active on the ATP synthase of Mycobacterium tuberculosis. Science 307:223–227. doi: 10.1126/science.1106753. [DOI] [PubMed] [Google Scholar]
- 3.Diacon AH, Pym A, Grobusch MP, Patientia R, Rustomjee R, Page-Shipp L, Pistorius C, Krause R, Bogoshi M, Churchyard G, Venter A, Allen J, Palomino JC, De Marez T, van Heeswijk RP, Lounis N, Meyvisch P, Verbeeck J, Parys W, de Beule K, Andries K, Mc Neeley DF. 2009. The diarylquinoline TMC207 for multidrug-resistant tuberculosis. N Engl J Med 360:2397–2405. doi: 10.1056/NEJMoa0808427. [DOI] [PubMed] [Google Scholar]
- 4.Diacon AH, Donald PR, Pym A, Grobusch M, Patientia RF, Mahanyele R, Bantubani N, Narasimooloo R, De Marez T, van Heeswijk R, Lounis N, Meyvisch P, Andries K, McNeeley DF. 2012. Randomized pilot trial of eight weeks of bedaquiline (TMC207) treatment for multidrug-resistant tuberculosis: long-term outcome, tolerability, and effect on emergence of drug resistance. Antimicrob Agents Chemother 56:3271–3276. doi: 10.1128/AAC.06126-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Diacon AH, Pym A, Grobusch MP, de los Rios JM, Gotuzzo E, Vasilyeva I, Leimane V, Andries K, Bakare N, De Marez T, Haxaire-Theeuwes M, Lounis N, Meyvisch P, De Paepe E, van Heeswijk RP, Dannemann B, TMC207-C208 Study Group. 2014. Multidrug-resistant tuberculosis and culture conversion with bedaquiline. N Engl J Med 371:723–732. doi: 10.1056/NEJMoa1313865. [DOI] [PubMed] [Google Scholar]
- 6.Pym A, Diacon A, Tang S-J, Conradie F, Danilovits M, Chuchottaworn C, Vasilyeva I, Andries K, Bakare N, De Marez T, Haxaire-Theeuwes M, Lounis N, Meyvisch P, Van Baelen B, van Heeswijk RP, Dannemann B, TMC207-C209 Study Group. 2016. Bedaquiline in the treatment of multi- and extensively drug-resistant tuberculosis. Eur Respir J 47:564–574. doi: 10.1183/13993003.00724-2015. [DOI] [PubMed] [Google Scholar]
- 7.Janssen Pharmaceutical. 2012. Prescribing information for Sirturo (bedaquiline) tablets. Janssen Pharmaceutical, Titusville, NJ: http://www.accessdata.fda.gov/drugsatfda_docs/label/2012/204384s000lbl.pdf. [Google Scholar]
- 8.Janssen-Cilag International NV. 2014. Sirturo (bedaquiline) tablets summary of product characteristics. Janssen-Cilag International NV, Beerse, Belgium: https://www.medicines.org.uk/emc/medicine/29644. [Google Scholar]
- 9.WHO. 2013. The use of bedaquiline in the treatment of multidrug-resistant tuberculosis. Interim policy guidance. WHO, Geneva, Switzerland: http://apps.who.int/iris/bitstream/10665/84879/1/9789241505482_eng.pdf. [PubMed] [Google Scholar]
- 10.Centers for Disease Control and Prevention. 2013. Provisional CDC guidelines for the use and safety monitoring of bedaquiline fumarate (Sirturo) for the treatment of multidrug-resistant tuberculosis. MMWR Recomm Rep 62:1–12. [PubMed] [Google Scholar]
- 11.Palomino JC, Martin A, Camacho M, Guerra H, Swings J, Portaels F. 2002. Resazurin microtiter assay plate: simple and inexpensive method for detection of drug resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother 46:2720–2722. doi: 10.1128/AAC.46.8.2720-2722.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martin A, Camacho M, Portaels F, Palomino JC. 2003. Resazurin microtiter assay plate testing of Mycobacterium tuberculosis susceptibilities to second-line drugs: rapid, simple, and inexpensive method. Antimicrob Agents Chemother 47:3616–3619. doi: 10.1128/AAC.47.11.3616-3619.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Andries K, Villellas C, Coeck N, Thys K, Gevers T, Vranckx L, Lounis N, de Jong BC, Koul A. 2014. Acquired resistance of Mycobacterium tuberculosis to bedaquiline. PLoS One 9:e102135. doi: 10.1371/journal.pone.0102135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hartkoorn RC, Upekar S, Cole ST. 2014. Cross-resistance between clofazimine and bedaquiline through upregulation of MmpL5 in Mycobacterium tuberculosis. Antimicrob Agents Chemother 58:2979–2981. doi: 10.1128/AAC.00037-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Almeida D, Loerger T, Tyagi S, Andries K, Mdluli K, Grosset J, Sacchettini J, Nuermberger E. 2015. Loss-of-function mutations in pepQ confer cross-resistance to bedaquiline and clofazimine in Mycobacterium tuberculosis. Abstr 46th Union World Conf Lung Health, abstr EP-140-04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coeck N, Villellas C, Meehan CJ, Lounis N, Niemann S, Rigouts L, de Jong B, Andries K. 2015. Unexpected high frequency of Rv0678 mutations in MDR-TB patients without documented prior use of clofazimine or bedaquiline. Abstr 46th Union World Conf Lung Health, p S45. [Google Scholar]
- 17.Clinical and Laboratory Institute Standards. 2012. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, 9th ed CLSI document M07-A9. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 18.Clinical and Laboratory Standards Institute. 2011. Susceptibility testing of mycobacteria, nocardiae, and other aerobic actinomycetes; approved standard, 2nd ed CLSI document M24-A2. Clinical and Laboratory Standards Institute, Wayne, PA. [PubMed] [Google Scholar]
- 19.Clinical and Laboratory Institute Standards. 2008. Development of in vitro susceptibility testing criteria and quality control parameters; approved guideline, 3rd ed CLSI document M23-A3. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 20.Kaniga K, Cirillo DM, Hoffner S, Ismail N, Metchock B, Pfyffer G, Sloutsky A, Venter A. 2015. Abstr 46th Union World Conf Lung Health, abstr PC-806-04. [Google Scholar]
- 21.Lounis N, Vranckx L, Gevers T, Kaniga K, Andries K. 2016. In vitro culture conditions affecting the minimal inhibitory concentration of bedaquiline against M. tuberculosis. Med Mal Infect 46:220–225. doi: 10.1016/j.medmal.2016.04.007. [DOI] [PubMed] [Google Scholar]
- 22.Canetti G, Rist N, Grosset J. 1963. Mesure de la sensibilité du bacille tuberculeux aux drogues antibacillaires par la méthode des proportions. Rev Tuberc Pneumol (Paris) 27:217–272. [PubMed] [Google Scholar]
- 23.Peñuelas-Urquides K, Villarreal-Treviño L, Silva-Ramírez B, Rivadeneyra-Espinoza L, Said-Fernández S, de León MB. 2013. Measuring of Mycobacterium tuberculosis growth. A correlation of the optical measurements with colony forming units. Braz J Microbiol 44:287–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.European Committee on Antimicrobial Susceptibility Testing. 2015. Breakpoint tables for interpretation of MICs and zone diameters, version 5.0, valid from 2015-01-01. http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_5.0_Breakpoint_Table_01.pdf.
- 25.WHO. 2008. Policy guidance on drug-susceptibility testing (DST) of second-line antituberculosis drugs. WHO, Geneva, Switzerland: http://www.who.int/tb/publications/2008/whohtmtb_2008_392. [PubMed] [Google Scholar]
- 26.Rothlauf MV, Brown GL, Blair EB. 1981. Isolation of mycobacteria from undecontaminated specimens with selective 7H10 medium. J Clin Microbiol 13:76–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Joloba ML, Johnson JL, Feng PJ, Bozeman L, Goldberg SV, Morgan K, Gitta P, Boom HW, Heilig CM, Mayanja-Kizza H, Eisenach KD. 2014. What is the most reliable solid culture medium for tuberculosis treatment trials? Tuberculosis (Edinb) 94:311–316. doi: 10.1016/j.tube.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 28.Cambau E, Viveiros M, Machado D, Raskine L, Ritter C, Tortoli E, Matthys V, Hoffner S, Richter E, Perez Del Molino ML, Cirillo DM, van Soolingen D, Böttger EC. 2015. Revisiting susceptibility testing in MDR-TB by a standardized quantitative phenotypic assessment in a European multicentre study. J Antimicrob Chemother 70:686–696. doi: 10.1093/jac/dku438. [DOI] [PubMed] [Google Scholar]
- 29.Keller PM, Hömke R, Ritter C, Valsesia G, Bloemberg GV, Böttger EC. 2015. Determination of MIC distribution and epidemiological cutoff values for bedaquiline and delamanid in Mycobacterium tuberculosis using the MGIT 960 system equipped with TB eXiST. Antimicrob Agents Chemother 59:4352–4355. doi: 10.1128/AAC.00614-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Torrea G, Coeck N, Desmaretz C, Van De Parre T, Van Poucke T, Lounis N, de Jong BC, Rigouts L. 2015. Bedaquiline susceptibility testing of Mycobacterium tuberculosis in an automated liquid culture system. J Antimicrob Chemother 70:2300–2305. doi: 10.1093/jac/dkv117. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.