Abstract
Background
LIM domain only 2 (LMO2, human gene) is a key transcription factor that regulates hematopoiesis and vascular development. However, its role in adult endothelial function has been incompletely characterized.
Methods and Results
In vitro loss‐ and gain‐of‐function studies on LMO2 were performed in human umbilical vein endothelial cells with lentiviral overexpression or short hairpin RNA knockdown (KD) of LMO2, respectively. LMO2 KD significantly impaired endothelial proliferation. LMO2 controls endothelial G1/S transition through transcriptional regulation of cyclin‐dependent kinase 2 and 4 as determined by reverse transcription polymerase chain reaction (PCR), western blot, and chromatin immunoprecipitation, and also influences the expression of Cyclin D1 and Cyclin A1. LMO2 KD also impaired angiogenesis by reducing transforming growth factor‐β (TGF‐β) expression, whereas supplementation of exogenous TGF‐β restored defective network formation in LMO2 KD human umbilical vein endothelial cells. In a zebrafish model of caudal fin regeneration, RT‐PCR revealed that the lmo2 (zebrafish gene) gene was upregulated at day 5 postresection. The KD of lmo2 by vivo‐morpholino injections in adult Tg(fli1:egfp) y1 zebrafish reduced 5‐bromo‐2′‐deoxyuridine incorporation in endothelial cells, impaired neoangiogenesis in the resected caudal fin, and substantially delayed fin regeneration.
Conclusions
The transcriptional factor LMO2 regulates endothelial proliferation and angiogenesis in vitro. Furthermore, LMO2 is required for angiogenesis and tissue healing in vivo. Thus, LMO2 is a critical determinant of vascular and tissue regeneration.
Keywords: angiogenesis, endothelial cell, LIM domain only 2, proliferation, regeneration, transcription factors
Subject Categories: Vascular Biology
Introduction
LIM domain only 2 (LMO2, human gene) is a critical transcription factor for initiation of yolk sac and definitive hematopoiesis.1 The homozygous LMO2 null mutation leads to failure of yolk sac erythropoiesis and embryonic lethality around E10.5 in mice.1 LMO2 overexpression (OE) impedes the differentiation of T progenitor cells, and aberrant activation of LMO2 by chromosomal translocations contributes to T‐cell acute lymphoblastic leukemia in human.2, 3 As a transcription factor, LMO2 does not directly bind DNA, but instead forms a DNA‐binding complex with its two LIM–zinc‐finger‐like protein interaction modules.4 LMO2 interacts with GATA‐1, TAL1, E2A, and Ldb1/NL1 to form a DNA‐binding complex that recognizes a bipartite DNA motif comprising an E‐box, CAGGTG followed 9 bp downstream by a GATA site.5
In addition, LMO2 plays an essential role in embryonic angiogenesis.4 In fact, Lmo2 (mouse gene) null embryonic stem cells in mouse chimeras showed marked disorganization of the vascular system.4 Lmo2 knockdown (KD) by morpholino (Mo) decreases the number of intersomitic vessel sprouts and axial vessel formation in zebrafish embryos.6 In endothelial cells (ECs), Fli1, Elf1, and Ets1 regulate LMO2 gene expression through direct binding to its proximal promoter.7 Unlike other transcription factors crucial for endothelial lineage development such as ETV2, LMO2 remains highly expressed in adult ECs, suggesting a key role for this transcription factor in adult EC function.
LMO2 has been suggested to play an important role in tumor angiogenesis.8 Indeed, it is a robust marker of endothelium in vascular neoplasms9 such as infantile hemangioma.10 In the EAhy926 cell line, an immortalized EC line, OE of LMO2 promotes angiogenesis and cell proliferation.10 However, it is unclear how LMO2 controls proliferation in the immortalized EC line, nor is it known whether normal adult endothelial function and vascular homeostasis are so regulated.
To assess the role of LMO2 in adult EC function, we performed in vitro loss‐ and gain‐of function experiments of LMO2 in human umbilical vein ECs (HUVECs) to assess major aspects of endothelial function such as proliferation, network formation, acetylated‐low density lipoprotein (acLDL) uptake, NO production, and cell surface marker expression. We further screened and identified the key molecular targets of LMO2, cyclin‐dependent kinase (CDK) 2 and CDK4, and related cyclins involved in endothelial proliferation and transforming growth factor‐β (TGF‐β1)–promoted angiogenesis. We examined the role of LMO2 in vivo in a caudal fin resection model in adult zebrafish. Our studies suggest that LMO2 is a critical determinant of angiogenesis and tissue regeneration.
Material and Methods
Chemicals and Antibodies
All chemicals were purchased from Sigma‐Aldrich (St. Louis, MO) unless otherwise stated. EGM‐2 medium and bullet kit was from Lonza (Walkersville, MD). Angiogenesis RT Profiler PCR Array and chromatin immunoprecipitation (ChIP) polymerase chain reaction (PCR) primers were from Qiagen (Valencia, CA). TGF‐β ELISA kits were from R&D Systems (Minneapolis, MN). Taqman primers, cell proliferation kit, AF594‐conjugated acLDL, CellTracker Red, and Griess kit was from Invitrogen (Carlsbad, CA). PE‐human CD31 antibody and APC‐human CD144 antibody was from BD Biosciences (San Jose, CA). ChIP kit was from Cell Signaling Technology (Beverly, MA). Anti‐human LMO2 antibody (clone SP51) was purchased from Spring Bioscience (Pleasanton, CA). Cyclin D1, p21, and horseradish peroxidase (HRP)–conjugated goat anti‐mouse/rabbit antibodies were from Santa Cruz Biotechnology Inc (Santa Cruz, CA). Cyclin A, Cyclin E, CDK2, and CDK4 antibodies were from Millipore (Billerica, MA). β‐Tubulin antibody and anti‐zebrafish lmo2 (zebrafish gene) antibody was from Abcam (Cambridge, UK). Lentiviral particles of control (CT) short hairpin RNA (shRNA) and LMO2 shRNA were from Santa Cruz Biotechnology Inc. CT noncoding and LMO2 open reading frame (ORF) OE lentiviral particles were from Applied Biological Materials Inc (Richmond, BC, Canada).
Zebrafish Husbandry
Tg(fli1:egfp) y1 fish were raised according to standard procedures11 and kept at 28°C under a 14/10‐hour light/dark cycle and fed with dry meal (Gemma Micro) twice per day. Animals were housed and all experiments were carried out in accordance with the recommendations of the Institutional Animal Care and Use Committee. All surgery procedures were performed under anesthesia.
Stable Cell Generation
HUVEC were cultured in EBM‐2 basal medium supplemented with EGM‐2 bullet kits. HUVECs at passage 4 were grown to 60% confluence before infection with lentiviral particles containing shRNA targeting LMO2 or ORF of LMO2. Scramble shRNA containing virus or virus without LMO2 ORF were used as controls. Stable KD or OE of LMO2 was obtained by puromycin (1 μg/mL) selection for consecutive 14 days after viral infection. LMO2 KD or OE was confirmed by gene expression and protein level analysis.
Protein Extraction and Western Blot Analysis
Cultured cells or zebrafish tissue samples were collected and solubilized in RIPA buffer (25 mmol/L Tris‐HCl pH 7.6, 150 mmol/L NaCl, 1% NP‐40, 1% sodium deoxycholate, and 0.1% SDS) supplemented with protease inhibitor cocktail. Protein concentration was measured using BCA assay. Samples (containing 50–100 μg protein) were subjected to polyacrylamide gel electrophoresis (4–12% gradient) and transferred onto nitrocellulose membranes. Blots were stained with 1% Ponceau S for loading controls. Blots were then blocked with 5% nonfat milk in PBST (PBS+0.1% Tween) for 1 hour at room temperature and probed with primary antibody overnight at 4°C. They were then washed 4 times with PBST for 10 minutes. HRP‐conjugated goat anti‐mouse or rabbit antibodies were incubated for 1 hour at room temperature. Blots were washed 4 times with PBST for 10 minutes. Antigen‐antibody complexes were then detected by exposure in FluorChem M system from ProteinSimple (San Jose, CA). β‐tubulin was used as loading control.
RNA Extraction and RT‐PCR
mRNA was extracted from cultured cells or homogenized zebrafish caudal fins using column purification.12 The mRNA was reverse transcribed into cDNA. Taqman primers targeting specific genes were then used for real‐time PCR (RT‐PCR; QuantStudio Real‐Time PCR System, Life Technologies, Carlsbad, CA). 18S was used as an internal control for HUVEC studies and β‐actin was used as an internal control for zebrafish studies. Angiogenesis RT Profiler PCR Array was performed according to the manufacturer's manual.
Endothelial Proliferation Assay, Network Formation Assay, and Griess Assay
Cell proliferation was examined by proliferation assay. HUVECs were seeded at the same starting number 5×104 cells in EGM‐2 medium and counted and subcultured every other day for 4 days. Angiogenesis was examined by matrigel network formation assay. Briefly, 24‐well plates were coated with Matrigel 300 μL (BD Biosciences), and incubated at 37°C for 30 minutes. In each well, 7×104 cells in 500 μL EGM‐2 medium was seeded. After 16 hours, cells were stained with CellTracker Red 5 μmol/L for 30 minutes and visualized under the fluorescence microscope. Griess assay was used for NO measurement based on the manufacturer's manual.
ELISA Assay
TGF‐β1 ELISA assay was performed following the manufacturer's manual.
ChIP‐PCR Assay
ChIP assay was performed following the manufacturer's manual. Briefly, DNA and protein were crosslinked by 1% formaldehyde. Chromatin was isolated and digested with micrococcal nuclease. The DNA‐protein complex was then precipitated with control IgG or antibodies against LMO2 overnight at 4°C and protein A/G–conjugated magnetic beads for 1 hour. Cross‐links were reversed. The extracted DNA was used as a template for PCR amplification of the targeted promoter region. The putative LMO2 complex binding sites were predicted using the DECODE (Decipherment of DNA Elements) database.
Vivo‐Mo Injection in Zebrafish
Suppression of lmo2 transcription factor was achieved by using antisense vivo‐Mo 5′‐GTAGAAGCCATTTTCAATATGATTC‐3′ (Gene Tools LLC, Philomath, OR) targeting the LMO2 mRNA translation initiation site. Vivo‐Mo is made by covalently binding a standard Mo to a synthetic scaffold containing guanidinium head groups as a delivery moiety.13 An antisense vivo‐Mo that targets human β‐globin intron mutation 5′‐CCTCTTACCTCATTACAATTTATA‐3 was used as control. Vivo‐Mo was injected into the retro‐orbital vein, as previously described.14 Briefly, zebrafish were anesthetized in tricaine 0.05 mg/mL, and 2 μL of 0.1 mmol/L vivo‐Mo solution was loaded in a glass capillary and injected with a standard microinjector (IM300 Microinjector; Narishige, Tokyo, Japan) on days 0, 2, 4, 6, 8, 10, 12, and 14.
BrdU Incorporation in Zebrafish
To label proliferating cells, a dose of 5 μL of 5‐bromo‐2′‐deoxyuridine (BrdU; 10 mmol/L in PBS) was injected into the retro‐orbital vein at days 11 and 13 after the day 0 injection of vivo‐MO. At day 15, fish were euthanized for enzymatic digestion in PBS containing trypsin 0.4%, 1 mmol/L EDTA, and collagenase P 0.05 mg/mL, and incubated for 45 minutes at 28.5°C with pipetting every 5 to 10 minutes to obtain single‐cell suspension for fluorescence‐activated cell sorter (FACS) analysis.
Caudal Fin Resection and Regeneration
Caudal fin resection in adult fishes was performed with sterile razor blades at day 4 after the day 0 injection of vivo‐Mo and allowed to regenerate. Total regeneration was measured as previously described.15 Briefly, fin images were collected before amputation and time points after amputation. The new tissue area (in pixels) of the caudal fin from the new distal fin edge to the amputation plane was quantified in each fish up to 5 days postresection using Image J software. The percentage of regeneration for each fin at each time point was defined as percentage of regeneration=100× (regenerated tissue area/original fin area amputated).
FACS Analysis
HUVECs were washed with PBS and trypsinized to generate single‐cell suspension. For cell surface marker detection, single‐cell suspension cells were stained with indicated antibody. For cell cycle analysis, single‐cell suspension was fixed with 70% EtOH at 4°C overnight. Cells were then stained using propidium iodide (PI)/RNase buffer for 30 minutes and immediately run on a BD LSR II machine (BD Biosciences). BrdU incorporation assay in zebrafish was performed as previously described.16 Data were analyzed by FlowJo software (FlowJo, LLC, Ashland, OR). Cell cycle distribution was analyzed using the cell cycle package of FlowJo.
Data Analysis
Statistical analysis was performed with Prism 7 software (GraphPad Software, Inc, La Jolla, CA). Results were expressed as mean±SEM. The Shapiro‐Wilk test was used to confirm the null hypothesis that the data follow a normal distribution. Statistical comparisons between the two groups were then performed via Student t test, and one‐way ANOVA test was used to analyze multiple groups. Bonferroni corrections test was applied for multiple comparisons. P<0.05 was considered significant.
Results
Generation of LMO2 KD HUVECs and LMO2 OE HUVECs
A stable line of LMO2 KD HUVECs was generated by infection with lentiviral particles containing shRNA targeting LMO2 followed by puromycin selection. A CT HUVEC line was generated using lentiviral scramble shRNA. Compared with CT cells, the LMO2 KD cells exhibited a 70% decrease in LMO2 gene expression detected by RT‐PCR (Figure 1A) and an 80% reduction in protein level by Western blot (Figure 1B). The morphology of LMO2 KD cells was similar to CT (Figure S1). A stable line of HUVECs overexpressing LMO2 was generated by lentiviral particles encoding the ORF of LMO2 followed by puromycin selection. The CT HUVEC line was generated using lentiviral particles without an LMO2 ORF. Compared with CT cells, the LMO2 OE stable cells had 8‐fold increases in LMO2 gene expression detected by RT‐PCR (Figure 1C) and 7‐fold increases in protein level by Western blot (Figure 1D).
Figure 1.
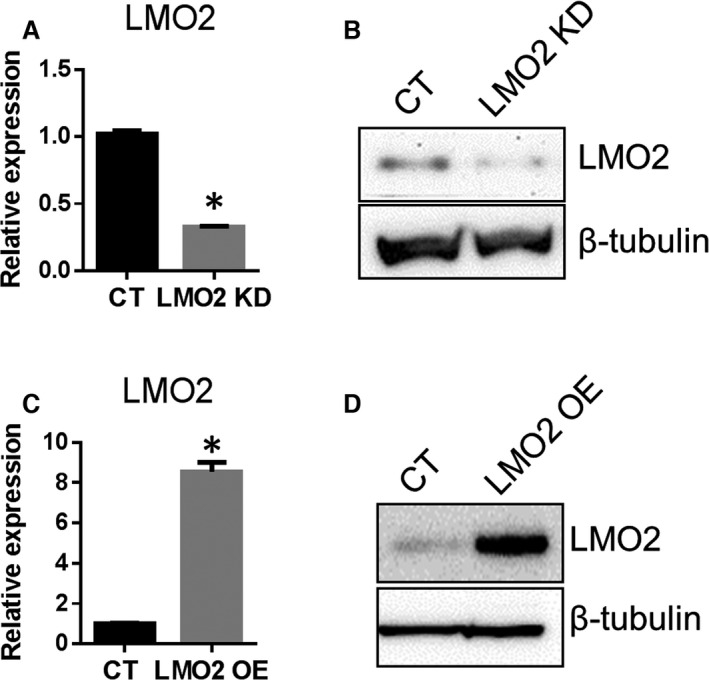
Generation of LMO2 knockdown (KD) or overexpressing cells. Human umbilical vein endothelial cells (HUVECs) passage 4 (P4) were infected with lentiviral scramble short hairpin RNA (shRNA) virus or LMO2 shRNA virus and selected with puromycin (1 μg/mL) for 14 consecutive days to generate stable KD cells. HUVECs P4 were infected with lentiviral particles encoding LMO2 open reading frame (ORF) or virus without the ORF and selected with puromycin to generate stable overexpression (OE) or control (CT) cell lines. A, reverse transcription polymerase chain reaction (RT‐PCR) examination of LMO2 mRNA level of KD cells. B, Western blot examination of LMO2 protein level in KD cells. C, RT‐PCR examination of LMO2 mRNA level in OE cells. D, Western blot examination of LMO2 protein level in OE cells. Data are presented as mean±SEM (n=3). * P<0.05 vs CT.
LMO2 KD Hindered G1/S Transition and Reduced Proliferation
Cell growth was attenuated in the LMO2 KD as shown by total cell numbers counted every other day for 4 days (Figure 2A). This effect was likely due to an impairment of cell proliferation, as cell cycle analysis by PI staining and FACS revealed that the fraction of cells in S phase was decreased (4.5% versus 9% in CT) and that in G1 phase was slightly increased (Figure 2B and 2C), indicating an impairment of G1/S transition. Further examination of G1/S transition‐related cell cycle genes by Western blot showed decreased levels of CDK4, CDK2, Cyclin D1, Cyclin A1, while Cyclin E and p21 expression were not significantly changed (Figure 2D). ENCODE database prediction suggests that both the CCGATAACGG region starting at +232 of CDK4 (Gene ID: 1019) and the CCCTCAGGTGGTG region starting at +2700 of CDK2 (Gene ID: 1017) could be potential LMO2 complex binding sites. ChIP assays confirmed that LMO2 complex binds to both regions (Figure 2E and 2F). In LMO2 KD HUVECs, LMO2 complex binding is reduced in the CDK2 gene (by 50%; Figure 2E) and in the CDK4 gene (by 75%; Figure 2F) consistent with a partial KD of LMO2 and downregulated gene expression of CDK2 and CDK4, respectively.
Figure 2.
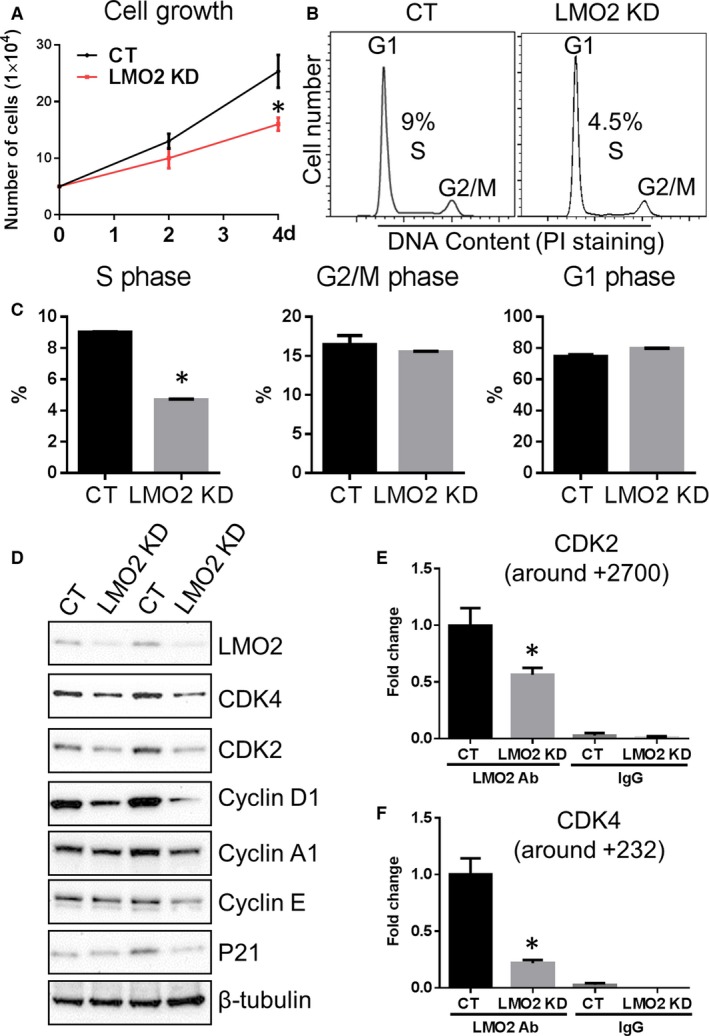
LMO2 knockdown (KD) reduced cell proliferation and hindered G1/S transition. A, Cell growth curve. B, Cell cycle analysis by propidium iodide (PI) staining. C, Quantification of cell fractions in G1, S, and G2/M phases. D, Western blot examination of G1/S transition‐related genes. E and F, chromatin immunoprecipitation polymerase chain reaction analysis of LMO2 complex binding to CDK2 gene (+2700) and CDK4 gene (+232) in both control (CT) and LMO2 KD cells. Data are presented as mean±SEM (n=3). *P<0.05 vs CT.
LMO2 OE Increased G1/S Transition but Not Proliferation
Cell cycle analysis showed that the fraction of cells in S phase were increased (14.7% versus 7.9% CT) and that in G1 phase decreased in LMO2 OE condition (Figure 3A and 3B). However, cell proliferation was not changed by LMO2 OE (Figure S2). Following LMO2 OE, Cyclin D1 mRNA level was increased, whereas CDK2 and CDK4 mRNA levels remained stable (Figure 3C). Interestingly, LMO2 OE increased only Cyclin D1 and Cyclin A1 protein levels but not that of CDK2, CDK4, Cyclin E, or p21 (Figure 3D).
Figure 3.
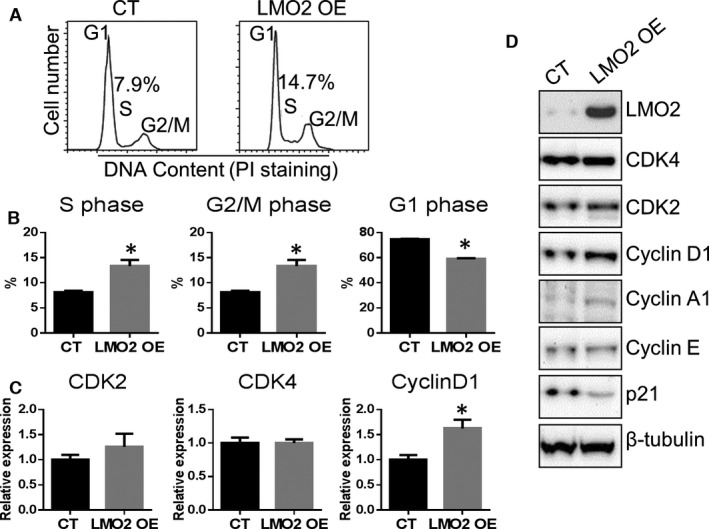
LMO2 overexpression promoted endothelial G1/S transition. A, Cell cycle analysis by propidium iodide (PI) staining. B, Quantification of cell fractions in G1, S, and G2/M phases. C, mRNA level of G1/S‐related genes. D, Western blot examination of G1/S‐related genes. Data are presented as mean±SEM (n=3). * P<0.05 vs control (CT).
LMO2 KD Impaired Endothelial Network Formation: Role of TGF‐β1
The LMO2 KD impaired endothelial network formation in matrigel (Figure 4A). Further analysis of 84 angiogenesis‐related genes using the Qiagen Angiogenesis RT Profiler PCR Array revealed that there was a substantial reduction in the expression of core angiogenesis genes (33 of 84; Figure 4B) in the LMO2 KD cells. Among them, TGF‐β1 promoter region is predicted to have LMO2 complex binding sites, indicating that it could be a direct transcriptional target of LMO2. We confirmed by RT‐PCR (Figure 4C) and ELISA (Figure 4D) that TGF‐β1 was downregulated in LMO2 KD cells. To determine the contribution of TGF‐β downregulation to impaired angiogenic processes in LMO2 KD cells, we determined whether network formation could be rescued by TGF‐β. Indeed, supplementation of the HUVEC LMO2 KD with exogenous TGF‐β restored defective network formation (Figure 4F). ENCODE database prediction suggests that two regions on TGF‐β1 (Gene ID: 7040) promoter could potentially have LMO2 complex binding. One region, CCGCAGCTGCTGC, starts at the proximal –901 position and the other region, CAGATAGGGG, starts at the distal –4030 position. The ChIP assay revealed that LMO2 complex binds to both regions (Figure 4G), confirming that TGF‐β1 is a direct transcriptional target of LMO2. Interestingly, in LMO2 KO cells, the decreased binding of LMO2 complex was only observed in the proximal –901 binding site, suggesting that this binding site is more sensitive to LMO2 downregulation. Notably, OE of LMO2 did not further increase TGF‐β1 level, probably because LMO2 binding to DNA involves the participation of other components of the binding complex (Figure 4E). This explanation is consistent with the observation that LMO2 OE did not further promote endothelial network formation (Figure S3).
Figure 4.
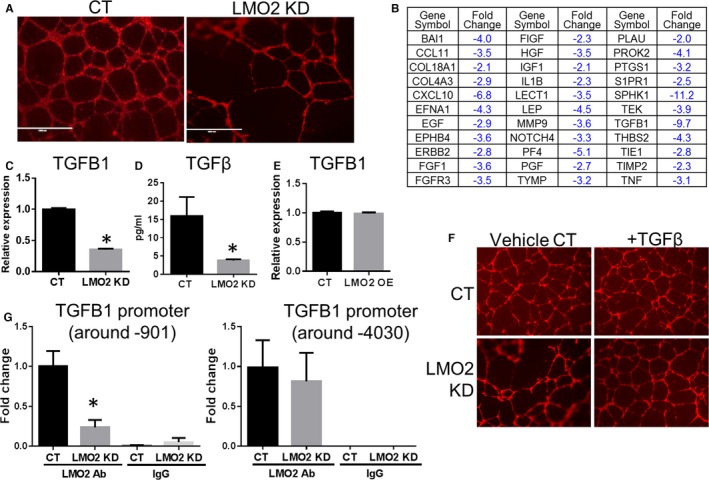
LMO2 knockdown (KD) impaired endothelial network formation, which can be rescued by exogenous transforming growth factor‐β1 (TGF‐β1). A, Network formation of control (CT) and LMO2 KD human umbilical vein endothelial cells (HUVECs). B, List of downregulated genes identified in LMO2 KD in comparison to CT HUVECs using angiogenesis polymerase chain reaction (PCR) array. C, Reverse transcription PCR (RT‐PCR) of TGF‐β1. D, ELISA for TGF‐β levels in culture medium of CT and LMO2 KD cells. E, RT‐PCR of TGF‐β1 in CT and LMO2 overexpression HUVECs. F, Network formation of CT and LMO2 KD HUVECs with or without supplementation of TGF‐β 10 ng/mL. G, Chromatin immunoprecipitation PCR analysis of LMO2 complex binding to TGF‐β1 promoter (–901 and –4030 region) in both CT and LMO2 KD cells. Data are presented as mean±SEM (n=3). * P<0.05 vs CT.
LMO2 KD Did Not Cause Global Loss of Endothelial Function
Because the LMO2 KD caused a severe impairment in angiogenic processes, we assessed other basic functions of ECs after LMO2 KD. We assessed the expression of endothelial surface markers, the uptake of acLDL, and the elaboration of NO. Compared with CT cells, LMO2 KD did not significantly alter acLDL uptake, the expression of CD31 and CD144 as assessed by FACS analysis, or the synthesis of NO as assessed by Griess assay (Figure S4).
Upregulation of lmo2 Gene Expression during Regeneration of the Caudal Fin
To test whether lmo2 is required for tissue regeneration, we assessed the gene expression of lmo2 and another early endothelial marker gene tie2 in caudal fin tissue at 0, 1, 2, and 5 days after caudal fin resection (Figure 5A). We observed that tie2 gene expression remained stable (Figure 5C) from day 0 to day 5 (at which time about 50% of the caudal fin was regenerated). Conversely, lmo2 gene expression level significantly increased by 50% at day 5 when about 50% of the caudal fin was regenerated (Figure 5B).
Figure 5.
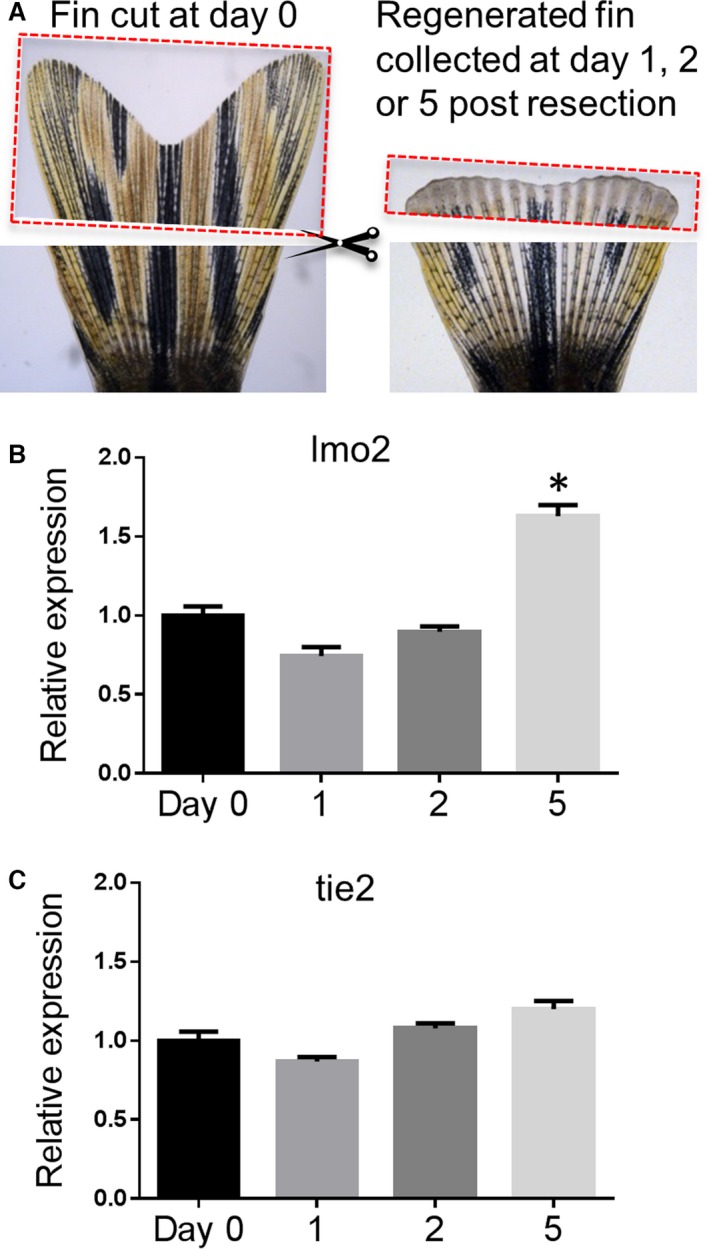
lmo2 gene expression increased following caudal fin resection and the regeneration process. A, To analyze changes in gene expression in the regenerating caudal fin, the amputated tissues from 15 fish were collected at day 0 (left panel) and regenerating distal tissues were resected and collected at 1, 2, or 5 days after the original resection (n=5 for each time point) (right panel, a fin at 2 days postresection). B, Gene expression of lmo2 in the regenerating fin. C, Gene expression of tie2 in the regenerating fin. Data are presented as mean±SEM (n=5). *P<0.05 vs control.
Adult Endothelial Proliferation Was Impaired with lmo2 KD
To test the hypothesis that LMO2 plays a key role in the proliferation of adult ECs in vivo, we injected vivo‐Mo targeting lmo2 translation start site in adult Tg(fli1:egfp) y1 zebrafish (2.5 months old) and quantified cell proliferation by BrdU incorporation (Figure 6A). The lmo2 vivo‐Mo significantly downregulated lmo2 expression compared with the CT group injected with a vivo‐Mo targeting human β‐globin, as shown by Western blot (Figure 6E). The BrdU incorporation rate of GFP+ ECs was 13.7% in the CT vivo‐Mo group, whereas it decreased to 7.2% in the lmo2 vivo‐Mo–injected group (Figure 6B and 6D). In contrast, the BrdU incorporation rate in GFP− cells was similar between the CT vivo‐Mo (1.77%) and lmo2 vivo‐Mo injection groups (1.85%) (Figure 6B and 6C).
Figure 6.
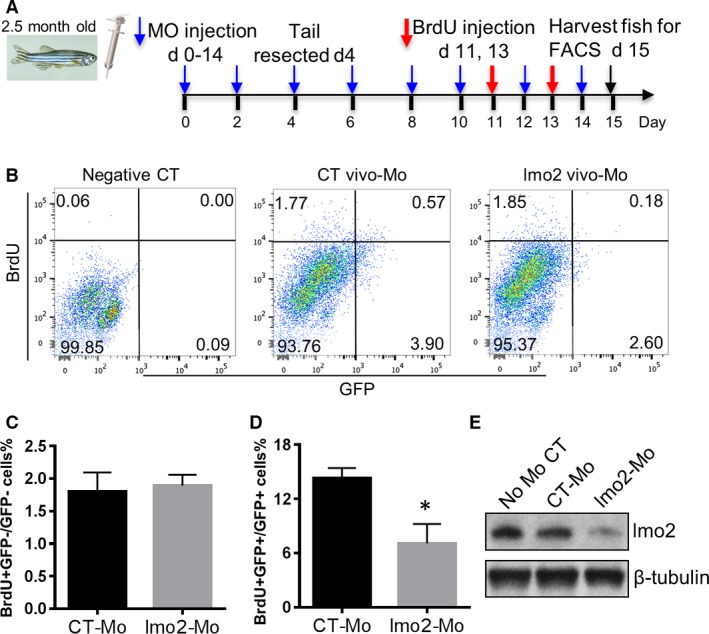
lmo2 knockdown (KD) by vivo‐morpholino (Mo) in zebrafish reduced proliferating GFP+ cells. A, Adult Tg(fli1:egfp) y1 zebrafish fish (2.5 months old) received a retro‐orbital injection of PBS solution containing lmo2 or control (CT) vivo‐Mo, then the caudal fin was resected and injected with 5‐bromo‐2′‐deoxyuridine (BrdU), as shown in the diagram. B, At day 15, whole fish were enzymatically digested and cell suspension was analyzed by flow cytometry. Negative control was Wik (non‐GFP) fish line not injected with BrdU. BrdU+ and GFP+ events are shown in CT and lmo2 vivo‐Mo–injected fish. C and D, Diagrams showing the percentage of BrdU+GFP−/GFP− cells and BrdU+GFP+/GFP+ cells. E, Western blot of lmo2 expression at day 10 in zebrafish without vivo‐Mo injection or injected with CT or lmo2 vivo‐Mo injection. Data are presented as mean±SEM (n=10). *P<0.05 vs CT.
Regeneration of the Vasculature and Parenchyma Was Impaired with lmo2 KD
The caudal fin was resected at 4 days after the first vivo‐Mo injection and its regeneration was followed up to day 15 (Figure 7A). At day 1 and 2 postresection, the percentage of fin regeneration was similar in lmo2 KD and CT fish (Figure 7A and 7B). However, at 5 days postresection, the percentage of caudal fin regeneration was only 28% in LMO2 KD fish compared with 44% in CT fish (Figure 7A and 7B). Furthermore, neoangiogenesis was impaired as the regenerated vasculature area was significantly smaller (0.47 mm2) in lmo2 KD fish compared with CTs (0.74 mm2) at day 5 postresection (Figure 7C and 7D). Interestingly, the morphology of the neovessels in the lmo2 KD fish were aberrant with tangled vessel complexes (Figure 7C), in comparison to the standard vascular branching observed in the CT fish.
Figure 7.
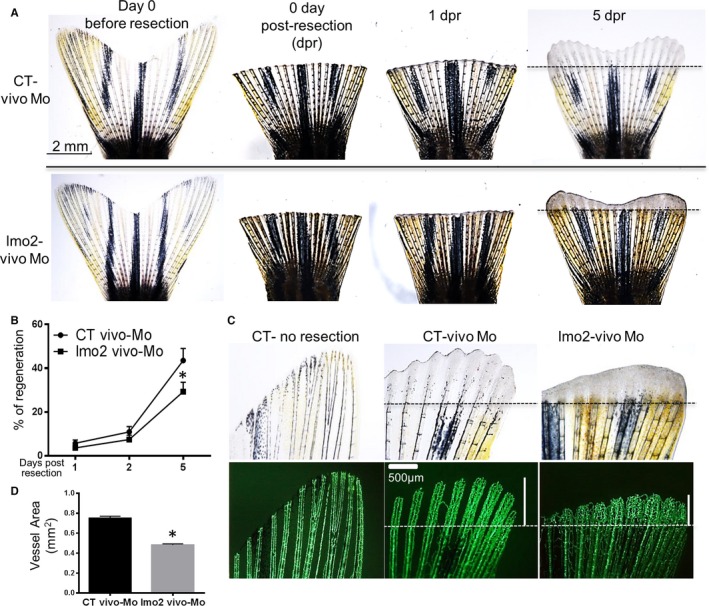
lmo2 KD by vivo‐morpholino (Mo) in zebrafish reduced caudal fin regeneration and impaired vascular regeneration. Adult Tg(fli1:egfp) y1 zebrafish fish (2.5 months old) were injected with lmo2 or control (CT) vivo‐Mo, then the caudal fin was resected and fin regeneration was observed. A, Representative caudal fin images before resection and then at 0, 1, 2, and 5 days postresection (dpr) in fish injected with CT or lmo2 vivo‐Mo. B, Quantification of the regeneration. The whole area of the caudal fin at day 0 before resection and the new fin tissue area in each fish from the new distal fin edge to the amputation plane were quantified at 0, 1, 2, and 5 dpr. Caudal fin regeneration for each fin at each time point was defined as percentage of regeneration=100× (regenerated tissue area/original fin area amputated). C, Representative image showing decreased vasculature length in the regenerated fin in lmo2 KD fish compared with CT. The vertical white bar represents the length of the regenerated vasculature. D, Quantification of the regenerated vascular area in CT at 5 dpr. Data are presented as mean±SEM (n=10). *P<0.05 vs CT.
Discussion
Transcriptional Factor LMO2
LMO2 is a key transcription factor that regulates hematopoiesis1 and embryonic vascular development.4 LMO2 is considered as an oncogene in T cells as aberrant activation of LMO2 by chromosomal translocations contributes to T‐cell acute lymphoblastic leukemia in humans.2, 3 Lmo2 transgenic mice develop T‐cell leukemia as Lmo2 OE impedes T‐cell differentiation.17, 18, 19 LMO2 expression in adult murine and human ECs has been reported.4 However, the role of LMO2 in adult angiogenesis has not been systematically studied. Our studies indicate that LMO2 is a critical factor in adult angiogenesis and endothelial proliferation, and in the response to injury, as it mediates the vascular regeneration required for tissue repair.
LMO2 and Adult Angiogenesis
To examine the role of LMO2 in endothelial function, we generated stable LMO2 KD cells using lentiviral shRNA in HUVECs. The LMO2 KD was associated with severely impaired angiogenesis as assessed by endothelial network formation. We hypothesized that LMO2 may modulate the expression of genes involved in angiogenesis. Accordingly, we used an angiogenesis PCR array for the network of core angiogenic genes and found that almost half of these genes (33 of 84), including TGF‐β1, were downregulated more than 2‐fold. TGF‐β1 is the most downregulated angiogenic cytokine detected in our experimental system. Further, ENCODE database prediction suggests one proximal (starting at –901) and one distal (starting at –4030) consensus sequence in the TGF‐β1 promoter, which was confirmed by our ChIP assays. TGF‐β is an angiogenic cytokine that regulates physiological and tumor angiogenesis.20, 21 We found that supplementation of TGF‐β could rescue the impaired network formation in the LMO2 KD, indicating that TGF‐β is a critical mediator of the angiogenic effects of LMO2. Subsequently, we used ChIP analysis to confirm that LMO2 binds to both the proximal and distal sites in the TGF‐β1 promoter. Notably, the proximal binding site seems to be more sensitive to a reduced level of LMO2. We further confirmed that KD of LMO2 reduces TGF‐β1 gene expression.
Additional analysis (Data S1) using the Gene Expression Omnibus (GEO) database ChIP‐seq data indicates that the LMO2 transcriptional complex may bind to more than 1 000 gene promoters in murine hemogenic endothelium22 and mouse hematopoietic progenitor cells (Figure S5A). Among these genes, only 627 Lmo2 downstream genes are identified in both hemogenic endothelium and hematopoietic progenitor cells, suggesting Lmo2 complex binding targets vary between cell types. This observation might be explained by the existence of differences between cell types in the prevalence of LMO2 binding partners. Interestingly, five genes from the GEO database that contain consensus sequences for the LMO2 binding complex (Figure S5B), specifically COL18A1, EPHB4, IGF1, PF4, and SPHK1, were also detected in our angiogenesis PCR array. These genes could potentially be direct transcriptional targets of LMO2 in adult ECs. Besides direct transcriptional regulation, the mechanism of LMO2‐mediated indirect regulation requires future elucidation.
EC Proliferation is Regulated by LMO2
Our data further suggest that in ECs, LMO2 is a key regulator of G1/S transition and cell proliferation. A recent bioinformatics analysis23 predicted binding of the LMO2 complex to promoter regions of cell cycle genes such as CDK2 and CDK4, suggesting a potential regulation by LMO2 of cell cycle progression. This prediction has not been experimentally verified until now. We found that LMO2 KD in HUVECs decreased cell proliferation (Figure 2A) and hindered the G1/S transition (Figure 2B). Master genes that regulate G1/S transition24 such as CDK2, CDK4, Cyclin D1, and Cyclin A1 were downregulated in LMO2 KD, while Cyclin E and p21 did not change. Consistent with a previous bioinformatics prediction report,23 our ChIP assay (Figure 2E and 2F) confirms the binding of LMO2 complex to CDK2 (CCCTCAGGTGGTG site at +2700) and CDK4 (CCGATAACGG at +232). We also confirmed that CDK4 is a direct transcriptional target of LMO2 (Figure S5B).
The OE of LMO2 increased the expression of Cyclin D1 and Cyclin A1 and was associated with an increased G1/S transition. Since the LMO2 complex does not have a putative binding site for the promoter regions of Cyclin D1 and Cyclin A1, its indirect regulation of these genes may be mediated by an effect of LMO2 on other transcription factors. Although OE of LMO2 in HUVECs promoted G1/S transition and increased S phase (Figure 3A), it did not further promote proliferation (Figure S2). Thus, while LMO2 is necessary for normal proliferation of mature ECs, it is not sufficient to accelerate their proliferation. This may be due to the fact that LMO2 binds to DNA with the help of other protein components of its DNA binding complex. As LMO2 forms protein‐protein interactions with a variety of known binding partners5, 25, 26 it may also regulate gene expression through nontranscriptional mechanisms.4, 27, 28 These other complex components may become rate limiting in the setting of LMO2 OE. This hypothesis would be consistent with the observation that the expression of CDK2 and CDK4 was not increased after LMO2 OE. In T cells where LMO2 is an oncogene, LMO2 OE does not cause increased cycling but does cause quiescence,29 suggesting that LMO2 functions differently in different cell types.
Notably, LMO2 appears to have specificity for regulation of angiogenic processes. We found that LMO2 KD did not cause a global impairment of endothelial function as it did not alter acLDL uptake, NO secretion, or the expression of specific EC surface markers. To summarize, our in vitro studies indicate that LMO2 is a critical and fairly specific regulator of endothelial proliferation and angiogenesis. Accordingly, we further investigated whether LMO2 is involved in the response to injury as endothelial proliferation and angiogenesis are required for tissue regeneration.
LMO2 Mediates Vascular and Tissue Regeneration After Injury
As a model for vascular and tissue regeneration, we chose the caudal fin resection model in zebrafish. Following amputation, lineage‐restricted mesenchymal progenitor cells form at the wound edge, presumably via dedifferentiation of mature somatic cells of the stump. ECs sprout into the avascular area.27 Angiogenic cytokines such as vascular endothelial growth factor have been shown to play a crucial role in this process. Other major signaling pathways that regulate fin regeneration include fibroblast growth factor, Hedgehog, bone morphogenetic protein, retinoic acid, Wnt/β‐catenin, insulin‐like growth factor, activin, Notch, mechanistic target of rapamycin complex 1, and calcineurin.30, 31
We first examined lmo2 gene expression during the regeneration of the resected caudal fin in zebrafish. We discovered that lmo2 gene expression is increased in regenerating caudal fin at day 5 postresection (Figure 5B), whereas the expression of tie2, an early endothelial marker32 known to promote vascular stability,33 was unchanged. This suggests that lmo2 upregulation is an early factor in vascular and tissue regeneration.
To further confirm the role of lmo2 in tissue repair in vivo, caudal fin regrowth was examined in adult zebrafish transgenic for fli1:egfp to mark ECs. In this model, ≈4% of the cells of the fish are GFP+ cells. This finding agrees with the range in humans, where ECs comprise about 3% to 6% of all cells in the body.34 In these animals, we injected the lmo2 KD Mo immediately prior to the caudal fin injury. We observed that the percentage of proliferating ECs, as assessed by BrdU incorporation, was drastically decreased by the Mo KD. By contrast, the proliferation rate in residual non‐ECs was not affected. This finding suggests that lmo2 is necessary for adult EC proliferation in vivo after an injury. Notably, the lmo2 KD also substantially limited tail regeneration. This observation would be consistent with impairment in neovascularization, as it is critical to restore perfusion in the regeneration of injured tissue.
Of note, the length and the morphology of the neovasculature in the regenerating fin was affected in the lmo2 KD. In addition to shorter branch lengths, the newly formed vessels were also aberrant in the lmo2 KD, forming tangled vascular complexes rather than the more linear morphology of the neovessels forming during fin regeneration in the CT animals.
Vascular regeneration in the caudal fin was severely impaired in lmo2 KD zebrafish, associated with a substantial reduction in fin regeneration. Thus, LMO2 appears to be a novel and potent player in vascular and tissue regeneration. The translational relevance of this observation is that OE of LMO2 might be useful in diseases characterized by ischemia and impaired angiogenesis.5, 25, 26 Alternatively, LMO2 expression in the vasculature of some tumors8, 9 suggests that disruption of LMO2 regulation might be useful in disorders characterized by pathological angiogenesis.35 To target T‐cell leukemia, antibodies or aptamers targeting LMO2 have been developed.36, 37 Although such agents may be effective anti‐tumor therapies, our studies suggest that such inhibitors might adversely affect adult EC function and normal regeneration. Thus, careful examination of the vascular effects of these inhibitors should be considered in their preclinical development and clinical trials.
Conclusions
Our studies provide the first evidence that LMO2 is a critical transcription factor that regulates adult EC proliferation and angiogenesis. In addition, LMO2 appears to be critically involved in the response to injury. By its regulation of vascular regeneration, LMO2 is required for normal tissue repair. These results indicate that LMO2 may be a good target for therapeutic modulation in tissue regeneration.
Sources of Funding
This work was supported in part by grants to Dr Cooke from the National Institutes of Health (U01 HL100397) and the Cancer Prevention and Research Institute (RP150611). Dr Meng is supported by a postdoctoral fellowship from the American Heart Association.
Disclosures
None.
Supporting information
Data S1. ChIP‐seq data analysis.
Figure S1. Representative bright field image of CT and LMO2 KD HUVECs.
Figure S2. LMO2 OE did not promote HUVEC proliferation.
Figure S3. LMO2 OE did not promote network formation on matrigel.
Figure S4. LMO2 knockdown in ECs did not interfere with acLDL uptake, NO secretion and cell surface marker levels. A, FACS analysis of AF594‐acLDL uptake in both CT and LMO2 KD HUVEC. B, Quantification of mean fluorescence intensity (MFI) of AF594‐acLDL. C, NO production measurement using Griess assay. D, FACS analysis of PE‐CD31 and APC‐CD144. E and F, Quantification of MFI of CD31 and CD144. Data are presented as mean±SEM (n=3).
Figure S5. ChIP analysis of the LMO2 transcriptional targets in hemogenic endothelium and hematopoietic progenitor cells. A, LMO2 transcriptional targets. B, LMO2 transcriptional targets that are also identified in adult ECs.
(J Am Heart Assoc. 2016;5:e004117 doi:10.1161/JAHA.116.004117)
References
- 1. Warren AJ, Colledge WH, Carlton MB, Evans MJ, Smith AJ, Rabbitts TH. The oncogenic cysteine‐rich LIM domain protein rbtn2 is essential for erythroid development. Cell. 1994;78:45–57. [DOI] [PubMed] [Google Scholar]
- 2. Boehm T, Foroni L, Kaneko Y, Perutz M, Rabbitts T. The rhombotin family of cysteine‐rich LIM‐domain oncogenes: distinct members are involved in T‐cell translocations to human chromosomes 11p15 and 11p13. Proc Natl Acad Sci USA. 1991;88:4367–4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Royer‐Pokora B, Loos U, Ludwig W. TTG‐2, a new gene encoding a cysteine‐rich protein with the LIM motif, is overexpressed in acute T‐cell leukaemia with the t (11; 14)(p13; q11). Oncogene. 1991;6:1887–1893. [PubMed] [Google Scholar]
- 4. Yamada Y, Pannell R, Forster A, Rabbitts TH. The oncogenic LIM‐only transcription factor Lmo2 regulates angiogenesis but not vasculogenesis in mice. Proc Natl Acad Sci USA. 2000;97:320–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wadman IA, Osada H, Grütz GG, Agulnick AD, Westphal H, Forster A, Rabbitts TH. The LIM‐only protein Lmo2 is a bridging molecule assembling an erythroid, DNA‐binding complex which includes the TAL1, E47, GATA‐1 and Ldb1/NLI proteins. EMBO J. 1997;16:3145–3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Patterson LJ, Gering M, Patient R. Scl is required for dorsal aorta as well as blood formation in zebrafish embryos. Blood. 2005;105:3502–3511. [DOI] [PubMed] [Google Scholar]
- 7. Landry J‐R, Kinston S, Knezevic K, Donaldson IJ, Green AR, Göttgens B. Fli1, Elf1, and Ets1 regulate the proximal promoter of the LMO2 gene in endothelial cells. Blood. 2005;106:2680–2687. [DOI] [PubMed] [Google Scholar]
- 8. Yamada Y, Pannell R, Forster A, Rabbitts TH. The LIM‐domain protein Lmo2 is a key regulator of tumour angiogenesis: a new anti‐angiogenesis drug target. Oncogene. 2002;21:1309–1315. [DOI] [PubMed] [Google Scholar]
- 9. Gratzinger D, Zhao S, West R, Rouse RV, Vogel H, Gil EC, Levy R, Lossos IS, Natkunam Y. The transcription factor LMO2 is a robust marker of vascular endothelium and vascular neoplasms and selected other entities. Am J Clin Pathol. 2009;131:264–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sun ZJ, Cai Y, Chen G, Wang R, Jia J, Chen XM, Zheng LW, Zhao YF. LMO2 promotes angiogenesis probably by up‐regulation of bFGF in endothelial cells: an implication of its pathophysiological role in infantile haemangioma. Histopathology. 2010;57:622–632. [DOI] [PubMed] [Google Scholar]
- 11. Westerfield M. The Zebrafish Book: A Guide for the Laboratory Use of Zebrafish (Danio rerio). Eugene: University of Oregon Press; 2000. [Google Scholar]
- 12. Gu Q, Yang X, Lin L, Li S, Li Q, Zhong S, Peng J, Cui Z. Genetic ablation of solute carrier family 7a3a leads to hepatic steatosis in zebrafish during fasting. Hepatology. 2014;60:1929–1941. [DOI] [PubMed] [Google Scholar]
- 13. Li YF, Morcos PA. Design and synthesis of dendritic molecular transporter that achieves efficient in vivo delivery of morpholino antisense oligo. Bioconjug Chem. 2008;19:1464–1470. [DOI] [PubMed] [Google Scholar]
- 14. Pugach EK, Li P, White R, Zon L. Retro‐orbital injection in adult zebrafish. J Vis Exp. 2009. pii: 1645. doi: 10.3791/1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Petrie TA, Strand NS, Yang CT, Rabinowitz JS, Moon RT. Macrophages modulate adult zebrafish tail fin regeneration. Development. 2014;141:2581–2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ergul AA, Halim DÖ, Adams MM. Bromodeoxyuridine (BrdU) labeling and immunohistochemical detection in adult zebrafish brain. Protoc Exch. 2013. doi:10.1038/protex.2013.087. [Google Scholar]
- 17. Larson RC, Osada H, Larson TA, Lavenir I, Rabbitts TH. The oncogenic LIM protein Rbtn2 causes thymic developmental aberrations that precede malignancy in transgenic mice. Oncogene. 1995;11:853–862. [PubMed] [Google Scholar]
- 18. Larson RC, Lavenir I, Larson TA, Baer R, Warren AJ, Wadman I, Nottage K, Rabbitts TH. Protein dimerization between Lmo2 (Rbtn2) and Tal1 alters thymocyte development and potentiates T cell tumorigenesis in transgenic mice. EMBO J. 1996;15:1021–1027. [PMC free article] [PubMed] [Google Scholar]
- 19. Neale GA, Rehg JE, Goorha RM. Disruption of T‐cell differentiation precedes T‐cell tumor formation in LMO‐2 (rhombotin‐2) transgenic mice. Leukemia. 1997;11(suppl 3):289–290. [PubMed] [Google Scholar]
- 20. Pepper MS. Transforming growth factor‐beta: vasculogenesis, angiogenesis, and vessel wall integrity. Cytokine Growth Factor Rev. 1997;8:21–43. [DOI] [PubMed] [Google Scholar]
- 21. Viñals F, Pouysségur J. Transforming growth factor β1 (TGF‐β1) promotes endothelial cell survival during in vitro angiogenesis via an autocrine mechanism implicating TGF‐α signaling. Mol Cell Biol. 2001;21:7218–7230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Goode DK, Obier N, Vijayabaskar MS, Lie ALM, Lilly AJ, Hannah R, Lichtinger M, Batta K, Florkowska M, Patel R, Challinor M, Wallace K, Gilmour J, Assi SA, Cauchy P, Hoogenkamp M, Westhead DR, Lacaud G, Kouskoff V, Gottgens B, Bonifer C. Dynamic gene regulatory networks drive hematopoietic specification and differentiation. Dev Cell. 2016;36:572–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lee S‐K, Choi YS, Cha J, Moon E‐J, Lee S‐W, Bae M‐K, Sohn T‐K, Won Y, Ma S, Bae Kong E. Identification of novel anti‐angiogenic factors by in silico functional gene screening method. J Biotechnol. 2003;105:51–60. [DOI] [PubMed] [Google Scholar]
- 24. Bertoli C, Skotheim JM, de Bruin RA. Control of cell cycle transcription during G1 and S phases. Nat Rev Mol Cell Biol. 2013;14:518–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Schmeichel KL, Beckerle MC. The LIM domain is a modular protein‐binding interface. Cell. 1994;79:211–219. [DOI] [PubMed] [Google Scholar]
- 26. Valge‐Archer VE, Osada H, Warren AJ, Forster A, Li J, Baer R, Rabbitts TH. The LIM protein RBTN2 and the basic helix‐loop‐helix protein TAL1 are present in a complex in erythroid cells. Proc Natl Acad Sci USA. 1994;91:8617–8621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Xu C, Hasan SS, Schmidt I, Rocha SF, Pitulescu ME, Bussmann J, Meyen D, Raz E, Adams RH, Siekmann AF. Arteries are formed by vein‐derived endothelial tip cells. Nat Commun. 2014;5:5758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Coma S, Allard‐Ratick M, Akino T, van Meeteren LA, Mammoto A, Klagsbrun M. GATA2 and Lmo2 control angiogenesis and lymphangiogenesis via direct transcriptional regulation of neuropilin‐2. Angiogenesis. 2013;16:939–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cleveland SM, Smith S, Tripathi R, Mathias EM, Goodings C, Elliott N, Peng D, El‐Rifai W, Yi D, Chen X, Li L, Mullighan C, Downing JR, Love P, Dave UP. Lmo2 induces hematopoietic stem cell‐like features in T‐cell progenitor cells prior to leukemia. Stem Cells. 2013;31:882–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wehner D, Weidinger G. Signaling networks organizing regenerative growth of the zebrafish fin. Trends Genet. 2015;31:336–343. [DOI] [PubMed] [Google Scholar]
- 31. Poss KD, Shen J, Nechiporuk A, McMahon G, Thisse B, Thisse C, Keating MT. Roles for Fgf signaling during zebrafish fin regeneration. Dev Biol. 2000;222:347–358. [DOI] [PubMed] [Google Scholar]
- 32. Foubert P, Matrone G, Souttou B, Lere‐Dean C, Barateau V, Plouet J, Le Ricousse‐Roussanne S, Levy BI, Silvestre JS, Tobelem G. Coadministration of endothelial and smooth muscle progenitor cells enhances the efficiency of proangiogenic cell‐based therapy. Circ Res. 2008;103:751–760. [DOI] [PubMed] [Google Scholar]
- 33. von Tell D, Armulik A, Betsholtz C. Pericytes and vascular stability. Exp Cell Res. 2006;312:623–629. [DOI] [PubMed] [Google Scholar]
- 34. Bianconi E, Piovesan A, Facchin F, Beraudi A, Casadei R, Frabetti F, Vitale L, Pelleri MC, Tassani S, Piva F, Perez‐Amodio S, Strippoli P, Canaider S. An estimation of the number of cells in the human body. Ann Hum Biol. 2013;40:463–471. [DOI] [PubMed] [Google Scholar]
- 35. Carmeliet P. Angiogenesis in health and disease. Nat Med. 2003;9:653–660. [DOI] [PubMed] [Google Scholar]
- 36. Nam CH, Lobato MN, Appert A, Drynan LF, Tanaka T, Rabbitts TH. An antibody inhibitor of the LMO2‐protein complex blocks its normal and tumorigenic functions. Oncogene. 2008;27:4962–4968. [DOI] [PubMed] [Google Scholar]
- 37. Appert A, Nam CH, Lobato N, Priego E, Miguel RN, Blundell T, Drynan L, Sewell H, Tanaka T, Rabbitts T. Targeting LMO2 with a peptide aptamer establishes a necessary function in overt T‐cell neoplasia. Cancer Res. 2009;69:4784–4790. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1. ChIP‐seq data analysis.
Figure S1. Representative bright field image of CT and LMO2 KD HUVECs.
Figure S2. LMO2 OE did not promote HUVEC proliferation.
Figure S3. LMO2 OE did not promote network formation on matrigel.
Figure S4. LMO2 knockdown in ECs did not interfere with acLDL uptake, NO secretion and cell surface marker levels. A, FACS analysis of AF594‐acLDL uptake in both CT and LMO2 KD HUVEC. B, Quantification of mean fluorescence intensity (MFI) of AF594‐acLDL. C, NO production measurement using Griess assay. D, FACS analysis of PE‐CD31 and APC‐CD144. E and F, Quantification of MFI of CD31 and CD144. Data are presented as mean±SEM (n=3).
Figure S5. ChIP analysis of the LMO2 transcriptional targets in hemogenic endothelium and hematopoietic progenitor cells. A, LMO2 transcriptional targets. B, LMO2 transcriptional targets that are also identified in adult ECs.


