Abstract
This study was undertaken to evaluate the utility of matrix-assisted laser desorption ionization–time of flight mass spectrometry with the Vitek MS Plus system for identifying Mycobacterium abscessus subspecies in order to facilitate more rapid and appropriate therapy. A total of 175 clinical M. abscessus strains were identified by whole-genome sequencing analysis: 139 Mycobacterium abscessus subsp. abscessus and 36 Mycobacterium abscessus subsp. massiliense. The research-use-only (RUO) Saramis Knowledge Base database v.4.12 was modified accordingly by adding 40 M. abscessus subsp. abscessus and 19 M. abscessus subsp. massiliense reference spectra to construct subspecies SuperSpectra. A blind test, used to validate the remaining 116 isolates, yielded 99.1% (n = 115) reliability and only 0.9% (n = 1) error for subspecies identification. Among the two subspecies SuperSpectra, two specific peaks were found for M. abscessus subsp. abscessus and four specific peaks were found for M. abscessus subsp. massiliense. Our study is the first to report differential peaks 3,354.4 m/z and 6,711.1 m/z, which were specific for M. abscessus subsp. massiliense. Our research demonstrates the capacity of the Vitek MS RUO Saramis Knowledge Base database to identify M. abscessus at the subspecies level. Moreover, it validates the potential ease and accuracy with which it can be incorporated into the IVD system for the identification of M. abscessus subspecies.
INTRODUCTION
Mycobacterium abscessus is one of the most common pathogens isolated from patients with cystic fibrosis and the second most prevalent, rapidly growing nontuberculous mycobacteria (NTM) species causing NTM pulmonary diseases (1, 2). Although it might not be the most virulent NTM pathogen, inherent antibiotic resistance makes it notoriously difficult to treat (3–7). Among multiple resistance mechanisms, resistance to macrolides is noteworthy (8). In this regard, acquired resistance to clarithromycin is always associated with a mutation in the 23S rRNA gene; intrinsic resistance is conferred mainly by the erythromycin methylase (erm) gene (3). The erm gene plays a significant role in the drug susceptibility and antibiotic resistance of M. abscessus. Mycobacterium abscessus is divided into three subspecies: Mycobacterium abscessus subsp. massiliense, Mycobacterium abscessus subsp. bolletii, and Mycobacterium. abscessus subsp. abscessus (9). The most meaningful genomic difference between the three subspecies is that M. abscessus subsp. massiliense possesses an incomplete and inactive erm gene, which is 276 bp in length and harbors deletion mutations at two positions (10–13). M. abscessus subsp. massiliense can acquire clarithromycin resistance as a consequence of a 23S rRNA mutation at position A2058 (12, 14). Due to an inactive erm gene, M. abscessus subsp. massiliense does not exhibit inducible resistance after exposure to macrolides. M. abscessus subsp. abscessus and M. abscessus subsp. bolletii, on the other hand, exhibit inducible resistance (9, 15). As a result, lung diseases caused by different M. abscessus subspecies exhibit contrasting responses to macrolide-based antibiotic therapy (1, 8, 11, 16–19). As such, identification of isolates at the subspecies level, combined with antimicrobial susceptibility testing, is very important for clinicians to make proper therapeutic decisions.
Accurate identification of M. abscessus at the subspecies level is still complicated. Although PCR can differentiate M. abscessus subsp. massiliense from M. abscessus subsp. abscessus and M. abscessus subsp. bolletii by identifying the fragmented erm(41) gene, it still requires sequence analysis of several housekeeping genes (20, 21). In addition, PCR methods that depend upon gene sequencing are costly. Indeed, whole-genome sequencing (WGS), the most reliable method of identification, is extremely expensive, and the data analysis is time-consuming. Moreover, WGS is not practical in many clinical microbiology laboratories. Therefore, an easier, more rapid, accurate, and cost-effective diagnostic tool is needed to distinguish between M. abscessus subspecies.
Reportedly, matrix-assisted laser desorption ionization−time of flight mass spectrometry (MALDI-TOF MS) offers an effective method for identifying the M. abscessus complex in clinical laboratories (22). However, improvements in MALDI-TOF MS are needed to identify very close taxa that include the M. abscessus subspecies (23). A number of studies used spectrum analysis to show that several peaks differentiated M. abscessus at the subspecies level; these findings, however, were inconsistent (23–27). Possibly, these inconsistencies in subspecies profiles were determined by geographic location and related to the horizontal transfer of genes during the evolution of M. abscessus worldwide. The analysis of mass spectra provides a new opportunity to increase the range of MALDI-TOF MS application.
In the present study, 175 clinical M. abscessus isolates were collected at the Shanghai Pulmonary Hospital and analyzed by WGS. Two subspecies were observed by using the unweighted pair group with arithmetic mean (UPGMA) method. These subspecies were used as references for MALDI-TOF MS identification. We propose that the two subspecies, M. abscessus subsp. abscessus and M. abscessus subsp. massiliense, can be directly and automatically identified by updating the spectral database using the Vitek MS Plus system (bioMérieux SA, Marcy l'Etoile, France). Considering the absence of M. abscessus subsp. bolletii in our collection, it was not covered in our present research but remains an important area for further study.
MATERIALS AND METHODS
Bacteria collection and DNA extraction. (i) Bacteria collection.
In this study, 175 clinical M. abscessus isolates were obtained from the Shanghai Pulmonary Hospital affiliated with Tongji University. Mycobacterial isolates were obtained from respiratory samples of sputum and bronchoalveolar lavage fluid. These isolates were previously identified as NTM by MGIT960 medium culture and the p-nitrobenzoic acid (PNB) test. All isolates were further identified as M. abscessus by sequencing the rpoB gene. The study was approved by the Ethics Committee of Tongji University and the Shanghai Pulmonary Hospital.
Respiratory samples were transferred to Lowenstein-Jensen (LJ) agar plates after alkali treatment with 4% NaOH. Bacterial smears prepared from the colonies that grew were stained and examined microscopically to identify acid-fast bacteria. The acid-fast positive colonies were isolated and cultured in LJ medium at 37°C for 3 to 7 days (14, 28) and then were further used for DNA extraction and MALDI-TOF MS identification.
(ii) DNA extraction.
DNA extraction was performed as described previously with slight modification (29). A 10-μl loopful of bacteria grown in LJ medium was transferred to a microcentrifuge tube that contained 1 ml of TE (10 mM Tris 10, 1 mM EDTA [pH 8.0]) buffer and 250 ml of 0.5-mm glass beads. After vortexing, a 400-μl aliquot of the suspension was mixed with 1 mg/ml lysozyme and incubated overnight at 37°C. Then, 70 μl of sodium dodecyl sulfate (10%) and 10 μl of proteinase K (10 mg/ml) were added, and the mixture was incubated at 65°C for 20 min. A 100-μl solution of 10% N-acetyl-N,N,N-trimethyl ammonium bromide (CTAB) and NaCl (0.7 M) was added followed by 100 μl of NaCl (0.5 M) alone, and the mixture was incubated at 65°C for 10 min. Afterward, 750 μl of chloroform-isoamylalcohol (24:1) was added, and the tube was centrifuged at 13,000 rpm for 5 min. A supernatant was generated by the addition of a 60% volume of isopropanol. The tube was then incubated at −20°C for 20 min and centrifuged at 13,000 rpm for 15 min. The supernatant was discarded, and the sediment was washed with 70% ethanol and then dissolved in 200 μl of TE buffer. The genomic DNA recovered in the sediment was quantified using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Inc., West Palm Beach, FL, USA). High-quality DNA samples (optical density at 260/280 nm (OD260/280) of 1.8 to 2.0, >6 μg) were used to construct 350- to 450-bp fragment libraries.
Library construction and Illumina HiSeq sequencing.
For Illumina pair-end sequencing, at least 3 μg of genomic DNA derived from each strain was used for sequencing library construction. Paired-end libraries with ∼400-bp insert sizes were prepared following Illumina's standard genomic DNA library preparation procedure. Purified genomic DNA was sheared into smaller fragments with a desired size using a Covaris focused ultrasonicator (Thermo Fisher Scientific); blunt ends were generated with T4 DNA polymerase. After addition of an A base to the 3′ end of the blunt phosphorylated DNA fragments, adapters were ligated to the ends. The desired fragments were purified by gel electrophoresis and then selectively enriched and amplified by PCR. The index tag was introduced into the adapter at the PCR stage as appropriate, and a library quality test was performed. The qualified Illumina paired-end library was used to conduct Illumina HiSeq sequencing (two 150-bp reads).
Genome assembly and evolution tree construction.
Before the SPAdes (v.3.6.0; http://bioinf.spbau.ru/en/spades) software default parameters were used to assemble the genome draft (30), SPAdes was combined with BayesHammer (31) in order to adjust the bases and the program carefully to correct sequence assembly errors and any incomplete insertions. QUAST (v.2.3; http://quast.bioinf.spbau.ru/) was used to evaluate the result of the assembly (32).
MASH (v.1.0.1; https://github.com/marbl/Mash) was used with reference genomes to calculate the distance of strains between subspecies (33). Phylogenetic trees of the subspecies were constructed using the UPGMA method. Established strains ATCC 19977 (M. abscessus subsp. abscessus), CIP108297 (M. abscessus subsp. massiliense), and CIP108541 (M. abscessus subsp. bolletii) served as reference controls (11, 14, 34, 35). H37Rv (M. tuberculosis) was used as a separate species reference control.
MALDI-TOF MS. (i) Sample preparation.
A protocol developed by bioMérieux and improved by Machen et al. (36) was used to prepare the samples. Colonies were transferred from an LJ agar plate to a microcentrifuge tube that contained 800 μl of 70% ethanol and 250 μl of 0.5-mm glass beads. The microcentrifuge tube was vortexed for 15 min and then incubated at room temperature for 10 min. Subsequently, the mycobacteria were suspended by vortexing 10 s. The suspension was transferred to an empty microcentrifuge tube and centrifuged at 13,000 rpm for 5 min. The supernatant was discarded, and the pellet was dried for 10 min, resuspended in 10 μl of 70% formic acid, and incubated for 2 to 5 min at room temperature. Acetonitrile (10 μl) was added, and the tube was then centrifuged at 10,000 rpm for 3 min. One microliter of supernatant was added to a spot on a disposable target plate. The spot was dried completely, covered with 1 μl of α-cyano-4-hydroxycinnamic acid (CHCA) matrix solution and dried again before loading into the MALDI-TOF MS.
(ii) MALDI-TOF MS acquisition.
MALDI-TOF MS measurement of the isolates was performed using the Vitek MS Plus system. The spectra were acquired in linear positive ion mode at a laser frequency of 50 Hz across m/z 2,000 to 20,000 Da. For each target slide, the Escherichia coli reference strain ATCC 8739 was used for instrument calibration according to the manufacturer's specifications. After spectrum acquisition, the data were transferred from the Vitek MS acquisition station to the Saramis analysis server. The data were reported with number of peaks and the highest level matches compared to those for the Saramis 4.12 research-use-only (RUO) database (bioMérieux SA).
(iii) Spectra database upgrading.
Spectral data of 175 clinical isolates were collected and analyzed. Subsequently, the spectra from 40 M. abscessus subsp. abscessus isolates and from 19 M. abscessus subsp. massiliense isolates were imported into the RUO Saramis database. New folders of subspecies were added under the original M. abscessus species in the spectral taxonomy tree, and then the imported spectra were pasted into the respective subspecies folders. SuperSpectra were generated by creating consensus spectra that contained the 40 main peaks, which were found with 100% frequency in each respective subspecies. SuperSpectra were then activated for subsequent automated identification at the subspecies level.
Validation.
The updated database included the 59 imported reference spectra and two new subspecies SuperSpectra. It was validated by a blind test of the remaining 116 consecutive collection isolates not used to construct the new SuperSpectra.
RESULTS
Reference identification.
NTM, which are resistant to PNB, grow well in LJ medium supplemented with PNB. Our clinical M. abscessus strains were first detected by growth in the presence of PNB and further confirmed by MALDI-TOF MS identification and sequencing of the rpoB gene. The DNA was extracted from all 175 strains for WGS.
Whole-genome sequencing results.
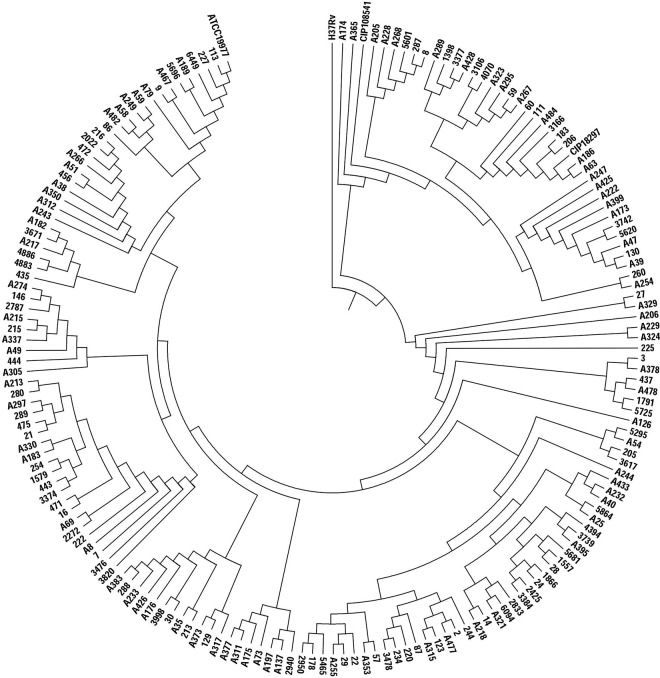
H37Rv (M. tuberculosis) was used as an exterior reference during cluster analysis, while ATCC 19977 (M. abscessus subsp. abscessus) (37), CIP108297 (M. abscessus subsp. massiliense), and CIP108541 (M. abscessus subsp. bolletii) served as the internal reference control strains. The WGS dendrogram revealed two clusters, perfectly congruent with the two subspecies. The 175 isolates consisted of 139 M. abscessus subsp. abscessus and 36 M. abscessus subsp. massiliense (Fig. 1).
FIG 1.
Using the UPGMA method, 175 strains of Mycobacterium abscessus were divided into two groups consisting of 139 subsp. abscessus isolates and 36 subsp. massiliense isolates. Established strains ATCC 19977 (M. abscessus subsp. abscessus), CIP108297 (M. abscessus subsp. massiliense), and CIP108541 (M. abscessus subsp. bolletii) served as reference controls; H37Rv (M. tuberculosis) was used as a separate species reference control.
MALDI-TOF MS results.
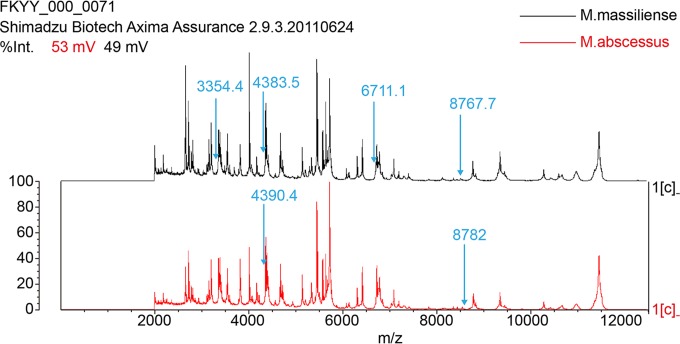
All 175 isolates were identified as M. abscessus (average identification confidence score of >95%) by comparing the collected spectra to the research-use-only (RUO) Saramis Knowledge Base database v.4.12. After expansion of the spectral database and upgrading of the taxonomy tree, the 116 blind-tested strains were differentiated into two subspecies. The identification confidence scores approached 99.9%; 99.1% of the isolates (n = 115/116) were correctly identified. The single error, sample A173, was a M. abscessus subsp. massiliense isolate that was misidentified because it exhibited a pattern with two peaks (4,390 m/z and 8,782 m/z) that were characteristic of M. abscessus subsp. abscessus. None of the strains used to construct the subspecies SuperSpectra was misidentified using our modified database.
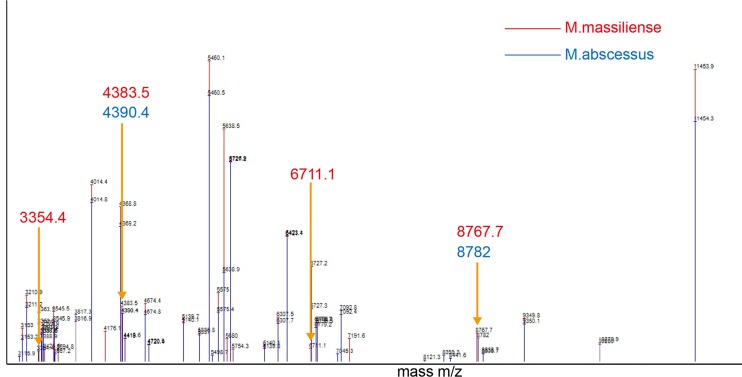
For each subspecies, 40 peaks that occurred with 100% frequency were selected for constructing the corresponding SuperSpectra (Fig. 2). Between the two subspecies SuperSpectra, 30 peaks overlapped and 10 peaks were unique. The 30 peaks that were superimposable showed a frequency of >98.3% among all 175 M. abscessus isolates; the difference of occurrence rates between the two subspecies was <2.8%. Fourteen of the misalignments exhibited a frequency difference between subspecies of <20.8%, while six peaks showed differences of >95.8%. The specific peak signals are shown in Fig. 3 and 4.
FIG 2.
SuperSpectra of M. abscessus subsp. massiliense and M. abscessus subsp. abscessus were generated using Saramis Premium software. Among a combined 40 peaks, 30 peaks overlap, while 10 peaks are unique for each subspecies. Six peaks are regarded as highly specific signals (3,354.4 m/z, 4,383.5 m/z, 4,390.4 m/z, 6,711.1 m/z, 8,767.7 m/z, and 8,782.0 m/z).
FIG 3.
Spectrometric profiles and specific M. abscessus subsp. massiliense and M. abscessus subsp. abscessus peaks were obtained by the Vitek MS Plus system and analyzed with the research-use-only (RUO) Saramis Knowledge Base database v.4.12.
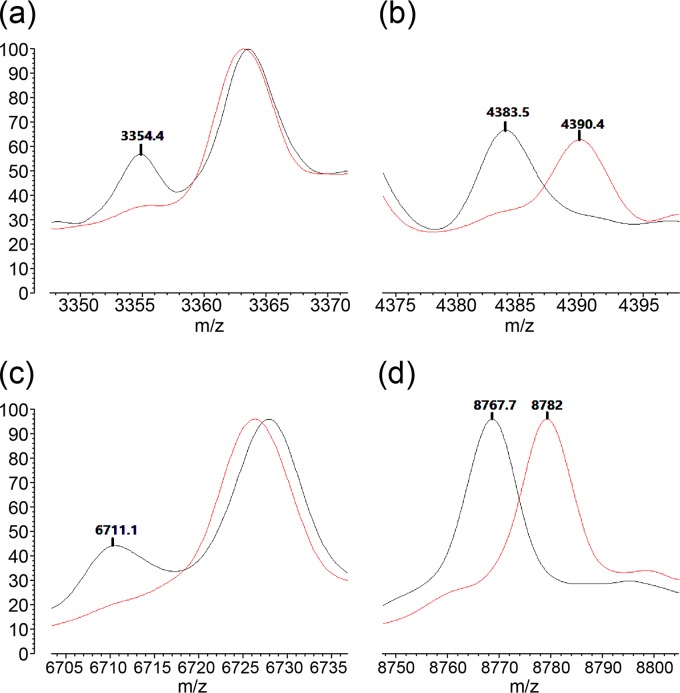
FIG 4.
Six peaks differentiate two M. abscessus subspecies. The signals at 3,354.4 m/z and 6,711.1 m/z are specific for M. abscessus subsp. massiliense (a and c). The signals at 4,383.5 m/z and 8,767.7 m/z are also specific for M. abscessus subsp. massiliense; the signals at 4,390.4 m/z and 8,782.0 m/z are specific for M. abscessus subsp. abscessus (b and d).
We found two specific peaks at 4,390.4 m/z and 8,782.0 m/z for M. abscessus subsp. abscessus and four specific peaks at 3,354.4 m/z, 4,383.5 m/z, 6,711.1 m/z, and 8,767.7 m/z for M. abscessus subsp. massiliense. Table 1 shows the differential peaks and their frequencies in the two subspecies. Peaks at 3,354.4 m/z and 6,711.1 m/z were only observed in M. abscessus subsp. massiliense. In detail, signals at 4,390.4 m/z and 8,782.0 m/z were found in 137/139 and 138/139, respectively, of the M. abscessus subsp. abscessus isolates; signals at 4,383.5 m/z and 8,767.7 m/z were found in 35/36 of the M. abscessus subsp. massiliense isolates. Signals at 3,354.4 m/z and 6,711.1 m/z were found in all M. abscessus subsp. massiliense (36/36); a signal at 3,354.4 m/z was detected in only two of the M. abscessus subsp. abscessus isolates. A signal at 6,711.1 m/z was not found among any of the M. abscessus subsp. abscessus isolates.
TABLE 1.
Six specific peaks analyzed and selected using Saramis Premium software
| Subspecies | Detection frequency (n [%]) at peak (m/z) of: |
|||||
|---|---|---|---|---|---|---|
| 3,354.4 | 4,383.5 | 4,390.4 | 6,711.1 | 8,767.7 | 8,782.0 | |
| M. abscessus subsp. massiliense (n = 36) | 36 (100) | 35 (97.2) | 1 (2.8) | 36 (100) | 35 (97.2) | 1 (2.8) |
| M. abscessus subsp. abscessus (n = 139) | 2 (1.4) | 2 (1.4) | 137 (98.6) | 0 (0) | 1 (0.7) | 138 (99.3) |
DISCUSSION
As an emerging proteomic tool for microbial identification, MALDI-TOF MS is superior in cost and speed. Previous reports showed >84% accuracy in mycobacteria species-level identification (22, 36, 38, 39) but found difficulty in distinguishing closely related (sub)species (40). The present study confirms the accuracy of MALDI-TOF MS in identifying M. abscessus at the species level (18, 22, 36, 39–41). Discriminating the subspecies correctly was the real challenge since a quick, reliable, and economic method is still not available (42). The study described herein was undertaken to determine the ability of MALDI-TOF MS to differentiate M. abscessus isolates at the subspecies level.
In the present study, all 175 isolates were precisely identified as M. abscessus at the species level using the RUO Saramis Knowledge Base. Next, the RUO Saramis Knowledge Base database was expanded with 59 spectra of clinical isolates (40 M. abscessus subsp. abscessus and 19 M. abscessus subsp. massiliense) previously identified using WGS and traditional identification methods (18).
Identifications with confidence scores of >90% obtained by activated subspecies-specific SuperSpectra were considered accurate at the subspecies level (43). Expansion of the Saramis Knowledge Base database enabled MALDI-TOF MS to discriminate between the abscessus and massiliense subspecies of M. abscessus. To optimize the accuracy of any MALDI-TOF MS identification system, it is critical to add sufficient spectral data to expand the database. This is required to address both unclaimed taxa and claimed taxa in which additional discrimination is desired. Subsequently, a blind test of 116 WGS genotyped isolates revealed 99.1% accuracy and 0.9% error. The single error occurred for isolate A173, M. abscessus subsp. massiliense, which was misidentified as M. abscessus subsp. abscessus. This misidentified isolate displayed specific signal patterns characteristic of both subspecies upon further analysis of the peak profiles and mass list, which might have occurred as the result of an ambiguous proteome caused by interspecies lateral gene transfer (44). Recently, this speculation was tested with a classification algorithm based upon 40 total strains composed of 24 M. abscessus subsp. abscessus, 10 M. abscessus massiliense subspecies, and 6 M, abscessus bolletii subspecies isolates and validated by 49 strains using the MALDI Biotyper (Bruker Daltonics, Bremen, Germany) (24). The improved algorithm reached 94% accuracy with 2% uncertainty and 4% error. Moreover, it accounted for the misidentification of strains that exhibited the biomarkers of both M. abscessus subsp. abscessus and M. abscessus subsp. massiliense (24). These hybrid allelic forms and associated proteomes displayed by several M. abscessus isolates were reported in previous studies (11, 24, 25, 44). In addition, misidentification of the M. abscessus subsp. massiliense isolate in the study conducted here may be due in part to the limited number of strains collected and used to construct the massiliense subspecies SuperSpectrum. It is obvious from the clinical data, that the incidence of the massiliense subspecies in Shanghai, China, is smaller than the incidence of the abscessus subspecies. Consequently, optimization of the M. abscessus subsp. massiliense SuperSpectrum may require the inclusion of more precise MS data. As suggested above, the percentage of massiliense subspecies determined among the total M. abscessus isolates seems to vary, depending upon geographic location. The ratio of M. abscessus subsp. massiliense to M. abscessus subsp. abscessus isolated was much lower in the United States and Europe (9, 23, 24, 45), while the rate was much higher in Japan, South Korea, and Taiwan (5, 14, 46–48). The results of the study presented herein show that 20.6% of M. abscessus identified in Shanghai, China, belonged to the massiliense subspecies. This rate is closer to that found in the United States and Europe than in other Asian locales.
Several previous studies indicated that, by aligning the specific peak signals, MALDI-TOF MS clearly differentiated M. abscessus subsp. massiliense from the other two subspecies (23–27). However, discrepancies in the specific peak signals of the three subspecies were reported. The studies of Teng et al. (26) and Suzuki et al. (25), for example, showed results similar to those reported here: specific peaks for M. abscessus subsp. abscessus at around 4,368 (4,368.24) m/z, 7,638 (7,639.70, 7,637.24) m/z, 8,782 (8,783.84, 8,781.77) m/z, and 9,475 (9,477.48, 9,473.82) m/z; and specific peaks for M. abscessus subsp. massiliense at around 4,385 (4,386.24, 4,385.05) m/z, 7,668 (7,669.20, 7,667.09) m/z, and 8,769 (8,771.73, 8,767.98) m/z. In contrast, the groups of Fangous et al. (24) and Panagea et al. (23) found three additional peaks at 2,081 m/z, 3,123 m/z, and 3,378 m/z for M. abscessus subsp. abscessus; peaks at 3,108 m/z and 3,378 m/z were observed for M. abscessus subsp. massiliense. M. abscessus subsp. bolletii, on the other hand, was rarely recovered in these studies due to its infrequent occurrence. Studies that described peaks for M. abscessus subsp. bolletii reported distinctly different results (24, 25). These studies suggested, however, that M. abscessus subsp. bolletii was so closely related to M. abscessus subsp. abscessus that discrimination by MALDI-TOF MS was difficult. In our WGS experiment, M. abscessus isolates were divided into two clusters in keeping with the observations of other investigators (18). Our results indicated that signals at 4,390.4 or 4,383.5 m/z and 8,782.0 or 8,767.7 m/z clearly differentiated M. abscessus subsp. massiliense from M. abscessus subsp. abscessus with almost 100% accuracy. This is consistent with the findings of other Asian researchers (25, 26). Peaks around 7,638 m/z or 7,668 m/z, described by Taiwanese and Japanese investigators, were not included in our SuperSpectra (26, 27). This suggests that the two peaks in our data set might be ambiguous and not specific enough for identification, which might be due in part to the nature of the culture conditions used. There is no clear evidence, however, that such conditions prominently interfere with MALDI-TOF spectra (18, 25).
M. abscessus isolates collected in Europe and the United States revealed unique peak patterns, i.e., specific signals at 2,081 m/z and 3,378 m/z (23, 24). Possibly these occurred as a function of horizontal gene transfer and genetic background differences that influence the evolution of M. abscessus worldwide (35, 49). This possibility concurs with the fact that our profiles are similar to others found in Asia but diverge from those found in Europe and the United States (25–27). The 3,354.4 m/z and 6,711.1 m/z peaks specific for M. abscessus subsp. massiliense described herein, however, have not been reported previously. More global MALDI-TOF MS experiments are needed to substantiate the effects of geographic distribution on M. abscessus subspecies type.
Mycobacterium abscessus is the most common, rapidly growing mycobacterial species that causes NTM pulmonary disease (11, 50–55). Different subspecies may indicate divergent treatment plans, especially for those plans that involve clarithromycin (5, 28, 46–49, 56). Consequently, development of a fast and accurate technique for the identification of M. abscessus subspecies is urgently needed. The present study demonstrated the potential use of the MALDI-TOF MS RUO Saramis Knowledge Base database to identify M. abscessus at the subspecies level. Upon verification of its ability to distinguish M. abscessus subspecies isolated worldwide, MALDI-TOF MS may be incorporated into the IVD system to ease use and increase diagnostic accuracy (26, 39). Once incorporated, MALDI-TOF MS will contribute significantly to the early treatment of diseases caused by M. abscessus subspecies.
ACKNOWLEDGMENTS
We thank David H. Pincus for his careful review and edit of the manuscript and Kevin Li for his useful suggestions and assistance.
We declare no conflict of interest.
This project was supported by grants from the National Natural Science Foundation of China (no. 811012310), the Medical Guide Program of the Shanghai Science and Technology Committee (no. 14411970500 and 14411962900), the Key Project of the Shanghai Municipal Health and Family Planning Commission (no. 201540367), and the Central Universities Basic Research Program (no.1511219024).
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.Bryant JM, Grogono DM, Greaves D, Foweraker J, Roddick I, Inns T, Reacher M, Haworth CS, Curran MD, Harris SR, Peacock SJ, Parkhill J, Floto RA. 2013. Whole-genome sequencing to identify transmission of Mycobacterium abscessus between patients with cystic fibrosis: a retrospective cohort study. Lancet 381:1551–1560. doi: 10.1016/S0140-6736(13)60632-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Olivier KN, Weber DJ, Wallace RJ Jr, Faiz AR, Lee JH, Zhang Y, Brown-Elliot BA, Handler A, Wilson RW, Schechter MS, Edwards LJ, Chakraborti S, Knowles MR, Nontuberculous Mycobacteria in Cystic Fibrosis Study Group. 2003. Nontuberculous mycobacteria. I: multicenter prevalence study in cystic fibrosis. Am J Respir Crit Care Med 167:828–834. [DOI] [PubMed] [Google Scholar]
- 3.Brown-Elliott BA, Vasireddy S, Vasireddy R, Iakhiaeva E, Howard ST, Nash K, Parodi N, Strong A, Gee M, Smith T, Wallace RJ Jr. 2015. Utility of sequencing the erm(41) gene in isolates of Mycobacterium abscessus subsp. abscessus with low and intermediate clarithromycin MICs. J Clin Microbiol 53:1211–1215. doi: 10.1128/JCM.02950-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choi GE, Min KN, Won CJ, Jeon K, Shin SJ, Koh WJ. 2012. Activities of moxifloxacin in combination with macrolides against clinical isolates of Mycobacterium abscessus and Mycobacterium massiliense. Antimicrob Agents Chemother 56:3549–3555. doi: 10.1128/AAC.00685-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim SY, Kim CK, Bae IK, Jeong SH, Yim JJ, Jung JY, Park MS, Kim YS, Kim SK, Chang J, Kang YA. 2015. The drug susceptibility profile and inducible resistance to macrolides of Mycobacterium abscessus and Mycobacterium massiliense in Korea. Diagn Microbiol Infect Dis 81:107–111. doi: 10.1016/j.diagmicrobio.2014.10.007. [DOI] [PubMed] [Google Scholar]
- 6.Köser CU, Ellington MJ, Peacock SJ. 2014. Whole-genome sequencing to control antimicrobial resistance. Trends Genet 30:401–407. doi: 10.1016/j.tig.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Renna M, Schaffner C, Brown K, Shang S, Tamayo MH, Hegyi K, Grimsey NJ, Cusens D, Coulter S, Cooper J, Bowden AR, Newton SM, Kampmann B, Helm J, Jones A, Haworth CS, Basaraba RJ, DeGroote MA, Ordway DJ, Rubinsztein DC, Floto RA. 2011. Azithromycin blocks autophagy and may predispose cystic fibrosis patients to mycobacterial infection. J Clin Invest 121:3554–3563. doi: 10.1172/JCI46095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koh WJ, Jeon K, Lee NY, Kim BJ, Kook YH, Lee SH, Park YK, Kim CK, Shin SJ, Huitt GA, Daley CL, Kwon OJ. 2011. Clinical significance of differentiation of Mycobacterium massiliense from Mycobacterium abscessus. Am J Respir Crit Care Med 183:405–410. doi: 10.1164/rccm.201003-0395OC. [DOI] [PubMed] [Google Scholar]
- 9.Zelazny AM, Root JM, Shea YR, Colombo RE, Shamputa IC, Stock F, Conlan S, McNulty S, Brown-Elliott BA, Wallace RJ Jr, Olivier KN, Holland SM, Sampaio EP. 2009. Cohort study of molecular identification and typing of Mycobacterium abscessus, Mycobacterium massiliense, and Mycobacterium bolletii. J Clin Microbiol 47:1985–1995. doi: 10.1128/JCM.01688-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choo SW, Wee WY, Ngeow YF, Mitchell W, Tan JL, Wong GJ, Zhao Y, Xiao J. 2014. Genomic reconnaissance of clinical isolates of emerging human pathogen Mycobacterium abscessus reveals high evolutionary potential. Sci Rep 4:4061. doi: 10.1038/srep04061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jeon SM, Lim NR, Kwon SJ, Shim TS, Park MS, Kim BJ, Kim SH. 2014. Analysis of species and intra-species associations between the Mycobacterium abscessus complex strains using pulsed-field gel electrophoresis (PFGE) and multi-locus sequence typing (MLST). J Microbiol Methods 104:19–25. doi: 10.1016/j.mimet.2014.05.024. [DOI] [PubMed] [Google Scholar]
- 12.Maurer FP, Ruegger V, Ritter C, Bloemberg GV, Bottger EC. 2012. Acquisition of clarithromycin resistance mutations in the 23S rRNA gene of Mycobacterium abscessus in the presence of inducible erm(41). J Antimicrob Chemother 67:2606–2611. doi: 10.1093/jac/dks279. [DOI] [PubMed] [Google Scholar]
- 13.Mougari F, Raskine L, Ferroni A, Marcon E, Sermet-Gaudelus I, Veziris N, Heym B, Gaillard JL, Nassif X, Cambau E. 2014. Clonal relationship and differentiation among Mycobacterium abscessus isolates as determined using the semiautomated repetitive extragenic palindromic sequence PCR-based DiversiLab system. J Clin Microbiol 52:1969–1977. doi: 10.1128/JCM.03600-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim HY, Kim BJ, Kook Y, Yun YJ, Shin JH, Kim BJ, Kook YH. 2010. Mycobacterium massiliense is differentiated from Mycobacterium abscessus and Mycobacterium bolletii by erythromycin ribosome methyltransferase gene (erm) and clarithromycin susceptibility patterns. Microbiol Immunol 54:347–353. doi: 10.1111/j.1348-0421.2010.00221.x. [DOI] [PubMed] [Google Scholar]
- 15.Ripoll F, Pasek S, Schenowitz C, Dossat C, Barbe V, Rottman M, Macheras E, Heym B, Herrmann JL, Daffe M, Brosch R, Risler JL, Gaillard JL. 2009. Non mycobacterial virulence genes in the genome of the emerging pathogen Mycobacterium abscessus. PLoS One 4:e5660. doi: 10.1371/journal.pone.0005660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adékambi T, Berger P, Raoult D, Drancourt M. 2006. rpoB gene sequence-based characterization of emerging non-tuberculous mycobacteria with descriptions of Mycobacterium bolletii sp. nov., Mycobacterium phocaicum sp. nov and Mycobacterium aubagnense sp nov. Int J Syst Evol Microbiol 56:133–143. doi: 10.1099/ijs.0.63969-0. [DOI] [PubMed] [Google Scholar]
- 17.Adékambi T, Reynaud-Gaubert M, Greub G, Gevaudan MJ, La Scola B, Raoult D, Drancourt M. 2004. Amoebal coculture of “Mycobacterium massiliense” sp. nov. from the sputum of a patient with hemoptoic pneumonia. J Clin Microbiol 42:5493–5501. doi: 10.1128/JCM.42.12.5493-5501.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leao SC, Tortoli E, Euzeby JP, Garcia MJ. 2011. Proposal that Mycobacterium massiliense and Mycobacterium bolletii be united and reclassified as Mycobacterium abscessus subsp. bolletii comb. nov., designation of Mycobacterium abscessus subsp. abscessus subsp. nov. and emended description of Mycobacterium abscessus. Int J Syst Evol Microbiol 61:2311–2313. doi: 10.1099/ijs.0.023770-0. [DOI] [PubMed] [Google Scholar]
- 19.Lyu J, Kim BJ, Kim BJ, Song JW, Choi CM, Oh YM, Lee SD, Kim WS, Kim DS, Shim TS. 2014. A shorter treatment duration may be sufficient for patients with Mycobacterium massiliense lung disease than with Mycobacterium abscessus lung disease. Respir Med 108:1706–1712. doi: 10.1016/j.rmed.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 20.Kim K, Hong SH, Kim BJ, Kim BR, Lee SY, Kim GN, Shim TS, Kook YH, Kim BJ. 2015. Separation of Mycobacterium abscessus into subspecies or genotype level by direct application of peptide nucleic acid multi-probe-real-time PCR method into sputa samples. BMC Infect Dis 15:325. doi: 10.1186/s12879-015-1076-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakanaga K, Sekizuka T, Fukano H, Sakakibara Y, Takeuchi F, Wada S, Ishii N, Makino M, Kuroda M, Hoshino Y. 2014. Discrimination of Mycobacterium abscessus subsp. massiliense from Mycobacterium abscessus subsp. abscessus in clinical isolates by multiplex PCR. J Clin Microbiol 52:251–259. doi: 10.1128/JCM.01327-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen JH, Yam WC, Ngan AH, Fung AM, Woo WL, Yan MK, Choi GK, Ho PL, Cheng VC, Yuen KY. 2013. Advantages of using matrix-assisted laser desorption ionization-time of flight mass spectrometry as a rapid diagnostic tool for identification of yeasts and mycobacteria in the clinical microbiological laboratory. J Clin Microbiol 51:3981–3987. doi: 10.1128/JCM.01437-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Panagea T, Pincus DH, Grogono D, Jones M, Bryant J, Parkhill J, Floto RA, Gilligan P. 2015. Mycobacterium abscessus complex identification with matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol 53:2355–2358. doi: 10.1128/JCM.00494-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fangous MS, Mougari F, Gouriou S, Calvez E, Raskine L, Cambau E, Payan C, Hery-Arnaud G. 2014. Classification algorithm for subspecies identification within the Mycobacterium abscessus species, based on matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol 52:3362–3369. doi: 10.1128/JCM.00788-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suzuki H, Yoshida S, Yoshida A, Okuzumi K, Fukusima A, Hishinuma A. 2015. A novel cluster of Mycobacterium abscessus complex revealed by matrix-assisted laser desorption ionization-time-of-flight mass spectrometry (MALDI-TOF MS). Diagn Microbiol Infect Dis 83:365−370. doi: 10.1016/j.diagmicrobio.2015.08.011. [DOI] [PubMed] [Google Scholar]
- 26.Teng SH, Chen CM, Lee MR, Lee TF, Chien KY, Teng LJ, Hsueh PR. 2013. Matrix-assisted laser desorption ionization-time of flight mass spectrometry can accurately differentiate between Mycobacterium massiliense (M. abscessus subspecies bolletti) and M. abscessus (sensu stricto). J Clin Microbiol 51:3113–3116. doi: 10.1128/JCM.01239-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tseng SP, Teng SH, Lee PS, Wang CF, Yu JS, Lu PL. 2013. Rapid identification of M. abscessus and M. massiliense by MALDI-TOF mass spectrometry with a comparison to sequencing methods and antimicrobial susceptibility patterns. Future Microbiol 8:1381–1389. doi: 10.2217/fmb.13.115. [DOI] [PubMed] [Google Scholar]
- 28.Luo L, Li B, Chu H, Huang D, Zhang Z, Zhang J, Gui T, Xu L, Zhao L, Sun X, Xiao H. 2016. Characterization of Mycobacterium abscessus subtypes in Shanghai of China: drug sensitivity and bacterial epidemicity as well as clinical manifestations. Medicine (Baltimore) 95:e2338. doi: 10.1097/MD.0000000000002338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Somerville W, Thibert L, Schwartzman K, Behr MA. 2005. Extraction of Mycobacterium tuberculosis DNA: a question of containment. J Clin Microbiol 43:2996–2997. doi: 10.1128/JCM.43.6.2996-2997.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nikolenko SI, Korobeynikov AI, Alekseyev MA. 2013. BayesHammer: Bayesian clustering for error correction in single-cell sequencing. BMC Genomics 14(Suppl 1):S7. doi: 10.1186/1471-2164-14-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gurevich A, Saveliev V, Vyahhi N, Tesler G. 2013. QUAST: quality assessment tool for genome assemblies. Bioinformatics 29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ondov BD, Treangen TJ, Mallonee AB, Bergman NH, Koren S, Phillippy AM. 2015. Mash: fast genome and metagenome distance estimation using MinHash. Genome Biol 20:132. doi: 10.1186/s13059-016-0997-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Davidson RM, Hasan NA, de Moura VC, Duarte RS, Jackson M, Strong M. 2013. Phylogenomics of Brazilian epidemic isolates of Mycobacterium abscessus subsp. bolletii reveals relationships of global outbreak strains. Infect Genet Evol 20:292–297. doi: 10.1016/j.meegid.2013.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Davidson RM, Hasan NA, Reynolds PR, Totten S, Garcia B, Levin A, Ramamoorthy P, Heifets L, Daley CL, Strong M. 2014. Genome sequencing of Mycobacterium abscessus isolates from patients in the united states and comparisons to globally diverse clinical strains. J Clin Microbiol 52:3573–3582. doi: 10.1128/JCM.01144-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Machen A, Kobayashi M, Connelly MR, Wang YF. 2013. Comparison of heat inactivation and cell disruption protocols for identification of mycobacteria from solid culture media by use of Vitek matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol 51:4226–4229. doi: 10.1128/JCM.02612-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Howard ST. 2013. Recent progress towards understanding genetic variation in the Mycobacterium abscessus complex. Tuberculosis 93(Suppl):S15–S20. doi: 10.1016/S1472-9792(13)70005-2. [DOI] [PubMed] [Google Scholar]
- 38.Boyle DP, Zembower TR, Qi C. 2015. Evaluation of Vitek MS for rapid classification of clinical isolates belonging to Mycobacterium avium complex. Diagn Microbiol Infect Dis 81:41–43. doi: 10.1016/j.diagmicrobio.2014.09.026. [DOI] [PubMed] [Google Scholar]
- 39.Dunne WM Jr, Doing K, Miller E, Miller E, Moreno E, Baghli M, Mailler S, Girard V, van Belkum A, Deol P. 2014. Rapid inactivation of Mycobacterium and Nocardia species before identification using matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol 52:3654–3659. doi: 10.1128/JCM.01728-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saleeb PG, Drake SK, Murray PR, Zelazny AM. 2011. Identification of mycobacteria in solid-culture media by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol 49:1790–1794. doi: 10.1128/JCM.02135-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seng P, Drancourt M, Gouriet F, La Scola B, Fournier PE, Rolain JM, Raoult D. 2009. Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin Infect Dis 49:543–551. doi: 10.1086/600885. [DOI] [PubMed] [Google Scholar]
- 42.Leung JM, Olivier KN. 2013. Nontuberculous mycobacteria: the changing epidemiology and treatment challenges in cystic fibrosis. Curr Opin Pulm Med 19:662–669. doi: 10.1097/MCP.0b013e328365ab33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mather CA, Rivera SF, Butler-Wu SM. 2014. Comparison of the Bruker Biotyper and Vitek MS matrix-assisted laser desorption ionization-time of flight mass spectrometry systems for identification of mycobacteria using simplified protein extraction protocols. J Clin Microbiol 52:130–138. doi: 10.1128/JCM.01996-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Macheras E, Konjek J, Roux AL, Thiberge JM, Bastian S, Leao SC, Palaci M, Sivadon-Tardy V, Gutierrez C, Richter E, Rusch-Gerdes S, Pfyffer GE, Bodmer T, Jarlier V, Cambau E, Brisse S, Caro V, Rastogi N, Gaillard JL, Heym B. 2014. Multilocus sequence typing scheme for the Mycobacterium abscessus complex. Res Microbiol 165:82–90. doi: 10.1016/j.resmic.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 45.Bastian S, Veziris N, Roux AL, Brossier F, Gaillard JL, Jarlier V, Cambau E. 2011. Assessment of clarithromycin susceptibility in strains belonging to the Mycobacterium abscessus group by erm(41) and rrl sequencing. Antimicrob Agents Chemother 55:775–781. doi: 10.1128/AAC.00861-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Harada T, Akiyama Y, Kurashima A, Nagai H, Tsuyuguchi K, Fujii T, Yano S, Shigeto E, Kuraoka T, Kajiki A, Kobashi Y, Kokubu F, Sato A, Yoshida S, Iwamoto T, Saito H. 2012. Clinical and microbiological differences between Mycobacterium abscessus and Mycobacterium massiliense lung diseases. J Clin Microbiol 50:3556–3561. doi: 10.1128/JCM.01175-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang YC, Liu MF, Shen GH, Lin CF, Kao CC, Liu PY, Shi ZY. 2010. Clinical outcome of Mycobacterium abscessus infection and antimicrobial susceptibility testing. J Microbiol Immunol Infect 43:401–406. doi: 10.1016/S1684-1182(10)60063-1. [DOI] [PubMed] [Google Scholar]
- 48.Lyu J, Jang HJ, Song JW, Choi CM, Oh YM, Lee SD, Kim WS, Kim DS, Shim TS. 2011. Outcomes in patients with Mycobacterium abscessus pulmonary disease treated with long-term injectable drugs. Respir Med 105:781–787. doi: 10.1016/j.rmed.2010.12.012. [DOI] [PubMed] [Google Scholar]
- 49.Hoefsloot W, van Ingen J, Andrejak C, Angeby K, Bauriaud R, Bemer P, Beylis N, Boeree MJ, Cacho J, Chihota V, Chimara E, Churchyard G, Cias R, Daza R, Daley CL, Dekhuijzen PN, Domingo D, Drobniewski F, Esteban J, Fauville-Dufaux M, Folkvardsen DB, Gibbons N, Gomez-Mampaso E, Gonzalez R, Hoffmann H, Hsueh PR, Indra A, Jagielski T, Jamieson F, Jankovic M, Jong E, Keane J, Koh WJ, Lange B, Leao S, Macedo R, Mannsaker T, Marras TK, Maugein J, Milburn HJ, Mlinko T, Morcillo N, Morimoto K, Papaventsis D, Palenque E, Paez-Pena M, Piersimoni C, Polanova M, Rastogi N, Richter E, et al. . 2013. The geographic diversity of nontuberculous mycobacteria isolated from pulmonary samples: an NTM-NET collaborative study. Eur Respir J 42:1604–1613. doi: 10.1183/09031936.00149212. [DOI] [PubMed] [Google Scholar]
- 50.Anonymous. 1997. Diagnosis and treatment of disease caused by nontuberculous mycobacteria. This official statement of the American Thoracic Society was approved by the Board of Directors, March 1997. Medical Section of the American Lung Association. Am J Respir Crit Care Med 156:S1–25. [DOI] [PubMed] [Google Scholar]
- 51.Benwill JL, Wallace RJ Jr. 2014. Mycobacterium abscessus: challenges in diagnosis and treatment. Curr Opin Infect Dis 27:506–510. doi: 10.1097/QCO.0000000000000104. [DOI] [PubMed] [Google Scholar]
- 52.Brown-Elliott BA, Wallace RJ Jr. 2002. Clinical and taxonomic status of pathogenic nonpigmented or late-pigmenting rapidly growing mycobacteria. Clin Microbiol Rev 15:716–746. doi: 10.1128/CMR.15.4.716-746.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Griffith DE, Aksamit T, Brown-Elliott BA, Catanzaro A, Daley C, Gordin F, Holland SM, Horsburgh R, Huitt G, Iademarco MF, Iseman M, Olivier K, Ruoss S, von Reyn CF, Wallace RJ Jr, Winthrop K, ATS Mycobacterial Diseases Subcommittee, American Thoracic Society, Infectious Disease Society of America. 2007. An official ATS/IDSA statement: diagnosis, treatment, and prevention of nontuberculous mycobacterial diseases. Am J Respir Crit Care Med 175:367–416. doi: 10.1164/rccm.200604-571ST. [DOI] [PubMed] [Google Scholar]
- 54.Medjahed H, Gaillard JL, Reyrat JM. 2010. Mycobacterium abscessus: a new player in the mycobacterial field. Trends Microbiol 18:117–123. doi: 10.1016/j.tim.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 55.O'Driscoll C, Konjek J, Heym B, Fitzgibbon MM, Plant BJ, Ni Chroinin M, Mullane D, Lynch-Healy M, Corcoran GD, Schaffer K, Rogers TR, Prentice MB. 2016. Molecular epidemiology of Mycobacterium abscessus complex isolates in Ireland. J Cyst Fibros 15:179–185. doi: 10.1016/j.jcf.2015.05.007. [DOI] [PubMed] [Google Scholar]
- 56.Simons S, van Ingen J, Hsueh PR, Van Hung N, Dekhuijzen PN, Boeree MJ, van Soolingen D. 2011. Nontuberculous mycobacteria in respiratory tract infections, eastern Asia. Emerg Infect Dis 17:343–349. doi: 10.3201/eid170310060. [DOI] [PMC free article] [PubMed] [Google Scholar]