Abstract
Background
Chitin, the second most abundant biopolymer on earth after cellulose, is found in probably all fungi, many animals (mainly invertebrates), several protists and a few algae, playing an essential role in the development of many of them. This polysaccharide is produced by type 2 glycosyltransferases, called chitin synthases (CHS). There are several contradictory classifications of CHS isoenzymes and, as regards their evolutionary history, their origin and diversity is still a matter of debate.
Results
A genome-wide analysis resulted in the detection of more than eight hundred putative chitin synthases in proteomes associated with about 130 genomes. Phylogenetic analyses were performed with special care to avoid any pitfalls associated with the peculiarities of these sequences (e.g. highly variable regions, truncated or recombined sequences, long-branch attraction). This allowed us to revise and unify the fungal CHS classification and to study the evolutionary history of the CHS multigenic family. This update has the advantage of being user-friendly due to the development of a dedicated website (http://wwwabi.snv.jussieu.fr/public/CHSdb), and it includes any correspondences with previously published classifications and mutants. Concerning the evolutionary history of CHS, this family has mainly evolved via duplications and losses. However, it is likely that several horizontal gene transfers (HGT) also occurred in eukaryotic microorganisms and, even more surprisingly, in bacteria.
Conclusions
This comprehensive multi-species analysis contributes to the classification of fungal CHS, in particular by optimizing its robustness, consensuality and accessibility. It also highlights the importance of HGT in the evolutionary history of CHS and describes bacterial chs genes for the first time. Many of the bacteria that have acquired a chitin synthase are plant pathogens (e.g. Dickeya spp; Pectobacterium spp; Brenneria spp; Agrobacterium vitis and Pseudomonas cichorii). Whether they are able to produce a chitin exopolysaccharide or secrete chitooligosaccharides requires further investigation.
Electronic supplementary material
The online version of this article (doi:10.1186/s12862-016-0815-9) contains supplementary material, which is available to authorized users.
Keywords: Chitin synthase, Bacteria, Evolution, Classification, Horizontal gene transfer, Fungi
Background
Chitin is a biological polymer consisting of carbohydrate molecules bonded together to form long chains of polysaccharides. Unlike starch and glycogen that are storage polysaccharides in plants and animals respectively, chitin is a structural polysaccharide organised as crystalline microfibrils and with enormous tensile strength. It contributes to the rigidity and integrity of cells, tissues or body surfaces in a wide range of organisms, protecting and giving them shape, as seen with cellulose or pectin in plant and algal cell walls. So far, the presence of this structural polysaccharide has been mainly demonstrated in the cell walls of mycota, the exoskeleton of hexapoda or crustacea, and in the radula or beak of mollusca [1], where it plays a major role in development and growth. In addition, the presence and subcellular location of chitin in invertebrate hemocytes suggests another role for this polysaccharide in the immune system of diverse animals [2]. It was generally thought that there was no chitin in vertebrates but this polymer has been described in several ray-finned fishes (Actinopterygi) [3, 4] and in some amphibians [5]. The role of endogenous chitin in the biology of these vertebrates remains elusive. Chitin has also been sporadically found in structures from a diverse range of eukaryotic microorganisms, such as the cell wall of a few chlorophyta (green microalgae), the cyst wall or lorica, of ciliophora (ciliated protozoans), the theca of choanoflagellida (flagellated protozoans), and the test or cyst wall, of amoebozoa (amoeboid protozoans) [6]. It is also present in the large family of heterokonta protists, for example in the cell wall of oomycota, the spines of diatomae and the stalk of chrysophyta [7].
Chitin is an hexosamine polymer composed of beta-(1,4)-linked linear chains of more than 5,000 N-acetylglucosamine residues that are highly cross-linked with hydrogen bonds. In insects, chitin is deposited exclusively on the apical sides of epithelial cells, facing the external environment (body surface, gut and tracheal lumen) [8]. In fungal cell walls, there is a common fibrillar core composed of branched beta-(1,3)-glucan to which chitin and other polysaccharides are covalently bound. Chitin accounts for 1-2% of the cell wall mass in yeasts and up to 30% in molds [9, 10]. Elongation of the chitin polymer is catalyzed by a highly conserved enzyme called chitin synthase, CHS (UDP-N-acetyl-D-glucosamine: chitin 4-beta-N-acetylglucosaminyl-transferase; EC 2.4.1.16). The CHS enzyme belongs to the GT2 family of processive polymerizing glycosyltransferases which includes the synthases for cellulose, callose, curdlan, mannan, hyaluronate and alginate polymers [11]. In fungi, chitin biosynthesis requires a set of multiple CHS isoenzymes that are encoded by a multigenic family. Although they share a common central catalytic domain, CHS isoenzymes from fungi and other species (metazoa, protists) can differ greatly in their N- and C-termini parts. All CHS utilize UDP-N-acetylglucosamine (UDP-GlcNAc) from the cytoplasm as a substrate and catalyze multiple transfers of the activated sugar donor to the non-reducing end of the growing chain. Multiple transmembrane domains are found in every CHS protein and these probably form a channel in the cell membrane through which linear chitin is extruded into the extracellular space as it is described for cellulose biosynthesis [12]. Nevertheless, the exact mechanism by which chitin is assembled into microfibrils and cross-linked with other components of the cell wall is still poorly understood.
The importance of chs genes in fungal biology has been extensively investigated by reverse genetics. Mutants have been constructed by disrupting or deleting particular chs genes in fungal species and no less than a hundred mutants have been made so far in more than twenty fungi (Additional file 1: Table S1). The multiplicity of chs genes in fungal genomes has necessitated their classification for comparative functional genomics, and fungal CHS isoenzymes have been classified into multiple divisions and classes according to protein similarities in their catalytic domain. More than 50 phylogenetic analyses have been published and differences, in the names and in the number of classes have complicated the situation (Additional file 2: Table S2). Initial attempts to classify CHS mainly involved the Ascomycota sequences (Table 1). Organising classes I, II and III into the division 1 and the classes IV and V into the division 2 was the result of working on a small number of fungal sequences [13–15]. Then, several other classes (V, VI, VII etc.) or a new division 3, were added but nomenclature was unfortunately decided heterogeneously between the different study groups [16–21]. More recently, CHS classification was extended to several basidiomycota species but by still following the same nomenclature used for Ascomycota species [22] (Table 1). Finally, a class VIII was recently proposed but it was for three completely distinct clades of putative CHS, resulting in an unusable classification [10, 23, 24]. Moreover, all these different classifications were initially based upon CHS sequences from Ascomycota and the actual classifications are not well suited to other fungi (e.g. Mucoromycotina).
Table 1.
Summarized history of chitin synthase protein classifications in fungi
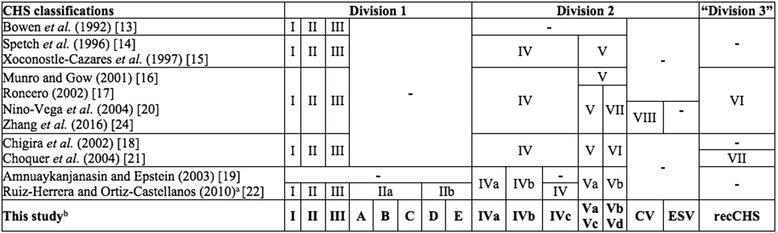
In bold, the chitin synthase classification issued from our study
aInversion between classes IVa and IVb and inversion between classes Va and Vb were corrected in Ortiz-Castellanos and Ruiz-Herrera [110]
bCV: Chlorovirus-like CHS class; ESV: Ectocarpus siliculosus-like CHS class; recCHS: recombined chitin synthase
In order to update and standardize CHS classifications, we performed a comparative multi-species analysis across many sequenced and annotated genomes. A databank of CHS proteins was generated from a similarity search of CHS Pfam domains in the complete proteomes. Maximum likelihood (ML) and Bayesian phylogenetic trees were constructed to provide a global view of the whole CHS family. Applying a rigorous method of phylogenetic analysis, we organised the fungal sequences and obtained a more robust classification. We extended the study to chitin synthases from other species in order to elucidate chs gene evolution. In particular, this genome-wide phylogenetic analysis confirms the occurrence of multiple gene losses, duplications and horizontal gene transfers within this family of glycosyltransferases. Surprisingly, this work provides, for the first time, evidence of a chitin synthase horizontal gene transfer from eukaryota to bacterial genomes.
Results and discussion
In silico detection of putative CHS in annotated and complete proteomes enabled us to identify more than eight hundreds of CHS sequences in about 120 eukaryota species. We also found CHS sequences from a few viruses and bacteria, but no CHS could be found in archeal species (Fig. 1; Additional file 3: Table S3). Most fungal chitin synthases fall into two distinct divisions (Fig. 2).
Fig. 1.
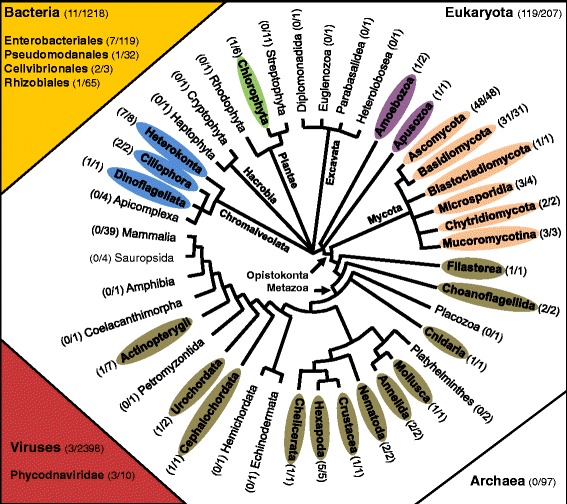
Distribution of chitin synthases among eukaryotes, bacteria and viruses. Eukaryotic species phylogeny was adapted from [100], with modifications for Hacrobia [101], Ciliophora [102], Coelacanthimorpha [103], and Amoebozoa, Apusozoa, Filasterea and Choanoflagellida [104]. Groups in which the presence of chitin synthase protein was detected are shaded in grey and written in bold. In each group, we include, in brackets, the number of species harboring chitin synthase proteins compared to the number of species analyzed. These species are listed in Additional file 3: Table S3 and Additional file 14: Table S7
Fig. 2.
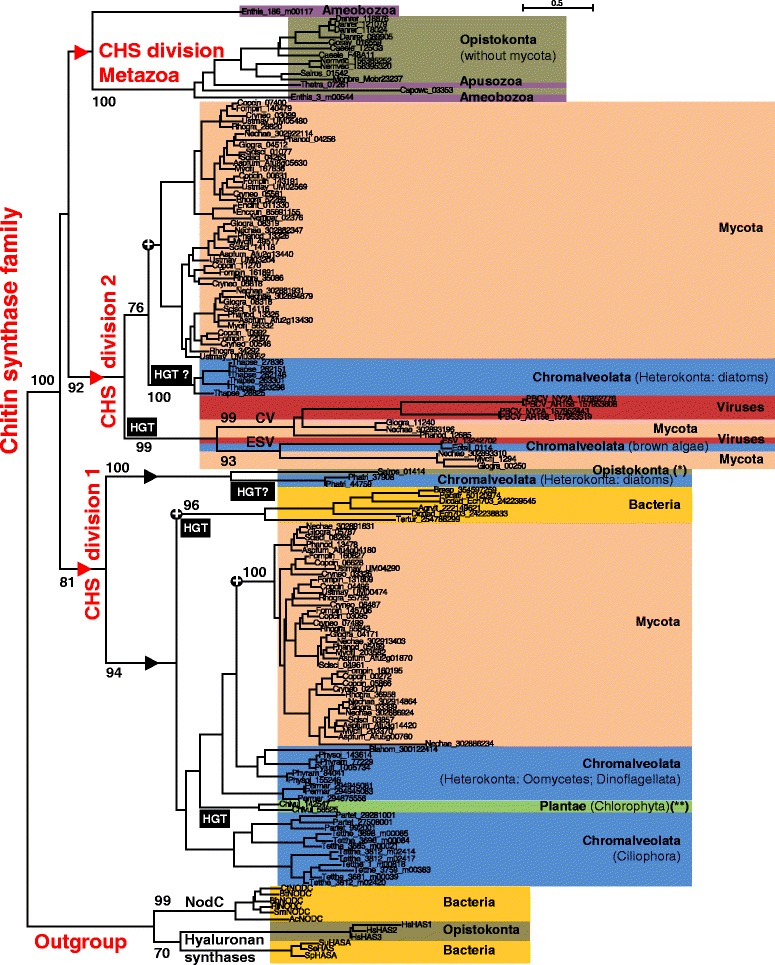
Evolution of chitin synthases (CHS). A ML phylogeny based on 222 amino acid alignment positions of 157 sequences was constructed with PhyML using NODC proteins and Hyaluronan synthases as outgroups. Bootstraps of interest ≥60 are shown above the branches. Black circles with a + correspond to root nodes of the subtrees detailed in another figure. Horizontal gene transfers (HGT) are indicated on the branches leading to transferred sequences. The Bayesian phylogenetic approach gave similar results (Additional file 15: Figure S7). The abbreviations used in sequence names are listed in Additional file 13: Table S6. (*) Acanthamoeba and Ichtyosporea sequences were also described in this group [43]. (**) Picochlorum sp. sequence is also present in this group [50]
Update of CHS fungi classification
We observed that in Dikarya fungi, Ascomycota and Basidiomycota, up to 15 CHS-encoding genes were found in the same species (e.g. Postia placenta; Additional file 3: Table S3). In Ascomycota yeasts, the number of CHS is usually lower than in filamentous fungi and ranges from one (e.g. Schizosaccharomyces pombe) to seven (Yarrowia lipolytica) [25]. In Hemiascomycota yeasts, the presence of seven CHS in Yarrowia lipolytica and only three in Saccharomyces cerevisiae can only be explained by several losses in this clade (Additional file 4: Figure S1). In Mucoromycotina, Chytridiomycota and Blastocladiomycota, 15 to 38 chs genes were found depending on the species suggesting an even larger expansion of the CHS family in these early-branching fungal lineages (Additional file 3: Table S3). Finally, none or only one, chs gene was found in four Microsporidia fungal genomes (Encephalitozoon cuniculi, Encephalitozoon intestinalis, Nematocida parisii and Nosema ceranae). This characteristic seems consistent with the extreme reduction and compaction that was observed in these particular genomes [26]. The large number of detected CHS, and their diversity, provided the opportunity to establish a more robust fungal classification for this protein family. In our definition, a class means a set of CHS sequences, from different Ascomycota, Basidiomycota and/or Mucoromycotina species, which forms a well-supported group in the trees constructed with two phylogenetic approaches (ML and Bayesian). For now, we consider the genome quality and number from Chytridiomycota and Blastocladiomycota species insufficient to accurately classify these sequences. Ascomycota yeast CHS sequences were excluded from most phylogenetic analyses in order to reduce the risk of long-branch attraction (LBA) as substantial artefact was described in the phylogeny of Saccharomyces species [27]. LBA could have contributed to erroneously cluster together long branches, irrespective of the true relationships of sequences [28]. Such exclusion is not a problem as classification of CHS from yeasts is easy (Additional file 4: Figure S1).
Fungal division 1 : five new classes A to E in addition to classes I, II and III
Fungal division 1 consists mainly of protein sequences with an N-terminal Chitin_synth_1N domain (PF08407), a conserved catalytic site and 6 predicted transmembrane domains in the C-terminal region (Fig. 3). In this division, different fungal lineage-specific classes were found with only one, corresponding to class III, which was common to Ascomycota and Basidimycota (Fig. 4). Previous attempts to group the classes A to E mainly with Ascomycota classes I or II resulted in different classifications as these groups were not well supported by the phylogenies (see references in Additional file 2: Table S2). A possible explanation for the observed phylogeny is that several duplications arose for this division at an early stage in fungal evolution and, thereafter, different gene losses might have occurred in the different lineages. More than forty fungal mutants impaired in a division 1 chs gene were generated in Ascomycota species and a few in Basidiomycota species (see references in Additional file 1: Table S1). Some differences in CHS activity in vitro, chitin content and conidiation were observed for chs mutants impaired in Ascomycota classes I or II. These classes probably have redundant roles as the single corresponding mutants were not affected in their growth and morphology. On the contrary, class III CHS seems to play a crucial role in the hyphal tip growth as several class III mutants were strongly affected in their morphology, with reduced growth resulting in small colonies and abnormal highly-branched hyphae (Additional file 1: Table S1). This class was lost early on during Saccharomycotina yeast evolution, as only Yarrowia lipolytica possess a class III chs gene (Additional file 4: Figure S1). Conversely, this class was expanded in some Pezizomycotina species as already mentioned for Fusarium sp. and Aspergillus sp. [23]. Some duplicated copies are fast evolving sequences, which contrasts with the strong sequence conservation observed in this class, and they might have acquired different roles (Additional file 5: Figure S2).
Fig. 3.
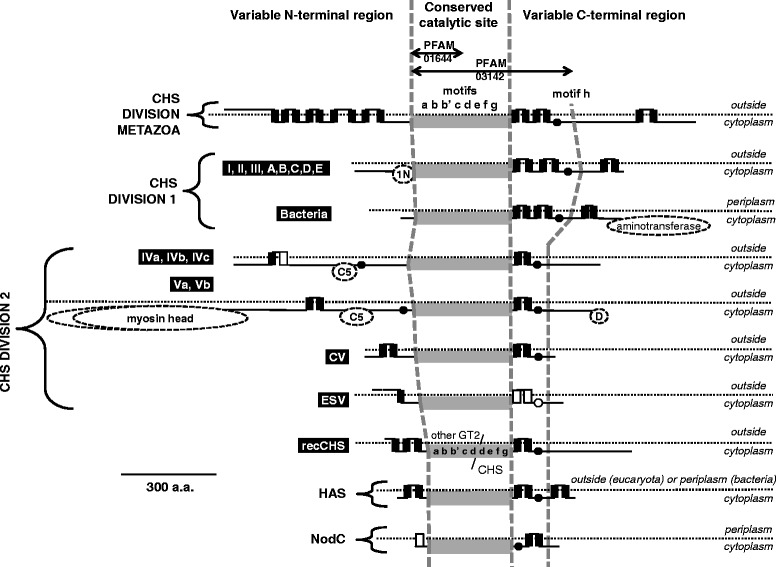
Structural features of the chitin synthase proteins. The core of CHS proteins is always composed of conserved motifs named a-h and playing a role in the active site of the enzyme (Additional file 7: Figure S3). The domains related to chitin synthase used for their detection, Chitin_synth_1 (PF01644) and Chitin_synth_2 (PF03142), correspond to the a-c and a-h regions respectively. Other domains from the Pfam library, 1N (Chitin_synth_1N; PF08407), Aminotransferase (DegT_DnrJ_EryC1; PF01041), Myosin head (Myosin_head; P00063), C5 (Cyt-b5; PF00173) and D (DEK_C; PF08766) were often detected in some CHS clades and are indicated by dashed boxes. The length variation of the myosin head domain in the clade V is represented by a dashed line in the dashed box of this domain. N-terminal and C-terminal regions of CHS are variable and transmembrane segments detected in almost all the sequences of one clade are shown with black solid squares. The less frequent additional segments are shown with white solid squares. Predicted outside or cytoplasmic localizations of eukaryota CHS protein segments are indicated thanks to a dotted line (or periplasmic and cytoplasmic localizations for bacterial CHS and HAS). Cytoplasmic localization of the conserved motifs a-h in homologous GT2 glycosyltransferases was confirmed with LacZ, PhoA and/or GFP reporter fusions [105–109]. According to these studies, some predicted transmembrane domains are, in fact, putative membrane domains that do not cross the membrane (black circles or white circles for the less frequent ones). The distant motif h might interact with the other conserved motifs as its cytoplasmic localization was confirmed
Fig. 4.
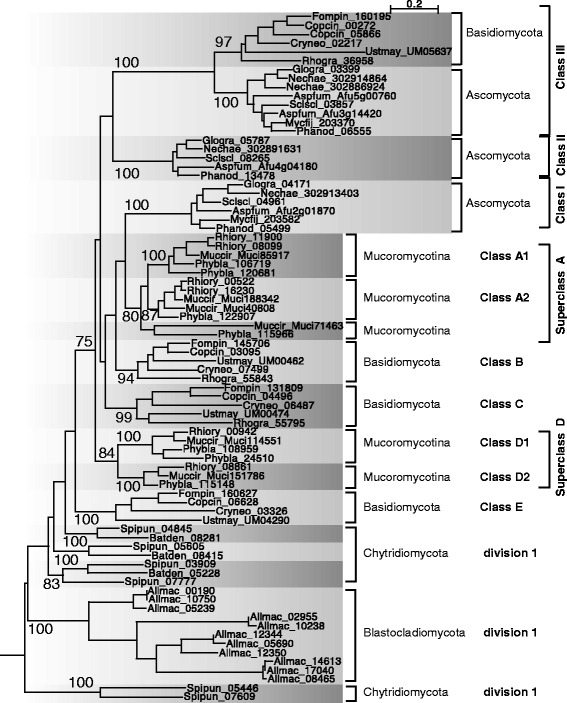
Evolution of fungal CHS belonging to division 1. A ML phylogeny, based on 489 amino acid alignment positions of 78 sequences, was constructed with PhyML. The root was placed according to the phylogeny in the Additional file 16: File S2. The Bayesian phylogenetic approach gave similar results (Additional file 17: Figure S8)
Fungal division 2 : two superclasses IV and V
Fungal division 2 is clearly divided into two monophyletic superclasses IV and V (Fig. 5). These are composed of protein sequences which contain, in addition to the chitin synthase catalytic site, one or two predicted transmembrane domains in the N-ter and C-ter regions plus a cytochrome-b5-like domain (PF00173) in the N-ter region, which has a proposed role as a binding site for lipid ligands [29] (Fig. 3). Sequences of the superclass V often have two additional domains: a myosin motor domain (PF00063) fused to their N-terminus extremity, involved in intracellular trafficking of CHS and site specificity of chitin secretion [30] (Fig. 3), and a DEK-C domain (PF08766), of unknown function, at their C-terminus. Each superclass was divided into several classes, most of which are conserved between Ascomycota, Basidiomycota and Mucoromycotina (Fig. 5). The class IVb was lost in Ascomycota and the ancestor of most Hemiascomycota yeasts lost all division 2 classes except the class IVa (Additional file 4: Figure S1). The class IVc was only found in Mucoromycotina, where an expansion of classes IVa, Va and Vb was also observed. Proteins from superclasses IV and V were also detected in the proteomes of Chytridiomycota and Blastocladiomycota whereas Microsporidia proteomes only contained one member of the superclass IV. Despite the fact that the superclass IV was found in the largest number of fungal species, the corresponding mutants do not usually exhibit any apparent phenotypic change compared to the wild-type strain (Additional file 1: Table S1). By contrast, mutants of class Va or Vb are usually strongly affected in their morphology (swelling, baloon formation, intrahyphal hyphae), in the response to cell wall stresses and in virulence [31] (Additional file 1: Table S1).
Fig. 5.
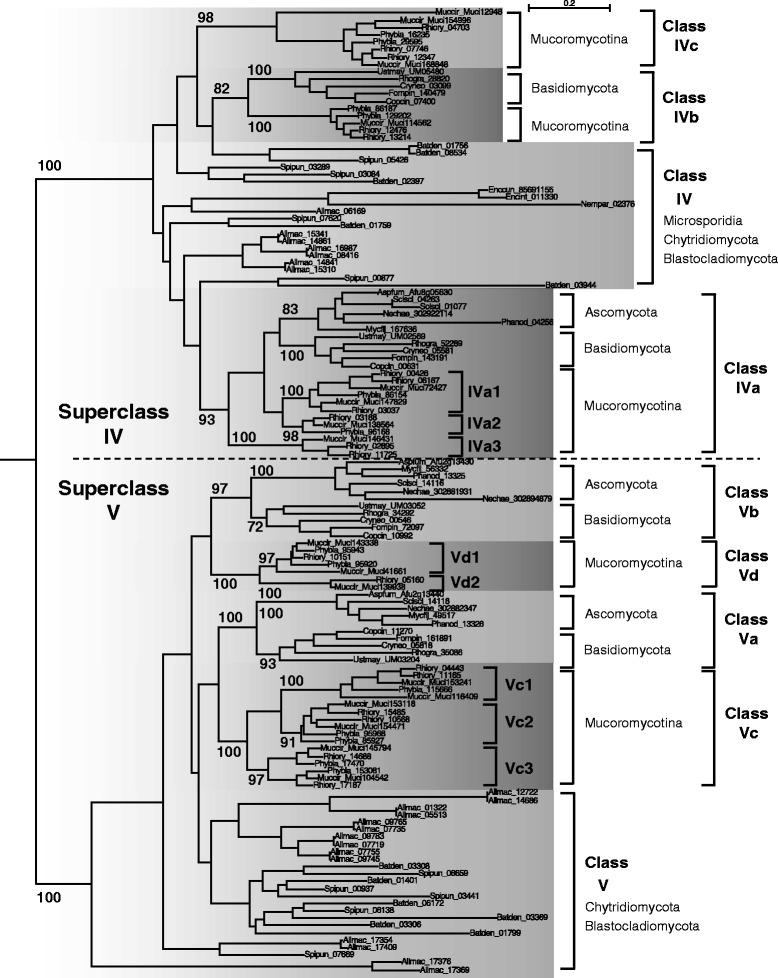
Evolution of fungal CHS belonging to division 2. A ML phylogeny, based on 485 amino acid alignment positions of 131 sequences, was constructed with PhyML. The root was placed according to the phylogeny in the Additional file 18: File S3. The Bayesian phylogenetic approach gave similar results (Additional file 19: Figure S9)
Fungal division 2: two virus-like classes
In fungal division 2, in addition to the two superclasses IV and V, two others classes were found (Fig. 2). These fungal classes, that we have called ESV (Ectocarpus siliculosus Virus-like CHS) and CV (Chloroviruses-like CHS), were found in proteomes of some Ascomycota (Sordariomycetes, Dothideomycetes and Eurotiomycetes), some Basidiomycota (Ustilaginomycetes only possessing a ESV CHS), a Chytridiomycota (only possessing a CV CHS) and some Phycodnaviridae giant algal-viruses (Additional file 6: Table S4). The CV class was recently proposed as a new class VIII [24] but we do not recommend this denomination as class VIII is used for other distinct groups of putative CHS [10, 23] (Additional file 2: Table S2). Independent horizontal transfers of these chs genes with their neighboring genes, in fungal genomes from chloroviruses and phaeoviruses respectively, have already been suggested [32]. In addition to the strong similarity between CV and ESV viral and fungal sequences, the corresponding genes are clustered on genomes with an UDP-N-acetylglucosamine 6-dehydrogenase gene (UNGD). This two-gene cluster, conserved in some fungal species and in viruses of distantly related algae, suggests that these two genes were probably transmitted together and that this gene pair might work in concert : UNGD family enzymes provide precursors for glycosyltransferase enzymes [32]. Moreover, the fungal and viral CV genes are also colocated with a polysaccharide deacetylase. These genetic material exchanges between fungi and algal-viruses are perplexing. However, as exemplified by the 600 million year old lichen-fossils, interactions betwen fungi and algae have long existed [33]. Furthermore, gene transfers from the ancestor of Dothideo/Sordariomycetes to the ancestor of the terrestrial alga Trebouxia decolorans have also been proposed [34]. There is currently no evidence for the functionality of the fungal proteins encoded by the genes from the ESV class. However, the CV class genes of Glomerella graminicola and Gibberella zeae were found to be differentially expressed during plant infection [23] and a deletion mutant in a CV class chs gene was recently described in Fusarium graminearum (called Fgchs8 gene) [24]. Disruption of this gene resulted in a reduced accumulation of chitin, decreased CHS activity, sensitivity to SDS and reduced pathogenicity. It has been suggested that the Fgchs8 gene is required for cell wall development in F. graminearum. The chs genes from the CV and ESV classes were duplicated in some Ascomycota lineages and they were lost in others [32] (Additional file 6: Table S4). They also show higher evolutionary rates than other fungal CHS (Fig. 2; Additional file 5: Figure S2). The virus containing a chs gene from the ESV class is, more precisely, a lysogenic phaeovirus. DNA from this virus, including the ESV chs gene, is integrated into the genome of the pluricellular brown alga Ectocarpus Siliculosus [35]. This integrated viral ESV chs gene is transcriptionally silent in the algae and is probably not functional [36]. Viruses with at least one chs gene from the CV class include some chloroviruses infecting the unicellular green alga Chlorella with a lytic infection style (e.g. Paramecium Bursaria Chlorella viruses). Some chloroviruses form chitin on the surface of infected cells which might protect virus-infected algae from uptake by other organisms [37]. Indeed, the heterologous expression of the chlorovirus CVK2 chs gene was performed in E. coli leading to the production of fibers by the bacterium [38].
Outside fungal divisions : a class of CHS probably emerging from a recombination event
In our analysis, a complete CHS class, previously called VI, VII (Table 1; Additional file 2: Table S2) or sometimes division 3 [39], was treated separately. We suspect a recombination event of being at the origin of this class (Fig. 3). Associated with a reduced taxonomic distribution in fungi, including some Ascomycota groups (Sordariomycetes, Dothideomycetes, Eurotiomycetes and Leotiomycetes) and a Mucoromycotina species (Additional file 6: Table S4), this class was also recently detected in a chromalveolate (Rhizaria), Plasmodiophora brassicae, an obligate biotrophic pathogen of crucifers [40]. In this study, we suggest that the corresponding protein sequences have characteristics associated with a rearrangement in their ancestor: a duplication of the QXXXY motif (motif d in Fig. 3 and Additional file 7: Figure S3) and a phylogenetic signal which seems to differ in the N- and C-termini sequence fragments located on either side of this duplication (Additional file 8: Figure S4). Indeed, while the C-terminal region appears to be similar to that in other CHS (Additional file 8: Figure S4A), the N-terminal region is closer to hyaluronan synthase proteins in the phylogenetic tree (Additional file 8: Figure S4B) and shares the same organization of transmembrane domains (Fig. 3). This observation could be explained by an ancient recombination between the ancestor’s sequences of two glycosytransferase family 2 proteins. An alternative explanation, that we can not completely exclude, is that the ancestor of this class underwent a period of an accelerated rate of evolution which blurred the phylogenetic signal. In both cases (recombination or transient high evolutionary rate), we excluded them from phylogenetic analyses as they might have provoked artefactual groups due to long-branch attractions. HGTs are probably at the origin of this CHS class in one Mucoromycotina (Mortierella verticillata), one Chytridiomycota (Batrachochytrium dendrobatidis) and one chromalveolata (Plasmodiophora brassicae) (Additional file 6: Table S4). In Ascomycota groups, where this class is present, the corresponding genes are probably functional as they are strictly fixed in one well-conserved copy (Additional file 3: Table S3; Additional file 5: Figure S2). However, their chitin synthase activity has not yet been proved so we recommend using the term recCHS (“recombined” CHS) for these sequences. Few fungal deletion mutants were obtained for this class and their phenotypes are divergent. RecCHS orthologs have probably evolved with different roles in these fungi during growth and development [41, 42].
Origin of CHS in eukaryotes
The phylogeny was obtained with a protein sequences sample representative of the entire set of chitin synthases detected in this study (Fig. 2). At least three chitin synthase genes were present in the ancestor genome of Opistokonta: one ancestral chs for the Metazoa division, one for the division 2 and one for the division 1 (see red triangles in Fig. 2). A previous study suggested four ancestral chs genes in the Last Opisthokonta Common Ancestor (LOCA) [43] but we propose that two of them belong to division 1 and they may have diverged from a common ancestral sequence in LOCA (see black triangles in Fig. 2). The chitin synthase genes probably appeared earlier, given the basal positions in the Metazoa division of one sequence of the Amoebozoa Entamoeba histolytica and the CHS of the apusomonad Thecamonas trahens. The distribution of taxa possessing a chs gene in Metazoa indicates that chs genes were lost several times independently in this phylum (Fig. 1; in Platyhelminthes, in Hemichordata, in some Actinopterygii etc.). On the other hand, family expansion occurred in some species, such as in the gastropod Lottia gigantea [44] and the amphioxus Branchiostoma floridae [45]. It is noteworthy that a second CHS sequence from Entamoeba histolytica did not significantly group with any of the chitin synthase divisions, which raises the question of its origin. However, heterologous expression of this protein in the yeast Saccharomyces cerevisiae confirmed its CHS activity [46]. The presence of chitin synthases belonging to different divisions in diverse chromalveolates, such as ciliates, diatoms, oomycetes and other protists, might be the result of a deeper evolutionary origin of CHS in the Last Common Eukaryotic Ancestor [40, 47] (Figs. 1 and 2; Additional file 3: Table S3). However, an alternative plausible model for the origin of chromalveolate CHS could imply several independent horizontal gene transfers at different times during the chromalveolate evolutionary history [48], as we suggested for the chs of the rhizaria Plasmodiophora brassicae (see above in Update of CHS fungi classification). Diatom chs genes from Thalassiosira pseudonana (division 2) and Phaeodactylum tricornutum (division 1), were probably acquired by an HGT from Opistokonta (fungi or metazoa; Fig. 2).
Other HGT must have occurred to explain the actual taxonomic distribution of chs genes and their sequence diversity. If the division 1 chs of the green algae (Trebouxiophyceae) Chlorella [49] (Fig. 2) and Picochlorum [50] were vertically inherited from the Plantae ancestor, it would imply that these genes were independently lost in many lineages of the plantae. As mentioned for the CV and ESV classes of division 2 (Update of CHS fungi classification), chs gene exchanges might have occurred between algae-related viruses, including chloroviruses and fungi. However, the Chlorella CHS belongs to division 1 and seems to resemble oomycete and ciliate CHS more than fungal ones (Fig. 2). Therefore, a possible source of the transfer could be a ciliate living in symbiosis with a green alga, such as Paramecium bursaria.
Hence, the evolutionary history of chitin synthase genes suggests different independent horizontal gene transfers among diverse eukaryotic microorganisms (Fig. 2). While viruses are known to be gene transfer agents, membrane vesicles, which are not impaired by receptor recognition in the way that viruses are, could be large spectrum transducing agents [51–53].
Bacterial chitin synthase genes
The analysis of bacterial proteomes gave unexpected results as it revealed a dozen of bacteria possessing at least one division 1 CHS whereas no bacterial CHS had been previously described (Figs. 1, 2 and 6; Additional file 3: Table S3). These bacteria correspond mainly to Gammaproteobacteria (7 Enterobacteriaceae, 2 Cellvibrionaceae and 1 Pseudomonadaceae) but there is also an Alphaproteobacteria (Rhizobiaceae). Finally, one chs gene was detected in the genome of the Cyanobacteria Tolypothrix campylonemoides (NCBI Reference Sequence: NZ_JXB01000008) but it was not included as it is split by a transposase insertion and is probably not functional. The well supported monophyletic group, formed by the corresponding CHS proteins, suggests a unique transfer of a eukaryotic division 1 chs gene to a bacterial genome and multiple HGT might have occurred then between bacterial species. Several cases of the lateral transfer of genes, protein domain-encoding fragments or repeat elements have been described, from animals to bacteria (reviewed in [54–56]) and from Plantae to bacteria [57–59]. However, the detected eukaryote to bacteria transfers are fewer than the bacteria to eukaryote ones [54, 60] for the following possible reasons: (i) the barrier formed by spliceosomal introns in eukaryotic genes [54, 61]; (ii) the smaller population size of eukaryotes which, therefore, offers a reduced pool of potential donors [61]; and (iii) the small number of eukaryotic genes that might present a selective advantage for bacteria and which could, thus, persist in their genome after being transferred [62]. Interestingly, the family 14 of carbohydrate-binding modules (CBM14s), which are short chitin-binding modules predominantly found in animal and fungal genomes, were also detected in seven bacterial genomes as putative HGT [63]. However, the bacterial species involved were different from those harboring chitin synthase genes. Most of the bacterial chs genes were co-localized with a set of co-oriented genes, in a classical bacterial operon organization (Fig. 6) including a sugar epimerase and hypothetical protein encoding-genes. This unusual pattern of gene order conservation between distantly related bacteria suggest that the corresponding genes were transferred and maintained together, possibly because they participate in a common function in these bacteria. Noticeably, two of the hypothetical proteins possess bacterial domains of unknown functions DUF1800 and DUF1501, that are predicted to be part of the same operon in OperonDB [64, 65]. Enterobacteriales species (Brenneria, Pectobacterium and Dickeya) clearly acquired these DUF- and CHS-encoding genes during the same transfer because close enterobacteriales relatives without chs have neither the DUF1501- nor the DUF1800-encoding genes (Additional file 9: Table S5). The alphaproteobacteria Agrobacterium vitis has two pairs of DUF1501-DUF1800 encoding genes organized in tandem. Phylogenetic trees revealed that the pair of proteins encoded by the genes co-localized with the chs are more closely related to their homologs in gammaproteobacteria than to their homologs in alphaproteobacteria (Additional file 10: Figure S5). Hence, these DUF-encoding genes were also transferred with the chs in A. vitis. Some transfers seem to be recent as the corresponding chs belongs to a genomic region with a different G + C content, compared with the neighboring genomic context (Additional file 11: Figure S6). This is the case for one of the two Dickeya_dadantii_Ech703 chs genes (second HGT of a chs in this genome) and the chs of Cedecea neteri and Pseudomonas cichorii, which are localized in regions with a lower G + C content. The secondary metabolism gene cluster, containing one chs in Teredinibacter turnereae, also appeared with a different G + C composition higher than the neighboring genomic context. Some bacterial chitin synthase genes have evolved additional features that differentiate them from eukaryotic division 1 CHS (Fig. 6). First, they have a short N-terminal region and they lack the Eukaryota Chitin_synt_1N domain (PF08407) (Fig. 3). Secondly, an accretion of an aminotransferase domain probably occurred in the ancestor of Pectobacterium and Brenneria (Fig. 3).
Fig. 6.
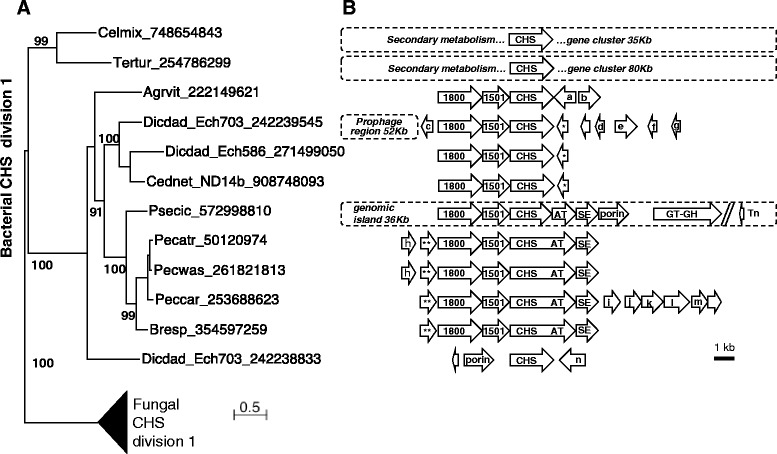
Phylogeny and genomic context of bacterial chitin synthases. a Phylogenetic relationships of bacterial chitin synthases. The ML tree is shown with numbers above the branches, indicating support in bootstrap analyses (100 replicates). The tree was rooted with division 1 fungal CHS from A. fumigatus and U. maydis. The Bayesian phylogenetic approach gave the exact same topology (Additional file 20: Figure S10). b Gene organization of each variable region containing chitin synthase. The limits of regions were obtained by comparison with ortholog regions in proximal species without the chs gene. Each represented region is aligned with the corresponding CHS in the phylogenetic tree. CHS in arrows is the chitin synthase domain and AT is the aminotransferase domain. 1800 and 1501: proteins with a domain of unknown function, DUF1800 (PF08811) and DUF1501 (PF07394) respectively; SE: sugar epimerase; a: transcriptional regulator with a sugar-binding domain; b: 2,4-dihydroxyhept-2-ene-1,7-dioic acid aldolase; c: OsmC family protein; d: DUF465; e: lipid A biosynthesis lauroyl (or palmitoleoyl) acyltransferase; f: cold-shock protein; g: DUF2511; h: efflux protein; i: gluconate-5-dehydrogenase; j: taurine dioxygenase; k: sulfotransferase NodH; l: aminotransferase; m: hydroxylase; and n: AraC family transcriptional regulator. The dotted rectangles correspond to regions detected as a prophage, a secondary metabolism cluster or a genomic island. Empty arrows and those with stars represent hypothetical proteins. Among them, groups of arrows with the same number of stars are orthologs
In a large variety of bacteria, type 2 glycosyltransferases (GT2) homologous to chitin synthases have been described. These transmembrane enzymes are localized at the inner membrane of Gram negative bacteria and synthesize different exopolysaccharides (EPS) into the periplasm. These EPS are made of cellulose, curdlan, alginate or hyaluronic acid [66]. In Pseudomonas aeruginosa, secretion of alginate through the peptidoglycan and the outer membrane is ensured by an envelope-spanning multiprotein complex including channel proteins [67]. We found that some bacterial chs genes are colocated with porin or efflux protein-encoding genes (Fig. 6). It is possible that these bacterial chs genes, and their neighbouring transporter encoding-genes, are involved in the production and secretion of an EPS made of chitin.
EPS are matrix components of bacterial biofilms which are known to play a major role in pathogenic or symbiotic interactions between bacteria and animals or plants [68]. It is noticeable that, among bacteria in which a chs gene was found, Pseudomonas cichorii and Agrobacterium vitis are plant-pathogens and secrete an EPS made of alginate and curdlan, respectively [69, 70]. Indeed, A. vitis is able to build biofilms on abiotic as well as on plant root surfaces, probably due to its EPS [71]. Other bacterial species displaying a chs gene are also plant pathogens: a Brenneria spp., three Pectobacterium species and two Dickeya dadantii isolates. The later two bacterial isolates, D. dadantii Ech586 and D. dadantii Ech703, should be reclassified as two different species, Dickeya zeae Ech586 and Dickeya paradisiaca Ech703, respectively [72]. It may be that the bacterial chitin synthase plays a role in the parasitic interaction between these bacteria and their plant hosts.
CHS activities in fungi or metazoa are usually described as producing long crystalline chains of chitin (fibers) but it is possible that a bacterial CHS secretes small soluble chitosaccharides, or eventually chitooligosaccharides (COS), instead. Indeed, the phytopathogenic oomycete Aphanomyces euteiches does not secrete chitin fibers although two chs genes are present in this species [73]. However, the encoded CHS activities of A. euteiches are thought to be active because a small soluble and noncrystalline glucan-chitosaccharide was detected in its cell wall [74]. Interestingly, one of the two chs genes in D. dadantii Ech703 and the chs gene of Pseudomonas cichorii are co-located and have the same orientation as a putative chitoporin-encoding gene (Fig. 6). In the marine bacterium Vibrio harveyi, chitoporin is a transporter of COS through the outer membrane [75]. COS are known to play a key role as signal molecules in multiple plant-microbe interactions. In parasitic interactions between plants and fungi, COS are released from the digestion of the fungal cell wall by secreted plant chitinases. The resulting COS are then recognized by plant receptors as Microbial Associated Molecular Patterns (MAMP) and they induce plant immunity [76]. In symbiotic interactions, modified COS, called lipochitooligosaccharides (LCOs), which include Myc and Nod factors, modulate plant host immunity. Myc factors are produced and secreted by arbuscular mycorrhizal fungi but the enzymes responsible for their biosynthesis have not yet been identified [77]. Nod factors are synthesized by NodC, a bacterial GT2 homologous to CHS, and they induce nodule formation during a symbiotic interaction between Rhizobiacea (nitrogen-fixing bacteria) and leguminous host plants [78]. Whether bacterial chs genes are able to synthesize COS implicated in plant interactions is still uncertain.
Several bacterial chs genes are fused with a putative aminotransferase domain and they are colocated with a putative sugar epimerase-encoding gene (Fig. 6). These two additional functions could be associated with modifications of chitin or COS. A gene of unknown functions, and composed of the DUF1501 domain, was also found colocated with several bacterial chs. The encoded DUF1501-containing protein carries a twin-arginine motif which would imply that it is exported by the twin-arginine translocation (Tat) pathway (http://www.compgen.org/tools/PRED-TAT/ [79]). The bacterial Tat system allows folded proteins to be moved across membranes without significant ion leakage [80]. The DUF1501 domain has also been described as forming part of a conserved machinery in compartimentalized species from the Planctomycetes, Verrucomicrobia and Chlamydiae (PVC) super-phylum [81]. These informations about DUF1501 proteins are hardly connectable with the biological role of known CHS. However, our hypothesis is that DUF1501, DUF1800 and CHS proteins might have a functional link in bacteria as they share a common conserved operon organization. It would be very interesting to study the function of bacterial CHS and also of the proteins that seem associated to them. Finally, in the particular cases of Teredinibacter turnerae and Cellvibrio mixtus subsp. mixtus, the chs genes are localized in the middle of secondary metabolism gene clusters (80 Kb and 35 Kb, respectively) and they could be involved in the production of a bioactive molecule [82].
Conclusions
Bacterial chitin synthase genes constitute a new example of genes acquired by a bacterium via horizontal transfer from a eukaryotic donor. The chitin synthase activity of the bacterial CHS and the possible selective advantage for the corresponding bacteria, often implied in plant interactions, needs further investigation. CHS-encoding genes have also been transferred between eukaryotic microorganisms. We recommend avoiding the use of this multigenic family to elucidate the phylogenetic relationships between the different eukaryotic species, especially since many duplications and losses are also observed in different lineages.
Fungal CHS are, unfortunately, not an exception and the difficulty of classifying these sequences has led to discordant classifications. We took advantage of the current study to determine which can be robustly classified and then to construct the most consensual classification possible. To facilitate the use of this new classification, any information that corresponds with the previously published versions is provided (Additional file 2: Table S2), together with the databank, in fasta format, of all the classified CHS sequences (Additional file 12: File S1). A website also permits blast queries to the databank (http://wwwabi.snv.jussieu.fr/public/CHSdb). This facilitates a search for the class of a previously identified CHS, even if the corresponding accession number has changed (as is often the case for fungal sequences). It is also an aid for the classification of CHS from species closely related to those analyzed.
Methods
Data collection
The 208 eukaryotic complete proteomes were downloaded from different databases (Additional file 13: Table S6). The main sources were: Ensembl (56 species; http://www.ensembl.org/), the DOE Joint Genome Institute (53 species; http://www.jgi.doe.gov/), the National Center for Biotechnology Information (31 species; http://www.ncbi.nlm.nih.gov/), and the Broad Institute of MIT and Harvard (29 species; http://www.broadinstitute.org/). The tested proteomes of 1218 bacteria, 97 archaea and 2398 viruses were also downloaded from the NCBI (Additional file 14: Table S7). The hyaluronan synthase and nodulation NodC protein sequences employed in this study were those used in [21] and [83].
Identification of chitin synthases
CHS protein sequences share several motifs (Fig. 3; Additional file 7: Figure S3), spanning the catalytic site and the C-terminal part of these enzymes, and two major related chitin synthase Pfam domains [84], Chitin_synth_1 (PF01644) and Chitin_synth_2 (PF03142). To identify the CHS sequences, similarity searches were first performed with these two domains in complete proteomes (see Methods – Data collection) using the RPS-BLAST program [85], with an E-value cutoff of 1e-05. This cutoff was judged sufficiently high to detect all chitin synthase proteins as some hits corresponded to other glycosyltransferase proteins of the GT2 family. Indeed, unlike the Chitin_synth_1 domain which is very specific to division 1 CHS sequences, the Chitin_synth_2 domain could be found in dozens of glycosyl transferase sequences that were not chitin synthases (e.g. hyaluronan synthases, cellulose synthases etc.). To eliminate these non-CHS sequences, phylogenetic trees were performed (see Methods – Sequences analysis) and only proteins forming a clear monophyletic group with known chitin synthase divisions, using hyaluronan synthase and nodulation NodC protein sequences as the outgroups, were considered as chitin synthase proteins. The presence of conserved motifs essential for enzymatic activity (D, D, D, QXXRW) was also checked as it guarantees that the proteins are potentially functional and could endow a chitin synthase activity. Proteins that do not possess these motifs were, thus, considered as dubious and disqualified from our study. For example, a recently proposed class VIII [10] was not retained due to the absence of the essential motif QXXRW.
Some CHS classes showed reduced taxonomic distribution (recCHS, ESV, CV, Bacterial CHS etc.). To gain a better understanding of the origin of these sequences, it was important to have a more precise idea of the species that possess the corresponding genes. In these cases, the analysis was completed by a BLASTP search, carried out at the NCBI, using the non-redundant protein sequences (nr) database.
Sequence analysis
All phylogenetic analyses were performed using the following procedure. First, amino acid sequences were aligned using MAFFT with the E-INSI algorithm and default settings [86]. Next, regions in the resulting multiple sequence alignments that were suitable for phylogenetic inference were selected using BMGE [87]. The BLOSUM60 and BLOSUM30 matrices were used, respectively, for alignments with sequences from a single division and alignments containing sequences from different divisions. This step removed any ambiguously aligned or highly variable regions in order to improve the overall performance of the phylogenetic reconstructions. Thirdly, phylogenetic inferences were obtained with two approaches. ML trees were constructed with PhyML 3.0 [88] using the following parameters: the LG model, with empirical amino acid frequencies, an estimated proportion of invariable sites, subtree pruning and regrafting (SPR) and five random starting trees added to the standard BioNJ starting tree. The support of the data for each internal branch of the phylogenies was estimated using non-parametric bootstraps, with 100 replicates. Bayesian inferences were obtained with MrBayes v3.2.6 [89] with a fixed WAG model of amino acid substitution and a gamma correction (four discrete categories plus a proportion of invariant sites). The program was run with four chains for 100 000 generations and trees were sampled every 100 generations. To construct the consensus tree, the first 250 trees were discarded “burnin” and posterior probabilities were used as support for internal branches. NJplot [90] and FigTree [91] were used for outputting ML and Bayesian trees respectively.
Hyaluronan synthase (HAS) and NodC sequences were used as the outgroup for the global CHS tree (Fig. 2) as they are distinct type 2 glycosyltransferases. All CHS sequences from the databank were classified using phylogenies. However, only a part of them was used in the phylogenetic trees presented here, so that the trees remain readable. They correspond to the CHS of a sample of species chosen to maximised the preservation of the global CHS tree topology observed with all the sequences.
The domains were annotated by searching protein sequences against the Pfam library of HMMs with pfam_scan.pl [92]. The transmembrane helices in proteins were predicted with TMHMM 2.0 [93].
Genomic context analysis
The genomic context of bacterial chitin synthases was analyzed with multiple genome alignments using Mauve 2.3.1 [94]. Gene organization around the chitin synthase encoding-genes was compared with that of ortholog regions in genome(s) from proximate species lacking the chitin synthase gene. Hence, four groups of genomes were compared. First, the Pectobacterium group comprised Pectobacterium carotovorum subsp. carotovorum WPP14, P. carotovorum subsp. brasiliensis PBR1692, Brenneria sp. EniD312, P. wasabiae WPP163, P. carotovorum subsp. carotovorum PC1 and P. atrosepticum SCRI1043. Second, a Dickeya group comprised Dickeya dadantii 3937, D. dadantii Ech586, D. dadantii Ech703 and D. zeae Ech1591. Third, an Agrobacterium group was composed of Agrobacterium vitis S4 and Agrobacterium radiobacter K84. Finally, a Pseudomonas group comprised P. cichorii JBC1, P. syringae pv. tomato DC300, P. syringae pv. syringae B728a and P. syringae pv. phaseolicola 1448A. In each case, this allowed us to localize the limits between the variable region containing the chitin synthase coding gene and the core surrounding regions shared by all proximate species (syntenic shared blocks). Predictions concerning genomic islands were retrieved from IslandViewer [95] and predicted prophages were sourced from Prophinder [96]. A G + C content analysis was performed with overlapping sliding windows of 1000 bp at a step of 30 bp using JaDis [97].
Acknowledgements
We are grateful to the D.S.I Pôle Calcul (University of Paris VI) for providing computational resources. We also thank Florence Hommais and Joel Pothier for helpful discussions.
Funding
This work was supported by the University Claude Bernard Lyon 1, the CNRS and Bayer SAS.
Availability of data and materials
The dataset(s) supporting the conclusions of this article is (are) included within the article (and its Additional files 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19 and 20).
Authors’ contributions
IRG and MC conceived and coordinated the study. IRG, SB, CS and NC acquired the sequence data and made the bioinformatic and phylogenetic analyses. MC and MCS acquired and compared the data on mutants. IRG and SB developped the website. MC, IRG, SG and MB interpreted the data and drafted the paper. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Abbreviations
- CHS
Chitin synthase(s)
- CV
Chlorovirus-like CHS class
- ESV
Ectocarpus siliculosus-like CHS class
- HAS
Hyaluronan synthase(s)
- HGT
Horizontal gene transfer(s)
- LBA
Long branch attraction
- LOCA
Last Opistokonta common ancestor
- ML
Maximum Likelihood
- recCHS
Recombined chitin synthase
- UNGD
UDP-N-acetylglucosamine 6-dehydrogenase gene
Additional files
Classification of fungal chitin synthase single mutants and comparison of their phenotype (1986–2016). In the first tab, phenotypes of mutants are summarized. In the second tab, phenotypes of mutants are detailed. In the third tab are the references of the single mutants publications. (XLSX 59 kb)
Correspondence between the published chitin synthase classifications (1992–2016) (first tab) and their references (second tab). (XLSX 29 kb)
Account of all chitin synthase encoding sequences in the genomes of species included in this study. (XLSX 81 kb)
Phylogeny of Hemiascomycota yeast CHS. A ML phylogeny based on 404 amino acid alignment positions of 42 sequences was constructed with PhyML. Bootstraps of interest and ≥60 are shown above the branches. (PDF 49 kb)
Nonsynonymous substitution rates (dN) estimated for pairwise comparisons between ascomycota chs from the different classes. Comparisons were restricted to a region spanning the catalytic domain from the c motif to the g motif (Fig. 3 and Additional file 7: Figure S3). The PAML [98] codeml program was used to estimate dN with the codon substitution model of Goldman and Yang [99]. (PDF 47 kb)
Account of species displaying recCHS, ESV or CV chitin synthases (tab 1) and account detail of recCHS, ESV and CV sequences in these species (tab 2). (XLSX 15 kb)
Eight conserved signatures in chitin synthases (adapted from [21]). (PDF 90 kb)
Phylogenies of CHS N- and C-termini regions. The N-terminal region, used for phylogeny, ends before the motif d of the catalytic domain and the C-terminal region starts after this motif (Fig. 3 and Additional file 7: Figure S3) The two corresponding alignments were obtained by extracting these regions from the alignement obtained with the full sequences. They were then filtered with BMGE prior to the tree reconstruction analysis. This resulted in 72 amino acid alignment positions of 119 CHS N-terminal region sequences and 148 amino acid alignment positions of 120 CHS C-terminal region sequences. Phylogenetic trees were constructed with PhyML (A-B) and MrBayes (C-D) Bootstraps ≥60 (PhyML) and posterior probabilities (MrBayes) of interest are shown above the branches. (PDF 133 kb)
Co-occurrence of DUF1501, DUF1800 and CHS encoding genes in bacterial genomes. (DOCX 60 kb)
Phylogenies of DUF1800 and DUF1501 bearing proteins. ML phylogenies based on (A) 383 amino acid alignment positions of 14 DUF1800 bearing sequences and (B) 335 amino acid alignment positions of 14 DUF1501 bearing sequences were constructed with PhyML. Bootstraps of interest and ≥60 are shown above the branches. (PDF 51 kb)
G + C content variation around the recently laterally transferred chitin synthase genes of D. dadantii Ech703, Cedecea neteri and Pseudomonas cichorii. The G + C content was computed in 1Kb sliding windows, with a step of 30 bp, along a genomic region containing the chs-like gene. Genes from the variable region are indicated with white arrows. Black arrows show the length and position of the surrounding genes. For each gene, the G + C content at the third codon position (GC3%) is indicated with a black dot. The horizontal line corresponds to the G + C content of the entire region. (PDF 178 kb)
Databank of all classified CHS in fasta format. (TXT 980 kb)
Eukaryotic species analyzed in this study. (XLSX 17 kb)
Archeae, Bacteria and Virus genomes analyzed in this study. (XLSX 124 kb)
Bayesian phylogeny of chitin synthases. (PDF 69 kb)
ML phylogeny used to place the root in Fig. 4. (TXT 5 kb)
Bayesian phylogeny of fungal CHS belonging to division 1. (PDF 189 kb)
ML phylogeny used to place the root in Fig. 5. (TXT 6 kb)
Bayesian phylogeny of fungal CHS belonging to division 2. (PDF 241 kb)
Bayesian phylogeny of bacterial chitin synthases. (PDF 71 kb)
Contributor Information
Isabelle R. Gonçalves, Phone: (33) 4 72 44 85 43, Email: isabelle.goncalves@univ-lyon1.fr
Sophie Brouillet, Email: sophie.brouillet@upmc.fr.
Marie-Christine Soulié, Email: marie-christine.soulie@upmc.fr.
Simonetta Gribaldo, Email: simonetta.gribaldo@pasteur.fr.
Catherine Sirven, Email: catherine.sirven@bayer.com.
Noémie Charron, Email: ncharron1@gmail.com.
Martine Boccara, Email: martine.boccara@upmc.fr.
Mathias Choquer, Email: mathias.choquer@univ-lyon1.fr.
References
- 1.Cauchie H-M. Chitin production by arthropods in the hydrosphere. Hydrobiologia. 2002;470:63–95. doi: 10.1023/A:1015615819301. [DOI] [Google Scholar]
- 2.Heath-Heckman EAC, McFall-Ngai MJ. The occurrence of chitin in the hemocytes of invertebrates. Zoology. 2011;114:191–198. doi: 10.1016/j.zool.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wagner GP, Lo J, Laine R, Almeder M. Chitin in the epidermal cuticle of a vertebrate (Paralipophrys trigloides, Blenniidae, Teleostei) Experientia. 1993;49:317–319. doi: 10.1007/BF01923410. [DOI] [Google Scholar]
- 4.Zaku SG, Emmanuel SA, Aguzue OC, Thomas SA. Extraction and characterization of chitin; a functional biopolymer obtained from scales of common carp fish (Cyprinus carpio L.): A lesser known source. Afr J Food Sci. 2011;5:478–483. [Google Scholar]
- 5.Tang WJ, Fernandez JG, Sohn JJ, Amemiya CT. Chitin is endogenously produced in vertebrates. Curr Biol. 2015;25:897–900. doi: 10.1016/j.cub.2015.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mulisch M. Chitin in protistan organisms: distribution, synthesis and deposition. Eur J Protistol. 1993;29:1–18. doi: 10.1016/S0932-4739(11)80291-9. [DOI] [PubMed] [Google Scholar]
- 7.Durkin CA, Mock T, Armbrust EV. Chitin in diatoms and its association with the cell wall. Eukaryot Cell. 2009;8:1038–1050. doi: 10.1128/EC.00079-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Merzendorfer H. The cellular basis of chitin synthesis in fungi and insects: Common principles and differences. Eur J Cell Biol. 2011;90:759–769. doi: 10.1016/j.ejcb.2011.04.014. [DOI] [PubMed] [Google Scholar]
- 9.Free SJ. Fungal cell wall organization and biosynthesis. Adv Genet. 2013;81:33–82. doi: 10.1016/B978-0-12-407677-8.00002-6. [DOI] [PubMed] [Google Scholar]
- 10.Mélida H, Sain D, Stajich JE, Bulone V. Deciphering the uniqueness of Mucoromycotina cell walls by combining biochemical and phylogenomic approaches: The unique features of Mucoromycotina cell walls. Environ Microbiol. 2015;17:1649–1662. doi: 10.1111/1462-2920.12601. [DOI] [PubMed] [Google Scholar]
- 11.Coutinho PM, Deleury E, Davies GJ, Henrissat B. An evolving hierarchical family classification for Glycosyltransferases. J Mol Biol. 2003;328:307–317. doi: 10.1016/S0022-2836(03)00307-3. [DOI] [PubMed] [Google Scholar]
- 12.Morgan JLW, Strumillo J, Zimmer J. Crystallographic snapshot of cellulose synthesis and membrane translocation. Nature. 2012;493:181–186. doi: 10.1038/nature11744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bowen AR, Chen-Wu JL, Momany M, Young R, Szaniszlo PJ, Robbins PW. Classification of fungal chitin synthases. Proc Natl Acad Sci. 1992;89:519–523. doi: 10.1073/pnas.89.2.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Specht CA, Liu Y, Robbins PW, Bulawa CE, Iartchouk N, Winter KR, et al. The chsD and chsE genes of Aspergillus nidulans and their roles in chitin synthesis. Fungal Genet Biol. 1996;20:153–167. doi: 10.1006/fgbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 15.Xoconostle-Cázares B, Specht CA, Robbins PW, Liu Y, León C, Ruiz-Herrera J. Umchs5, a gene coding for a class IV chitin synthase in Ustilago maydis. Fungal Genet Biol. 1997;22:199–208. doi: 10.1006/fgbi.1997.1014. [DOI] [PubMed] [Google Scholar]
- 16.Munro CA, Gow NAR. Chitin synthesis in human pathogenic fungi. Med Mycol. 2001;39:41–53. doi: 10.1080/mmy.39.1.41.53. [DOI] [PubMed] [Google Scholar]
- 17.Roncero C. The genetic complexity of chitin synthesis in fungi. Curr Genet. 2002;41:367–378. doi: 10.1007/s00294-002-0318-7. [DOI] [PubMed] [Google Scholar]
- 18.Chigira Y, Abe K, Gomi K, Nakajima T. chsZ, a gene for a novel class of chitin synthase from Aspergillus oryzae. Curr Genet. 2002;41:261–267. doi: 10.1007/s00294-002-0305-z. [DOI] [PubMed] [Google Scholar]
- 19.Amnuaykanjanasin A, Epstein L. A class V chitin synthase gene, chsA is essential for conidial and hyphal wall strength in the fungus Colletotrichum graminicola (Glomerella graminicola) Fungal Genet Biol. 2003;38:272–285. doi: 10.1016/S1087-1845(02)00563-7. [DOI] [PubMed] [Google Scholar]
- 20.Niño-vega GA, Carrero L, San-Blas G. Isolation of the CHS4 gene of Paracoccidioides brasiliensis and its accommodation in a new class of chitin synthases. Med Mycol. 2004;42:51–57. doi: 10.1080/1369378031000153811. [DOI] [PubMed] [Google Scholar]
- 21.Choquer M, Boccara M, Gonçalves IR, Soulié M-C, Vidal-Cros A. Survey of the Botrytis cinerea chitin synthase multigenic family through the analysis of six euascomycetes genomes. Eur J Biochem. 2004;271:2153–2164. doi: 10.1111/j.1432-1033.2004.04135.x. [DOI] [PubMed] [Google Scholar]
- 22.Ruiz-Herrera J, Ortiz-Castellanos L. Analysis of the phylogenetic relationships and evolution of the cell walls from yeasts and fungi: Fungal wall evolution. FEMS Yeast Res. 2010;10:225–243. doi: 10.1111/j.1567-1364.2009.00589.x. [DOI] [PubMed] [Google Scholar]
- 23.Li M, Jiang C, Wang Q, Zhao Z, Jin Q, Xu J-R, et al. Evolution and functional insights of different ancestral Orthologous clades of chitin synthase genes in the fungal tree of life. Front Plant Sci. 2016;7:37. doi: 10.3389/fpls.2016.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Y-Z, Chen Q, Liu C-H, Liu Y-B, Yi P, Niu K-X, et al. Chitin synthase gene FgCHS8 affects virulence and fungal cell wall sensitivity to environmental stress in Fusarium graminearum. Fungal Biol. 2016;120:764–774. doi: 10.1016/j.funbio.2016.02.002. [DOI] [PubMed] [Google Scholar]
- 25.Sheng W, Yamashita S, Ohta A, Horiuchi H. Functional differentiation of chitin synthases in Yarrowia lipolytica. Biosci Biotechnol Biochem. 2013;77:1275–1281. doi: 10.1271/bbb.130111. [DOI] [PubMed] [Google Scholar]
- 26.Peyretaillade E, El Alaoui H, Diogon M, Polonais V, Parisot N, Biron DG, et al. Extreme reduction and compaction of microsporidian genomes. Res Microbiol. 2011;162:598–606. doi: 10.1016/j.resmic.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 27.Fares MA, Byrne KP, Wolfe KH. Rate asymmetry after genome duplication causes substantial long-branch attraction artifacts in the phylogeny of Saccharomyces species. Mol Biol Evol. 2006;23:245–253. doi: 10.1093/molbev/msj027. [DOI] [PubMed] [Google Scholar]
- 28.Felsenstein J. Cases in which parsimony or compatibility methods will be positively misleading. Syst Biol. 1978;27:401–410. doi: 10.1093/sysbio/27.4.401. [DOI] [Google Scholar]
- 29.Mifsud W, Bateman A. Membrane-bound progesterone receptors contain a cytochrome b5-like ligand-binding domain. Genome Biol. 2002;3:5. doi: 10.1186/gb-2002-3-12-research0068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schuster M, Treitschke S, Kilaru S, Molloy J, Harmer NJ, Steinberg G. Myosin-5, kinesin-1 and myosin-17 cooperate in secretion of fungal chitin synthase. EMBO J. 2012;31:214–227. doi: 10.1038/emboj.2011.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fernandes C, Gow NAR, Gonçalves T. The importance of subclasses of chitin synthase enzymes with myosin-like domains for the fitness of fungi. Fungal Biol Rev. 2016;30:1–14. doi: 10.1016/j.fbr.2016.03.002. [DOI] [Google Scholar]
- 32.Chen Z, Martinez DA, Gujja S, Sykes SM, Zeng Q, Szaniszlo PJ, et al. Comparative genomic and transcriptomic analysis of Wangiella dermatitidis, a major cause of Phaeohyphomycosis and a model black yeast human pathogen. G3 (Bethesda) 2014;4:561–578. doi: 10.1534/g3.113.009241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yuan X, Xiao S, Taylor TN. Lichen-like symbiosis 600 million years ago. Science. 2005;308:1017–1020. doi: 10.1126/science.1111347. [DOI] [PubMed] [Google Scholar]
- 34.Beck A, Divakar PK, Zhang N, Molina MC, Struwe L. Evidence of ancient horizontal gene transfer between fungi and the terrestrial alga Trebouxia. Org Divers Evol. 2014;15:235–248. doi: 10.1007/s13127-014-0199-x. [DOI] [Google Scholar]
- 35.Delaroque N, Müller DG, Bothe G, Pohl T, Knippers R, Boland W. The complete DNA sequence of the Ectocarpus siliculosus virus EsV-1 genome. Virology. 2001;287:112–132. doi: 10.1006/viro.2001.1028. [DOI] [PubMed] [Google Scholar]
- 36.Cock JM, Sterck L, Rouzé P, Scornet D, Allen AE, Amoutzias G, et al. The Ectocarpus genome and the independent evolution of multicellularity in brown algae. Nature. 2010;465:617–621. doi: 10.1038/nature09016. [DOI] [PubMed] [Google Scholar]
- 37.Kawasaki T, Tanaka M, Fujie M, Usami S, Sakai K, Yamada T. Chitin synthesis in Chlorovirus CVK2-infected chlorella cells. Virology. 2002;302:123–131. doi: 10.1006/viro.2002.1572. [DOI] [PubMed] [Google Scholar]
- 38.Yamada T, Kawasaki T. Microbial synthesis of hyaluronan and chitin: New approaches. J Biosci Bioeng. 2005;99:521–528. doi: 10.1263/jbb.99.521. [DOI] [PubMed] [Google Scholar]
- 39.James TY, Pelin A, Bonen L, Ahrendt S, Sain D, Corradi N, et al. Shared signatures of parasitism and Phylogenomics Unite Cryptomycota and Microsporidia. Curr Biol. 2013;23:1548–1553. doi: 10.1016/j.cub.2013.06.057. [DOI] [PubMed] [Google Scholar]
- 40.Schwelm A, Fogelqvist J, Knaust A, Jülke S, Lilja T, Bonilla-Rosso G, et al. The Plasmodiophora brassicae genome reveals insights in its life cycle and ancestry of chitin synthases. Sci Rep. 2015;5:11153. doi: 10.1038/srep11153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Traeger S, Nowrousian M. Functional analysis of developmentally regulated genes chs7 and sec22 in the Ascomycete Sordaria macrospora. G3 (Bethesda) 2015;5:1233–1245. doi: 10.1534/g3.115.017681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morcx S, Kunz C, Choquer M, Assie S, Blondet E, Simond-Côte E, et al. Disruption of Bcchs4, Bcchs6 or Bcchs7 chitin synthase genes in Botrytis cinerea and the essential role of class VI chitin synthase (Bcchs6) Fungal Genet Biol. 2013;52:1–8. doi: 10.1016/j.fgb.2012.11.011. [DOI] [PubMed] [Google Scholar]
- 43.Torruella G, de Mendoza A, Grau-Bové X, Antó M, Chaplin MA, del Campo J, et al. Phylogenomics reveals convergent evolution of lifestyles in close relatives of animals and fungi. Curr Biol. 2015;25:2404–2410. doi: 10.1016/j.cub.2015.07.053. [DOI] [PubMed] [Google Scholar]
- 44.Zakrzewski A-C, Weigert A, Helm C, Adamski M, Adamska M, Bleidorn C, et al. Early divergence, broad distribution, and high diversity of animal chitin synthases. Genome Biol Evol. 2014;6:316–325. doi: 10.1093/gbe/evu011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guerriero G. Putative chitin synthases from Branchiostoma floridae show extracellular matrix-related domains and mosaic structures. Genomics Proteomics Bioinformatics. 2012;10:197–207. doi: 10.1016/j.gpb.2012.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Van Dellen KL, Bulik DA, Specht CA, Robbins PW, Samuelson JC. Heterologous expression of an Entamoeba histolytica chitin synthase in Saccharomyces cerevisiae. Eukaryot Cell. 2006;5:203–206. doi: 10.1128/EC.5.1.203-206.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cavalier-Smith T, Chao EE-Y. Phylogeny of choanozoa, apusozoa, and other protozoa and early eukaryote megaevolution. J Mol Evol. 2003;56:540–563. doi: 10.1007/s00239-002-2424-z. [DOI] [PubMed] [Google Scholar]
- 48.Blank CE. An expansion of age constraints for microbial clades that lack a conventional fossil record using phylogenomic dating. J Mol Evol. 2011;73:188–208. doi: 10.1007/s00239-011-9467-y. [DOI] [PubMed] [Google Scholar]
- 49.Blanc G, Duncan G, Agarkova I, Borodovsky M, Gurnon J, Kuo A, et al. The Chlorella variabilis NC64A genome reveals adaptation to photosymbiosis, coevolution with viruses, and cryptic sex. Plant Cell. 2010;22:2943–2955. doi: 10.1105/tpc.110.076406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Foflonker F, Price DC, Qiu H, Palenik B, Wang S, Bhattacharya D. Genome of the halotolerant green alga Picochlorum sp. reveals strategies for thriving under fluctuating environmental conditions. Environ Microbiol. 2015;17:412–426. doi: 10.1111/1462-2920.12541. [DOI] [PubMed] [Google Scholar]
- 51.Kulp A, Kuehn MJ. Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Annu Rev Microbiol. 2010;64:163–184. doi: 10.1146/annurev.micro.091208.073413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Biller SJ, Schubotz F, Roggensack SE, Thompson AW, Summons RE, Chisholm SW. Bacterial vesicles in marine ecosystems. Science. 2014;343:183–186. doi: 10.1126/science.1243457. [DOI] [PubMed] [Google Scholar]
- 53.Leung KF, Dacks JB, Field MC. Evolution of the multivesicular body ESCRT machinery; retention across the eukaryotic lineage. Traffic. 2008;9:1698–1716. doi: 10.1111/j.1600-0854.2008.00797.x. [DOI] [PubMed] [Google Scholar]
- 54.Dunning Hotopp JC. Horizontal gene transfer between bacteria and animals. Trends Genet. 2011;27:157–163. doi: 10.1016/j.tig.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Anderson MT, Seifert HS. Opportunity and means: horizontal gene transfer from the human host to a bacterial pathogen. mBio. 2011;2:e00005-11-e00005-11. [DOI] [PMC free article] [PubMed]
- 56.Patrick S, Blakely GW. Crossing the eukaryote-prokaryote divide: A ubiquitin homolog in the human commensal bacterium Bacteroides fragilis. Mob Genet Elem. 2012;2:149–151. doi: 10.4161/mge.21191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kajava AV, Anisimova M, Peeters N. Origin and evolution of GALA-LRR, a new member of the CC-LRR subfamily: from plants to bacteria? PLoS One. 2008;3:e1694. doi: 10.1371/journal.pone.0001694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Arias MC, Danchin EGJ, Coutinho P, Henrissat B, Ball S. Eukaryote to gut bacteria transfer of a glycoside hydrolase gene essential for starch breakdown in plants. Mob Genet Elem. 2012;2:81–87. doi: 10.4161/mge.20375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Miyashita H, Kuroki Y, Kretsinger RH, Matsushima N. Horizontal gene transfer of plant-specific leucine-rich repeats between plants and bacteria. Nat Sci. 2013;5:580–598. [Google Scholar]
- 60.Soanes D, Richards TA. Horizontal gene transfer in eukaryotic plant pathogens. Annu Rev Phytopathol. 2014;52:583–614. doi: 10.1146/annurev-phyto-102313-050127. [DOI] [PubMed] [Google Scholar]
- 61.Keeling PJ, Palmer JD. Horizontal gene transfer in eukaryotic evolution. Nat Rev Genet. 2008;9:605–618. doi: 10.1038/nrg2386. [DOI] [PubMed] [Google Scholar]
- 62.Ochman H, Moran NA. Genes lost and genes found: evolution of bacterial pathogenesis and symbiosis. Science. 2001;292:1096–1099. doi: 10.1126/science.1058543. [DOI] [PubMed] [Google Scholar]
- 63.Chang T-C, Stergiopoulos I. Inter- and intra-domain horizontal gene transfer, gain-loss asymmetry and positive selection mark the evolutionary history of the CBM14 family. FEBS J. 2015;282:2014–2028. doi: 10.1111/febs.13256. [DOI] [PubMed] [Google Scholar]
- 64.Ermolaeva MD, White O, Salzberg SL. Prediction of operons in microbial genomes. Nucleic Acids Res. 2001;29:1216–1221. doi: 10.1093/nar/29.5.1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pertea M, Ayanbule K, Smedinghoff M, Salzberg SL. OperonDB: a comprehensive database of predicted operons in microbial genomes. Nucleic Acids Res. 2009;37:D479–D482. doi: 10.1093/nar/gkn784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schmid J, Sieber V, Rehm B. Bacterial exopolysaccharides: biosynthesis pathways and engineering strategies. Front Microbiol. 2015;6:496. doi: 10.3389/fmicb.2015.00496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hay ID, Wang Y, Moradali MF, Rehman ZU, Rehm BHA. Genetics and regulation of bacterial alginate production. Environ Microbiol. 2014;16:2997–3011. doi: 10.1111/1462-2920.12389. [DOI] [PubMed] [Google Scholar]
- 68.Killiny N, Martinez RH, Dumenyo CK, Cooksey DA, Almeida RPP. The exopolysaccharide of Xylella fastidiosa is essential for biofilm formation, plant virulence, and vector transmission. Mol Plant-Microbe Interact. 2013;26:1044–1053. doi: 10.1094/MPMI-09-12-0211-R. [DOI] [PubMed] [Google Scholar]
- 69.Fett WF, Wijey C. Yields of alginates produced by fluorescent pseudomonads in batch culture. J Ind Microbiol. 1995;14:412–415. doi: 10.1007/BF01569959. [DOI] [Google Scholar]
- 70.McIntosh M, Stone BA, Stanisich VA. Curdlan and other bacterial (1 → 3)-β-d-glucans. Appl Microbiol Biotechnol. 2005;68:163–173. doi: 10.1007/s00253-005-1959-5. [DOI] [PubMed] [Google Scholar]
- 71.Abarca-Grau AM, Penyalver R, López MM, Marco-Noales E. Pathogenic and non-pathogenic Agrobacterium tumefaciens, A. rhizogenes and A. vitis strains form biofilms on abiotic as well as on root surfaces. Plant Pathol. 2011;60:416–425. doi: 10.1111/j.1365-3059.2010.02385.x. [DOI] [Google Scholar]
- 72.Marrero G, Schneider KL, Jenkins DM, Alvarez AM. Phylogeny and classification of Dickeya based on multilocus sequence analysis. Int J Syst Evol Microbiol. 2013;63:3524–3539. doi: 10.1099/ijs.0.046490-0. [DOI] [PubMed] [Google Scholar]
- 73.Badreddine I, Lafitte C, Heux L, Skandalis N, Spanou Z, Martinez Y, et al. Cell wall Chitosaccharides are essential components and exposed patterns of the Phytopathogenic Oomycete Aphanomyces euteiches. Eukaryot Cell. 2008;7:1980–1993. doi: 10.1128/EC.00091-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nars A, Lafitte C, Chabaud M, Drouillard S, Mélida H, Danoun S, et al. Aphanomyces euteiches cell wall fractions containing novel glucan-chitosaccharides induce defense genes and nuclear calcium oscillations in the plant host Medicago truncatula. PLoS One. 2013;8:e75039. doi: 10.1371/journal.pone.0075039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Suginta W, Chumjan W, Mahendran KR, Schulte A, Winterhalter M. Chitoporin from Vibrio harveyi, a channel with exceptional sugar specificity. J Biol Chem. 2013;288:11038–11046. doi: 10.1074/jbc.M113.454108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sanchez-Vallet A, Mesters JR, Thomma BPHJ. The battle for chitin recognition in plant-microbe interactions. FEMS Microbiol Rev. 2015;39:171–183. doi: 10.1093/femsre/fuu003. [DOI] [PubMed] [Google Scholar]
- 77.Sun J, Miller JB, Granqvist E, Wiley-Kalil A, Gobbato E, Maillet F, et al. Activation of symbiosis signaling by Arbuscular Mycorrhizal fungi in legumes and rice. Plant Cell. 2015;27:823–838. doi: 10.1105/tpc.114.131326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Geremia RA, Mergaert P, Geelen D, Van Montagu M, Holsters M. The NodC protein of Azorhizobium caulinodans is an N-acetylglucosaminyltransferase. Proc Natl Acad Sci. 1994;91:2669–2673. doi: 10.1073/pnas.91.7.2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bagos PG, Nikolaou EP, Liakopoulos TD, Tsirigos KD. Combined prediction of Tat and Sec signal peptides with hidden Markov models. Bioinforma Oxf Engl. 2010;26:2811–2817. doi: 10.1093/bioinformatics/btq530. [DOI] [PubMed] [Google Scholar]
- 80.Berks BC. The twin-arginine protein translocation pathway. Annu Rev Biochem. 2015;84:843–864. doi: 10.1146/annurev-biochem-060614-034251. [DOI] [PubMed] [Google Scholar]
- 81.Kamneva OK, Knight SJ, Liberles DA, Ward NL. Analysis of genome content evolution in PVC bacterial super-phylum: assessment of candidate genes associated with cellular organization and lifestyle. Genome Biol Evbol. 2012;4:1375–1390. doi: 10.1093/gbe/evs113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yang JC, Madupu R, Durkin AS, Ekborg NA, Pedamallu CS, Hostetler JB, et al. The complete genome of Teredinibacter turnerae T7901: an intracellular Endosymbiont of marine wood-boring bivalves (Shipworms). Ahmed N, editor. PLoS ONE. 2009;4:e6085. [DOI] [PMC free article] [PubMed]
- 83.Mohammed Ali AM, Kawasaki T, Yamada T. Genetic rearrangements on the Chlorovirus genome that switch between hyaluronan synthesis and chitin synthesis. Virology. 2005;342:102–110. doi: 10.1016/j.virol.2005.07.026. [DOI] [PubMed] [Google Scholar]
- 84.Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, et al. Pfam: the protein families database. Nucleic Acids Res. 2014;42:D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Criscuolo A, Gribaldo S. BMGE (Block Mapping and Gathering with Entropy): a new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol Biol. 2010;10:210. doi: 10.1186/1471-2148-10-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 89.Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinforma Oxf Engl. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 90.Perrière G, Gouy M. WWW-query: An on-line retrieval system for biological sequence banks. Biochimie. 1996;78:364–369. doi: 10.1016/0300-9084(96)84768-7. [DOI] [PubMed] [Google Scholar]
- 91.Rambaud A, Drummond A. FigTree v1.3.1. 2009. [Google Scholar]
- 92.Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, et al. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2016;44:D279–D285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sonnhammer EL, von Heijne G, Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc Int Conf Intell Syst Mol Biol. 1998;6:175–182. [PubMed] [Google Scholar]
- 94.Darling ACE, Mau B, Blattner FR, Perna NT. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Langille MGI, Brinkman FSL. IslandViewer: an integrated interface for computational identification and visualization of genomic islands. Bioinformatics. 2009;25:664–665. doi: 10.1093/bioinformatics/btp030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lima-Mendez G, Van Helden J, Toussaint A, Leplae R. Prophinder: a computational tool for prophage prediction in prokaryotic genomes. Bioinformatics. 2008;24:863–865. doi: 10.1093/bioinformatics/btn043. [DOI] [PubMed] [Google Scholar]
- 97.Gonçalves I, Robinson M, Perrière G, Mouchiroud D. JaDis: computing distances between nucleic acid sequences. Bioinformatics. 1999;15:424–425. doi: 10.1093/bioinformatics/15.5.424. [DOI] [PubMed] [Google Scholar]
- 98.Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci CABIOS. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- 99.Goldman N, Yang Z. A codon-based model of nucleotide substitution for protein-coding DNA sequences. Mol Biol Evol. 1994;11:725–736. doi: 10.1093/oxfordjournals.molbev.a040153. [DOI] [PubMed] [Google Scholar]
- 100.Piednoël M, Gonçalves IR, Higuet D, Bonnivard E. Eukaryote DIRS1-like retrotransposons: an overview. BMC Genomics. 2011;12:621. doi: 10.1186/1471-2164-12-621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Okamoto N, Chantangsi C, Horák A, Leander BS, Keeling PJ. Molecular phylogeny and description of the novel katablepharid Roombia truncata gen. et sp. nov., and establishment of the Hacrobia taxon nov. PLoS One. 2009;4:e7080. doi: 10.1371/journal.pone.0007080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Keeling PJ, Burger G, Durnford DG, Lang BF, Lee RW, Pearlman RE, et al. The tree of eukaryotes. Trends Ecol Evol. 2005;20:670–676. doi: 10.1016/j.tree.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 103.Amemiya CT, Alföldi J, Lee AP, Fan S, Philippe H, Maccallum I, et al. The African coelacanth genome provides insights into tetrapod evolution. Nature. 2013;496:311–316. doi: 10.1038/nature12027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Paps J, Medina-Chacón LA, Marshall W, Suga H, Ruiz-Trillo I. Molecular phylogeny of unikonts: new insights into the position of apusomonads and ancyromonads and the internal relationships of opisthokonts. Protist. 2013;164:2–12. doi: 10.1016/j.protis.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Heldermon C, DeAngelis PL, Weigel PH. Topological organization of the hyaluronan synthase from Streptococcus pyogenes. J Biol Chem. 2001;276:2037–2046. doi: 10.1074/jbc.M002276200. [DOI] [PubMed] [Google Scholar]
- 106.Karnezis T. Topological characterization of an inner membrane (1- > 3)- beta-D-glucan (curdlan) synthase from Agrobacterium sp. strain ATCC31749. Glycobiology. 2003;13:693–706. doi: 10.1093/glycob/cwg093. [DOI] [PubMed] [Google Scholar]
- 107.Oglesby LL, Jain S, Ohman DE. Membrane topology and roles of Pseudomonas aeruginosa Alg8 and Alg44 in alginate polymerization. Microbiology. 2008;154:1605–1615. doi: 10.1099/mic.0.2007/015305-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Remminghorst U, Hay ID, Rehm BHA. Molecular characterization of Alg8, a putative glycosyltransferase, involved in alginate polymerisation. J Biotechnol. 2009;140:176–183. doi: 10.1016/j.jbiotec.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 109.Dorfmueller HC, Ferenbach AT, Borodkin VS, van Aalten DMF. A structural and biochemical model of processive chitin synthesis. J Biol Chem. 2014;289:23020–23028. doi: 10.1074/jbc.M114.563353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ortiz-Castellanos L, Ruiz-Herrera J. Phylogenetic relationships of the wall-synthesizing enzymes of Basidiomycota confirm the phylogeny of their subphyla. Folia Microbiol (Praha) 2015;60:143–150. doi: 10.1007/s12223-014-0354-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The dataset(s) supporting the conclusions of this article is (are) included within the article (and its Additional files 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19 and 20).


