Abstract
Background:
A dinucleotide variant rs368234815 in interferon lambda 4 (IFNL4) gene was recently found to be associated with the hepatitis C virus (HCV) treatment response. This study aimed to assess the impact of IFNL4 rs368234815 polymorphism on treatment response to pegylated-IFN alpha (Peg-IFN-α) and ribavirin (RBV) in hemophilic patients with chronic hepatitis C (CHC).
Materials and Methods:
In this retrospective study, 92 hemophilic patients with CHC who were treated with Peg-IFN-α/RBV were investigated. Single-nucleotide polymorphisms (SNPs) in IFNL genomic region including rs368234815, rs12979860, and rs8099917 were analyzed by DNA sequencing.
Results:
Of the 92 patients, 63 (68.5%) achieved sustained virological response (SVR). Of the 43 patients with rs368234815 TT/TT genotype, 36 (83.7%) achieved SVR, while in 49 patients with non-TT/TT genotypes, 27 (55.1%) achieved SVR. Other pretreatment parameters predicted SVR were patients’ body mass index, HCV genotype, rs12979860, and rs8099917 SNPs. In multivariate analysis, all above-mentioned parameters except rs8099917 remained as predictors of SVR. IFNL4 rs368234815 was a strong predictor of SVR; however, the prediction power of this SNP was the same as that of rs12979860 SNP in the patients of the current study.
Conclusion:
IFNL4 rs368234815 SNP can be considered for decision-making in the treatment of HCV-infected patients.
Keywords: Genetic polymorphism, hepatitis C, human interferon lambda 4 protein
INTRODUCTION
An estimated 150–200 million people worldwide are infected with hepatitis C virus (HCV) which can lead to cirrhosis and/or liver cancer.[1,2] Hepatitis C is a major cause of morbidity and mortality in hemophilic patients who received clotting factor concentrates before the availability of virus-inactivated clotting factors in the mid-1980s. Some factors including host and viral parameters can affect the treatment response to antiviral therapy among patients with chronic hepatitis C (CHC).[3] In recent years, single-nucleotide polymorphisms (SNPs) located upstream of interferon lambda 3 (IFNL3) gene were identified as predictors of HCV spontaneous and treatment-induced clearance.[4,5,6] More recently, Prokunina-Olsson et al.[7] reported a novel transiently induced region upstream of IFNL3 (IL28B) on chromosome 19 that harbors dinucleotide variant rs368234815 (TT/ΔG), which was in strong linkage disequilibrium (LD) with rs12979860, a genetic marker strongly associated with sustained virological response (SVR) of HCV after pegylated-IFN (Peg-IFN) plus ribavirin (RBV) combination treatment. The perfect correlation of these two genetic variants in Caucasian patients was reported previously.[8]
This study aimed to evaluate the role of IFNL4 rs368234815 polymorphism on response to Peg-IFN-α/RBV in hemophilic patients with CHC.
PATIENTS AND METHODS
In this retrospective study, a total of 92 hemophilic patients were selected randomly from more than three hundred hemophilic cases with CHC who referred from the Iranian Hemophilia Foundation and Evaluated in Tehran Hepatitis Clinic (Tehran, Iran) from 2011 to 2013. All the patients were above 18 years of age with quantifiable HCV RNA (>25 IU/mL) in serum for more than 6 months before the study and had no previous history of antiviral therapy for CHC. The patients were treated according to the label, once-weekly injections of 180 μg of Peg-IFN-α-2a (Pegasys, Roche, Basel, Switzerland) or 1.5 μg/kg of PegIFN-α-2b (PegIntron, Schering-Plough, Las Piedras, Puerto Rico, USA) and weight-based RBV (Copegus, Roche or Rebetol, Schering-Plough) was given orally at an 800–1200 mg/day for 24–72 weeks according to the HCV genotype and on-treatment virological response. Informed consent was obtained from all patients who participated in this study. The study protocol conforms to the ethical guidelines of the 1975 Declaration of Helsinki. The goal of treatment was SVR, defined as undetectable serum HCV RNA 24 weeks after cessation of therapy.
HCV RNA was quantified in all patients by the COBAS TaqMan HCV Test (Roche Diagnostics). Genotyping of rs368234815, rs12979860, and rs8099917 SNPs was performed by DNA sequencing as previously described.[9]
The liver biopsy for determination of liver histology is not obligatory before starting HCV treatment and also, the liver biopsy procedure is life threatening in hemophilic patients. As a result, liver fibrosis and cirrhosis were assessed by transient elastography using FibroScan 502 machine (EchoSense) for a proportion of patients and the results were defined as F0–F4, the result of >F3 or >12.5 Kpa were considered as severe fibrosis or cirrhosis. For instances which FibroScan was not accessible, evidence of liver cirrhosis was defined based on clinical and imaging evidence.
Fisher's exact test was used for analysis of categorical variables and t-test for continuous variables. Hardy–Weinberg Equilibrium (HWE) was assessed for SNPs and the LD between these SNPs was calculated. All baseline variables that had a P < 0.2 in univariate analysis were entered to logistic regression models. To prevent multicollinearity resulted by high LD between the SNPs, different logistic regression models with the inclusion of a single SNP were considered.[10] P < 0.05 was considered to be statistically significant. Statistical analysis was performed using SPSS version 20 (IBM SPSS).
RESULTS
The patients’ characteristics and on-treatment response by SVR are shown in Table 1. The distribution of rs368234815, rs12979860, and rs8099917 genotypes were in HWE (P = 0.37, P = 0.37, and P = 0.86, respectively). The distribution of rs12979860 and rs368234815 were similar which resulted in strong LD (D’ =1.0, r2 = 1.0) between them. The LD between both rs368234815 and rs12979860 with rs8099917 was moderate (D’ =1.0, r2 = 0.51). Sixty-three (68.5%) reached SVR with antiviral therapy. Among patients’ baseline characteristics, body mass index (BMI) <25, HCV genotype-3, rs12979860 CC, rs8099917 TT, and rs368234815 TT/TT were associated with achievement of SVR. Of the 43 patients with rs368234815 TT/TT genotype, 36 (83.7%) achieved SVR, while in 49 patients with non-TT/TT genotypes, 27 (55.1%) achieved SVR. In patients with HCV genotype-1 infection, 76.0% of patients with rs368234815 TT/TT genotype achieved SVR, while 48.6% of ΔG carriers achieved SVR (P = 0.038). In HCV genotype-3 infection, 94.4% and 75.0% of patients with rs368234815 TT/TT and non-TT/TT genotypes achieved SVR, respectively (P = 0.274).
Table 1.
Patients’ characteristics by achievement of sustained virological response
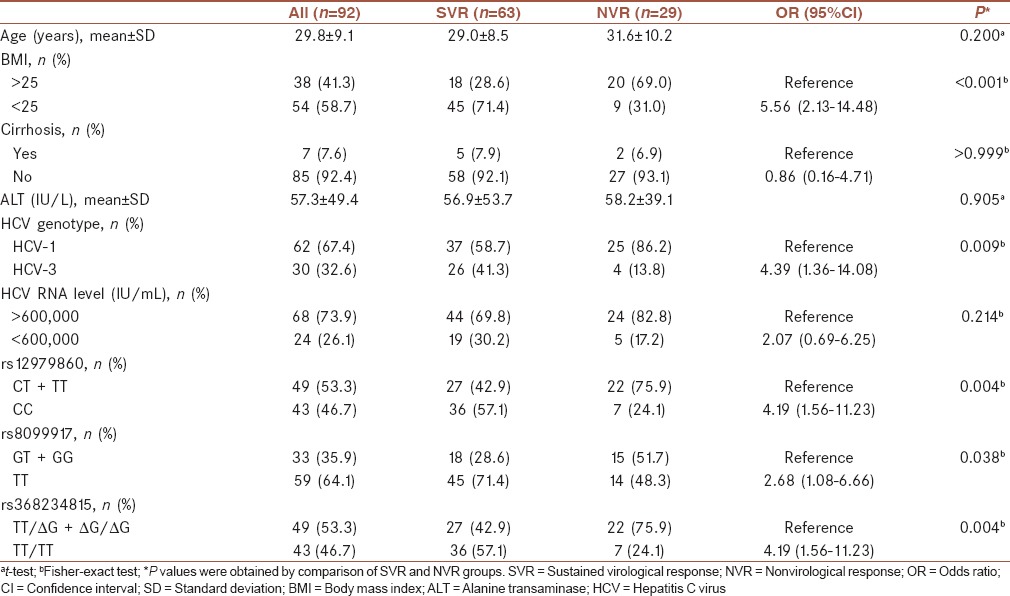
In multivariate analysis of baseline parameters, BMI <25, HCV genotype-3, rs12979860 CC (logistic regression model 1), and rs368234815 TT/TT (logistic regression model 3) were remained as predictors of SVR [Table 2].
Table 2.
Multivariate analysis of baseline predictors of sustained virological response

DISCUSSION
In a cohort of hemophilic patients with CHC, the presence of TT/TT at rs368234815 SNP was shown to confer a higher chance of SVR after antiviral therapy compared to SVR rate in the patients who carried ΔG allele which was similar to the observations in nonhemophilic patients.[11,12] Previous studies showed that SVR rate was doubled in hemophilic cases with CC genotype at rs12979860 than in those with T allele.[13,14] A former study from Iran found 61% SVR rate among 367 Iranian hemophilic patients with CHC who were treated with Peg-IFN-α-2a and RBV.[15]
Prokunina-Olsson et al.[7] showed that in the Asian population, the rs368234815 SNP may provide no more information on the grounds of the haplotype structure in which the IFNL4/IL28B SNPs were tightly linked to each other, whereas determination of IFNL4 rs368234815 genotype in patients with African ancestry might be superior to determination of other SNPs in IFNL genomic region. We found strong LD between the rs368234815 and rs12979860 SNPs which was similar to that in the previous studies.[11,12,16] Although some studies emphasized that there was no superiority in additional testing of IFNL4 rs368234815 for treatment prediction in Caucasian patients, a recent study concluded that the determination of this variant may be superior to that of known IL28B variants for patient management using IFN-based regimens.[17] Furthermore, if we accept the argument that IFNL4 rs368234815 is the functional variant in the process of HCV spontaneous and treatment-induced clearance, then it would seem to make sense to base clinical decisions on rs368234815 SNP, rather than a correlated variant such as rs12979860 SNP, even if the correlation is high.[8]
The response to IFN-based regimens for HCV infection varies considerably according to the host and viral factors and the presence or absence of an early response during treatment. Moreover, IFN and RBV treatment are associated with a number of side effects. New direct-acting antivirals which were approved for the treatment of CHC lead to high SVR rate with minimal side effects. It is likely that in the future the high efficacy of the new medications will overwhelm the predictive value of variables such as IFNL4 genotype; however, it can be considered that these new drugs might not be indicated for all patients categories (such as patients with HCV genotype-3 infection) and they may not be affordable in low-income communities for cost-effectiveness reason. Furthermore, since the notably proportion of patients with both HCV genotype-1 and -3 infections who harbored favorable rs368234815 TT/TT genotype achieved SVR, it is rational to personalize the treatment decision for HCV-infected patients according to rs368234815 marker. It means that the patients with rs368234815 TT/TT genotype can benefit from the cost-effective treatment regimen (Peg-IFN/RBV) which can decrease the burden of liver disease in the community. The main limitation of the present study was the small number of the patients which limited the power of the study. Another limitation was the different methods for the assessment of liver cirrhosis.
CONCLUSION
Our results indicated that the rs368234815 marker shows equivalent performance in prediction of SVR to the rs12979860 variant in Iranian hemophilic patients with CHC. Although there was a strong LD between both genetic variants, given rs368234815 seems to be the functional variant in the process of HCV treatment clearance, it can be considered as a replacement for rs12979860 in clinical practice. Furthermore, the rs368234815 marker can be considered for decision making in the treatment of HCV-infected patients.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
AUTHORS’ CONTRIBUTION
MK contributed in the conception of the work, conducting the study, revising the draft, approval of the final version of the manuscript, and agreed for all aspects of the work. SMA contributed in the conducting the study, approval of the final version of the manuscript, and agreed for all aspects of the work. BB contributed in the conducting the study, revising the draft, approval of the final version of the manuscript, and agreed for all aspects of the work. AP contributed in the conducting the study, approval of the final version of the manuscript, and agreed for all aspects of the work. HS contributed in the conception of the work, conducting the study, revising the draft, approval of the final version of the manuscript, and agreed for all aspects of the work.
REFERENCES
- 1.Gravitz L. Introduction: A smouldering public-health crisis. Nature. 2011;474:S2–4. doi: 10.1038/474S2a. [DOI] [PubMed] [Google Scholar]
- 2.Mohd Hanafiah K, Groeger J, Flaxman AD, Wiersma ST. Global epidemiology of hepatitis C virus infection: New estimates of age-specific antibody to HCV seroprevalence. Hepatology. 2013;57:1333–42. doi: 10.1002/hep.26141. [DOI] [PubMed] [Google Scholar]
- 3.Sharafi H, Alavian SM. IL28B polymorphism, explanation for different responses to therapy in hepatitis C patients. Hepat Mon. 2011;11:958–9. doi: 10.5812/kowsar.1735143X.794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sharafi H, Alavian SM, Behnava B, Pouryasin A, Keshvari M. The impact of IFNL4 rs12979860 polymorphism on spontaneous clearance of hepatitis C; a case-control study. Hepat Mon. 2014;14:e22649. doi: 10.5812/hepatmon.22649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haj-Sheykholeslami A, Keshvari M, Sharafi H, Pouryasin A, Hemmati K, Mohammadzadehparjikolaei F. Interferon-lambda polymorphisms and response to pegylated interferon in Iranian hepatitis C patients. World J Gastroenterol. 2015;21:8935–42. doi: 10.3748/wjg.v21.i29.8935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kalantari H, Tavakoli T, Bagherpour B, Honarmand S. Interleukin-28B (rs12979860) gene variation and treatment outcome after peginterferon plus ribavirin therapy in patients with genotype 1 of hepatitis C virus. J Res Med Sci. 2014;19:1062–7. [PMC free article] [PubMed] [Google Scholar]
- 7.Prokunina-Olsson L, Muchmore B, Tang W, Pfeiffer RM, Park H, Dickensheets H, et al. A variant upstream of IFNL3 (IL28B) creating a new interferon gene IFNL4 is associated with impaired clearance of hepatitis C virus. Nat Genet. 2013;45:164–71. doi: 10.1038/ng.2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keshvari M, Pouryasin A, Behnava B, Sharafi H, Hajarizadeh B, Alavian SM. Letter: The rs12979860 and ss469415590 polymorphisms of IFNL4 gene are in strong linkage disequilibrium in Caucasian patients with chronic hepatitis C. Aliment Pharmacol Ther. 2014;39:343. doi: 10.1111/apt.12589. [DOI] [PubMed] [Google Scholar]
- 9.Sharafi H, Pouryasin A, Alavian SM, Behnava B, Keshvari M, Mehrnoush L, et al. Development and validation of a simple, rapid and inexpensive PCR-RFLP method for genotyping of common IL28B polymorphisms: A useful pharmacogenetic tool for prediction of hepatitis C treatment response. Hepat Mon. 2012;12:190–5. doi: 10.5812/hepatmon.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keshvari M, Sharafi H, Hajarizadeh B, Alavian SM. Can we include genetic variants with high linkage disequilibrium into a multiple logistic model? Liver Int. 2014;34:964. doi: 10.1111/liv.12464. [DOI] [PubMed] [Google Scholar]
- 11.Stättermayer AF, Strassl R, Maieron A, Rutter K, Stauber R, Strasser M, et al. Polymorphisms of interferon-lambda4 and IL28B – Effects on treatment response to interferon/ribavirin in patients with chronic hepatitis C. Aliment Pharmacol Ther. 2014;39:104–11. doi: 10.1111/apt.12547. [DOI] [PubMed] [Google Scholar]
- 12.Real LM, Neukam K, Herrero R, Guardiola JM, Reiberger T, Rivero-Juarez A, et al. IFNL4 ss469415590 variant shows similar performance to rs12979860 as predictor of response to treatment against hepatitis C virus genotype 1 or 4 in Caucasians. PLoS One. 2014;9:e95515. doi: 10.1371/journal.pone.0095515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mancuso ME, Linari S, Aghemo A, Bartolozzi D, Santagostino E, Rumi MG, et al. Interferon lambda 3 rs12979860 polymorphism in patients with haemophilia and HCV infection: A predictor of spontaneous viral clearance and sustained virological response. Thromb Haemost. 2014;111:1067–76. doi: 10.1160/TH13-11-897. [DOI] [PubMed] [Google Scholar]
- 14.Maor Y, Morali G, Bashari D, Pénaranda G, Schapiro JM, Martinowitz U, et al. Genetic polymorphism of IL28B in hepatitis C-infected haemophilia patients in Israel. Haemophilia. 2013;19:150–6. doi: 10.1111/j.1365-2516.2012.02932.x. [DOI] [PubMed] [Google Scholar]
- 15.Alavian SM, Tabatabaei SV, Keshvari M, Behnava B, Miri SM, Elizee PK, et al. Peginterferon alpha-2a and ribavirin treatment of patients with haemophilia and hepatitis C virus infection: A single-centre study of 367 cases. Liver Int. 2010;30:1173–80. doi: 10.1111/j.1478-3231.2010.02296.x. [DOI] [PubMed] [Google Scholar]
- 16.Ochi H, Miki D, Hayes CN, Abe H, Hayashida Y, Kubo M, et al. IFNL4/IL-28B haplotype structure and its impact on susceptibility to hepatitis C virus and treatment response in the Japanese population. J Gen Virol. 2014;95(Pt 6):1297–306. doi: 10.1099/vir.0.060103-0. [DOI] [PubMed] [Google Scholar]
- 17.Covolo L, Bibert S, Donato F, Bochud PY, Lagging M, Negro F, et al. The novel ss469415590 variant predicts virological response to therapy in patients with chronic hepatitis C virus type 1 infection. Aliment Pharmacol Ther. 2014;39:322–30. doi: 10.1111/apt.12568. [DOI] [PubMed] [Google Scholar]


